Abstract
Selective suicide inhibitors represent a seductively attractive approach for inactivation of therapeutically relevant enzymes since they are generally devoid of off-target toxicity in vivo. While most suicide inhibitors are converted to reactive species at enzyme active sites, theoretically bioactivation can also occur in ectopic (secondary) sites that have no known function. Here, we report an example of such an “ectopic suicide inhibition”, an unprecedented bioactivation mechanism of a suicide inhibitor carried out by a non-catalytic site of thioredoxin glutathione reductase (TGR). TGR is a promising drug target to treat schistosomiasis, a devastating human parasitic disease. Utilizing hits selected from a high throughput screening campaign, time-resolved X-ray crystallography, molecular dynamics, mass spectrometry, molecular modeling, protein mutagenesis and functional studies, we find that 2-naphtholmethylamino derivatives bound to this novel ectopic site of Schistosoma mansoni (Sm)TGR are transformed to covalent modifiers and react with its mobile selenocysteine-containing C-terminal arm. In particular, one 2-naphtholmethylamino compound is able to specifically induce the pro-oxidant activity in the inhibited enzyme. Since some 2-naphtholmethylamino analogues show worm killing activity and the ectopic site is not conserved in human orthologues, a general approach to development of novel and selective anti-parasitic therapeutics against schistosoma is proposed.
Keywords: Schistosomiasis, Redox metabolism, Suicide inhibitor, Quinone methide, Secondary site
1. Introduction
Thioredoxin reductases (TrxR) head the thioredoxin (Trx)-related pathways both in archaea and in higher eukaryotes. Trx-related pathways are central in redox signaling, defense against oxidative stress, and DNA synthesis. Given their central role in metabolism, these enzymes are targets both of drugs already used in the clinic to fight cancer and inflammatory diseases in humans [1] and of several ongoing drug-design projects aimed at finding new drugs against parasitic pathogens. TrxR are obligate homodimers that shuttle electrons from NADPH to oxidized Trx, utilizing a FAD cofactor, a pair of cysteines, situated at the si-face of the flavin. The transfer of electrons from NADPH to the FAD/Cys redox site is also shared by the closely related glutathione reductases (GRs), with glutathione disulfide reduction occurring at this site. TrxRs from higher eukaryotes utilize a mobile, redox active C-terminal arm of the symmetrical subunit to shuttle electrons from FAD to the protein surface where Trx reduction occurs. In mammalian TrxRs, a selenocysteine (Sec, U) occupies the penultimate position of the C-terminus and is capable of redox cycling with an adjacent conserved cysteine.
Thioredoxin glutathione reductase (TGR) is a TrxR-like enzyme, which is a natural chimeric protein being a fusion of a glutaredoxin (Grx) domain to the N-terminus of a Sec-containing TrxR domain [2]. The presence of the Grx domain confers on this enzyme the unique capability to simultaneously reduce both oxidized glutathione and Trx [3]. TGR is the lone enzyme responsible for maintaining redox homeostasis in Schistosoma and related parasites that lack independent TrxR and GR proteins. For this reason, TGR is one of the most promising drug targets against schistosomiasis, a devastating neglected disease, and other related infections such as taeniasis and echinococcosis [4,5]. Schistosomiasis affects more than 200 million people worldwide and only one drug, praziquantel, is available for treatment, which is administered to tens of millions of people each year [6]. Less sensitive parasite strains are emerging due to this massive drug administration, making the identification of new therapies an urgent need.
Sec-containing TrxR-like enzymes, including TGR from S. mansoni (SmTGR), were recently subjected to several high throughput screening (HTS) campaigns [7–9]. Due to the presence of highly nucleophilic Sec and the low pKa Cys coupled to redox active cofactors, they are particularly prone to inhibition by electrophiles, metal-containing compounds, oxidants, and redox cyclers [10]. Indeed, quinones and more general Michael acceptors are often identified as good inhibitors of this protein family. Improving their efficacy utilizing a combinatorial chemistry approach is, in general, an unfeasible task due their high inherent reactivity coupled with a low specificity [1]. Many chemotypes identified as hits in HTS are pan-assay interference compounds (PAINS); they are often found as active hits against different targets in multiple HTS campaigns and are regarded as poor drug leads [11]. In contrast, hits that are not electrophiles or oxidants, but can be converted to such reactive compounds in situ, hold greater promise as drug leads. This is especially true if the reactive moiety is revealed only upon reaching the final biological target, as occurs in some suicide inhibitors.
Quinone methides (QMs) are covalent modifiers of biological macromolecules via Michael addition, showing pronounced biological activity in vivo [12]. Furthermore, they have been implicated as the ultimate metabolites responsible for the activity of several antitumor drugs, antibiotics, and DNA alkylators [13]. They can be generated in situ by different mechanisms, including photoactivation and elimination of good leaving groups (for a comprehensive review see Refs. [14,15]) and efforts are directed towards the possibility of combining their intrinsic reactivity with site-directing features to create affinity reagents [16]. Considering that (i) QMs are known to inhibit human TrxR by forming covalent adducts with the nucleophilic centers of the enzyme [17] and (ii) inhibition of TrxR by suicide inhibitors has been recently demonstrated as a feasible strategy [18], the utilization of a masked QM, activated by the TrxR itself, represents a promising approach to inhibit selectivity this challenging drug-target family.
In the present work, we show that 2-naphtholmethylamino compounds (2NAMOs, Table 1), found in a quantitative HTS against SmTGR (GenBank: AAK85233.1; bioassay AID 485364), are covalent modifiers of this enzyme upon their transformation to a QM form. In particular, 1-[(dimethylamino)methyl]-2-naphthol (compound 4 in Table 1), binds to a solvent-accessible secondary site (Fig. 1), not implicated in the canonical enzyme function, and the specific amino acid environment present therein facilitates the transformation of the initial molecule into 1-methylene-1,2-dihydronaphthalene-2-one, an oQM, by a base-catalyzed elimination mechanism. We were able to follow the progression of this acid-base catalysis by time resolved crystallography, using the freeze-trapping method and molecular dynamics. We demonstrated that SmTGR is inhibited by 4 faster than an SmTGR mutant in which amino acid residues proposed to be involved in the adventitious QM transformation were modified. Furthermore, the oQM generated from the 2NAMO precursors is demonstrated by site-directed mutagenesis, functional studies and mass spectrometry to covalently modify functional amino acids of the protein inhibiting its enzymatic activity. Given that (i) some 2NAMO analogues show activity against ex vivo worms, displaying low toxicity in animal models [19] and (ii) in human TrxR and GR the 2NAMO binding site is not conserved, we suggest that these findings may impact the search for selective inhibitors against TGR crucial in schistosomiasis.
Table 1.
2NAMO Compounds investigated in this work.
| Compound | Chemical formula | CID | Apparent IC50 (μM) | pka of the leaving group upon oQM formation | |||
|---|---|---|---|---|---|---|---|
| TGR | Rat TrxRa | Human GR | GSTO1b | ||||
| 1 | 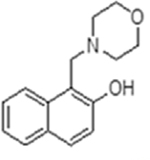 |
33839 | 0.61 ± 0.09 | 10 | 70.97 ± 11.66 | inactive | 8.3 (morpholine) |
| 2 | 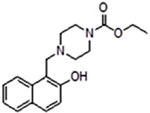 |
224037 | 0.65 ± 0.12 | 12.6 | 35% inhibition @ 2 mM | inactive | 7.8 (ethyl piperazine-1-carboxylate) |
| 3 | 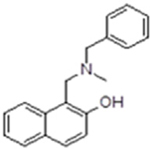 |
577695 | 0.84 ± 0.03 | inactive | 20% inhibition @ 2 mM | inactive | 9.4 (N-benzylmethylamine) |
| 4 | 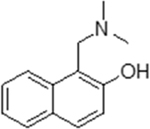 |
223180 | 11.2 ± 1.9 | 31.6 | 1,833 ± 73 | inactive | 11 (dimethylamine) |
| 5 | 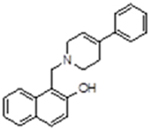 |
2408384 | 11.87 ± 3.57 | inactive | inactive | inactive | 9.6 (4-phenyl-1,2,3,6-tetrahydropyridine) |
| 6 | 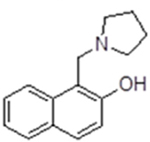 |
194653 | 28.83 ± 5.37 | inactive | inactive | inactive | 11.2 (pyrolidine) |
| 7 |  |
21436 | 57.63 ± 4.57 | inactive | inactive | n.d.c | 11.2 (piperidine) |
From bioassay AID 588453.
From bioassay AID 66188991.
Not determined. The compounds are ordered starting from the most potent. The pKas of compounds 2, 3 and 5 were calculated (http://chemicalize.com).
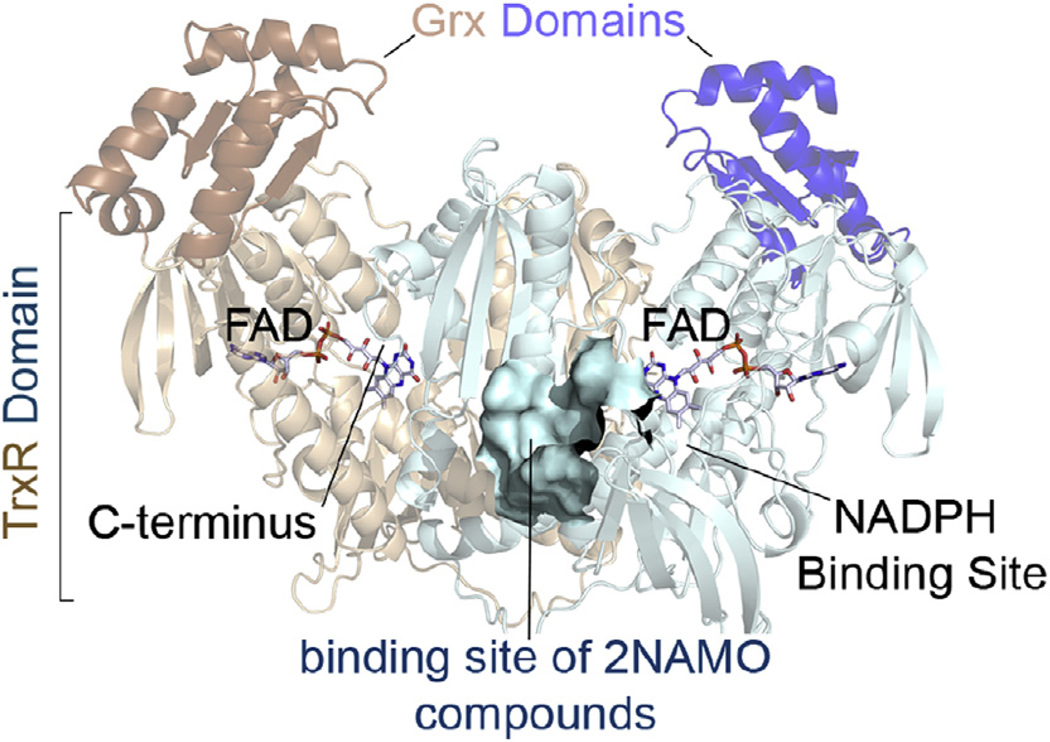
Fig. 1.
The physiological dimer of SmTGR in ribbons and the localization of the binding site of 2NAMOs relatively to the main active sites of the enzyme, is shown. The TrxR and Grx domains of a subunit are respectively colored in cyan and blue, while those belonging to the symmetrical one are in light-orange and orange. The binding site of 2NAMOs is shown as solvent-exposed surface. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2. Results and discussion
Covalent modification of proteins is of major interest due to the diversity of its applications that span from the search for chemical probes to new therapeutics. The major concern for the use of covalent inhibitors as drugs is their high reactivity coupled to low selectivity; however, an improvement of the latter could be possible with the development of suicide inhibitors [20]. This strategy relies on the binding and transformative capacity of biological macromolecules to activate a reagent’s masked reactivity in situ. QMs and related electrophiles can be generated through a range of strategies to alkylate various targets and are considered particularly suitable to be employed in such approaches [13]. Here, the unprecedented observation of the transformation of a suicide inhibitor carried out by a secondary site of a drug-target endowed with the appropriate chemical environment to generate a covalent inhibitor of the same protein is reported. Considering the unique mechanism of action, i.e. bioactivation of suicide inhibitors at a protein secondary site, we name this mechanism “ectopic suicide inhibition”. Moreover, we demonstrate the possibility to modulate the reactivity of masked oQM by changing the nature of the leaving group and implicate Sec as one the primary sites of alkylation by this class of compounds.
2.1. 2NAMOs are inhibitors of SmTGR and are able to form oQM species in solution
Several 2NAMO derivatives are present as hits in the quantitative high-throughput screen against SmTGR (bioassay AID 485364). We confirmed their apparent IC50 using 15 min preincubation (Table 1). All the compounds can efficiently inhibit the enzyme only in the presence of NADPH, and the inhibition appears to be irreversible even upon high dilution of the inhibited protein, with the measured IC50 representing a kinetic property rather than an equilibrium thermodynamic parameter [21].
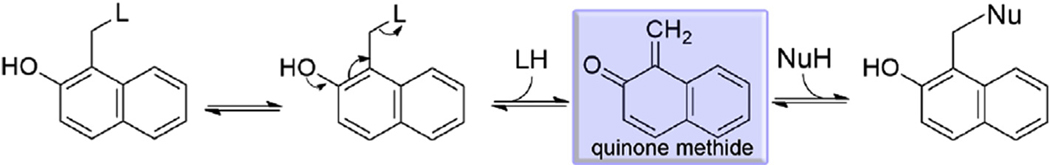
2NAMO compounds are Mannich adducts and are potentially capable of generating an oQM via loss of the amino group through an acid/ base-catalyzed elimination mechanism (Scheme 1 [15]). It is interesting to note that the IC50, determined after 15 min incubation against SmTGR, shows some correlation with the pKa of the amino group, which is a measure of the leaving group ability [22]. The lack of a stronger correlation with the pKa could be due to differences in the hydrophobic properties of the amino leaving groups. Once formed, oQMs are electrophilic Michael acceptors able to react with nucleophiles as shown in Scheme 1.
Scheme 1.
Elimination reaction of 2NAMO compounds followed by nucleophilic addition onto the electrophilic sp2 carbon of quinone methide. L is the leaving group and Nu is the nucleophile.
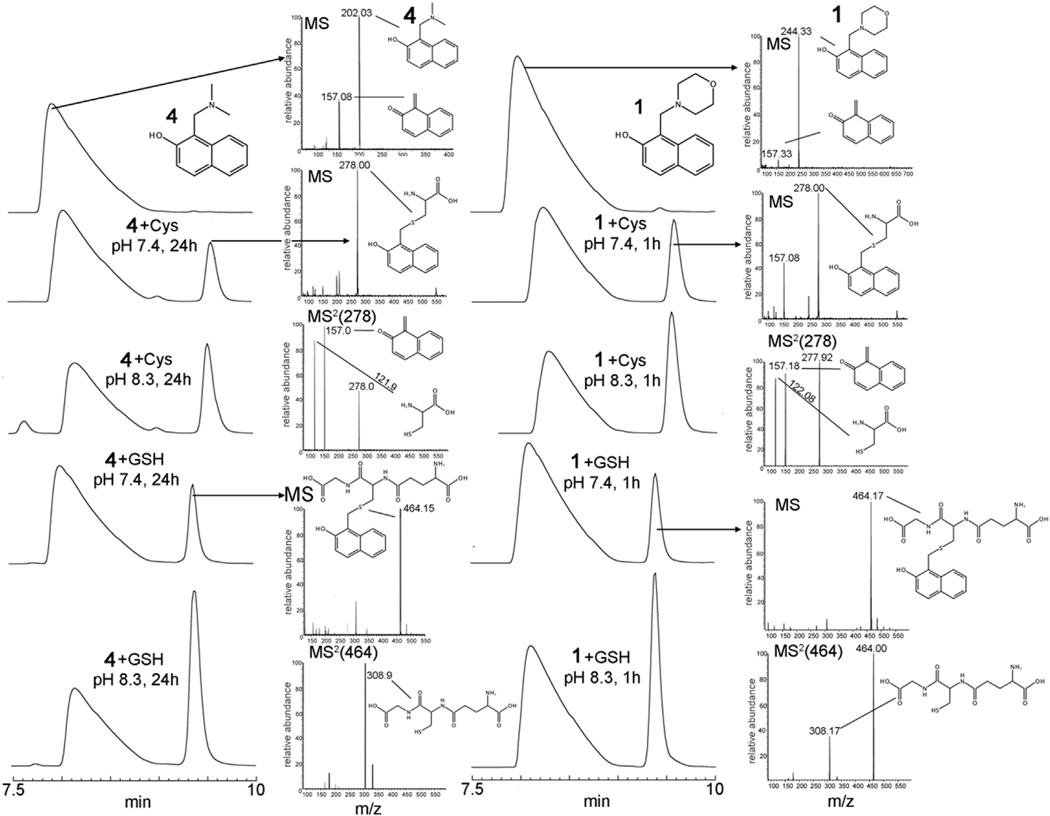
To test this potential mechanism, 1 and 4 were incubated with free Cys, Sec or glutathione (GSH). We followed the time course of the reaction by RP-HPLC and analyzed the separated peaks by tandem mass spectrometry (Fig. 2 and Supplemental Fig. 1). Compound 4 incubated at pH 7.4 and at ambient temperature with both GSH and Cys gives rise to a new detectable species after several hours. On the other hand, in 1 h at pH 7.4, compound 1 generates the same species in amounts compatible to those generated by 4 in 24 h. Both peaks in the RP-HPLC of the initial compounds were confirmed by mass spectrometry to belong to 1 and 4 of Table 1 ([1H]+ = 244 and [4H]+ = 202, see Fig. 2). However, when they are injected by direct infusion into the ESI source, a peak corresponding to the oQM species ([oQMH]+ = 157) is detected, the intensity of which is reduced if the capillary voltage and temperature of the ESI source are lowered, indicating an intrinsic lability of the precursors in the vapor phase. The new peaks formed upon incubation of 1 and 4 with both Cys and GSH were subjected to collision-induced dissociation (CID) tandem mass spectrometry analysis confirming that they are the products of reaction of oQM with thiol nucleophiles (see Fig. 2). 2NAMOs undergo an addition reaction with Sec at pH 7.4 as well. Sec (pKa = 5.2) is 100-times more nucleophilic than Cys [23]. The observed increased rate of QM formation for 1 with respect to 4, at pH 7.4, is likely due to the lower basicity and increased leaving group ability, as also observed for other QMs precursors [24]. Thus, in principle, it would be possible to modulate the reactivity of a 2NAMO compound just by modulating the leaving group basicity. Compound 1 was incubated with Sec in presence of 6 equivalents of TCEP for 1 h, bubbling nitrogen into the solution in order to maintain Sec in a reduced state. The adduct between Sec and oQM is clearly formed as demonstrated by mass spectrometry (Supplemental Fig. 1). The reaction between 4 and Sec was not carried out due to the difficulty in maintaining anaerobic conditions for longer times.
Fig. 2.
RP-HPLC and MS spectra of 4 and 1 alone (left and right up), after incubation with Cys and GSH at different pH values and the relative mass spectra (MS and MS2) of the peaks. Compound 1 is able to generate oQM faster than 4: at 1 h, 1 produces the adducts between oQM and thiols in similar amounts to those produced by 4 in 24 h.
To get further insights into the base-catalyzed elimination mechanism reported in Scheme 1, the same experiments between 2NAMOs and nucleophiles were carried out at a higher pH. The increased rate of oQM-adduct formation observed at pH 8.3 for both 1 and 4 in presence of thiols (Fig. 2) can be mostly due to a higher degree of deprotonation of the 2-naphthol moiety [(the pKa of the naphtholic oxygen of 2NAMOs should be somewhat lower than that of the 2-naphthol (≈9.6; [25], due to the intra-molecular H-bond between the oxygen and the positively charged amino group as observed in the crystal structure of compound 1 [26]], rather than an increased concentration of the active nucleophiles, i.e. the thiolate of GSH and cysteine (pKa ≈ 8.3 [23]). Accordingly, as demonstrated for other QM precursors, deprotonation of the OH is the starting event for the electronic rearrangement that ends with elimination of the leaving group and formation of reactive QM[27].
2.2. Time-resolved structural studies of oQM formation on an adventitious SmTGR pocket
We were able to observe the formation of oQM in a solvent-exposed pocket of SmTGR by means of time-resolved X-ray studies. This was possible due to the high solubility of the inactive precursor allowing the use of high concentrations in soaking experiments, which favors rapid diffusion of the compound through the crystal lattice with respect to the slow acid-base reaction (see below). We tested 1, 3, 4 and 7 by X-ray crystallography. Crystals soaked with 1 and 3 immediately underwent cracking and co-crystallization trials with all the compounds failed due to partial protein precipitation during incubation. We solved the structures of SmTGR-U597C at different soaking times [1 h PDB ID: 6RTJ; 2 h PDB ID: 6RTM and 4 h PDB ID: 6RTO] using 25 mM of 4 in the absence of NADPH. The structures were solved at a resolution ranging from 2.0 Å to 2.3 Å (See Supplemental Table 1), thus below 2.5 Å, which is recognized as the resolution limit to reliably determine the position of the atoms of a ligand in an electron density map [28]. We could not soak for longer times because the crystals cracked resulting in low resolution data and disordered diffraction patterns, possibly due to cysteine alkylation, an event that we cannot detect in the crystal structures.
At 1 h soaking time, 4 binds to a solvent-exposed pocket that localizes almost halfway between the NADPH binding pocket and the mobile C-terminal arm of the same subunit (Figs. 1 and 3). Hydrophobic residues constitute the bottom of the pocket while three carboxylic acid residues, E330, D334 and E337, lie on the rim cavity. E337 forms a salt-bridge with K345. The positive nitrogen of the dimethylamine group of 4, which lies out of the aryl plane, forms an electrostatic interaction with E330 (4.5 Å) and an H-bond (2.6 Å) between the carboxyl group of D334 and the OH of the naphthol is present. Compound 4 is also engaged in an edge-to-face pi-pi interaction with F343 and in a series of hydrophobic contacts with L320, V316, F343 and with the alkyl chain of K354. In the difference electron density map at 1 h, the arm containing the amine group is clearly present and exposed to the solvent (see Fig. 3, Panel A and D). The RSCC (real space correlation coefficient) of 4 is 0.89, indicating a good fit into electron density [29] and its averaged B-factor is slightly higher than that of the contacting residues. At 2 h soaking time, a flat electron density is present in the same position of that observed at 1 h (Fig. 3, Panel B and E), compatible with the oQM form of 4 (RSCC = 0.92); this electron density is still branched in proximity to D334, accounting for the H-bond between the oQM’s oxygen and the aspartate. However, a small blob of positive electron density is present in the same position of the dimethylamino group of 4, as found in Panels A and D of Fig. 3. After structure refinement and fitting of the oQM into the crude electron density, the 2F0-Fc map contoured at 0.5σ, which in general can detect less occupied conformers [30], indicates that some oQM precursors are still not transformed (see the insert of Panel E in Fig. 3). This is in agreement with other time-resolved studies carried out by the freeze-trapping method in which it is not possible to perfectly synchronize all the molecules in a crystal for a given chemical reaction [31]. At 4 h, the electron density is completely flat, the averaged B-factor of 4 is not significantly higher with respect to that of the compounds found at 1 h and 2 h of soaking time, despite an initial apparently less defined positive electron density (see Fig. 3, Panel C and F).
Fig. 3.
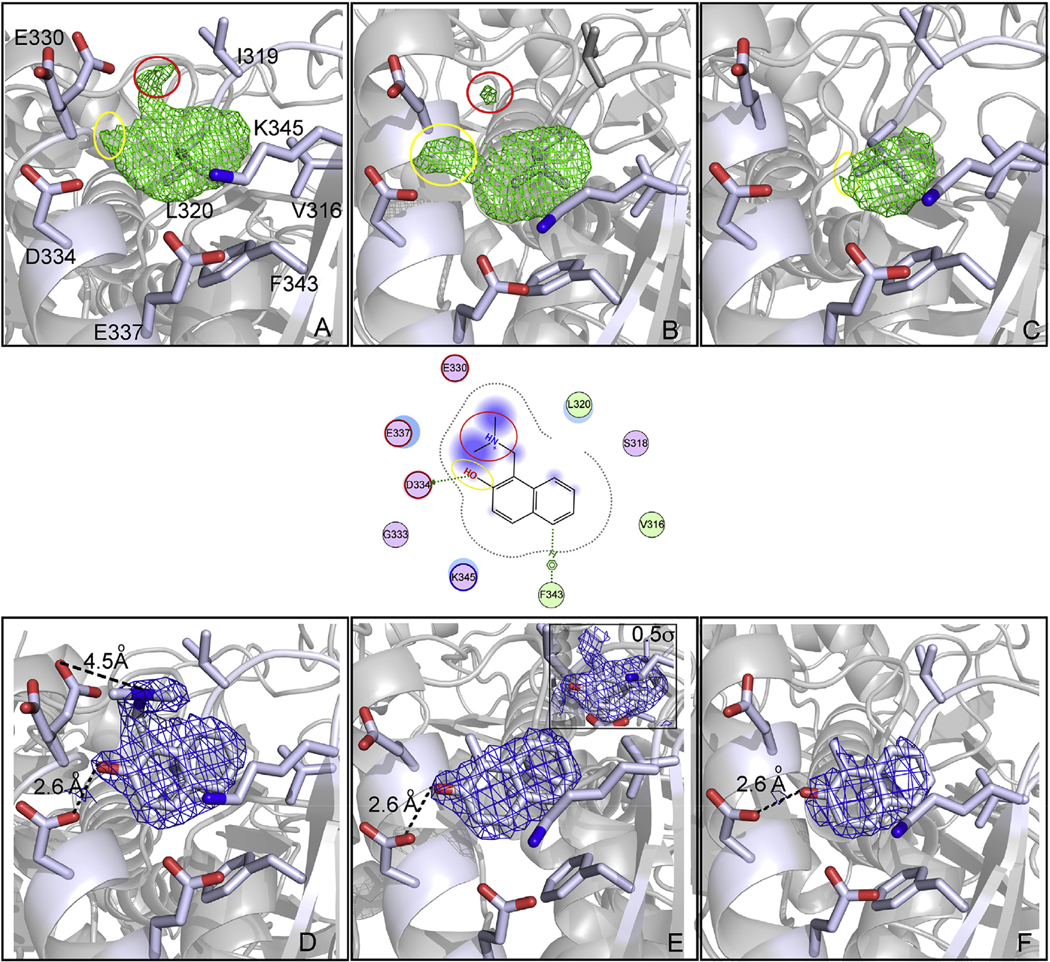
Snapshots of the transformation of 4 into a oQM in a secondary pocket of SmTGR. Panels represent the crystal structure of SmTGR in complex with 4 after 1 h soaking (A and D), 2 h soaking (B and E) and 4 h soaking (C and F). Polar interactions and H-bonds are displayed as dotted-lines. A two-dimensional depiction of the environment of the ligand is shown in the middle of the figure. The green electron density maps represent the Fo − Fc omit map contoured at 3σ and the blue electron density maps represent the 2Fo − Fc omit map contoured at 1σ, unless where it is differently indicated. At 1 h the electron densities are doubled branched indicating the prominent presence of the intact 4; after 2 h the density is flat, except for a small positive electron density spot localized in the same position of the dimethylamino group of 4 at 1 h. In the insert of Panel E, the 2Fo-Fc electron density, contoured at 0.5σ, indicates that few molecules of 4 are still not transformed in the crystal lattice. After 4 h, the density is completely flat without any branched portion that protrudes out of the aryl plane, indicating completion of the reaction. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The carboxylic group of D334 is well positioned to serve as a base for the OH of the naphthol, while E330 may be involved in the binding of the positive amino substituent of the aryl. These interactions, together with the pH dependence observed for the 2NAMOs transformation (Fig. 2), allow us to propose that the binding site of 4 facilitates oQM formation as suggested by the electron density changes observed in the crystal structures solved at different soaking times.
To obtain further preliminary insights into this mechanism we ran molecular dynamics simulations for the complex between SmTGR and compound 4 and conducted a distance analysis of the key residues in the 2NAMO binding site (Supplemental Fig. 2). In SmTGR, E330 is largely responsible for electrostatic interactions with the amino group of 4. It maintains consistent average distance of 5.64 Å between the carbon atom of the carboxyl group and the amine of the ligand throughout the simulation, which is indicative of a stable salt bridge. Although the average distances between D334 and E337 and the amino group of 4 are 7.28 and 7.15 Å, respectively, compound 4 adopt two almost equally populated and occurring simultaneously distinct conformations with the average distances 5.5 Å and 8.9 Å between the amino group of 4 and E330 and 6 Å and 8 Å between the amino group of 4 and E337, indicating that 4 oscillates between two stable conformation with (5.5 Å and 6 Å) and without (8.9 Å and 8 Å) salt bridge with these residues.
E337 is largely in salt bridge with K345 with an average distance of 3.96 Å, while E330 and D334 and K345 maintain an average distance of 10.5 and 8.97 Å, respectively. Throughout the simulations, D334 is either in direct or indirect contact via one water molecule hydrogen bond with the hydroxyl group of the ligand 4. The average distance between E330, D334, and E337 and the hydroxyl group of 1 is 6.38, 5.09, and 5.19 Å, respectively. The distance between D334 and the hydroxyl group of 4 remains stable with only minor oscillation around 5 Å. Unlike D334, E330 and E337 and the hydroxyl group of compound 4 form two stable conformations with an average distances 4.7 and 8.5 Å and 3.7 and 6.5 Å, respectively (Supplemental Fig. 2). The patterns of the interactions suggest that the hydroxyl group of compound 4 forms either hydrogen bond with E330 or with E337, while maintaining a stable hydrogen bond with D334. The average distance between the acidic sidechains of E330 and D334 and D334 and E337 remain stable at an average distance 7.97 and 6.9 Å, respectively. Altogether these observations suggest that the pKa of the three acidic sidechains of E330, D334, and E337 located on the same side of the helix and in proximity to each other is affected for at least one of them. We hypothesize that the pKa of D334 is higher than what is typically expected for the carboxyl group in solution at physiological pH exposed for aspartic acids in proteins because D334 is located between E330 and E337 and there are no other residues next to D334 capable of stabilizing the negative charge. If the pKa of D334 is, indeed, higher, D334 may deprotonate the hydroxyl group of 2NAMOs more effectively compared to what is expected in solution, leading to an accelerated rate of oQM formation in the 2NAMO binding site of SmTGR. The distance between the positively charged amino group of K345 and C9 and the amino group of ligand 4 is at an average of 4.9 and 5.56 Å. K345 is located directly above the aromatic ring of ligand 4 and is expected to form cation-π bond and stabilize the negative charge on the hydroxyl group of ligand 1 upon deprotonation required for oQM formation. On the other hand, K345 is close enough to the amino group of compound 4, facilitating the proton transfer also necessary for oQM formation.
Overall, these data suggest that the hydroxyl and amino groups of 4, the polar sidechains of D334, E337, K345, and the bridging water molecules may form a continuous flow of electrostatic and hydrogen bonds interactions. These concerted interactions are typically observed for proton transfer over multiple functional groups and water molecules, a.k.a. “proton wires”, in biomolecular systems [32] and, in our case, can facilitate seamless protonation/deprotonation events necessary for the formation of oQM.
2.3. The binding site of 2NAMOs of SmTGR facilitates their transformation into oQM
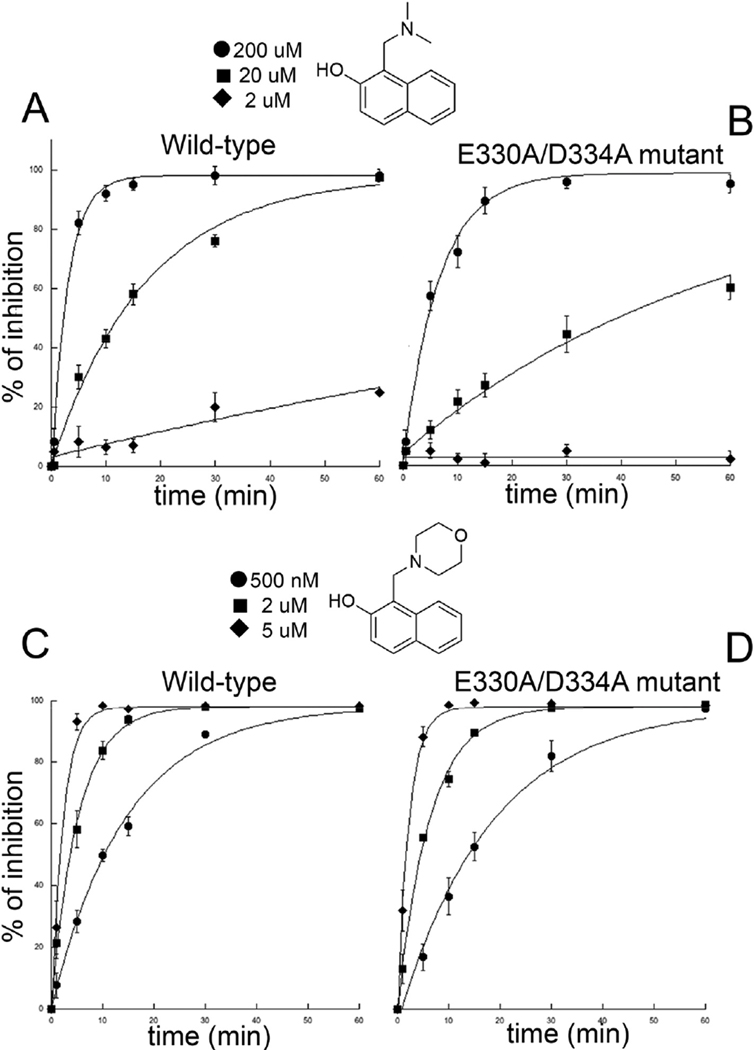
Molecular dynamic simulations together with the 3D structures derived from SmTGR crystals frozen at different incubation times with 4 suggest that its base-promoted transformation into an oQM is carried out in the 2NAMO binding site. In support of this, we produced a double mutant E330A/D334A, where the two acidic residues relevant for binding and catalysis are mutated. The steady state parameters of the double mutant, measured at variable NADPH concentration and constant DTNB [KM(NADPH) = 20 μM; kcat = 31 s−1], are close to those previously found for the wild type enzyme [33,34]. The double mutant is still inhibited by 4, but a 4-fold higher concentration of the inhibitor is required to achieve the same degree of inhibition observed for the WT enzyme (apparent IC50 at 15’ = 44.0 ± 4.9 μM). Therefore, it is likely that the observed inhibition is due to spontaneously formed oQM, as indicated by the results reported in Fig. 2. The time-dependent inactivation of the WT and the E330A/D334A mutant by 4 was determined. It was found that WT is inhibited 3 times faster than the E330A/D334 A at 20 μM, but the difference increases at lower concentrations of compound 4. Indeed, at 2 μM the pseudo first-order inhibition rate of E330A/D334A is almost zero [kapp(SmTGR-E330A/D334A) = 1.3 10−5 min−1], while it is still significant for the WT [kapp(SmTGR WT) = 4.7 10−3 min−1]. With 4 at 2 μM, the difference between the two protein forms is about 360-fold, demonstrating a clear catalytic role for the 2NAMO binding site in oQM production (Fig. 4). The global fitting of the observed kinetics yields the second order rate constants reported in the legend of Fig. 4. Enzyme inhibition occurs in the minute time window and it is due to oQM formation (see below). This time course is apparently not in agreement with the time required for oQM formation observed in crystallo. However, it is well-known that reaction in the crystal lattice can be slower by as much as 200-fold compared to those in solution due to the decreased thermal motions of the molecules in the solid state [31,35]. At higher concentrations of 4, differences in the inactivation rates between SmTGR variants become smaller, and at 2 mM (results not shown) disappear due to the large amount of spontaneously generated oQM. Indeed, considering that the pKa of 4 should be lower than that of the of 2-naphthol, i.e. 9.6, due to the intra-molecular H-bond between the oxygen and the amino group observed in some 2NAMO compounds [26], the deprotonated species competent for oQM formation is more than 12 μM at pH 7.4, when 2 mM concentrations of 4 are tested. This amount is more than enough to completely modify the 40–100 nM SmTGR present in the assays. Accordingly, when WT and mutant are incubated with 1, which is highly reactive and capable of rapidly and spontaneously forming an oQM in solution, only a slight increase in the inhibition rate for the WT is observed (Fig. 4).
Fig. 4.
Time dependent inactivation of SmTGR WT and E330/D334 by compounds 4 and 1. The calculated second order rate constants for the inhibition of SmTGR WT and E330/D334 by 4 are respectively: 2.75 × 10−3 μM−1 min−1 and 8.81 × 10−4 μM−1 min−1; while the second order inhibition rates of SmTGR WT and E330/D334 by 1 are: 0.11 μM−1 min−1 and 0.08 μM−1 min−1.
2.4. Proposed mechanism of SmTGR inhibition by 2NAMOs
SmTGR-U597C mutant is inhibited by 4 and 1 with apparent IC50s of 2 mM and 100 μM at 15 min, respectively, 200-fold higher than those determined for the WT enzyme. This indicates a primary role for the U597 in the mechanism of oQM alkylation and, indeed, selenolates are about 2 orders of magnitude more nucleophilic than thiols at pH around 7 [23]. Furthermore, at pH 6.4 SmTGR WT is still inhibited in the low micromolar range by 4 (not shown), indicating that Sec (pKa = 5.2) or a low pKa cysteine is involved.
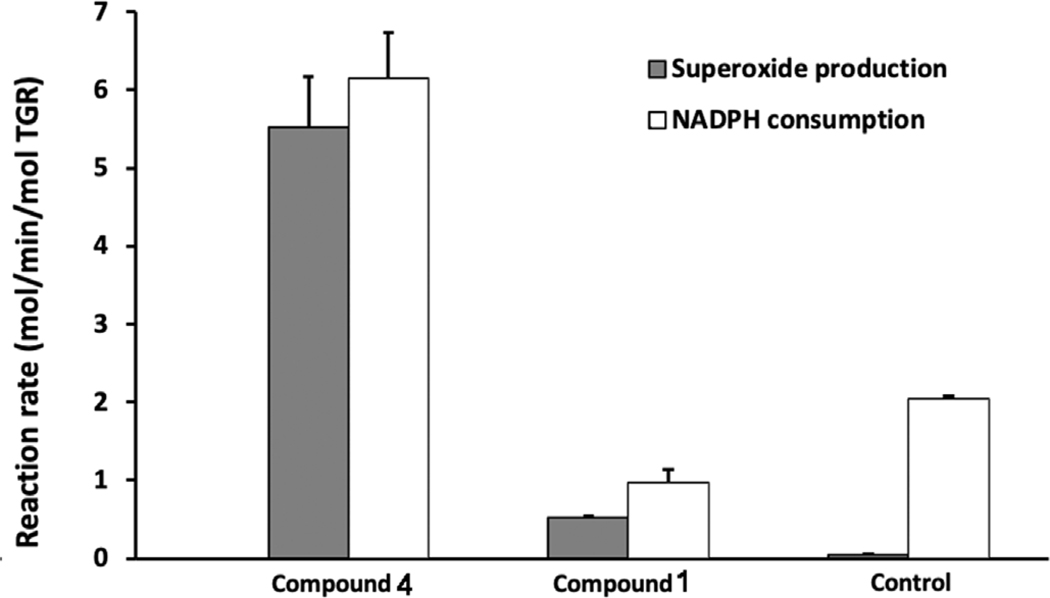
Another indication that the C-terminal redox center is implicated in the mechanism of action of 2NAMOs is the acquisition by the inhibited enzyme of the NADPH oxidase activity, i.e. the capability to transfer reducing equivalents to oxygen generating hydrogen peroxide or superoxide. This is a hallmark of the TrxR-like enzymes once the C-terminal redox center (C596–U597) is covalently modified and thus unable to undergo redox cycling [7]. The NADPH oxidase activity was tested using the pyrogallol method (Fig. 5 [36]). Compound 4 is able to induce this activity in the WT enzyme, while compound 1 is not. This difference can be rationalized by taking into account the capacity of 4 to generate reduced amounts of oQM (see Fig. 2) in the time window of the experiment, which can selectively alkylate the more nucleophilic Sec inducing the pro-oxidant activity of SmTGR, while the greater amount of oQM produced by 1 may alkylate other nucleophilic sites once the C-terminus has been saturated, a situation not compatible with NADPH oxidase activity and already observed for other alkylating agents, capable of reacting in a non-specific manner with Sec and Cys of SmTGR [37].
Fig. 5.
Superoxide production and NADPH consumption by TGR. TGR (500 nM) was incubated with NADPH (100 μM) and compounds 1 and 4 for 30 min. Samples were desalted to remove unreacted compounds. NADPH consumption and superoxide production using pyrogallol red of the inhibited enzymes were determined. Addition of 10 units of SOD abolished oxidation of pyrogallol red (not shown).
Moreover, a short MD simulation on SmTGR WT model shows that the oQM generated into the 2NAMO binding site could be directly sequestered by the C-terminal arm, thus accounting for the observed specificity of compound 4. The two charged residues in the C-terminal tail, K585 and K586, are the last tightly attached to the rest of the protein [38], whereas the C-terminal tail consisting of S(587)GVSPIV-SGCUG(598) is flexible and can swing to shuttle electrons amongst Cys154/159 (the FAD redox site), Cys28/31 (the Grx active site), and the Trx substrate. An analysis of the amino acid composition of SmTGR shows that there are only few regions that do not contain acidic or basic residues: the Sec-containing catalytic arm, the FAD and the Grx redox sites and the Trx docking site. Considering that the Sec597 arm has to travel tens of angstroms to find these interaction sites [38], it appears that the hydrophobic areas were designed by the evolution to improve the efficiency of the SmTGR catalytic machinery. Interestingly, the 2NAMO binding site is also among the hydrophobic areas that can be reached by the Sec597 catalytic tail. In fact, the distance between K585 and K586, which probably serve as a hinge for the flexible hydrophobic C-terminal arm, and the 2NAMO binding site is comparable to 25–30 Å distances between K585/596 and Cys28/31, Cys154/159, and Trx docking site (Supplemental Fig. 3).
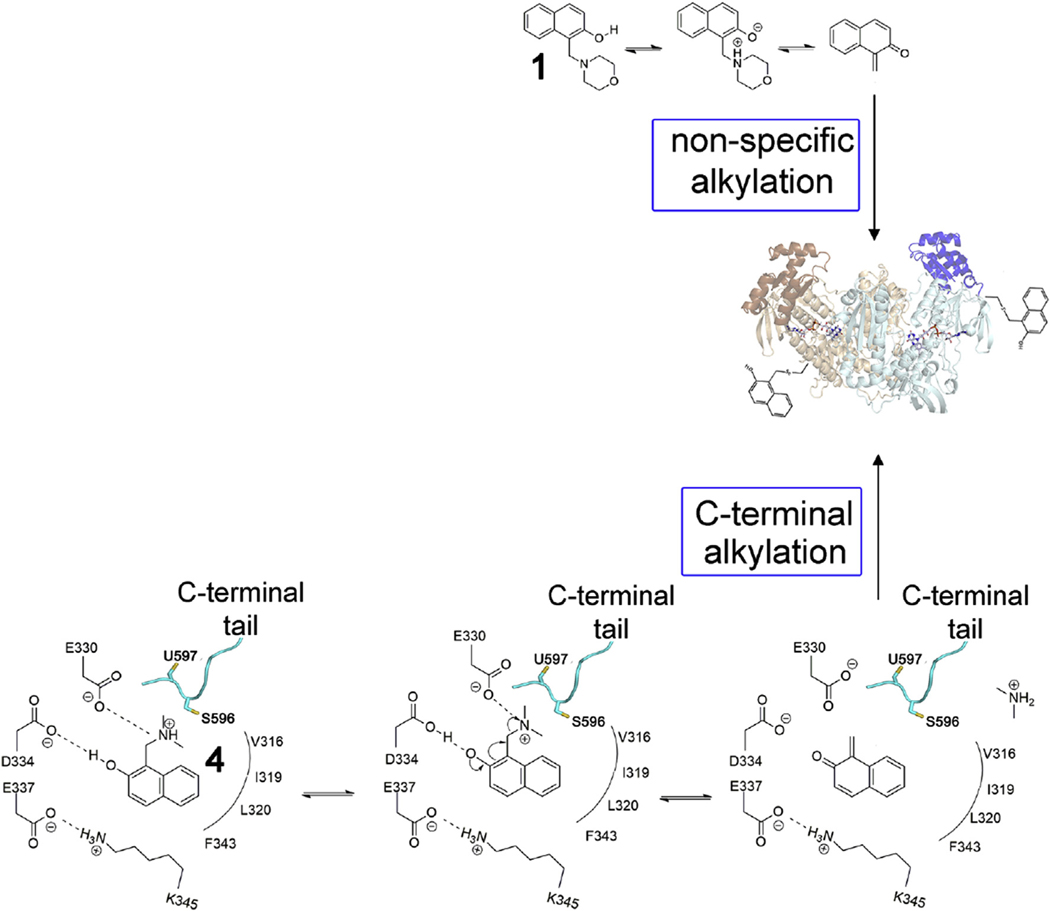
In summary, 2NAMO compounds vary in their intrinsic rate to form oQM in solution depending on their leaving group ability. The 2NAMOs, which are slower in generating oQM, such as 4, may utilize a secondary site in SmTGR, where the amino acidic environment facilitates this transformation. Once the reactive species is generated it leaves the site (likely due to diminished affinity caused by the loss of the positively charged amino group) and reacts primarily with the nucleophilic Sec, leading to inhibition of enzymatic activity of SmTGR (Fig. 6) and induction of the pro-oxidant activity. Compound 1, which is capable to spontaneously generate a high amount of oQM in solution, results in a non-specific alkylation of SmTGR (Fig. 6).
Fig. 6.
Proposed mechanism of SmTGR inhibition by 2NAMOs. The major routes of SmTGR inhibition by 1 and 4 are displayed. The formation of oQM by compounds 1 and 4 followed by covalent modification of the protein is in accordance with irreversible and NADPH-dependent inhibition mechanism observed for this class of compounds. The oQM, generated by 4 in the 2NAMO binding site, can be sequestered by the mobile reduced C-terminal arm.
2.5. 2NAMO compounds are able to kill S. mansoni worms
At 50 μM, 4 has no activity against ex vivo cultured worms while 1 and 3 kill 13% and 24% of the worms after 144 h, respectively (Table 2). The lack of activity against ex vivo worms reflects the decreased capacity of 4 both to form oQM in solution and, likely, to penetrate the worm membrane in comparison to more lipophilic 1 and 3. Other 2NAMOs potentially capable of forming oQM have been found to have selective worm-killing activity. For example, several very lipophilic steroid-containing 2NAMO derivatives [19] efficiently kill worms and display low toxicity against human cells. Therefore, we suggest that these 2NAMO analogues exert, at least in part, their pharmacological activity through inhibiting SmTGR by means of a QM-based mechanism, as demonstrated for the first time in the present work.
Table 2.
Activity of compounds against live, cultured worms. Compounds were added to media and worm survival was quantitated at indicated times.
| Compound | IC50 (μM) | Adult S. mansoni (144 h) | Larval S. mansoni(120 h) | |
|---|---|---|---|---|
| 50 μM | 10 μM | 10 μM | ||
| 1 | 0.61 ± 0.09 | 13% dead | 8% dead | n.d.a |
| 2 | 0.65 ± 0.12 | 5% dead | n.d.a | n.d.a |
| 3 | 0.84 ± 0.03 | 24% dead | 0% dead | 19% dead |
| 4 | 11.2 ± 1.9 | 0% dead | 0% dead | n.d.a |
| 6 | 57.63 ± 4.57 | 0% dead | n.d.a | n.d.a |
n.d. – not determined.
2.6. Interaction of 2NAMO compounds with orthologues and model Cys-containing enzymes
An analysis by Wang and collaborators [39] shows that compounds 1 and 4 are active in only a small percentage (4 was active in ≈1.5%; 10 HTS out of over 655 HTS) of screens reported in PubChem. This low hit rate indicates a degree of specificity for this class of compounds. Indeed, when they are tested against enzymes that contain low pKa cysteines crucial for catalysis, such as Rat norvegicus TrxR (RnTrxR), human glutathione S-transferase Ω−1 (GSTO1), and human GR, they do not have equivalent activities (Table 1). The potency of compounds 4 and 1 against RnTrxR are 31 μM and 10 μM, respectively, whereas their activity against SmTGR is 11.2 and 0.61 μM, respectively (Table 1). Considering that the C-terminus, including Sec, the FAD redox center, and most of reactive cysteines are conserved both in the Trx domain of SmTGR and in RnTrxR [40], the difference in specificity observed for compounds 1 and 4 should be sought in differences in the 2NAMO binding site. RnTrxR has a putative 2NAMO binding site size similar to that of SmTGR. An analysis of acidic and basic residues SmTGR and RnTrxR, which are likely to be responsible for catalysis and stabilization of the intermediates, shows somewhat minor but important differences (see Supplemental Fig. 4).
To gain additional insights into the differences in selectivity between SmTGR, RnTrxR, and hGR, we compared the 2NAMO binding site in smTGR with the putative 2NAMO binding sites in RnTrxR and hGR by means of molecular dynamic simulations. It was found that loss of 1 from the 2NAMO binding site occurs at a higher frequency in RnTrxR than in SmTGR. A comparison of the RnTrxR and SmTGR ligand-protein complex geometry during the simulations suggests that E330 in SmTGR plays a key role in binding of the ligand via ionic interaction with its amino group and restricting the escape of the ligand from the binding site. N234 in TrxR in the equivalent position of E330 is unable to fulfill this function because, while it still can form a hydrogen bond, it lacks the charge and is shorter and less bulky than E330. In the absence of the third acidic sidechain in TrxR, E238 (equivalent to D334 in SmTGR) and E241 (equivalent to E337 in SmTGR) maximize their distance and likely retain the pKa expected for an isolated acid side chain in water. The position and orientation of E241 is further restricted by its interaction with more directional guanidino sidechain of R249. The same interaction between E337 and K345 in SmTGR is much less directional and is easier to maintain throughout the simulation. The much larger and planar guanidino sidechain of R249 in RnTrxR limits the ensemble of binding modes available to 4. In fact, due to the large size and planarity of the guanidino portion of the sidechain of R249, its deviation from a co-planar orientation with the ring portion of 4 is expected to cause dissociation of the ligand from the putative 2NAMO binding site in RnTrxR. Overall, the network of the electrostatic and hydrogen bond interactions in RnTrxR is disorganized, probably leading to less efficient catalysis of oQM formation. Although these studies do not allow quantitative assessment of the degree of the difference between SmTGR and TrxR, they do support the higher propensity of the 2NAMO binding site in SmTGR to catalyze formation of oQM qualitatively.
The GSTO1 inhibition by 1 and 4 is not detected in the conditions tested. It is known that this enzyme is less prone to react with hydrophobic electrophilic substrates as compared with other GSTs, due to a more hydrophilic environment surrounding the catalytic low pKa Cys [41]. Thus, the lack of inhibition of GSTO1 may be attributed to a low capability of the functional site of this enzyme to host the hydrophobic oQM. Compounds 1 and 4 are much weaker inhibitors of hGR with respect to SmTGR (see Table 1), with similar values to those found for the SmTGR-U597C mutant. Modeling of hGR shows that the area corresponding to the putative 2 NAMO binding site in hGR is too shallow to accommodate any of the compounds in Table 1 in an effective fashion. hGR lacks the C-terminal redox center and the 2NAMO binding site (see Supplemental Fig. 4), but it retains the catalytic cysteine couple close to the FAD cofactor. We speculate that, in this case, the weaker inhibition is due to the alkylation of these two cysteines, which are less prone to interact with oQM due to their intrinsic less nucleophilic character with respect to Sec.
3. Conclusions
A novel unique site in SmTGR catalyzing bioactivation of 2NAMO derivatives to reactive oQMs was identified using structural, functional, and molecular dynamics studies. Our time-resolved X-ray experiments show elimination of the amino group of 2NAMO in compound 4 and formation of the corresponding oQM. To the best of our knowledge this is the first example of a suicide inhibitor activation carried out by a secondary/ectopic site of a drug-target with no function in the enzymatic mechanism, which we call here “ectopic suicide inhibition”. The bioactivation via a novel ectopic suicide inhibition mechanism followed by enzyme alkylation at a crucial redox site is consistent with molecular dynamics simulations and further supported by the in vitro functional characterization of different SmTGR variants. Moreover, considering that: (i) this ectopic site is not conserved in human orthologs, (ii) it is possible to modulate the reactivity of the 2NAMOs by changing the nature of the leaving group and (iii) 2NAMOs show activity against ex vivo worms, the design of novel anti-parasitic agents, characterized by a longer half-life and an increased selectivity over human enzymes is attainable. Overall, the discovery of the “ectopic suicide inhibition” mechanism offers a novel avenue for development of novel and safe therapeutics for schistosomiasis.
4. Materials and methods
Materials and reagents –
C18 column (Knauer). Tris(hydroxymethyl) aminomethane (Tris), potassium iodide (KI), 1,4-Dithiothreitol (DTT), β-mercaptoethanol (βme), dimethyl sulfoxide (DMSO), 1-[(Dimethylamino)methyl]-2-naphthol (4; CID:223180), 1-(Morpholin-4-ylmethyl)-2-naphthol (1; CID:33839), 5, 5-dithio-bis-(2-nitrobenzoic acid) (DTNB), sodium phosphate, sodium chloride (NaCl), selenocystine, Tris(2-carboxyethyl)phosphine hydrochloride were from Sigma-Aldrich. Nicotinamide adenine dinucleotide phosphate reduced (NADPH) was from Sigma and Cayman Chemicals. Polyethylene glycol 3350 (PEG 3350), bis(2-hydroxyethyl)aminotris(hydroxymethyl)methane (bis-Tris) were from Molecular Dimension. EDTA was from Euroclone.
4.1. Protein expression and purification
SmTGR wildtype (WT), SmTGR-Sec597Cys and SmTGR-E330A/ D334A mutant –
SmTGR WT protein was expressed in BL21 cells co-transformed with pSUABC and purified as previously described [33]. A form of WT that was replete with Sec was prepared as described [42,43]. The SmTGR-Sec597Cys mutant was prepared as described [34]. The SmTGR-E330A/D334A mutant was generated using NEBaseChanger and the Q5 Site-Directed Mutagenesis Kit. The DNA of the WT was used as a template and the sequences of the DNA primers used in polymerase chain reactions (PCRs) were 5′-AAGGTTGGTGcCTATA TGGAG-3′ (forward primer) and 5′-TGCAGCCATTTGTTGATC -3′ (reverse primer). Amplified DNA was purified with a PureYield™ Plasmid Miniprep System (Promega Corporation) and sequenced. SmTGR-E330A/D334A mutant was expressed and purified following the same procedure of the WT [33,42]. Protein concentrations were determined by FAD absorption at 463 nm (ε463 = 11.3 mM−1 cm−1). All the proteins were stored at −20 °C adding 50% glycerol.
Human GR –
The codon-optimized sequence for human cytoplasmic GR was synthesized by GenScript (Piscataway, NJ) in pET-15b. For protein expression the plasmid was transferred to Escherichia coli strain BL21(DE3). An overnight culture was diluted 1:100 in LB plus ampicillin (34 μg/ml) and cultured at 37 °C to OD600 = 0.6. At this point IPTG and riboflavin were added to 1 mM and 20 μg/ml, respectively, and the culture continued for 3 h, after which cells were collected by centrifugation. The cell pellets were frozen in dry ice ethanol and thawed at room temperature three times. The pellets were suspended in buffer 1 (50 mM potassium phosphate, pH 7.4, 500 mM NaCl) plus 30 mM imidazole, 1 mg/ml lysozyme, 100 μM FAD, and 1 mM phenylmethylsulfonyl fluoride. The suspension was sonicated and then centrifuged at 16,000 rpm for 60 min. The supernatant was filtered through a 0.45 μm filter and applied to pre-equilibrated nickel-affinity column (His-Trap Chelating, GE Healthcare). The column was washed with buffer 1 plus 30 mM imidazole, and then with buffer 1 plus 100 mM imidazole. The protein was eluted with buffer 1 plus 500 mM imidazole. Ultrafiltration (Amicon Ultra) was used to remove imidazole and to concentrate protein against buffer 1. The protein concentration was determined from FAD absorption at 463 nm (ε463 = 11.3 mM−1 cm−1). The protein was stored in buffer 1 plus 50% glycerol at −20 °C.
4.2. Functional studies
Steady state characterization of the SmTGR WT and E330A/D334A –
All the functional assays were carried out using 40 nM of WT and of the SmTGR-E330A/D334A mutant. Steady state experiments were carried out in 50 mM sodium phosphate (pH 7.4), 10 mM EDTA and 3 mM 5,5-dithio-bis-(2-nitrobenzoic acid) (DTNB) varying NADPH concentrations from 10 μM to 2 mM. Reaction was monitored by DTNB reduction at 412 nm (ε412nm = 13.6 mM−1cm−1).
IC50 measurements –
Assays were performed at room temperature in 100 mM potassium phosphate (pH 6.4 and pH 7.4) and 10 mM EDTA. SmTGR WT (40 nM), SmTGR-E330A/D334A (40 nM), SmTGR-Sec597Cys (400 nM) and human GR (10 nM) were incubated with 100 μM NADPH and different concentration of the compounds (or DMSO as control) at room temperature for 15 min. Then the TrxR activity inhibition of TGR was determined performing a DTNB assay started by addition of a second aliquot of NADPH (100 μM) and DTNB (3 mM). Reaction was monitored by absorbance increase at 412 nm by 2-thio-5-nitrobenzoic acid formation. GR activity was determined by addition of a second aliquot of NADPH (100 μM) and GSSG (100 μM) and the reaction was monitored by NADPH consumption at 340 nm. Both of the assays were performed on Thermo Multiskan Spectrum plate reader. All assays were done in triplicate.
Reversibility of SmTGR inhibition by 1 and 4 –
SmTGR WT(500 nM) was incubated with NADPH (100 μM) and 50 μM inhibitors for 30 min. After incubation samples were desalted using 7.0 kDa-cutoff spin Zeba desalting column (Thermo Scientific) and the DTNB assay was performed.
Time dependent inactivation –
SmTGR-E330A/D334A and SmTGR WT (40–100 nM) were incubated at room temperature with 1 and 4 at concentrations ranging from 0.020 μM to 2000 μM in presence of NADPH (100 μM) in 50 mM sodium phosphate pH 7.4, 10 mM EDTA plus 100 mM NaCl. The residual enzyme activities were reported as percentage of the controls, in which the compounds were replaced by DMSO. Reactions were monitored, taking aliquots at different time intervals, by the addition of fresh NADPH (100 μM) and DTNB (3 mM) in disposable plastic cuvettes. Reduction of DTNB was followed for 120s by absorbance increase at 412 nm. The data presented in Fig. 4 are the average of 3 or 9 independent measurements carried out with three or more different enzyme preparations. Global data analysis was carried out using the package Octave on an Intel based PC running the Linux operating system. The least squares minimization routine was written in house and makes use of the precompiled subprogram “fminunc”. The routine can minimize a pseudo-first order equation or a two exponentials sequential reaction scheme in which the first process is pseudo-first order. Analysis according to two sequential steps essentially gave the same result, the second step being much faster than the first one and poorly determined.
Detection of superoxide –
The pyrogallol red method was used [36,44]. SmTGR WT(500 nM) was incubated with NADPH (100 μM) and 50 μM inhibitors for 30 min. Samples were desalted using 7.0 kDa-cutoff spin Zeba desalting column (Thermo Scientific). For the detection of superoxide production and NADPH consumption, 100 μl of a pyrogallol red (50 μM) and NADPH (300 μM) solution was mixed with 100 μl of each sample above and then monitored at 340 nm and 540 nm simultaneously for 2 h for NADPH or pyrogallol red oxidation, respectively. The same reactions were performed with addition of 10 units of SOD to each well. NADPH consumption was determined using ε340nm = 6.22 mM−1cm−1 and superoxide production was determined by consumption of pyrogallol red by using ε540nm = 23.9 mM−1cm−1 with a stoichiometric relation of 1 molecule pyrogallol red react with 2 molecules superoxide.
Reversed phase HPLC –
Solutions of pure 1 and 4 at 1 mM or incubated with both 1 mM Cys and GSH, in 5 mM sodium phosphate (pH 7.4 and pH 8.3), were injected at different time intervals into an AZURA HPLC system (Knauer) equipped with a C18 column and equilibrated with MQ water plus 0.1% formic acid. For the reaction with Sec, 0.5 mM of selenocysteine was firstly reduced with 3 mM of TCEP in nitrogen atmosphere at pH 7.4 and then 1 mM of 1 was added; the mixture was incubated for 1 h and then injected into the HPLC system, as for the other reactions with thiols. Elution was performed using a gradient from 0 to 100% of CH3CN + 0.1% formic acid. The reaction outcomes were monitored by UV detection at 278 nm. All the peaks were manually collected.
Mass spectrometry–
Peaks from RP-HPLC were analyzed in a LTQ Orbitrap XL Discovery equipped with a ESI source (Thermo Fisher Scientific) in positive ion mode. For the direct-infusion experiments, a flow of 15 μL/min was provided by a syringe pump using a 250 μL syringe (Hamilton, Reno, NV, USA). The following settings were applied: capillary voltage of −4.0 kV, mass range of m/z 125 to 2,000 and capillary temperature of 260 °C. Calibration was performed with standard calibration mixture containing caffeine (m/z 195), MRFA peptide (m/z 524) and Ultramark 1600 (m/z 1,022–1,922).
4.3. Structural studies
Protein crystallization and soaking experiments –
SmTGR-Sec597Cys crystals were obtained as previously reported [42]. Briefly, the protein is equilibrated in 20 mM Tris pH7.4, concentrated to 4.5 mg/ml and mixed with equal amounts of a solution containing PEG3350 20%, 0.2 M KI, 100 mM BisTris 7.0 and DTT 5 mM. Several crystals were washed twice in a well solution where the reducing agent was removed and then incubated with 25 mM of 4. Crystals were picked up at 1 h, 2 h and 4 h and flash frozen in liquid nitrogen. After 5–6 h of soaking time, crystals began to crack yielding a poor diffraction pattern when subjected to synchrotron light source.
X-ray data collections and refinement –
Crystals derived from different soaking contact times were subjected to the same data collection procedure: 100 K, 400 frames, 0.5° of oscillation range, 1 Å wavelength and using the same X-ray dose at Elettra XRD1 (Trieste, Italy). All the crystals belong to the C2 space group, presenting more or less the same dimensions (a = 142, b = 102, c = 58. β = 112.89), as reported in Supplemental Table 1. Crystals diffracted from 2.05 to 2.3 Å of resolution. Data were processed with XDS [45] and reduced with CCP4 [46]. Structures were solved by the molecular replacement method (Phaser [47]) using the structure of the oxidized SmTGR (pdb code: 2V6O [2]) as a search model. Model building and refinement were performed using Coot [48] and PHENIX/Refine [49]. The ligands were modeled into the electron density only after convergence of the refinement procedure.
Molecular dynamics simulations –
Coordinates of X-ray model of TGR (PDB ID:2X99; 6RTJ), RnTrxR (PDB ID:3EAN), human GR (PDB ID:3DJG) were downloaded from the PDB. All the structure preparation steps were performed in Molecular Operating Environment (MOE, [50]). The proteins were subjected to the “structure preparation” procedure. Hydrogen atoms were added using the Protonate 3D algorithm. The monomer of TGR or RnTrxR were solvated in a periodic box with water and 0.1 M KCl using MOE “Solvate” module. The water box was extended at least 10 Å from the protein. The energy of the resulting structure was minimized with periodic boundary conditions (PBC) enabled and utilizing AMBER14:EHT forcefield in MOE [51,52] until the RMS gradient reached 0.01 kcal/mol/Å2. The “Molecular Dynamics” module in MOE was used to prepare the resulting structure for MD simulations using NAMD software, version 13, Linux-x86_64-multicore-CUDA (http://www.ks.uiuc.edu/Research/namd/ [53]). The MD trajectories were analyzed in VMD, version 1.9.3 [54]. The final plots with the distance data were prepared in Mathematica [55]. Each solvated complex was heated to 300 K for 1000 ps, equilibrated for 5000 ps in the NVT ensemble at 1 atm and 300 K. The non-bonded interactions were switched at 8 Å and zero smoothly at 10 Å (cutoff 10 Å, switchdist 8 Å, nonbondedScaling 1, pairlistdist 11.5 Å, limitdist 0.5 Å). The temperature was maintained using of Langevin dynamics with a damping coefficient of 5.0 ps−1. In PBC, the wrapAll parameter for used to calculate all the coordinates around periodic boundaries. Electrostatic interactions in PBC were treated using the Particle Mesh Ewald (PME) method and PME GridSpacing set at 1.0 Å. Covalent bonds with hydrogen atoms were kept rigid using ShakeH with the following parameters rigid bonds - all, rigid tolerance – 10−6 Å, and water molecules were kept rigid using Settle algorithm. The time step size for integration of each step of the simulation was 2 fs. All the other parameters were unchanged from the default settings in the NAMD software. The final production run was obtained from the last 20 ns of the simulation. In the analysis of the simulations, the pairwise distances were calculated between the pairs of Glu330 - N-LIG, Asp334 - N-LIG, Glu337 - N-LIG, Asp334 - O-LIG, Glu337 - O-LIG, Lys345 - C5-LIG, Glu337 - Lys345, Glu330 - Asp334, Asp334 - Glu337, where N-LIG and O-LIG correspond to the nitrogen and oxygen atoms in the substituents attached to C1 and C2 and C9-LIG – to the C9 carbon atom of the naphthyl ring in compound 4, respectively. In the amino acids, the distances were measured using the carbon atoms in the carboxyl groups of the sidechains in E330, D334, and E337 and nitrogen atom in the sidechain of K345. The distance plots were prepared in Mathematica [55] and are provided in Supplemental Fig. 2.
4.4. Ex vivo experiments with Adult worms and Schistosomula
Adult worms were isolated from infected mice as described [42] and cultured in RPMI medium + 10% fetal calf serum, 10 mM glutamine, and 1 × penicillin/streptomycin for 24 h before compound addition. Larval worms were prepared by mechanical transformation and Percoll gradient isolation from cercariae isolated from infected Biomphalaria glabrata as described [42]. Larval worms were cultured in M-199 + 10% fetal calf serum for 24 h before the addition of compounds. Worms were observed daily and scored as dead when no movement was seen after several minutes’ observation.
Accession numbers.
PBB ID: 6RTJ, SmTGR in complex with compound 4 at 1 h soaking; PBB ID: 6RTM, SmTGR in complex with compound 4 at 2 h soaking; PBB ID: 6RTO, SmTGR in complex with compound 4 at 4 h soaking.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health (NIH, USA) - National Institute of Allergy and Infectious Diseases (NIAID): Nai grant R33AI127635 to GRT, PAP, DLW, and FA and by Italian Ministry of Education, University and Research (MIUR) PRIN 20154JRJPP to AB. MA has been supported by MIUR under the national project FSE/FESR - PON Ricerca e Innovazione 2014–2020 ( (N° AIM1887574). We are grateful to Maurizio Polentarutti and Nicola Demitri (Elettra synchrotrone Trieste, Italy) for data collection support. B. glabrata snails were provided by the NIAID Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD) through NIH-NIAID Contract HHSN272201000005I for distribution through BEI Resources. NAMD and VMD were developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign. We thank the developers for the academic licenses for this software. We thank Elias Arnér, Karolinska Institutet (Sweden) for the kind gift of SmTGR in pABC2.
Abbreviations:
- FAD
flavin adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate reduced
- HEPE
1-(2-hydroxyethyl)piperazine functional group
- GR
glutathione reductase
- TrxR
high molecular weight mammalian thioredoxin reductase
- TGR
thioredoxin glutathione reductase
- GSH
glutathione
- Trx
thioredoxin
- Sec
selenocysteine
- Grx
glutaredoxin
- qHTS
quantitative high throughput screening
- oQM
ortho quinone methide
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.freeradbiomed.2019.12.019.
References
- [1].Zhang J, Zhang B, Li X, Han X, Liu R, Fang J, Small molecule inhibitors of mammalian thioredoxin reductase as potential anticancer agents: an update, Med. Res. Rev. 39 (2019) 5–39. [DOI] [PubMed] [Google Scholar]
- [2].Angelucci F, Miele AE, Boumis G, Dimastrogiovanni D, Brunori M, Bellelli A, Glutathione reductase and thioredoxin reductase at the crossroad: the structure of Schistosoma mansoni thioredoxin glutathione reductase, Proteins 72 (2008) 936–945. [DOI] [PubMed] [Google Scholar]
- [3].Alger HM, Williams DL, The disulfide redox system of Schistosoma mansoni and the importance of a multifunctional enzyme, thioredoxin glutathione reductase, Mol. Biochem. Parasitol. 121 (2002) 129–139. [DOI] [PubMed] [Google Scholar]
- [4].Prast-Nielsen S, Huang HH, Williams DL, Thioredoxin glutathione reductase: its role in redox biology and potential as a target for drugs against neglected diseases, Biochim. Biophys. Acta 1810 (2011) 1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Williams DL, Bonilla M, Gladyshev VN, Salinas G, Thioredoxin glutathione reductase-dependent redox networks in platyhelminth parasites, Antioxidants Redox Signal. 19 (2013) 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Global Health Estimates, Deaths by Cause, Age, Sex, by Country and by Region, 2000–2016, World Health Organization, Geneva, 2016 2018. [Google Scholar]
- [7].Stafford WC, Peng X, Olofsson MH, Zhang X, Luci DK, Lu L, Cheng Q, Trésaugues L, Dexheimer TS, Coussens NP, Augsten M, Ahlzén HM, Orwar O, Östman A, Stone-Elander S, Maloney DJ, Jadhav A, Simeonov A, Linder S, Arnér ESJ, Irreversible inhibition of cytosolic thioredoxin reductase 1 as a mechanistic basis for anticancer therapy, Sci. Transl. Med. 10 (428) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Simeonov A, Jadhav A, Sayed AA, Wang Y, Nelson ME, Thomas CJ, Inglese J, Williams DL, Austin CP, Quantitative high-throughput screen identifies inhibitors of the Schistosoma mansoni redox cascade, PLoS Neglected Trop. Dis. 2 (2008) e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li T, Ziniel PD, He PQ, Kommer VP, Crowther GJ, He M, Liu Q, Van Voorhis WC, Williams DL, Wang MW, High-throughput screening against thioredoxin glutathione reductase identifies novel inhibitors with potential therapeutic value for schistosomiasis, Infect Dis Poverty 4 (2015) 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Saccoccia F, Angelucci F, Boumis G, Carotti D, Desiato G, Miele AE, Bellelli A, Thioredoxin reductase and its inhibitors, Curr. Protein Pept. Sci. 15 (6) (2014) 621–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Baell JB, Nissink JWM, Seven year itch: pan-assay interference compounds (PAINS) in 2017-utility and limitations, ACS Chem. Biol. 13 (2018) 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dunlap T, Chandrasena RE, Wang Z, Sinha V, Wang Z, Thatcher GR, Quinone formation as a chemoprevention strategy for hybrid drugs: balancing cytotoxicity and cytoprotection, Chem. Res. Toxicol. 20 (2007) 1903–1912. [DOI] [PubMed] [Google Scholar]
- [13].Dufrasne F, Gelbcke M, Neve J, Kiss R, Kraus JL, Quinone methides and their prodrugs: a subtle equilibrium between cancer promotion, prevention, and cure, Curr. Med. Chem. 18 (2011) 3995–4011. [DOI] [PubMed] [Google Scholar]
- [14].Toteva MM, Richard JP, The generation and reactions of Quinone Methides, Adv. Phys. Org. Chem. 45 (2011) 39–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bolton JL, Quinone methide bioactivation pathway: contribution to toxicity and/ or cytoprotection? Curr. Org. Chem. 18 (2014) 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhou Q, Rokita SE, A general strategy for target-promoted alkylation in biological system, Proc. Natl. Acad. Sci. 26 (2003) 15452–15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Citta A, Folda A, Bindoli A, Pigeon P, Top S, Vessières A, Salmain M, Jaouen G, Rigobello MP, J. Med. Chem. 57 (2014) 8849–8859. [DOI] [PubMed] [Google Scholar]
- [18].Li X, Hou Y, Meng X, Ge C, Ma H, Li J, Fang J, Selective activation of a prodrug by thioredoxin reductase providing a strategy to target cancer cells, Angew Chem. Int. Ed. Engl. 57 (2018) 6141–6145. [DOI] [PubMed] [Google Scholar]
- [19].Krieg R, Jortzik E, Goetz AA, Blandin S, Wittlin S, Elhabiri M, Rahbari M, Nuryyeva S, Voigt K, Dahse HM, Brakhage A, Beckmann S, Quack T, Grevelding CG, Pinkerton AB, Schönecker B, Burrows J, Davioud-Charvet E, Rahlfs S, Becker K, Arylmethylamino steroids as antiparasitic agents, Nat. Commun. 8 (2017) 14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Johnson DS, Weerapana E, Cravatt BF, Strategies for discovering and derisking covalent, irreversible enzyme inhibitors, Future Med. Chem. 2 (2010) 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bellelli A, Carey J, Reversible Ligand Binding: Theory and Experiment, John Wiley and sons inc., Hoboken, NJ, USA, 2018. [Google Scholar]
- [22].Boyd DB, Leaving group ability and pKa in elimination reactions, J. Org. Chem. 50 (1985) 885–886. [Google Scholar]
- [23].Reich HJ, Hondal RJ, Why nature chose selenium, ACS Chem. Biol. 11 (4) (2016) 821–841. [DOI] [PubMed] [Google Scholar]
- [24].Weinert EE, Dondi R, Colloredo-Melz S, Frankenfield KN, Mitchell CH, Freccero M, Rokita SE, Substituents on quinone methides strongly modulate formation and stability of their nucleophilic adducts, J. Am. Chem. Soc. 128 (2006) 11940–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rosenberg JL, Brinn I, Excited state dissociation rate constants in naphthols, J. Phys. Chem. 76 (1972) 3558–3562. [Google Scholar]
- [26].Ma SS, Zhang MJ, Yuan DY, Qi ZB, 1-Morpholinomethyl-2-naphthol, Acta Crystallogr. E61 (2005) o1370–o1371. [Google Scholar]
- [27].Driscoll JP, Kornecki K, Wolkowski JP, Chupak L, Kalgutkar AS, O’Donnell JP, Bioactivation of phencyclidine in rat and human liver microsomes and recombinant P450 2B enzymes: evidence for the formation of a novel quinone methide intermediate, Chem. Res. Toxicol. 20 (2007) 1488–1497. [DOI] [PubMed] [Google Scholar]
- [28].Deller MC, Rupp B, Models of protein-ligand crystal structures: trust, but verify, J. Comput. Aided Mol. Des. 29 (2015) 817–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Smart OS, Horský V, Gore S, Svobodová Vařeková R, Bendová V, Kleywegt GJ, Velankar S, Validation of ligands in macromolecular structures determined by X-ray crystallography, Acta Crystallogr D Struct Biol 74 (2018) 228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lang PT, Ng HL, Fraser JS, Corn JE, Echols N, Sales M, Holton JM, Alber T, Automated electron-density sampling reveals widespread conformational polymorphism in proteins, Protein Sci. 19 (2010) 1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hajdu J, Neutze R, Sjögren T, Edman K, Szöke A, Wilmouth RC, Wilmot CM, Analyzing protein functions in four dimensions, Nat. Struct. Biol. 7 (2000) 1006–1012. [DOI] [PubMed] [Google Scholar]
- [32].Hammes-Schiffer S, Stuchebrukhov AA, Theory of coupled electron and proton transfer reactions, Chem. Rev. 110 (2010) 6939–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kuntz AN, Davioud-Charvet E, Sayed AA, Califf LL, Dessolin J, Arnér ES, Williams DL, Thioredoxin glutathione reductase from Schistosoma mansoni: an essential parasite enzyme and a key drug target, PLoS Med. 4 (6) (2007) e206. Erratum in: PLoS Med. 4, e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huang HH, Day L, Cass CL, Ballou DP, Williams CH Jr., Williams DL, Investigations of the catalytic mechanism of thioredoxin glutathione reductase from Schistosoma mansoni, Biochemistry 50 (2011) 5870–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W, Watching DNA polymerase η make a phosphodiester bond, Nature 487 (2012) 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cortés-Ríos J, Torres MJ, Campos-Bustamante MP, Romero-Parra J, Letelier ME, Pessoa-Mahana D, Chung H, Faúndez M, NADPH oxidase activity: spectrophotometric determination of superoxide using pyrogallol red, Anal. Biochem. 536 (2017) 96–100. [DOI] [PubMed] [Google Scholar]
- [37].Lyu H, Petukhov PA, Banta PR, Jadhav A, Lea WA, Cheng Q, Arnér ESJ, Simeonov A, Thatcher GRJ, Angelucci F, Williams DL, Characterization of lead compounds targeting the selenoprotein thioredoxin glutathione reductase for treatment of schistosomiasis, ACS Infectious Diseases (2019) Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Angelucci F, Dimastrogiovanni D, Boumis G, Brunori M, Miele AE, Saccoccia F, Bellelli A, Mapping the catalytic cycle of Schistosoma mansoni thioredoxin glutathione reductase by X-ray crystallography, J. Biol. Chem. 285 (2010) 32557–32567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang Y, Cheng T, Bryant SH, PubChem BioAssay: a decade’s development toward open high-throughput screening data sharing, SLAS Discov 22 (2017) 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Angelucci F, Sayed AA, Williams DL, Boumis G, Brunori M, Dimastrogiovanni D, Miele AE, Pauly F, Bellelli A, Inhibition of Schistosoma mansoni thioredoxin-glutathione reductase by auranofin: structural and kinetic aspects, J. Biol. Chem. 284 (2009) 28977–28985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Whitbread AK, Masoumi A, Tetlow N, Schmuck E, Coggan M, Board PG, Characterization of the omega class of glutathione transferases, Methods Enzymol. 401 (2005) 78–99. [DOI] [PubMed] [Google Scholar]
- [42].Silvestri I, Lyu H, Fata F, Boumis G, Miele AE, Ardini M, Ippoliti R, Bellelli A, Jadhav A, Lea WA, Simeonov A, Cheng Q, Arnér ESJ, Thatcher GRJ, Petukhov PA, Williams DL, Angelucci F, Fragment-based discovery of a regulatory site in thioredoxin glutathione reductase acting as “doorstop” for NADPH entry, ACS Chem. Biol. 13 (2018) 2190–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cheng Q, Arnér ES, Selenocysteine insertion at a predefined UAG codon in a release factor 1 (RF1)-depleted Escherichia coli host strain bypasses species barriers in recombinant selenoprotein translation, J. Biol. Chem. 292 (2017) 5476–5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Faúndez M, Rojas M, Bohle P, Reyes C, Letelier ME, Aliaga ME, Speisky H, Lissi E, López-Alarcón C, Pyrogallol red oxidation induced by superoxide radicals: application to evaluate redox cycling of nitro compounds, Anal. Biochem. 419 (2) (2011) 284–291. [DOI] [PubMed] [Google Scholar]
- [45].Kabsch W, Xds, Acta crystallogr., Sect. D: Biol. Crystallogr. 66 (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, Overview of the CCP4 suite and current developments, Acta Crystallogr. D Biol. Crystallogr. 67 (2011) 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, J. Appl. Crystallogr. 40 (2007) 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallogr. D Biol. Crystallogr. 60 (2004) 2126–2132. [DOI] [PubMed] [Google Scholar]
- [49].Echols N, Moriarty NW, Klei HE, Afonine PV, Bunkoczi G, Headd JJ, McCoy AJ, Oeffner RD, Read RJ, Terwilliger TC, Adams PD, Automating crystallographic structure solution and refinement of protein-ligand complexes, Acta Crystallogr. Sect. D Biol. Crystallogr. 70 (2014) 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Molecular Operating Environment (MOE), Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2019.
- [51].Gerber PR, Muller K, MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry, J. Comput. Aided Mol. Des. 9 (1995) 251–268. [DOI] [PubMed] [Google Scholar]
- [52].Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C, ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB, J. Chem. Theory Comput. 11 (2015) 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K, Scalable molecular dynamics with NAMD, J. Comput. Chem. 26 (2005) 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Humphrey W, Dalke A, Schulten K, VMD: visual molecular dynamics, J. Mol. Graph. 14 (33–38) (1996) 27–38. [DOI] [PubMed] [Google Scholar]
- [55].Mathematica, Wolfram Research, Inc, (2019) Champaign, Illinois.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.