Abstract
Access of molecular oxygen to the respiratory electron transport chain at the mitochondria costs in the generation of reactive oxygen-derived species (ROS). ROS induces progressive damage to macromolecules in all living cells, hence, rapid defense mechanisms to maintain cellular redox homeostasis are vital. NEDD8/Rub1 is a highly conserved ubiquitin-like modifier that has recently been identified as a key regulator of cellular redox homeostasis. In this review, I will present NEDD8/Rub1, its modification cascade of enzymes, substrates and hydrolases. After introduction, I will show that the NEDD8/Rub1 pathway is linked with mitochondria physiology, namely, oxidative stress. In the rest of the review, I will approach the Ascomycota phylum of the kingdom fungi instrumentally, to present existing links between NEDD8/Rub1 vitality and the aerobic lifestyle of model species belonging to three subphyla: Saccharomycotina (S. cerevisiae and C. albicans), Pezizomycotina (A. nidulans and N. crassa), and Taphrinomycotina (S. pombe).
Keywords: NEDD8, Rub1, Cullin-RING E3 ligase, COP9 signalosome, NEDP1, Ascomycota, Mitochondria
Abbreviations: Ub, ubiquitin; Ubl, ubiquitin-like; SUMO, Small Ubiquitin-like Modifier; ULP1, Ub Like Protease 1; NEDD8, Neural precursor cell Expressed Developmentally Downregulated gene 8; Rub1, Related ub 1; CRLs, cullin-RING E3 ligases; WHB, winged-helix B; CSN, COP9 signalosome; SR, substrate receptor; UCH, ubiquitin carboxyl terminal hydrolase; HIF-α1, Hypoxia-inducible factor α-1; PCNA, proliferating cell nuclear antigen; mtDNA, mitochondrial DNA; ETC, electron transfer respiratory chain; ROS, reactive oxygen species; PARP1, Poly ADP-Ribose Polymerase 1
Graphical abstract
Highlights
-
•
NEDD8/Rub1 is a key regulator of cellular redox homeostasis.
-
•
Ascomycota species that produce mitochondria-derived ROS during glycolysis require NEDD8/Rub1for viability.
-
•
NEDD8/Rub1 essentiality correlates with the existence of NEDP1 in the organism genome.
1. Introduction
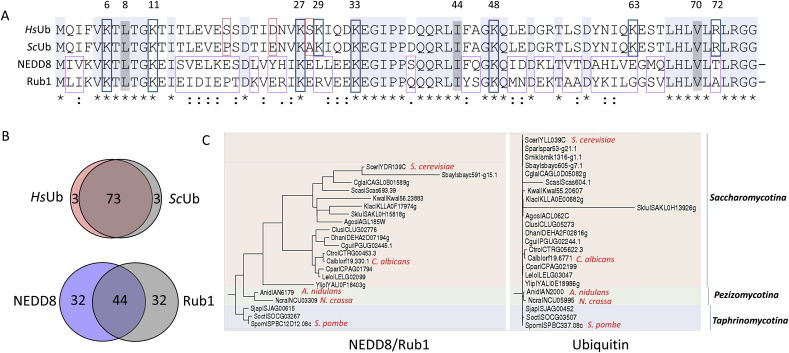
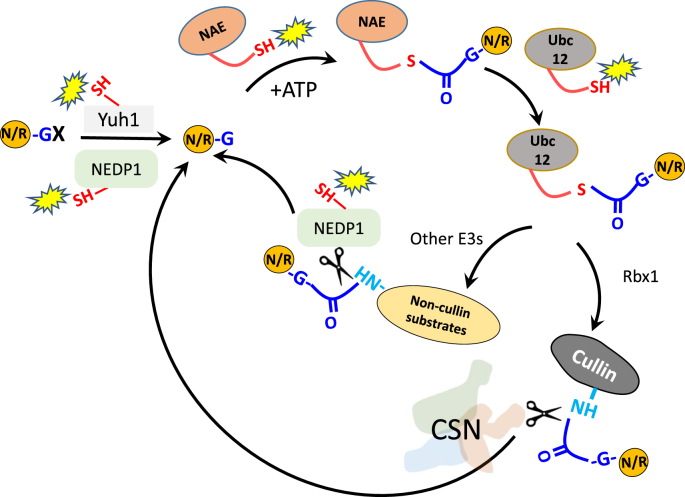
The eukaryotic family of ubiquitin-like (Ubl) polypeptides is a class of evolutionary conserved reversible protein modifiers that regulate a variety of fundamental cellular processes [1,2]. The first discovered and most widely studied member of this family is ubiquitin (Ub) [3]. Ub is a most conserved polypeptide, showing only 3:76 amino acid difference between the Saccharomyces cerevisiae and human orthologs (Fig. 1). Ub modifies thousands of targets in processes that require hundreds of enzymes, involving the recognition, covalent modification and release of Ub to and from substrates [4]. The Ubl family comprises additional members, among which, small ubiquitin-like modifiers (SUMOs) and neural precursor cell expressed developmentally downregulated gene 8 (NEDD8) are the most widely studied to date. Each of the Ubl modifiers requires a cognate cascade of enzymes for the covalent attachment to substrates (Fig. 2). NEDD8 is the closest paralog of Ub; however, it shows a higher variety in sequence then Ub, with a diversity of as many as 32 out of its 76 amino acids between the human NEDD8 and its S. cerevisiae ortholog, Related Ub 1 (Rub1) (Fig. 1). Unlike the expended Ub pathway, the NEDD8/Rub1 modification pathway includes numerous enzymes and the list of substrates is much shorter, with extensive reports on a single family of conserved protein targets, namely “cullins” (in S. cerevisiae Cdc53/yCul1, yCul3 and Rtt101/yCul4; in humans the typical Cul1-5, and atypical Cul 7 and Cul 9/PARC) [[5], [6], [7]]. Each cullin serves as a platform for the construction of modular multi-subunit Ub ligases, belonging to the family of cullin-RING E3 ligases (CRLs). Similar to ubiquitination, the conjugation of NEDD8/Rub1 to cullin substrates (known as “NEDDylation”) requires a cascade of enzymes (Fig. 3). Both NEDD8 and Rub1 orthologs are synthesized as precursors that first need to be trimmed by C-terminal hydrolases [8,9]. The subsequent conjugation of NEDD8/Rub1 to a precise Lys residue of each cullin is mediated by the NEDDylation cascade of enzymes, starting with a dimeric NEDD8 E1 activating enzyme (NAE1), consisting of Ula 1/Uba3 in S. cerevisiae and APPBP1/UBA3 in humans, followed by an E2 conjugating enzyme (Ubc12 in S. cerevisiae; UbE2M or UbE2F in humans) [[10], [11], [12]] and the RING E3 subunit of CRLs (Hrt1 in S. cerevisiae; RBX1 and RBX2 in humans), together with the co-E3, DCN1 that stimulates the NEDDylation reaction [13,14]. Cullin NEDDylation enhances CRL activity in vitro probably by facilitating the recruitment of the Ub-charged E2 enzyme [15]. The NEDDylation site is located at the C-terminal “winged-helix B” (WHB) inhibitory domain of the cullin [16]. Recent structural studies of CRL1 (a.k.a. the SCF) through cryo-electron microscopy have revealed that covalent attachment of NEDD8 to the WHB domain of Cul1 leads to extensive rearrangements of the CRL components, which could explain the enhanced ubiquitination activity [17]. Cullin modification is reversed by the COP9 signalosome (CSN), a multi-subunit cullin-NEDD8/Rub1-specific deNEDDylase [18,19] (Fig. 3). Over time, studies undertaken by multiple laboratories have identified additional substrates for NEDD8 [[20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30]]. These non-cullin substrates are conjugated to NEDD8 monomers either directly, or indirectly through poly-NEDD8 or mixed Ub–NEDD8 chains. These studies have led to the finding of an additional deNEDDylase, named NEDP1 (a.k.a. Ub-like protease (ULP)8, SENP8, DEN1) that detaches NEDD8 from non-cullin substrates [[31], [32], [33]] (Fig. 4). While the NEDDylation cascade of enzymes is highly conserved across phyla, deNEDDylases are more diverse in sequence, complexity and even in their existence. This review analyzes the knowledge on Ascomycota lifestyle instrumentally to present a link between NEDD8/Rub1 functionality, vitality and cell metabolic programming (Fig. 5).
Fig. 1.
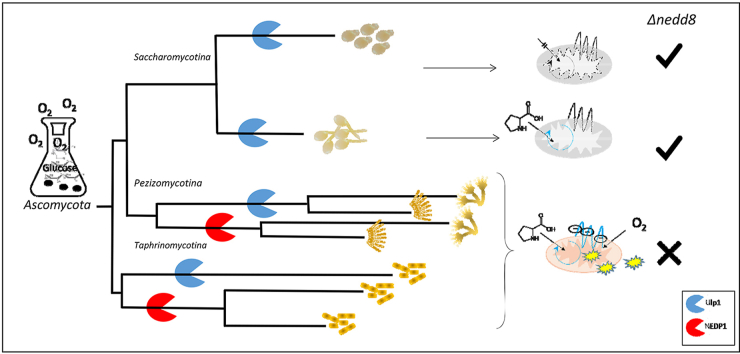
Orthology and paralogy between ubiquitin and NEDD8/Rub1 modifiers. (A) multiple sequence alignment (Clustal W) of the modifiers from Saccharomyces cerevisiae (Sc) and Homo sapiens (Hs). The hydrophobic patch residues of Ub (L8, I44, and V70) are in grey. Conserved residues are shaded in light grey; lysine residues in a blue frame; non-conserved residues are framed in red (ubiquitin) or purple (NEDD8/Rub1). (B) Venn diagram representing the number of conserved distinct amino acids between Homo sapiens and S. cerevisiae orthologues. (C) Orthologues of Ubiquitin and NEDD8/Rub1 within the Ascomycota phylum. The tree shows clearly that Ubiquitin is much more conserved across the Ascomycota phylum and NEDD8/Rub1 conservation is higher within sub-phyla than cross sub-phyla. The described model species are in red. Figures display information according to the Fungal Orthogroups Repository website: http://www.broadinstitute.org/regev/orthogroups/. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Illustration of the NEDD8/Rub1 modification cycle. The precursor form of NEDD8/Rub1 (-GX) is processed by at least two hydrolases: NEDP1 and Yuh1, each of them trims the modifier into the matured form (-G). Subsequently, the Gly residue forms a reactive intermediate thioester with a cysteine of NAE in an ATP-dependent manner before the trans-thiolation to Ubc12. The four enzymes of NEDP1, Yuh1, NAE and Ubc12 include a catalytic cysteine that might be prone to oxidation (yellow star). Next, NEDD8/Rub1 is transferred from Ubc12 to a specific lysine on the substrate through an E3 ligase. For cullin NEDDylaiton, the E3 is the Rbx1 subunit CRLs. Other substrates might be targeted for NEDDylaiton by other E3 ligases. DeNEDDylases of CSN (for cullins) and NEDP1 (for other substrates) deconjugate NEDD8 from the substrates. The terminology in this figure is mostly based on S. cerevisiae. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
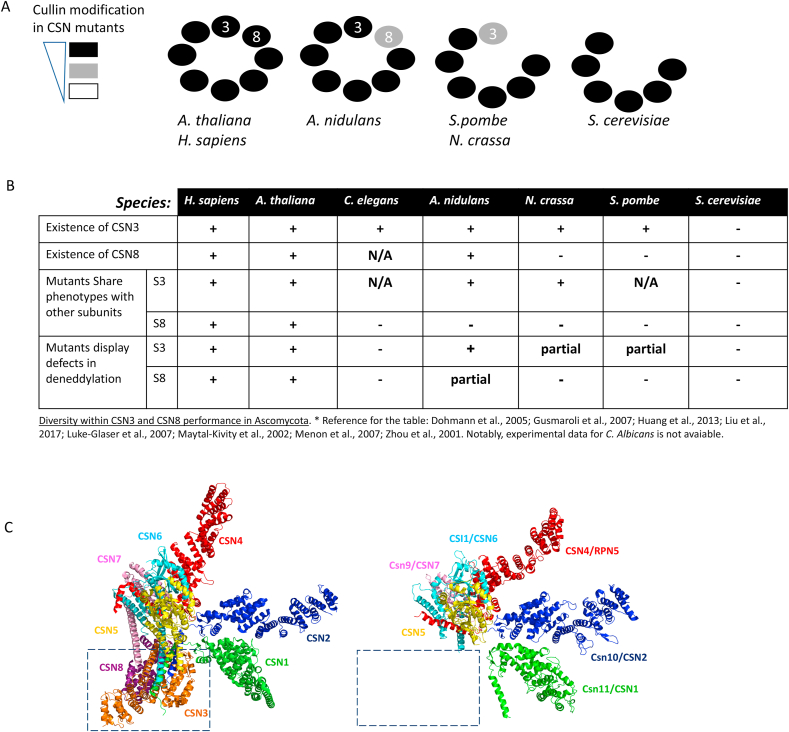
Plasticity within CSN assemblages. (A) CSN complexes do not always bear the CSN3 and CSN8 subunits. (B) The table shows that CSN3 and CSN8 are not always fully committed to the deNEDDylation activity (“partial”). The data is according to Dohmann et al., 2005; Gusmaroli et al., 2007; Huang et al., 2013; Liu et al., 2017; Luke-Glaser et al., 2007; Maytal-Kivity et al., 2002; Menonet al., 2007; Zhou et al., 2001 [56,[140], [141], [142], [143], [144], [145]]. (C) 3D structure of human and S. Cerevisiae CSN complexes. Both complexes present a highly conserved deNEDDylase core complex. Yet, the S. cerevisiae CSN complex lacks the CSN3/8 module (dashed box). Human CSN structure is according to Lingaraju et al. 2014 [19]. Empirical model for the S. cerevisiae CSN is according to Sinha et al. 2020 [61].
Fig. 4.
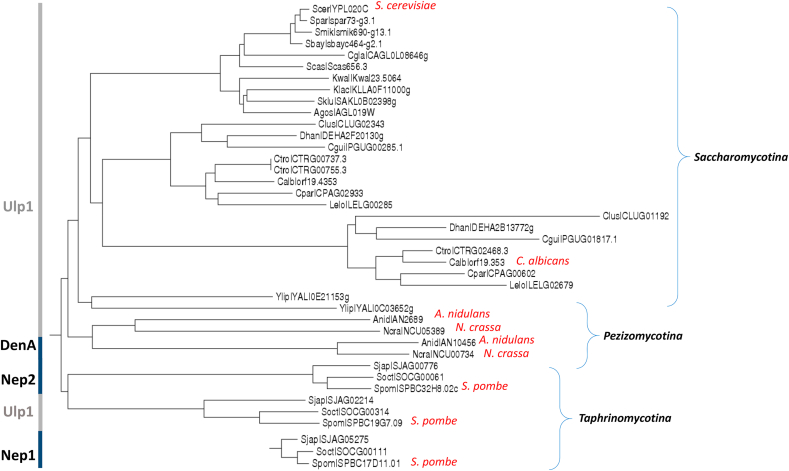
Ascomycota Orthologs of Ulp1 and their paralogy with DENP1. The subphylum of Saccharomycotina includes a single copy of Ulp1. Species of the Pesisomycotina subphyla includes a Ulp1 paralog that was identified experimentally as DENP1 (DenA). Notably, Taphrinomycotina species include 2 copies of NEDP1 (Nep1 and Nep2). Nep1 and Nep2 are differ in the position of the active site cysteine relative to the rest of the protein. The heterogeneity in molecular mass and active site position is typical of this family of proteases [146,147], thus, the branch of Nep1 is not connected to the genetree. The figure produced through the Fungal Orthogroups Repository website: http://www.broadinstitute.org/regev/orthogroups/.
Fig. 5.
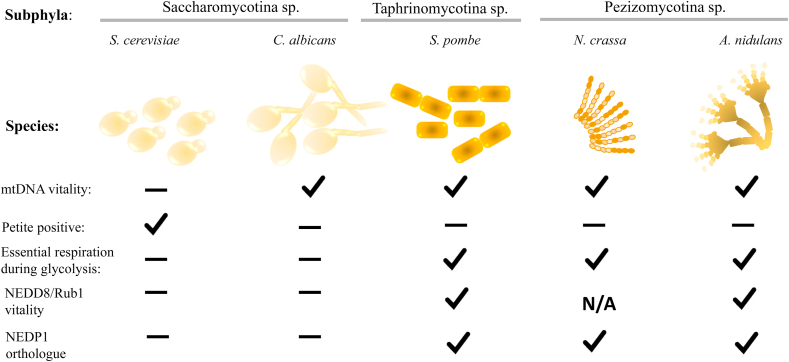
NEDD8 vitality and mitochondria respiration lifestyle of Ascomycota model species. The five species shown belong to three subphyla of Ascomycota. The mitochondrial metabolic programming of these fungi is variable. As discussed in the text: essentiality of mitochondrial respiration derived ROS during glycolysis correlates with the vitality of NEDD8/Rub1.
2. Crossroads between ubiquitin and NEDD8 pathways
The NEDDylation pathway intersects with ubiquitination at several junctions. The first intersecting enzyme is the ubiquitin C-terminal hydrolase 3 ([UCHL3], or, Yuh1 in S. cerevisiae), which exposes the conjugating residue, Gly76 of both, Ub and NEDD8 (Fig. 2). Similarly, all documented NEDD8/Rub1 E3s are also Ub ligases [34]. Apparently, the Ub and NEDD8/Rub1 pathways split into distinct cascades for the E1 and E2 activities. Notably, residue 72 within each modifier is the key recognition site for each the E1 enzymes: ubiquitin activating enzyme (UAE) and NAE [[35], [36], [37]]. NAE exhibits specificity to NEDD8/Rub1 due to an arginine residue near the catalytic site of UBA3/Uba3 that could possibly clash with the highly conserved Arg 72 residue of Ub [[35], [36], [37], [38]]. In contrast, UAE prefers Ub over Rub1 [39]. Notably, in certain circumstances, NEDD8/Rub1 approaches UAE to enter the ubiquitinome [[39], [40], [41], [42]]. This can be achieved if most of the Ub bulk is incorporated into chains due to a pharmacological treatment with proteasome inhibitors, or in response to stress [[40], [41], [42], [43], [44]]. In such circumstances, NEDD8/Rub1 is activated by UAE, transferred to Ub-E2s and eventually incorporated into Ub chains. It had been suggested that introducing the yeast Rub1 into the ubiquitinome decreases the average length of heterologous mixed Rub1-Ub chains relative to homogenous poly-Ub chains [45]. The mammalian NEDD8 has been reported to form in vivo homogenous polymers and tripeptides in cellular stresses, including DNA damage and oxidative stress, respectively [[46], [47], [48]]. The spillover of NEDD8/Rub1 into the Ub pathway and the ability to form homogenous chains could indicate other function(s) of NEDD8/Rub1 apart from activating CRLs.
3. The NEDD8/Rub1 deconjugating enzymes
3.1. The CSN complex
3.1.1. Evolutionary conservation of the CSN
The CSN is a multi-subunit cullin-deNEDDylating enzyme harboring a JAMM/MPN+ metalloprotease motif within the catalytic subunit Csn5. The CSN shows high paralogy to the 19S proteasome lid, which contributes deubiquitination activity to the 26S holocomplex through the metalloprotease subunit Rpn11 [49,50]. The proteasome lid and the CSN complexes are required for viability of multicellular organisms. Yet, unlike the high conservation of the proteasome lid across all eukaryotic phyla, the CSN shows more diversity, especially in fungal species [[51], [52], [53]]. For example, S. cerevisiae proteasome lid subunits can be substituted with Arabidopsis thaliana orthologs. In contrast, S. cerevisiae CSN subunits are not interchangeable with orthologs of higher organisms [54,55]. The CSN had been studied in model organism species belonging to the three monophyletic subphyla of the fungal phylum Ascomycota: Saccharomycotina (including S. cerevisiae and Candida albicans), Pezizomycotina (including Aspergillus nidulans and Neurospora crassa), and Taphrinomycotina (including Schizosaccharomyces pombe). In these species, the CSN is not vital, although mutants sometimes lead to physiological and morphological alterations, among them, defects in cell-cycle progression, vacuole morphology, circadian clock or fruit body formation [[56], [57], [58], [59], [60], [61]].
3.1.2. Conserved and diverged CSN functions
The specificity of the CSN complex to cullin-NEDD8 conjugates stems from the JAMM/MPN+ motif in Csn5/Jab1, which becomes functional only upon interaction of the holocomplex with a CRL. Indeed, the complex-incorporated Csn5 is autoinhibited at the Glu 104 residue in the autoinhibitory loop (Ins-1). Interaction of the complex with a NEDDylated CRL leads to a series of conformational change events in Csn2, Csn4 and Csn7, triggering rearrangements in the Csn5–Csn6 dimer, resulting in Csn5 activation by priming the Csn5 JAMM/MPN+ motif for deNEDDylation [19,62]. In addition to intrinsic deNEDDylation activity, the CSN also possesses two kinds of deubiquitination activity: one that deconjugates ubiquitin from mono-ubiquitinated substrates and is mediated by Csn5 [42,63]; and the other that depolymerizes polyubiquitin chains through CSN-associated deubiquitinating enzymes: Usp 48 in humans, Ubp12/Usp 15 in S. pombe, A. nidulans and humans, and Ubp3 in S. cerevisiae [54,64,65]. Both deNEDDylation and deubiquitination functions of the CSN inhibit CRLs in vitro [[65], [66], [67]]. Additionally, the CSN also controls CRL activity in two non-enzymatic manners: inhibition of cullin NEDDylation through the binding of Csn2 and Csn4 to the CRL-RING subunit Rbx1 [19,62,68,69]; and hindrance of the target binding site in the substrate receptor (SR) of CRLs by the Csn3 and Csn8 subunits [19,70]. The overall negative activity of the CSN is believed to protect CRL ligases in vivo from auto-ubiquitination of self-components. Consequently, the CSN is considered a positive regulator of CRL-mediated physiological functions. Consistent with this, in the absence of CSN, many SRs are auto-ubiquitinated, and hence are destabilized [71,72]. Indeed, a direct interaction between the CSN and CRL-SRs has been described in many organisms; in each of them, dissociation of CSN subunits results in the loss of BTB and F box protein stability [[71], [72], [73], [74], [75], [76], [77]]. The CRL SR interacting components, Csn3 and Csn8, are two atypical Proteasome, COP9, Initiation factor 3 (PCI)-containing subunits [50]. In canonical CSN complexes, these subunits co-interact through their N-terminal repeats, and are considered as a module [19]. Interestingly, both Csn3 and Csn8 exhibit diverged phylogenetic performance in the Ascomycota species by not always participating in CSN assemblages [54,[78], [79], [80]] (Fig. 3a and b), even if a direct ortholog gene exists in their genome [50,81]. Indeed, orthologs of these subunits exist and participate in CSN assemblages of A. nidulans (Csn3, Csn8), N. crassa (Csn3) and S. pombe (Csn3), yet their mutants are distinguished from other csn mutants by showing a slighter CSN characteristic biochemical phenotype of accumulated cullin-NEDD8 conjugates [56,67,71,82] (Fig. 3b). The minor accumulation of NEDDylated cullins could be explained if these subunits are not part of the core deNEDDylase complex, and instead are primarily involved in the regulation of substrate ubiquitination and turnover [53] (Fig. 3c). Evidently, S. cerevisiae CSN complexes lacking the Csn3/Csn8 module harbor a highly conserved deNEDDylase activity, but do not affect the turnover of typical CRL substrates such as p27/Sic 1 or other key cell-cycle regulators [83,84]. Up-to-date, the turnover of a single CRL substrate was found to be regulated by the S. cerevisiae CSN complex Mth 1, a negative regulator of the glucose-sensing signal transduction pathway and a substrate of SCFGrr1 [84]. It should be noted that in higher organisms, the Csn3/8 module is involved in redox regulation, since conditional perturbation of Cullin-deNEDDylation by cardiomyocyte-restricted knockout of mouse CSN8 has been found to increase myocardial oxidation [85,86]. Altogether, this suggests that the requirement of the CSN for vitality and/or functionality exhibits a great phylogenetic diversity.
3.2. Other deNEDDylases
Identification of the CSN as a cullin-specific deNEDDylase indicates a requirement for a more general deNEDDylase to facilitate the exposure of the conjugation residue Gly76 of NEDD8/Rub1 and to release NEDD8 from non-cullin substrates. These distinct functions could be achieved either by a difunctional enzyme similar to the deSUMOylating enzyme ULP1 [87], or by two distinct enzymes, with each one bearing one of the functions. In fact, UCHL3/Yuh1 catalyzes the initial processing of NEDD8/Rub1 by hydrolyzing the peptide bond at the carboxyl terminus of both Ub and NEDD8/Rub1 (Fig. 2). UCHL3/Yuh1 is highly conserved in eukaryotes, including the three Ascomycota subphyla. Nevertheless, Ub is probably a better substrate of UCHL3/Yuh1 as the inhibitory constant (Ki) for NEDD8/Rub1 is 20-fold higher compared to Ub [8]. Many eukaryotes harbor an additional deNEDDylase, namely NEDP1. NEDP1 is a NEDD8-specific hydrolase from the ULP family that plays a dual role by both processing the carboxyl terminus of NEDD8 and also releasing covalently attached NEDD8 molecules from multiple substrates [32,33,[88], [89], [90]]. NEDP1 is a direct paralog of Ulp1 (Fig. 4). Orthologs of NEDP1 are included in Pezizomycotina and Taphrinomycotina, but not in Saccharomycotina species. Interestingly, the S. pombe genome includes two orthologs of NEDP1 known as Nep1 and Nep2 [90] (Fig. 4). NEDP1 deletion mutants of A. nidulans (ΔdenA) and S. pombe (Δnep1/Δnep2) are viable [90,91]. Notably, the functional redundancy in S. pombe could be connected with the essential nature of NEDD8 conjugation in fission yeast.
4. Essentiality of NEDD8 and the cullin-NEDDylation site in Ascomycota
If the regulation of CRL activity was the only important function of NEDD8, it would be expected that the NEDDylation and deNEDDylation enzymes would be similarly required for organism viability. Yet, non-essentiality of the CSN in Ascomycota species does not constantly conform with the requirement of NEDD8/Rub1 for vitality. Indeed, NEDD8/Rub1 is essential for the viability of most studied Ascomycota model species, except for S. cerevisiae and C. albicans [92,93]. S. cerevisiae strains, mutated in the NEDDylation modification site of either Cdc53/Cul1 (K760R) or Rtt101 (K791R) are viable and show moderated phenotypes of altered ergosterol quantity and sensitivity to a DNA single-strand break, respectively [61,94]. Similarly, the C. albicans mutant of Rub1 (rub1−/−) exhibits mild growth defects, including a pseudohypha-like phenotype associated with the pathogenic morphology of this fungus. The phenotype is shared with the non-NEDDylated point mutant of Cul1 (K699R), but not with csn5−/− [92]. The non-essentiality of NEDD8/Rub1 in both, S. cerevisiae and C. albicans is correlated with the absence of DENP1-encoding gene in their genomes.
Although belonging to the same phylum, the fission yeast S. pombe is only distantly related to S. cerevisiae and is believed to have separated more than 350 million years ago [95]. Indeed, S. pombe NEDD8/Rub1 is required for organism viability and the NEDDylation site of the only essential S. pombe cullin, Cul1/Pcu1 (K713) is also vital [96]. Unexpectedly, the overexpression of pcu1Δ720–767, a C-terminal truncated mutant lacking the autoinhibitory WHB domain of Cul1/Pcu1, rescues pcu1 K713/R unviability [46,97,98]. These findings could be explained if increased flexibility of the WHB free mutant suppresses the necessity of NEDD8/Rub1 for SCF activation [17]. Surprisingly, NAE is still essential in this mutant, indicating the vital NEDDylation of the non-cullin substrate(s) in S. pombe.
5. Cullin-free NEDD8 and the response to mitochondrial oxidative stress
Oxidation is the strongest stress inducer of the formation of non-cullin conjugates of NEDD8 [43]. A growing number of studies identify a direct link between mitochondrial-derived oxidation and the performance of non-cullin NEDD8 conjugates. For example, mice flavoproteins of the mitochondrial electron transfer chain (ETC) are stabilized by NEDDylation and are rapidly degraded upon deficiency in NEDDylation cascade enzymes, leading to pathologies related to fatty acid oxidation disorders [99]. The modification of hypoxia-inducible factor (HIF)-α1 protein by NEDD8 has a great impact on cell survival under circumstances where hypoxia and oxidative stress coexist [100]. Stressed mammalian cells accumulate poly-NEDD8 chains [43], and the treatment with hydrogen peroxide inactivates NEDP1, resulting in an accumulation of unanchored NEDD8 trimers [47]. These NEDD8 trimers bind and inhibit poly-ADP ribose polymerase (PARP)1 to prevent the activation of apoptosis. In correlation with these data, the proliferating cell nuclear antigen (PCNA) is modified by NEDD8 and stabilized upon hydrogen peroxide treatment due to inhibition of NEDP1 [101].
The link between NEDD8/Rub1 and oxidative stress is highly conserved. Indeed, the loss of NEDD8/Rub1 in C. albicans promotes sensitivity to oxidative stress [92]. Likewise, Δrub1, a null mutant of NEDD8/Rub1 in S. cerevisiae, exhibits altered mitochondrial morphology and decreased ergosterol quantity, a well-known phenotype of cells with a complete loss of mitochondrial DNA (mtDNA ρ0) [45,61,102]. Altogether, this suggests a highly conserved link between the biology of NEDD8/Rub1 and mitochondria-derived oxidation.
6. NEDD8/Rub1 and the respiration lifestyle of various fungal species
The mitochondria ETC is a central source of cellular reactive oxygen species (ROS) [[103], [104], [105]]. Mitochondrial ROS is considered as a key pathogenic trigger of diseases such as inflammation, cancer, cardiac, neurodegeneration and other aging disorders [[106], [107], [108]]. Accordingly, it will not be surprising if the diverged existence or essentiality of NEDD8 and NEDP1 in Ascomycota is linked with aerobic lifestyle and essentiality of the mitochondria. The stated Ascomycota groups have the ability to ferment glucose in the presence of oxygen and to proliferate under anaerobic conditions, and thereby employ the mitochondria for respiration by different metabolic programs. Unfortunately, with the exception of the information on these popular species, reliable data concerning respiration strategies of other Ascomycota species are poorly known.
6.1. The respiration lifestyle of S. cerevisiae
Much of our knowledge in this area is based on research of S. cerevisiae due to biotechnological interest in this organism, which began in the 19th century following the identification of glucose fermentation by Louis Pasteur. S. cerevisiae cells approach anaerobic metabolism even under aerobic conditions in the presence of oxygen. This phenomenon is known as “aerobic glycolysis” or the “Crabtree effect.” Aerobic glycolysis in Crabtree-positive organisms such as S. cerevisiae enables a higher rate of ATP production at the fermentation growth phase, usually without accumulating ROS [109,110]. As a Crabtree-positive organism, S. cerevisiae cells convert glucose to ethanol and carbon dioxide through alcoholic fermentation, as long as glucose is available during the logarithmic growth phase and regardless of the presence of oxygen. During this phase, the oxidation of carbohydrates in the mitochondrial ETC is suppressed [111]. Towards the end of the fermentative phase, under a critical dilution rate of glucose, S. cerevisiae cells undergo a physiological transition from anaerobic glycolysis to mitochondrial respiration, which is accompanied by a high production of ROS. This critical point is known as the “diauxic shift,” and is characterized by de novo transcription of metabolic and antioxidant genes [[112], [113], [114]]. In fact, S. cerevisiae cells can survive without mtDNA (ρ0) as long as they are grown on a fermentative carbon source [115]. Moreover, ρ0 yeast cells, which do not have respiratory capacity, form a characteristically reduced colony size, termed “petite” [115].
6.2. S. cerevisiae mitochondria and NEDD8/Rub1
We asked how the respiration lifestyle of S. cerevisiae might be linked with NEDD8/Rub1 functionality. Recent studies present a dramatic loss in the S. cerevisiae yCul1/Cdc53 NEDDylation status at the diauxic shift [84,110]. Indeed, the S. cerevisiae NEDDylation cascade of enzymes is sensitive to a natural increase in metabolic ROS during mitochondrial respiration, leading to the loss of Ubc12~NEDD8 thioester forms and eventually in Cdc53/yCul1 NEDDylation status. Correspondingly, elevation of ROS in mutants lacking a proper antioxidant machinery, or in wild-type cells treated by uncouplers of the mitochondria ETC, leads to a decrease in Cdc53/yCul1 NEDDylation status. This phenomenon is evolutionarily conserved, as induction of ROS in human cells by low concentrations of hydrogen peroxide blocks the NEDDylation enzymes and the transfer of NEDD8 to cullins [116,117]. Conversely, rpn11-m1, a proteasome mutant strain that does not accumulate ROS due to a high capacity of antioxidants, exhibits continuous Ubc12~NEDD8 thioester formation in all growth phases [110,118]. On the other hand, CSN metalloprotease activity is stable during oxidation (diauxic and post-diauxic phases) [84,110,119]. Inhibition of the S. cerevisiae NEDDylation cascade by cumulative mitochondrial and exogenous ROS supplemented by continuous CSN activity, results in the loss of cullin NEDDylation and the appearance of cullin-free NEDD8/Rub1molecules.
6.3. C. albicans mitochondria and NEDD8/Rub1
C. albicans is an mtDNA ρ+ Crabtree-positive organism, harboring effective mitochondria during the pre-diauxic anaerobic growth phase [120]. This attribute might appear to conflict with the suggestion that NEDD8/Rub1 essentiality correlates with mitochondrial respiratory activity. However, the lifestyles of C. albicans and S. cerevisiae are quite dissimilar. The mitochondria of C. albicans have a central role in fungal virulence, even at the fermentation phase [121]. C. albicans is a pleomorphic pathogenic fungus with two distinct morphologies: yeast-like and filamentous-like. The filamentous morphology could be further dissected into a pseudo-hyphae form and a pathogenic invasive-hyphae form [122]. The most potent inducers of the invasive form are amino acids such as arginine and proline, which are catabolized to glutamate [123]. These amino acids enter the tricarboxylic acid (TCA) cycle in the mitochondria through α-ketoglutarate. The catabolism of proline in the mitochondria leads to increased levels of intracellular ATP, which subsequently promotes the switch between the yeast and hyphal morphologies, a key strategy to escape from degradation in macrophages [124]. Interestingly, similar to S. cerevisiae cells, when C. albicans is grown aerobically in a high-glucose medium complemented with proline, mitochondria are no longer required [124]. Moreover, even without the addition of proline, cells at the pre-diauxic phase exhibit a rapid induction of oxidative stress-resistant genes and are less susceptible to oxidative stress [125]. This could be explained as survival tactics of pathogens, which are attracted by neutrophils that try to kill them by oxidative burst [126]. Accordingly, the non-essentiality of NEDD8/Rub1 in C. albicans can be explained by the function of mitochondria in the aerobic fermentation phase mainly for the metabolism of amino acids and not for the purpose of respiration.
6.4. S. pombe mitochondria and NEDD8/Rub1
Similar to C. albicans, the fission yeast S. pombe is also an mtDNA ρ+ Crabtree-positive organism with partial active mitochondria at the fermentation growth phase. However, unlike species of the Saccharomycotina sub-phylum, S. pombe cells respire a particular amount of oxygen during the fermentation growth phase [127]. S. pombe cells treated at the fermentation phase with antimycin A, a potent ETC inhibitor, have a lower biomass, consume more glucose and produce more ethanol than control cells, indicating that the mitochondrial respiratory loss is compensated for by increased alcoholic fermentation. The above suggests that the oxidative phosphorylation of ETC is active during fermentation and that energy gained in the respiration process is required for yeast proliferation and increase of biomass [128]. Indeed, Malecki et al. [128] identified 154 genes in S. pombe that were found to be important for respiration but did not have orthologs in S. cerevisiae, suggesting that the genetic basis for respiratory growth is remarkably distinct between these yeast species. The increased respiration rate at the fermentation phase in S. pombe is associated with low activity of a sole enzyme, pyruvate kinase 1 (Pyk1), a protein that forms a homo-tetramer during glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic respiration (TCA cycle). Accelerated Pyk1 activity restricts the respiration at the fermentation phase and leads to changes in cellular metabolism and physiology, most notably sensitivity to oxidative stress [129]. Mitochondrial respiration accompanied by the formation of ROS in the fermentation phase of S. pombe, correlates with the requirement of NEDD8/Rub1 for viability.
6.5. Mitochondria of the Aspergillus and Neurospora species
A. nidulans and N. crassa are Crabtree-negative filamentous fungal species. They predominantly oxidize pyruvate to carbon dioxide through the TCA cycle, and show little or no repression of the TCA cycle in glucose-rich growth conditions. As a result, mitochondria are essential all the way through [130]. While the CSN and CRLs have been extensively studied in these species, little is known about NEDD8/Rub1 per se [60,71,80,82,131,132]. NEDD8 and the NEDDylation cascade enzymes are required for the viability of A. nidulans [91]. To date, experimental data on NEDD8/Rub1 in N. crassa are not available. However, considering the biology of the CSN/NEDD8 axis in this organism and its aerobic lifestyle, NEDD8 is probably essential for vitality [80].
7. Concluding remarks and perspectives
Access of molecular oxygen to the mitochondria ETC leads to the generation of ROS [[103], [104], [105]]. ROS induces progressive damage to macromolecules, cells and tissues. The cumulative damage could trigger diseases such as inflammation, cancer, cardiac, neurodegeneration and other aging disorders [[106], [107], [108]]. Ascomycota model species offer an opportunity to identify a highly conserved link between metabolic oxidative stress and NEDD8/Rub1essentiality. These species are facultative anaerobic organisms that utilize both anaerobic and aerobic lifestyles for optimal growth and have fundamental adaptations to ecological niches and environmental pressures to which they may be exposed. The vitality of NEDD8/Rub1 in Ascomycota species is consistent with the presence of functional mitochondrial respiration that accompanies ROS production along all stages of growth (Fig. 5).
The NEDDylation cascade mediates the conjugation of NEDD8/Rub1 to cullin and non-cullin substrates. As expected, vitality of the conjugation pathway enzymes correlates with that of NEDD8/Rub1. Of note, orthologs of the ROS-sensitive deNEDDylase NEDP1 appear only in genome of organisms with vital mtDNA and NEDD8/Rub1. Conversely, subunits of the CSN, the only cullin deNEDDylase eraser, are not essential in all three Ascomycota sub-phyla, regardless of the species’ respiration lifestyle. Hence, the seemingly strict function of CSN in regulation of CRL substrate turnover is less conserved than imagined. Why the CSN exists in species of the three sub-phyla if it is not vital and does not significantly regulate CRL activity is unknown. It is possible that the most conserved function of the CSN is to release NEDD8/Rub1 from the cullin “reservoirs” to support the non-cullin function of this modifier. Indeed, CSN activity is constitutive and not sensitive to ROS [119]. Furthermore, the accumulation of cullin-free NEDD8/Rub1 molecules during oxidation is conserved from S. cerevisiae to humans [47,110]; therefore, this function could be considered as a most conserved character of NEDD8/Rub1. In fact, the anaerobic flagellated protozoan parasite Giardia has a relic mitochondrion (mitosome) that lacks mtDNA and has a genome lacking CSN subunits and a clear ortholog for NEDD8/Rub1 [54,133,134]. Most interestingly, reports of recent genome sequencing of an anaerobic microbe Monocercomonoides sp. PA203 (now termed Monocercomonoides exilis), present a compelling case for the complete lack of mitochondrial organelles [135]. The genome of this organism includes a single gene of tri-Ub (AAW22168.1a), but lacks an ortholog for NEDD8/Rub1. This is a first clear case of an organism that has Ub but lacks NEDD8/Rub1.
Despite the considerable research in the field of NEDD8/Rub1, many questions remain unanswered. For example, there is still no clear understanding of how the various biochemical activities attributed to NEDD8/Rub1 translate into its physiological functions in different organisms, particularly mammals. One interesting point is the increased production of ROS in many cancers, which originate from mitochondrial dysfunction [136]. It has been shown that various cancer cells deal with ROS through a metabolic switch between glycolytic and oxidative metabolism in a reversible fashion, analogous to the Crabtree effect, known as the “Warburg effect” [137]. Indeed, information on glucose uptake, glycolytic activity and downregulation of mitochondrial metabolism in Crabtree-positive species corresponds to the analogous Warburg effect in cancer cells. Given the growing interest in the use of NEDD/Rub1 as a therapeutic target for cancer, and the development of MLN-4924 (pevonedistat), a potent inhibitor of NAE [117,138,139], understanding the role of NEDD8/Rub1 in mitochondria metabolism could potentially lead to a better prediction of cancer types, developmental stages, and tumor progression that could potentially be targeted by this inhibitor.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
I would like to thank the ISF grant number 162/2017 for funding the studies in my lab.
References
- 1.Cappadocia L., Lima C.D. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 2017 doi: 10.1021/acs.chemrev.6b00737. 10.1021/acs.chemrev.6b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershko A., Eytan E., Ciechanover A., Haas A.L. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells: relationship to the breakdown of abnormal proteins. J. Biol. Chem. 1982;257:3964–13970. [PubMed] [Google Scholar]
- 4.Smalle J., Vierstra R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 5.Hotton S.K., Callis J. Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- 6.Mahon C., Krogan N.J., Craik C.S., Pick E. Cullin E3 ligases and their rewiring by viral factors. Biomolecules. 2014;4:897–930. doi: 10.3390/biom4040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skaar J.R., Florens L., Tsutsumi T., Arai T., Tron A., Swanson S.K., Washburn M.P., DeCaprio J.A. PARC and CUL7 form atypical cullin RING ligase complexes. Canc. Res. 2007;67:2006–2014. doi: 10.1158/0008-5472.CAN-06-3241. [DOI] [PubMed] [Google Scholar]
- 8.Johnston S.C., Riddle S.M., Cohen R.E., Hill C.P. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linghu B., Callis J., Goebl M.G. Rub1p processing by Yuh1p is required for wild-type levels of Rub1p conjugation to Cdc53p. Eukaryot. Cell. 2002;1:491–494. doi: 10.1128/EC.1.3.491-494.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osaka F., Kawasaki H., Aida N., Saeki M., Chiba T., Kawashima S., Tanaka K., Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hori T., Osaka F., Chiba T., Miyamoto C., Okabayashi K., Shimbara N., Kato S., Tanaka K. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 12.Zhou W., Xu J., Tan M., Li H., Li H., Wei W., Sun Y. UBE2M is a stress-inducible dual E2 for neddylation and ubiquitylation that promotes targeted degradation of UBE2F. Mol. Cell. 2018;70:1008–1024. doi: 10.1016/j.molcel.2018.06.002. e1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott D.C., Monda J.K., Grace C.R., Duda D.M., Kriwacki R.W., Kurz T., Schulman B.A. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott D.C., Monda J.K., Bennett E.J., Harper J.W., Schulman B.A. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawakami T., Chiba T., Suzuki T., Iwai K., Yamanaka K., Minato N., Suzuki H., Shimbara N., Hidaka Y., Osaka F. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duda D.M., Borg L.A., Scott D.C., Hunt H.W., Hammel M., Schulman B.A. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baek K., Krist D.T., Prabu J.R., Hill S., Klugel M., Neumaier L.M., von Gronau S., Kleiger G., Schulman B.A. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature. 2020;578:461–466. doi: 10.1038/s41586-020-2000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei N., Serino G., Deng X.W. The COP9 signalosome: more than a protease. Trends Biochem. Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Lingaraju G.M., Bunker R.D., Cavadini S., Hess D., Hassiepen U., Renatus M., Fischer E.S., Thoma N.H. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 20.Jones J., Wu K., Yang Y., Guerrero C., Nillegoda N., Pan Z.Q., Huang L. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J. Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loftus S.J., Liu G., Carr S.M., Munro S., La Thangue N.B. NEDDylation regulates E2F-1-dependent transcription. EMBO Rep. 2012;13:811–818. doi: 10.1038/embor.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu J., Liu C., Wei R., Xie P., He S., Zhang L. Nedd8 targets ubiquitin ligase Smurf2 for neddylation and promote its degradation. Biochem. Biophys. Res. Commun. 2016;474:51–56. doi: 10.1016/j.bbrc.2016.04.058. [DOI] [PubMed] [Google Scholar]
- 23.Xie P., Zhang M., He S., Lu K., Chen Y., Xing G., Lu Y., Liu P., Li Y., Wang S. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat. Commun. 2014;5:3733. doi: 10.1038/ncomms4733. [DOI] [PubMed] [Google Scholar]
- 24.Mahata B., Sundqvist A., Xirodimas D.P. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene. 2012;31:3060–3071. doi: 10.1038/onc.2011.482. [DOI] [PubMed] [Google Scholar]
- 25.Sundqvist A., Liu G., Mirsaliotis A., Xirodimas D.P. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xirodimas D.P. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 27.Xirodimas D.P., Saville M.K., Bourdon J.C., Hay R.T., Lane D.P. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 28.Xirodimas D.P., Sundqvist A., Nakamura A., Shen L., Botting C., Hay R.T. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9:280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hakenjos J.P., Bejai S., Ranftl Q., Behringer C., Vlot A.C., Absmanner B., Hammes U., Heinzlmeir S., Kuster B., Schwechheimer C. ML3 is a NEDD8- and ubiquitin-modified protein. Plant Physiol. 2013;163:135–149. doi: 10.1104/pp.113.221341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogl A.M., Phu L., Becerra R., Giusti S.A., Verschueren E., Hinkle T.B., Bordenave M.D., Adrian M., Heidersbach A., Yankilevich P. Global site-specific neddylation profiling reveals that NEDDylated cofilin regulates actin dynamics. Nat. Struct. Mol. Biol. 2020;27:210–220. doi: 10.1038/s41594-019-0370-3. [DOI] [PubMed] [Google Scholar]
- 31.Schwechheimer C. NEDD8-its role in the regulation of Cullin-RING ligases. Curr. Opin. Plant Biol. 2018;45:112–119. doi: 10.1016/j.pbi.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 32.Coleman K.E., Bekes M., Chapman J.R., Crist S.B., Jones M.J., Ueberheide B.M., Huang T.T. SENP8 limits aberrant neddylation of NEDD8 pathway components to promote cullin-RING ubiquitin ligase function. Elife. 2017;6 doi: 10.7554/eLife.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan-Erdene T., Nagamalleswari K., Yin L., Wu K., Pan Z.-Q., Wilkinson K.D. Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 2003;278:28892–28900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 34.Enchev R.I., Schulman B.A., Peter M. Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohnsack R.N., Haas A.L. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J. Biol. Chem. 2003;278:26823–26830. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 36.Walden H., Podgorski M.S., Huang D.T., Miller D.W., Howard R.J., Minor D.L., Jr., Holton J.M., Schulman B.A. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 37.Souphron J., Waddell M.B., Paydar A., Tokgoz-Gromley Z., Roussel M.F., Schulman B.A. Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8's E1. Biochemistry. 2008;47:8961–8969. doi: 10.1021/bi800604c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walden H., Podgorski M.S., Schulman B.A. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 39.Whitby F.G., Xia G., Pickart C.M., Hill C.P. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 40.Hjerpe R., Thomas Y., Chen J., Zemla A., Curran S., Shpiro N., Dick L.R., Kurz T. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 2012;441:927–936. doi: 10.1042/BJ20111671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hjerpe R., Thomas Y., Kurz T. NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. J. Mol. Biol. 2012;421:27–29. doi: 10.1016/j.jmb.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 42.Singh R.K., Zerath S., Kleifeld O., Scheffner M., Glickman M.H., Fushman D. Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system. Mol. Cell. Proteomics. 2012;11:1595–1611. doi: 10.1074/mcp.M112.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leidecker O., Matic I., Mahata B., Pion E., Xirodimas D.P. The ubiquitin E1 enzyme Ube 1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle. 2012;11:1142–1150. doi: 10.4161/cc.11.6.19559. [DOI] [PubMed] [Google Scholar]
- 44.Maghames C.M., Lobato-Gil S., Perrin A., Trauchessec H., Rodriguez M.S., Urbach S., Marin P., Xirodimas D.P. NEDDylation promotes nuclear protein aggregation and protects the Ubiquitin Proteasome System upon proteotoxic stress. Nat. Commun. 2018;9:4376. doi: 10.1038/s41467-018-06365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sylvia Zerath Gurevich A.S., Joseph Longworth, Singh Rajesh K., Lemma Betsegaw E., Thakur Anita, Popp Oliver, Kornitzer Daniel, Reis Noa, Scheffner Martin, Dittmar Gunnar, Pick Elah, Fushman David, Glickman Michael H. Rub1/NEDD8, a ubiquitin-like modifier, is also a ubiquitin modifier. bioRxiv. 2020 doi: 10.1101/2020.06.18.159145. [DOI] [Google Scholar]
- 46.Girdwood D., Xirodimas D.P., Gordon C. The essential functions of NEDD8 are mediated via distinct surface regions, and not by polyneddylation in Schizosaccharomyces pombe. PloS One. 2011;6 doi: 10.1371/journal.pone.0020089PONE-D-11-05772. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keuss M.J., Hjerpe R., Hsia O., Gourlay R., Burchmore R., Trost M., Kurz T. Unanchored tri-NEDD8 inhibits PARP-1 to protect from oxidative stress-induced cell death. EMBO J. 2019;38 doi: 10.15252/embj.2018100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailly A.P., Perrin A., Serrano-Macia M., Maghames C., Leidecker O., Trauchessec H., Martinez-Chantar M.L., Gartner A., Xirodimas D.P. The balance between mono- and NEDD8-chains controlled by NEDP1 upon DNA damage is a regulatory module of the HSP70 ATPase activity. Cell Rep. 2019;29:212–224. doi: 10.1016/j.celrep.2019.08.070. e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pick E., Pintard L. A journey to the land of the rising sun with the Cop9/Signalosome and related Zomes. EMBO Rep. 2009;10:343–348. doi: 10.1038/embor.2009.27. Epu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pick E., Hofmann K., Glickman M.H. PCI complexes: beyond the proteasome, CSN, and eIF3 Troika. Mol. Cell. 2009;35:260–264. doi: 10.1016/j.molcel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 51.Braus G.H., Irniger S., Bayram O. Fungal development and the COP9 signalosome. Curr. Opin. Microbiol. 2010;13:672–676. doi: 10.1016/j.mib.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 52.Serino G., Pick E. Duplication and familial promiscuity within the proteasome lid and COP9 signalosome kin complexes. Plant Sci. 2013;203–204:89–97. doi: 10.1016/j.plantsci.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 53.Pick E., Golan A., Zimbler J.Z., Guo L., Sharaby Y., Tsuge T., Hofmann K., Wei N. The minimal deneddylase core of the COP9 signalosome excludes the Csn6 MPN(-) domain. PloS One. 2012;7 doi: 10.1371/journal.pone.0043980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Z., Kleifeld O., Lande-Atir A., Bsoul M., Kleiman M., Krutauz D., Book A., Vierstra R.D., Hofmann K., Reis N. Dual function of Rpn5 in two PCI complexes, the 26S proteasome and COP9 signalosome. Mol. Biol. Cell. 2011;22:911–920. doi: 10.1091/mbc.E10-08-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu H., Reis N., Lee Y., Glickman M.H., Vierstra R.D. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maytal-Kivity V., Piran R., Pick E., Hofmann K., Glickman M.H. COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Rep. 2002;12:1215–1221. doi: 10.1093/embo-reports/kvf235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Busch S., Schwier E.U., Nahlik K., Bayram O., Helmstaedt K., Draht O.W., Krappmann S., Valerius O., Lipscomb W.N., Braus G.H. An eight-subunit COP9 signalosome with an intact JAMM motif is required for fungal fruit body formation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8089–8094. doi: 10.1073/pnas.0702108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosel D., Kimmel A.R. The COP9 signalosome regulates cell proliferation of Dictyostelium discoideum. Eur. J. Cell Biol. 2006;85:1023–1034. doi: 10.1016/j.ejcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 59.Mundt K.E., Porte J., Murray J.M., Brikos C., Christensen P.U., Caspari T., Hagan I.M., Millar J.B., Simanis V., Hofmann K. The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr. Biol. 1999;9:1427–1430. doi: 10.1016/s0960-9822(00)80091-3. [DOI] [PubMed] [Google Scholar]
- 60.He Q., Cheng P., He Q., Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 2005;19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinha A., Israeli R., Cirigliano A., Gihaz S., Trabelcy B., Braus G.H., Gerchman Y., Fishman A., Negri R., Rinaldi T. The COP9 signalosome mediates the Spt23 regulated fatty acid desaturation and ergosterol biosynthesis. Faseb. J. 2020 doi: 10.1096/fj.201902487R. 10.1096/fj.201902487R. [DOI] [PubMed] [Google Scholar]
- 62.Cavadini S., Fischer E.S., Bunker R.D., Potenza A., Lingaraju G.M., Goldie K.N., Mohamed W.I., Faty M., Petzold G., Beckwith R.E. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature. 2016;531:598–603. doi: 10.1038/nature17416. [DOI] [PubMed] [Google Scholar]
- 63.Rabut G., Le Dez G., Verma R., Makhnevych T., Knebel A., Kurz T., Boone C., Deshaies R.J., Peter M. The TFIIH subunit Tfb3 regulates cullin neddylation. Mol. Cell. 2011;43:488–495. doi: 10.1016/j.molcel.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dubiel W., Chaithongyot S., Dubiel D., Naumann M. The COP9 signalosome: a multi-DUB complex. Biomolecules. 2020;10 doi: 10.3390/biom10071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A.F., Tanaka K., Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 66.Yang X., Menon S., Lykke-Andersen K., Tsuge T., Di X., Wang X., Rodriguez-Suarez R.J., Zhang H., Wei N. The COP9 signalosome inhibits p27(kip 1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr. Biol. 2002;12:667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- 67.Zhou C., Wee S., Rhee E., Naumann M., Dubiel W., Wolf D.A. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol. Cell. 2003;11:927–938. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 68.Enchev R.I., Scott D.C., da Fonseca P.C., Schreiber A., Monda J.K., Schulman B.A., Peter M., Morris E.P. Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2012 doi: 10.1016/j.celrep.2012.08.019. 10.1016/j.celrep.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mosadeghi R., Reichermeier K.M., Winkler M., Schreiber A., Reitsma J.M., Zhang Y., Stengel F., Cao J., Kim M., Sweredoski M.J. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. Elife. 2016;5 doi: 10.7554/eLife.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bornstein G., Ganoth D., Hershko A. Regulation of neddylation and deneddylation of cullin 1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11515–11520. doi: 10.1073/pnas.0603921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Z., Wang Y., Cai G., He Q. Neurospora COP9 signalosome integrity plays major roles for hyphal growth, conidial development, and circadian function. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt M.W., McQuary P.R., Wee S., Hofmann K., Wolf D.A. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol. Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peth A., Berndt C., Henke W., Dubiel W. Downregulation of COP9 signalosome subunits differentially affects the CSN complex and target protein stability. BMC Biochem. 2007;8:27. doi: 10.1186/1471-2091-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wee S., Geyer R.K., Toda T., Wolf D.A. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 2005;7:387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- 75.Cope G.A., Deshaies R.J. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Denti S., Fernandez-Sanchez M.E., Rogge L., Bianchi E. The COP9 signalosome regulates Skp 2 levels and proliferation of human cells. J. Biol. Chem. 2006;281:32188–32196. doi: 10.1074/jbc.M604746200. [DOI] [PubMed] [Google Scholar]
- 77.Fischer E.S., Scrima A., Bohm K., Matsumoto S., Lingaraju G.M., Faty M., Yasuda T., Cavadini S., Wakasugi M., Hanaoka F. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 78.Luke-Glaser S., Roy M., Larsen B., Le Bihan T., Metalnikov P., Tyers M., Peter M., Pintard L. CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo. Mol. Cell Biol. 2007;27:4526–4540. doi: 10.1128/MCB.01724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu C., Powell K.A., Mundt K., Wu L., Carr A.M., Caspari T. Cop9/signalosome subunits and Pcu 4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 2003;17:1130–1140. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J., Hu Q., Chen H., Zhou Z., Li W., Wang Y., Li S., He Q. Role of individual subunits of the Neurospora crassa CSN complex in regulation of deneddylation and stability of cullin proteins. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilmes G.M., Bergkessel M., Bandyopadhyay S., Shales M., Braberg H., Cagney G., Collins S.R., Whitworth G.B., Kress T.L., Weissman J.S. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beckmann E.A., Kohler A.M., Meister C., Christmann M., Draht O.W., Rakebrandt N., Valerius O., Braus G.H. Integration of the catalytic subunit activates deneddylase activity in vivo as final step in fungal COP9 signalosome assembly. Mol. Microbiol. 2015;97:110–124. doi: 10.1111/mmi.13017. [DOI] [PubMed] [Google Scholar]
- 83.Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D.A., Wei N., Shevchenko A., Deshaies R.J. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 84.Zemla A., Thomas Y., Kedziora S., Knebel A., Wood N.T., Rabut G., Kurz T. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat. Commun. 2013;4:1641. doi: 10.1038/ncomms2628. [DOI] [PubMed] [Google Scholar]
- 85.Su H., Li J., Osinska H., Li F., Robbins J., Liu J., Wei N., Wang X. The COP9 signalosome is required for autophagy, proteasome-mediated proteolysis, and cardiomyocyte survival in adult mice. Circ Heart Fail. 2013;6:1049–1057. doi: 10.1161/CIRCHEARTFAILURE.113.000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao P., Wang C., Li J., Su H., Yang L., Wu P., Lewno M.T., Liu J., Wang X. COP9 signalosome suppresses RIPK1-RIPK3-mediated cardiomyocyte necroptosis in mice. Circ Heart Fail. 2020;13 doi: 10.1161/CIRCHEARTFAILURE.120.006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li S.-J., Hochstrasser M. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 2003;160:1069–1082. doi: 10.1083/jcb.200212052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mendoza H.M., Shen L.-n., Botting C., Lewis A., Chen J., Ink B., Hay R.T. NEDP1, a highly conserved cysteine protease that deNEDDylates cullins. J. Biol. Chem. 2003;278:25637–25643. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 89.Mergner J., Kuster B., Schwechheimer C. DENEDDYLASE1 protein counters automodification of neddylating enzymes to maintain NEDD8 protein homeostasis in Arabidopsis. J. Biol. Chem. 2017;292:3854–3865. doi: 10.1074/jbc.M116.767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O'Donoghue J.E., Bech-Otschir D., Larsen I.B., Wallace M., Hartmann-Petersen R., Gordon C. Nedd8 processing enzymes in Schizosaccharomyces pombe. BMC Biochem. 2013;14:8. doi: 10.1186/1471-2091-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Christmann M., Schmaler T., Gordon C., Huang X., Bayram O., Schinke J., Stumpf S., Dubiel W., Braus G.H. Control of multicellular development by the physically interacting deneddylases DEN1/DenA and COP9 signalosome. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sela N., Atir-Lande A., Kornitzer D. Neddylation and CAND1 independently stimulate SCF ubiquitin ligase activity in Candida albicans. Eukaryot. Cell. 2012;11:42–52. doi: 10.1128/EC.05250-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Whitby F.G., Xia G., Pickart C.M., Hill C.P. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 94.Buser R., Kellner V., Melnik A., Wilson-Zbinden C., Schellhaas R., Kastner L., Piwko W., Dees M., Picotti P., Maric M. The replisome-coupled E3 ubiquitin ligase Rtt101Mms22 counteracts Mrc1 function to tolerate genotoxic stress. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoffman C.S., Wood V., Fantes P.A. An ancient yeast for young geneticists: a primer on the Schizosaccharomyces pombe model system. Genetics. 2015;201:403–423. doi: 10.1534/genetics.115.181503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Osaka F., Saeki M., Katayama S., Aida N., Toh-E A., Kominami K., Toda T., Suzuki T., Chiba T., Tanaka K. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yamoah K., Oashi T., Sarikas A., Gazdoiu S., Osman R., Pan Z.Q. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Furukawa K., Mizushima N., Noda T., Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 2000;275:7462–7465. doi: 10.1074/jbc.275.11.7462. [DOI] [PubMed] [Google Scholar]
- 99.Zhang X., Zhang Y.L., Qiu G., Pian L., Guo L., Cao H., Liu J., Zhao Y., Li X., Xu Z. Hepatic neddylation targets and stabilizes electron transfer flavoproteins to facilitate fatty acid beta-oxidation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2473–2483. doi: 10.1073/pnas.1910765117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ryu J.H., Li S.H., Park H.S., Park J.W., Lee B., Chun Y.S. Hypoxia-inducible factor alpha subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J. Biol. Chem. 2011;286:6963–6970. doi: 10.1074/jbc.M110.188706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guan J., Yu S., Zheng X. NEDDylation antagonizes ubiquitination of proliferating cell nuclear antigen and regulates the recruitment of polymerase eta in response to oxidative DNA damage. Protein Cell. 2018;9:365–379. doi: 10.1007/s13238-017-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cirigliano A., Macone A., Bianchi M.M., Oliaro-Bosso S., Balliano G., Negri R., Rinaldi T. Ergosterol reduction impairs mitochondrial DNA maintenance in S. cerevisiae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:290–303. doi: 10.1016/j.bbalip.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 103.Starkov A.A. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N. Y. Acad. Sci. 2008;1147:37–52. doi: 10.1196/annals.1427.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lushchak V.I. Adaptive response to oxidative stress: bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011;153:175–190. doi: 10.1016/j.cbpc.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 105.McCord J.M. The evolution of free radicals and oxidative stress. Am. J. Med. 2000;108:652–659. doi: 10.1016/s0002-9343(00)00412-5. [DOI] [PubMed] [Google Scholar]
- 106.Benhar M., Engelberg D., Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3:420–425. doi: 10.1093/embo-reports/kvf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Devine M.J., Plun-Favreau H., Wood N.W. Parkinson's disease and cancer: two wars, one front. Nat. Rev. Canc. 2011;11:812–823. doi: 10.1038/nrc3150. [DOI] [PubMed] [Google Scholar]
- 108.Chen Z., Zhong C. Decoding Alzheimer's disease from perturbed cerebral glucose metabolism: implications for diagnostic and therapeutic strategies. Prog. Neurobiol. 2013;108:21–43. doi: 10.1016/j.pneurobio.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 109.Pfeiffer T., Morley A. An evolutionary perspective on the Crabtree effect. Front Mol Biosci. 2014;1:17. doi: 10.3389/fmolb.2014.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bramasole L., Sinha A., Gurevich S., Radzinski M., Klein Y., Panat N., Gefen E., Rinaldi T., Jimenez-Morales D., Johnson J. Proteasome lid bridges mitochondrial stress with Cdc53/Cullin1 NEDDylation status. Redox Biol. 2019;20:533–543. doi: 10.1016/j.redox.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Postma E., Verduyn C., Scheffers W.A., Van Dijken J.P. Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1989;55:468–477. doi: 10.1128/AEM.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brauer M.J., Saldanha A.J., Dolinski K., Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maris A.F., Assumpcao A.L., Bonatto D., Brendel M., Henriques J.A. Diauxic shift-induced stress resistance against hydroperoxides in Saccharomyces cerevisiae is not an adaptive stress response and does not depend on functional mitochondria. Curr. Genet. 2001;39:137–149. doi: 10.1007/s002940100194. [DOI] [PubMed] [Google Scholar]
- 114.Soontorngun N. Reprogramming of nonfermentative metabolism by stress-responsive transcription factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 2016 doi: 10.1007/s00294-016-0609-z. 10.1007/s00294-016-0609-z. [DOI] [PubMed] [Google Scholar]
- 115.Day M. Yeast petites and small colony variants: for everything there is a season. Adv. Appl. Microbiol. 2013;85:1–41. doi: 10.1016/B978-0-12-407672-3.00001-0. [DOI] [PubMed] [Google Scholar]
- 116.Kumar A., Wu H., Collier-Hyams L.S., Kwon Y.M., Hanson J.M., Neish A.S. The bacterial fermentation product butyrate influences epithelial signaling via reactive oxygen species-mediated changes in cullin-1 neddylation. J. Immunol. 2009;182:538–546. doi: 10.4049/jimmunol.182.1.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Downs C.A., Kumar A., Kreiner L.H., Johnson N.M., Helms M.N. H2O2 regulates lung epithelial sodium channel (ENaC) via ubiquitin-like protein Nedd8. J. Biol. Chem. 2013;288:8136–8145. doi: 10.1074/jbc.M112.389536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Livnat-Levanon N., Kevei E., Kleifeld O., Krutauz D., Segref A., Rinaldi T., Erpapazoglou Z., Cohen M., Reis N., Hoppe T. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014;7:1371–1380. doi: 10.1016/j.celrep.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 119.Bramasole L., Sinha A., Harshuk D., Cirigliano A., Gurevich S., Yu Z., Carmeli R.L., Glickman M.H., Rinaldi T., Pick E. The proteasome lid triggers COP9 signalosome activity during the transition of Saccharomyces cerevisiae cells into quiescence. Biomolecules. 2019;9 doi: 10.3390/biom9090449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Niimi M., Kamiyama A., Tokunaga M. Respiration of medically important Candida species and Saccharomyces cerevisiae in relation to glucose effect. J. Med. Vet. Mycol. 1988;26:195–198. [PubMed] [Google Scholar]
- 121.Sun N., Parrish R.S., Calderone R.A., Fonzi W.A. Unique, diverged, and conserved mitochondrial functions influencing Candida albicans respiration. mBio. 2019;10 doi: 10.1128/mBio.00300-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sudbery P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 123.Land G.A., McDonald W.C., Stjernholm R.L., Friedman T.L. Factors affecting filamentation in Candida albicans: relationship of the uptake and distribution of proline to morphogenesis. Infect. Immun. 1975;11:1014–1023. doi: 10.1128/IAI.11.5.1014-1023.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Silao F.G.S., Ward M., Ryman K., Wallstrom A., Brindefalk B., Udekwu K., Ljungdahl P.O. Mitochondrial proline catabolism activates Ras 1/cAMP/PKA-induced filamentation in Candida albicans. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1007976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Van Ende M., Wijnants S., Van Dijck, Sugar P. Sensing and Signaling in Candida albicans and Candida glabrata. Front. Microbiol. 2019;10:99. doi: 10.3389/fmicb.2019.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rodaki A., Bohovych I.M., Enjalbert B., Young T., Odds F.C., Gow N.A., Brown A.J. Glucose promotes stress resistance in the fungal pathogen Candida albicans. Mol. Biol. Cell. 2009;20:4845–4855. doi: 10.1091/mbc.E09-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Heslot H., Goffeau A., Louis C. Respiratory metabolism of a "petite negative"yeast Schizosaccharomyces pombe 972h. J. Bacteriol. 1970;104:473–481. doi: 10.1128/JB.104.1.473-481.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Malecki M., Bitton D.A., Rodriguez-Lopez M., Rallis C., Calavia N.G., Smith G.C., Bahler J. Functional and regulatory profiling of energy metabolism in fission yeast. Genome Biol. 2016;17:240. doi: 10.1186/s13059-016-1101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kamrad S., Grossbach J., Rodriguez-Lopez M., Mulleder M., Townsend S., Cappelletti V., Stojanovski G., Correia-Melo C., Picotti P., Beyer A. Pyruvate kinase variant of fission yeast tunes carbon metabolism, cell regulation, growth and stress resistance. Mol. Syst. Biol. 2020;16 doi: 10.15252/msb.20199270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chambergo F.S., Bonaccorsi E.D., Ferreira A.J., Ramos A.S., Ferreira Junior J.R., Abrahao-Neto J., Farah J.P., El-Dorry H. Elucidation of the metabolic fate of glucose in the filamentous fungus Trichoderma reesei using expressed sequence tag (EST) analysis and cDNA microarrays. J. Biol. Chem. 2002;277:13983–13988. doi: 10.1074/jbc.M107651200. [DOI] [PubMed] [Google Scholar]
- 131.von Zeska Kress M.R., Harting R., Bayram O., Christmann M., Irmer H., Valerius O., Schinke J., Goldman G.H., Braus G.H. The COP9 signalosome counteracts the accumulation of cullin SCF ubiquitin E3 RING ligases during fungal development. Mol. Microbiol. 2012 doi: 10.1111/j.1365-2958.2012.07999.x. 10.1111/j.1365-2958.2012.07999.x. [DOI] [PubMed] [Google Scholar]
- 132.Busch S., Eckert S.E., Krappmann S., Braus G.H. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2003;49:717–730. doi: 10.1046/j.1365-2958.2003.03612.x. [DOI] [PubMed] [Google Scholar]
- 133.Fort P., Kajava A.V., Delsuc F., Coux O. Evolution of proteasome regulators in eukaryotes. Genome Biol Evol. 2015;7:1363–1379. doi: 10.1093/gbe/evv068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jedelsky P.L., Dolezal P., Rada P., Pyrih J., Smid O., Hrdy I., Sedinova M., Marcincikova M., Voleman L., Perry A.J. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PloS One. 2011;6 doi: 10.1371/journal.pone.0017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Karnkowska A., Treitli S.C., Brzon O., Novak L., Vacek V., Soukal P., Barlow L.D., Herman E.K., Pipaliya S.V., Panek T. The oxymonad genome displays canonical eukaryotic complexity in the absence of a mitochondrion. Mol. Biol. Evol. 2019;36:2292–2312. doi: 10.1093/molbev/msz147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ahmad I.M., Aykin-Burns N., Sim J.E., Walsh S.A., Higashikubo R., Buettner G.R., Venkataraman S., Mackey M.A., Flanagan S.W., Oberley L.W. Mitochondrial O2*- and H2O2 mediate glucose deprivation-induced stress in human cancer cells. J. Biol. Chem. 2005;280:4254–4263. doi: 10.1074/jbc.M411662200. [DOI] [PubMed] [Google Scholar]
- 137.Diaz-Ruiz R., Rigoulet M., Devin A. The Warburg and Crabtree effects: on the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim. Biophys. Acta. 2011;1807:568–576. doi: 10.1016/j.bbabio.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 138.Oladghaffari M., Islamian J.P., Baradaran B., Monfared A.S. MLN4924 therapy as a novel approach in cancer treatment modalities. J. Chemother. 2016;28:74–82. doi: 10.1179/1973947815Y.0000000066. [DOI] [PubMed] [Google Scholar]
- 139.Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., Brownell J.E., Burke K.E., Cardin D.P., Critchley S. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 140.Dohmann E.M., Kuhnle C., Schwechheimer C. Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell. 2005;17:1967–1978. doi: 10.1105/tpc.105.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gusmaroli G., Feng S., Deng X.W. The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell. 2004;16:2984–3001. doi: 10.1105/tpc.104.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang H., Quint M., Gray W.M. The eta 7/csn3-3 auxin response mutant of Arabidopsis defines a novel function for the CSN3 subunit of the COP9 signalosome. PloS One. 2013;8 doi: 10.1371/journal.pone.0066578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu Q., Zhou Y., Tang R., Wang X., Hu Q., Wang Y., He Q. Increasing the unneddylated Cullin1 portion rescues the csn phenotypes by stabilizing adaptor modules to drive SCF assembly. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou C., Seibert V., Geyer R., Rhee E., Lyapina S., Cope G., Deshaies R.J., Wolf D.A. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2001;2:7. doi: 10.1186/1471-2091-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Menon S., Tsuge T., Dohmae N., Takio K., Wei N. Association of SAP130/SF3b-3 with Cullin-RING ubiquitin ligase complexes and its regulation by the COP9 signalosome. BMC Biochem. 2008;9:1. doi: 10.1186/1471-2091-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Watts F.Z. SUMO modification of proteins other than transcription factors. Semin. Cell Dev. Biol. 2004;15:211–220. doi: 10.1016/j.semcdb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 147.Zhou L., Watts F.Z. Nep1, a Schizosaccharomyces pombe deneddylating enzyme. Biochem. J. 2005;389:307–314. doi: 10.1042/BJ20041991. [DOI] [PMC free article] [PubMed] [Google Scholar]