Abstract
Transcription of human papillomavirus (HPV) genes proceeds unidirectionally from multiple promoters. Direct profiling of transcription start sites (TSSs) by Cap Analysis Gene Expression (CAGE) is a powerful strategy for examining individual HPV promoter activity. The objective of this study was to evaluate alterations of viral promoter activity during infection using CAGE technology. We used CAGE-based sequencing of 46 primary cervical samples, and quantitatively evaluated TSS patterns in the HPV transcriptome at a single-nucleotide resolution. TSS patterns were classified into two types: early promoter-dominant type (Type A) and late promoter-dominant type (Type B). The Type B pattern was more frequently found in CIN1 and CIN2 lesions than in CIN3 and cancer samples. We detected transcriptomes from multiple HPV types in five samples. Interestingly, in each sample, the TSS patterns of both HPV types were the same. The viral gene expression pattern was determined by the differentiation status of the epithelial cells, regardless of HPV type. We performed unbiased analyses of TSSs across the HPV genome in clinical samples. Visualising TSS pattern dynamics, including TSS shifts, provides new insights into how HPV infection status relates to disease state.
Subject terms: Infectious-disease diagnostics, Virology, Cervical cancer, Biotechnology, Cancer, Computational biology and bioinformatics, Microbiology, Oncology
Introduction
Uterine cervical cancer is the second most commonly diagnosed cancer, and the third leading cause of mortality among women in developed countries1. Persistent infection with high-risk human papillomaviruses (HR-HPVs) is the main cause of cancer development2–4. During the last two decades, HPV-induced carcinogenesis has been extensively studied. The HPV-derived E6 and E7 oncoproteins inactivate the p53 and pRb tumour suppressor proteins, respectively, which results in resistance to apoptosis and promotion of cell proliferation. Continuous high expression of E6 and E7 is the most important factor in cervical cancer progression5,6.
HPV transcription is unidirectional and generates numerous viral transcripts via differential RNA splicing. At least 13 transcripts from eight HPV genes were identified in HPV-16–infected W12E cells7. These transcripts overlap, complicating the evaluation of expression levels of each transcript. There are two major promoters in the HPV genome: the early promoter, located in the long control region (LCR); and the late promoter, located in the E7 gene downstream of the early promoter. The early promoter controls the expression of E6 and E7, while the late promoter regulates the expression of E1, E2, E4, and E5 (which are important for cell differentiation and viral replication), as well as the expression of L1 and L2 capsid protein genes8. The activity of these promoters is regulated by cellular transcription factors, or by the epigenetic alteration of the viral genome.
A transcriptome profiling method known as Cap Analysis Gene Expression (CAGE)9 can be used to determine the 5′-terminal sequence of RNA, allowing for promoter detection and quantitative measurement of promoter activity. The two main CAGE protocols currently used are no-amplification non-tagging (nAnTi)-CAGE10, which does not involve PCR amplification, and nanoCAGE11, which is designed to process samples that yield nanograms of RNA. nanoCAGE is based on PCR amplification, with the PCR bias removed through the use of unique molecular identifiers12. We previously reported that nAnTi-CAGE may be used to identify precise transcription start sites (TSSs) in the HPV genome, and have used this technology to quantify the activity of multiple promoters in three cell lines and one patient sample13. Direct evaluation of TSSs may represent a novel diagnostic strategy to assess HPV infection status and disease progression.
HPV genes are differentially expressed in parallel with the differentiation programme of the cervical epithelium. At the initial stage of HPV infection, the copy number of the viral genome in cells in the basal layer of the cervical epithelium is very low. Viral DNA replication proceeds along with epithelial differentiation14,15. In the upper epithelial layers, the viral late genes L1 and L2 are expressed to allow viral capsid assembly, packaging, and shedding from the superficial layer of the epithelium. As the viral late gene expression is promoted, E2 suppresses the activity of the early promoter by binding to the E2 binding sites (E2BS) of the LCR16–18. Thus, in the late stages of epithelial differentiation, HPV early promoter activity is relatively suppressed. As the severity of cervical intraepithelial neoplasia (CIN) increases, sustained high expression of E6 and E7 is driven by the early promoter, and, conversely, L1 gene expression is suppressed19. Several methods have been devised to evaluate the expression of late genes, such as L1 or E4, as biomarkers for CIN progression19–25. Precise evaluation of the late gene expression patterns could support their use as novel biomarkers for cervical cancer progression.
In this context, we propose that a quantitative assessment of promoter activity, by evaluating TSS activity, would allow for classification of HPV status, as well as CIN severity. In the present study, we developed a novel approach for the evaluation of differences of viral promoter activity at the single-nucleotide level using CAGE technology in clinical HPV samples.
Results
HPV TSS patterns of cervical lesions
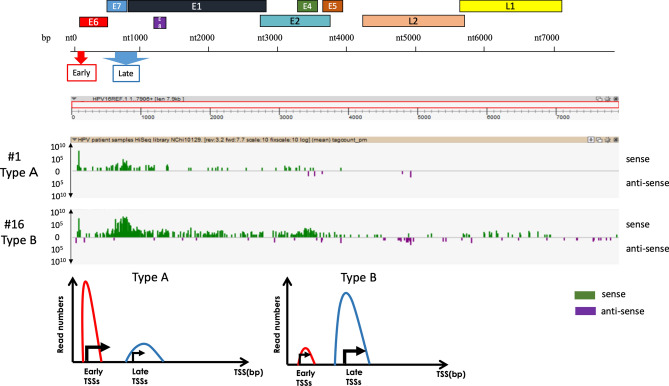
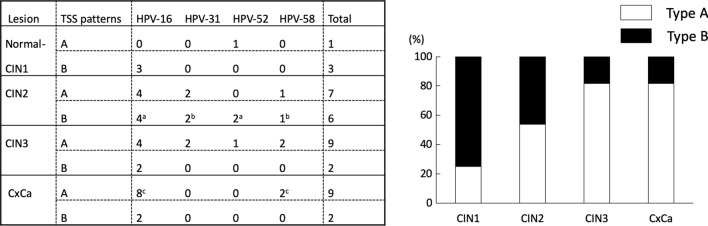
Forty-six cervical lesions, from normal and cancerous lesions, were analysed by nAnTi-CAGE or nanoCAGE (9 for nAnTi-CAGE and 37 for nanoCAGE). As the principle of both nAnTi-CAGE and nanoCAGE is highly similar, we first performed nAnTi-CAGE analysis for 9 samples, and we used 37 samples for nanoCAGE analysis, which is a novel technology developed after nAnTi-CAGE to meaningfully observe TSS pattern dynamics with CAGE analysis. The HPV TSS patterns were classified into broad TSS types. First, we visualised TSS activity at a single-nucleotide level using ZENBU software26. We identified two TSS patterns when focusing on the most activated TSS clusters: the early promoter-activated pattern and the late promoter-activated pattern, which were designated Type A and Type B, respectively. To analyse multiple HPV subtypes in parallel, we defined broad windows containing the early and late promoters in any HPV genome: from nucleotide 80 to 110, and from nucleotide 600 to 950, respectively. We discovered TSS patterns indicative of the early and late promoters, and we subsequently refined the TSS pattern definitions so that Type A included samples where one-third of early promoter activity ≥ late promoter activity; Type B, one-third of early promoter activity < late promoter activity (Fig. 1). The cervical lesion grades and corresponding HPV TSS types are summarised in Fig. 2 and Table 1. Type B was more common in CIN2 or CIN1 than in other samples, while CIN3 or cancerous lesions were predominantly Type A (chi-square test, p = 0.0224), and the observed frequency of Type B decreased with CIN progression (Cochran–Armitage test, p = 0.0208).
Figure 1.
nAnTi-CAGE and nanoCAGE TSSs of HPV-positive cervical lesions. Forty-six cervical lesions, representing both normal and cancerous lesions, were analysed by nAnTi-CAGE or nanoCAGE (9 for nAnTi-CAGE and 39 for nanoCAGE). The HPV TSS patterns were investigated and classified by the prominent TSS types. Regardless of the HPV type, early and late promoter activity was defined by the numbers of TSSs in each transcriptome that started either between nucleotides 80 and 110, or between nucleotides 600 and 950. The TSS patterns were defined as follows: Type A, one-third of early promoter activity ≥ late promoter activity; Type B, one-third of early promoter activity < late promoter activity. To visualise TSS levels at the single-nucleotide level, nanoCAGE data were visualised using ZENBU software. Representative data for each TSS pattern are shown. TSS transcription start site.
Figure 2.
nAnTi-CAGE and nanoCAGE TSSs of HPV-positive cervical lesions. Thirty-nine cervical lesions with HPV-16, HPV-31, HPV-52, and/or HPV-58 infections, isolated from normal and cancerous lesions, were analysed by nAnTi-CAGE or nanoCAGE. The HPV TSS patterns were investigated and classified according to the prominent TSS types. Regardless of the HPV strain, early and late promoter activity was defined by the numbers of TSSs in each transcriptome that started either between nucleotides 80 and 110, or between nucleotides 600 and 950. The TSS patterns were defined as follows: Type A, one-third of early promoter activity ≥ late promoter activity; Type B, one-third of early promoter activity < late promoter activity. The TSS patterns of HPV-positive cervical lesions along with HPV type and cervical lesion status are summarised. aTwo samples were co-infected with HPV-16 and HPV-52. bOne sample was co-infected with HPV-31 and HPV-58. cOne sample was co-infected with HPV-16 and HPV-58. HPV human papillomavirus; TSS transcription start site; CIN cervical intraepithelial neoplasia; CxCa cervical cancer.
Table 1.
Summary of clinical information for patients with cervical lesions.
| Sample ID | Age (yr) | Stage | 1st HPV | 2nd HPV | TSS pattern | Early | Late | Early | Late |
|---|---|---|---|---|---|---|---|---|---|
| #34 | 37 | NILM | 52 | Type A | 1048 | 48 | – | – | |
| #40 | 49 | NILM | – | – | – | – | – | – | |
| #25 | 28 | NILM | 16 | Type B | 2 | 3 | – | – | |
| #13 | 29 | CIN1 | 16 | Type B | 2 | 3 | – | – | |
| C1072_ATG | 37 | CIN1 | 16 | Type B | 441 | 17,349 | – | – | |
| #4 | 36 | CIN1-2 | 31 | Type A | 71 | 1 | – | – | |
| #30 | 43 | CIN1-2 | 52 | 16 | Type B | 1497 | 1355 | 0 | 6 |
| #6 | 32 | CIN2 | 31 | 58 | Type B | 1299 | 1422 | 33 | 41 |
| #7 | 36 | CIN2 | 31 | Type A | 28 | 5 | – | – | |
| #8 | 37 | CIN2 | 58 | Type A | 987 | 130 | – | – | |
| #9 | 34 | CIN2 | 16 | Type A | 200 | 9 | – | – | |
| #16 | 27 | CIN2 | 16 | Type B | 635 | 7107 | – | – | |
| #23 | 40 | CIN2 | 16 | Type A | 314 | 18 | – | – | |
| #24 | 32 | CIN2 | 16 | Type B | 157 | 303 | – | – | |
| #26 | 36 | CIN2 | – | – | – | – | – | – | |
| #29 | 38 | CIN2 | 31 | Type B | 895 | 352 | – | – | |
| #33 | 39 | CIN2 | 16 | Type A | 204 | 23 | – | – | |
| #41 | 45 | CIN2 | 16 | Type A | 879 | 37 | – | – | |
| C1072_ACG | 43 | CIN1-2 | 52 | 16 | Type B | 439 | 12,827 | 9 | 75 |
| #20 | 42 | CIN2-3 | – | – | – | – | – | – | |
| #37 | 44 | CIN2-3 | 16 | Type B | 417 | 1447 | – | – | |
| #1 | 36 | CIN3 | 16 | Type A | 832 | 124 | – | – | |
| #3 | 30 | CIN3 | 31 | Type A | 17 | 0 | – | – | |
| #12 | 66 | CIN3 | 67 | 58 | Type A | 558 | 25 | 54 | 0 |
| #17 | 47 | CIN3 | – | – | – | – | – | – | |
| #18 | 43 | CIN3 | 52 | Type A | 614 | 42 | – | – | |
| #19 | 42 | CIN3 | 58 | Type A | 533 | 38 | – | – | |
| #21 | 35 | CIN3 | 31 | Type A | 80 | 7 | – | – | |
| #22 | 42 | CIN3 | 16 | Type A | 310 | 27 | – | – | |
| #39 | 78 | CIN3 | 16 | Type A | 1932 | 167 | – | – | |
| #42 | 61 | CIN3 | 16 | Type A | 238 | 31 | – | – | |
| C1072_GCT | 30 | CIN3 | 16 | Type B | 218 | 269 | – | – | |
| #2 | 31 | AIS | 18 | Type B | 7 | 26 | – | – | |
| #27 | 28 | CxCa | 16 | 58 | Type A | 6 | 2 | 5 | 0 |
| #11 | 74 | CxCa | 16 | Type A | 643 | 99 | – | – | |
| #5 | 33 | CxCa | 16 | Type A | 277 | 40 | – | – | |
| #32 | 37 | CxCa | 18 | Type B | 77 | 58 | – | – | |
| #14 | 62 | CxCa | 58 | Type A | 6684 | 198 | – | – | |
| #38 | 32 | CxCa | 16 | Type B | 288 | 112 | – | – | |
| #43 | 55 | CxCa | – | – | – | – | – | – | |
| C1059_ACC | 63 | CxCa | 16 | Type A | 635 | 114 | – | – | |
| C1059_ATG | 44 | CxCa | 16 | Type A | 4939 | 126 | – | – | |
| C1059_ACG | 59 | CxCa | 16 | Type B | 3384 | 2224 | – | – | |
| C1065_CAC | 33 | CxCa | 16 | Type A | 3253 | 117 | – | – | |
| C1065_GCG | 35 | CxCa | 16 | Type A | 1855 | 152 | – | – | |
| C1065_ATG | 67 | CxCa | 16 | Type A | 1097 | 18 | – | – |
Forty-six cervical lesions, from normal and cancerous lesions, were analyzed by nAnTi-CAGE or nanoCAGE (9 for nAnTi-CAGE and 37 for nanoCAGE). The HPV TSS patterns were classified into broad TSS types: the early promoter-activated pattern and the late promoter-activated pattern, which were designated Type A and Type B, respectively. We defined broad windows containing the early and late promoters in any HPV genome: from nucleotide 80 to 110, and from nucleotide 600 to 950, respectively. TSS patterns were defined as follows: Type A, one-third of early promoter activity ≥ late promoter activity: and Type B, one-third of early promoter activity < late promoter activity.
AIS adenocarcinoma in situ; CIN cervical intraepithelial neoplasia; CxCa cervical cancer; NILM negative for intraepithelial lesion malignancy; TSS transcription start site.
We then investigated whether the initial observation of multiple TSS patterns would be supported by a more systematic approach. We fitted Gaussian mixture models27 to investigate the accuracy of the classified HPV-derived TSS types. Among 37 samples analyzed by nano-CAGE, 33 samples of which HPV-derived TSS was detected were included in this study. Of them, 2 samples were co-infected with two HPV genotypes. Thirty-five HPV-derived TSS types were classified by Gaussian mixture models, and compared to the types of HPV-derived TSSs classified according to the averaged difference in expression between the early and late promoters, defined as (early – late)/(early + late). The model with the highest likelihood was univariate, with two components and unequal variance: this corresponded closely to Type A and the union of Type B, since only one Type A sample (sample #27) was classified as Type B (Fig. 3).
Figure 3.
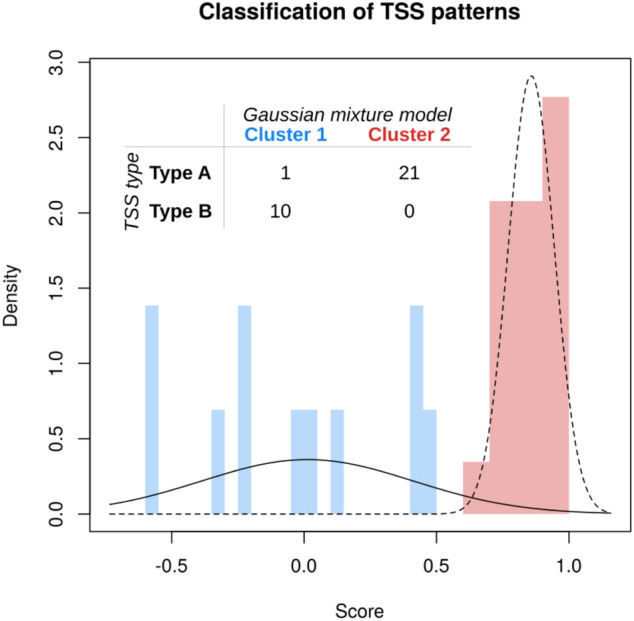
Classification of TSS patterns by Gaussian mixture models. The score was defined as the average difference between the expression levels of the early and late promoters, and a Gaussian mixture model was fitted to these scores. The model with the highest likelihood was univariate, with two components and unequal variance. The TSS patterns were classified according to the averaged difference in expression between the early and late promoters, defined as (early − late)/(early + late). TSS transcription start site.
TSS patterns in multiple infections
We detected transcriptomes of multiple HPV types in five samples in the current study (Table 1). The following co-infections were observed: HPV-16 and HPV-52 (samples C1072_ACG and #30); HPV-31 and HPV-58 (sample #6); HPV-16 and HPV-58 (sample #27); and HPV-67 and HPV-58 (sample #12). Interestingly, the TSS patterns of both detected HPV types were the same in each sample (Fig. 4). Furthermore, a dominant HPV type was apparent in each case of co-infection (Fig. 4 and Table 1).
Figure 4.
TSS patterns of cervical lesions with HPV co-infection. (a) nAnTi-CAGE analysis of a CIN sample co-infected with HPV-16 and HPV-52. To visualise TSS activity at a single-nucleotide level, nAnTi-CAGE data were visualised using ZENBU software. (b) nanoCAGE analysis of a CIN sample co-infected with HPV-67 and HPV-58. To visualise TSS activity at a single-nucleotide level, nanoCAGE data were visualised using ZENBU software. HPV human papillomavirus; TSS transcription start site; CIN cervical intraepithelial neoplasia.
Assessment of small promoters by nAnTi-CAGE technology
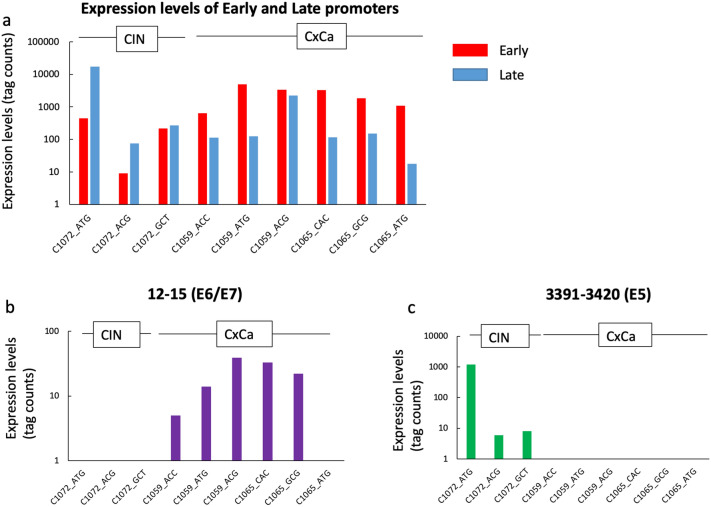
In a previous study, we identified numerous HPV-derived TSS clusters in a CIN cell line and a CIN sample13. In the present study, we used nAnTi-CAGE to detect small HPV16-derived TSS clusters, as well as the prominent early and late promoters, in clinical samples (Table 2). One of the small TSS clusters was found to be for the E8^E2 gene, and is located at nt1125-1148. We identified the E8^E2 TSS in 3 of 6 cancer samples and 2 of 3 CIN samples. Another small TSS cluster found to be for the E5 gene, located at nt3391-34207, was identified in all CIN samples. Furthermore, we also identified a cluster located at nt12-15 in all cervical cancer samples. Focusing on the early and late promoters, as well as the cluster located at nt12-15 for the E6/E7 genes and nt3391-3420 for the E5 gene, there are changes in gene expression according to the usage of each viral promoter (Fig. 5).
Table 2.
Summary of the HPV-16-derived tag numbers of cervical samples by nAnTi-CAGE.
| Start of cluster (HPV-16 nt) | End of cluster (HPV-16 nt) | Strand | C1072_ATG | C1072_ACG | C1072_GCT | C1059_ACC | C1059_ATG | C1059_ACG | C1065_CAC | C1065_GCG | C1065_ATG |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 15 | + | · | · | · | 5 | 14 | 39 | 33 | 22 | · |
| 90 | 97 | + | 425 | 9 | 214 | 616 | 4896 | 3315 | 3177 | 1801 | 1085 |
| 670 | 672 | + | 230 | · | · | 6 | · | 30 | · | 11 | · |
| 710 | 713 | + | 536 | · | · | · | · | 47 | · | · | · |
| 714 | 717 | + | 425 | · | · | · | · | 45 | · | · | · |
| 741 | 798 | + | 12,965 | 58 | 11 | 39 | 32 | 1217 | 26 | 50 | 6 |
| 930 | 932 | + | 16 | · | · | · | 5 | · | · | · | · |
| 952 | 955 | + | 128 | · | · | · | 17 | · | · | · | · |
| 972 | 974 | + | 6 | · | · | · | · | · | · | · | · |
| 997 | 999 | + | 7 | · | · | · | · | · | · | · | · |
| 1120 | 1122 | + | · | · | · | · | · | 8 | · | · | · |
| 1125 | 1148 | + | 29 | · | 6 | 29 | · | 116 | · | 8 | · |
| 1225 | 1228 | + | · | · | · | · | · | 7 | · | · | · |
| 1234 | 1236 | + | · | · | · | · | · | · | · | · | · |
| 1259 | 1262 | + | 20 | · | · | · | · | · | · | · | · |
| 1398 | 1400 | + | · | · | · | · | · | · | · | · | · |
| 1509 | 1512 | + | · | · | · | · | 43 | · | · | · | · |
| 1563 | 1566 | + | · | · | · | · | 5 | · | · | · | · |
| 1570 | 1573 | + | · | · | · | · | 5 | · | · | · | · |
| 1675 | 1678 | + | 11 | · | · | · | 17 | · | · | · | · |
| 2009 | 2011 | + | · | · | · | · | · | · | · | · | · |
| 2057 | 2060 | + | · | · | · | · | · | · | · | · | · |
| 3357 | 3361 | + | 25 | · | · | · | · | · | · | · | · |
| 3391 | 3420 | + | 1197 | 6 | 8 | · | · | · | · | · | · |
| 3441 | 3443 | + | 11 | · | · | · | · | · | · | · | · |
| 3444 | 3446 | + | 55 | · | · | · | · | · | · | · | · |
| 3495 | 3497 | + | 64 | · | · | · | · | · | · | · | · |
| 3585 | 3589 | + | 162 | · | · | · | · | · | · | · | · |
| 3633 | 3638 | + | · | · | · | · | · | · | · | · | · |
| 4022 | 4024 | + | · | · | · | · | · | · | · | · | · |
| 5661 | 5665 | + | · | · | · | · | · | · | · | · | · |
| 7684 | 7687 | + | · | · | · | · | · | 8 | · | · | · |
| 7782 | 7785 | + | · | · | · | · | · | · | · | · | · |
| 7852 | 7856 | + | · | · | · | · | · | · | · | · | · |
| 592 | 652 | − | 13 | · | · | · | · | · | · | · | · |
| 945 | 950 | − | · | · | · | · | · | · | · | · | · |
| 1311 | 1328 | − | · | · | · | · | · | · | · | · | · |
| 1884 | 1891 | − | · | · | · | · | · | · | · | · | · |
| 2850 | 2893 | − | 13 | · | · | · | · | · | · | · | · |
| 4852 | 4933 | − | · | · | · | · | · | · | · | · | · |
| 7561 | 7632 | − | 9 | · | · | · | · | · | · | · | · |
CAGE tag 5′-coordinates were used for Paraclu clustering with the following parameters: (i) minimum five tags per cluster; (ii) (maximum density/baseline density) ≥ 2; and (iii) 100-bp maximum cluster length. Tag numbers < five were designated as negative for each TSS cluster.
nt12-15, nt1125-1148, and nt3391-3420 are highlighted in bold.
“·” indicates negative for each TSS cluster (tag numbers < five).
Figure 5.
HPV-16–derived tag numbers of cervical samples by nAnTi-CAGE. (a) Expression levels of early and late promoters across the cervical samples. (b) The cluster located at nt12-15 for E6/E7 genes. (c) The cluster located at nt3391-3420 for E5 gene. HPV human papillomavirus; TSS transcription start site; CIN cervical intraepithelial neoplasia; CxCa cervical cancer.
Discussion
In the present study, we noted that the TSS patterns in the HPV genome may reflect the lesion stage of infected tissue. In all cancer samples and in several CIN samples, the prominent TSS patterns corresponded to the early promoter, while in low-grade CIN samples, the dominant TSS clusters had shifted from the early to the late promoter. Furthermore, in lesions with multiple infections, the prominent TSS patterns were the same, regardless of HPV type.
Quantitative visualisation of TSS activation had previously been difficult to capture. Previous studies revealed the presence of two major promoters on the HPV genome: the early and late promoters. The activation of these promoters has been extensively investigated in reporter assays using cultured cells. However, quantitative evaluation of promoter activity in clinical samples remained challenging. Furthermore, overlapping transcripts complicated the quantitative evaluation of the individual transcripts. CAGE technology facilitates this by enabling the detection of precise TSSs and the quantitative evaluation of their activity. In the present study, we quantitatively assessed TSS activation in clinical samples using CAGE, and determined the occurrence of at least two TSS patterns in clinical samples: Type A and Type B. Considering that the expression of late genes is up-regulated in later stages of the viral life cycle, which is coordinated with epithelial differentiation, the Type B TSS pattern could represent a normal viral life cycle and correlate with lower CIN grades. As shown in Fig. 2, most of the samples with lower CIN grades (i.e. CIN1 and CIN2) showed the Type B pattern, which could represent a normal viral life cycle, while the Type A pattern accounted for a larger proportion in CIN3 and cancer samples.
Another important finding was the detection of weak TSS cluster activity, such as that of the TSS clusters at nt12-15, nt1125-1148, and nt3391-3420. In particular, in our cohort, a weak E6/E7 TSS cluster, nt12-15, was only detected in cervical cancer samples; however, an E5 TSS cluster, nt3391-3420, was only detected in CIN samples. In addition to the TSS patterns, the expression of these weak TSS clusters could serve as diagnostic biomarkers for cervical cancer progression. In the present study, we also identified the cluster at nt1125-1148, a TSS cluster of E8^E228, which is regulated by E1 and E2. The E8^E2 protein plays an important role in regulating viral genome replication during the course of infection28–31, and E8^E2 expression inhibits the proliferation of cancer cells32. However, until now, there has been no evidence for the existence of E8^E2 in clinical samples. After a direct evaluation of TSSs, we report here for the first time the identification and quantitation of an activated E8 promoter in clinical samples in three of seven cervical cancers and two of three CIN samples. Further analysis and clinical follow-up of specific patients are required to elucidate the association between E8^E2 expression and cancer progression.
We then demonstrated that the TSS pattern was the same in co-infected samples, regardless of the HPV types involved. It is plausible that viral gene expression changes in parallel with the differentiation of the infected epithelial cells33,34. The viral gene expression pattern may thus be determined by the differentiation status of the epithelial cells, regardless of HPV type. In well-differentiated superficial cells, the HPV late promoter is activated33,34. In contrast, in high-grade CIN samples, lack of epithelial differentiation may be associated with a stable expression of HPV early genes, such as genes encoding the E6 and E7 oncoproteins.
We originally defined the A and B TSS patterns based on visual inspection of the data, and defined Type A as having early > late promoter activity, and Type B as having late > early promoter activity. Independent classification based on Gaussian mixture models suggested that these definitions could be refined using machine learning. Nevertheless, we demonstrated the feasibility of a novel method for the evaluation of altered HPV promoter activity during disease progression in clinical samples. This constitutes a proof-of-principle for the utility of TSS patterns as a diagnostic marker for CIN severity or progression. An extended CAGE study with more samples would allow for further assessment of the possibility of linking TSS patterns to disease state. Such a study would need to balance the requirement for screening a large number of samples with the requirement for sequencing a sufficiently high number of reads from each sample. Using the classification proposed in the present study, distinguishing between Type A and B patterns required at least 16 tags (in total) for the early and late promoters. The HPV transcriptome represents only a fraction of the available sequence libraries, varying roughly between 1 per million and 1 per cent. Therefore, either new samples should be sequenced at a depth of 10 to 20 million reads, or an enrichment method should be developed to address this issue. A limitation of the present study was that the number of clinical samples was not sufficient to allow statistical validation of the association between different TSS patterns and the severity of CIN lesions, or the differentiation status of the epithelium.
In conclusion, in this study we demonstrated the feasibility to analyse TSS activity at the single-nucleotide level using CAGE technology in clinical HPV samples. Further work on a larger cohort following the same patients over time will be needed for determining the sensitivity and specificity of the quantification of dynamic changes of TSS patterns as a biomarker of disease progression.
Methods
Patients and clinical samples
HPV-infected cervical tissues were obtained from biopsy or surgery samples. Diagnosis was confirmed by experienced pathologists and gynaecological oncologists through pathological and colposcopic examination at the University of Tokyo Hospital. HPV-infected cervical tissues were also examined by H&E staining, and the extent of dysplasia was evaluated. Cervical intraepithelial neoplasia was categorised as grade 1, 2, and 3 (CIN1, CIN2, and CIN3) depending upon the proportion of abnormal cell thickness. Then, experienced gynaecological oncologists confirmed the biopsied samples as CIN or a cervical cancer lesion. The samples for CAGE analysis were taken from the same area that met the diagnostic criteria of a cervical lesion. All experimental procedures were approved by the institutional review board at The University of Tokyo (approval number G0637-6), and signed informed consent for the use of the tissues and genomic data was obtained from each participant. Preparation of nanoCAGE libraries at RIKEN was approved by the institutional review board at the Yokohama Campus (approval number H26-26). For the analysis, RNA (5 µg for nAnTi-CAGE and 500 ng for nanoCAGE) was extracted from each sample using an miRNeasy kit (Qiagen, Hilden, Germany). RNA quality was assessed using a Bioanalyzer (Agilent) and standardised to an RNA integrity number (RIN) of > 7.0 for nAnTi-CAGE or > 5.0 for nanoCAGE. The purity of RNA samples was assessed using NanoDrop analysis, which confirmed that the A260/A290 and A260/A230 ratios were > 1.7.
Ethical considerations
This study was approved by the institutional review board at The University of Tokyo (approval number G0637-6) in accordance with the Declaration of Helsinki. All patients provided written informed consent for study participation.
nAnTi-CAGE library construction
First-strand cDNA was transcribed to include the 5′-end of capped RNA, and CAGE ‘barcode’ tags were attached as previously described35. The sequenced CAGE tags were mapped to the HPV-16 and HPV-52 genome based on the infected HPV genotypes using BWA software (v0.5.9), discarding ribosomal or non-A/C/G/T base-containing RNAs. For the HPV-16 genes, CAGE tag 5′-coordinates were used for Paraclu clustering36 with the following parameters: (i) minimum five tags per cluster; (ii) (maximum density/baseline density) ≥ 2; and (iii) 100-bp maximum cluster length. Tag numbers < 5 were designated as negative for each TSS cluster.
nanoCAGE library construction
nanoCAGE libraries were constructed from isolated RNA as previously described12, with some modifications. The reverse-transcription products were eluted in 40 µL, and qPCR was conducted using the SYBR Premix Ex Taq kit (TaKaRa). Cycle numbers were estimated as Ct + 4 cycles, and PCR was conducted to generate cDNA using the Ex Taq enzyme (TaKaRa). PCR products were eluted in 30 μL of sterile distilled water after purification, and 0.3 ng of each sample was tagmented individually at 55 °C for 5 min. The extension time of the final PCR was 30 s, and the final purification was achieved using one volume of AMPure reagent (Beckman Coulter, Inc), with the products eluted in 25 μL of reaction mixture. The multiplexed libraries were then paired-end sequenced in five lanes of a HiSeq 2000 sequencer (Illumina) and aligned to human genome version hg38 supplemented with all the HPV genomes available on the Papillomavirus Episteme database37 on 5 Sep, 2016, using the CAGEscan pipeline v3.0 (https://gitlab.com/mcfrith/cagescan-pipeline, Kratz et al., in preparation), which assembles overlapping pairs originating from the same molecule and maps them to the genome using the LAST aligner38.
Statistics
The association between cervical lesion grades and TSS types was evaluated by the chi-square test and Cochrane–Armitage test using JMP Pro version 12.2.0 (SAS Institute, Cary, NC, USA). A p value < 0.05 was considered statistically significant. The ‘densityMclust’ function in the R package mclust v5.427 was used to compare the likelihood of different Gaussian mixtures. After defining a score as the average difference between the expression levels of the early and late promoters, we fitted Gaussian mixture models to these scores. The TSS patterns were classified according to the averaged difference in expression between the early and late promoters, calculated as (early—late)/(early + late). The minimum number of samples required for achieving confidence < 0.25 (i.e. to distinguish between Types A and B) was determined using the ‘ciss.wald’ function in the R package binomSamSize v0.1–5 (Fig. 3).
Acknowledgements
The authors are grateful to Dr Lawrence Banks and Dr Miranda Thomas (Tumour Virology Laboratory, International Centre for Genetic Engineering and Biotechnology, Italy) for valuable comments on the manuscript. We thank Bogumil Kaczkowski for critical comments, and Genome Network Analysis Service Support Facility (GeNAS) at RIKEN for the sequencing of CAGE libraries. This work was supported by a Grant-in-Aid for Scientific Research (KN and CP) from the Ministry of Education, Science, and Culture (Japan) and a grant to RIKEN CLST (DGT) from the MEXT, Japan. This study was also partially supported by Grants-in-Aid for Practical Research for Innovative Cancer Control (KK, Grant Number: 15656298) and J-PRIDE (AT, Grant number: 19fm0208013h0003) from the Japan Agency for Medical Research and Development (AMED).
Author contributions
Conception and design: A.T., K.N. Acquisition of data: A.T., K.N., H.N., Y.K., S.K. Analysis and interpretation of data: A.T., C.P., K.N., K.K., K.H., T.N., I.K. Writing, review, and/or revision of the manuscript: A.T., K.N., C.P., K.O., I.K. Study supervision: K.O., K.K., P.C., Y.O., T.F.
Data availability
Demultiplexed sequence files are being submitted to the Japanese Genotype–Phenotype Archive (JGA).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Lowy DR, Schiller JT. Reducing HPV-associated cancer globally. Cancer Prev. Res. (Phila.) 2012;5:18–23. doi: 10.1158/1940-6207.CAPR-11-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Human papillomavirus-associated cancers—United States, 2004–2008. Morb. Mortal. Wkly. Rep. 2012;61:258–261. [PubMed] [Google Scholar]
- 4.Yim E-K, Park J-S. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res. Treat. 2005;37:319–324. doi: 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeon S, Allen-Hoffmann BL, Lambert PF. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995;69:2989–2997. doi: 10.1128/JVI.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBride AA, Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017;13:e1006211. doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milligan SG, Veerapraditsin T, Ahamet B, Mole S, Graham SV. Analysis of novel human papillomavirus type 16 late mRNAs in differentiated W12 cervical epithelial cells. Virology. 2007;360:172–181. doi: 10.1016/j.virol.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doorbar J, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 9.Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murata M, et al. Detecting expressed genes using CAGE. Methods Mol. Biol. 2014;1164:67–85. doi: 10.1007/978-1-4939-0805-9_7. [DOI] [PubMed] [Google Scholar]
- 11.Plessy C, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat. Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulain S, et al. NanoCAGE: a method for the analysis of coding and noncoding 5′-capped transcriptomes. Methods Mol. Biol. 2017;1543:57–109. doi: 10.1007/978-1-4939-6716-2_4. [DOI] [PubMed] [Google Scholar]
- 13.Taguchi A, et al. Characterization of novel transcripts of human papillomavirus type 16 using cap analysis gene expression technology. J. Virol. 2015;89:2448–2452. doi: 10.1128/JVI.03433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doorbar J. The papillomavirus life cycle. J. Clin. Virol. 2005;32(Suppl 1):7–15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Nagasaka K, Kawana K, Osuga Y, Fujii T. PDZ domains and viral infection: versatile potentials of HPV-PDZ interactions in relation to malignancy. BioMed Res. Int. 2013;2013:369712. doi: 10.1155/2013/369712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rapp B, et al. Cell-type-specific separate regulation of the E6 and E7 promoters of human papillomavirus type 6a by the viral transcription factor E2. J. Virol. 1997;71:6956–6966. doi: 10.1128/JVI.71.9.6956-6966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steger G, Corbach S. Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J. Virol. 1997;71:50–58. doi: 10.1128/JVI.71.1.50-58.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stubenrauch F, Colbert AM, Laimins LA. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 1998;72:8115–8123. doi: 10.1128/JVI.72.10.8115-8123.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, et al. Expression of HPV L1 capsid protein in cervical specimens with HPV infection. Diagn. Cytopathol. 2008;36:864–867. doi: 10.1002/dc.20922. [DOI] [PubMed] [Google Scholar]
- 20.Mehlhorn G, et al. HPV L1 detection discriminates cervical precancer from transient HPV infection: a prospective international multicenter study. Mod. Pathol. 2013;26:967–974. doi: 10.1038/modpathol.2012.233. [DOI] [PubMed] [Google Scholar]
- 21.Rauber D, Mehlhorn G, Fasching PA, Beckmann MW, Ackermann S. Prognostic significance of the detection of human papilloma virus L1 protein in smears of mild to moderate cervical intraepithelial lesions. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008;140:258–262. doi: 10.1016/j.ejogrb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Griesser H, Sander H, Walczak C, Hilfrich RA. HPV vaccine protein L1 predicts disease outcome of high-risk HPV+ early squamous dysplastic lesions. Am. J. Clin. Pathol. 2009;132:840–845. doi: 10.1309/AJCPCU0HBFFFGDTV. [DOI] [PubMed] [Google Scholar]
- 23.Negri G, et al. p16 ink4a and HPV L1 immunohistochemistry is helpful for estimating the behavior of low-grade dysplastic lesions of the cervix uteri. Am. J. Surg. Pathol. 2008;32:1715–1720. doi: 10.1097/PAS.0b013e3181709fbf. [DOI] [PubMed] [Google Scholar]
- 24.Choi YS, et al. Human papillomavirus L1 capsid protein and human papillomavirus type 16 as prognostic markers in cervical intraepithelial neoplasia 1. Int. J. Gynecol. Cancer. 2010;20:288–293. doi: 10.1111/IGC.0b013e3181cd184c. [DOI] [PubMed] [Google Scholar]
- 25.Griffin H, et al. Stratification of HPV-induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM. Mod. Pathol. 2015;28:977–993. doi: 10.1038/modpathol.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severin J, et al. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat. Biotechnol. 2014;32:217–219. doi: 10.1038/nbt.2840. [DOI] [PubMed] [Google Scholar]
- 27.Scrucca L, Fop M, Murphy TB, Raftery AE. McLust 5: clustering, classification and density estimation using Gaussian finite mixture models. R. J. 2016;8:289–317. doi: 10.32614/RJ-2016-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lace MJ, Anson JR, Thomas GS, Turek LP, Haugen TH. The E8^E2 gene product of human papillomavirus type 16 represses early transcription and replication but is dispensable for viral plasmid persistence in keratinocytes. J. Virol. 2008;82:10841–10853. doi: 10.1128/JVI.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fertey J, et al. Interaction of the papillomavirus E8^E2C protein with the cellular CHD6 protein contributes to transcriptional repression. J. Virol. 2010;84:9505–9515. doi: 10.1128/JVI.00678-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Straub E, Dreer M, Fertey J, Iftner T, Stubenrauch F. The viral E8^E2C repressor limits productive replication of human papillomavirus 16. J. Virol. 2014;88:937–947. doi: 10.1128/JVI.02296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dreer M, et al. Interaction of NCOR/SMRT repressor complexes with papillomavirus E8^E2C proteins inhibits viral replication. PLoS Pathog. 2016;12:e1005556. doi: 10.1371/journal.ppat.1005556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fertey J, et al. Growth inhibition of HeLa cells is a conserved feature of high-risk human papillomavirus E8^E2C proteins and can also be achieved by an artificial repressor protein. J. Virol. 2011;85:2918–2926. doi: 10.1128/JVI.01647-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grassmann K, Rapp B, Maschek H, Petry KU, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 1996;10:2339–2349. doi: 10.1128/JVI.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozbun MA, Meyers C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 1997;71:5161–5172. doi: 10.1128/JVI.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Y, et al. Tor signaling regulates transcription of amino acid permeases through a GATA transcription factor Gaf1 in fission yeast. PLoS ONE. 2015;10:e0144677. doi: 10.1371/journal.pone.0144677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frith MC, et al. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Doorslaer K, et al. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. 2017;45:D499–D506. doi: 10.1093/nar/gkw879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Demultiplexed sequence files are being submitted to the Japanese Genotype–Phenotype Archive (JGA).