SUMMARY
Dengue virus (DENV) infects an estimated 390 million people each year worldwide. As tetravalent DENV vaccines have variable efficacy against DENV serotype 2 (DENV2), we evaluated the role of genetic diversity within the pre-membrane (prM) and envelope (E) proteins of DENV2 on vaccine performance. We generated a recombinant DENV2 genotype variant panel with contemporary prM and E isolates that are representative of global genetic diversity. The DENV2 genotype variants differ in growth kinetics, morphology, and virion stability. Importantly, the DENV2 genotypic variants are differentially neutralized by monoclonal antibodies, polyclonal serum neutralizing antibodies from DENV2-infected human subjects, and vaccine-elicited antibody responses from the TV003 NIH DENV2 monovalent and DENV tetravalent vaccines. We conclude that DENV2 prM and E genetic diversity significantly modulates antibody neutralization activity. These findings have important implications for dengue vaccines, which are being developed under the assumption that intraserotype variation has minimal impact on neutralizing antibodies.
Graphical Abstract
In Brief
Martinez et al. demonstrate that dengue virus serotype 2 (DENV2) genetic variation modulates neutralizing antibody activity from infection and vaccination. This observation underlines that genotypic variation impacts dengue virus 2 evasion from humoral immunity, suggesting that intraserotype genotypic variation should be considered in designing dengue vaccines.
INTRODUCTION
Dengue virus (DENV) is one of the most significant arthropod-borne viruses (arbovirus) with respect to disease burden in humans, causing ~390 million new infections each year worldwide (Bhatt et al., 2013). DENV is primarily spread by the female mosquitoes Aedes aegypti and Aedes albopictus, which are two vectors characterized by rapidly expanding geographic distributions throughout the world (Kraemer et al., 2019; Lambrechts et al., 2010, 2011). As a consequence of human activities and environmental changes that favor mosquito vectors, over the next 30–50 years, DENV infection incidence rates are also projected to increase in frequency in locations where it is currently endemic and also spread to new parts of the world (Kraemer et al., 2019). DENV is classified into four serotypes, which include DENV serotype 1 (DENV1), DENV2, DENV3, and DENV4. Primary human infections with any of the DENV serotypes are most often asymptomatic; however, disease cases can lead to high fevers, severe joint and muscle pain, and rash (Simmons et al., 2012). Secondary DENV infection with a heterologous serotype increases the risk of developing severe dengue hemorrhagic fever (DHF) or dengue hemorrhagic shock (DHS) (Simmons et al., 2012). The high global disease burden of DENV, coupled with the potential for disease enhancement after a secondary infection, has stimulated the development of multiple candidate vaccines, including killed, recombinant vectored and live-attenuated tetravalent vaccines (Bauer et al., 2015; Biswal et al., 2019; Hadinegoro et al., 2015; Kirkpatrick et al., 2016; Schmidt et al., 2017).
Recently, the World Health Organization (WHO) and the U.S. Food and Drug Administration (FDA) approved a live-attenuated tetravalent DENV vaccine, Dengvaxia (developed by Sanofi Pasteur), in people with preexisting immunity acquired via natural infections. The vaccine is approved for use in seropositive individuals, because vaccine efficacy was high in this group (Sridhar et al., 2018). In seronegative children, overall vaccine efficacy was low, and vaccination increased the risk of developing severe disease upon wild-type DENV infection (Sridhar et al., 2018). Moreover, the vaccine efficacy of Dengvaxia is highly variable for the four DENV serotypes, with a vaccine efficacy of 74%, 75%, and 63% against DENV4, DENV3, and DENV1, respectively. Yet Dengvaxia performs remarkably poorly against DENV2 with an overall efficacy of 39% (Juraska et al., 2018), and in select populations less than 10% efficacy as shown in Thai children (Sabchareon et al., 2012). The immunologic or virologic basis for the poor performance of Dengvaxia against DENV2 has not been well defined. Likely explanations include the poor replication of the DENV2 vaccine component as has been described previously (Torresi et al., 2017) as well as genetic and antigenic differences between the DENV2 vaccine component and circulating DENV2 strains.
It is well known that within each DENV serotype there is genetic and antigenic diversity resulting in multiple distinct genotypes (Shrivastava et al., 2018; Waman et al., 2016; Weaver and Vasilakis, 2009). However, genetic and antigenic differences between DENVs belonging to the same serotype have not been considered significant enough to impact long-term protective immunity. DENV vaccines have been formulated under the assumption that the envelope (E) proteins from a single strain will stimulate broadly protective antibody responses to all genotypes within a serotype. Several recent studies challenge this assumption. Previously our group and other groups have demonstrated that natural antigenic variation in the E protein of DENV strains within serotypes 1, 3, and 4 can have a large impact on the efficiency of neutralization by monoclonal antibodies (mAbs) and immune sera from people exposed to DENV infections or vaccines (Brien et al., 2010; Gallichotte et al., 2018a; Messer et al., 2012; Wahala et al., 2010). There are also documented cases of individuals being reinfected with homologous serotypes (Forshey et al., 2016). Recent analysis of samples and data from DENV vaccine trials suggest that intraserotype antigenic variation has an impact on vaccine performance. In Dengvaxia clinical trials, a genetic sieve analysis of viral sequences from the vaccine and placebo arms demonstrated that the vaccine was more efficacious against a vaccine-matched strain of DENV4 compared to a vaccine mismatched strain (Juraska et al., 2018). These observations highlight the need to further study how intraserotype E protein variation impacts antibody neutralization and protective immunity.
DENV2 is consistently responsible for large epidemics in Asia and Latin America. Despite the fact that multiple DENV2 genotypes currently circulate in different parts of the world, few studies have attempted to systematically evaluate the impact of genotype-specific genetic/antigenic variation on antibody binding and neutralization. A genetic sieve analysis of DENV2 isolates from one Dengvaxia clinical trial demonstrated differences in efficacy against different genotypes (Rabaa et al., 2017), but further studies are needed to define the overall impact on vaccine efficacy and the specific epitopes and residues responsible for vaccine escape.
Consequently, the goal of this study was to evaluate the impact of DENV2 genotypic variation on humoral neutralizing responses in natural and experimental infections and following TV003 DENV2 monovalent and tetravalent live-attenuated vaccination. To this end, we generated an isogenic panel of recombinant DENV2 viruses encoding different pre-membrane (prM) and E glycoprotein genes derived from different DENV2 genotypes, which included circulating isolates from several parts of the world. Using this panel of reagents, we evaluated the neutralization profiles of different genotypic variants to monoclonal and human clinical samples. Our data demonstrate that natural genetic variation harbored in genotype variants significantly modulates the neutralization activity of antibodies, especially in monovalent and tetravalent vaccinated subjects. These data highlight the importance of considering genetic and antigenic diversity within DENV2 when studying human epidemics and evaluating vaccine responses.
RESULTS
Design and Generation of a DENV2 Genotype Panel Representative of Global Genetic Diversity
The four distinct dengue virus serotypes have closely related but distinct genomes that lead to both genetic and antigenic convergence and divergence (Bell et al., 2019). However, the impact of genetic diversity within each serotype, particularly dengue virus serotype 2, on neutralizing responses and vaccine performance is less clear. As the majority of human neutralizing antibodies target the viral E proteins (Rey et al., 2018; Tsai et al., 2017), we examined the genetic relationships within the prM and E sequences from contemporary DENV2 isolates representing different genotypes that have circulated in different parts of the world over the past five decades (Table S1). While the DENV2 isolates segregated into six distinct genotypes, a neighbor-joining tree analysis indicated that isolates within each of the American, Asian-American, Asian I, Asian II, Cosmopolitan, and Sylvatic DENV2 genotypes clustered closely together, reflecting minimal genetic variation within each genotype (Figure 1A). In contrast, each of the six DENV2 genotypes exhibited considerable differences within their prM (9.1%; range, 4.5%–20.2%) and E glycoprotein (8.5%; range, 5.0%–16.4%) with respect to the other genotypes, consistent with earlier reports (Shrivastava et al., 2018). Despite the overall genetic differences among the genotypes, both the WHO Asian I – Thailand – 1974 DENV2 reference strain and Dengvaxia Thailand PUO-218 1980 DENV2 vaccine strain clustered relatively close to contemporary circulating Asian I isolates, with an E % differences of 3.8% and 4% for Dengvaxia Thailand PUO-218 and the WHO Asian I – Thailand – 1974 reference strain compared to other Asian I genotypic variants, respectively. Similarly, the NIH DENV2 component of the tetravalent TV003 vaccine, based on the New Guinea C1980 strain from the Asian II genotype, closely clustered with contemporary circulating DENV2 strains, with a 2.7% E % difference compared to other Asian II genotypic variants (Figure 1A). To assess how contemporary circulating DENV2 strains from distinct parts of the world antigenically differ from one another, we selected representative DENV2 genotype variants from each branch to generate a panel of recombinant viruses that comprehensively represent global diversity within this serotype (Figure 1A). Using our DENV reverse genetics platform described previously (Gallichotte et al., 2018b), we generated an isogenic panel of eleven DENV2 genotype variants as represented in the phylogenetic tree (Figure 1A). Each genotype variant contained their endogenous prM and E sequences and utilized the WHO S16803 reference strain as the receiving backbone strain (Figure 1B).
Figure 1. DENV2 Genotypic Variant Panel that Is Representative of Global Genetic Diversity.
(A) Phylogenetic relationships of the prM and E sequences among the six dengue virus serotype 2 genotypes: American, Asian I, Asian II, Asian-American, Cosmopolitan, and Sylvatic. Colored circles represent DENV2 genotypic isolates that were produced recombinantly. Red arrow represents the dengue virus 2 NIH vaccine strain, and the blue arrow shows the DENV2 Dengvaxia vaccine strain.
(B) Schematic representation of recombinant DENV2 prM and E viral constructs on the S16803 WHO reference strain backbone.
See also Table S1.
Phenotypic Characteristics and Growth Kinetics of DENV2 Genotypic Variants from Distinct Parts of the World
We recovered the DENV2 genotypic global panel recombinant viruses in C6/36 insect cells and characterized their growth kinetics and stability. The DENV2 genotypic variants had distinct foci morphologies, with three distinct phenotypes with respect to their foci size: small foci (1–4.99 1e−3 mm2), medium size foci (5–9.99 1e−3 mm2), and larger and more diffuse foci (10–15 1e−3 mm2). The Asian-American – Puerto Rico – 2003 had small foci (Figure 2A). The American – Tonga – 1974, Cosmopolitan – India – 2001, Sylvatic Asian – Malaysia – 2008, and Asian II – California – 2009, Cosmopolitan – Singapore – 2009, Cosmopolitan – Sri Lanka – 2004, Asian I – Thailand – 2016, Cosmopolitan – Sri Lanka – 2016, and Asian I (WHO ref) – Thailand – 1974 isolates had medium size foci. Finally, the Sylvatic African – Guinea – 1981 had large foci (Figure 2B). Moreover, the DENV2 genotype variants induced differential cytopathic effects (CPEs) in C6/36 cells. The American – Tonga – 1974, Sylvatic – African Guinea – 1981, Sylvatic – Asian Malaysia – 2008, Asian II – California – 2009, and Cosmopolitan – Sri Lanka – 2009 induced low levels of CPE compared to mock-infected C6/36 cells at 4 days post-infection at a multiplicity of infection (MOI) of 0.01 (Figure S1). In contrast, the Cosmopolitan – India – 2001, Asian I – Thailand – 2016, and the Asian I (WHO ref) – Thailand – 1974, induced medium levels of CPE relative to the mock-infected C6/36 cells. Finally, the Asian-American – Puerto Rico – 2003, Cosmopolitan – Singapore – 2009, and Cosmopolitan – Sri Lanka – 2016 induced high levels of CPE compared to mock.
Figure 2. Virologic Characteristics of DENV2 Genotypic Global Panel.
(A) DENV2 genotype variant foci size in Vero-81 cells at 48 h post-infection.
(B) Quantification of foci size (1e−3 mm2).
(C) Multistep growth curve kinetics of dengue virus 2 genotypic variants in C6/36 insect cells. Cells were infected with a multiplicity of infection of 0.01. p values displayed are from a Kruskal-Wallis test.
(D) Stability of the DENV2 genotype panel at 4°C, 28°C, 37°C, and 40°C. Mean percentage of infection is shown from two independent repeats. Bars denote the range.
We examined the growth kinetics of the DENV2 genotypic variants in a multistep growth curve started at a MOI of 0.01 in C6/36 and in Vero cells. Both in C6/36 cells and in Vero cells, the DENV2 genotype variants all differed in their growth kinetics at the sampled time points (days 1, 2, 3, 4, 5, 6, and 7) (Figures 2C and S1; Table S2). In C6/36 cells, the heavily tissue-culture-adapted WHO reference strain Asian I – Thailand – 1974, the American – Tonga – 1974, and the Sylvatic African – Guinea – 1981 isolates grew to highest infectious viral titers by day 5 post-infection and remained the highest among the isolates until day 7 post-infection. In Vero cells, both Sylvatic African – Guinea – 1981 and Cosmopolitan – India – 2001 grew to the highest titers by day 2 and remained the highest by day 7. In contrast, both the Cosmopolitan – Singapore – 2009 and Cosmopolitan – Sri Lanka – 2004 isolates grew to the lowest infectious viral titers by day 3 and remained the lowest by day 7 in C636 cells, whereas the Asian – Thailand – 2016 variant grew to the lowest titers in Vero cells (Figure S1; Table S2).
To define the thermostability of the DENV2 genotypic variants, equivalent focus-forming units (FFUs) of each recombinant virus were incubated at 4°C, 28°C, 37°C, and 40°C for 8 h before measuring infectivity in a Vero focus-forming assay. We observed variability in the thermostability with the Cosmopolitan – India – 2001 and Sylvatic Asian – Malaysia – 2009 prM and E isolates retaining the highest infectivity when exposed to 28°C and 37°C temperatures (Figure 2D). However, both of these relatively stable isolates lost most of their relative infectivity at 40°C. In contrast, the Sylvatic African – Guinea – 1981 prM and E isolate exhibited high viral infectivity when exposed to 28°C and 37°C and remained the most infectious when exposed to 40°C, suggesting this Sylvatic African – Guinea – 1981 isolate was more stable than other DENV2 genotypic isolates in the global panel (Figure 2D).
The Genetic Diversity within the prM and E of DENV2 Genotype Variants
The genetic diversity within DENV serotypes can exceed 6% (Shrivastava et al., 2018). While studies in mice have found that DENV2 genotypic variation can impact neutralizing mAb activity (Sukupolvi-Petty et al., 2010), it is unclear if this genetic diversity within the DENV2 localizes within key epitopes on the viral E proteins and impacts infection-derived and vaccine-elicited antibodies in humans. To examine DENV2 genotypic variation within prM and E, we performed a sequence alignment of these proteins and compared their genetic diversity relative to the Asian I – Thailand – 1974 WHO reference strain (Figure 3). DENV2 isolates differed at 25 amino acid residues within the prM compared to the WHO reference strain (Figure 3A), with both the Sylvatic African – Guinea – 1981 and Sylvatic Asian – Malaysia – 2008 showing the greatest divergence, differing at 13 distinct amino acid residues from the reference sequence. The DENV2 genotypic variant global panel also exhibited considerable genetic diversity within the E protein, a key target of neutralizing antibodies (Figure 3B). DENV2 genotypic variants included in this study differed at 32 amino acid residues within E domain I (EDI) and II (EDII) and had 12 distinct amino acid residues within EDIII with respect to the WHO reference strain (Figures 3B and S2). Again, both sylvatic genotypic variants exhibited the greatest genetic diversity within EDI, EDII, and EDIII compared to other human isolates in the DENV2 genotypic variant panel. Some amino acid residue positions within E were highly variant, with several DENV2 genotype variants differing at several amino acid residues. Amino acid residue positions 71, 129, 141, and 390 all differed in at least five genotypic variants (Figures 3C and S2), potentially implicating these E protein amino acid residue sites as critical for evading neutralizing antibody responses and could be sites of immune pressure. While we utilized the same strain to illustrate the amino acid change across the DENV2 E protein (Figure S2), it is likely that the distinct DENV2 genotypic variant structures could also have slight differences in the E protein conformation.
Figure 3. PrM and E Genetic Variation among DENV2 Genotypic Variants.
(A) Amino acid variation within the prM.
(B) Amino acid variation within the EDI, EDII, and EDIII among distinct DENV2 genotypes from different parts of the world.
(C) Amino acid residue variation on the E protein surface. Amino acid residue variation hotspots relative to S16803 WHO reference strain.
See also Figure S2.
The Impact of DENV2 Genetic Variation on the Neutralization Activity of Human-Derived mAbs
To determine the impact of DENV2 E protein diversity on modulating the neutralization activity of human antibodies, we tested human mAbs with diverse specificities that were derived from plasmablasts or memory B cells isolated from patients during early and late convalescent stages of DENV2 infections: 1662, 1671-1, 1671-4, 1615, 1628, 1660, 2D22, and 3F9. Both 2D22 and 3F9 target distinct and complex quaternary epitopes that are only expressed on whole virions (Fibriansah et al., 2015; Gallichotte et al., 2018b; Smith et al., 2014). DENV2-specific mAb 3F9 targets a complex quaternary epitope within EDI, whereas DENV2-specific mAb 2D22 targets a complex quaternary epitope that spans EDIII (Gallichotte et al., 2018b). All plasma-blast-derived mAbs (1,662, 1,671-1, 1,671-4 1,615, 1,628, and 1,660) bound to both the TV003 and American – Tonga –1974 DENV2 strains and to the chimeric DENV2/4 EDIII virus, but not DENV1, DENV3, DENV4 virions or recombinant E or EDIII, suggesting these mAbs may target a 2D22-like quaternary epitope that spans into EDIII (Figure S3) (Nivarthi et al., 2019).
To test if these distinct type-specific and cross-reactive DENV-specific IgG mAbs differentially neutralize DENV2, we measured the neutralization activity of these mAbs against the DENV2 genotype global panel. Both 2D22 and 3F9 strongly neutralized nearly all strains of the DENV2 genotype panel (Figures 4A, 4B, and 4I). The one exception to this pattern was the strain with the Asian-American – Puerto Rico – 2003 prM and E proteins, which was 10- to 100-fold less efficiently neutralized by mAb 3F9. The plasmablast-derived mAbs 1662, 1671-1, 1615, 1628, and 1660 neutralized the DENV2 panel less potently than 2D22 and 3F9 derived from memory B cells. The plasmablast-derived mAbs also displayed greater variability in neutralization between genotypic variants in the panel (Figures 4C–4H). In fact, mAbs 1662, 1671-1, 1671-4, and 1615 all had greater than 2 logs difference in their IC50 (inhibitory concentration at which a 50% reduction in focus forming units is observed) against the DENV2 genotypic variants (Figure 4I). Together these data suggest that the neutralization potency of DENV2-type-specific and strongly neutralizing human mAbs such as 2D22 and 3F9 are relatively insensitive to DENV2 genotypic variation. In contrast, neutralizing mAbs derived from the acute stage plasmablasts of individuals experiencing DENV2 infections are strongly influenced by genotypic variation between DENV2 strains.
Figure 4. The Neutralization Activity of Monoclonal Antibodies (mAbs) against Distinct DENV2 Genotypic Variants.
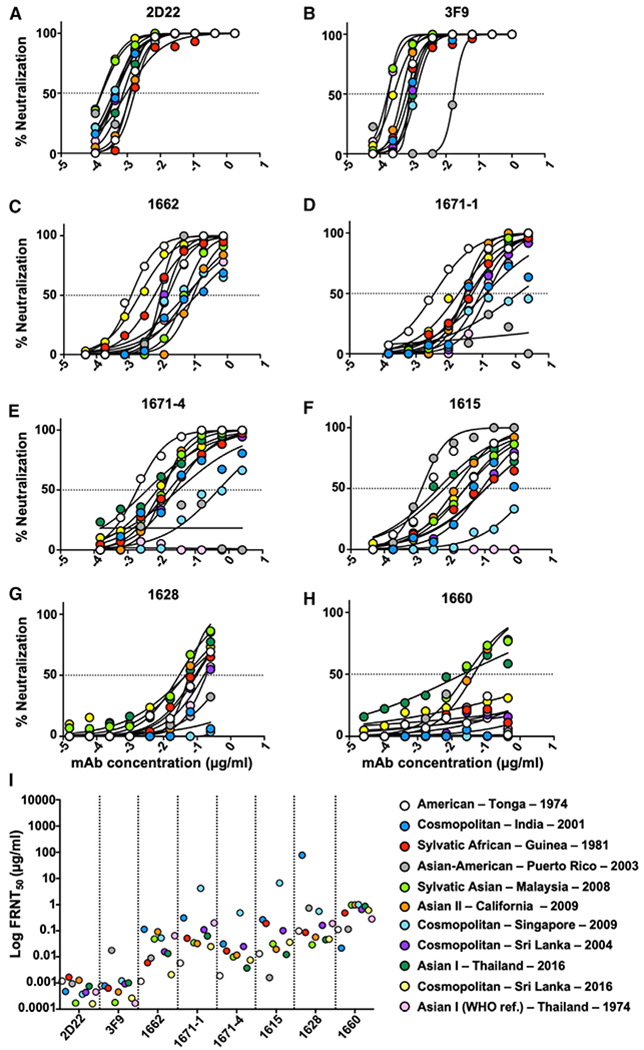
(A and B) Memory-B-cell-derived mAbs 2D22 and 3F9.
(C–H) Plasmablast-derived mAbs 1662, 1671-1, 1671-4, 1615, 1628, and 1660.
(I) mAb neutralization IC50 values against DENV2 recombinant viruses. See also Figure S3.
Polyclonal Genotype-Specific Neutralizing Profiles after Naturally Occurring and Experimental DENV2 Human Infection
To examine if naturally occurring DENV2 genotype variation influences neutralization by polyclonal serum antibodies, we determined the IC50 focus reduction neutralization test (FRNT) neutralization endpoints for each DENV2 genotype using convalescent serum after primary DENV2 infections from (1) a naturally infected traveler cohort and (2) experimental human challenge studies (Kirkpatrick et al., 2016). After a natural primary infection, we observed significant differences in the neutralization activity against distinct DENV2 genotype variants, with a wide range of neutralization activity against diverse strains, which was heavily driven by sylvatic strains (Figures 5A and S4). From the DENV2 genotype panel, both the Sylvatic African – Guinea – 1981 and Sylvatic Asian – Malaysia – 2008 were the most neutralization sensitive to neutralizing antibodies from naturally infected patients with a primary DENV2 infection (Figure 5A and S4; Table S3). Some strains exhibited greater than 10-fold variation in neutralization sensitivity. Among these, DENV2 Asian-American – Puerto Rico – 2003 and the American – Tonga – 1974 recombinant variants were the most uniformly neutralization-resistant strains to convalescent polyclonal sera from natural DENV2 infection.
Figure 5. Variable Neutralization Activity of Polyclonal Serum Responses from Individuals after a Primary Infection against DENV2 Genotypic Variants.
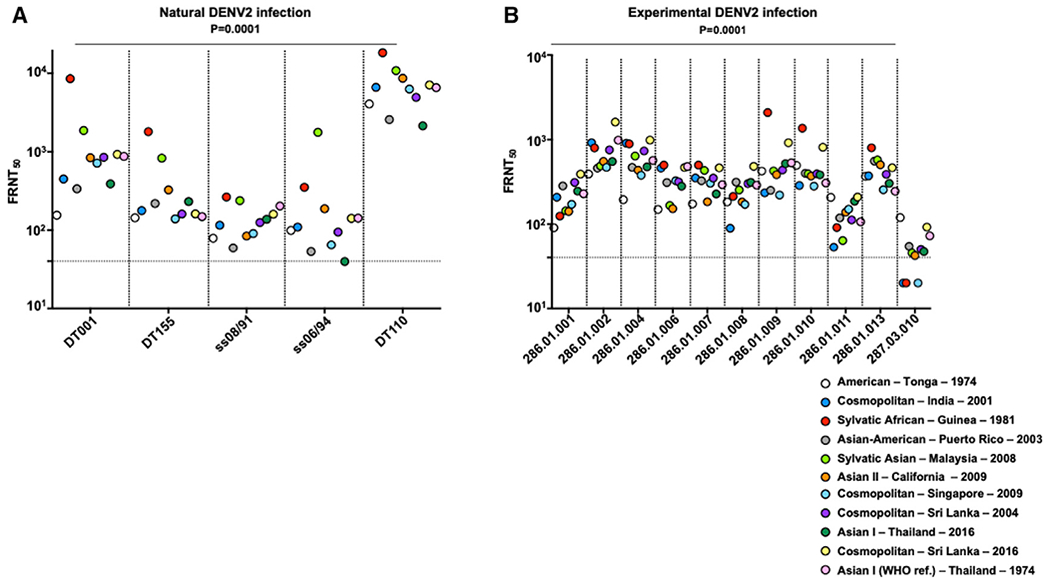
(A) The neutralization activity of naturally infected individuals. Horizontal dashed line denotes the starting dilution tested.
(B) The neutralization activity of experimentally infected patients against dengue virus 2 variants. p values are from a Friedman’s test.
See also Figures S4 and S5 and Table S3.
At 180 days after experimental infection, many individuals had greater than a 4-fold difference in neutralization titers. We observed differences in neutralization among the DENV2 genotype global panel when tested against polyclonal serum derived from experimentally rDEN2Δ30 American – Tonga – 1974 DENV2-infected volunteers (Figures 5B and S5). Experimentally infected individuals did not elicit the highest levels of neutralizing antibodies against the challenge American – Tonga – 1974 virus, rather both the Cosmopolitan – Sri Lanka – 2016 and the Sylvatic African – Guinea – 1981 genotypic variants were generally neutralized the most efficiently (Figures 5B and S5; Table S3). Altogether, these results suggest that host-elicited polyclonal neutralizing antibodies from an experimental DENV2 infection differentially neutralize DENV2 genotypic variants, and that some DENV2 strains are more resistant to neutralization than other strains. Previous controlled human infection models (CHIMs) utilized the American – Tonga – 1974 strain (Kirkpatrick et al., 2016). It is interesting that the American – Tonga – 1974 variant was relatively more neutralization resistant, relative to other DENV2 genotypes, to human sera from an experimental infection (Figure 5B). Utilizing more neutralization-resistant dengue strains in the setting of a CHIM provides a more rigorous experimental setting to test dengue vaccines, placing a higher level of confidence in dengue vaccine efficacy. Moreover, this observation may also have implications in vaccine design. The experimentally infected individuals included in this study were from the placebo arm of the study and thus dengue seronegative at the time of infection, it is encouraging that higher levels of neutralizing antibodies were raised against diverse DENV2 genotypes relative to the challenge American – Tonga – 1974 strain, suggesting that this genotype can elicit high-quality neutralizing antibody responses.
Polyclonal Genotype-Specific Neutralizing Profiles after NIH DENV2 Monovalent and NIH DENV Tetravalent TV003 Experimental Vaccination
To examine if the DENV2 prM and E genetic variation had an impact on vaccine-elicited neutralizing antibody responses, we tested the neutralization profiles of the genotype panel against human polyclonal sera from the TV003 NIH DENV2 monovalent and the DENV tetravalent vaccinated volunteers on day 180 post-vaccination. We examined the neutralizing activity in DENV2 monovalent and DENV tetravalent vaccinees that were seronegative at baseline. Monovalent vaccination resulted in similar overall neutralization titers compared to those elicited after American – Tonga – 1974 DENV2 challenge, although the range in variation was greater. DENV2 monovalent vaccinees exhibited greater than 4-fold variable neutralization activity against distinct DENV2 genotypic variants (Figures 6A and S6). While the sylvatic strains were generally the most sensitive strains to neutralization, the circulating contemporary DENV2 genotypic variants in the global panel also had considerable differences in their neutralization sensitivity to vaccine-elicited neutralizing antibodies after monovalent vaccination (Figures 6A and S6; Table S3).
Figure 6. The Neutralization Activity of Vaccine-Elicited Antibody Responses in TV003 NIH Experimental Vaccinees against DENV2 Genotypic Variants.
(A) The neutralization activity in individuals immunized with the TV003 NIH DENV2 monovalent vaccine. Horizontal dashed line denotes the starting dilution tested.
(B) The neutralization activity in individuals immunized with theTV003 NIH DENV tetravalent vaccine against DENV2 viruses. p values are from a Friedman’s test.
See also Figures S6 and S7 and Table S3.
On the other hand, the majority of the TV003 NIH DENV tetravalent vaccinees best neutralized both the Cosmopolitan – Sri Lanka – 2016 and the Cosmopolitan – Indian – 2001 strains (Figures 6B and S7). While we did not include the TV003 NIH vaccine strain, Asian II New Guinea C, in our DENV2 genotype panel, the Asian II – California – 2009 variant is closely related to the NIH vaccine strain (Figure 1A). The Asian II – California – 2009 variant was neutralized with medium potency by vaccine-elicited antibodies from both the TV003 NIH DENV2 monovalent vaccine and NIH DENV tetravalent vaccine (Figures 6A and 6B). Both TV003 NIH DENV2 monovalent and DENV tetravalent experimental vaccinees weakly neutralized the Cosmopolitan – Singapore – 2009 and Asian-American – Puerto Rico – 2003 strains (Figures 6A and 6B). Four of 10 individuals from the tetravalent vaccine group mounted low to nonmeasurable responses to several contemporary strains. Similar results were reported after DENV4 monovalent and DENV tetravalent vaccination (Gallichotte et al., 2018a). Altogether, these findings suggest that individual responses differ by >10-fold in our neutralizing assay (FRNT) and that some DENV vaccinated individuals did not produce measurable neutralizing titers against several contemporary DENV2 genotypic variants that are circulating in DENV-endemic regions.
Genotype Variation Impacts DENV2-Type-Specific Neutralizing Antibody Responses after Experimental Infection
The CYD-TDV DENV tetravalent vaccine elicited low levels of type-specific neutralizing antibody responses against DENV2 compared to other serotypes (Hadinegoro et al., 2015; Henein et al., 2017; Sabchareon et al., 2012). To examine if DENV2-type-specific neutralizing antibodies are impacted by genotypic variation, we measured the neutralization activity of type-specific DENV2-specific immunoglobulin G (IgG) neutralizing antibodies. We depleted cross-reactive antibodies and measured neutralizing responses against the distinct DENV2 genotypic variants. In individuals experimentally infected with the American – Tonga – 1974 DENV2 strain, the majority of the neutralization activity against the DENV2 genotypic variants was retained following the depletion of DENV1, DENV3, and DENV4 cross-reactive antibody responses (Figures 7A and 7B). The Asian II – California – 2009 variant was the most neutralization sensitive before and after depletion in individuals that were experimentally infected with the American – Tonga – 1974 variant. We observed a 94% DENV2-type-specific response following depletion of cross-reactive antibody responses, suggesting that in primary infected individuals, the majority of the neutralizing antibody response is type specific (Figure 7C; Table S4). However, we still observed statistically significant differences in neutralization activity following depletion of DENV-cross-reactive antibodies across the panel, suggesting that type-specific neutralizing antibodies elicited in the setting of experimental infection are impacted by DENV2 genotypic variation within the prM and E.
Figure 7. The Neutralization Activity of Human Vaccine-Elicited Antibody Responses following Cross-Reactive Antibody Depletion against DENV2 Genotypic Variants.
(A) The neutralization activity of BSA-depleted DENV2-specific IgG neutralizing antibodies from experimentally infected individuals.
(B) The neutralization activity of cross-reactive antibody-depleted DENV2-specific IgG neutralizing antibodies in experimentally infected individuals.
(C) Percent DENV2-serotype-specific neutralizing antibodies in experimentally infected individuals following cross-reactive antibody depletion. Bars denote mean and standard deviation.
See also Table S4.
DISCUSSION
The development of an effective DENV vaccine represents one of the great challenges of the 21st century for the global health community (Halstead, 2018). In addition to solving fundamental problems like variable replication and poorly balanced immunity to the four DENV serotypes, investigators also need to consider intraserotype strain variation and breadth of vaccine-induced immunity (Bäck and Lundkvist, 2013; Dejnirattisai et al., 2010; Gallichotte et al., 2018a). Not only is genotype replacement a recurring theme during successive DENV outbreaks (Lambrechts et al., 2012), but also DENV2 includes highly virulent genotypes and is the most prominent serotype globally. Consequently, defining the molecular mechanisms of DENV2 antigenic variability and vaccine performance is a clear priority for identifying improved correlates of protective immunity.
Among the three live-attenuated tetravalent DENV vaccines, DENV2 neutralizing responses were low after Dengvaxia vaccination and high following Takeda’s TAK-003 and the TV003 NIH vaccination (Biswal et al., 2019; Kirkpatrick et al., 2015; Whitehead et al., 2017). Most importantly, TV003 NIH tetravalent vaccination followed by DENV2 American – Tonga – 1974 challenge was 100% efficacious in human volunteers, although some had boosted neutralizing responses, which is suggestive of low level and/or abortive infections (Kirkpatrick et al., 2016). A recent study reported higher frequencies of DENV2 genotypes in breakthrough infections from Dengvaxia-vaccinated individuals, suggesting that DENV2 genotypic diversity may modulate vaccine protection and breakthrough infection (Rabaa et al., 2017). However, experimental data demonstrating that the genotypic diversity observed in DENV2 can modulate differential neutralization activity of vaccine-elicited antibodies is lacking to date. A potential reason why these data may be lacking is because of the difficulty in assessing the impact of genotypic diversity on immunity given that both laboratory and clinical isolates differ across their entire genomes, and these differences can lead to vastly different growth properties. Our DENV2 reverse genetics system circumvents these limitations by isolating the genetic variability to the prM and E, making our virus panel ideal tool for evaluating the impact of genotypic diversity of host immunity. The goal of this study was to evaluate DENV2 genotype sensitivity to neutralizing antibodies elicited after natural and experimental infection as well as DENV2 monovalent and tetravalent vaccination of volunteers.
To gain a better understanding of the impact of the genetic diversity within key neutralizing antibody epitopes on host humoral immune evasion, we designed and generated a panel of contemporary DENV2 genotype variants that capture the genetic diversity in circulating strains globally. Interestingly, we observed considerable genetic diversity within DENV2 isolates, with sylvatic DENV2 strains clustering farther apart from more frequently circulating and endemic genotypes (Figure 1A). Despite the genetic distance of the sylvatic isolates, these strains were potently neutralized by natural, experimental, and vaccine sera supporting earlier studies that the TV003 NIH vaccine will protect the public from sylvatic strain emergence events (Durbin et al., 2013). Our analyses suggest that the contemporary DENV2 genotypic isolates differ at key targets of host-elicited neutralizing antibodies in the E glycoprotein (Figure 3). For some DENV2-type-specifc and neutralizing human mAbs, more than 100-fold difference in the IC50 was observed against the distinct DENV2 genotypic variants (Figure 4), underlining that the previously unappreciated intraserotypic genetic diversity can modulate the neutralization activity of many human mAbs. For example, mAbs 2D22 and 3F9, which were isolated from an individual infected with a DENV2 Cosmopolitan Sri Lanka strain, mostly had good neutralization coverage against the DENV2 genotypic variants (Figure 4I), with only a few neutralization-resistant variants relative to the Cosmopolitan – Sri Lanka – 2004 strain. In contrast, the plasmablast-derived mAbs 1662, 1671-1, 1671-4, 1615, 1628, and 1660, which were derived from individuals infected with an American – Tonga – 1974 strain, were highly sensitive to prM and E genotype variation (Figure 4I).
Several vaccinated individuals had little if any neutralizing antibody titers against several genotype strains, suggesting that they may be more vulnerable to infection via genotype-mediated natural variation. For example, naturally infected individual DT001, whom was likely infected with a DENV2 Cosmopolitan Sri Lankan genotype based on the country and date of infection at the time of travel, neutralized the majority of viruses in the panel with respect to both Cosmopolitan variants in our panel except for American – Tonga – 1974 and the Asian-American – Puerto Rico – 2003 variants (Figure 5A). For naturally infected patient DT155, whom was likely infected with an Asian-American genotype given the country and date of the infection at the time of travel, many of the DENV2 strains were neutralized with respect to the Asian-American – Puerto Rico – 2003 strain, except for the American-Tonga – 1974, Cosmopolitan – India – 2001, and Cosmopolitan – Singapore – 2009 strains (Figure 5A). Many of the naturally infected patients, but not experimentally infected patients, generally neutralized both the Sylvatic African – Guinea – 1981 and the Sylvatic Asian – Malaysia – 2008 with the greatest potency, suggesting that these strains may be more neutralization sensitive to antibodies derived from wild-type DENV2 infection. Only two of the 11 American – Tonga – 1974 experimentally infected patients (i.e., 286.01.011 and 287.03.010) poorly neutralized the American – Tonga – 1974 strain, suggesting that these patients had less coverage against DENV2 genotypic variants (Figure 5B). Almost half experimentally infected subjects (i.e., 286.01.001, 286.01.002, 286.01.004, 286.01.006, and 286.01.007) had broad DENV2 genotypic neutralizing responses, with all of the variants neutralized with greater potency relative to the American – Tonga – 1974 experimental infection strain (Figure 5B). In contrast, in both the DENV2 monovalent and DENV tetravalent vaccinated subjects, the neutralization breadth was narrow, and fewer DENV2 genotypic variants were neutralized (Figure 6). For DENV2 monovalent subjects 250.01.03, 250.01.07, 250.01.11, 250.01.15, and 250.01.17, the majority of DENV2 strains were more neutralization resistant with respect to the Asian II – California – 2009 genotype, which is the same genotype as the TV003 DENV2 vaccine (Figure 6A). Similarly, in the overwhelming majority of TV003 DENV tetravalent subjects, most DENV2 genotypic variants were more neutralization resistant relative to the vaccine-matched Asian II – California – 2009 genotype, suggesting that the tetravalent TV003 may also induce narrow neutralizing antibody repertoires against DENV2 strains. While it is not yet clear why the neutralization breadth of the DENV2 monovalent and DENV tetravalent vaccines is narrower compared to the neutralizing response in the setting of experimental infection, viral replication differences in the TV003 vaccine strain compared to the American – Tonga – 1974 challenge strain may partially explain this observation.
DENV2 natural variation can influence neutralization activity by at least three different mechanisms, including (1) prM and E-mediated maturation status and epitope presentation, (2) immune-mediated evolution in key E glycoprotein epitopes, or a combination of both, and (3) breathing differences that are modulated by the prM and E genetic diversity. Likewise, some strains appear to be uniformly neutralization sensitive to the many human antibodies included in our study. One potential explanation is that the most neutralization sensitive DENV2 genotypic variants were generally the most stable, which may be indicative of less virion breathing, and thus more easily neutralized by antibodies. Consistent with this hypothesis, the Sylvatic Asian – Malaysia – 2008 had among the highest infectivity at 28°C and 37°C, and similarly, the Sylvatic African – Guinea – 1981 had the highest infectivity in stability assays at 40°C (Figure 2D), and both Sylvatic variants were generally the most neutralization sensitive to natural infection and vaccination sera (Figures 5 and 6). This observation potentially suggests that stability may be related to “breathing” and as a result epitope exposure in some DENV2 genotypic variants. Particularly, based on these observations, some DENV2 variants that are more stable may breathe less and may be more susceptible to neutralizing antibodies. Thus, it is possible that stability may be related to epitope exposure and therefore neutralization sensitivity, which is clinically relevant as virion stability could have an impact on DENV vaccine design. This hypothesis would be consistent with a previous study that found that DENV virus breathing could alter antibody recognition and epitope accessibility (Dowd et al., 2015).
The Takeda-003 (TAK-003) tetravalent vaccine showed a 97% efficacy against DENV2 shortly after the second dose of the vaccination schedule (Biswal et al., 2019), yet DENV2 breakthrough infections still occurred. While breakthrough DENV2 infections were rare, it will be important to evaluate the role for DENV2 genotypic variation of the circulating strains at TAK-003 clinical sites on potentially modulating these rare breakthrough infections. Thus, a key question elicited from these studies is whether natural DENV2 genotypic variation will circumvent the robust type-specifc neutralizing responses that are elicited following TAK-003 vaccination. Secondary and vaccine breakthrough infections provide an opportunity for increased hospitalization rates, as was observed at later stages of the trial in young children that received Sanof’s Dengvaxia (Hadinegoro et al., 2015), especially given that this vaccine does not perform equally well against DENV1, DENV3, and DENV4 (Biswal et al., 2019).
While our data demonstrate that prM and E glycoprotein natural genetic variation impacts the magnitude of the human neutralizing antibody responses, defining the mechanism(s) regulating neutralization escape will require future studies. As the DENV2 global genotype panel is comprised of chimeric prM and E viruses incorporated into the S16803 WHO reference backbone strain, these reagents will be useful in dissecting the role of these genes on DENV2 maturation status and neutralization. While polyclonal vaccine-elicited responses by DENV tetravalent vaccines are complex and may be difficult to map at the amino acid residue resolution (Swanstrom et al., 2018), the large panel of variants provides an opportunity to link common amino acid signatures associated with polyclonal escape (Georgiev et al., 2013). Moreover, the immunologically well-characterized DENV2 genotype panel provides a robust tool for evaluating antibody responses in the setting of phase 3 efficacy studies. Our findings suggest that the current DENV nomenclature and properties of the variants within each serotype may be misleading. Our data argue that the distinct genotypes within DENV2 should be considered distinct from one another in terms of their neutralization properties and provide a paradigm shift in thinking how the genotypic variants within a serotype modulate immunity. These findings have major implications in dengue vaccine design and point to a role in genotypic variation being a factor in potentially driving vaccine efficacy and failure. Moreover, our data argue for targeted surveillance of circulating DENV strains in dengue endemic regions where experimental vaccines are undergoing clinical efficacy trials. This targeted surveillance of endemic dengue genotypic variants should inform the design of next-generation dengue vaccines with improved coverage against genetically diverse strains to ultimately improve vaccine efficacy in humans. As DENV vaccines are iteratively improved, genotypic variation should be considered in designing vaccines.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David R. Martinez (david.rafael.martinez@gmail.com).
Materials Availability
Plasmids generated in this study are available upon request.
Data and Code Availability
Not applicable.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells and media
Insect C6/36 (CRL-1660) cells were obtained from the American Type Culture Collection (ATCC). C6/36 cells were grown in minimal essential medium (MEM) media supplemented with 5% fetal bovine serum (FBS) and 100 U/mL penicillin, 100 mg/mL streptomycin and incubated at 32°C with 5% CO2. Vero-81 (CCL-81) cells were obtained ATCC. Vero cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) Ham’s F12 50/50 media 5% fetal bovine serum (FBS), 100 U/mL penicillin, 100 mg/mL streptomycin, 1% GlutaMax, 1% Sodium Bicarbonate, 1% non-essential amino acids, and incubated at 37°C with 5% CO2, as previously described by our group (Gallichotte et al., 2018a).
Human subjects
Five human serum samples from a primary dengue virus 2 infection were obtained from a dengue Traveler Cohort enrolled at the University of North Carolina Chapel Hill (IRB #08-0895). Eleven human serum samples from 180 days post primary infection were obtained from the control arm of a human dengue virus challenge model in which patients were infected with the DENV2 American – Tonga – 1974 strain (Kirkpatrick et al., 2016). Study subjects from the control arm of the human challenge study were healthy flavivirus naive males with a media age of 30 years (Kirkpatrick et al., 2016). Eight human serum samples from 180 days post vaccination were obtained from the NIH DENV2 (rDEN2/4D30) monovalent TV003 experimental vaccine, and twelve human serum samples from 180 days post vaccination were obtained from the NIH tetravalent (rDEN1D30, rDEN2/4D30, rDEN3D30/31, and rDEN4D30) TV003 experimental vaccine (Kirkpatrick et al., 2016; Lindow et al., 2013; Whitehead et al., 2003). Monovalent and tetravalent vaccine study subjects were healthy adult males and non-pregnant females between ages of 18-50 years, flavivirus seronegative, and without known chronic conditions and have been previously described (Lindow et al., 2013). Human natural infection and vaccine serum samples were received as de-identified material and were deemed as “not human subject research” by the University of North Carolina Chapel Hill Institutional Review Board.
METHOD DETAILS
Generation of the phylogenetic tree
Dengue virus 2 (DENV2) sequences from distinct parts of the world were downloaded from the Virus Pathogen Database and Analysis Resource (ViPR). DENV2 pre-membrane (prM) and envelope (E) sequences were extracted and used to generate the phylogenetic tree, which was constructed in Geneious using the Neighbor-Joining method using a Jukes–Cantor genetic distance model with global alignment and free end gaps. The radial phylogram was visualized in CLC Sequence Viewer 8 and prepared for publication in Adobe Illustrator CC 2019.
Virus design and recovery
Recombinant DENV2 chimeric viruses were designed by inserting the endogenous prM and E sequences from the distinct isolates into the DENV2 Thailand – Asian I – 1974 (S16803) WHO reference strain as described previously (Gallichotte et al., 2018a). Briefly DENV2 prM and E sequences were synthetically derived and introduced into a four-plasmid infectious clone system utilizing the DENV2 WHO reference strain: S16803 subclone. DENV2 sequences were then digested and ligated together and DENV2 full-length genomic RNA was generated using T7 RNA polymerase. Infectious genome-length DENV2 RNA transcripts were electroporated into C6/36 (CRL-1660) cells and four days later, supernatants were collected and used to establish passage 0 viruses to make working stocks. All DENV2 genotypic recombinant viruses were grown to passage 2 and the prM and E genes were then sequenced to confirm sequence identity.
Dengue virus 2 titration and immunostaining
Vero cells were plated 1 day prior to infection at a density of 20,000 cells per well in 96-well flat-bottom plates. Virus stocks were thawed on ice for 1 hour and were then serially diluted 10-fold and pipette tips being exchanged after each dilution. Media was then removed from plates and serially diluted viruses were added to cells and incubated for 1 hour at 37°C. After incubation, 125 μL of methylcellulose (1% methylcellulose Opti-MEM supplemented with 2% FBS, 100 U/ml penicillin, 100 mg/ml streptomycin) was overlaid and cells were incubated for 48 hours at 37°C with 5% CO2. Media was then removed from wells and washed with 1X PBS and fixed with 10% Formalin for 20 min at room temperature. Fixed cell membranes were then permeabilized by washing twice with 100 μL with 1X Permeabilization buffer (ThermoFisher). Fixed and permeabilized cells were then blocked for 10 minutes at room temperature with blocking buffer (1X Permeabilization buffer containing 5% non-fat dry milk). Fixed and permeabilized cells were stained with 1:1000 diluted anti-prM hybridoma, 2H2, and anti-E hybridoma, 4G2, in blocking buffer for 30 min at 37°C. Fixed and permeabilized cells were washed three times with 1X PBS and were stained with an anti-mouse horseradish peroxidase (HRP)-labeled secondary antibody for 30 min at 37°C. After washing three times with 1X PBS, foci were developed with TrueBlue substrate and were visualized with an ImmunoSpot C.T.L. Analyzer using BioSpot 7. 0.
Dengue virus 2 growth curves
C6/36 and Vero cells were seeded onto 6-well plates 1 day prior to infection. The DENV2 genotype strains were diluted at a multiplicity of infection (MOI) of 0.01 and added to cells for 1 hour at 32°Cfor C6/36 cells or at 37°C for Vero cells. The virus inoculum was removed, and C6/36 cells were then washed 3 times with fresh media and the media was replaced after the third wash. Media was sampled at 24, 48, 72, 96, 120, 144, and 168 hours post infection and media was immediately frozen after each sampling at −80°C. Viruses were titered and immunostained as described above.
Thermostability assay
DENV2 genotypic variants were diluted to an MOI of 1 and incubated at 4°C, 28°C, 37°C, or 40°C for 8 hr. Heat blocks (Benchmark Scientific) set at 4°C, 28°C, 37°C, or 40°C with two separate thermometers to accurately monitor temperature were used to carry out the thermostability experiments. The thermostability experiments on the DENV2 genotypic variants were conducted at the same time and on the same day. At the end of the 8-hour incubation, DENV2 viruses were transferred to 4°C for 15 min and the same volume of DENV2 viruses was added in duplicate on Vero cells to obtain a titer. Thermostability assays were repeated at least two times. Viruses were titered and immunostained as described above. The thermostability was calculated by dividing the titer obtained at the test temperature (i.e., 40°C) over the 4°C titer times 100 and expressed as a percent. A Kruskal-Wallis test was used to compare viral growth kinetics from different time points (P values shown in Table S2).
Patient serum samples and monoclonal antibodies
Monoclonal antibodies 1662, 1671-1, 1671-4, 1615, 1628 and 1660 were derived from plasmablasts from patients experimentally infected with the American – Tonga – 1974 DENV2 strain as described previously (Nivarthi et al., 2019). Briefly, PBMCs were stained with plasmablast-specific markers and IgG + plasmablasts were single cell sorted by fluorescence-activated cell sorting (FACS) into 96-well plates containing RNAlater. The IgG+ plasmablast markers were: CD3−CD14−IgA−IgM− CD19 + CD20low/−CD27 + CD38hi. Plasmablasts RNA was then reverse transcribed, and VDJ gene PCR, barcode assignment, sequence assembly, and V(D)J assignment was performed as described previously (Tan et al., 2014). Antibody genes were then expressed in mammalian expression vectors in frame with a human IgG1 Fc region and were generated in HEK293 cells for small scale transfection as described previously (Nivarthi et al., 2019). Plasmablast-derived mAbs were screened against DENV2 antigens by ELISA as described below.
Antibody depletion studies
Dynabeads M-280 tosylactivated were covalently bound to anti-DENV E mAb 1M7 overnight at 37°C. Bead:mAb complex was blocked with 1% BSA in PBS at 37°C, and then washed with 0.1 M 2-(N-morpholino) ethane sulfonic acid (MES) buffer. Beads were then incubated with BSA (control) or purified antigens for 1 hr at 37°C, and then washed three times with PBS. Bead:mAb:-DENV complex was fixed with 2% paraformaldehyde in PBS for 20 min, and then washed four times with PBS. DENV-specific antibodies were depleted from sera by incubating beads with sera diluted 1:10 in PBS for 1 hr at 37°C with end-over-end mixing for at least three sequential rounds of depletions. For all the sera, BSA coated beads were used for the control depleted condition to estimate the baseline neutralization titers before depletion. For estimating the DENV2 TS Abs, beads conjugated with a mixture of the WHO reference strain dengue virus antigens: DENV1 (West Pac 1974), DENV3 (CH53489), and DENV4 (TVP-376) were used to deplete sera. As a homologous control, where all the DENV2 specific antibodies (TS and CR) were depleted was also performed for each sample to serve as a background control. Removal of DENV antibodies to the depleting antigen was confirmed by DENV ELISA as previously described by our group (Swanstrom et al., 2019). All the depleted sera were further tested in a Vero-81 based FRNT assays against the panel of the DENV2 genotypic variants. We calculated IC50 values by using the sigmoidal dose response (variable slope) via Prism 7 (GraphPad Software, San Diego, CA, USA). Reported values were required to have an R2 > 0.75, a hill slope > 0.5, and an IC50 within the range of the dilutions. Using the (neutralization 50%) Neut50 titers the percentage of type-specific (TS) nAbs against DENV2 genotypic variants were calculated using the following formulas.
Binding ELISAs
Non-chimeric viruses used in the ELISA screens were grown in Vero-81 cells at 37 °C, as previously described (Nivarthi et al., 2017). Recombinant envelope protein (recE) (80% of E protein) from WHO reference DENV2 S16803 strain was produced in our laboratory or purchased from Hawaii Biotech, Inc.(Modis et al., 2004) Recombinant EDIII protein from DENV2 was produced in-house as described previously (Wahala et al., 2009). Recombinant viruses were constructed using a four-cDNA cloning strategy as described previously (Gallichotte et al., 2015). Briefly, the rDENV2/4 was created by introducing the envelope protein (E) domain III (EDIII) residues from DENV2 (Nicaragua 694) into the DENV4 A subclone (Sri Lanka 1992; GenBank KJ160504.1) and replacing E nucleotides 900 to 1179 with the corresponding nucleotides encoding variant DENV2 amino acids. The parental viruses (DENV2 Nicaraguan 694 IC, DENV4 Sri Lanka 1992) were also tested alongside binding ELISAs with the rDENV2/4 (EDIII) virus. Only samples that bound to DENV2 Nicaraguan 694 IC were considered positive for binding to rDENV2/4(EDIII).
For detecting Dengue-specific IgG reactive to whole DENV virions (DENV1 TV003, DENV2 TV003, DENV3 TV003, DENV4 TV003, DENV2 American-Tonga74, rDENV2/4 EDIII, DENV2 Nicaraguan IC, DENV4 SriLanka1992), the viral stock of each serotype was titrated separately by ELISA using a fixed amount of highly-cross reactive serum sample (DT000) to achieve OD405 = 1.0 to normalize among serotypes and among batches in ELISA assays. Equivalent quantities (volume of each viral stock equivalent to an OD of 1.0) of DENV viruses were captured individually by plate-bound mouse anti-E mAb 4G2 overnight at 4 °C. Recombinant E proteins were directly coated (rE - 100 ng/well; rEDIII - 200 ng/well) on ELISA plates overnight at 4 °C (Gallichotte et al., 2015). Plates were blocked with 3% (vol/vol) normal goat serum (Thermo, USA), in Tris-buffered saline (TBS) containing 0.05% (vol/vol) Tween 20 (blocking buffer). Plasmablast-derived IgG1 mAbs were tested at a fixed concentration of 20 μg/ml. Following 3X washes, alkaline-phosphatase conjugated secondary anti-human IgG antibodies were used to detect binding of primary antibodies with p-nitrophenyl phosphate substrate, and OD405 reaction color changes were quantified by spectrophotometry.
Focus reduction neutralization test
Vero-81 cells (ATCC: CCL-81) were seeded at 200,000 cells per well the day before the focus reduction neutralization test (FRNT) in 96-well plates. Monoclonal antibody and serum samples were diluted to 1:40 in Vero infection media and were serially diluted at 1:4. Dengue virus was then diluted to 3200 focus forming units per milliliter (FFU/ml) and 30 μL and was diluted with 30 μL of serially diluted antibody sample fora 1:1 ratio and incubated at 37°C for 1 hour. After the 1-hour incubation, the media was removed from Vero-81 cells and 50 μL of antibody-virus mixture was added and incubated for 1 hour at 37°C. Following the incubation, cells were overlaid with 125 μL of methylcellulose and incubated at 37°C for 48 hours.
QUANTIFICATION AND STATISTICAL ANALYSIS
Dengue virus envelope structures were visualized and rendered using PyMOL - The PyMOL Molecular Graphics System Version 2.2.2. All statistical tests were run on GraphPad Prism Version 7.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| DENV2-specific IgG 1662 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1671-1 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1671-4 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1615 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1652 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1585 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1591 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1660 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1636 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1606 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 1678 | Nivarthi et al., 2019 | N/A |
| DENV2-specific IgG 2D22 | Gallichotte et al., 2018 | N/A |
| DENV2-specific IgG 3F9 | Gallichotte et al., 2018 | N/A |
| DENV-4G2 (hybridoma-produced Ig) | ATCC HB-112 | Cat# D1-4G2-4-15 |
| DENV-2H2 (hybridoma-produced Ig) | ATCC HB-114 | Cat# D3-2H2-9-21 |
| Anti-Mouse IgG (H+L) Antibody, Human Serum Adsorbed and Peroxidase-Labeled | SeraCare | Cat# 5450-0011 |
| Bacterial and Virus Strains | ||
| DENV2 prM and E: Tonga, 1974, American | This manuscript | GenBank ID: AY744147 |
| DENV2 prM and E: Thailand, 1974, Asian I | This manuscript | GenBank ID: GU289914 |
| DENV2 prM and E: Thailand, 2016 Asian I | This manuscript | GenBank ID: LC410185 |
| DENV2 prM and E: California, 2009, Asian II | This manuscript | GenBank ID: HQ541798 |
| DENV2 prM and E: Puerto Rico, 2003, Asian-American | This manuscript | GenBank ID: EU687235 |
| DENV2 prM and E: India, 2001, Cosmopolitan | This manuscript | GenBank ID: DQ448237 |
| DENV2 prM and E: Sri Lanka, 2004, Cosmopolitan | This manuscript | GenBank ID: GQ252677 |
| DENV2 prM and E: Singapore, 2009, Cosmopolitan | This manuscript | GenBank ID: KM279605 |
| DENV2 prM and E: Sri Lanka, 2016, Cosmopolitan | This manuscript | GenBank ID: KY495803.1 |
| DENV2 prM and E: Guinea, 1981, Sylvatic | This manuscript | GenBank ID: EF105378 |
| DENV2 prM and E: Malaysia, 2008, Sylvatic | This manuscript | GenBank ID: FJ467493 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fetal Bovine Serum, ultra-low IgG | Thermo Fisher Scientific | Cat#16250078 |
| 100x Penicillin Streptomycin Glutamine | Thermo Fisher Scientific | Cat#10378016 |
| DMEM, high glucose, GlutaMAX Supplement | Thermo Fisher Scientific | Cat#10566-024 |
| Dulbecco’s Phosphate-Buffered Saline, 1X withcalcium and magnesium | Corning Life Sciences | Cat#21–030-CM |
| Dubelcco’s modified Eagle’s/Ham’s F-12 50/50 Mix | GIBCO | 10-092-CV |
| Opti-MEM I | GIBCO | 31985-070 |
| Non-fat dry milk | Bio-Rad | Cat#1706404 |
| Recombinant DNA | ||
| Primer: DENV2_forw_1 5′-GCA GGC ATG ATC ATT ATG TTG ATT C | This manuscript | N/A |
| Primer: DENV2_forw_2 5′-CAC GAA CTG AAA CAT GGA TGT CAT C | This manuscript | N/A |
| Primer: DENV2_forw_3 5′-GCT GCC CAA CAC AAG GAG AAC CTA G | This manuscript | N/A |
| Primer: DENV2_forw_4 5′-GAA AAT AAA GCT TGG CTG GTG CAC A | This manuscript | N/A |
| Primer: DENV2_forw_5 5′-GTT CTC CAT GTA AGA TCC CTT TTG A | This manuscript | N/A |
| Primer: DENV2_forw_6 5′-CAC CAA GTT TTC GGA GCA ATC TAT G | This manuscript | N/A |
| Primer: DENV2_reve_1 5′-CTC AAG ATC CAA GTT TCA ATT CTC T | This manuscript | N/A |
| Primer: DENV2_reve_2 5′-CAG ACG AAC CTT TTG TCC TGC TCT TC | This manuscript | N/A |
| Primer: DENV2_reve_3 5′-CAA TTT GAT CCT TGT GTG TCC GCT C | This manuscript | N/A |
| Primer: DENV2_reve_4 5′-GTA ATC AGG CGA CCT AAA ACA TGT C | This manuscript | N/A |
| Primer: DENV2_reve_5 5′-CTA TCC ATG TGA TAA TGA CTC CTA TG | This manuscript | N/A |
| Primer: DENV2_reve_6 5′-CTC CTG TCA TGA TAG TTA ACT TCA C | This manuscript | N/A |
| Experimental Models: Cell Lines | ||
| Vero | ATCC | CCL-81 |
| C6/36 cells | ATCC | CRL-1660 |
| Critical Commercial Assays | ||
| QIAamp Viral RNA Mini Kit | QIAGEN | Cat# 52904 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| mMESSAGE mMACHINE T7 Transcription Kit | Thermo Fisher Scientific | Cat# AM1344 |
| Deposited Data | ||
| N/A | N/A | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7.2 | GraphPad Software Inc. | https://www.graphpad.com |
| PyMOL | Schrödinger, Inc. | https://www.schrodinger.com |
| Other | ||
| Immunospot | C.T.L. | N/A |
Highlights.
Dengue virus serotype 2 (DENV2) genotypic variants have distinct characteristics
DENV2 genotypic variants exhibit variation in proteins prM and E
DENV2 prM and E variation modulates the neutralization activity of human mAbs
DENV2 genotypes are differentially neutralization-susceptible to vaccine antibodies
ACKNOWLEDGMENTS
D.R.M. is funded by National Institutes of Health (NIH) grant F32 AI152296 and a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award and was previously supported by NIH NIAID grant T32 AI007151. This work was funded by NIH NIAID grants R01 AI125198, R01 AI106695, and R01 AI107731 (awarded to A.M.d.S. and R.S.B.). The conclusions and opinions expressed in this article are those of the authors and do not necessarily reflect those of the NIH or U.S. Department of Health and Human Services.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108226.
DECLARATION OF INTERESTS
A.M.d.S. has consulted with Takeda, Merck, and GSK on developing DENV vaccine concepts. R.S.B. has consulted with Takeda on DENV and norovirus vaccine concepts. R.S.B. has had a sponsored program on norovirus vaccine development by Takeda. A.M.d.S. and R.S.B. are inventors on patents on DENV vaccines.
REFERENCES
- Bäck AT, and Lundkvist A (2013). Dengue viruses: an overview. Infect. Ecol. Epidemiol 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer K, Esquilin IO, Cornier AS, Thomas SJ, Quintero Del Rio AI, Bertran-Pasarell J, Morales Ramirez JO, Diaz C, Carlo S, Eckels KH, et al. (2015). A phase II, randomized, safety and immunogenicity trial of a rederived, live-attenuated dengue virus vaccine in healthy children and adults living in Puerto Rico. Am. J. Trop. Med. Hyg 93, 441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SM, Katzelnick L, and Bedford T (2019). Dengue genetic divergence generates within-serotype antigenic variation, but serotypes dominate evolutionary dynamics. eLife 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. (2013). The global distribution and burden of dengue. Nature 496, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal S, Reynales H, Saez-Llorens X, Lopez P, Borja-Tabora C, Kosalaraksa P, Sirivichayakul C, Watanaveeradej V, Rivera L, Espinoza F, et al. ; TIDES Study Group (2019). Efficacy of a Tetravalent Dengue Vaccine in Healthy Children and Adolescents. N. Engl. J. Med 381, 2009–2019. [DOI] [PubMed] [Google Scholar]
- Brien JD, Austin SK, Sukupolvi-Petty S, O’Brien KM, Johnson S, Fremont DH, and Diamond MS (2010). Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J. Virol 84, 10630–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, et al. (2010). Cross-reacting antibodies enhance dengue virus infection in humans. Science 328, 745–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, DeMaso CR, and Pierson TC (2015). Genotypic differences in dengue virus neutralization are explained by a single amino acid mutation that modulates virus breathing. MBio 6, e01559–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AP, Mayer SV, Rossi SL, Amaya-Larios IY, Ramos-Castaneda J, Eong Ooi E, Jane Cardosa M, Munoz-Jordan JL, Tesh RB, Messer WB, et al. (2013). Emergence potential of sylvatic dengue virus type 4 in the urban transmission cycle is restrained by vaccination and homotypic immunity. Virology 439, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM, et al. (2015). DENGUE VIRUS. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshey BM, Reiner RC, Olkowski S, Morrison AC, Espinoza A, Long KC, Vilcarromero S, Casanova W, Wearing HJ, Halsey ES, et al. (2016). Incomplete protection against dengue virus type 2 re-infection in Peru. PLoS Negl. Trop. Dis 10, e0004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Widman DG, Yount BL, Wahala WM, Durbin A, Whitehead S, Sariol CA, Crowe JE Jr., de Silva AM, and Baric RS (2015). A new quaternary structure epitope on dengue virus serotype 2 is the target of durable type-specific neutralizing antibodies. MBio 6, e01461–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Baric TJ, Nivarthi U, Delacruz MJ, Graham R, Widman DG, Yount BL, Durbin AP, Whitehead SS, de Silva AM, and Baric RS (2018a). Genetic variation between dengue virus type 4 strains impacts human antibody binding and neutralization. Cell Rep. 25, 1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Baric TJ, Yount BL Jr., Widman DG, Durbin A, Whitehead S, Baric RS, and deSilva AM (2018b). Human dengue virus serotype 2 neutralizing antibodies target two distinct quaternary epitopes. PLoS Pathog. 14, e1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. (2013). Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756. [DOI] [PubMed] [Google Scholar]
- Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, et al. ; CYD-TDV Dengue Vaccine Working Group (2015). Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N. Engl. J. Med 373, 1195–1206. [DOI] [PubMed] [Google Scholar]
- Halstead SB (2018). Which dengue vaccine approach isthe most promising, and should we be concerned about enhanced disease after vaccination? There is only one true winner. Cold Spring Harb. Perspect. Biol 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henein S, Swanstrom J, Byers AM, Moser JM, Shaik SF, Bonaparte M, Jackson N, Guy B, Baric R, and de Silva AM (2017). Dissecting antibodies induced by a chimeric yellow fever-dengue, live-attenuated, tetravalent dengue vaccine (CYD-TDV) in naive and dengue-exposed individuals. J. Infect. Dis 215, 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juraska M, Magaret CA, Shao J, Carpp LN, Fiore-Gartland AJ, Benkeser D, Girerd-Chambaz Y, Langevin E, Frago C, Guy B, et al. (2018). Viral genetic diversity and protective efficacy of a tetravalent dengue vaccine in two phase 3 trials. Proc. Natl. Acad. Sci. USA 115, E8378–E8387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick BD, Durbin AP, Pierce KK, Carmolli MP, Tibery CM, Grier PL, Hynes N, Diehl SA, Elwood D, Jarvis AP, et al. (2015). Robust and balanced immune responses to all 4 dengue virus serotypes following administration of a single dose of a live attenuated tetravalent dengue vaccine to healthy, flavivirus-naive adults. J. Infect. Dis 212, 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick BD, Whitehead SS, Pierce KK, Tibery CM, Grier PL, Hynes NA, Larsson CJ, Sabundayo BP, Talaat KR, Janiak A, et al. (2016). The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci. Transl. Med 8, 330ra36. [DOI] [PubMed] [Google Scholar]
- Kraemer MUG, Reiner RC Jr., Brady OJ, Messina JP, Gilbert M, Pigott DM, Yi D, Johnson K, Earl L, Marczak LB, et al. (2019). Publisher Correction: Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat. Microbiol 4, 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L, Scott TW, and Gubler DJ (2010). Consequences of the expanding global distribution of Aedes albopictus for dengue virus transmission. PLoS Negl. Trop. Dis 4, e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L, Paaijmans KP, Fansiri T, Carrington LB, Kramer LD, Thomas MB, and Scott TW (2011). Impact of daily temperature fluctuations on dengue virus transmission by Aedes aegypti. Proc. Natl. Acad. Sci. USA 108, 7460–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts L, Fansiri T, Pongsiri A, Thaisomboonsuk B, Klungthong C, Richardson JH, Ponlawat A, Jarman RG, and Scott TW (2012). Dengue-1 virus clade replacement in Thailand associated with enhanced mosquito transmission. J. Virol 86, 1853–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow JC, Durbin AP, Whitehead SS, Pierce KK, Carmolli MP, and Kirkpatrick BD (2013). Vaccination of volunteers with low-dose, live-attenuated, dengue viruses leads to serotype-specific immunologic and virologic profiles. Vaccine 31, 3347–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer WB, Yount B, Hacker KE, Donaldson EF, Huynh JP, de Silva AM, and Baric RS (2012). Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS Negl. Trop. Dis 6, e1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, and Harrison SC (2004). Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319. [DOI] [PubMed] [Google Scholar]
- Nivarthi UK, Kose N, Sapparapu G, Widman D, Gallichotte E, Pfaff JM, Doranz BJ, Weiskopf D, Sette A, Durbin AP, et al. (2017). Mapping the human memory B cell and serum neutralizing antibody responses to dengue virus serotype 4 infection and vaccination. J. Virol 91, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivarthi UK, Tu HA, Delacruz MJ, Swanstrom J, Patel B, Durbin AP, Whitehead SS, Pierce KK, Kirkpatrick BD, Baric RS, et al. (2019). Longitudinal analysis of acute and convalescent B cell responses in a human primary dengue serotype 2 infection model. EBioMedicine 41, 465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaa MA, Girerd-Chambaz Y, Duong Thi Hue K, Vu Tuan T, Wills B, Bonaparte M, van der Vliet D, Langevin E, Cortes M, Zambrano B, et al. (2017). Genetic epidemiology of dengue viruses in phase III trials of the CYD tetravalent dengue vaccine and implications for efficacy. eLife 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Stiasny K, Vaney MC, Dellarole M, and Heinz FX (2018). The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. 19, 206–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, et al. (2012). Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380, 1559–1567. [DOI] [PubMed] [Google Scholar]
- Schmidt AC, Lin L, Martinez LJ, Ruck RC, Eckels KH, Collard A, De La Barrera R, Paolino KM, Toussaint JF, Lepine E, et al. (2017). Phase 1 randomized study of a tetravalent dengue purified inactivated vaccine in healthy adults in the United States. Am. J. Trop. Med. Hyg 96, 1325–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S, Tiraki D, Diwan A, Lalwani SK, Modak M, Mishra AC, and Arankalle VA (2018). Co-circulation of all the four dengue virus serotypes and detection of a novel clade of DENV-4 (genotype I) virus in Pune, India during 2016 season. PLoS ONE 13, e0192672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CP, Farrar JJ, Nguyen V, and Wills B (2012). Dengue. N. Engl. J. Med 366, 1423–1432. [DOI] [PubMed] [Google Scholar]
- Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, and Crowe JE Jr. (2014). Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J. Virol 88, 12233–12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S, Luedtke A, Langevin E, Zhu M, Bonaparte M, Machabert T, Savarino S, Zambrano B, Moureau A, Khromava A, et al. (2018). Effect of dengue serostatus on dengue vaccine safety and efficacy. N. Engl. J. Med 379, 327–340. [DOI] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, et al. (2010). Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J. Virol 84, 9227–9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom JA, Henein S, Plante JA, Yount BL, Widman DG, Gallichotte EN, Dean HJ, Osorio JE, Partidos CD, de Silva AM, and Baric RS (2018). Analyzing the human serum antibody responses to a live attenuated tetravalent dengue vaccine candidate. J. Infect. Dis 217, 1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom JA, Nivarthi UK, Patel B, Delacruz MJ, Yount B, Widman DG, Durbin AP, Whitehead SS, De Silva AM, and Baric RS (2019). Beyond neutralizing antibody levels: the epitope specificity of antibodies induced by national institutes of health monovalent dengue virus vaccines. J. Infect. Dis 220, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YC, Kongpachith S, Blum LK, Ju CH, Lahey LJ, Lu DR, Cai X, Wagner CA, Lindstrom TM, Sokolove J, and Robinson WH (2014). Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol. 66, 2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torresi J, Richmond PC, Heron LG, Qiao M, Marjason J, Starr-Spires L, van der Vliet D, Jin J, Wartel TA, and Bouckenooghe A (2017). Replication and excretion of the live attenuated tetravalent dengue vaccine CYD-TDV in a flavivirus-naive adult population: assessment of vaccine viremia and virus shedding. J. Infect. Dis 216, 834–841. [DOI] [PubMed] [Google Scholar]
- Tsai WY, Lin HE, and Wang WK (2017). Complexity of human antibody response to dengue virus: implication for vaccine development. Front. Microbiol 8, 1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Kraus AA, Haymore LB,Accavitti-Loper MA, and deSilva AM (2009). Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology 392, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, and de Silva AM (2010). Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog. 6, e1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waman VP, Kolekar P, Ramtirthkar MR, Kale MM, and Kulkarni-Kale U (2016). Analysis of genotype diversity and evolution of Dengue virus serotype 2 using complete genomes. PeerJ 4, e2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, and Vasilakis N (2009). Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect. Genet. Evol 9, 523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead SS, Hanley KA, Blaney JE Jr., Gilmore LE, Elkins WR, and Murphy BR (2003). Substitution of the structural genes of dengue virus type 4 with those of type 2 results in chimeric vaccine candidates which are attenuated for mosquitoes, mice, and rhesus monkeys. Vaccine 21, 4307–4316. [DOI] [PubMed] [Google Scholar]
- Whitehead SS, Durbin AP, Pierce KK, Elwood D, McElvany BD, Fraser EA, Carmolli MP, Tibery CM, Hynes NA, Jo M, et al. (2017). In a randomized trial, the live attenuated tetravalent dengue vaccine TV003 is well-tolerated and highly immunogenic in subjects with flavivirus exposure prior to vaccination. PLoS Negl. Trop. Dis 11, e0005584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.


