Abstract
Background
Epigenome-wide association studies using peripheral blood have identified specific sites of DNA methylation associated with risk of various cancers and may hold promise to identify novel biomarkers of risk; however, few studies have been performed for pancreatic cancer and none using a prospective study design.
Methods
Using a nested case-control study design, incident pancreatic cancer cases and matched controls were identified from participants who provided blood at baseline in 3 prospective cohort studies. DNA methylation levels were measured in DNA extracted from leukocytes using the Illumina MethylationEPIC array. Average follow-up period for this analysis was 13 years.
Results
Several new genomic regions were identified as being differentially methylated in cases and controls; the 5 strongest associations were observed for CpGs located in genes TMEM204/IFT140, MFSD6L, FAM134B/RETREG1, KCNQ1D, and C6orf227. For some CpGs located in chromosome 16p13.3 (near genes TMEM204 and IFT140), associations were stronger with shorter time to diagnosis (eg, odds ratio [OR] = 5.95, 95% confidence interval [CI] = 1.52 to 23.12, for top vs bottom quartile, for <5 years between blood draw and cancer diagnosis), but associations remained statistically significantly higher even when cases were diagnosed over 10 years after blood collection. Statistically significant differences in DNA methylation levels were also observed in the gastric secretion pathway using Gene Set Enrichment Analysis (GSEA) analysis.
Conclusions
Changes in DNA methylation in peripheral blood may mark alterations in metabolic or immune pathways that play a role in pancreatic cancer. Identifying new biological pathways in carcinogenesis of pancreatic cancer using epigenome-wide association studies approach could provide new opportunities for improving treatment and prevention.
Pancreatic cancer accounts for the third highest number of cancer-related deaths in the United States, after lung and colorectal cancers, and is expected to surpass colorectal cancer in the next decade (1). Pancreatic cancer is highly lethal, with 93% of pancreatic cancer patients succumbing to their disease within 5 years of diagnosis, largely because of most cases being diagnosed at late stages. Screening for pancreatic cancer is not currently recommended for asymptomatic adults because existing screening methods have not been shown to reduce mortality (2). Improving sensitivity of tests is important, but for screening to be effective, patients need to be identified at early stages of the disease.
Some studies aimed at identifying early detection biomarkers have focused on measuring DNA methylation of promoter regions in known cancer genes using peripheral cell-free DNA in bloods of patients (3, 4). These methods are promising, but it is unclear whether they would identify cancers at early stages of the disease, when treatment would be most effective. Alternatively, high-dimensional arrays now provide the opportunity to agnostically interrogate hundreds of thousands of biomarkers, including quantification of DNA methylation throughout the genome. Using these technologies, studies have begun to examine how DNA methylation levels in peripheral leukocytes vary by cancer status (5–7).
Several case-control studies on pancreatic cancer have compared DNA methylation in blood leukocytes of pancreatic cancer patients with healthy controls (8–10). However, these studies were retrospective case-control studies, and they cannot differentiate between recent changes to those that occurred months or years prior to diagnosis. Conducting prospective nested case-control studies using blood samples that were obtained from healthy individuals many years prior to diagnosis provides a unique opportunity to identify biomarkers that may be linked to disease progression. As DNA methylation levels can change over time to impact gene expression, identifying changes decades prior to disease could assist in uncovering pathways that contribute to the development of cancer, and novel genomic regions identified using these methods could potentially be targeted for prevention or drug intervention. To our knowledge, this study presents results from the first epigenome-wide association study (EWAS) on pancreatic cancer risk using prediagnostic peripheral blood leukocytes.
Methods
Study Populations
The primary EWAS was conducted using prediagnostic bloods of pancreatic cancer cases and matched controls selected from the Nurses’ Health Study (NHS), the Physicians’ Health Study (PHS), and the Health Professionals Follow-up Study (HPFS).
For this analysis, 403 incident cases were confirmed to have pancreatic cancer among the participants who provided blood samples prior to cancer diagnosis. A control subject was matched to each case on cohort (which also matches on sex), age (+/− 1 year), date of blood draw (month 3+/− and year), smoking (never, former, current), and race (white or other). Because of low DNA concentrations in some of the samples and samples removed after data processing of the arrays (see Supplementary Methods, available online), the initial 1:1 matching was not always conserved and resulted in some cases and controls with no matched pair. The final dataset consisted of 393 cases and 431 controls.
DNA Methylation Measurements
DNA extracted from buffy coats was bisulfite-treated, and DNA methylation was measured with the Illumina Infinium MethylationEPIC BeadChip array (Illumina, Inc, CA, USA). Details on DNA methylation measurements and data processing are provided in the Supplementary Methods (available online).
Statistical Analyses
All statistical analyses were performed in R (version 3.5.1). The dataset of cases and controls from this nested case-control study was randomly divided into a training set (n = 577) and testing set (n = 247) to replicate findings and reduce chance findings; however, to maximize power, analyses were also conducted using all data combined. We initially conducted the EWAS analysis using a series of unconditional multivariable logistic regression models to estimate odds ratios (ORs) and 95% confidence intervals (CIs) for the CpG-specific DNA methylation levels and pancreatic cancer risk. Unconditional logistic regression models were used to estimate odds ratios and 95% confidence intervals to maximize power by including cases and controls without matched pairs (Ptrend based on continuous variable of median value for each quartile). All models were adjusted for age at blood draw, cohort, race (white or nonwhite), smoking status (never, former, current), date of blood draw (continuous), body mass index (BMI), and cell composition (except in the cell-specific models) (11, 12), given the potential for confounding by cell composition (13). Models were fit to the training and testing sets separately, with the intent to compare statistically significant results; models were also fit on the whole dataset. All P values were adjusted for multiple comparisons using the false discovery rate (FDR) method. The statistical method section for the risk prediction score analysis is provided in the Supplementary Methods (available online).
We used the DMRcate Bioconductor R package (14) to identify differentially methylated regions (DMRs) associated with pancreatic cancer risk. DMRcate was applied independently to the training and testing sets using the following settings: region length 2000 bp; min of 2 statistically significant CpGs; FDR P value < .05 (adjusting for the same covariates as in the single CpG analyses). Statistically significant regions (after correction for multiple comparisons) were compared between the training and testing sets; CpGs in regions identified in both datasets were further evaluated by using the combined dataset to test associations with pancreatic cancer using CpG quartiles (based on methylation levels in the controls). Associations were also examined by cohort (using cohort-specific cut points for the quartiles) and by time from blood draw to diagnosis. Finally, to increase power, we also conducted the DMRcate analysis on the combined (training and testing) dataset, and the top 5 most statistically significant regions were evaluated for patterns by cohort study and by time to diagnosis.
Because different cell types might exhibit differing associations between CpG-specific DNA methylation and pancreatic case-control status, an additional analysis was conducted to identify CpGs that might be unique to different cell types in blood (Supplementary Methods, available online).
To identify biological pathways and gene ontologies associated with pancreatic cancer, we used the methylGSA Bioconductor package (15) to perform gene set analyses. Analyses were performed separately on the training and testing sets, and the top genesets (rank-ordered based on P value) were compared between the training and testing sets to look for consistent pathways and/or ontologies.
All P values are based on 2-sided tests, and statistical significance was based on a P value of less than .05.
Results
Population Characteristics
The population characteristics for the cases and controls overall, and by the training and testing sets, are presented in Table 1. Participants in the nested case-control study were diagnosed with pancreatic cancer an average of 13 years (range 6 months to 26 years) after providing a blood sample; 44.8% cases and 45.0% of controls were women (selected from the NHS cohort), and the remaining cases and controls were men from the PHS (18.1% and 20.0%, respectively) and HPFS (37.2% and 35.0%, respectively) cohorts. Baseline characteristics were evenly distributed between the training and testing datasets (Table 1).
Table 1.
Baseline characteristicsa for subjects in 3 prospective cohort studies (NHS, HPFS, PHS) included in the nested case-control analysis
| Baseline characteristics | Total (n = 824) |
Training (n = 577) |
Testing (n = 247) |
|||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |
| Total No. | 393 | 431 | 276 | 301 | 117 | 130 |
| Cohort, No. (%) | ||||||
| NHS | 176 (44.8) | 194 (45.0) | 126 (45.7) | 137 (45.5) | 50 (42.7) | 57 (43.8) |
| HPFS | 146 (37.2) | 151 (35.0) | 103 (37.3) | 105 (34.9) | 43 (36.8) | 46 (35.4) |
| PHS | 71 (18.1) | 86 (20.0) | 47 (17.0) | 59 (19.6) | 24 (20.5) | 27 (20.8) |
| Mean age at blood draw (SD), y | 60.6 (7.9) | 60.2 (7.7) | 61.0 (8.1) | 60.4 (7.8) | 59.8 (7.5) | 59.8 (7.5) |
| Mean time before diagnosis (SD), yb | 13.0 (6.2) | 13.2 (6.2) | 12.4 (6.2) | |||
| Female, No. (%) | 176 (44.8) | 194 (45.0) | 126 (45.7) | 137 (45.5) | 50 (42.7) | 57 (43.8) |
| White, No. (%)c | 346 (94.0) | 406 (94.9) | 244 (93.8) | 284 (95.3) | 102 (94.4) | 122 (93.8) |
| Smoking, No. (%)d | ||||||
| Never | 159 (40.9) | 173 (40.3) | 111 (40.5) | 118 (39.5) | 48 (41.4) | 55 (42.6) |
| Former | 174 (44.7) | 190 (44.3) | 120 (43.8) | 134 (44.8) | 54 (46.6) | 56 (43.4) |
| Current | 57 (14.6) | 65 (15.2) | 43 (15.7) | 47 (15.7) | 14 (12.1) | 18 (14.0) |
| BMI, kg/m2, No. (%)e | 26.0 (4.3) | 25.6 (3.9) | 26.1 (4.3) | 25.5 (3.9) | 25.9 (4.1) | 25.9 (4.0) |
| Underweight and normal | 183 (47.3) | 215 (50.8) | 132 (48.7) | 155 (52.7) | 51 (44.0) | 60 (46.5) |
| Overweight | 144 (37.2) | 151 (35.7) | 91 (33.6) | 100 (34.0) | 53 (45.7) | 51 (39.5) |
| Obese | 60 (15.5) | 57 (13.5) | 48 (17.7) | 39 (13.3) | 12 (10.3) | 18 (14.0) |
| Diabetes, No. (%)f | 19 (4.8) | 11 (2.6) | 14 (5.1) | 10 (3.3) | 5 (4.3) | 1 (0.8) |
Covariates based on questionnaires closest to time of blood draw. BMI = body mass index; HPFS = Health Professionals Follow-up Study; NHS = Nurses’ Health Study; PHS = Physicians’ Health Study.
11 missing values.
28 missing values.
6 missing values.
14 missing values.
1 missing value.
Single CpG EWAS Analysis
The single CpG EWAS analyses performed in the training and testing sets (conducted separately) identified no statistically significant CpGs after a Bonferroni correction. We reduced our number of comparisons by including only CpGs with intraclass correlation coefficient (ICC) greater than 0.5 (over a 1-year period) from our pilot study (n = 199 719 CpGs) (16), but still no CpGs were statistically significant using this restrictive number of tests. Combining the 2 datasets to conduct an overall EWAS did not result in the identification of any single CpG that was statistically significant after multiple comparison correction (top 1000 most statistically significant CpGs prior to adjustment are provided in Supplementary Table 1, available online).
Polymethylomic Risk Score
Using the training dataset, we identified 99 CpGs using our cut points for mean difference and statistical significance using a volcano plot analysis (Supplementary Figure 1, available online). The lasso regression model reduced that number of CpGs to 92. Before developing a polymethylomic risk score, we evaluated whether the mean difference in methylation in these CpGs was consistent in the training and testing datasets. Unfortunately, the mean difference was in different directions for 42 out of 92 CpGs, leading us to conclude that the polymethylomic risk score would not be able to discriminate between cases and controls in the testing set. Using only cases who had given blood within 5 years of diagnosis did not improve our ability to develop a risk prediction score.
DMR Analysis
To explore alternative approaches, we applied a method (DMRcate) designed to identify DMRs by case and control status (14). The DMRcate methodology was developed to identify regions of chromosomes that are differentially methylated based on some phenotype or exposure, rather than focusing on the identification of single CpGs. By focusing on regions and combining information from multiple nearby methylation sites, this approach provides a more powerful statistical tool to identify changes in methylation that are associated with disease outcomes. Statistically significant regions were identified in each of the training and testing sets, but 1 of the regions was statistically significant (and in the same direction) in both datasets; this region, located on chromosome 16 (1583391–1584516; overlapping promoters for IFT140 and TMEM204), consisted of 13 CpGs that differed coordinately between the cases and controls in the 2 datasets (all were P < .05 FDR adjusted) and 3 additional CpGs in the testing dataset (Supplementary Table 2, available online). Of the 16 CpGs identified, only 12 remained statistically significant after combining the 2 datasets to examine overall associations with pancreatic cancer risk (Supplementary Table 3, available online).
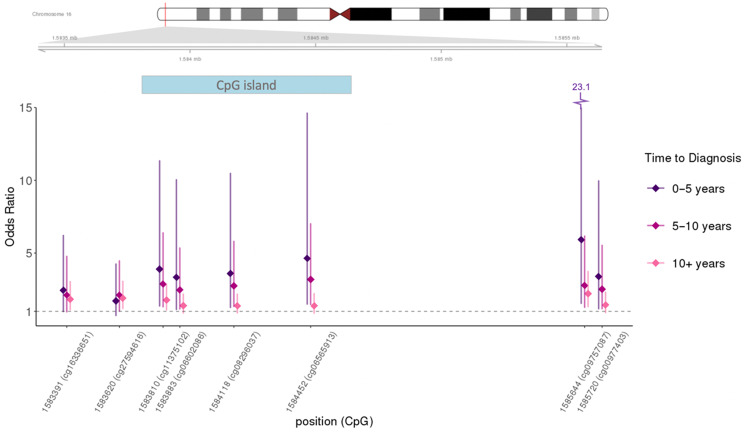
When we examined how these CpGs were associated by time between blood draw and cancer diagnosis, there was a striking pattern of higher risk associated with time more proximal to diagnosis evident in many of the CpGs; results are presented in Table 2 (top 12 statistically significant CpGs are included) and Figure 1 (the top 8 CpGs with visible time trends from Table 2 were included in the figure). The strongest association was noted for cg09757087 (Table 3); a greater than twofold increase in risk was identified for individuals in the highest quartile of methylation level, compared with the lowest quartile, and a close to sixfold increase was observed in subjects who provided blood no more than 5 years prior to cancer diagnosis (OR = 5.95, 95% CI = 1.52 to 23.12). This CpG was also identified in the top 1000 most statistically significant CpGs in the single CpG EWAS (P < .001; Supplementary Table 1, available online). For cg09757087, trends were statistically significant across quartiles in all strata of time to diagnosis and in all but the PHS cohort study (Table 3). Of note, ICCs for the CpGs in this region calculated at 2 time points 1 year apart in a previous pilot study (16) were between 0.88 and 0.98.
Table 2.
Top 12 CpGs in differentially methylated region (DMR) on chromosome 16 identified in training and testing datasets and combined for overall analysis
| CpG | Annotation |
Methylation beta value |
OR (95% CI)b |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Position | Relation to island | Cases | Controls | SD | Difference/SD | Overall | Time to diagnosis |
|||
| ≤5 years | 5–10 years | >10 years | ||||||||
| cg09757087a | 1585644 | S_Shore | 0.718 | 0.698 | 0.089 | 0.220 | 2.31 (1.46 to 3.66) | 5.92 (1.52 to 23.12) | 2.78 (1.25 to 6.19) | 2.21 (1.30 to 3.76) |
| cg11375102 | 1583810 | Island | 0.242 | 0.230 | 0.063 | 0.187 | 2.12 (1.37 to 3.26) | 3.90 (1.34 to 11.36) | 2.87 (1.28 to 6.42) | 1.76 (1.07 to 2.91) |
| cg27594616 | 1583620 | N_Shore | 0.218 | 0.205 | 0.081 | 0.160 | 1.89 (1.25 to 2.88) | 1.71 (0.69 to 4.28) | 2.11 (0.99 to 4.49) | 1.90 (1.17 to 3.09) |
| cg06565913 | 1584452 | Island | 0.368 | 0.352 | 0.086 | 0.184 | 1.86 (1.22 to 2.84) | 4.64 (1.47 to 14.64) | 3.19 (1.45 to 7.04) | 1.37 (0.84 to 2.24) |
| cg00977403a | 1585720 | N_Shore | 0.367 | 0.348 | 0.090 | 0.207 | 1.89 (1.21 to 2.94) | 2.45 (0.96 to 6.24) | 2.12 (0.93 to 4.80) | 1.82 (1.08 to 3.07) |
| cg16336651 | 1583391 | N_Shore | 0.542 | 0.531 | 0.056 | 0.197 | 1.75 (1.16 to 2.64) | 3.60 (1.24 to 10.50) | 2.76 (1.31 to 5.83) | 1.38 (0.86 to 2.21) |
| cg08296037 | 1584118 | Island | 0.393 | 0.372 | 0.095 | 0.215 | 1.75 (1.15 to 2.66) | 2.66 (1.00 to 7.07) | 2.43 (1.10 to 5.37) | 1.55 (0.96 to 2.51) |
| cg06602086 | 1583883 | Island | 0.297 | 0.278 | 0.111 | 0.173 | 1.74 (1.14 to 2.67) | 3.39 (1.15 to 9.99) | 2.53 (1.15 to 5.56) | 1.44 (0.88 to 2.34) |
| cg07341220 | 1583899 | Island | 0.204 | 0.192 | 0.075 | 0.163 | 1.72 (1.13 to 2.62) | 3.33 (1.10 to 10.06) | 2.47 (1.13 to 5.38) | 1.38 (0.86 to 2.21) |
| cg10465839 | 1584050 | Island | 0.259 | 0.246 | 0.079 | 0.165 | 1.70 (1.12 to 2.60) | 2.06 (0.77 to 5.49) | 2.58 (1.18 to 5.62) | 1.45 (0.89 to 2.36) |
| cg02193187 | 1583630 | N_Shore | 0.176 | 0.164 | 0.069 | 0.167 | 1.65 (1.09 to 2.51) | 1.85 (0.75 to 4.59) | 2.28 (1.02 to 5.06) | 1.53 (0.94 to 2.47) |
| cg07639376 | 1584516 | Island | 0.416 | 0.396 | 0.113 | 0.173 | 1.51 (1.00 to 2.28) | 1.85 (0.75 to 4.59) | 2.16 (1.02 to 4.57) | 1.33 (0.83 to 2.15) |
Only identified in testing dataset. CI = confidence interval; OR = odds ratio.
Q4 vs Q1. Model adjusted for age at blood draw, race, date of blood draw, cohort study, smoking status, body mass index, and cell proportions.
Figure 1.
Odds ratio (95% confidence interval) by time to diagnosis for top 8 CpGs in differentially methylated region (DMR) on chromosome 16. Diamonds represent the odds ratio (OR); bars through diamonds represent 95% confidence interval (CI).
Table 3.
Associations between cg09757087 (TMEM204/IFT140) and risk of pancreatic cancer across each quartile, overall, by time to diagnosis, and by cohort study
| Cg09757087 quartiles | Model 1a |
Model 2b |
||
|---|---|---|---|---|
| No. cases/controls | OR (95% CI) | No. cases/controls | OR (95% CI) | |
| Overall (n = 824)c | ||||
| Q1 | 63/108 | 1.00 (referent) | 63/107 | 1.00 (referent) |
| Q2 | 96/107 | 1.60 (1.05 to 2.44) | 93/103 | 1.61 (1.05 to 2.46) |
| Q3 | 104/108 | 1.81 (1.17 to 2.79) | 99/104 | 1.76 (1.13 to 2.74) |
| Q4 | 130/108 | 2.34 (1.49 to 3.68) | 129/107 | 2.31 (1.46 to 3.66) |
| Ptrend | <.001 | <.001 | ||
| ≤5 years to diagnosisc | ||||
| Q1 | 3/108 | 1.00 (referent) | 3/107 | 1.00 (referent) |
| Q2 | 10/107 | 3.35 (0.88 to 12.71) | 10/103 | 3.39 (0.89 to 12.94) |
| Q3 | 20/108 | 7.47 (2.07 to 26.97) | 19/104 | 7.47 (2.04 to 27.30) |
| Q4 | 17/108 | 6.01 (1.55 to 23.24) | 17/107 | 5.92 (1.52 to 23.12) |
| Ptrend | .004 | .006 | ||
| 5–10 years to diagnosisc | ||||
| Q1 | 15/108 | 1.00 (referent) | 15/107 | 1.00 (referent) |
| Q2 | 20/107 | 1.58 (0.74 to 3.38) | 20/103 | 1.54 (0.71 to 3.31) |
| Q3 | 14/108 | 1.07 (0.47 to 2.45) | 13/104 | 0.95 (0.40 to 2.22) |
| Q4 | 34/108 | 2.96 (1.35 to 6.48) | 33/107 | 2.78 (1.25 to 6.19) |
| Ptrend | .01 | .03 | ||
| >10 years to diagnosisc | ||||
| Q1 | 41/108 | 1.00 (referent) | 41/107 | 1.00 (referent) |
| Q2 | 62/107 | 1.60 (0.98 to 2.60) | 59/103 | 1.60 (0.97 to 2.62) |
| Q3 | 69/108 | 1.93 (1.17 to 3.18) | 66/104 | 1.91 (1.15 to 3.19) |
| Q4 | 77/108 | 2.16 (1.28 to 3.65) | 77/107 | 2.21 (1.30 to 3.76) |
| Ptrend | .004 | .004 | ||
| NHS (n = 370) d | ||||
| Q1 | 27/49 | 1.00 (referent) | 27/49 | 1.00 (referent) |
| Q2 | 47/48 | 1.90 (1.01 to 3.60) | 47/48 | 1.94 (1.02 to 3.68) |
| Q3 | 35/48 | 1.41 (0.72 to 2.77) | 33/47 | 1.37 (0.69 to 2.72) |
| Q4 | 67/49 | 2.44 (1.22 to 4.90) | 67/49 | 2.49 (1.24 to 5.00) |
| Ptrend | .04 | .04 | ||
| HPFS (n = 297)d | ||||
| Q1 | 21/38 | 1.00 (referent) | 20/37 | 1.00 (referent) |
| Q2 | 39/37 | 2.02 (0.99 to 4.14) | 38/34 | 2.23 (1.06 to 4.66) |
| Q3 | 44/38 | 2.41 (1.16 to 5.00) | 40/34 | 2.63 (1.22 to 5.65) |
| Q4 | 42/38 | 2.43 (1.13 to 5.23) | 41/37 | 2.67 (1.20 to 5.95) |
| Ptrend | .03 | .02 | ||
| PHS (n = 157)d | ||||
| Q1 | 16/22 | 1.00 (referent) | 16/22 | 1.00 (referent) |
| Q2 | 13/21 | 0.98 (0.36 to 2.70) | 13/21 | 0.90 (0.32 to 2.53) |
| Q3 | 25/21 | 1.71 (0.68 to 4.33) | 25/21 | 1.51 (0.59 to 3.89) |
| Q4 | 17/22 | 1.47 (0.49 to 4.40) | 17/22 | 1.36 (0.44 to 4.16) |
| Ptrend | .28 | .38 | ||
Model 1: Adjusted for age at blood draw, race, date of blood draw, cohort study, and cell proportions. CI = confidence interval; HPFS = Health Professionals Follow-up Study; NHS = Nurses’ Health Study; OR = odds ratio; PHS = Physicians’ Health Study.
Model 2: Additional adjusted for smoking status and body mass index.
Cut offs (overall): Q1: ≤0.624; Q2: 0.625–0.703; Q3: 0.704–0.764; Q4: ≥0.765.
Cut offs (study specific): Q1: ≤0.624 for NHS, ≤0.625 for HPFS and ≤0.622 for PHS; Q2: 0.625–0.703 for NHS, 0.626–0.703 for HPFS and 0.626–0.701 for PHS; Q3: 0.704–0.764 for NHS, 0.704–0.765 for HPFS and 0.705–0.764 for PHS; Q4: ≥0.765 for NHS, ≥0.766 for HPFS and ≥0.766 for PHS.
To examine whether we missed important regions by splitting our dataset into 2 independent sets (reducing overall power), we combined the dataset to conduct an overall DMRcate analysis. We identified 3 regions that were more statistically significant (overall) than the chromosome 16 region (TMEM204/IFT140—ranking fourth overall) and several more regions that were also statistically significant (Supplementary Table 4, available online). The top 3 regions were located in genes MFSD6L, FAM134B, and KCNQ1DN. For each of these regions, as well as for the fifth strongest region, we selected the CpGs with the strongest associations with pancreatic cancer risk for comparison by cohort and time to diagnosis (Table 4). For each of these CpGs, direction of associations was robust across each of the cohorts and was statistically significant for almost all CpGs. However, in contrast to the TMEM204/IFT140 region, associations with pancreatic cancer in these regions were similar by time to diagnosis.
Table 4.
Associations between the strongest CpGsa in each of the top regions identified using DMRcate on combined datasets (excluding region 4 TMEM204/IFT140 presented in Tables 2 and 3) and risk of pancreatic cancer across each quartile, overall, by time to diagnosis, and by cohort study
| Quartiles for CpGs | Region 1 (MFSDL6) |
Region 2 (FAM134b) |
Region 3 (KCNQ1DN) |
Region 5 (C6orf227) |
|---|---|---|---|---|
| cg24203800 | cg04851848 | cg04457979 | cg05602975 | |
| OR (95% CI)b | OR (95% CI)b | OR (95% CI)b | OR (95% CI)b | |
| Overall (n = 824) | ||||
| Q1 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Q2 | 0.99 (0.67 to 1.45) | 1.32 (0.86 to 2.04) | 1.64 (1.06 to 2.52) | 1.49 (0.99 to 2.26) |
| Q3 | 0.87 (0.59 to 1.28) | 1.73 (1.13 to 2.67) | 2.15 (1.40 to 3.29) | 1.23 (0.81 to 1.88) |
| Q4 | 0.41 (0.26 to 0.63) | 2.25 (1.43 to 3.55) | 2.16 (1.40 to 3.33) | 1.95 (1.30 to 2.93) |
| Ptrend | <.001 | <.001 | <.001 | .004 |
| ≤5 years to diagnosis | ||||
| Q1 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Q2 | 0.91 (0.43 to 1.95) | 1.08 (0.42 to 2.78) | 1.22 (0.48 to 3.11) | 1.83 (0.79 to 4.22) |
| Q3 | 0.40 (0.16 to 1.02) | 0.96 (0.35 to 2.59) | 2.17 (0.89 to 5.30) | 0.93 (0.37 to 2.39) |
| Q4 | 0.42 (0.16 to 1.06) | 1.39 (0.52 to 3.70) | 1.38 (0.54 to 3.57) | 1.04 (0.41 to 2.68) |
| Ptrend | .02 | .55 | .30 | .69 |
| 5–10 years to diagnosis | ||||
| Q1 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Q2 | 0.76 (0.38 to 1.54) | 1.82 (0.76 to 4.34) | 1.69 (0.69 to 4.16) | 1.24 (0.55 to 2.79) |
| Q3 | 1.06 (0.55 to 2.03) | 2.83 (1.20 to 6.69) | 2.86 (1.23 to 6.66) | 1.00 (0.45 to 2.21) |
| Q4 | 0.47 (0.22 to 1.02) | 2.74 (1.11 to 6.77) | 3.41 (1.46 to 7.96) | 2.28 (1.10 to 4.72) |
| Ptrend | .15 | .02 | .002 | .03 |
| >10 years to diagnosis | ||||
| Q1 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Q2 | 1.08 (0.70 to 1.67) | 1.32 (0.80 to 2.18) | 1.70 (1.04 to 2.79) | 1.59 (0.98 to 2.60) |
| Q3 | 0.83 (0.53 to 1.30) | 1.71 (1.04 to 2.81) | 1.94 (1.18 to 3.17) | 1.48 (0.90 to 2.44) |
| Q4 | 0.37 (0.22 to 0.62) | 2.36 (1.41 to 3.97) | 2.00 (1.21 to 3.29) | 2.26 (1.41 to 3.63) |
| Ptrend | <.001 | <.001 | .009 | .002 |
| NHS (n = 370) | ||||
| Q1 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Q2 | 0.96 (0.54 to 1.73) | 1.77 (0.91 to 3.43) | 1.90 (0.98 to 3.68) | 1.10 (0.59 to 2.06) |
| Q3 | 1.00 (0.56 to 1.82) | 2.10 (1.07 to 4.10) | 2.23 (1.15 to 4.33) | 1.30 (0.69 to 2.43) |
| Q4 | 0.47 (0.24 to 0.91) | 2.73 (1.38 to 5.41) | 2.76 (1.43 to 5.33) | 1.91 (1.05 to 3.50) |
| Ptrend | .05 | .005 | .003 | .03 |
| HPFS (n = 297) | ||||
| Q1 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Q2 | 0.55 (0.28 to 1.08) | 1.16 (0.57 to 2.36) | 1.97 (0.92 to 4.22) | 1.83 (0.91 to 3.66) |
| Q3 | 0.77 (0.40 to 1.46) | 1.09 (0.52 to 2.30) | 2.59 (1.22 to 5.47) | 0.86 (0.41 to 1.79) |
| Q4 | 0.30 (0.14 to 0.63) | 2.11 (0.98 to 4.55) | 2.82 (1.31 to 6.06) | 1.73 (0.88 to 3.40) |
| Ptrend | .008 | .07 | .009 | .36 |
| PHS (n = 157) | ||||
| Q1 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Q2 | 1.29 (0.53 to 3.13) | 1.16 (0.35 to 3.82) | 0.61 (0.21 to 1.77) | 2.66 (0.90 to 7.83) |
| Q3 | 0.68 (0.26 to 1.77) | 3.09 (1.08 to 8.79) | 1.79 (0.69 to 4.64) | 2.26 (0.77 to 6.57) |
| Q4 | 0.26 (0.08 to 0.81) | 3.46 (1.12 to 10.69) | 0.66 (0.24 to 1.82) | 2.96 (1.02 to 8.60) |
| Ptrend | .01 | .007 | .98 | .10 |
Based on P values and consistency across cohort studies. CI = confidence interval; HPFS = Health Professionals Follow-up Study; NHS = Nurses’ Health Study; OR = odds ratio; PHS = Physicians’ Health Study.
Adjusted for age at blood draw, race, date of blood draw, cohort study, smoking status, body mass index, and cell proportion.
We conducted stratified analyses to examine whether our results were modified by BMI or smoking status. Associations for some of the CpGs were stronger in overweight and obese subjects in regions 3 and 4 and among current smokers in regions 1, 3, and 4 (Supplementary Table 5, available online). Results were similar among whites only and those who did not report having diabetes at baseline (data not shown).
Cell-Specific Analyses of CpGs in Top 5 DMRs
We conducted additional analyses to identify CpGs within the identified DMRs that might be unique to different immune cell types in blood (17). Our results suggest that the methylation differences between cases and controls varied by cell type for regions 4 and 5; we observed stronger associations for most of the CpGs in regions 4 and 5 in CD4 T cells, CD8 T cells, and natural killer cells than in neutrophils or monocytes (Table 5; not all CpGs are shown). For CpGs in regions 1, 2, and 3, the differences by cell types were not as marked (Table 5).
Table 5.
Top CpGs (3 per region based on P values) for each immune cell type in the top 5 differentially methylated regions (DMR)a
| Region and CpG | CD4T |
CD8T |
NK |
Neutrophil |
Monocytes |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI)b | P | OR (95% CI)b | P | OR (95% CI)b | P | OR (95% CI)b | P | OR (95% CI)b | P | |
| 1 | ||||||||||
| cg04259560 | 0.39 (0.20 to 0.74) | .004 | 0.44 (0.25 to 0.76) | .003 | 0.30 (0.17 to 0.53) | <.001 | 0.49 (0.32 to 0.75) | .001 | 0.42 (0.26 to 0.70) | <.001 |
| cg24203800 | 0.47 (0.27 to 0.81) | .006 | 0.51 (0.31 to 0.84) | .008 | 0.43 (0.27 to 0.70) | <.001 | 0.47 (0.31 to 0.73) | <.001 | 0.37 (0.23 to 0.60) | <.001 |
| cg11685316 | 0.33 (0.15 to 0.73) | .006 | 0.46 (0.16 to 1.30) | .143 | 0.29 (0.13 to 0.63) | .002 | 0.63 (0.42 to 0.94) | .02 | 0.52 (0.34 to 0.79) | .002 |
| 2 | ||||||||||
| cg04851848 | 2.77 (1.52 to 5.04) | <.001 | 2.78 (1.44 to 5.37) | .002 | 2.08 (0.77 to 5.64) | .15 | 2.04 (1.32 to 3.14) | .001 | 2.74 (1.52 to 4.93) | <.001 |
| cg22728178 | 1.72 (1.12 to 2.64) | .01 | 2.03 (1.28 to 3.20) | .002 | 2.06 (0.60 to 7.11) | .25 | 1.72 (1.13 to 2.60) | .01 | 2.06 (1.22 to 3.48) | .007 |
| cg20376277 | 1.53 (1.00 to 2.34) | .05 | 1.96 (1.03 to 3.73) | .039 | 2.07 (0.85 to 5.06) | .11 | 1.94 (1.27 to 2.97) | .002 | 1.82 (1.16 to 2.86) | .009 |
| 3 | ||||||||||
| cg14582642 | 1.70 (1.08 to 2.68) | .02 | 1.98 (0.99 to 3.95) | .05 | 2.64 (0.86 to 8.14) | .09 | 1.41 (0.94 to 2.11) | .09 | 1.77 (1.13 to 2.78) | .01 |
| cg05290058 | 2.19 (0.99 to 4.84) | .05 | 2.14 (1.13 to 4.03) | .02 | 2.06 (0.80 to 5.33) | .14 | 1.70 (1.12 to 2.60) | .01 | 1.66 (1.10 to 2.50) | .02 |
| cg17239974 | 4.83 (1.51 to 15.5) | .008 | 1.88 (0.94 to 3.75) | .07 | 1.81 (0.78 to 4.18) | .17 | 1.17 (0.77 to 1.77) | .47 | 1.56 (0.84 to 2.91) | .16 |
| 4 | ||||||||||
| cg11375102 | 1.94 (1.07 to 3.52) | .03 | 3.00 (1.15 to 7.80) | .02 | 1.56 (0.35 to 6.84) | .56 | 1.44 (0.94 to 2.19) | .09 | 1.54 (0.94 to 2.50) | .08 |
| cg00463982 | 2.22 (1.11 to 4.34) | .02 | 2.06 (0.97 to 4.36) | .06 | 1.56 (0.95 to 2.58) | .08 | 1.47 (0.97 to 2.24) | .07 | 1.19 (0.71. 2.01) | .51 |
| cg09757087 | 2.72 (0.78 to 9.41) | .12 | 1.24 (0.38 to 4.00) | .72 | 1.84 (1.17 to 2.88) | .008 | 1.95 (1.28 to 2.97) | .002 | 2.42 (1.47 to 3.97) | <.001 |
| 5 | ||||||||||
| cg08301503 | 2.53 (1.40 to 4.57) | .002 | 3.00 (1.15 to 7.80) | .02 | 3.41 (1.44 to 8.07) | .005 | 1.69 (1.09 to 2.60) | .02 | 1.34 (0.81 to 2.24) | .26 |
| cg06289138 | 2.21 (1.17 to 4.16) | .03 | 3.17 (1.09 to 9.18) | .03 | 1.96 (1.18 to 3.26) | .01 | 1.26 (0.85 to 1.88) | .27 | 1.39 (0.81 to 2.40) | .24 |
| cg00536532 | 1.89 (0.85 to 4.21) | .12 | 2.06 (0.97 to 4.36) | .06 | 2.07 (1.02 to 4.21) | .04 | 1.27 (0.85 to 1.88) | .25 | 1.68 (1.09 to 2.59) | .02 |
Results for B cells were similar (not shown). CI = confidence interval; NK = natural killer; OR = odds ratio.
Adjusted for age at blood draw, race, date of blood draw, cohort study, smoking status, and body mass index.
GSEA Analysis
Using a gene set enrichment analysis (GSEA) (15), we identified 225 pathways that overlapped in the 2 datasets (training and testing). After multiple comparisons adjustments, 4 pathways were borderline statistically significant in the training dataset (P = .056), and 14 pathways were statistically significant in the testing dataset (P < .05) (Supplementary Table 6, available online). The 2 overlapping pathways between the top findings in 2 datasets were gastric acid secretion and melanogenesis. The gastric acid secretion pathway includes 72 genes; the normalized enrichment score was 1.258 (testing) and 1.127 (training). The melanogenesis pathway includes 100 genes; the normalized enrichment score was 1.175 (testing) and 1.100 (training).
Using cell-specific analyses, we examined pathways using GSEA in all cases and controls; none of the cell-specific pathways were statistically significant after correction for multiple comparisons, with the exception of neutrophils (Supplementary Table 7, available online). For neutrophils, 5 pathways were statistically significant (P < .05) after a Bonferroni adjustment; type I diabetes, acute myeloid leukemia, and endometrial cancer pathways were among those. Of note, pancreatic cancer pathways were identified in natural killer cells (P = .007 prior to multiple comparison correction).
Discussion
In this large study, pooling pancreatic cancer cases from 3 cohort studies, we report associations between several new genomic regions and pancreatic cancer risk; genes in these regions included TMEM204/IFT140, MFSD6L, FAM134b/RETREG1, KCNQ1D, and C6orf227. Associations between DNA methylation and pancreatic cancer risk in CpGs located in the TMEM204/IFT140 region were stronger in lymphocyte subtypes, specifically CD4 and CD8T cells, than in myeloid cells, suggesting that this could result from shifts in the percentages of smaller activated subtypes of lymphocytes. We also identified 2 pathways where methylation levels were related to risk of pancreatic cancer, namely, gastric acid secretion and melanogenesis.
For the TMEM204/IFT140 region, we observed, on average, higher DNA methylation levels in subjects who had blood drawn closer to time of cancer diagnosis, suggesting that methylation, as assessed in the whole blood array platform, increases as disease progresses. In contrast, this was not observed for the other top regions identified, possibly because methylation levels in those regions are more stable over time and less influenced by factors that impact progression or reflect disease onset. These results may have different implications regarding the role of the genes on etiology vs disease onset.
Several of the probes in the DMR region on chromosome 16 (TMEM204/IFT140 region) that we identified have been previously identified as biologically significant in carcinogenesis, including in pancreatic cancer. In The Cancer Genome Atlas data, DNA methylation levels of probe cg08296037 were strongly inversely correlated with expression of TMEM204 in peripheral blood of acute myeloid leukemia patients [r = −0.79; P = 2.3 × 10–39 (18)]. TMEM204 is expressed in all cancers with low specificity, but mean levels are higher in pancreatic cancer tissue than any other cancer tissue (RNAseq The Cancer Genome Atlas data); low TMEM204 expression has been associated with poor liver cancer outcome but improved survival in melanoma (19).
The DMRcate analysis conducted on all subjects combined identified potential regions of interest that will need to be replicated in other populations. Two of the top regions identified in this analysis included genes that have been linked to cancer in other studies. One region includes gene FAM134B, a tumor suppressor gene previously associated with colorectal and esophageal squamous cell cancers (20). Promoter hypermethylation of FAM134B (in tumor tissue) has been associated with poor prognosis in colorectal cancer (21). Another region includes long noncoding RNA KCNQ1DN, which has been associated with Wilms tumors (22) and gastric cancer (23). DNA methylation levels in these regions did not change with shorter time to diagnosis, suggesting they may represent genes that impact susceptibility to cancer risk.
The findings from the GSEA analysis suggest that methylation changes in the gastric secretion pathway may play a role in pancreatic cancer. Ulcers have been linked to pancreatic cancer in several studies (24–26), especially for gastric ulcers, and although these associations increase as the time to diagnosis is shorter, they are present up to 10 years prior to cancer diagnosis (25). It has also been suggested that stomach acid secretion may alter bacterial landscape in the stomach and influence nitrosamine levels that, jointly, may contribute to pancreatic carcinogenesis (27). Identification of changes in DNA methylation in the gastric acid secretion pathways may provide new insights into biological mechanisms; other studies will have to confirm and further examine these findings.
Positive results from this study should be interpreted with caution given that some findings may have been due to chance; however, we tried to minimize chance findings by splitting our data into a training set and a testing set and emphasizing the results that were consistent in both sets. On the other hand, differences in methylation levels between cases and controls in single CpGs might have been missed because of the large number of corrections for multiple comparisons (>800 000). Our multivariate models were adjusted for age, sex, race, smoking, and BMI to control for potential confounding, but we did not adjust for medical conditions that are known to increase the risk of pancreatic cancer, such as chronic pancreatitis, because we did not have those data available in our dataset. Finally, because we had a limited number of non-Caucasian participants in this study, our findings may not be generalizable to other populations.
To our knowledge, this is the first study to conduct an EWAS on pancreatic cancer risk using peripheral blood collected prior to cancer diagnosis. We identified several regions that were differentially methylated in participants who later developed pancreatic cancer. We also observed that genes involved in gastric secretion were differentially methylated in cases compared with controls. Findings from this study suggest that changes in methylation levels that occur more than 10 years prior to cancer diagnosis can influence risk and may be markers of altered pathways involved in carcinogenesis. For some CpGs in the region including genes TMEM204/IFT140, methylation differences increased in magnitude closer to the time of diagnosis, perhaps reflecting changes in metabolic systems, a pattern similar to those observed between diabetes and pancreatic cancer. Although differences in methylation levels are modest, it is important to remember the progress made and insights gained from genome-wide association studies, even with small effects sizes (28, 29). Additional studies will unquestionably provide critical insights into biological pathways that could lead to major breakthroughs in clinical settings.
Funding
The research reported in this publication was primarily supported by the National Institutes of Health (NIH)/National Cancer Institute grant R01 CA207110. In addition, other NIH funds contributed to the support of the investigators: P30 CA168525 and the Kansas IDeA Network of Biomedical Research Excellence Bioinformatics Core, supported in part by the National Institute of General Medical Science (NIGMS) Award P20GM103418.
Notes
Role of the funder: The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclosures: The authors have no conflicts of interests.
Author Contributions: Conception and design: D.S. Michaud, K.T. Kelsey. Development of methodology: D.S. Michaud, K.T. Kelsey. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): D.S. Michaud, K.T. Kelsey. Analysis and interpretation of data (eg, statistical analysis, biostatistics, computational analysis): D.S. Michaud, M. Ruan, D.C. Koestler, D. Pei, C.J. Marsit, I. De Vivo, K.T. Kelsey. Writing, review, and/or revision of the manuscript: D.S. Michaud, M. Ruan, D.C. Koestler, D. Pei, C.J. Marsit, I. De Vivo, K.T. Kelsey. Administrative, technical, or material support (ie, reporting or organizing data, constructing databases): D.S. Michaud, M. Ruan. Study supervision: D.S. Michaud, K.T. Kelsey.
Data Availability Statement: All data from this study have been deposited in dbGAP and will be available on January 3, 2020 [“DNA Methylation Markers and Pancreatic Cancer Risk in 3 Cohort Studies (NHS, PHS, HPFS)” phs001917.v1.p1]. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001917.v1.p1
Supplementary Material
References
- 1. Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. [DOI] [PubMed] [Google Scholar]
- 2. Owens DK, Davidson KW, Krist AH, et al. Screening for pancreatic cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. JAMA. 2019;322(5):438–444. [DOI] [PubMed] [Google Scholar]
- 3. Eissa MAL, Lerner L, Abdelfatah E, et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin Epigenetics. 2019;11(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liggett T, Melnikov A, Yi QL, et al. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer. 2010;116(7):1674–1680. [DOI] [PubMed] [Google Scholar]
- 5. Barth SD, Schulze JJ, Kuhn T, et al. Treg-mediated immune tolerance and the risk of solid cancers: findings from EPIC-Heidelberg. J Natl Cancer Inst. 2015;107(11):djv224. [DOI] [PubMed] [Google Scholar]
- 6. Bartlett AH, Liang JW, Sandoval-Sierra JV, et al. Longitudinal study of leukocyte DNA methylation and biomarkers for cancer risk in older adults. Biomark Res. 2019;7(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grieshober L, Graw S, Barnett MJ, et al. Methylation-derived neutrophil-to-lymphocyte ratio and lung cancer risk in heavy smokers. Cancer Prev Res. 2018;11(11):727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pedersen KS, Bamlet WR, Oberg AL, et al. Leukocyte DNA methylation signature differentiates pancreatic cancer patients from healthy controls. PLOS One. 2011;6(3):e18223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dauksa A, Gulbinas A, Barauskas G, et al. Whole blood DNA aberrant methylation in pancreatic adenocarcinoma shows association with the course of the disease: a pilot study. PLOS One. 2012;7(5):e37509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neale RE, Clark PJ, Fawcett J, et al. Association between hypermethylation of DNA repetitive elements in white blood cell DNA and pancreatic cancer. Cancer Epidemiol. 2014;38(5):576–582. [DOI] [PubMed] [Google Scholar]
- 11. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salas LA, Koestler DC, Butler RA, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adalsteinsson BT, Gudnason H, Aspelund T, et al. Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PLOS One. 2012;7(10):e46705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peters TJ, Buckley MJ, Statham AL, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ren X, Kuan PF. methylGSA: a Bioconductor package and Shiny app for DNA methylation data length bias adjustment in gene set testing. Bioinformatics. 2019;35(11):1958–1959. [DOI] [PubMed] [Google Scholar]
- 16. Zaimi I, Pei D, Koestler DC, et al. Variation in DNA methylation of human blood over a 1-year period using the Illumina MethylationEPIC array. Epigenetics. 2018;13(10-11):1056–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rahmani E, Schweiger R, Rhead B, et al. Cell-type-specific resolution epigenetics without the need for cell sorting or single-cell biology. Nat Commun. 2019;10(1):3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng M, Brägelmann J, Kryukov I, Saraiva-Agostinho N, Perner S. FirebrowseR: an R client to the Broad Institute's Firehose Pipeline. Database (Oxford). 2017;2017:baw160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):660. 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 20. Islam F, Gopalan V, Lam AK. RETREG1 (FAM134B): a new player in human diseases: 15 years after the discovery in cancer. J Cell Physiol. 2018;233(6):4479–4489. [DOI] [PubMed] [Google Scholar]
- 21. Islam F, Gopalan V, Pillai S, et al. Promoter hypermethylation inactivate tumor suppressor FAM134B and is associated with poor prognosis in colorectal cancer. Genes Chromosomes Cancer. 2018;57(5):240–251. [DOI] [PubMed] [Google Scholar]
- 22. Xin Z, Soejima H, Higashimoto K, et al. A novel imprinted gene, KCNQ1DN, within the WT2 critical region of human chromosome 11p15.5 and its reduced expression in Wilms’ tumors. J Biochem. 2000;128(5):847–853. [DOI] [PubMed] [Google Scholar]
- 23. Hu Z, Yang D, Tang Y, et al. Five-long non-coding RNA risk score system for the effective prediction of gastric cancer patient survival. Oncol Lett. 2019;17(5):4474–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo J, Nordenvall C, Nyren O, et al. The risk of pancreatic cancer in patients with gastric or duodenal ulcer disease. Int J Cancer. 2007;120(2):368–372. [DOI] [PubMed] [Google Scholar]
- 25. Bao Y, Spiegelman D, Li R, et al. History of peptic ulcer disease and pancreatic cancer risk in men. Gastroenterology. 2010;138(2):541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bosetti C, Lucenteforte E, Bracci PM, et al. Ulcer, gastric surgery and pancreatic cancer risk: an analysis from the International Pancreatic Cancer Case-Control Consortium (PanC4). Ann Oncol. 2013;24(11):2903–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Risch HA. Pancreatic cancer: helicobacter pylori colonization, N-nitrosamine exposures, and ABO blood group. Mol Carcinog. 2012;51(1):109–118. [DOI] [PubMed] [Google Scholar]
- 28. Visscher PM, Wray NR, Zhang Q, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101(1):5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–1701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.