Abstract
Objective:
To evaluate the consistency and objectivity of deep neural networks in embryo scoring and making disposition decisions for biopsy and cryopreservation in comparison to grading by highly trained embryologists.
Design:
Prospective double-blind study using retrospective data.
Setting:
U.S.-based large academic fertility center.
Patients:
Not applicable.
Intervention(s):
Embryo images (748 recorded at 70 hours postinsemination [hpi]) and 742 at 113 hpi) were used to evaluate embryologists and neural networks in embryo grading. The performance of 10 embryologists and a neural network were also evaluated in disposition decision making using 56 embryos.
Main Outcome Measures:
Coefficients of variation (%CV) and measures of consistencies were compared.
Results:
Embryologists exhibited a high degree of variability (%CV averages: 82.84% for 70 hpi and 44.98% for 113 hpi) in grading embryo. When selecting blastocysts for biopsy or cryopreservation, embryologists had an average consistency of 52.14% and 57.68%, respectively. The neural network outperformed the embryologists in selecting blastocysts for biopsy and cryopreservation with a consistency of 83.92%. Cronbach’s α analysis revealed an α coefficient of 0.60 for the embryologists and 1.00 for the network.
Conclusions:
The results of our study show a high degree of interembryologist and intraembryologist variability in scoring embryos, likely due to the subjective nature of traditional morphology grading. This may ultimately lead to less precise disposition decisions and discarding of viable embryos. The application of a deep neural network, as shown in our study, can introduce improved reliability and high consistency during the process of embryo selection and disposition, potentially improving outcomes in an embryology laboratory.
Keywords: Convolutional neural networks, deep-learning, embryo assessment, machine learning
Abstract
Objetivo:
evaluar la consistencia y objetividad de redes neuronales profundas en la valoración de embriones y toma de decisiones de biopsia y criopreservación en comparación con la valoración por parte de embriólogos altamente entrenados.
Diseño:
Estudio prospectivo doble ciego utilizando datos retrospectivos.
Escenario:
Centro de fertilidad académico de Estados Unidos.
Pacientes:
No aplica.
Intervenciones:
Imágenes de embriones (748 registradas a las 70 horas post inseminación (hpi) y 724 a las 113 horas post hpi) fueron utilizadas para evaluar a los embriólogos y a las redes neuronales profundas en la valoración de embriones. El desempeño de 10 embriólogos y de redes neuronales profundas también fue evaluado en la toma de decisiones utilizando 56 embriones.
Medidas de resultado principal:
Se compararon coeficientes de variación (%CV) y medidas de consistencia.
Resultados:
Los embriólogos presentaron un alto grado de variabilidad (promedio de %CV: 82.84% y 44.98%) en la valoración de los embriones. Al seleccionar los blastocistos para biopsia o criopreservación, los emrbriólogos tuvieron un promedio de consistencia de 52.14% y 57.68%, respectivamente. La red neuronal tuvo un desempeño superior al de los embriólogos al seleccionar blastocistos para biopsia o criopreservación con una consistencia de 83.92%. Un análisis de Cronbach reveló un coeficiente α de 0.60 para los emrbriólogos y 1.00 para la red.
Conclusiones:
Los resultados de nuestro estudio muestran un alto grado de variabilidad intra e inter embri_ologo al valorar los embriones, probablemente debido a la naturaleza subjetiva de la valoración morfológica tradicional. Esto podría llevar a decisiones menos precisas y a descartar embriones viables. La aplicación de una red neuronal profunda, como se muestra en nuestro estudio, puede introducer una fiabilidad mejorada y alta consistencia durante el proceso de selección de embriones y disposición, potencialmente mejorando los resultados en un laboratorio de embriología.
In vitro fertilization techniques involve culturing embryos for multiple days in controlled and monitored environmental conditions and are assessed at different stages of embryo development to validate their quality. Highly trained embryologists make visual assessments of embryo morphologies, and such evaluations are used to make decisions on the fate of these embryos. However, these conventional visual evaluations of embryo morphology have been observed to possess high intervariability and intravariability, and the lack of consistency in such assessments leads to variability in decision making that is detrimental to both patient care and advancement of the field overall (1–4).
Computer-assisted evaluations could help minimize variability among embryologists in embryo scoring and decision-making processes. Rule-based computer vision image analysis approaches have focused on measuring specific expert-defined parameters such as zona pellucida thickness variation, number of blastomeres, degree of cell symmetry and cytoplasmic fragmentation, in embryo images collected using highly controlled imaging systems (5–7). Commercially available, time-lapse imaging (TLI) systems, for example, make use of such computer vision-based image analysis methods in evaluating embryos, and have enabled regular and automated data acquisition of embryo development under controlled environments, along with identifying objective morphokinetic parameters. These highly expensive and bulky instruments, which are used by less than 20% of fertility centers in the United States, have helped to improve objectivity of the decision-making processes in embryology laboratories but are unable to improve consistency among embryologists, owing to their reliance on measurements that require human interference (3, 8, 9). Furthermore, these methods are limited to evaluating specific parameters and require embryologists to dedicate more time in evaluating their measures to meet their own laboratory specific criteria (10).
Fully automated assessments of embryos, where quality grades are assigned without user interventions from an image analysis standpoint, is challenging, partially due to the complexity of embryo morphologies. However, advances in machine learning have enabled objective and accurate image classification in both medical and nonmedical fields without the need for manual feature engineering (11–15). Unsurprisingly, there has been significant interest in the research community on using machine learning-based frameworks for embryo analysis (16–26). The focus of most research in this domain primarily revolves around the use of various machine learning concepts toward the identification of the best-quality embryo based on their developmental grade or potential for implantation.
Machine learning, a concept of artificial intelligence, focuses on techniques that enable computer systems to improve with data and experience. The classical machine learning techniques require hand-crafted features (human input) for efficient performance. Others have used a variety of classical machine learning techniques such as support vector machines, logistic regression models, random forests, and Bayesian classifiers for embryo assessment (16). Deep-learning, which is a subset of machine learning, takes a different approach through representation learning: namely, the system identifies features directly and does not rely on hand-crafted features. One of the most popular deep-learning concepts that has seen numerous implementations in the field of assisted reproduction is convolutional neural networks (CNN) (19–22). Such systems are “trained” using large datasets of embryo images, where they learn the required parameters associated with specific qualities by themselves over time. Furthermore, unlike rule-based computer vision algorithms, these networks are “black boxes,” and their decision-making process is not easy to interpret. These systems have been evaluated for their ability to correctly classify between different qualities of embryos; however, their abilities in consistency have not been evaluated (20–22).
In this study, we report the performance of embryologists in terms of consistency in evaluating embryo morphology as part of their routine clinical tasks, and compared their results with the results obtained using a CNN-based framework. First, we intended to study the variability of embryo scoring between embryologists and the variability associated with CNN. We evaluated neural networks trained with embryo images recorded at 70 hours postinsemination (hpi) and 113 hpi in embryo scoring and compared its performance with that of embryologists. Although embryo scoring is an important task performed by embryologists that is used toward clinical decision-making, it does not fully capture the implications of variability in decision making in a clinical setting. We studied the consistency of 10 embryologists in performing routine clinical tasks such as selecting embryos for biopsy and cryopreservation with the alternative of discarding embryos, and compared their results with the results obtained through the CNN-based approach. The criteria for embryo selection for biopsy and cryopreservation at the participating fertility center are the same (>3CC blastocyst at 113 hpi). Our results suggest that these networks far exceed human performance in terms of consistency in embryo scoring and decision making.
MATERIALS AND METHODS
Embryo Dataset and Neural Network Training
We collected 3469 embryo images recorded at 70 and 113 hpi from the Massachusetts General Hospital (MGH) fertility center in Boston, Massachusetts, under institutional review board approval (IRB#2017P001339 and IRB#2019P002392). These embryo images were annotated by embryologists with a minimum of 5 years’ experience. Only images of embryos that were completely nondiscernible were removed from the study as part of the data cleaning procedure. The networks reported in this study were trained using embryo image data recorded with an Embryoscope at 70 hpi and 113 hpi as reported previously (20, 21, 27). Briefly, the CNN was developed using the dataset of embryo images collected at 70 and 113 hpi from 3469 embryos. Only embryos that fertilized normally by 18 hpi were used in training and validating the neural network. A five-class categorization system based on the blastocyst quality at 113 hpi was employed, and was used to train the neural network in both prediction and classification tasks. The network “learns” by identifying patterns in images that are associated with the respective classes of embryos. Data augmentations through flips and rotations were performed during training to ensure the robustness of the trained algorithm. Embryos evaluated in this study were independent of the training set; that is, the CNN was evaluated using embryo image data that it had not seen during training.
Embryo Scoring Assessments
A total of 10 embryologists participated in our study, with 8 at the senior level with more than 5 years of experience and 2 at the junior level with less than 2 years of experience (Table S1). The embryos were categorized into 5 classes based on their morphological features (1 = poor, 2 = fair, 3 = good, 4 = great, and 5 = excellent) at 2 timepoints of 70 hpi and 113 hpi. Eight embryologists evaluated 748 embryo images acquired at 70 hpi, and 7 embryologists evaluated 742 embryo images acquired at 113 hpi. Neural networks trained with both 70 hpi and 113 hpi embryo images were tested three times to measure their variability. The neural network was trained using the annotated data based on the developmental stage grading system used at the fertility center. The grading system used was consolidated into five measures of quality (classes 1–5) in this study. The grades provided by the system and the embryologists for each embryo were used in measuring their overall repeatability and similarity. In cases in which the embryologists were uncertain about the quality/grade of the embryo, they were allowed to report a score of zero. Coefficients of variation were calculated to describe the variability between embryologists on their scoring tendencies (interobserver variability). An intraclass correlation coefficient (ICC) analysis was performed to evaluate the agreement between the embryologists in scoring embryos.
Embryo Biopsy and Cryopreservation Consistency Assessments
Ten embryologists participated in the experiments for selecting embryos for biopsy and cryopreservation. The criteria for embryo selection in both cases were the same (>3CC blastocysts). Both groups (i.e., embryologists and CNN) were presented with a blinded set of 56 blastocysts imaged at 113 hpi, which they were tasked with deciding between biopsy or discard. Similarly, they were also asked to decide between cryopreservation or discard of the same set of blinded embryos. In both tasks, each embryo was also rotated 90°, 180°, and 270° and presented to the embryologists at random with the original set of images (n = 224). The embryologists were blinded to both their decisions and the indices of duplicate images. We compared the decisions made, with the developmental grades available for each embryo as part of the historical clinical data. Each embryologist also evaluated the 56 embryos to assign a developmental grade to each embryo based on the grading system used by the fertility center, which evaluated the degree of blastocoel expansion, inner cell mass, and trophectoderm. We also analyzed the consistency of the CNN in embryo assessment (discard or cryopreservation/biopsy; n = 224) and compared its results with the consistency of the embryologists. Consistency here is defined as the percentage of cases in which their decisions were invariant to rotations. In addition, we measured the internal consistency between each task for every individual embryologist (intraobserver variability). A mode was used to calculate the final decisions of every embryologist for each embryo in the two tasks, and the modes (their decisions) were compared along with their own developmental grade assessment through the Cronbach’s α statistical analysis (intraobserver variability).
Statistical Analysis
Coefficient of variation estimations, interclass correlation coefficient calculations, Cronbach’s α tests for internal consistency, and one-sample t-tests were performed using Medcalc 14.8.1. Coefficient of variation was used to describe the relative dispersion of scoring tendencies observed among embryologists and the neural network for the same set of embryos, by calculating the ratio of the standard deviation to the mean of categorization frequencies of embryologists. An interclass correlation coefficient was also calculated taking into consideration absolute agreement to understand the reliability of a typical embryologist (28). Similarly, Cronbach’s α was used to compare the reliability in the network’s and embryologists’ decisions by comparing their decisions across the three tasks: namely, embryo scoring, selection for biopsy, and cryopreservation. One-sample t-tests were performed to compare the consistency and α values of the embryologists and the network (29).
RESULTS
System Evaluation in Embryo Scoring Against Trained Embryologists
The consistency of embryologists in scoring embryo morphologies at 70 hpi was assessed based on a five-grade system (1 = poor, 2 = fair, 3 = good, 4 = great, and 5 = excellent). Eight embryologists performed visual assessments of embryo quality using 748 embryo images recorded at 70 hpi. The frequency of each grade among the embryologists was compared by calculating the coefficient of variation (%CV) across the eight embryologists. The embryologists classified embryos (n = 748) as poor, fair, good, great, and excellent with %CVs of 109.11%, 49.15%, 49.86%, 44.30%, and 161.79%, respectively (Figure 1a, Figure S1). A high degree of variability (%CV average-82.84%) was observed among embryologists when assessing embryos at 70 hpi. An ICC analysis revealed a single-measure absolute agreement rating of 0.2036 with a 95% confidence interval (CI) ranging from 0.1509 to 0.2577. Similarly, seven embryologists classified embryo images (n = 742) recorded at 113 hpi as poor, fair, good, great, and excellent using embryo images with %CVs of 51.45%, 43.69%, 52.58%, 21.41% and 55.75%, respectively (Figure 1b, Figure S1). Although the degree of variability between the embryologists was still high (%CV average-44.98%), they had a relatively better agreement in terms of assessing embryo images recorded at 113 hpi compared to when they assessed data recorded at 70 hpi. The results of these experiments showed that there was significant variation among the embryologists in terms of embryo quality assessments. Furthermore, ICC analysis revealed a relatively higher single-measure absolute agreement rating of 0.4010 (confidence interval [CI] 0.3626–0.4399). In comparison, neural networks perform with 100% repeatability for classifying embryo images recorded at 70 hpi and 113 hpi as poor, fair, good, great, and excellent.
FIGURE 1.
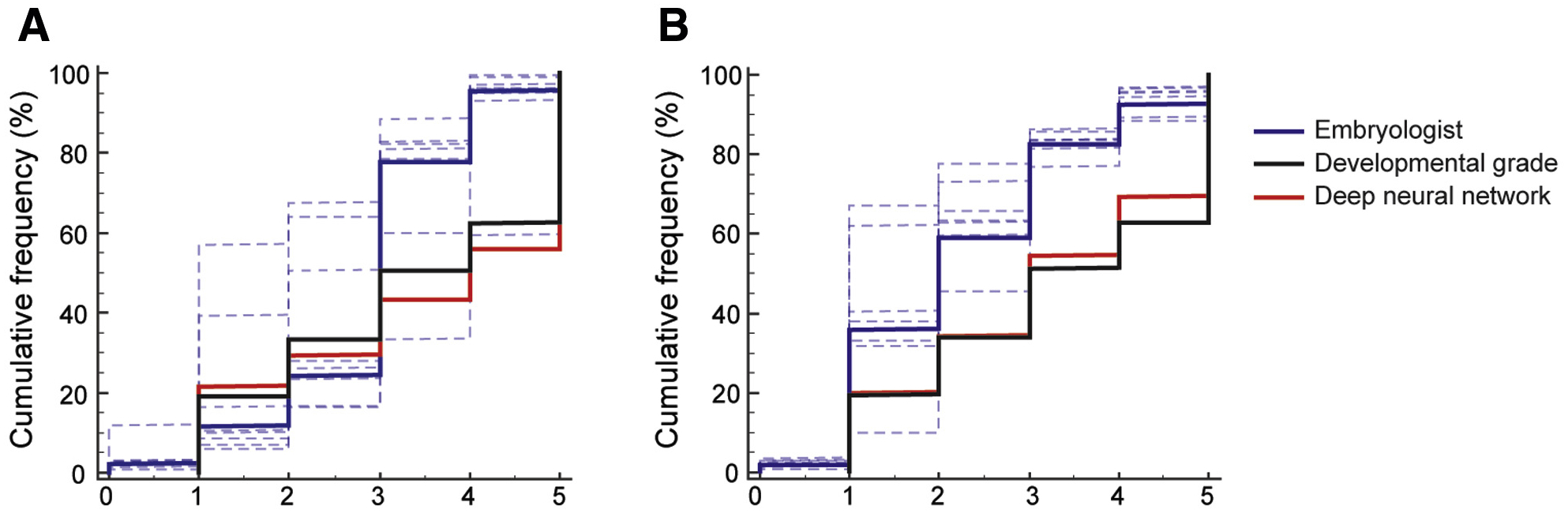
Objectivity and consistency among embryologists in embryo assessments. The cumulative frequency charts show the distribution of embryos based on quality scores assigned by embryologists along with the distribution of their developmental grades, and scores generated by the deep-neural network using embryo images recorded at (A) 70 hpi (n = 748) and (B) 113 hpi (n = 742). Dotted lines represent the individual distributions, and bold lines represent the consensus. The scores 1–5 represent the different embryo quality grades in increasing order; score 0 represents categorization when an embryologist was uncertain about the quality grade of an embryo.
System Evaluation for Consistency in Biopsy and Cryopreservation Decisions Compared to Trained Embryologist Decisions
For the task of selecting embryos for biopsy, we observed that the embryologists (n = 56, 10 embryologists) performed on average with a consistency of 52.14% (CI 40.99%–63.29%), whereas the CNN outperformed (P < .05; t-test) the embryologists with a consistency of 83.92% (Figure 2A). Similarly, in selecting embryos for cryopreservation (n = 56, 10 embryologists), we observed that the embryologists performed with an average consistency of 57.68% (CI 47.39%– 67.97%), whereas the CNN outperformed the embryologists with a consistency of 83.92% (P < .05; t-test) (Figure 2B). The comparison of the overall decisions made by the embryologists for each task along with their own developmental grade scores through a Cronbach’s α analysis revealed an average α value of 0.60 (CI 47.39–67.97). In contrast, the system performed significantly better (P < .05; t-test) with an excellent α of 1.00 (lower CI 1.00) (Figure 2C).
FIGURE 2.
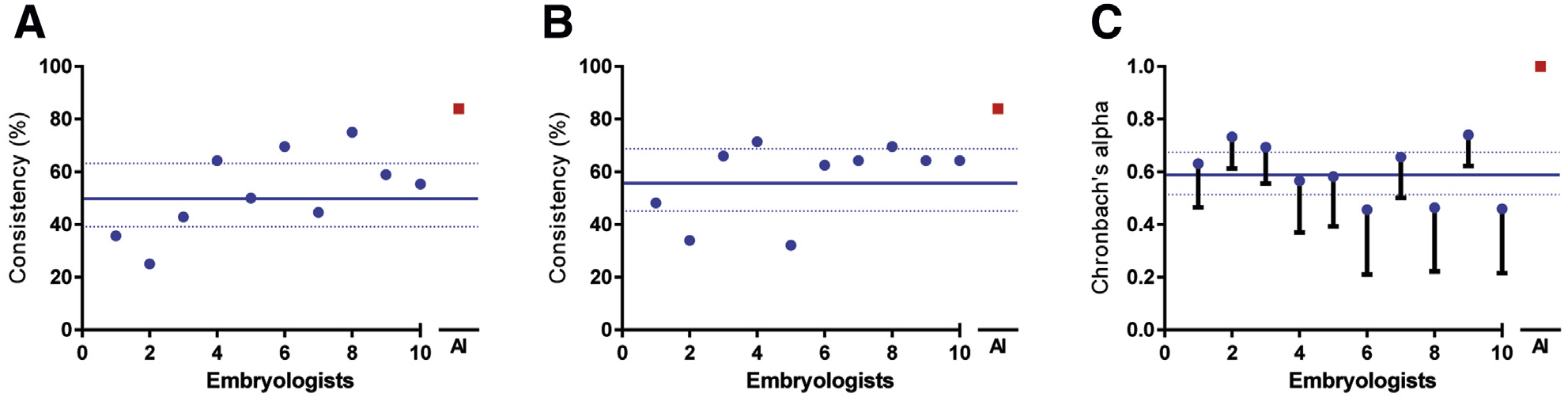
Comparison of consistency and reliability in decision making among embryologists and the deep-neural network. (A) and (B) represent the consistency among embryologists and the deep neural network in disposition decision making when presented with the task of either discarding or selecting embryos for biopsy and the task of either discarding or selecting embryos for cryo-preservation (n = 56). (C) Measure of reliability for each individual and the deep neural network, through Cronbach’s α analysis of internal consistency among the tasks of biopsy, cryopreservation, and developmental stage grading (>3CC or <3CC). The criterion for both biopsy and cryopreservation of embryos was the exhibition of a >3CC developmental stage.
DISCUSSION
Embryo scoring is a routine clinical task performed by embryologists to gauge the quality of embryos. Clinically, embryo scoring influences decisions made as part of patient care. In research, the validity of many interventional studies is estimated with embryo score improvements. Embryologists are required to perform such important evaluations without support systems, on top of their original set of responsibilities. The lack of consistency among embryologists is of significant concern, as variable decisions can lead to the increase in potentially discarding viable embryos and decreasing cumulative pregnancy rates.
In our studies, embryologists had a significant variability when evaluating embryos based on the developmental grade and their overall quality. We observed that the embryologists scored embryos at 70 hpi and 113 hpi with high variability and poor reliability. Furthermore, the embryologists were inconsistent in classifying embryos based on the perceived quality, whereas the neural network remained close to its original developmental-grade–based categorization. Similarly, even embryo images oriented at different angles led to laboratory staff making final embryo disposition decisions (perform embryo biopsy, cryopreserve, discard) with high variability. In such simulated clinical decision–making experiments, the average consistency of embryologists in making disposition decisions was 0.60, as revealed by the Cronbach’s α test. The recommended internal consistency range should be higher than 0.9 α coefficient in clinical settings (30). The results from our study confirm those of other independent studies that show high variability among embryologists (1–4). Thus far, there has never been a scalable solution available for this important problem in embryology.
However, our evaluations with deep-neural networks indicate that such systems can be used to improve objectivity and consistency of embryo assessment. The evaluated CNN performed significantly better compared to embryologists in all tested tasks and with a high consistency of 84%, and can be further improved (robustness to perspective shift) through additional training. Such a neural network–based system does not rely on an embryologist’s input and thus is not affected by fatigue or other human factors. Such systems can potentially be helpful in relieving the burden of mundane routine tasks from embryologists, freeing them for more important responsibilities; however the efficacy still needs to be tested in clinical settings through additional prospective studies. Currently, the system is suitable for use as a decision support system, which can speed up certain tasks and act as a safety net by automatically highlighting possible erroneous classifications/discards that may have a detrimental effect on patient care.
Recently, multiple independent studies focused on deep learning–based applications in embryo scoring have been reported (6, 18, 20, 21). These studies suggest improved objectivity and consistency with the use of deep learning; however, it is important to understand that such results cannot be generalized across any and all neural network–based approaches or for all datasets. For example, if these networks “overfit” (i.e., memorize) during training, they tend to perform poorly in terms of consistency. Furthermore, most neural networks do not adapt well to different imaging systems and are limited to systems that were used in gathering the training data. Although CNN-based approaches provide an alternative to current methods of embryo scoring, limitations to their performance, among other factors, are dependent on the dataset used in training and the methodology involved in training such systems. Additionally, the use of time lapse data is still of interest to the assisted reproduction community, and in such cases, multi-dimensional neural networks and recurrent neural networks, are generally used. However, these forms of networks have similar limitations, and it is important to emphasize that the generalizability of neural networks stems from the features learned by the network and is not inherently applicable to neural networks. Studies on the robustness of a neural network’s performance are needed to better understand each trained model, because feature learning is done by the system, and a system’s learning is greatly affected by the training methodologies used.
Advances in artificial intelligence (AI) have paved the way for a new direction of computer-assisted decision support systems. Our AI-based approach requires no handcrafted feature selection, and the algorithm automatically learns and identifies important embryo features for automated assessments. These systems do not need expensive hardware for their operation. These algorithms are easily adaptable to different imaging platforms and are not limited to expensive time-lapse imaging platforms. Previous studies on using deep-neural networks have shown their portability to inexpensive hardware such as portable computers (<$100) and smartphones (<$5) (12, 22). The underlying costs of the technology associated with developing these networks promises easier and increased adoption of the technology from a scalability standpoint. The approach shows significant promise and warrants prospective randomized control trials to study its effectiveness in clinical practice.
Supplementary Material
Acknowledgments:
The authors would like to thank embryology staff from Massachusetts General Hospital for participating in this study. This work was supported by the Brigham and Women’s Hospital (BWH) Precision Medicine Developmental Award (BWH Precision Medicine Program) and Partners Innovation Discovery Grant (Partners Health-care). It was also partially supported through 1R01AI118502 and R01AI138800 Awards (National Institutes of Health).
C.L.B., M.K.K., P.T., and H.S. have a patent (WO2019068073A1) pending. H.S. has received grants from National Institutes of Health (1R01AI118502 and R01AI138800), during the conduct of the study.
Footnotes
C.L.B. has nothing to disclose; P.T. has nothing to disclose; M.K.K. has nothing to disclose; H.K. has nothing to disclose; I.S. has nothing to disclose; I.D. has nothing to disclose; R.G. has nothing to disclose; R.G. has nothing to disclose; R.P. has nothing to disclose; H.S. has nothing to disclose.
Discuss: You can discuss this article with its authors and other readers at https://www.fertstertdialog.com/users/16110-fertility-and-sterility/posts/56966-28655
REFERENCES
- 1.Storr A, Venetis CA, Cooke S, Kilani S, Ledger W. Inter-observer and intraobserver agreement between embryologists during selection of a single day 5 embryo for transfer: a multicenter study. Hum Reprod 2017;32: 307–14. [DOI] [PubMed] [Google Scholar]
- 2.Baxter Bendus AE, Mayer JF, Shipley SK, Catherino WH. Interobserver and intraobserver variation in day 3 embryo grading. Fertil Steril 2006;86: 1608–15. [DOI] [PubMed] [Google Scholar]
- 3.Paternot G, Devroe J, Debrock S, D’Hooghe TM, Spiessens C. Intra- and inter-observer analysis in the morphological assessment of early-stage embryos. Reprod Biol Endocrinol 2009;7:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tunis SR, Clarke M, Gorst SL, Gargon E, Blazeby JM, Altman DG, et al. Improving the relevance and consistency of outcomes in comparative effectiveness research. J Comp Effect Res 2016;5:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocha JC, Passalia FJ, Matos FD, Takahashi MB, Maserati MP Jr, Alves MF, et al. Automatized image processing of bovine blastocysts produced in vitro for quantitative variable determination. Sci Data 2017;4:170192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocha JC, Passalia FJ, Matos FD, Takahashi MB, Ciniciato DdS, Maserati MP, et al. A method based on artificial intelligence to fully automatize the evaluation of bovine blastocyst images. Sci Rep 2017;7:7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filho ES, Noble JA, Wells D. A review on automatic analysis of human embryo microscope images. Open Biomed Eng J 2010;4:170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundvall L, Ingerslev HJ, Breth Knudsen U, Kirkegaard K. Inter- and intraobserver variability of time-lapse annotations. Hum Reprod 2013;28: 3215–21. [DOI] [PubMed] [Google Scholar]
- 9.Dolinko AV, Farland LV, Kaser DJ, Missmer SA, Racowsky C. National survey on use of time-lapse imaging systems in IVF laboratories. J Assist Reprod Genet 2017;34:1167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y-G, Lazzaroni-Tealdi E, Wang Q, Zhang L, Barad DH, Kushnir VA, et al. Different effectiveness of closed embryo culture system with time-lapse imaging (EmbryoScope™) in comparison to standard manual embryology in good and poor prognosis patients: a prospectively randomized pilot study. Reprod Biol Endocrinol 2016;14:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potluri V, Kathiresan PS, Kandula H, Thirumalaraju P, Kanakasabapathy MK, Pavan SKS, et al. An inexpensive smartphone-based device for point-of-care ovulation testing. Lab Chip 2019;19:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436. [DOI] [PubMed] [Google Scholar]
- 14.Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med 2019;25: 24–9. [DOI] [PubMed] [Google Scholar]
- 15.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25:44–56. [DOI] [PubMed] [Google Scholar]
- 16.Curchoe CL, Bormann CL. Artificial intelligence and machine learning for human reproduction and embryology presented at ASRM and ESHRE 2018. J Assist Reprod Genet 2019;36:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran D, Cooke S, Illingworth PJ, Gardner DK. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum Reprod 2019;34:1011–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khosravi P, Kazemi E, Zhan Q, Malmsten JE, Toschi M, Zisimopoulos P, et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit Med 2019;2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thirumalaraju P, Bormann CL, Kanakasabapathy M, Doshi F, Souter I, Dimitriadis I, et al. Automated sperm morpshology testing using artificial intelligence. Fertil Steril 2018;110:e432. [Google Scholar]
- 20.Dimitriadis I, Bormann CL, Thirumalaraju P, Kanakasabapathy M, Gupta R, Pooniwala R, et al. Artificial intelligence-enabled system for embryo classification and selection based on image analysis. Fertil Steril 2019;111:e21. [Google Scholar]
- 21.Thirumalaraju P, Hsu JY, Bormann CL, Kanakasabapathy M, Souter I, Dimitriadis I, et al. Deep learning-enabled blastocyst prediction system for cleavage stage embryo selection. Fertil Steril 2019;111:e29. [Google Scholar]
- 22.Kanakasabapathy M, Dimitriadis I, Thirumalaraju P, Bormann CL, Souter I, Hsu J, et al. An inexpensive, automated artificial intelligence (AI) system for human embryo morphology evaluation and transfer selection. Fertil Steril 2019;111:e11. [Google Scholar]
- 23.Thirumalaraju P, Bormann CL, Kanakasabapathy MK, Kandula H, Shafiee H. Deep learning-enabled prediction of fertilization based on oocyte morphological quality. Fertil Steril 2019;112:e275. [Google Scholar]
- 24.Kanakasabapathy MK, Thirumalaraju P, Gupta R, Pooniwala R, Kandula H, Souter I, et al. Improved monitoring of human embryo culture conditions using a deep learning-derived key performance indicator (KPI). Fertil Steril 2019;112:e70–1. [Google Scholar]
- 25.Kanakasabapathy MK, Thirumalaraju P, Bormann CL, Kandula H, Sai Pavan SK, Yarravarapu D, et al. Deep learning-enabled smartphone-based system for automated embryo assessments and evaluation. Fertil Steril 2019;112:e285–6. [Google Scholar]
- 26.Hariton E, Dimitriadis I, Kanakasabapathy MK, Thirumalaraju P, Gupta R, Pooniwala R, et al. A deep learning framework outperforms embryologists in selecting day 5 euploid blastocysts with the highest implantation potential. Fertil Steril 2019;112:e77–8. [Google Scholar]
- 27.Thirumalaraju P, Kanakasabapathy MK, Gupta R, Pooniwala R, Kandula H, Souter I, et al. Automated quality assessment of individual embryologists performing ICSI using deep learning-enabled fertilization and embryo grading technology. Fertil Steril 2019;112:e71.31623745 [Google Scholar]
- 28.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull 1979;86:420–8. [DOI] [PubMed] [Google Scholar]
- 29.Cronbach LJ. Coefficient alpha and the internal structure of tests. Psychometrika 1951;16:297–334. [Google Scholar]
- 30.Bland JM, Altman DG. Cronbach’s alpha. BMJ (Clin Research Ed) 1997;314: 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


