Abstract
Background
We aimed at predicting fatigue after breast cancer treatment using machine learning on clinical covariates and germline genome-wide data.
Methods
We accessed germline genome-wide data of 2799 early-stage breast cancer patients from the Cancer Toxicity study (NCT01993498). The primary endpoint was defined as scoring zero at diagnosis and higher than quartile 3 at 1 year after primary treatment completion on European Organization for Research and Treatment of Cancer quality-of-life questionnaires for Overall Fatigue and on the multidimensional questionnaire for Physical, Emotional, and Cognitive fatigue. First, we tested univariate associations of each endpoint with clinical variables and genome-wide variants. Then, using preselected clinical (false discovery rate < 0.05) and genomic (P < .001) variables, a multivariable preconditioned random-forest regression model was built and validated on a hold-out subset to predict fatigue. Gene set enrichment analysis identified key biological correlates (MetaCore). All statistical tests were 2-sided.
Results
Statistically significant clinical associations were found only with Emotional and Cognitive Fatigue, including receipt of chemotherapy, anxiety, and pain. Some single nucleotide polymorphisms had some degree of association (P < .001) with the different fatigue endpoints, although there were no genome-wide statistically significant (P < 5.00 × 10−8) associations. Only for Cognitive Fatigue, the predictive ability of the genomic multivariable model was statistically significantly better than random (area under the curve = 0.59, P = .01) and marginally improved with clinical variables (area under the curve = 0.60, P = .005). Single nucleotide polymorphisms found to be associated (P < .001) with Cognitive Fatigue belonged to genes linked to inflammation (false discovery rate adjusted P = .03), cognitive disorders (P = 1.51 × 10−12), and synaptic transmission (P = 6.28 × 10−8).
Conclusions
Genomic analyses in this large cohort of breast cancer survivors suggest a possible genetic role for severe Cognitive Fatigue that warrants further exploration.
Fatigue is one of the most common and distressing long-term side effects experienced by breast cancer survivors after treatment (1). During active treatment, the vast majority of patients experience some fatigue, which typically improves over the first year after primary treatment completion, although around 30% of patients continue to report severe fatigue for many years (2–4). Several studies suggested that the intensity and duration of fatigue experienced by cancer patients are statistically significantly greater than those of healthy controls, with substantial evidence that cancer-related fatigue affects patients’ social and work lives and has a substantial negative impact on quality of life and daily activities (2,3,5–10).
Cancer-related fatigue is a complex, multidimensional, and heterogeneous symptom, involving physical, emotional, and cognitive dimensions (11). Previous research has pointed at age, preexisting depression and fatigue, early stress, comorbidities, physical inactivity, and specific treatment classes as clinical risk factors for onset and persistence of cancer-related fatigue (12,13). In addition, there is growing evidence for associations of cancer-related fatigue and biological factors, including genetic factors. Particularly, prior data suggested that activation of chronic inflammation pathways might contribute to posttreatment fatigue through central nervous system signaling. One of the most solid hypotheses is that fatigue is associated with single nucleotide polymorphisms (SNPs) related to genes coding for proinflammatory cytokines, including polymorphisms in TNFα, IL8, IL6, IL1β, and IL1RN (14,15). Other biological factors that may be implicated in cancer-related fatigue include hypothalamic-pituitary-adrenal axis deregulation, five-hydroxyl-tryptophan deregulation, and alterations in adenosine triphosphate and muscle metabolism (1,14–20). Nevertheless, evidence supporting these associations is inconsistent, and findings were not always validated because of several study limitations, including the focus on the most acute effects of cancer treatments, not accounting for different fatigue dimensions, small sample sizes, or retrospective or cross-sectional designs. Particularly, comprehensive genome-wide association studies (GWAS) have not been previously performed, which limits our understanding of fatigue after breast cancer treatment (2,8,10,12).
Although conventional GWAS have provided insights for many human complex traits (21), effect sizes of common SNPs are usually small, and adjustment for multiplicity leads to underpowered analyses (22). Machine learning methodologies emerged as an alternative data-driven approach that seeks to identify joint contributions of multiple SNPs to complex traits, eventually aiming for a prediction model that can aid clinical decision-making. Recently, preconditioned random forest regression (PRFR), proposed by Oh et al. (23) as means to prioritize the SNPs with predictive benefits, led to discovery of SNP panels of high relevance to radiotherapy-related toxicities (24).
To address the limitations of the previous studies on fatigue in breast cancer survivors, we applied a machine learning approach on data from the Cancer Toxicity (CANTO) cohort, consisting of large prospective, longitudinal, clinical, patient-reported outcomes and genomic data of survivors of early-stage breast cancer, to search the genome for a panel of fatigue-associated SNPs that could help predict severe fatigue 1 year after completion of primary breast cancer therapy and potentially suggest putative biological mechanisms of cancer-related fatigue.
Methods
Study Procedures
The CANTO study (NCT01993498) is a prospective cohort study that enrolled 12 012 patients between 2012 and 2018.
Patients were evaluated at diagnosis (baseline) and then for 5 years following completion of primary treatment, including surgery, adjuvant chemotherapy, or radiation therapy, whichever came last. For this study, data on diagnosis and 1 year after completion of primary treatment were used. Clinical data were prospectively collected by dedicated nurse practitioners. Socioeconomic characteristics and validated patient-reported outcome data including European Organization for Research and Treatment of Cancer quality-of-life questionnaires (EORTC QLQ-C30 and EORTC-QLQ-FA12 [fatigue-specific module]) (11,25), Global Physical Activity Questionnaire-16,12 and Hospital Anxiety and Depression Scale were also collected (23). Blood samples were collected at diagnosis for the purpose of DNA extraction from whole blood lymphocytes (26). The study was approved by the National Regulatory Authorities and Ethics Committee (ID-RCB: 2011-A01095-36, 11–039). All patients enrolled in the study provided written informed consent, including consent for the biological data collection.
Fatigue Endpoints
As a primary endpoint, severe fatigue was defined at 1 year after the end of primary treatment using the EORTC QLQ-C30 Overall Fatigue subscale and the EORTC QLQ-FA12 Physical, Emotional, and Cognitive Fatigue subscales. The continuum of scores for each endpoint was dichotomized into an event or nonevent endpoint variable to define severe or nonsevere fatigue, respectively. Patients were considered to have severe fatigue if they reported a fatigue score higher than the quartile 3 in the fatigue score distribution at 1 year after primary treatment completion. This cutoff was determined qualitatively to isolate patients with higher fatigue scores as seen from the distribution of score changes (Supplementary Figure 1, available online).
Study Cohort
Clinical Cohort
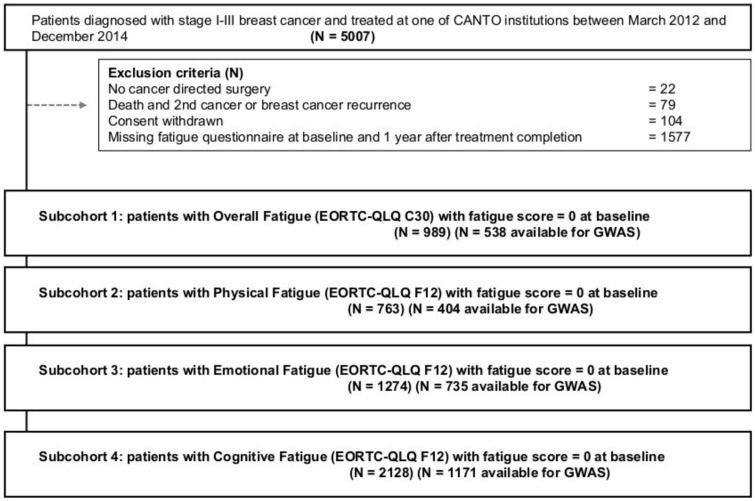
We accessed clinical data from 5007 patients enrolled in CANTO between March 2012 and December 2014. The main exclusion criteria to define a study group for each fatigue domain included absence of cancer-directed surgery to include only patients treated with curative intent; death, secondary cancer, or breast cancer recurrence to focus on a population disease free; withdrawn consent; missing baseline or follow-up scores for each fatigue endpoint; and nonzero baseline scores for the respective fatigue domain, because we were interestedin isolating the fatigue events that developed after breast cancer diagnosis and therefore more likely associated with treatment (Figure 1). The resulting clinical sample sizes were 989, 763, 1274, and 2128 for Overall, Physical, Emotional, and Cognitive Fatigues, respectively (Supplementary Tables 1 and 2, available online, detail cohort characteristics).
Figure 1.
Consolidated Standards of Reporting Trial (CONSORT) diagram of study population. Patients with no fatigue scores available had Overall more missing information in most baseline characteristics and other patient-reported outcomes. In selected characteristics, we recorded statistically significant differences between the 2 groups of patients. Missing fatigue score correlated with education, income, and TNM stage (Supplementary Table 4, available online). CANTO = Cancer Toxicity study; EORTC-QLQ = European Organization for Research and Treatment quality of life; GWAS = genome-wide association studies.
GWAS Data
By July 2018, 3895 patients from the entire CANTO cohort were genotyped at study inclusion and had available information for 687 572 germline SNPs (Illumina InfiniumExome24 version 1.1 and Illumina GSA24 v1.0). Standard quality control (14) was applied, filtering 1) 68 individuals with high genetic similarity, non-European origin, and low X chromosome heterozygosity (<0.15); and 2) 177 746 SNPs due to minor allele frequency less than 0.01, missing rate greater than 0.05, and Hardy-Weinberg Equilibrium P less than 10–5. Finally, 2 patients with an SNP missing rate greater than 0.05 were removed. Thus, 3825 patients with 509 826 SNPs passed the quality control. The genomic study cohort was defined by an overlap between the 3825 genotyped patients and the clinical overall cohorts as described above (N = 2799). The resulting sample size per fatigue endpoint was 538, 404, 735, and 1171 for Overall, Physical, Emotional, and Cognitive Fatigue, respectively (Figure 1; Supplementary Table 1, available online).
Statistical Analysis
We hypothesized that fatigue has genetic determinants that differ by fatigue domain, and cancer-related fatigue can be best predicted by combining genomic and clinical data.
Univariate Analyses of Clinical and Genomic Variables
First, we investigated univariate associations between each fatigue endpoint and clinical variables selected on the basis of clinical judgment. The Benjamini-Hochberg procedure was applied to the P values to identify statistically significantly associated variables (false discovery rate < 0.05) (27). Then a genome-wide association scan was performed to test associations between each SNP and the fatigue endpoints. The association was tested using the χ2 test under the additive model while adjusting for the first 3 principal components for ancestry.
Multivariate Modeling of Fatigue Using Genetic and Clinical Variables
Using machine-learning techniques, predictive modeling on severe fatigue at 1 year after primary treatment completion was built based on patterns in patients’ permutations of SNPs. To this end, a multivariable prediction model, based on PRFR methods, was built as described by Oh et al. (23).
First, the data were randomly split into the training and validation setswith matching event rate and distribution for the clinical variables with statistically significant univariate associations (Table 2). The PRFR model was built and validated separately on these 2 disjoint subsets (a holdout approach) (28). To reduce modeling computational complexity, an independent screening (29,30) was performed on the GWAS training data to filter likely irrelevant predictors: the SNPs with univariate correlation (P < .001), as determined empirically by previous studies (23,24), were selected for further predictive modeling. Missing genotypes (<5%) in the training set were imputed with the most frequent value.
Table 2.
Predictive performance of PRFR in the validation dataset with respect to the Overall, Physical, Emotional, and Cognitive fatiguea
| Fatigue category or endpoint | No. of samples (train/test) | Event rate, % | No. of SNPs with P < .001 | PRFR performance |
|||
|---|---|---|---|---|---|---|---|
| SNP only |
SNP + clinical |
||||||
| AUC | P b | AUC | P b | ||||
| Overall (EORTC-QLQ-C30) | 377/161 | 12.5 | 309 | 0.42 | .89 | NA | — |
| Fatigue domains (EORTC-QLQ-12) | |||||||
| Physical | 283/121 | 19.1 | 277 | 0.44 | .78 | NA | — |
| Emotional | 515/220 | 20.8 | 257 | 0.42 | .96 | 0.42 | .96 |
| Cognitive | 820/351 | 17.0 | 299 | 0.59 | .01 | .60 | .005 |
EORTC QLQ = European Organization for Research and Treatment of Cancer Quality of Life; NA = not applicable; PRFR = preconditioned random forest regression; SNP = single-nucleotide polymorphism.
P value was estimated using Mason and Graham’s test and was 2-sided.
The predictive performance of PRFR in the validation cohort was measured using the area under the curve (AUC) metric. Using Mason and Graham’s test (31), statistical significance of the AUC was tested under the null hypothesis of AUC not higher than random (0.5). For the endpoints that were predicted by genomic profiles with AUC greater than 0.5, contribution of the clinical variables to predictive performance was also investigated. The PRFR model was retrained with additional predictors from the clinical domain with statistically significant univariate association. The resulting risk model’s goodness of risk calibration was performed by 1) grouping the patients in the validation set by 3 equally sized high, intermediate, and low predicted risk bins and 2) calculating actual prevalence of fatigue within each bin.
For comparison with other conventional multivariable methods, least absolute shrinkage and selection operator and conventional random forest models were also built using the same training and validation sets as the PRFR model. Also, to preclude the possibility that the genomic model merely reflects ancestry differences confounding the outcomes, the comparison included a logistic regression model using only the first 3 principal components of genotypes as predictors.
Biological Interpretation of the Prediction Models
We performed an additional statistical analysis on the predictive modeling results to uncover the potential biomarkers and biological processes that might contribute to posttherapy fatigue. To this end, the PRFR ranked relative importance of predictors, also known as variable importance measure (VIM). To control for the effects of the clinical variables, we used the VIM from the PRFR model that was built with the genomic and clinical variables combined. The SNPs with the highest 50% VIM were taken to the following steps for biological interpretation. The SNPs were mapped within 50 000 base pairs of proximity according to the genome build 19 (hg19). The resulting gene list was analyzed for enrichment of previously known biological processes, pathways, or biomarker groups for certain diseases. For comparison, the enrichment analysis was also done using the initial SNP set with univariate correlation P less than .001 without the VIM-based filtering. In addition, an interactome analysis searched for a network of genes that are connected through previously known interactions. MetaCore (Thompson Reuters, New York, NY) was used for the enrichment and interactome analyses.
Analyses were performed using SAS (v.9.4) and R (v.3.6.0) packages GenABEL (21). All statistical tests were 2-sided.
Results
Baseline clinical characteristics are represented in Supplementary Table 2 (available online).
Univariate Analyses of Clinical and Genomic Variables
No statistically significant clinical variables were found for Overall and Physical fatigue endpoints. In contrast, anxiety (P = 4.34 × 10–6, odds ratio [OR] = 1.90 and 95% confidence interval [CI] = 1.34 to 2.67; for doubtful, OR = 2.22, 95% CI = 1.45 to 3.35 for certain vs absent) and pain (P = 7.78 × 10–5, OR = 1.02, 95% CI = 1.01 to 1.02 for unit pain score increase) were statistically significantly associated with increased risk for Emotional Fatigue. For Cognitive Fatigue, anxiety (P = 1.41 × 10–4, OR = 1.64, 95% CI = 1.24 to 2.17 for doubtful, OR = 1.62, 95% CI = 1.19, 2.2 for certain vs absent), depression (P = 4.29 × 10–4, OR = 1.87, 95% CI = 1.11 to 3.08, for doubtful, OR = 3.07, 95% CI = 1.31 to 6.86 for certain vs absent), and pain (P = 2.98 × 10–8, OR = 1.02, 95% CI = 1.01 to 1.02 for unit score increase) were statistically significantly associated (Table 1). No genome-wide significant SNPs (P < 5.00 × 10–8) were found to be associated with any of the endpoints. There was no notable genomic inflation for any of the 4 endpoints (Supplementary Figure 2, available online). These results were consistent for the genome-wide scan within the training subcohorts. The number of SNPs from the genome-wide scan within the training set with some degree of association (P < .001) was 309 for Overall, 277 for Physical, 257 for Emotional, and 299 for Cognitive Fatigue.
Table 1.
Statistical significance of association between clinical covariates and 4 fatigue endpoints
| Variable | Fatigue categories/endpoints |
|||||||
|---|---|---|---|---|---|---|---|---|
| Overall (N = 989) |
Physical (N = 763) |
Emotional (N = 1274) |
Cognitive (N = 2128) |
|||||
| Odds ratio (95% CI) | P | Odds ratio (95% CI) | P | Odds ratio (95% CI) | P | Odds ratio (95% CI) | P | |
| Sociodemographic | ||||||||
| Age, continuous | 0.99 (0.97 to 1.01) | .16 | 0.99 (0.97 to 1) | .14 | 0.99 (0.98 to 1) | .06 | 0.98 (0.97 to 0.99) | .003 |
| Education | ||||||||
| College or higher (referent) | 1.00 (Ref) | .79 | 1.00 (Ref) | .70 | 1.00 (Ref) | .38 | 1.00 (Ref) | .02 |
| High school | 0.86 (0.55 to 1.35) | 1.15 (0.74 to 1.78) | 1.24 (0.9 to 1.71) | 1.36 (1.04 to 1.77) | ||||
| Primary school | 0.93 (0.52 to 1.63) | 1.24 (0.69 to 2.19) | 1.21 (0.78 to 1.85) | 1.55 (1.07 to 2.23) | ||||
| Monthly household incomeb (euros) | ||||||||
| 1500 (Referent) | 1.00 (Ref) | .50 | 1.00 (Ref) | .21 | 1.00 (Ref) | .35 | 1.00 (Ref) | .22 |
| 1500-3000 | 0.7 (0.37 to 1.39) | 0.61 (0.35 to 1.12) | 0.83 (0.53 to 1.33) | 0.77 (0.52 to 1.14) | ||||
| 3000 | 0.72 (0.39 to 1.43) | 0.67 (0.37 to 1.23) | 1.03 (0.66 to 1.64) | 0.92 (0.63 to 1.38) | ||||
| Employment statusb | ||||||||
| Nonactive (Referent) | 1.00 (Ref) | .03 | 1.00 (Ref) | .05 | 1.00 (Ref) | .03 | 1.00 (Ref) | 6.95x10−4 |
| Active | 1.54 (1.04 to 2.3) | 1.48 (1.01 to 2.17) | 1.37 (1.03 to 1.82) | 1.5 (1.18 to 1.91) | ||||
| Marital statusb | ||||||||
| Not married (Referent) | 1.00 (Ref) | .80 | 1.00 (Ref) | .51 | 1.00 (Ref) | .82 | 1.00 (Ref) | .65 |
| Married | 0.83 (0.53 to 1.34) | 0.85 (0.54 to 1.34) | 0.95 (0.68 to 1.34) | 1.08 (0.81 to 1.46) | ||||
| Clinical | ||||||||
| Hormonal statusb | ||||||||
| Premenopause (Referent) | 1.00 (Ref) | .83 | 1.00 (Ref) | .29 | 1.00 (Ref) | .08 | 1.00 (Ref) | .004 |
| Postmenopause | 0.93 (0.61 to 1.47) | 0.79 (0.53 to 1.2) | 0.76 (0.57 to 1.03) | 0.7 (0.55 to 0.89) | ||||
| Smoking statusb | ||||||||
| Smoker (Referent) | 1.00 (Ref) | .26 | 1.00 (Ref) | .58 | 1.00 (Ref) | .16 | 1.00 (Ref) | .005 |
| Ex-smoker | 0.97 (0.49 to 1.97) | 0.74 (0.37 to 1.48) | 0.82 (0.5 to 1.34) | 0.74 (0.5 to 1.09) | ||||
| Nonsmoker | 0.71 (0.4 to 1.32) | 0.77 (0.44 to 1.39) | 0.69 (0.46 to 1.06) | 0.6 (0.44 to 0.84) | ||||
| Alcohol statusb,d | ||||||||
| No (Referent) | 1.00 (Ref) | 1.00 | 1.00 (Ref) | .49 | 1.00 (Ref) | .08 | 1.00 (Ref) | .10 |
| Yes | 1.01 (0.52 to 1.82) | 1.24 (0.71 to 2.11) | 1.43 (0.95 to 2.13) | 0.72 (0.48 to 1.05) | ||||
| Physical activity (GPAQ 16)a | ||||||||
| Q1 (Referent) | 1.00 (Ref) | .20 | 1.00 (Ref) | .06 | 1.00 (Ref) | .03 | 1.00 (Ref) | .36 |
| Q2 | 1.47 (0.84 to 2.58) | 1.19 (0.67 to 2.13) | 0.9 (0.61 to 1.32) | 0.77 (0.55 to 1.07) | ||||
| Q3 | 0.96 (0.54 to 1.7) | 0.83 (0.47 to 1.45) | 0.68 (0.45 to 1.01) | 0.85 (0.61 to 1.18) | ||||
| Q4 | 0.87 (0.49 to 1.55) | 0.98 (0.57 to 1.69) | 0.59 (0.39 to 0.89) | 0.79 (0.57 to 1.11) | ||||
| Charlson comorbidity score, continuous | 1.17 (0.98 to 1.4) | .09 | 1 (0.83 to 1.22) | .97 | 0.97 (0.83 to 1.14) | .82 | 0.96 (0.83 to 1.11) | .59 |
| Depression (HADS)b | ||||||||
| Absent (Referent) | 1.00 (Ref) | .59 | 1.00 (Ref) | .06 | 1.00 (Ref) | .06 | 1.00 (Ref) | 4.29 x 10−4c |
| Doubtful | 1.4 (0.51 to 3.29) | 0.87 (0.21 to 2.65) | 2.33 (0.94 to 5.45) | 1.87 (1.11 to 3.08)c | ||||
| Certain | 1.47 (0.36 to 4.53) | 6.48 (0.74 to 78.18) | NA | 3.07 (1.31 to 6.86)c | ||||
| Anxiety (HADS)b | ||||||||
| Absent (Referent) | 1.00 (Ref) | .86 | 1.00 (Ref) | .31 | 1.00 (Ref) | 4.34 x 10−6c | 1.00 (Ref) | 1.41 x 10−4c |
| Doubtful | 0.88 (0.53 to 1.43) | 1.39 (0.88 to 2.18) | 1.9 (1.34 to 2.67)c | 1.64 (1.24 to 2.17)c | ||||
| Certain | 0.93 (0.56 to 1.53) | 1.18 (0.7 to 1.97) | 2.22 (1.45 to 3.35)c | 1.62 (1.19 to 2.2)c | ||||
| Symptoms and quality of life | ||||||||
| Hot flashesb | ||||||||
| No (Referent) | 1.00 (Ref) | .27 | 1.00 (Ref) | .12 | 1.00 (Ref) | .006 | 1.00 (Ref) | .003 |
| Yes | 1.28 (0.84 to 1.94) | 1.4 (0.91 to 2.12) | 1.53 (1.13 to 2.06) | 1.46 (1.14 to 1.88) | ||||
| Pain (EORTC QLQ-C30),b continuous | 1.02 (1.01 to 1.04) | .006 | 1.02 (1 to 1.03) | .008 | 1.02 (1.01 to 1.02)c | 7.78 x 10−5c | 1.02 (1.01 to 1.02)c | 2.98 x 10−8c |
| Insomnia (EORTC QLQ-C30,)b continuous | 1.01 (1 to 1.01) | .11 | 1 (1 to 1.01) | .49 | 1 (1 to 1.01) | .07 | 1.01 (1 to 1.01) | 6.71 x 10−4 |
| Tumor characteristics | ||||||||
| Tumor grade | ||||||||
| 1 (Referent) | 1.00 (Ref) | .29 | 1.00 (Ref) | .25 | 1.00 (Ref) | .09 | 1.00 (Ref) | .70 |
| 2 | 1.49 (0.87 to 2.67) | 1.06 (0.64 to 1.79) | 1.47 (0.98 to 2.24) | 1.13 (0.81 to 1.58) | ||||
| 3 | 1.5 (0.82 to 2.84) | 1.45 (0.84 to 2.55) | 1.59 (1.02 to 2.53) | 1.15 (0.8 to 1.67) | ||||
| Tumor subtype | ||||||||
| HR+HER2+(Referent) | 1.00 (Ref) | .71 | 1.00 (Ref) | .02 | 1.00 (Ref) | .31 | 1.00 (Ref) | .29 |
| HR+HER2- | 0.95 (0.49 to 1.98) | 0.43 (0.25 to 0.78) | 0.95 (0.59 to 1.56) | 0.74 (0.52 to 1.07) | ||||
| HR-HER2+ | 1.49 (0.42 to 4.85) | 0.64 (0.19 to 1.95) | 1.61 (0.69 to 3.67) | 0.71 (0.33 to 1.44) | ||||
| HR-HER2- | 0.77 (0.26 to 2.18) | 0.62 (0.26 to 1.44) | 0.75 (0.34 to 1.59) | 0.91 (0.53 to 1.56) | ||||
| Tumor stage, AJCC | ||||||||
| I (Referent) | 1.00 (Ref) | .02 | 1.00 (Ref) | .31 | 1.00 (Ref) | .31 | 1.00 (Ref) | .01 |
| II | 1.77 (1.17 to 2.67) | 1.34 (0.9 to 2) | 1.21 (0.89 to 1.63) | 1.36 (1.06 to 1.75) | ||||
| III | 1.26 (0.55 to 2.64) | 1.21 (0.54 to 2.53) | 1.33 (0.78 to 2.21) | 1.57 (1.04 to 2.33) | ||||
| Treatment | ||||||||
| Chemotherapye | ||||||||
| No (Referent) | 1.00 (Ref) | .22 | 1.00 (Ref) | .04 | 1.00 (Ref) | .002 | 1.00 (Ref) | .002 |
| Yes | 1.28 (0.87 to 1.9) | 1.5 (1.02 to 2.2) | 1.56 (1.18 to 2.08) | 1.45 (1.14 to 1.84) | ||||
| Trastuzumab | ||||||||
| No (Referent) | 1.00 (Ref) | .14 | 1.00 (Ref) | .004 | 1.00 (Ref) | .05 | 1.00 (Ref) | .34 |
| Yes | 1.59 (0.85 to 2.84) | 2.3 (1.26 to 4.07) | 1.55 (0.99 to 2.4) | 1.2 (0.83 to 1.7) | ||||
| Endocrine therapy | ||||||||
| No (Referent) | 1.00 (Ref) | .16 | 1.00 (Ref) | .59 | 1.00 (Ref) | .40 | 1.00 (Ref) | .81 |
| Yes | 1.52 (0.88 to 2.78) | 1.17 (0.72 to 1.96) | 1.2 (0.82 to 1.78) | 1.05 (0.77 to 1.45) | ||||
| Breast surgery | ||||||||
| Breast conservation (Referent) | 1.00 (Ref) | .59 | 1.00 (Ref) | .05 | 1.00 (Ref) | .03 | 1.00 (Ref) | .44 |
| Mastectomy | 1.16 (0.72 to 1.81) | 1.55 (0.99 to 2.41) | 1.46 (1.05 to 2.01) | 1.12 (0.85 to 1.47) | ||||
| Lymphadenectomy | ||||||||
| No (Referent) | 1.00 (Ref) | .01 | 1.00 (Ref) | .04 | 1.00 (Ref) | .40 | 1.00 (Ref) | .006 |
| Axillary | 0.33 (0.05 to 3.78) | 1.91 (0.23 to 88.67) | Inf (0.87 to Inf) | |||||
| Sentinel lymph node biopsy | 0.26 (0.04 to 2.95) | 1.36 (0.16 to 63.1) | Inf (0.62 to Inf) | |||||
| Radiotherapy | ||||||||
| No (Referent) | 1.00 (Ref) | .78 | 1.00 (Ref) | 1.00 | 1.00 (Ref) | .55 | 1.00 (Ref) | .10 |
| Yes | 1.17 (0.58 to 2.6) | 1.04 (0.52 to 2.28) | 0.84 (0.51 to 1.43) | 1.53 (0.94 to 2.61) | ||||
AJCC = American Joint Committee on Cancer; CI = confidence interval; EORTC QLQ = European Organization for Research and Treatment of Cancer Quality of Life; GPAQ 16 = Global Physical Activity Questionnaire 16; HADS = Hospital Anxiety and Depression Scale; HER2 = human epidermal growth factor receptor 2; HR = hormone receptor; Q = quartile; Referent = reference level.
Assessed at baseline.
Statistical significance at Benjamini-Hochberg false discovery rate of 5%.
At least 1 drink per day.
In each subcohort, at least 86% of patients who received chemotherapy were treated with anthracycline and taxane combinations, mainly fluorouracil plus epirubicin plus cyclophosphamide followed by a taxane (docetaxel or paclitaxel) (see Supplementary Table 2, available online). In this setting, most patients received 6 cycles every 3 weeks with standard dose.
Predictive Performance of Genomic and Clinical Models
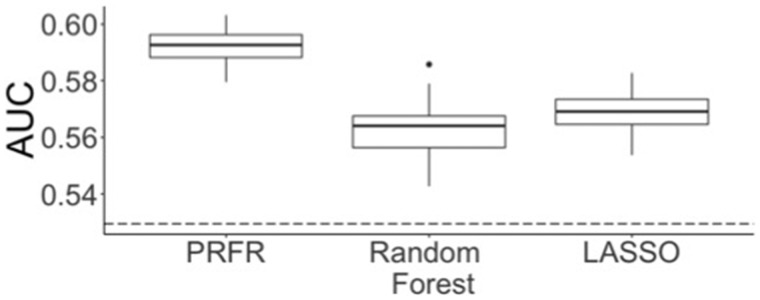
Only for the Cognitive Fatigue, the genomic-only model was validated with an AUC statistically significantly larger than 0.5 (AUC = 0.59, P = .01) (Table 2), which was marginally (not statistically significantly) improved to 0.60 (P = .005) by adding the aforementioned statistically significant clinical variables. The resulting clinico-genomic model for the Cognitive Fatigue showed good calibration (Figure 2); the predicted risk curve with respect to the 3 risk bins did not statistically significantly deviatefrom the actual severe fatigue occurrence (Hosmer-Lemeshow P = .09). The predictive performance of other conventional methods on the hold-out set was lower than for PRFR (Figure 3).
Figure 2.
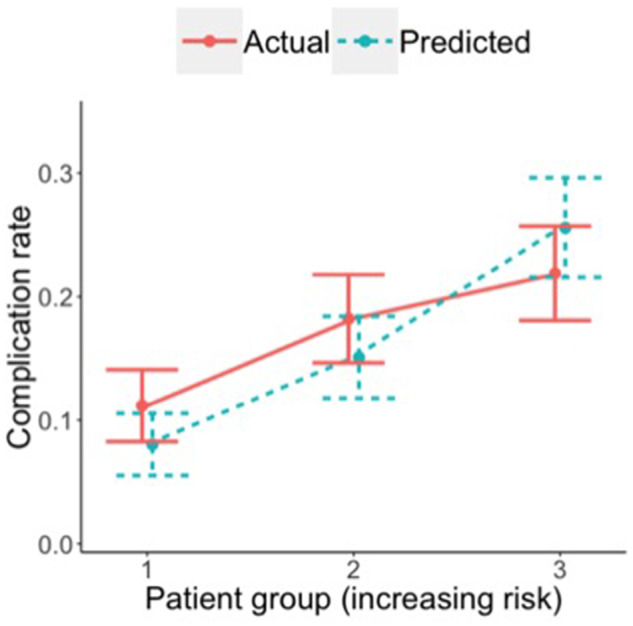
Risk calibration curve for the clinico-genomic Cognitive Fatigue prediction model.
Figure 3.
Comparison of the area under curve (AUC) in predicting Cognitive Fatigue between the preconditioned random forest regression (PRFR) method and other conventional multivariable regression methods. Confidence intervals on validation AUC were obtained by repeating the training process using randomly selected 80% of the training data. Dotted line = prediction AUC when the first 3 principal components for ancestry were used as the only predictors. LASSO = least absolute shrinkage and selection operator.
Biological Interpretation of the Genomic Models
Only the PRFR model for the Cognitive endpoint yielded an AUC with a P less than .05 and thus was analyzed for biological interpretability. The highest VIM was recorded for rs4742675 (VIM = 2.00 × 10–3, minor allele frequency = 0.24), which is located in an intergenic region in chromosome 9. In comparison, the clinical variable with the highest VIM was pain (VIM = 1.26 × 10–4, ranking = 101). The rest of the clinical variables scored relatively low compared with genomic variables. Out of 200 SNPs with top 50% VIM, 137 SNPs were annotated with at least 1 gene. The gene set enrichment analysis was performed using the 89 genes that were annotated to the 137 SNPs. Statistically significant enrichments in genes that are involved in cognitive and mood disorders (false discovery rate, P = 1.51 × 10−12) or inflammation or complementary system(P = .03) were observed from the selected SNPs but not from the original SNP set without VIM filtering (Table 3). Regardless of the filtering results, a biological process pertinent to synaptic transmission (P = 6.8 × 10−8) showed a high degree of enrichment. From the selected SNP list, Metacore analysis also identified a cluster of 4 gene products (Supplementary Figure 3, available online) consisting of Insulin-like Growth Factor (IGF)-1 receptor, Growth Factor Receptor Bound Protein 14 (GRB14), Fibroblast Growth Factor Receptor 1 (FGFR1), and Dual Leucine zipper Kinase (DLK). Supplementary Table 3 (available online) includes the VIM for all SNPs and clinical predictors for the Cognitive Fatigue model as well as annotation information for the SNP predictors.
Table 3.
The most statistically significantly enriched gene groups from the genes associated with the SNPs with GWAS scan P less than .001 (unfiltered) and the subset of those SNPs with top 50% VIM for the Cognitive Fatigue endpoint, obtained using the bioinformatics tool GeneGOa
| Pathway maps |
Biological process |
||||||
|---|---|---|---|---|---|---|---|
| Unfiltered |
Top 50% VIM |
Unfiltered |
Top 50% VIM |
||||
| Name | FDR | Name | FDR | Name | FDR | Name | FDR |
| Nociception or pronociceptive action of nociception in spinal cord at low doses | 2.48 × 10−4 | Immune response or lectin-induced complement pathway | 0.01 | Regulation of transport | 1.52 × 10−8 | Regulation of transport | 4.03 × 10−9 |
| O-glycan biosynthesis | 0.002 | Development/ oligodendrocyte differentiation from adult stem cells | 0.01 | Synaptic transmission, glutamatergic | 4.63 × 10−7 | Chemical synaptic transmission | 4.03 × 10−9 |
| Gamma-secretase proteolytic targets | 0.004 | Role of integrins in eosinophil degranulation in asthma | 0.01 | Regulation of localization | 8.25 × 10−7 | Anterograde trans-synaptic signaling | 4.03 × 10−8 |
| Calcium-dependent regulation of normal and asthmatic smooth muscle contraction | 0.02 | Degranulation of lung mast cells | 0.02 | Regulation of metal ion transport | 8.33 × 10−7 | Trans-synaptic signaling | 6.28 × 10−8 |
|
Complement pathway disruption in thrombotic microangiopathy |
0.02 |
Alternative complement cascade disruption in age-related macular degeneration |
0.02 |
Response to alkaloid |
8.33 × 10−7 |
Synaptic transmission glutamatergic |
6.28 × 10−8 |
|
Process networks |
Diseases (by biomarkers) |
||||||
|
Unfiltered |
Top 50% VIM |
Unfiltered |
Top 50% VIM |
||||
|
Name |
FDR |
Name |
FDR |
Name |
FDR |
Name |
FDR |
| Neurophysiological processor transmission of nerve impulse | 0.004 | Inflammation and complement system | 0.03 | Schizophrenia | 2.74 × 10−10 | Huntington disease | 1.26 × 10−12 |
| Development, neurogenesis, or synaptogenesis | 0.004 | — | — | Schizophrenia spectrum and other psychotic disorders | 2.74 × 10−10 | Chorea | 1.41 × 10−12 |
| — | — | — | — | Head and neck neoplasm | 6.11 × 10−10 | Brain ischemia | 1.51 × 10−12 |
| — | — | — | — | Digestive system diseases | 7.80 × 10−9 | Depressive disorders | 1.51 × 10−11 |
| — | — | — | — | Colorectal neoplasms | 1.08 × 10−8 | Cognition disorders | 1.51 × 10−10 |
Only the groups with FDR < 0.05 are shown. FDR = false discovery rate; GWAS = genome-wide association studies; SNP = single nucleotide polymorphism; VIM = variable importance measure.
Discussion
In this large multicentric, prospective, clinico-genomic longitudinal dataset of breast cancer survivors, we deployed machine learning techniques to investigate if high-dimensional genomic data could be used to build and validate a predictive model for the different known dimensions of fatigue. Although the ability of our models to identify clinic and genomic contributors of fatigue differed by fatigue domain, a group of SNPs and clinical variables was suggested to be associated with the cognitive domain.
Cancer-related fatigue is known to be complex in etiology, with possibly many clinical, bio-behavioral, and genetic contributors (1). Prior studies had several limitations. First, comprehensive integration of clinical, behavioral, and genetic information was lacking. Second, prior studies focused on candidate gene approaches mostly targeting proinflammatory cytokine activity that were largely not independently validated. Moreover, longitudinal design that follows patients from pretreatment into the survivorship period has not been implemented. Last, there has been lack of evaluation of the different dimensions of fatigue (1). In this study we tried to address all these limitations.
Our approach used machine learning to identify a group of SNPs and clinical information that may be associated with breast cancer–related Cognitive Fatigue. Several genes that were associated with the identified SNP were in alignment with prior knowledge of cancer-related fatigue. In the same way, the clinical predictors found in this data, including anxiety, depression, and pain, were previously shown to be associated with fatigue and cognitive dysfunction (12,13).
In the last decade, several studies highlighted a possible role of inflammation in cancer-related fatigue (14,15). In this context, our study suggested a link between enrichment in inflammatory complement system and onset of severe fatigue, which supports prior findings by Rajeevan et al. (32), who had reported associations between single nucleotide variations in complement activation pathway genes and chronic fatigue syndrome. Moreover, our interactome analysis uncovered the 4 gene products that are connected via previously known interactions or associations. The cluster included 2 growth-related proteins, IGF)-IR and FGFR1, which were previously named as potential biomarkers for cancer-related fatigue (33–35). In addition, the PRFR approach revealed new mechanisms that have not been previously explored in the scope of cancer-related fatigue. Among these, alteration of synaptic activity through glutamate was consistently discovered regardless of VIM filtering, which has been shown as an important pathway to chronic fatigue (36).
The predictive performance of the cognitive fatigue model was modest, only marginally improved by adding clinical variables, and was not statistically significantly different from the SNP-only model. The PRFR model made prediction predominantly using the genomic information, which was also reflected in the VIM distribution where baseline pain was the only variable in the top 50% of VIM. This could indicate information overlap: both SNPs and statistically significant clinical variables including pain, anxiety, and depression pertain to cognitive functions and behaviors. Notably, the agreement between the clinical and genomic factors might stress the close relation between a neurocognitive domain and cancer-related fatigue, which was also suggested by Van Dyk et al. (13). Also, there might exist a complex interplay between the genomic and baseline clinical characteristics that may have not been fully captured by the current algorithm.
Our study has important strengths, including its prospective and longitudinal design and the use of validated fatigue multidimensional questionnaires. In addition, patients in our study were treated with contemporary therapy protocols, and our models accounted for a number of sociodemographic, clinical, tumor, and treatment variables with low missing rates. Nevertheless, this study has some limitations. First, we set cutoffs to define our fatigue endpoint that we acknowledge as arbitrary. Second, limited sample size might have led to suboptimal predictive performance for the majority of the endpoints. This was partially due to exclusion of patients with nonzero baseline fatigue. However, this minimized confounding effects of heterogeneous baseline characteristics. Without this exclusion, the prediction would be dominated by clinical variables with minimal genomic impact (data not shown). Third, we excluded patients with missing fatigue questionnaires at baseline and follow-up. Specific populations with less education, lower income, or greater tumor stage might be underrepresented in this study (Supplementary Table 4, available online), which deserves future research. Fourth, aggressive filtering of genomic predictors was necessary in the attempt to reduce bias in permutation-based VIM in high dimensionality (37). Fifth, the study included individuals only of European origin, and thus the results are generalizable only to this population. Last, we acknowledge that the methodology and results reported in this article are mainly exploratory. Particularly, it is important to stress that the predictive power of the genomic variants identified as associated with fatigue is not sufficient to justify their use in clinical decision-making. Importantly, although our data point at pathways that may be worthy of further investigation, external validation of our findings is needed.
This study analyzed combined clinical and GWAS data from a large group of breast cancer survivors, suggesting a small genetic role for development of Cognitive Fatigue. This study broadens our understanding of cancer-related cognitive fatigue and informs further studies focused on identifying those patients with high risk of cognitive fatigue. Also, it explores the feasibility of machine learning techniques in predicting cancer-related fatigue, which deserves further investigation.
Funding
This study was supported by a Career Catalyst Research grant from Susan G. Komen (CCR17483507) to Ines Vaz-Luis, Odyssea to Ines Vaz-Luis, and Fondation ARC pour la recherche sur le cancer grant (CANTO-WORK-programme labillisé) to Ines Vaz-Luis and Gwenn Menvielle, Foundation Gustave Roussy to Ines Vaz-Luis. This research was supported by the French government under the “Investment for the Future” program managed by the National Research Agency (ANR), grant no ANR-10-COHO-0004. This research was supported by the Laboratory of Excellence GENMED (Medical Genomics) grant no. ANR-10-LABX-0013 managed by the ANR part of the Investment for the Future program. In addition, it also received support by National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748, U01 CA083178, R01 CA097397, R01 CA114236, P01 CA19656, and Breast Cancer Research Foundation grant BCRF-17–193.
Notes
Role of the funders: The study sponsors had no role in the design, collection, analysis, data interpretation, writing of the manuscript or decision to submit the manuscript.
Disclosures: J. O. Deasy has research contracts with Varian Medical Systems and Philips and is a shareholder in Paige.AI. A. Di Meglio reports personal fees from Thermo Fisher and grants from European Society for Medical Oncology (ESMO), fellowship support, outside the submitted work. O. Tredan reports personal fees from Roche, MSD, Novartis, Lilly, Astra Zeneca, and grants from Roche MSD, BMS, outside the submitted work. S. Michiels reports personal fees from Statistical advice: IDDI, Belgium and Janssen Cilag, France; Independent Data Monitoring Committee member: Hexal, J&J, Genticel, Mabxience, Steba, IQVIA, Roche, Sensorion, Biophytis, Servier, outside the submitted work. F. Andre reports grants from AstraZeneca, Pfizer, Lilly, Roche, Novartis, Daichii, outside the submitted work. I. Vaz-Luis reports personal fees from AstraZeneca, Kephren, Amgen, and Novartis, outside the submitted work. No other author reported disclosures.
Previous presentations: Results of this study were partly presented as an oral communication during the American Society of Clinical Oncology (ASCO) Congress 2019 (Chicago). DOI: 10.1200/JCO.2019.37.15_suppl.11515 Journal of Clinical Oncology 37, no. 15_suppl (May 20 2019) 11515–11515.
Supplementary Material
References
- 1. Bower JE. Cancer-related fatigue–mechanisms, risk factors, and treatments. Nat Rev Clin Oncol. 2014;11(10):597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abrahams HJ, Gielissen MF, Schmits IC, Verhagen CA, Rovers MM, Knoop H. Risk factors, prevalence, and course of severe fatigue after breast cancer treatment: a meta-analysis involving 12 327 breast cancer survivors. Ann Oncol. 2016;27(6):965–974. [DOI] [PubMed] [Google Scholar]
- 3. Bower JE, Ganz PA, Desmond KA, Rowland JH, Meyerowitz BE, Belin TR. Fatigue in breast cancer survivors: occurrence, correlates, and impact on quality of life. J Clin Oncol. 2000;18(4):743–753. [DOI] [PubMed] [Google Scholar]
- 4. Bower JE. Management of cancer-related fatigue. Clin Adv Hematol Oncol. 2006;4(11):828–829. [PubMed] [Google Scholar]
- 5. Curt G, Johnston PG. Cancer fatigue: the way forward. Oncologist. 2003;8(S1):27–30. [DOI] [PubMed] [Google Scholar]
- 6. Broeckel JA, Jacobsen PB, Horton J, Balducci L, Lyman GH. Characteristics and correlates of fatigue after adjuvant chemotherapy for breast cancer. J Clin Oncol. 1998;16(5):1689–1696. [DOI] [PubMed] [Google Scholar]
- 7. Dow KH, Ferrell BR, Leigh S, Ly J, Gulasekaram P. An evaluation of the quality of life among long-term survivors of breast cancer. Breast Cancer Res Treat. 1996;39(3):261–273. [DOI] [PubMed] [Google Scholar]
- 8. Curt GA, Breitbart W, Cella D, et al. Impact of cancer-related fatigue on the lives of patients: new findings from the fatigue coalition. Oncologist. 2000;5(5):353–360. [DOI] [PubMed] [Google Scholar]
- 9. Andrykowski MA, Curran SL, Lightner R. Off-treatment fatigue in breast cancer survivors: a controlled comparison. J Behav Med. 1998;21(1):1–18. [DOI] [PubMed] [Google Scholar]
- 10. Cella D, Lai JS, Chang CH, Peterman A, Slavin M. Fatigue in cancer patients compared with fatigue in the general United States population. Cancer. 2002;94(2):528–538. [DOI] [PubMed] [Google Scholar]
- 11. Weis J, Tomaszewski KA, Hammerlid E, et al. International psychometric validation of an EORTC quality of life module measuring cancer related fatigue (EORTC QLQ-FA12). J Natl Cancer Inst. 2017;109(5):djw273. [DOI] [PubMed] [Google Scholar]
- 12. Donovan KA, Small BJ, Andrykowski MA, Munster P, Jacobsen PB. Utility of a cognitive-behavioral model to predict fatigue following breast cancer treatment. Health Psychol. 2007;26(4):464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Dyk K, Bower JE, Crespi CM, Petersen L, Ganz PA. Cognitive function following breast cancer treatment and associations with concurrent symptoms. NPJ Breast Cancer. 2018;4(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bower JE, Ganz PA, Irwin MR, Arevalo JM, Cole SW. Fatigue and gene expression in human leukocytes: increased NF-kappaB and decreased glucocorticoid signaling in breast cancer survivors with persistent fatigue. Brain Behav Immun. 2011;25(1):147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bower JE, Ganz PA, Irwin MR, Kwan L, Breen EC, Cole SW. Inflammation and behavioral symptoms after breast cancer treatment: do fatigue, depression, and sleep disturbance share a common underlying mechanism? J Clin Oncol. 2011;29(26):3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bower JE. Cancer-related fatigue: links with inflammation in cancer patients and survivors. Brain Behav Immun. 2007;21(7):863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Collado-Hidalgo A, Bower JE, Ganz PA, Cole SW, Irwin MR. Inflammatory biomarkers for persistent fatigue in breast cancer survivors. Clin Cancer Res. 2006;12(9):2759–2766. [DOI] [PubMed] [Google Scholar]
- 18. Bower JE, Ganz PA, Aziz N, Fahey JL. Fatigue and proinflammatory cytokine activity in breast cancer survivors. Psychosom Med. 2002;64(4):604–611. [DOI] [PubMed] [Google Scholar]
- 19. Bower JE, Ganz PA, Tao ML, et al. Inflammatory biomarkers and fatigue during radiation therapy for breast and prostate cancer. Clin Cancer Res. 2009;15(17):5534–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Orre IJ, Reinertsen KV, Aukrust P, et al. Higher levels of fatigue are associated with higher CRP levels in disease-free breast cancer survivors. J Psychosom Res. 2011;71(3):136–141. [DOI] [PubMed] [Google Scholar]
- 21. Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187(2):367–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oh JH, Kerns S, Ostrer H, Powell SN, Rosenstein B, Deasy JO. Computational methods using genome-wide association studies to predict radiotherapy complications and to identify correlative molecular processes. Sci Rep. 2017;7(1):43381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee S, Kerns S, Ostrer H, Rosenstein B, Deasy JO, Oh JH. Machine learning on a genome-wide association study to predict late genitourinary toxicity after prostate radiation therapy. Int J Radiat Oncol Biol Phys. 2018;101(1):128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.EORTC. Manuals EORTC – quality of life. https://qol.eortc.org/manuals/. Published 2019. Accessed June 20, 2019.
- 26. Vaz-Luis I, Cottu P, Mesleard C, et al. UNICANCER: French prospective cohort study of treatment-related chronic toxicity in women with localised breast cancer (CANTO). ESMO Open. 2019;4(5):e000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 28. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): The TRIPOD Statement. Br J Surg. 2015;102(3):148–158. [DOI] [PubMed] [Google Scholar]
- 29. Fan J, Lv J. Sure independence screening for ultrahigh dimensional feature space. J R Stat Soc Ser B (Stat Methodol). 2008;70(5):849–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei Z, Wang W, Bradfield J, et al. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet. 2013;92(6):1008–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mason SJ, Graham NE. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: statistical significance and interpretation. Q J R Meteorol Soc. 2002;128(584):2145–2166. [Google Scholar]
- 32. Rajeevan MS, Dimulescu I, Murray J, Falkenberg VR, Unger ER. Pathway-focused genetic evaluation of immune and inflammation related genes with chronic fatigue syndrome. Hum Immunol. 2015;76(8):553–560. [DOI] [PubMed] [Google Scholar]
- 33. Thornton LM, Andersen BL, Blakely WP. The pain, depression, and fatigue symptom cluster in advanced breast cancer: covariation with the hypothalamic-pituitary-adrenal axis and the sympathetic nervous system. Health Psychol. 2010;29(3):333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hamre H, Zeller B, Kanellopoulos A, et al. Serum cytokines and chronic fatigue in adults surviving after childhood leukemia and lymphoma. Brain Behav Immun. 2013;30:80–87. [DOI] [PubMed] [Google Scholar]
- 35. Saligan LN, Olson K, Filler K, et al. Multinational Association of Supportive Care in Cancer Fatigue Study Group–Biomarker Working Group. The biology of cancer-related fatigue: a review of the literature. Support Care Cancer. 2015;23(8):2461–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ronnback L, Hansson E. On the potential role of glutamate transport in mental fatigue. J Neuroinflammation. 2004;1(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Winham SJ, Colby CL, Freimuth RR, et al. SNP interaction detection with Random Forests in high-dimensional genetic data. BMC Bioinformatics. 2012;13(1):164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.