Abstract
Significance: S-Persulfidation generates persulfide adducts (RSSH) on both small molecules and proteins. This process is believed to be critical in the regulation of biological functions of reactive sulfur species such as H2S, as well as in signal transduction. S-Persulfidation also plays regulatory roles in human health and diseases.
Recent Advances: Some mechanisms underlying the generation of low-molecular-weight persulfides and protein S-persulfidation in living organisms have been uncovered. Some methods for the specific delivery of persulfides and the detection of persulfides in biological systems have been developed. These advances help to pave the road to better understand the functions of S-persulfidation.
Critical Issues: Persulfides are highly reactive and unstable. Currently, their identification relies on trapping them by S-alkylation, but this is not always reliable due to rapid sulfur exchange reactions. Therefore, the presence, identity, and fates of persulfides in biological environments are sometimes difficult to track.
Future Directions: Further understanding the fundamental chemistry/biochemistry of persulfides and development of more reliable detection methods are needed. S-Persulfidation in specific protein targets is essential in organismal physiological health and human disease states. Besides cardiovascular and neuronal systems, the roles of persulfidation in other systems need to be further explored. Contradictory results of persulfidation in biology, especially in cancer, need to be clarified.
Keywords: S-persulfidation, persulfides, reactive sulfur species, chemical reaction, chemical probes, detection, diseases
Introduction
S-Persulfidation (also named as S-perthiolation or S-sulfhydration) leads to the formation of persulfides (RSSH) from thiols (RSH), in which R can be both small molecules and proteins. Persulfides belong to the family of reactive sulfur species, which play regulatory roles in biology. In mammals, the amounts of low molecular weight (LMW) persulfides and S-persulfidated proteins are unexpectedly high. For instance, up to 100 μM of LMW persulfides (such as glutathione persulfide [GSSH]) were found in the animal tissues (45). Ten percent to 25% of the proteins in mouse liver lysate, including actin, tubulin, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), are S-persulfidated under physiological conditions. Protein S-persulfidation is a recently defined oxidative post-translational modification that leads to the alteration of protein structures and functions (32, 48, 128). This concept was formulized along with the identification of H2S as a NO-like signaling molecule. For example, S-persulfidation augments GAPDH activity and enhances actin polymerization (80). Additionally, S-persulfidated proteins are rich in pancreatic MIN6 cells, especially under the condition of endoplasmic reticulum (ER) stress (36).
Given the importance of S-persulfidation in biology, it is crucial to understand some fundamental knowledge of this process, as well as difficulties associated with persulfidation research. This review summaries several key aspects about S-persulfidation, which include (i) pathways accounting for endogenous persulfidation; (ii) in vitro methods for protein S-persulfidation; (iii) fundamental chemistry and reactions of persulfides that are important for understanding biological functions of persulfidation; (iv) reagents that can specifically release persulfides; (v) methods for the detection/quantification of persulfidation; and (vi) the links between persulfidation and either organismal physiological health, such as vasorelaxation, learning and memory, or various human disorder states, such as cardiovascular diseases, neurodegenerative diseases, metabolic/immune disorders, and cancer. The readers are also encouraged to read some excellent reviews on this topic published recently (32, 65, 128).
Endogenous S-Persulfidation
S-Persulfidation is believed to be involved in H2S-based signal transduction. From the chemistry perspective, H2S cannot directly react with protein cysteine residues (-SH) to form persulfides. However, H2S could react with the oxidative forms of cysteine, such as disulfides (-S-S-), sulfenic acid (-S-OH), nitrosothiol (-SNO), to form persulfides. It should be noted that in aqueous solutions, the major form of H2S is its anion (HS−). The reactions of H2S are mainly performed through HS−. The reaction between H2S and LMW disulfides (such as cystine and GSSG) is a slow and reversible process, usually yielding a mixture of products (35). Such a reaction could be significantly accelerated in protein cases. For instance, the reaction between H2S and active disulfide of sulfide quinone oxidoreductase (SQR) protein is increased by ∼106-fold with respect to free cystine (78). It should be noted that in the cytosol, the concentrations of LMW and protein disulfides are quite low and the formed persulfides should be very limited. Hence, this reaction may be more relevant in ER and under oxidizing conditions (32).
Intracellular persulfide levels were found to increase by the treatment with H2O2, while to decrease by inhibiting cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE) (24). The phenomenon could be explained by two sequential reactions, that is, (H2O2 + P-SH → P-SOH + H2O) and (H2S + P-SOH → P-SSH + H2O). Importantly, this process can potentially protect against H2O2-mediated oxidative injury in cells. It is known that P-SOH can be further oxidized to sulfinic acids (P-SO2H) and sulfonic acids (P-SO3H) (117, 118), which are irreversible oxidized products of the original protein cysteine adducts. However, S-persulfidated proteins (P-SSH) would produce P-SSO2H and P-SSO3H under similar oxidative challenges. Both P-SSO2H and P-SSO3H can eventually be reduced back to P-SH and recover their normal function in the cells.
S-Nitrosation (RSNO) is another oxidative post-translational modification of protein cysteine residues (15, 69, 94). Similar to S-persulfidation, S-nitrosation is also involved in the regulation of protein functions, although sometimes functions of these two modifications are different or even opposite (113). In one study, 36% overlap of S-persulfidation and S-nitrosation proteomes was noted (36). In theory, RSNO can react with H2S to form persulfides and HNO. However, this reaction was found to be thermodynamically unfavored (+26 kJ/mol) (63). Nevertheless, this possibility should not be completely ruled out as it may be facilitated by protein microenvironments (32).
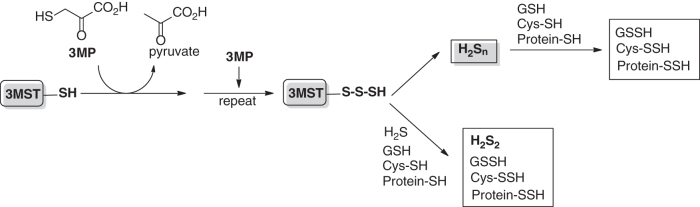
Polysulfide species such as H2Sn or RSnSH have been suggested by some researchers to be important regulating molecules in sulfur signaling while some other researchers disagree with this assessment (32). Polysulfides belong to the sulfane sulfur family and can transfer sulfur atom to cysteine residues to cause protein S-persulfidation. Such a process has been demonstrated with proteins including transient receptor potential (TRP) A1 and phosphatase and tensin homolog (PTEN) (37, 58). A series of studies by Kimura et al. suggested that endogenous H2Sn are produced mainly from 3-mercaptopyruvate by 3-mercaptopyruvate sulfurtransferase (3MST) (59). They also found that H2Sn could be generated from H2S by 3MST and rhodanese, and 3MST also produced cysteine- and GSH-persulfides (CysSSH and GSSH) (57). These reactions are described in Scheme 1, with persulfidated 3MST (3MST-SSnH) as the proposed key intermediate.
Scheme 1
Besides 3MST, other sulfur transfer enzymes may also be responsible for the formation of endogenous persulfide species (both LMW and protein substrates). These include rhodanese, SQR, CSE, CBS, cysteine desulfurase, and so on. Ida et al. found high levels (up to 100 μM) of LMW persulfides (CysSSH/GSSH) exist in cells and tissues, and these molecules were suggested to be the main biologically relevant persulfidation reagents (45, 88). A very recent work by Akaike et al. found that prokaryotic and mammalian cysteinyl-tRNA synthetases (CARSs) could effectively catalyze the production of cysteine persulfide (CysSSH) and polysulfides (CysSnSH) from cysteine (1). CARS was also found to integrate cysteine polysulfides into proteins during translational process. These LMW or protein-bound polysulfides can act as the precursors of H2Sn, and they may work collectively to induce endogenous protein S-persulfidation.
A recent work by Vitvitsky et al. found that H2S could reduce cytochrome c (Cyt C) via electron transfer process (114). This Cyt C-dependent H2S oxidation leads to HS radical formation. The resultant reactive sulfur species can further react with intracellular protein thiols to form persulfides, therefore enhancing persulfidation. This process is speculated to be particularly important in ischemia/reperfusion (I/R)-related injury as it protects proteins from reactive oxygen species (ROS)-induced hyperoxidation during the reperfusion phase.
In Vitro Methods for Protein S-Persulfidation
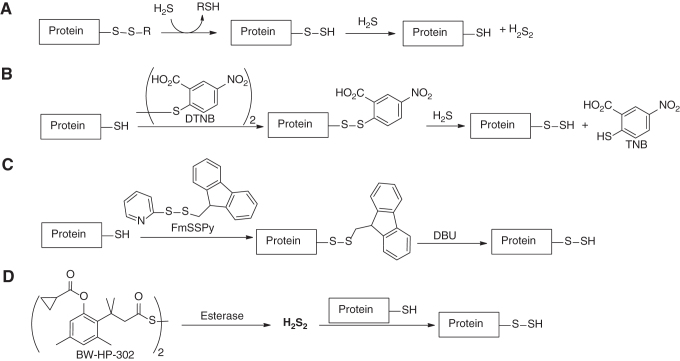
Protein persulfides are a class of important biological persulfides. While they are endogenously generated, relatively pure forms of protein persulfides are sometime needed for understanding their functions. To this end, several methods for the preparation of protein persulfides have been developed (Scheme 2):
Scheme 2
(i) Using papain as the model protein, Fukuto and colleagues showed that protein disulfides (P-SSR) could react with H2S to form persulfides (P-SSH) (35). Interestingly, excess H2S could further react protein persulfides to resume the original protein (P-SH), presumably with hydrogen persulfide (H2S2) as the byproduct.
(ii) 5,5′-Dithiobis-(2-nitrobenzoic acid) (DTNB, also known as Ellman's reagent) was used to react with protein-SH to form mixed disulfides of the protein, which could then react with H2S to form protein persulfides. In the latter step, a highly colored species, thionitrobenzoate (λmax = 412 nm), was formed and could be easily detected. As such, it provided a way to quantify protein persulfides. This method was validated on papain (35) and glutathione peroxidase (GPx) 3 (89).
(iii) A H2S-free method to prepare protein-SSH utilized 9-fluorenylmethyl pyridinyl disulfide (FmSSPy) to selectively block protein-SH (92). Then, the mixed protein disulfide was treated with DBU to release the 9-fluorenylmethyl protecting group and form protein persulfide.
(iv) Wang and colleagues developed a donor of H2S2 (BW-HP-302) that was activated by esterases (124). The liberated H2S2 could act as an electrophile to transfer the sulfur atom to protein-SH and furnish the persulfide. The authors tested GAPDH, and the persulfidated GADPH was analyzed by the tag-switch assay. All these methods provide indisputable syntheses of protein-SSH and open access to study the roles of protein persulfidation.
Fundamental Chemistry of Persulfides
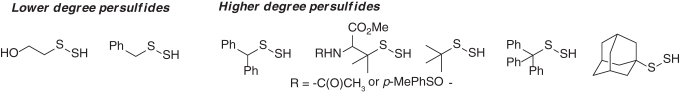
The chemistry and reactivity profiles of persulfides (R-SSH) are best studied using small-molecule substrates. Presently, a limited number of model persulfide substrates have been prepared, isolated, and fully characterized (5, 6, 40). These comprise a small fraction of lower degree (primary) persulfides such as 2-hydroxyethane persulfide [HO(CH2)2SSH] and benzyl persulfide (Bn-SSH), and the major pool of known persulfide substrates that include higher degree (secondary and tertiary) persulfides such as diphenylmethane-, tert-butyl- (tBu-), trityl- (Trt-), adamantyl (Ad-) persulfides, and derivatives of penicillamine persulfides (Scheme 3). Persulfides are inheritably unstable species, and trends of their stabilities are similar to those observed for S-nitrosothiols and sulfenic acids (4, 28, 127); where higher degree substituted persulfides are less prone to decomposition than lower degree substituted persulfides (5, 40). In most cases, the products of persulfide decomposition are polysulfides and elemental sulfur, that is, S8 (5).
Scheme 3
Persulfides are the sulfur equivalents of organic hydroperoxides with opposite reduction/oxidation (redox) properties. However, they share similar polarization across the O-O and S-S bond and exhibit electrophilic properties. Both sulfurs in the persulfide moiety are reactive electrophilic sites. Density functional theory and experimental data suggest difference in the regioselectivity of the addition of nucleophiles. The regioselectivity of the reaction of thiols and persulfides is dictated by the steric hindrance of the substituents. When the substituent on the persulfide is not sterically constrained, for instance a methyl group, nucleophiles preferentially react with the internal sulfur of persulfides (R-SSH), whereas sterically hindered persulfides such as t-BuSSH and TrtSSH are prone to be attacked by nucleophiles at the external sulfur (R-SSH) (5, 131).
Persulfides are weak acids (pKa ∼6.2), and the acid dissociation of -SSH is favored under normal physiological conditions, establishing the perthiolate anion (-SS−) as the major form in the biological medium. Therefore, persulfides are potential biological nucleophiles and prone to nucleophilic additions with biological electrophiles. Notably, the chemical reactions that perthiolate anions undergo with electrophiles are similar to those observed for R-SH. However, products derived from these reactions differ. For instance, in biological systems, persulfides can react with sulfur-containing electrophiles such as RSSR, RSOH, and H2O2 to yield trisulfides, polysulfides, and perthiosulfinate, respectively. Reactions with carbon-based biological electrophiles such as 8-nitroguanosine 3′,5′-cyclic monophosphate (8-nitro-cGMP) yield RSS-cGMP products, which in turn regulate signal transduction (45).
For decades, significant interest has been paid to the single electron chemistry of reactive sulfur species, especially hydrogen and electron transfer reactions of RSH to produce thiyl radicals (RS•) (27, 100). The formation of thiyl radicals can arise from exogenous stimuli, such as photolysis and radiolysis, and also by the action of oxygenated radicals, such as hydroxyl radical or superoxide, and electron transfer reactions with Fe-containing proteins. Implications of these sulfur radicals, for example, in proteins, are detrimental as they can lead to protein damage or altered function (16, 100). Compared with thiols (RSH), persulfides (RSSH) are more effective hydrogen donors as well as electron donors, making RSSH efficient radical scavengers. Persulfides are known to produce perthiyl radicals (20). In contrast to RSH, the weak bond dissociation enthalpy of the S-H bond in RSSH and stability of the perthiyl radical (RSS•) favor the formation of perthiyl radicals from the reaction with Fe(III) or the transfer of a hydrogen atom with other radical species, including peroxyl (ROO•), thiyl (RS•), alkoxyl (RO•), and alkyl radicals (R•) (8, 20). Compared with thiyl radical (RS•), RSS• are more stable and in consequence, less reactive and less toxic, due to the α-effect. Tertiary substituted substrates have mainly been utilized to study the chemistry of perthiyl radicals. Nonetheless, the formation of these radicals in proteins has been suggested. For instance, perthiyl radicals in Escherichia coli ribonucleotide reductase and pyruvate formate lyase were detected by electron spin resonance and proven to be vital for the inactivation of these two enzymes (23, 91). The major role elicited by perthiyl radicals is their ability to scavenge oxidants, suggesting their pivotal roles in the cell antioxidant defense (31).
Biocompatible Persulfide Donors
Persulfide donors are reagents that are capable of delivering RSSH payloads upon stimulation. The mechanism to trigger the release of persulfide should be unique and compatible to biological systems. Several strategies have been developed in this regard and can be classified in the following four categories.
Thiol-activated RSSH donors
Diallyl disulfide (DADS) and dially trisulfide (DATS) have been considered as H2S donors activated by cellular thiols (Cys and glutathione [GSH]). The mechanism of H2S release from DATS and DADS was recently revisited, and it was found that DATS is a rapid H2S donor via the intermediacy of S-allyl persulfide (68). The authors confirmed the generation of S-allyl persulfide and other polypersulfides (RSSnSH) derived from the reaction between GSH and DATS by intercepting the intermediates with an alkylating agent 9-bromobimane and liquid chromatography–mass spectrometry analysis. The products allyl-SS-bimane, allyl-SSS-bimane, and allyl-SSSS-bimane were confirmed. In contrast, similar experiments performed with DADS only produced trace amount of allyl-SS-bimane, and no detectable signal for allyl-SSS-bimane and allyl-SSSS-bimane. As such, DADS is not a useful H2S donor under the treatment of GSH. Previous discovery of DADS as a H2S donor is likely due to other polysulfide impurities in DADS samples. Notably, the formation of allyl-SS-bimane demonstrates that DATS is a potential persulfide donor. According to DFT calculations, the nucleophilic reaction of biological thiols to DATS is kinetically and thermodynamically driven toward the peripheral sulfur atom, which in turn produces the S-allyl perthiolate anion (18) rather than previously described SN2 reaction on the α-carbon. In addition, the calculations confirmed a significantly slower reaction for DADS when compared with DATS. These combined data provided direct evidence about the predominant persulfide donors in garlic, and new insights into the mechanism of H2S generation from garlic.
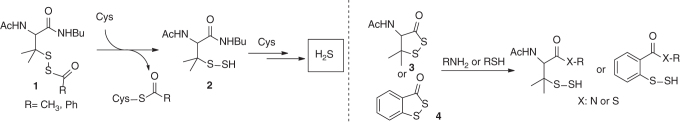
Our group reported a series of S-acyl penicillamine substrates 1 (S-acyl disulfides) as donors of H2S (134). These are also activated by nucleophilic thiols, such as Cys, through an S-acyl transfer reaction, which in turn reveals the penicillamine persulfide (Scheme 4). In this study, the penicillamine persulfide was not isolated. Instead, excess Cys triggered the decomposition of the persulfide and emanated H2S. Although this class of persulfide/H2S donors was found effective to repair myocardial I/R injury, it still remains an open question of whether the observed activity is attributed to the direct action of H2S or through some regulatory mechanisms associated with persulfides. More recently, we extended this class of molecules to a cyclic framework (51). From a chemistry standpoint, the cyclic framework was sought to exert different reactivity profiles when compared with acyclic structures as the tertiary α-carbon of the disulfide bond can induce some ring strain in compound 3. Biological reactive nucleophiles such as amines and thiols were tested, and both liberated the persulfide intermediate. Other persulfidated molecules such as benzodithiolane 4, and its selenyl surrogate yielded persulfide and selenylsulfide, respectively. Interestingly, the reactivity profiles of 3 and 4 in the presence of Cys and GSH significantly differed as H2S generation from 3 was ∼8-fold higher than 4 after 1 h. These results suggested a distinct reactivity and stability between the persulfidated species generated from 3 and 4 (e.g., tertiary-SSH vs. aryl-SSH). Such diverse reactivity widens the scope of the current library of S-acyl disulfide-based persulfide donors.
Scheme 4
Another class of S-acylated disulfide are those derived from dithioperoxyanhydrides (97) (Scheme 5). These compounds can release persulfide intermediates and subsequently H2S by the action of Cys or GSH. Mechanistically, an S-acyl transfer drives the formation of the persulfides. The profile of H2S release was illustrated in various aqueous medium including Dulbecco's phosphate-buffered saline buffer and cell lysate. An ex vivo vasorelaxation in rat aorta model demonstrated the efficacy of this class of donors, especially methyldithioperoxy anhydride [CH3(C = O)SS(C = O)CH3], to relax aortic rings in a dose-dependent manner.
Scheme 5
pH-activated RSSH donors
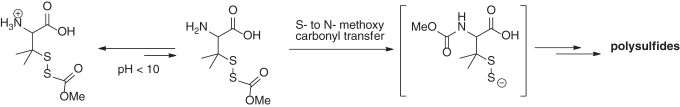
The ideal platform to release an active molecule is directly by aqueous environments and the variations in pH. The work by Galardon and colleagues demonstrated a slow and sustained production of H2S from an S-acylated disulfide scaffold (dithioperoxyanhydrides derivatives) at physiological pH 7.4 (97). Additional studies by this group led to a refined structure, which is a protected disulfide penicillamine derivative (Scheme 6) (3). In aqueous buffers, the –NH2 group triggered an S- to N-methoxy carbonyl transfer (analogous to native chemical ligation) to free up a perthiolate anion intermediate. Not surprisingly, a rapid release was attained at higher pH values (8.5) in contrast to lower pH values (6.5) as dissociation of the α-amino group increases as function of pH. It is worth noting that this persulfide donor was unable to produce H2S. Instead, a mixture polysulfides were identified as the end products. This persulfide donor may be suitable for studies centered in disseminating regulation of protein S-persulfidation in a H2S-free environment.
Scheme 6
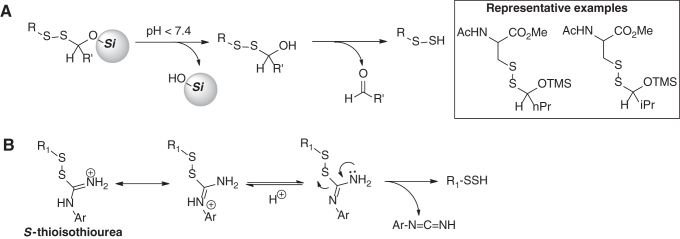
In this category, our group recently developed a general scaffold for the release of persulfides (52). The scaffold involves O-silyl protected unsymmetrical disulfides (Scheme 7A). This design combines the acid labile nature of the O-silyl group and the instability of hemithioacetals toward hydrolysis to unmask the persulfide under aqueous conditions. Several substrates were studied to explore the effects of the R-group and the lability of the O-silyl group. Substrates containing O-trimethylsilyl (-OTMS) were able to produce the corresponding persulfides at a wide range of pH (5.0, 6.0, and 7.4). For substrates containing O-triethylsilyl group (-OTES), the persulfide formation was much slower. These donors may serve as a framework to develop tools to deliver persulfides in cell organelles where pH is slightly acidic such as the lysosome.
Scheme 7
Another class of pH-activated RSSH donors was reported by Khodade and Toscano (56). The design of this new class of donors employed a urea moiety as part of unsymmetric disulfides to yield the S-thioisothiourea functionality (Scheme 7B). At pH 7.4, these donors produced persulfides as demonstrated in alkylation experiments with N-ethyl maleimide (NEM). The major byproduct was found to be phenyl cyanamide. The mechanism of RSSH release is shown in Scheme 7B. With some donors examined in this study, the generation of RSSH was accompanied by generation of RSOH. The concomitant formation of RSOH seemed to be directly controlled by the electronic properties of the N-aromatic group of the thiourea. Electron-donating groups appeared to increase the formation of RSOH, whereas electron-withdrawing groups inhibited its formation. In addition, RSSH release of these donors was pH dependent as the release was slower at lower pH.
Enzyme-activated RSSH donors
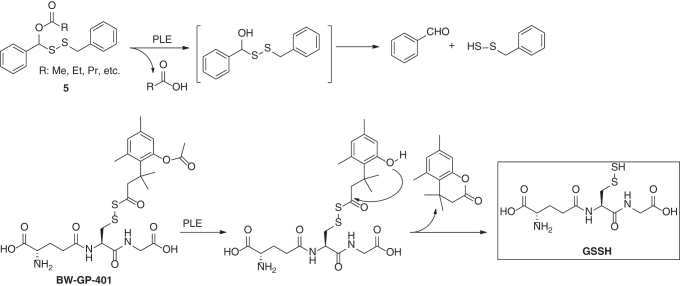
Enzyme-catalyzed release of bioactive payloads is paramount for prodrug design, and this strategy has been applied in the design of persulfide donors. In 2017, Wang and colleagues reported the first esterase-triggered RSSH donors 5 (Scheme 8) (137). Porcine liver esterase (PLE) could promote deacylation of 5 and led to the formation of benzyl persulfide. Later, the same group extended this strategy for the development of a GSSH donor BW-GP-401 (126). Upon PLE-catalyzed hydrolysis, an intramolecular lactonation produced a lactone and released GSSH. Direct evidence of the formation of GSSH was confirmed by capturing GSSH with DNFB. However, it is unclear how specific this donor is toward esterases, as similar acyl disulfides have been demonstrated to react with thiols via S-acyl transfer (3, 97, 134). Such a reaction may also lead to GSSH formation.
Scheme 8
ROS-activated RSSH donors
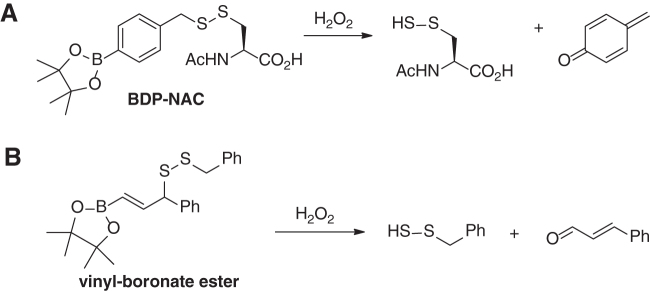
One possible way of generating protein-SSH is the reaction of H2S with protein sulfenic acids (P-SOH). Since protein-SOH are produced via the action of cysteine thiols and H2O2, it suggests that persulfides may coexist with oxidants like H2O2. Recently, Matson and colleagues reported a disulfide scaffold bpin-disulfide prodrug-N-acetyl cysteine (BDP-NAC) (Scheme 9A) that released cysteine persulfide in response to H2O2 (93). The scaffold contains a boronic ester, which is proven to selectively react with H2O2. The following self-degradation releases persulfide. Another work by Chakrapani and colleagues showed that vinyl-boronate ester-based disulfides could act as H2O2-triggered RSSH donors too (Scheme 9B) (13). It should be noted that persulfides are known to be highly reactive toward H2O2. Therefore, whether the released persulfides can be useful for the following applications is uncertain.
Scheme 9
Detection of S-Persulfidation
Cyanolysis
The first method that was used in the identification of protein persulfides is cyanolysis. As shown in Scheme 10, cyanide anion (CN−) can react with persulfides to form thiocyanate (SCN−), which can then be determined spectroscopically following the reaction with a ferric reagent. CN− could also react with other sulfane sulfur species like polysulfides and therefore causing false positives. This method is usually used for purified proteins. It cannot be used for identifying persulfide-containing proteins in biological samples such as cell lysates.
Scheme 10
Persulfide-derivatization-based methods
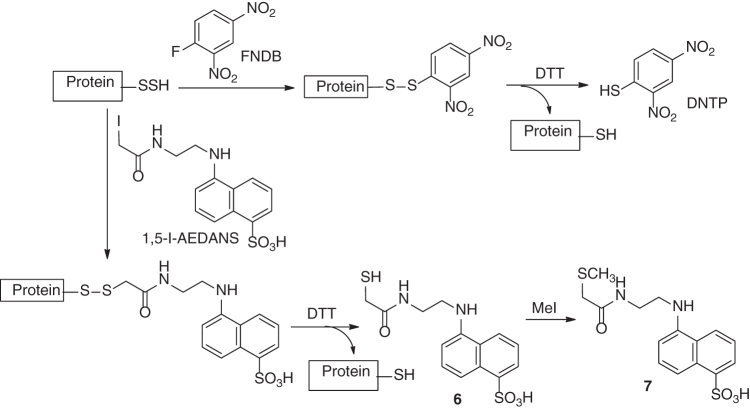
Due to the strong nucleophilicity of persulfides, certain electrophiles can be used to derivertize persulfides and applied for their detection. In one example, 1-fluoro-2,4-dinitrobenzene (FDNB) reacted with protein-SSH to form dinitrobenzene disulfide, which was next reduced by dithiothreitol (DTT) to release DNTP (Scheme 11) (99). The original concentration of persulfide could be determined by measurement of DNTP via spectroscopic methods. In another example, N-(iodoacetaminoethyl)-1-naphthylamine-5-sulfonic acid (1,5-I-AEDANS) was used to identify NifS-bound persulfide (135). The resulted NIfS-bound disulfide was cleaved by DTT to form NifS-SH and thiol 6. Methylation of 6 gave a stable thioether 7, which could be easily quantified. Again, these persulfide derivatization methods are limited to individual and purified proteins.
Scheme 11
Modified biotin-switch methods
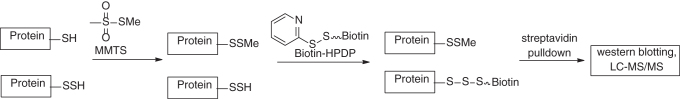
The first method that was used to identify persulfidated proteins in cell lysates is a modified biotin-switch method reported by Snyder and colleagues in 2009 (80). The rationale of this method is illustrated in Scheme 12. It was believed that methyl methanethiosulfonate (MMTS), a thiol-blocking reagent, could specifically block protein thiols (–SH) while leaves persulfides (–SSH) untouched. Then, the persulfide residues were tagged by a biotinylating agent, biotin-HPDP (N-[(6-biotinamido) hexyl]-3′-(2′-pyridyldithio) propionamide). With this method, it was found that 25%–50% hepatic proteins in mouse liver were persulfidated. However, the specificity of this method has been questioned as the selectivity of MMTS for –SH versus –SSH is hard to verify. Normally, –SSH are believed to be stronger nucleophiles than –SH. Thiol-blocking reagents such as iodoacetamide (IAM), NEM, MMTS, DTNB, and N-acetylcysteine pyridyldisulfide have been found to effectively block persulfides (89).
Scheme 12
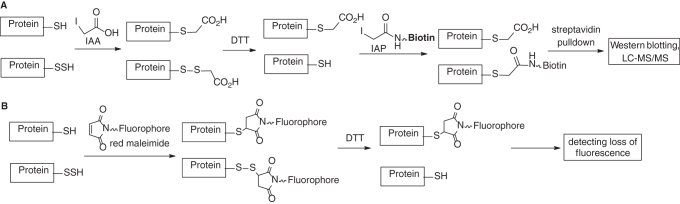
In another work by Tonks and colleagues, a different biotin-switch method was used in determining S-persulfidated proteins (Scheme 13A) (64): Both P–SH and P–SSH were first blocked by iodoacetic acid (IAA). Then, persulfide-derived disulfides were reduced by DTT to give free –SH. Subsequently, the –SH was labeled by iodoacetamide-linked biotin (IAP) for further isolation and analysis. Using this method, it was found that protein tyrosine phosphatase 1B could be reversibly inactivated by H2S and led to ER stress response. A similar method employing fluorophore-linked maleimide to detect protein persulfides was reported by Snyder and colleagues (Scheme 13B) (101). The protein mixtures were treated with fluorescent dye-linked maleimide to block both –SH and –SSH. DDT reduction should remove the fluorescent payload from –SSH-derived conjugates. The decrease of fluorescence could be quantified on a polyacrylamide gel and used to identify the original persulfides.
Scheme 13
Tag-switch method
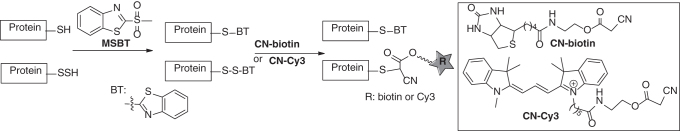
In 2014, our laboratory developed a tag-switch assay, which could be used for proteomic study of protein persulfidation (45, 129). This method employs two reagents to selectively label protein persulfides in a stepwise “tag-switch” process (Scheme 14). In step 1, both P–SH and P–SSH are blocked by methylsulfonyl benzothiazole (MSBT) to give the corresponding thioether (P-S-BT) and disulfide (P-SS–BT) adducts. Since benzothiazole-containing disulfides (P-S-SBT) are reactive toward certain carbon-based nucleophiles such as cyanoacetate, in step 2 reporting molecules can be introduced to P–S-SBT to form stable conjugates, indicating the original sites of protein persulfides. This process is selective for P–SSH as MSBT-blocked adducts from thiols (P-SH) do not react with carbon nucleophiles. Several cyanoacetate derivatives such as CN-biotin and CN-Cy3 have been developed as “tag-switch” labeling reagents. S-Persulfidated proteins identified by this method include protein disulfide isomerase, heat shock proteins, aldo-keto reductase, GAPDH, enolase, and phosphoglycerate kinase.
Scheme 14
Other biotin-labeling-based persulfide detections
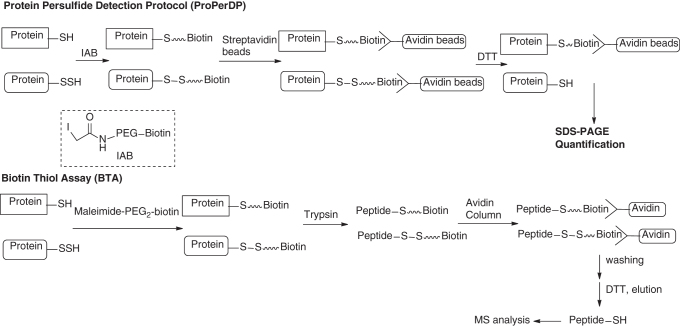
In 2016, Nagy and colleagues reported an easy-to-use and highly specific method for the detection/identification of protein persulfides, named ProPerDP (29). This method can be used in intact cells and tissue samples. With this method, the authors were able to quantify 1.52 and 11.6 μg/mg protein steady-state protein persulfide concentrations in HEK293 cells and mouse liver, respectively. The chemistry mechanism of the protocol is shown in Scheme 15: in the first step, protein-SH and –SSH groups are alkylated by iodoacetyl-PEG2-biotin (IAB). The resultant biotin-linked proteins are then pulled down from the protein mixture using streptavidin-coated magnetic beads. Persulfide-derived proteins can be further reduced by DTT to form protein-SH and released from the beads, whereas thiol (-SH)-derived proteins will remain bound to the beads. The released proteins can be quantified or visualized on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), which should reveal the original identity of persulfidated proteins.
Scheme 15
In almost the same time ProPerDP was developed, Hatzoglou and colleagues reported a biotin-thiol assay that was able to identify persulfide proteins based on a very similar mechanism (36). As shown in Scheme 15, maleimide-linked biotin is first used to label both protein –SH and –SSH residues. Then, proteins are trypsinized. Biotin-conjugated peptides (via thioether linkage or disulfide linkage) are immobilized by streptavidin beads. The disulfide-based peptides can be reduced by DTT and eluted from the beads for subsequent mass spectrometry analysis. These peptides are originally derived from protein persulfides so their identity/quantification can be used to deduce the information of persulfides.
Another similar persulfide-labeling approach (qPerS-SID) was combined with the stable isotope labeling with amino acid in culture method (73). This method allowed for quantitative persulfide proteomics analysis.
Fluorescent sensors for persulfides
Fluorescence-based methods have been widely used in the detection of biomolecules due to their rapid sensitive responses and spatiotemporal resolution capability. Up to date, fluorescent sensors that can specifically imagine protein persulfides have not been reported. However, several sensors have been developed for the detection of small-molecule persulfides. These works are summarized below.
SSP probes
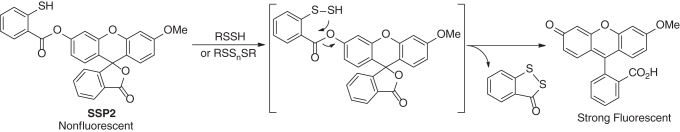
Persulfides belong to the sulfane sulfur family, in which molecules contain a sulfur atom with six valence electrons but no charge (known as S0). A unique reactivity of sulfane sulfurs is that they can react with certain nucleophiles, for instance, reacting with cyanide ion (CN−) to form thiocyanate (SCN−). Based on this reactivity, sulfane sulfur probes (such as SSP2) have been developed and were used to detect persulfides (Scheme 16). These probes employ a thiophenol to react with sulfane sulfurs to form an Ar-S-SH intermediate, which subsequently undergoes an intramolecular cyclization to release the pendant fluorophore. While these probes are selective for sulfane sulfurs including persulfides, they cannot differentiate each member in the sulfane sulfur family, for example, persulfides versus polysulfides.
Scheme 16
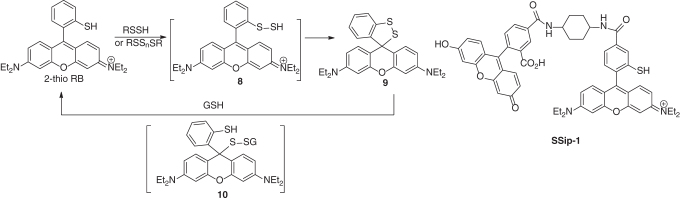
SSip-1
The reaction between persulfides and thiolate has also been used to develop reversible fluorescent sensors for persulfides. Urano and colleagues found that 2-thio rhodamine B (2-thio RB, Scheme 17) could react with persulfides or polysulfides to form 8, which then formed spirocycle 9, leading to fluorescence decrease at 560 nm (108). Interestingly, GSH could restore the fluorescence, via the reduction of the disulfide bond to form 10 and then elimination of GSSH to reform 2-thio RB. Based on this reactivity, a fluorescence resonance energy transfer (FRET)-based sensor SSip-1 was developed. SSip-1 could reversibly visualize sulfane sulfurs including persulfides in living cells.
Scheme 17
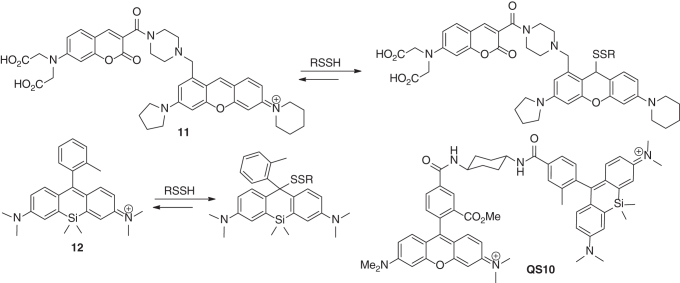
In another work by Ojida and colleagues, the strong nucleophilicity of persulfides was employed in the design of reversible ratiometric fluorescent sensors (54). For instance, sensor 11 contained a coumarin as the FRET donor, which is conjugated to a pyronine unit as the FRET acceptor. It showed two distinct emissions at 479 and 584 nm (excited at 410 nm) due to the coumarin and pyronine units. Upon addition of persulfides (using Na2S2 as the model), the dual-emission spectrum changed dramatically in a seesaw manner, with a large decrease at 584 nm and the concomitant increase at 479 nm. The ratio value R (F479nm/F584nm) could be used to determine the concentrations of persulfides. The sensing mechanism involves a nucleophilic addition of the persulfides with the pyronine unit, which modulates the intramolecular FRET efficiency. In addition, the nucleophilic addition was found to be reversible, and therefore, the sensor could also detect the decreases of persulfide levels. Very recently, Urano and colleagues found silicon-derived rhodamines could also undergo reversible nucleophilic addition with persulfides (110). For example, compound 12 showed high sensitivity and selectivity for persulfides (>10,000-fold over GSH). This template was used to develop QS10, a ratiometric sensor for persulfides.
Scheme 18
S-Persulfidation and Diseases
S-Persulfidation in cardiovascular and related health and diseases
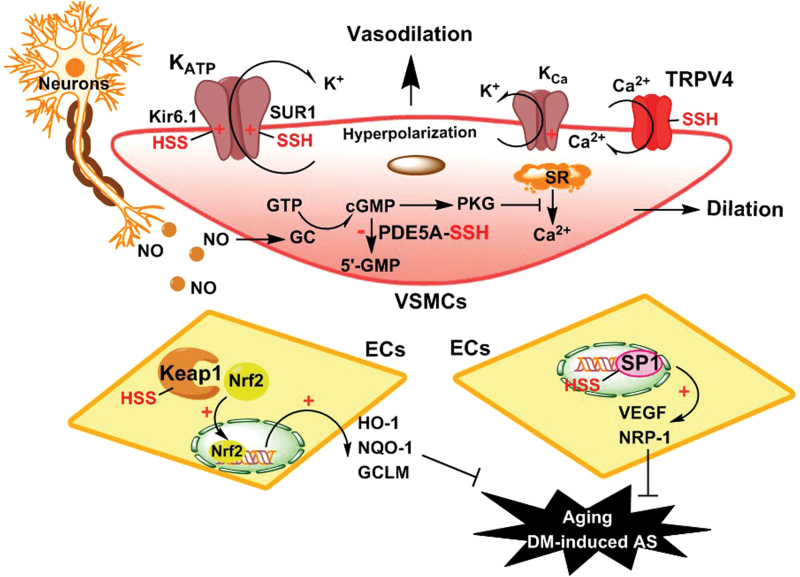
Vasorelaxation is one of the most important functions of H2S, and the underlying mechanisms have been investigated since the report of Kimura and colleagues (41). Several related proteins, such as ion channels and second messengers, are involved in H2S-induced vasorelaxation (Fig. 1) (7, 60, 81, 105, 109, 133). Among ion channels, ATP-sensitive potassium channel (KATP) is well documented (21, 47, 81). Classical KATP channels are composed of Kir6.x and SURx subunits, alongside additional components (102). Physiologically, KATP channels are controlled by intracellular ATP, as well as ADP and nucleotides. Many reports have shown that KATP channels can be regulated by H2S (47, 116, 133). For example, in aorta and mesenteric artery, H2S-induced vasorelaxation is mainly mediated by KATP channels (21, 133). Jiang et al. found that H2S-induced activation of KATP channels depends on SUR1 subunit's S-persulfidation at Cys6 and Cys26 residues (47). However, Mustafa et al. considered that S-persulfidation of Kir6.1 subunit at Cys43 site might be crucial to H2S-induced hyperpolarization and vasorelaxation (81).
FIG. 1.
Possible signaling roles of S-persulfidation in vasorelaxation. Neurons and ECs-released NO dilates VSMCs. In VSMCs, enhancement of K+ efflux through KATP or KCa channel hyperpolarizes cell membrane and induces vasodilation. The red P-SSH in KATP channel Kir6.1 and SUR1 subunits and in TRPV4 channel represents S-persulfidation of these channel proteins. The red cross (+) represents opening of the channels. The activation of GC/cGMP/PKC pathway inhibits SRs Ca2+ release and induces vasodilation. In ECs, S-persulfidation-induced Keap1 activation releases Nrf2, which undergoes nuclear translocation and induces the expression of HO-1, NQO-1, and GCLM. S-Persulfidation-induced SP1 activation promotes VEGF and NRP-1 expression, thus eliminating aging and DM-induced AS. AS, atherosclerosis; ECs, endothelial cells; GCLM, glutamate-cysteine ligase modifier; HO, heme oxygenase; KATP, ATP-sensitive potassium channel; Kca, calcium-activated potassium channel; Keap1, Kelch-like ECH-associated protein 1; NQO, NADPH quinone oxidoreductase; Nrf2, nuclear factor-like 2; NRP, neuropilin; PKC, protein kinase C; PKG, cGMP-dependent protein kinase; SP1, specificity protein 1; SR, sarcoplasmic reticulum; TRP, transient receptor potential; VEGF, vascular endothelial growth factor; VSMCs, vascular smooth muscle cells. Color images are available online.
Besides KATP channels, another kind of K+ channel can be controlled by intracellular Ca2+ ion content, also known as Ca2+ ion-activated K+ (KCa) channels. It has been shown that Ca2+ ion influx mediated by TRPV4 channels can activate KCa channels, thereby resulting in endothelial hyperpolarization and subsequent vasodilation (33). It is worth noting that H2S can S-persulfidate TRPV4 channel proteins at specific cysteine residues, as supported by a DTT assay (82). Apart from activating K+ channels, normal Ca2+ ion influx can cause contraction of muscle cells, including vascular smooth muscle cells. Therefore, lowering intracellular Ca2+ content will consequently lead to vasorelaxation. An important example is the signaling of cGMP, a second messenger, which has been demonstrated to reduce cytoplasmic Ca2+ content (10). Endogenous cGMP usually exerts its vasodilator action through the activation of cGMP-dependent protein kinase (PKG) (34). In cells, PKG has a wide range of functions to dilate blood vessels. For example, PKG can induce sarcoplasmic reticulum (SR) to store Ca2+, while it inhibits the release of Ca2+ by SR, thereby lowering cytoplasmic Ca2+ content and dilating smooth muscle cells. It is known that endogenous phosphodiesterase type 5 (PDE5) can hydrolyze intracellular cGMP, thereby balancing the physiological content of cGMP. However, if intracellular cGMP is deficient, low Ca2+-induced vasodilation and penile cavernous relaxation will be impaired, which may result in hypertension and erectile dysfunction. Therefore, inhibition of PDE5 is an important strategy of these dysfunctions through raising intracellular cGMP content. Interestingly, the increase in endogenous H2S amount or its exogenous application can inhibit PDE5A dimerization and resultant activation via S-persulfidation (105), thereby inducing vasodilation and penile cavernous relaxation. The effect mediated by S-persulfidation is similar to the effect of Sildenafil, a clinically used PDE5 inhibitor (12).
Besides vasorelaxation, H2S maintains vascular function and health through upregulation of vascular endothelial growth factor receptor 2 and neuropilin-1 (Fig. 1). The regulatory effects of H2S on these two proteins mainly attribute to S-persulfidation of a transcription factor, specificity protein 1 (SP1). It has been shown that S-persulfidation of Cys68 and Cys755 can enhance SP1 transcriptional activity (98).
Under conditions of oxidative lesion, S-persulfidation of redox-related proteins is distinctively involved in blood vessel protection. One of the most important examples is Kelch-like ECH-associated protein 1 (Keap1)/nuclear factor-like (Nrf)2 signaling pathway. The combination of Keap1 with Nrf2 usually activates the ubiquitin-proteasome pathway-mediated degradation, leading to a decrease in cytoplasm Nrf2 content. Once modified by electrophiles, such as 8-nitro-cGMP, the Keap1-Nrf2 heterodimer will release Nrf2. The translocated Nrf2 will bind to antioxidant response element in nuclear DNA and regulate the expression of redox-related genes, such as heme oxygenase (HO)-1, NADPH quinone oxidoreductase, and glutamate-cysteine ligase modifier (Fig. 1). As a nucleophile, H2S can directly react with some electrophiles, such as 8-nitro-cGMP, and thus abolish their oxidative injury (85). More importantly, H2S can also induce S-persulfidation of Keap1, indirectly relieving nucleophiles-induced oxidative lesions (42, 119, 123). Furthermore, H2S-mediated S-persulfidation of Keap1 at Cys151 residue significantly suppresses vascular atherosclerosis accelerated by diabetes (119). In addition, the aging of endothelial cells (ECs) and fibroblasts is of great importance to vascular dysfunction. In fact, the repair of DNA damages can retard the process and delay cellular senescence. It is reported that H2S-indcued Cys341 S-persulfidation of MEK1 enhances ERK1/2 phosphorylation and nuclear translocation, thereby activating PARP-1 and initiating DNA ligase III-mediated DNA damage repair (132).
Summarily, in blood vessels, under physiological condition, S-persulfidation of KATP channels, TRPV4 and PDE5 causes vasorelaxation. Under physiopathological conditions, S-persulfidation of Keap1 exerts antioxidative effects and attenuates vascular dysfunction induced by diabetes and aging.
Endogenous redox imbalance is also a major risk for heart disease. Intracellular accumulation of 8-nitro-cGMP is a main cause of redox imbalance. Usually, 8-nitro-cGMP derives from reaction between reactive nitrogen species and guanine nucleotide. It was reported that 4-week myocardial infarction and 6-week pressure overload could significantly increase 8-nitro-cGMP generation in mouse hearts (85). Persistent 8-nitro-cGMP injury eventually leads to cardiac disorder or even chronic heart failure. It has been well documented that 8-nitro-cGMP can induce S-guanylation of special cysteine residues and weaken the activity of target proteins, such as Keap1, PKG, H-Ras, and soluble N-ethylmaleimide-sensitive factor attachment receptors (2, 84, 96). Through S-persulfidation, cysteines (both small molecules and proteins) can be converted into persulfides (RSSH). Notably, RSSH has much enhanced ability to eliminate 8-nitro-cGMP comparing with reduced cysteines (RSH), thereby preventing specific proteins from S-guanylation induced by 8-nitro-cGMP (84, 85). Apart from the LMW CysSSH, S-persulfidation of proteins at special cysteine residues is essential to the redox homeostasis. For instance, treatment of cardiomyocytes with GYY4137 (a H2S donor) attenuates spontaneous hypertension-induced myocardial hypertrophy. The mechanism underlies H2S-medicated inhibition of Kruppel-like factor 5 (KLF5) transcription activity via S-persulfidation of SP1 at Cys664 (76). Additionally, in I/R-induced myocardial injury, H2S treatment during the reperfusion obviously lowers infarct size, thus improving myocardial contractile function. Further study showed that the levels of S-persulfidation and S-nitrosylation are significantly increased during the process (104). In summary, these studies suggest that S-persulfidation of LMW cysteine and cysteine residues of target proteins is essentially involved in cardioprotection (Fig. 2).
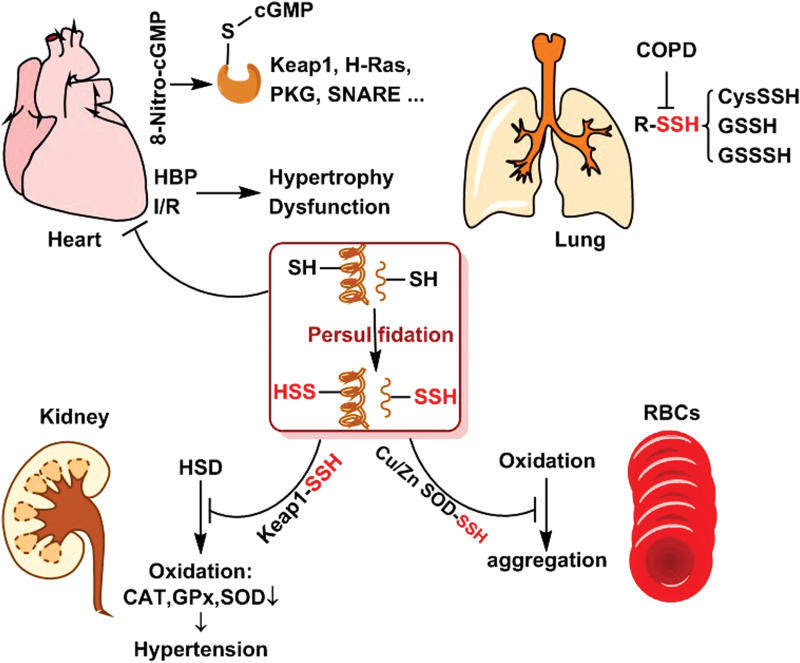
FIG. 2.
Roles of S-persulfidation in the indicated organs. In hearts, S-persulfidation blocks 8-nitro-cGMP-mediated target protein S-guanylation and inhibits HBP and I/R-induced hypertrophy and dysfunction. In kidneys, S-persulfidation of Keap1 prevents HSD and oxidation-induced HBP. In RBCs, S-persulfidation of Cu/Zn SOD attenuates oxidation-induced aggregation. In lungs, COPD condition reduces LMW S-persulfide contents. 8-nitro-cGMP, 8-nitroguanosine 3′,5′-cyclic monophosphate; COPD, chronic obstructive pulmonary disease; HBP, hypertension; HSD, high-salt diet; I/R, ischemia/reperfusion; LMW, low molecular weight; RBCs, red blood cells; SOD, superoxide dismutase. Color images are available online.
Apart from the heart and blood vessel, normal maintenance of cardiovascular function closely depends on other organs in the body, such as the lungs, kidneys, and even blood cells. Once these organs or cells are damaged or dysfunctional, secondary cardiovascular lesions will be initiated. In human chronic obstructive pulmonary disease (COPD), oxidative stress has been accepted as a major etiological factor driving the process of COPD (95). It was reported that the amounts of reactive persulfides, including CysSSH and GSSH, were distinctively lowered in lung cells and epithelial lining fluid of the patients with COPD (86). Significantly, the persulfide amounts have a positive correlation with the degree of airflow limitation (86) (Fig. 2).
In human chronic kidney disease, ROS appear to be responsible for the high incidence of cardiovascular events. It has been found that high-salt diet triggers renal excessive oxidation, and resultant functional and structural injury, evidenced by the increased levels of serum creatinine and urea, and decreased levels of 24-h urine protein. Meanwhile, the cardiovascular function is abnormal showing as secondary hypertension. Importantly, the donation of H2S improves renal function and structure and enhances urine volume, thereby alleviating the secondary hypertension of the patients with chronic kidney disease. Similar to the effects on other organs, the improvement of H2S on renal function is still associated with S-persulfidation of Keap1 and, subsequent Nrf2 disassociation and nuclear translocation. These effects will consequently trigger the expression of antioxidant genes, such as CAT, GPx, and superoxide dismutase (SOD) (43) (Fig. 2).
The blood system is a special flowable organ in the body, which consists of liquid plasma and tangible blood cells. The altered plasma components and dysfunctional blood cells can trigger secondary vascular lesions. As documented, S-persulfidation exists in red blood cells (RBCs) (14). When Cu/Zn SOD isolated from RBCs reacts with H2S, S-persulfidation will be generated. Further study shows that the modification occurs at Cys111 residue of Cu/Zn SOD protein. The S-persulfidated Cu/Zn SOD protein exhibits a slower acid-induced unfolding and is more resistant to oxidation-induced aggregation, such as copper and H2O2 (14, 26) (Fig. 2). S-Persulfidation of Cu/Zn SOD may be conducive to endothelial protection and decrease the risk of venous thrombosis.
S-Persulfidation in neural health and disease
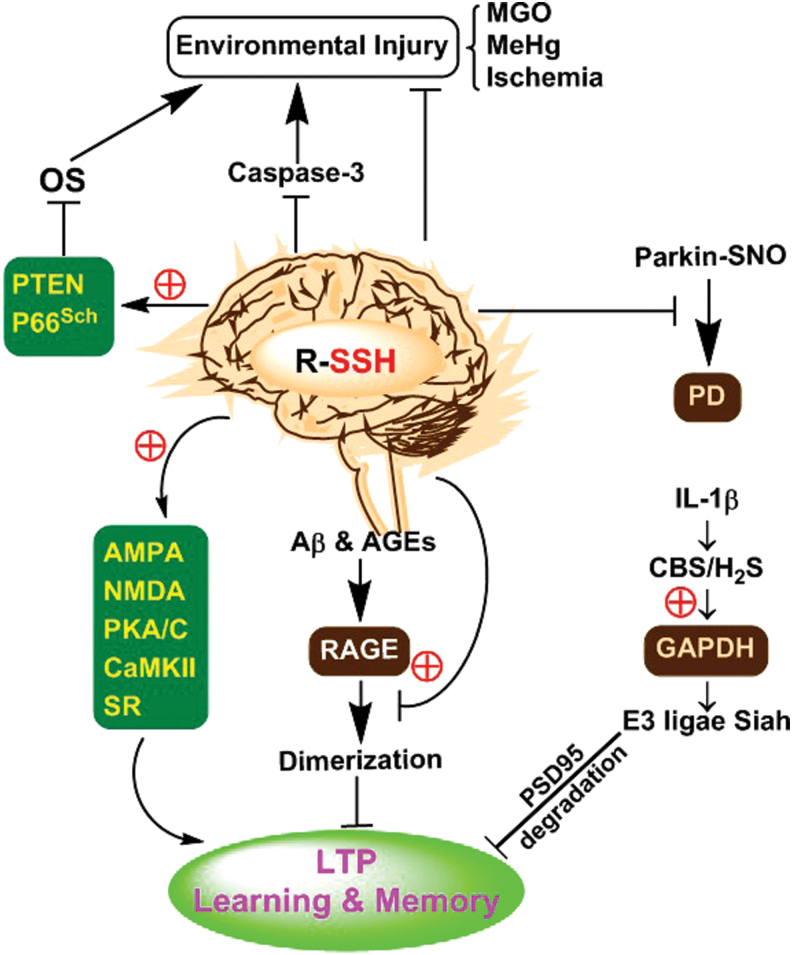
As a gaseous neuromodulator, physiological concentration of H2S is essential for learning and memory by affiliating hippocampus long-term potentiation (LTP), which is one of the most important forms of synaptic plasticity (74). During the development of synaptic plasticity, glutamate receptor-mediated postsynaptic excitation, including LTP, plays a very important role. Among those glutamate receptors, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor and N-methyl-d-aspartate (NMDA) receptor have been well documented. It was found that these receptors could be S-persulfidated during the process of synaptic plasticity (67, 74). These S-persulfidated proteins are seen in NR2A subunit of NMDA glutamate receptors and in postsynaptic proteins (67, 74). The postsynaptic proteins include protein kinase A (PKA), protein kinase C, and Ca2+/calmodulin-dependent protein kinases II, and they usually regulate the phosphorylation of glutamate receptors. In addition, D-serine in the brain acts as a neuronal signaling molecule and activates NMDA glutamate receptors, contributing to synaptic LTP generation. Endogenous D-serine is mainly produced due to the catalyzation of serine racemase (90). It has been shown that exogenous supplement of H2S or boost of H2S signal overtly restores S-persulfidation levels of serine racemase protein, followed by the improvement of age-related LTP deficit and hippocampus-dependent memory impairment (66). However, in inflammation-induced memory impairment, proinflammatory cytokine interleukin (IL)-1β can upregulate endogenous CBS and H2S; nevertheless, they seem to exert adverse effects. It has been reported that H2S can induce S-persulfidation of GAPDH in neuron dendrites. S-Persulfidation enhances GAPDHs binding ability to ubiquitin-protein ligase E3 (Siah protein). Then, E3 ligase Siah will link PSD95 synaptic scaffolding molecule, leading to PSD95s ubiquitination-dependent degradation and consequent memory impairment (77) (Fig. 3).
FIG. 3.
Possible signaling roles of S-persulfidation in the nervous system. S-Persulfidation of glutamate AMPA and NMDA receptors, postsynaptic proteins (PKA/C, CaMKII) and serine racemase is involved in normal learning and memory. S-Persulfidation of RAGE blunts Aβ and AGEs-impaired learning and memory. Proinflammatory cytokine IL-1β-mediated S-persulfidation of GAPDH harms learning and memory through degradation of PSD95. S-Persulfidation of parkin blocks its S-nitrosylation and inactivation and thus eliminates PD lesions. S-Persulfidation increases PTEN and P66Sch activity and decreases caspase-3 activity, thereby reducing environmental stimuli-induced neuronal lesions. Aβ, Abeta 1–42 peptide; AGE, advanced glycation end product; AMPA, α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid; CaMKII, calmodulin-dependent protein kinases II; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IL, interleukin; NMDA, N-methyl-d-aspartate; PD, Parkinson's disease; PKA, protein kinase A; PTEN, phosphatase and tensin homolog; RAGE, receptor for advanced glycation end product. Color images are available online.
Apart from brain, spinal cord is another important part of central neural system. A very recent study showed that breathed H2S could prevent ischemia-induced paraplegia through inhibition of inflammation and apoptosis. The mechanism was associated with induction of nuclear factor (NF)-κB p65 subunit S-persulfidation from S-nitrosylation (50).
In summary, S-persulfidation of proteins is significant in learning and memory of central neuronal system. It is not only involved in the development of normal learning and memory but also mediates inflammation-induced learning impairment.
In chronic neurodegenerative diseases, S-persulfidation of specific proteins is also of great significance (Fig. 3). Alzheimer's disease (AD) starts slowly and worsens over time, characterized by a gradual loss of short-term memory. In AD cellular models, the treatment of human SH-SY5Y neuroblastoma cells with Abeta 1–42 peptide (Aβ) or advanced glycation end products induces receptor for advanced glycation end products (RAGEs) dimerization and cellular senescence. It has been suggested that H2S can induce S-persulfidation of RAGE at Cys259/301 residues. By this mechanism, H2S blocks RAGEs dimer formation and persistent oxidation-induced neural lesions (138). Unlike the impaired memory of AD, Parkinson's disease (PD) mainly causes lesions in motor nervous system. It has been demonstrated that the mutation of parkin gene is one of the leading causes of PD. Normally, parkin protein acts as an ubiquitin E3 ligase, and its activity becomes low or even lost in the pathogenic process of inherited and sporadic PD (25). Notably, S-nitrosylation is a common reason for parkin's loss of ubiquitin ligase activity, which weakens its neuroprotective function (22). Furthermore, parkin's S-persulfidation abundance is significantly decreased in PD patients' brains. Interestingly, through upregulation of S-persulfidation, H2S is able to restore parkin's catalytic activity, consequently inhibiting PD-related lesions (113).
Akin to parkin, PTEN and P66Shc proteins can also be S-persulfidated, which is useful for the prevention of nerve cells against oxidation related injury. It is known that H2O2 and/or NO-induced modifications of specific cysteine residues of PTEN and P66Shc abolish the enzymatic activity. However, CBS/H2S can shift PTENs S-nitrosylation to S-persulfidation at Cys71 and Cys124 sites, followed by the improvement of enzymatic activity and downstream signaling in SH-SY5Y cells (Fig. 3) (38, 87, 120).
Sometimes, S-persulfidation of proteins may have inhibitory effects on enzymatic activities. For instance, in ischemic murine primary cortical neurons, due to S-persulfidation at Cys163, the apoptotic effector protein caspase-3 will lose activity and apoptotic death will decrease (75). In addition, under the condition of neuronal carbonyl stress induced by methylglyoxal (MGO), persulfide (Na2S2) as well as polysulfides (Na2S3 and Na2S4) exert significant neuroprotection by reducing the formation of MGO-modified proteins in SH-SY5Y cells (61, 62). It has also been found that induction of endogenous H2S suppresses MGO-induced peripheral neural injury (130).
In environmental electrophile-related neurotoxicity, exposure of cerebellar granule neurons to methyl mercury (MeHg) triggers S-guanylation and activation of H-Ras. However, MeHg-mediated neurotoxicity can be dramatically eliminated by the pretreatment with persulfide (46). Overall, in neurodegeneration and environmental stimuli-related lesions, S-persulfidation of proteins and LMW persulfides usually have significant protective effects.
S-Persulfidation and digestive system diseases
In I/R-induced gastric epithelial cell injury, the increased ROS levels significantly cause inflammatory response through NF-κB and mitogen-activated protein kinases signaling pathways. As aforementioned, Keap1-Nrf2 is essential to endogenous antioxidation. H2S-induced S-persulfidation of Keap1 leads to Nrf2 disassociation and activation, thereby inducing antioxidation gene expressions. The effects of H2S may have potential therapeutic value in acute gastric mucosal inflammatory lesions (39, 122).
In murine gastric smooth muscles, exogenous H2S can trigger S-persulfidation in voltage-dependent potassium channel (KV) protein, instead of KATP channel protein, and enhance the tonic contraction, which can be abolished by DTT (a reducing agent) (71). Oppositely, in colonic smooth muscles, H2S exhibits inhibitory effects on contraction, due to S-persulfidation-mediated inhibition of RhoA/Rho kinase (83). Additionally, in experimental colitis model, H2S apparently S-persulfidates KATP channel SUR2B subunit at Cys24/1455 sites, and such an effect attenuates NO-induced tyrosine nitration in another KATP channel subunit Kir6.1 (53). In summary, S-persulfidation has effects on epithelial cells, as well as smooth muscles, which seems to be tissue specific.
S-Persulfidation and bone health
Under physiological conditions, normal amount of H2S is a critical regulator of bone homeostasis. Bone marrow-derived mesenchymal stem cells (MSCs) are a common precursor of both adipocytes and osteoblasts. In fact, endogenously produced H2S in MSCs affects osteogenic differentiation and MSCs self-renewal. The effects of H2S are associated with the activation of TRPV3/V6/M4 channels-induced Ca2+ influx. H2S-induced channel protein activation is linked to S-persulfidation of cysteine residues. The increased Ca2+ content in cells consequently activates Wnt/beta-catenin signaling and in turn enhances osteogenic differentiation (72).
In addition, during bone fracture healing, CSE/H2S is capable of activating runt-related transcription factor 2 (RUNX2) and improves the fracture healing. Further study showed that H2S-mediated activation of RUNX2 relied on S-persulfidation at Cys123 and Cys132 (136). However, if the critical cysteine residues are S-nitrosylated, for instance, in peroxisome proliferator-activated receptor (PPAR)γ, the activation of target proteins will be inhibited, which is harmful for osteoblast differentiation (19). Under such conditions, S-persulfidation of specific proteins may have potential therapeutic effects.
S-Persulfidation in metabolic disorder and diabetes
Endogenous H2S possesses diverse roles in metabolisms of nutrients, such as glucose, lipid, and protein, in an insulin-dependent or -independent manner. H2S can regulate mitochondrial biogenesis via ATP synthase (121). Such effects rely on S-persulfidation on specific proteins. For example, in the liver, overexpressed CSE and exogenously applied H2S enhance the activity of pyruvate carboxylase (PC) and gluconeogenesis, through S-persulfidation of Cys265 in PC proteins (49). Beyond gluconeogenesis, H2S is able to facilitate hepatic glucose production from glucogen breakdown and thus increase blood glucose levels. The effects of H2S on glucogen breakdown are associated with S-persulfidation-mediated activation of glucose-6-phosphatase and fructose-1,6-bisphosphatase (112).
In addition to the regulation of nutrient metabolisms, the liver plays a role in alcohol metabolism. It has been known that drinking too much alcohol increases the risk of cancer. Disulfiram can be used to treat chronic alcoholism by inducing unpleasant effects when even small amounts of alcohol are consumed. The mechanism is due to the inhibition of aldehyde dehydrogenase (ALDH). A recent study by Iciek et al. showed that some reactive sulfur species (including GSSH, K2SX, and garlic-derived allyl trisulfide) also effectively inhibited the activity of ALDH. This inhibition could be easily reversed by reducing agents such as DTT and GSH, indicating that S-persulfidation mediates the regulation of ALDH activity (44).
In hepatocellular cells, H2S can improve the activity of ATP synthase through S-persulfidation on Cys244 and Cys294, thereby boosting mitochondrial ATP generation. It was pointed out that S-persulfidation-mediated activation was significant to maintain physiological ATP synthase activity and mitochondrial bioenergetics (79). However, during mitochondrial biogenesis and bioenergetics, CARSs can catalyze a reaction from cysteine to CysSSH. Importantly, this S-persulfidation occurs in cysteine residues of dynamin-related protein 1 (Drp1) and also in free form cysteines used for Drp1 protein translation (1). As a mediator of mitochondrial fission, the inactivation of Drp1 induced by S-persulfidation may recover mitochondrial function and ATP synthesis.
Besides livers, adipose tissues are another important organ related to nutrient metabolisms. It was reported that CSE/H2S promotes glucose uptake and lipid storage in adipocytes of fatty tissues, and the action relies on S-persulfidation of Cys139 in PPARγ (17). Actually, many functions in both the livers and fatty tissues, including metabolisms of glucose and lipid, are controlled by insulin from pancreatic islets. It has been shown that persulfides, such as CysSSH, cause tRNAs methylthiolation and stimulate insulin secretion in vitro and in vivo to regulate glucose and lipid homeostasis (107).
S-Persulfidation in immune system and inflammation
Besides barrier function, ECs in vascular lumen regulate the process of inflammation (125). It has been shown that CSE/H2S-induced S-persulfidation of RNA binding protein HuR at Cys13 prevents HuRs homodimerization and activity, thereby downregulating CD62E and cathepsin S. Through these effects, H2S can inhibit monocyte adherence and inflammatory response, thus delaying the atherogenesis process in ApoE−/− mice (9). Sirtuin-1, a histone deacetylase, exerts antiatherogenic effects by regulating the acetylation of proteins, such as P65, P53, and sterol response element binding protein, thereby reducing endothelial and macrophage inflammation. Notably, endogenous CSE/H2S can directly S-persulfidate Sirtuin-1 and enhance its activity (30), thus eliminating inflammation. In macrophages, H2S attenuates oxidative stress-induced mitochondrial ROS production and NLRP3 inflammasome activation by S-persulfidating c-Jun's Cys269 (70). On the condition of bacterial infection, the infected macrophages can remove the bacteria through generation of 8-nitro-cGMP in an autophagy-dependent manner. However, the smart bacteria, such as Salmonella, have a powerful ability to release reactive persulfides to block the antibacterial autophagy of macrophages, ensuring the bacteria's survival (55). Additionally, it was reported that endogenous CSE expression and H2S production were decreased in the allergic asthma of young patients. The decrease may induce type-2 immunoreaction. Notably, H2S-induced S-persulfidation of GATA3 protein attenuates type-2 immunoreaction, therefore eliminating allergic asthma symptom (115).
S-Persulfidation and cancer
Numerous studies have shown that H2S plays roles in cancer development and progression, through stimulation of angiogenesis, regulation of intracellular signaling, and cellular bioenergetics, while some reports are controversial (32). However, the available studies on the roles of S-persulfidation in cancer are very limited. In colonic carcinoma, endogenous CBS expression and H2S production are distinctively upregulated. The upregulation can enhance cellular bioenergetics and tumor growth. Lactate dehydrogenase A (LDHA) is a direct regulator of glycolysis and bioenergetics. Like the expression profile of CBS/H2S in colonic carcinoma, endogenous LDHA is overexpressed and can also enhance tumor growth. Treatment of human colon cancer (HCT116) cells with H2S boosts mitochondrial function and glycolysis, characterized by increased oxygen consumption and ATP production. Further study revealed that the effects of H2S were associated with LDHAs S-persulfidation of Cys163 and consequent activation (111).
In lung carcinoma, endogenous H2S is essential to tumor cell growth through repairing mitochondrial DNA damage. In fact, the induction of DNA damage is common action of chemotherapeutic drugs. Tumor cells repair the damaged DNA usually relying on exo/endonuclease gene (EXOG). It was found that H2S can activate EXOG, thereby enhancing tumor cells resistance to chemotherapeutic drugs. Further study uncovered that H2S-induced activation of EXOG is related to S-persulfidation at Cys76 residue (106).
Interestingly, a recent study showed that H2S could inhibit the efflux of doxorubicin (Dox) from liver cancer cells and enhance Doxs chemotherapy activity. The authors found that the treatment of H2S induced the S-persulfidation of retinoid X receptor (RXR) β and then inhibited the heterodimer formation between RXR β and liver X receptor α. Such effects eventually weakened ABCA1- and ABCG8-mediated efflux transportation of chemotherapeutic agents, thereby reversing drug resistance of live cancer cells (103).
Conclusions
The discovery of S-persulfidation in living organisms and its regulatory roles in redox biology are very attractive. S-Persulfidation leads to the formation of both small-molecule-based persulfides (RSSH) and protein persulfides. These species are highly reactive and thus quite unstable. Their presence or fates in biological environments are sometimes difficult to track. This review presents current understandings of persulfides (and S-persulfidation) chemistry, biochemistry, and their links with health and disease. In current studies, researchers commonly use thiol-blocking reagents, such as IAM, NEM, and monobromobimane, to convert unstable persulfides to stable derivatives for further analysis. However, these methods are not always reliable. As recently discovered by Nagy and colleagues, sulfur alkylation can alter sulfur speciation and sometimes can even change the identity of alkylated products (especially for polysulfide derivatives) (11). As such, the actual readouts do not necessarily represent true original (such as intracellular) speciation of sulfur species.
As discussed in this review, the specific target proteins of S-persulfidation are essential in organismal physiological health and human disease states (Table 1). Current studies are mainly focused on the cardiovascular system and neuronal system. Studies in other systems or organs are still limited. In addition, some reports are still contradictory, for instance, the roles of S-persulfidation in cancer growth. Apparently, further clarifications on these issues are needed, perhaps with the help of better understanding persulfide chemistry/biochemistry and more reliable detection methods. There is little doubt that the general area of investigation of S-persulfidation will be a topic of significant research interest in the coming years.
Table 1.
Representative Persulfidated Proteins and Their Functions
| Target proteins | Persulfide residues | Affected biological functions |
|---|---|---|
| Cardiovascular system | ||
| SUR1 subunit | C6/26 | Activates KATP channels and dilates blood vessels |
| Kir6.1 subunit | C43 | Activates KATP channels and dilates blood vessels |
| TRPV4 | N/A (Not detected) | Enhances KCa channel-dependent vasodilation |
| PDE5A | N/A (Not detected) | Inhibits cGMP degradation and induces cavernous relaxation |
| SP1 | C68/755 | Enhances VEGFR2- and neuropilin1-dependent anti-aging |
| C664 | Inhibits KLF5 activation and secondary myocardial hypertrophy | |
| Keap1 | C151 | Activates Nrf2 pathway and attenuates oxidative injuries |
| MEK1 | C341 | Activates ERK1/2 pathway and repairs damaged DNA |
| Cu/Zn-SOD | C111 | Inhibits RBCs aggregation and venous thrombosis |
| Neural system | ||
| NR2A | C87/320 (Speculated) | Activates NMDA glutamate receptors and improves memory |
| Serine racemase | C113 | Prevents age-related LTP deficit and memory impairment |
| GAPDH | C150 | Enhances Siah E3 ligase's binding, PSD95 degradation, and memory impairment |
| RAGE | C259/301 | Blocks RAGEs dimerization and persistent oxidation-induced neural lesions |
| Parkin | C95/182/C59 | Inhibits Parkinson's disease-related lesions |
| PTEN | C71/124 | Prevents from oxidative injury in nerve cells |
| Caspase-3 | C163 | Attenuates apoptotic death |
| Digestive system | ||
| SUR2B subunit | C24/1455 | Attenuates KATP channel Kir6.1's tyrosine nitration |
| Bone | ||
| RUNX2 | C123/132 | Improves fracture healing |
| Metabolic disorder and diabetes | ||
| Pyruvate carboxylase | C265 | Enhances gluconeogenesis |
| ATP synthase | C244/294 | Boosts ATP generation in liver cells |
| PPARγ | C139 | Promotes lipid storage |
| Inflammation and cancer | ||
| NF-κB p65 subunit | C38 | Increases NF-κB activity and attenuates macrophage apoptosis |
| HuR | C13 | Reduces monocyte adherence and inflammation |
| c-Jun | C269 | Inhibits ROS production and NLRP3 inflammasome activation |
| LDHA | C163 | Enhances colonic carcinoma energy synthesis and growth |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; KATP, ATP-sensitive potassium channel; Keap1, Kelch-like ECH-associated protein 1; LDHA, lactate dehydrogenase A; LTP, long-term potentiation; NF, nuclear factor; NMDA, N-methyl-d-aspartate; Nrf2, nuclear factor-like 2; PPAR, peroxisome proliferator-activated receptor; PTEN, phosphatase and tensin homolog; RAGE, receptor for advanced glycation end product; RBC, red blood cell; ROS, reactive oxygen species; RUNX2, runt-related transcription factor 2; SOD, superoxide dismutase; SP1, specificity protein 1; VEGF, vascular endothelial growth factor.
Abbreviations Used
- 3MP
3-mercaptopyruvate
- 3MST
3-mercaptopyruvate sulfurtransferase
- 8-nitro-cGMP
8-nitroguanosine 3′,5′-cyclic monophosphate
- Aβ
Abeta 1–42 peptide
- AD
Alzheimer's disease
- ALDH
aldehyde dehydrogenase
- BDP-NAC
bpin-disulfide prodrug-N-acetyl cysteine
- CARS
cysteinyl-tRNA synthetase
- CBS
cystathionine β-synthase
- COPD
chronic obstructive pulmonary disease
- CSE
cystathionine γ-lyase
- DADS
diallyl disulfide
- DATS
dially trisulfide
- Dox
doxorubicin
- Drp1
dynamin-related protein 1
- DTNB
5,5′-dithiobis-(2-nitrobenzoic acid)
- DTT
dithiothreitol
- ECs
endothelial cells
- ER
endoplasmic reticulum
- EXOG
exo/endonuclease gene
- FRET
fluorescence resonance energy transfer
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GPx
glutathione peroxidase
- GSH
glutathione
- GSSH
glutathione persulfide
- HO
heme oxygenase
- IAA
iodoacetic acid
- IAM
iodoacetamide
- IAP
iodoacetamide-linked biotin
- IL
interleukin
- I/R
ischemia/reperfusion
- KATP
ATP-sensitive potassium channel
- KCa
calcium-activated potassium channel
- Keap1
Kelch-like ECH-associated protein 1
- LDHA
lactate dehydrogenase A
- LMW
low molecular weight
- LTP
long-term potentiation
- MeHg
methyl mercury
- MGO
methylglyoxal
- MMTS
methyl methanethiosulfonate
- MSBT
methylsulfonyl benzothiazole
- MSCs
mesenchymal stem cells
- NEM
N-ethyl maleimide
- NF
nuclear factor
- NMDA
N-methyl-d-aspartate
- Nrf2
nuclear factor-like 2
- PC
pyruvate carboxylase
- PD
Parkinson's disease
- PDE5
phosphodiesterase type 5
- PKA
protein kinase A
- PKG
cGMP-dependent protein kinase
- PPAR
peroxisome proliferator-activated receptor
- PTEN
phosphatase and tensin homolog
- RAGE
receptor for advanced glycation end product
- RBC
red blood cell
- redox
reduction/oxidation
- ROS
reactive oxygen species
- RUNX2
runt-related transcription factor 2
- RXR
retinoid X receptor
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SILAC
stable isotope labeling with amino acid in culture
- SOD
superoxide dismutase
- SP1
specificity protein 1
- SQR
sulfide quinone oxidoreductase
- SR
sarcoplasmic reticulum
- TRP
transient receptor potential
- VEGF
vascular endothelial growth factor
Funding Information
This work was supported by the NIH (R01GM125968, R21DA046386) to M.X.; Natural Science Foundation of Guangdong Province Grant (2017A030313892) to C.-t.Y.
References
- 1. Akaike T, Ida T, Wei FY, Nishida M, Kumagai Y, Alam MM, Ihara H, Sawa T, Matsunaga T, Kasamatsu S, Nishimura A, Morita M, Tomizawa K, Nishimura A, Watanabe S, Inaba K, Shima H, Tanuma N, Jung M, Fujii S, Watanabe Y, Ohmuraya M, Nagy P, Feelisch M, Fukuto JM, and Motohashi H. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat Commun 8: 1177, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akaike T, Nishida M, and Fujii S. Regulation of redox signalling by an electrophilic cyclic nucleotide. J Biochem 153: 131–138, 2013 [DOI] [PubMed] [Google Scholar]
- 3. Artaud I and Galardon E. A persulfide analogue of the nitrosothiol SNAP: formation, characterization and reactivity. Chembiochem 15: 2361–2364, 2014 [DOI] [PubMed] [Google Scholar]
- 4. Baez NO, Reisz JA, and Furdui CM. Mass spectrometry in studies of protein thiol chemistry and signaling: opportunities and caveats. Free Radic Biol Med 80: 191–211, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bailey TS and Pluth MD. Reactions of isolated persulfides provide insights into the interplay between H2S and persulfide reactivity. Free Radic Biol Med 89: 662–667, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bailey TS, Zakharov LN, and Pluth MD. Understanding hydrogen sulfide storage: probing conditions for sulfide release from hydrodisulfides. J Am Chem Soc 136: 10573–10576, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beltowski J and Jamroz-Wisniewska A. Hydrogen sulfide and endothelium-dependent vasorelaxation. Molecules 19: 21183–21199, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bianco CL, Chavez TA, Sosa V, Saund SS, Nguyen QNN, Tantillo DJ, Ichimura AS, Toscano JP, and Fukuto JM. The chemical biology of the persulfide (RSSH)/perthiyl (RSS.) redox couple and possible role in biological redox signaling. Free Radic Biol Med 101: 20–31, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bibli SI, Hu J, Sigala F, Wittig I, Heidler J, Zukunft S, Tsilimigras DI, Randriamboavonjy V, Wittig J, Kojonazarov B, Schurmann C, Siragusa M, Siuda D, Luck B, Abdel Malik R, Filis KA, Zografos G, Chen C, Wang DW, Pfeilschifter J, Brandes RP, Szabo C, Papapetropoulos A, and Fleming I. Cystathionine gamma lyase sulfhydrates the RNA binding protein HuR to preserve endothelial cell function and delay atherogenesis. Circulation 139: 101–114, 2019 [DOI] [PubMed] [Google Scholar]
- 10. Blatter LA and Wier WG. Nitric oxide decreases [Ca2+]i in vascular smooth muscle by inhibition of the calcium current. Cell Calcium 15: 122–131, 1994 [DOI] [PubMed] [Google Scholar]
- 11. Bogdandi V, Ida T, Sutton TR, Bianco C, Ditroi T, Koster G, Henthorn HA, Minnion M, Toscano JP, van der Vliet A, Pluth MD, Feelisch M, Fukuto JM, Akaike T, and Nagy P. Speciation of reactive sulfur species and their reactions with alkylating agents: do we have any clue about what is present inside the cell? Br J Pharmacol 176: 646–670, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boolell M, Gepi-Attee S, Gingell JC, and Allen MJ. Sildenafil, a novel effective oral therapy for male erectile dysfunction. Br J Urol 78: 257–261, 1996 [DOI] [PubMed] [Google Scholar]
- 13. Bora P, Chauhan P, Manna S, and Chakrapani H. A vinyl-boronate ester-based persulfide donor controllable by hydrogen peroxide, a reactive oxygen species (ROS). Org Lett 20 7916–7920, 2018 [DOI] [PubMed] [Google Scholar]
- 14. Briggs RG and Fee JA. Sulfhydryl reactivity of human erythrocyte superoxide dismutase. On the origin of the unusual spectral properties of the protein when prepared by a procedure utilizing chloroform and ethanol for the precipitation of hemoglobin. Biochim Biophys Acta 537: 100–109, 1978 [DOI] [PubMed] [Google Scholar]
- 15. Broniowska KA and Hogg N. The chemical biology of S-nitrosothiols. Antioxid Redox Signal 17: 969–980, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burkey T, Hawari J, Lossing F, Lusztyk J, Sutcliffe R, and Griller D. The tert-butylperthiyl radical. J Org Chem 50: 4966–4967, 1985 [Google Scholar]
- 17. Cai J, Shi X, Wang H, Fan J, Feng Y, Lin X, Yang J, Cui Q, Tang C, Xu G, and Geng B. Cystathionine gamma lyase-hydrogen sulfide increases peroxisome proliferator-activated receptor gamma activity by sulfhydration at C139 site thereby promoting glucose uptake and lipid storage in adipocytes. Biochim Biophys Acta 1861: 419–429, 2016 [DOI] [PubMed] [Google Scholar]
- 18. Cai YR and Hu CH. Computational study of H2S release in reactions of diallyl polysulfides with thiols. J Phys Chem B 121: 6359–6366, 2017 [DOI] [PubMed] [Google Scholar]
- 19. Cao Y, Gomes SA, Rangel EB, Paulino EC, Fonseca TL, Li J, Teixeira MB, Gouveia CH, Bianco AC, Kapiloff MS, Balkan W, and Hare JM. S-Nitrosoglutathione reductase-dependent PPARgamma denitrosylation participates in MSC-derived adipogenesis and osteogenesis. J Clin Invest 125: 1679–1691, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chauvin JR, Griesser M, and Pratt DA. Hydropersulfides: H-Atom transfer agents Par excellence. J Am Chem Soc 139: 6484–6493, 2017 [DOI] [PubMed] [Google Scholar]
- 21. Cheng Y, Ndisang JF, Tang G, Cao K, and Wang R. Hydrogen sulfide-induced relaxation of resistance mesenteric artery beds of rats. Am J Physiol Heart Circ Physiol 287: H2316–H2323, 2004 [DOI] [PubMed] [Google Scholar]
- 22. Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, and Dawson TM. S-Nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science 304: 1328–1331, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Coves J, Le Hir de Fallois L, Le Pape L, Decout JL, and Fontecave M. Inactivation of Escherichia coli ribonucleotide reductase by 2'-deoxy-2'-mercaptouridine 5'-diphosphate. Electron paramagnetic resonance evidence for a transient protein perthiyl radical. Biochemistry 35: 8595–8602, 1996 [DOI] [PubMed] [Google Scholar]
- 24. Cuevasanta E, Lange M, Bonanata J, Coitino EL, Ferrer-Sueta G, Filipovic MR, and Alvarez B. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J Biol Chem 290: 26866–26880, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dawson TM and Dawson VL. The role of parkin in familial and sporadic Parkinson's disease. Mov Disord 25(Suppl 1): S32–S39, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Beus MD, Chung J, and Colon W. Modification of cysteine 111 in Cu/Zn superoxide dismutase results in altered spectroscopic and biophysical properties. Protein Sci 13: 1347–1355, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Denes F, Pichowicz M, Povie G, and Renaud P. Thiyl radicals in organic synthesis. Chem Rev 114: 2587–2693, 2014 [DOI] [PubMed] [Google Scholar]
- 28. Devarie-Baez NO, Zhang D, Li S, Whorton AR, and Xian M. Direct methods for detection of protein S-nitrosylation. Methods 62: 171–176, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doka E, Pader I, Biro A, Johansson K, Cheng Q, Ballago K, Prigge JR, Pastor-Flores D, Dick TP, Schmidt EE, Arner ES, and Nagy P. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci Adv 2: e1500968, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du C, Lin X, Xu W, Zheng F, Cai J, Yang J, Cui Q, Tang C, Cai J, Xu G, and Geng B. Sulfhydrated sirtuin-1 increasing its deacetylation activity is an essential epigenetics mechanism of anti-atherogenesis by hydrogen sulfide. Antioxid Redox Signal 30: 184–197, 2019 [DOI] [PubMed] [Google Scholar]
- 31. Everett SA and Wardman P. Perthiols as antioxidants: radical-scavenging and prooxidative mechanisms. Methods Enzymol 251: 55–69, 1995 [DOI] [PubMed] [Google Scholar]
- 32. Filipovic MR, Zivanovic J, Alvarez B, and Banerjee R. Chemical biology of H2S signaling through persulfidation. Chem Rev 118: 1253–1337, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Filosa JA, Yao X, and Rath G. TRPV4 and the regulation of vascular tone. J Cardiovasc Pharmacol 61: 113–119, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Francis SH, Busch JL, Corbin JD, and Sibley D. cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol Rev 62: 525–563, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Francoleon NE, Carrington SJ, and Fukuto JM. The reaction of H2S with oxidized thiols: generation of persulfides and implications to H2S biology. Arch Biochem Biophys 516: 146–153, 2011 [DOI] [PubMed] [Google Scholar]
- 36. Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, Wang B, Bebek G, Evans CR, Fox PL, Gerson SL, Hoppel CL, Liu M, Arvan P, and Hatzoglou M. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife 4: e10067, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Greiner R and Dick TP. Real-time assays for monitoring the influence of sulfide and sulfane sulfur species on protein thiol redox states. Methods Enzymol 555: 57–77, 2015 [DOI] [PubMed] [Google Scholar]
- 38. Greiner R, Palinkas Z, Basell K, Becher D, Antelmann H, Nagy P, and Dick TP. Polysulfides link H2S to protein thiol oxidation. Antioxid Redox Signal 19: 1749–1765, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo C, Liang F, Shah Masood W, and Yan X. Hydrogen sulfide protected gastric epithelial cell from ischemia/reperfusion injury by Keap1 S-sulfhydration, MAPK dependent anti-apoptosis and NF-kappaB dependent anti-inflammation pathway. Eur J Pharmacol 725: 70–78, 2014 [DOI] [PubMed] [Google Scholar]
- 40. Heimer NE, Field L, and Waites JA. Organic disulfides and related substances. 44. Preparation and characterization of 1-adamantyl hydrodisulfide as a stable prototype of the series. J Org Chem 50: 4164–4166, 1985 [Google Scholar]
- 41. Hosoki R, Matsuki N, and Kimura H. The possible role of hydrogen sulfide as an endogenous smooth muscle relaxant in synergy with nitric oxide. Biochem Biophys Res Commun 237: 527–531, 1997 [DOI] [PubMed] [Google Scholar]
- 42. Hourihan JM, Kenna JG, and Hayes JD. The gasotransmitter hydrogen sulfide induces Nrf2-target genes by inactivating the Keap1 ubiquitin ligase substrate adaptor through formation of a disulfide bond between Cys-226 and Cys-613. Antioxid Redox Signal 19: 465–481, 2013 [DOI] [PubMed] [Google Scholar]
- 43. Huang P, Shen Z, Liu J, Huang Y, Chen S, Yu W, Wang S, Ren Y, Li X, Tang C, Du J, and Jin H. Hydrogen sulfide inhibits high-salt diet-induced renal oxidative stress and kidney injury in Dahl rats. Oxid Med Cell Longev 2016: 2807490, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Iciek M, Gorny M, Bilska-Wilkosz A, and Kowalczyk-Pachel D. Is aldehyde dehydrogenase inhibited by sulfur compounds? In vitro and in vivo studies. Acta Biochim Pol 65: 125–132, 2018 [DOI] [PubMed] [Google Scholar]
- 45. Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, and Akaike T. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci U S A 111: 7606–7611, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ihara H, Kasamatsu S, Kitamura A, Nishimura A, Tsutsuki H, Ida T, Ishizaki K, Toyama T, Yoshida E, Abdul Hamid H, Jung M, Matsunaga T, Fujii S, Sawa T, Nishida M, Kumagai Y, and Akaike T. Exposure to electrophiles impairs reactive persulfide-dependent redox signaling in neuronal cells. Chem Res Toxicol 30: 1673–1684, 2017 [DOI] [PubMed] [Google Scholar]
- 47. Jiang B, Tang G, Cao K, Wu L, and Wang R. Molecular mechanism for H2S-induced activation of KATP channels. Antioxid Redox Signal 12: 1167–1178, 2010 [DOI] [PubMed] [Google Scholar]
- 48. Ju Y, Fu M, Stokes E, Wu L, and Yang G. H2S-mediated protein S-sulfhydration: a prediction for its formation and regulation. Molecules 22: 1334, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ju Y, Untereiner A, Wu L, and Yang G. H2S-induced S-sulfhydration of pyruvate carboxylase contributes to gluconeogenesis in liver cells. Biochim Biophys Acta 1850: 2293–2303, 2015 [DOI] [PubMed] [Google Scholar]
- 50. Kakinohana M, Marutani E, Tokuda K, Kida K, Kosugi S, Kasamatsu S, Magliocca A, Ikeda K, Kai S, and Sakaguchi M. Breathing hydrogen sulfide prevents delayed paraplegia in mice. Free Radic Biol Med 131: 243–250, 2019 [DOI] [PubMed] [Google Scholar]
- 51. Kang J, Ferrell AJ, Chen W, Wang D, and Xian M. Cyclic acyl disulfides and acyl selenylsulfides as the precursors for persulfides (RSSH), selenylsulfides (RSeSH), and hydrogen sulfide (H2S). Org Lett 20: 852–855, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kang J, Xu S, Radford MN, Zhang W, Kelly SS, Day JJ, and Xian M. O—>S relay deprotection: a general approach to controllable donors of reactive sulfur species. Angew Chem Int Ed Engl 57: 5893–5897, 2018 [DOI] [PubMed] [Google Scholar]
- 53. Kang M, Hashimoto A, Gade A, and Akbarali HI. Interaction between hydrogen sulfide-induced sulfhydration and tyrosine nitration in the KATP channel complex. Am J Physiol Gastrointest Liver Physiol 308: G532–G539, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kawagoe R, Takashima I, Uchinomiya S, and Ojida A. Reversible ratiometric detection of highly reactive hydropersulfides using a FRET-based dual emission fluorescent probe. Chem Sci 8: 1134–1140, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Khan S, Fujii S, Matsunaga T, Nishimura A, Ono K, Ida T, Ahmed KA, Okamoto T, Tsutsuki H, Sawa T, and Akaike T. Reactive persulfides from Salmonella typhimurium downregulate autophagy-mediated innate immunity in macrophages by inhibiting electrophilic signaling. Cell Chem Biol: 1403–1413, 2018 [DOI] [PubMed] [Google Scholar]
- 56. Khodade VS and Toscano JP. Development of S-substituted thioisothioureas as efficient hydropersulfide precursors. J Am Chem Soc 140: 17333–17337, 2018 [DOI] [PubMed] [Google Scholar]
- 57. Kimura Y, Koike S, Shibuya N, Lefer D, Ogasawara Y, and Kimura H. 3-Mercaptopyruvate sulfurtransferase produces potential redox regulators cysteine- and glutathione-persulfide (Cys-SSH and GSSH) together with signaling molecules H2S2, H2S3 and H2S. Sci Rep 7: 10459, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kimura Y, Mikami Y, Osumi K, Tsugane M, Oka J, and Kimura H. Polysulfides are possible H2S-derived signaling molecules in rat brain. FASEB J 27: 2451–2457, 2013 [DOI] [PubMed] [Google Scholar]
- 59. Kimura Y, Toyofuku Y, Koike S, Shibuya N, Nagahara N, Lefer D, Ogasawara Y, and Kimura H. Identification of H2S3 and H2S produced by 3-mercaptopyruvate sulfurtransferase in the brain. Sci Rep 5: 14774, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kiss L, Deitch EA, and Szabo C. Hydrogen sulfide decreases adenosine triphosphate levels in aortic rings and leads to vasorelaxation via metabolic inhibition. Life Sci 83: 589–594, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koike S, Kayama T, Yamamoto S, Komine D, Tanaka R, Nishimoto S, Suzuki T, Kishida A, and Ogasawara Y. Polysulfides protect SH-SY5Y cells from methylglyoxal-induced toxicity by suppressing protein carbonylation: a possible physiological scavenger for carbonyl stress in the brain. Neurotoxicology 55: 13–19, 2016 [DOI] [PubMed] [Google Scholar]
- 62. Koike S, Nishimoto S, and Ogasawara Y. Cysteine persulfides and polysulfides produced by exchange reactions with H2S protect SH-SY5Y cells from methylglyoxal-induced toxicity through Nrf2 activation. Redox Biol 12: 530–539, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koppenol WH and Bounds PL. Signaling by sulfur-containing molecules. Quantitative aspects. Arch Biochem Biophys 617: 3–8, 2017 [DOI] [PubMed] [Google Scholar]
- 64. Krishnan N, Fu C, Pappin DJ, and Tonks NK. H2S-induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci Signal 4: ra86, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lau N and Pluth MD. Reactive sulfur species (RSS): persulfides, polysulfides, potential, and problems. Curr Opin Chem Biol 49: 1–8, 2019 [DOI] [PubMed] [Google Scholar]
- 66. Li YL, Wu PF, Chen JG, Wang S, Han QQ, Li D, Wang W, Guan XL, Li D, Long LH, Huang JG, and Wang F. Activity-dependent sulfhydration signal controls N-methyl-D-aspartate subtype glutamate receptor-dependent synaptic plasticity via increasing D-serine availability. Antioxid Redox Signal 27: 398–414, 2017 [DOI] [PubMed] [Google Scholar]
- 67. Li YL, Zhou J, Zhang H, Luo Y, Long LH, Hu ZL, Chen JG, Wang F, and Wu PF. Hydrogen sulfide promotes surface insertion of hippocampal AMPA receptor GluR1 subunit via phosphorylating at serine-831/serine-845 sites through a sulfhydration-dependent mechanism. CNS Neurosci Ther 22: 789–798, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liang D, Wu H, Wong MW, and Huang D. Diallyl trisulfide is a fast H2S donor, but diallyl disulfide is a slow one: the reaction pathways and intermediates of glutathione with polysulfides. Org Lett 17: 4196–4199, 2015 [DOI] [PubMed] [Google Scholar]
- 69. Lima B, Forrester MT, Hess DT, and Stamler JS. S-Nitrosylation in cardiovascular signaling. Circ Res 106: 633–646, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lin Z, Altaf N, Li C, Chen M, Pan L, Wang D, Xie L, Zheng Y, Fu H, Han Y, and Ji Y. Hydrogen sulfide attenuates oxidative stress-induced NLRP3 inflammasome activation via S-sulfhydrating c-Jun at Cys269 in macrophages. Biochim Biophys Acta Mol Basis Dis 1864: 2890–2900, 2018 [DOI] [PubMed] [Google Scholar]
- 71. Liu DH, Huang X, Meng XM, Zhang CM, Lu HL, Kim YC, and Xu WX. Exogenous H2S enhances mice gastric smooth muscle tension through S-sulfhydration of KV 4.3, mediating the inhibition of the voltage-dependent potassium current. Neurogastroenterol Motil 26: 1705–1716, 2014 [DOI] [PubMed] [Google Scholar]
- 72. Liu Y, Yang R, Liu X, Zhou Y, Qu C, Kikuiri T, Wang S, Zandi E, Du J, Ambudkar IS, and Shi S. Hydrogen sulfide maintains mesenchymal stem cell function and bone homeostasis via regulation of Ca2+ channel sulfhydration. Cell Stem Cell 15: 66–78, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Longen S, Richter F, Kohler Y, Wittig I, Beck KF, and Pfeilschifter J. Quantitative persulfide site identification (qPerS-SID) reveals protein targets of H2S releasing donors in mammalian cells. Sci Rep 6: 29808, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Luo H, Wu PF, Han QQ, Cao Y, Deng SL, Wang J, Deng Q, Wang F, and Chen JG. Reactive sulfur species emerge as gliotransmitters to support memory via sulfuration-dependent gating of NR2A-containing N-methyl-D-aspartate subtype glutamate receptor function. Antioxid Redox Signal 30: 1880–1899, 2019 [DOI] [PubMed] [Google Scholar]
- 75. Marutani E, Yamada M, Ida T, Tokuda K, Ikeda K, Kai S, Shirozu K, Hayashida K, Kosugi S, Hanaoka K, Kaneki M, Akaike T, and Ichinose F. Thiosulfate mediates cytoprotective effects of hydrogen sulfide against neuronal ischemia. J Am Heart Assoc 4: e002125, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Meng G, Xiao Y, Ma Y, Tang X, Xie L, Liu J, Gu Y, Yu Y, Park CM, Xian M, Wang X, Ferro A, Wang R, Moore PK, Zhang Z, Wang H, Han Y, and Ji Y. Hydrogen sulfide regulates Kruppel-Like Factor 5 transcription activity via Specificity Protein 1 S-sulfhydration at Cys664 to prevent myocardial hypertrophy. J Am Heart Assoc 5: e004160, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mir S, Sen T, and Sen N. Cytokine-induced GAPDH sulfhydration affects PSD95 degradation and memory. Mol Cell 56: 786–795, 2014 [DOI] [PubMed] [Google Scholar]
- 78. Mishanina TV, Yadav PK, Ballou DP, and Banerjee R. Transient kinetic analysis of hydrogen sulfide oxidation catalyzed by human sulfide quinone oxidoreductase. J Biol Chem 290: 25072–25080, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Modis K, Ju Y, Ahmad A, Untereiner AA, Altaany Z, Wu L, Szabo C, and Wang R. S-Sulfhydration of ATP synthase by hydrogen sulfide stimulates mitochondrial bioenergetics. Pharmacol Res 113: 116–124, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, and Snyder SH. H2S signals through protein S-sulfhydration. Sci Signal 2: ra72, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mustafa AK, Sikka G, Gazi SK, Steppan J, Jung SM, Bhunia AK, Barodka VM, Gazi FK, Barrow RK, Wang R, Amzel LM, Berkowitz DE, and Snyder SH. Hydrogen sulfide as endothelium-derived hyperpolarizing factor sulfhydrates potassium channels. Circ Res 109: 1259–1268, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Naik JS, Osmond JM, Walker BR, and Kanagy NL. Hydrogen sulfide-induced vasodilation mediated by endothelial TRPV4 channels. Am J Physiol Heart Circ Physiol 311: H1437–H1444, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nalli AD, Wang H, Bhattacharya S, Blakeney BA, and Murthy KS. Inhibition of RhoA/Rho kinase pathway and smooth muscle contraction by hydrogen sulfide. Pharmacol Res Perspect 5: e00343, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nishida M, Nishimura A, Matsunaga T, Motohashi H, Kasamatsu S, and Akaike T. Redox regulation of electrophilic signaling by reactive persulfides in cardiac cells. Free Radic Biol Med 109: 132–140, 2017 [DOI] [PubMed] [Google Scholar]
- 85. Nishida M, Sawa T, Kitajima N, Ono K, Inoue H, Ihara H, Motohashi H, Yamamoto M, Suematsu M, Kurose H, van der Vliet A, Freeman BA, Shibata T, Uchida K, Kumagai Y, and Akaike T. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat Chem Biol 8: 714–724, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Numakura T, Sugiura H, Akaike T, Ida T, Fujii S, Koarai A, Yamada M, Onodera K, Hashimoto Y, Tanaka R, Sato K, Shishikura Y, Hirano T, Yanagisawa S, Fujino N, Okazaki T, Tamada T, Hoshikawa Y, Okada Y, and Ichinose M. Production of reactive persulfide species in chronic obstructive pulmonary disease. Thorax 72: 1074–1083, 2017 [DOI] [PubMed] [Google Scholar]
- 87. Ohno K, Okuda K, and Uehara T. Endogenous S-sulfhydration of PTEN helps protect against modification by nitric oxide. Biochem Biophys Res Commun 456: 245–249, 2015 [DOI] [PubMed] [Google Scholar]
- 88. Ono K, Akaike T, Sawa T, Kumagai Y, Wink DA, Tantillo DJ, Hobbs AJ, Nagy P, Xian M, Lin J, and Fukuto JM. Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic Biol Med 77: 82–94, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pan J and Carroll KS. Persulfide reactivity in the detection of protein S-sulfhydration. ACS Chem Biol 8: 1110–1116, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Panatier A, Theodosis DT, Mothet JP, Touquet B, Pollegioni L, Poulain DA, and Oliet SH. Glia-derived D-serine controls NMDA receptor activity and synaptic memory. Cell 125: 775–784, 2006 [DOI] [PubMed] [Google Scholar]
- 91. Parast CV, Wong KK, Kozarich JW, Peisach J, and Magliozzo RS. Electron paramagnetic resonance evidence for a cysteine-based radical in pyruvate formate-lyase inactivated with mercaptopyruvate. Biochemistry 34: 5712–5717, 1995 [DOI] [PubMed] [Google Scholar]
- 92. Park CM, Johnson BA, Duan J, Park JJ, Day JJ, Gang D, Qian WJ, and Xian M. 9-Fluorenylmethyl (Fm) disulfides: biomimetic precursors for persulfides. Org Lett 18: 904–907, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Powell CR, Dillon KM, Wang Y, Carrazzone RJ, and Matson JB. A persulfide donor responsive to reactive oxygen species: insights into reactivity and therapeutic potential. Angew Chem Int Ed Engl 57: 6324–6328, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Qian Q, Zhang Z, Orwig A, Chen S, Ding WX, Xu Y, Kunz RC, Lind NRL, Stamler JS, and Yang L. S-Nitrosoglutathione reductase dysfunction contributes to obesity-associated hepatic insulin resistance via regulating autophagy. Diabetes 67: 193–207, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rahman I. Oxidative stress and gene transcription in asthma and chronic obstructive pulmonary disease: antioxidant therapeutic targets. Curr Drug Targets Inflamm Allergy 1: 291–315, 2002 [DOI] [PubMed] [Google Scholar]
- 96. Raines KW, Cao GL, Lee EK, Rosen GM, and Shapiro P. Neuronal nitric oxide synthase-induced S-nitrosylation of H-Ras inhibits calcium ionophore-mediated extracellular-signal-regulated kinase activity. Biochem J 397: 329–336, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Roger T, Raynaud F, Bouillaud F, Ransy C, Simonet S, Crespo C, Bourguignon MP, Villeneuve N, Vilaine JP, Artaud I, and Galardon E. New biologically active hydrogen sulfide donors. Chembiochem 14: 2268–2271, 2013 [DOI] [PubMed] [Google Scholar]
- 98. Saha S, Chakraborty PK, Xiong X, Dwivedi SK, Mustafi SB, Leigh NR, Ramchandran R, Mukherjee P, and Bhattacharya R. Cystathionine beta-synthase regulates endothelial function via protein S-sulfhydration. FASEB J 30: 441–456, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sawahata T and Neal RA. Use of 1-fluoro-2,4-dinitrobenzene as a probe for the presence of hydrodisulfide groups in proteins. Anal Biochem 126: 360–364, 1982 [DOI] [PubMed] [Google Scholar]
- 100. Schoneich C. Mechanisms of protein damage induced by cysteine thiyl radical formation. Chem Res Toxicol 21: 1175–1179, 2008 [DOI] [PubMed] [Google Scholar]
- 101. Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, and Snyder SH. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell 45: 13–24, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Stephan D, Winkler M, Kuhner P, Russ U, and Quast U. Selectivity of repaglinide and glibenclamide for the pancreatic over the cardiovascular KATP channels. Diabetologia 49: 2039–2048, 2006 [DOI] [PubMed] [Google Scholar]
- 103. Stokes E, Shuang T, Zhang Y, Pei Y, Fu M, Guo B, Parissenti A, Wu L, Wang R, and Yang G. Efflux inhibition by H2S confers sensitivity to doxorubicin-induced cell death in liver cancer cells. Life Sci: 116–125, 2018 [DOI] [PubMed] [Google Scholar]
- 104. Sun J, Aponte AM, Menazza S, Gucek M, Steenbergen C, and Murphy E. Additive cardioprotection by pharmacological postconditioning with hydrogen sulfide and nitric oxide donors in mouse heart: S-sulfhydration vs. S-nitrosylation. Cardiovasc Res 110: 96–106, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sun Y, Huang Y, Yu W, Chen S, Yao Q, Zhang C, Bu D, Tang C, Du J, and Jin H. Sulfhydration-associated phosphodiesterase 5A dimerization mediates vasorelaxant effect of hydrogen sulfide. Oncotarget 8: 31888–31900, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Szczesny B, Marcatti M, Zatarain JR, Druzhyna N, Wiktorowicz JE, Nagy P, Hellmich MR, and Szabo C. Inhibition of hydrogen sulfide biosynthesis sensitizes lung adenocarcinoma to chemotherapeutic drugs by inhibiting mitochondrial DNA repair and suppressing cellular bioenergetics. Sci Rep 6: 36125, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Takahashi N, Wei FY, Watanabe S, Hirayama M, Ohuchi Y, Fujimura A, Kaitsuka T, Ishii I, Sawa T, Nakayama H, Akaike T, and Tomizawa K. Reactive sulfur species regulate tRNA methylthiolation and contribute to insulin secretion. Nucleic Acids Res 45: 435–445, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Takano Y, Hanaoka K, Shimamoto K, Miyamoto R, Komatsu T, Ueno T, Terai T, Kimura H, Nagano T, and Urano Y. Development of a reversible fluorescent probe for reactive sulfur species, sulfane sulfur, and its biological application. Chem Commun (Camb) 53: 1064–1067, 2017 [DOI] [PubMed] [Google Scholar]
- 109. Tang G, Yang G, Jiang B, Ju Y, Wu L, and Wang R. H2S is an endothelium-derived hyperpolarizing factor. Antioxid Redox Signal 19: 1634–1646, 2013 [DOI] [PubMed] [Google Scholar]
- 110. Umezawa K, Kamiya M, and Urano Y. A reversible fluorescent probe for real-time live-cell imaging and quantification of endogenous hydropolysulfides. Angew Chem Int Ed Engl 57: 9346–9350, 2018 [DOI] [PubMed] [Google Scholar]
- 111. Untereiner AA, Olah G, Modis K, Hellmich MR, and Szabo C. H2S-induced S-sulfhydration of lactate dehydrogenase a (LDHA) stimulates cellular bioenergetics in HCT116 colon cancer cells. Biochem Pharmacol 136: 86–98, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Untereiner AA, Wang R, Ju Y, and Wu L. Decreased gluconeogenesis in the absence of cystathionine gamma-lyase and the underlying mechanisms. Antioxid Redox Signal 24: 129–140, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Vandiver MS, Paul BD, Xu R, Karuppagounder S, Rao F, Snowman AM, Ko HS, Lee YI, Dawson VL, Dawson TM, Sen N, and Snyder SH. Sulfhydration mediates neuroprotective actions of parkin. Nat Commun 4: 1626, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Vitvitsky V, Miljkovic JL, Bostelaar T, Adhikari B, Yadav PK, Steiger AK, Torregrossa R, Pluth MD, Whiteman M, Banerjee R, and Filipovic MR. Cytochrome C reduction by H2S potentiates sulfide signaling. ACS Chem Biol 13: 2300–2307, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang P, Wu L, Ju Y, Fu M, Shuang T, Qian Z, and Wang R. Age-dependent allergic asthma development and cystathionine gamma-lyase deficiency. Antioxid Redox Signal 27: 931–944, 2017 [DOI] [PubMed] [Google Scholar]
- 116. Wang R. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev 92: 791–896, 2012 [DOI] [PubMed] [Google Scholar]
- 117. Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW, Kim K, and Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science 300: 653–656, 2003 [DOI] [PubMed] [Google Scholar]
- 118. Wood ZA, Poole LB, and Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300: 650–653, 2003 [DOI] [PubMed] [Google Scholar]
- 119. Xie L, Gu Y, Wen M, Zhao S, Wang W, Ma Y, Meng G, Han Y, Wang Y, Liu G, Moore PK, Wang X, Wang H, Zhang Z, Yu Y, Ferro A, Huang Z, and Ji Y. Hydrogen sulfide induces Keap1 S-sulfhydration and suppresses diabetes-accelerated atherosclerosis via Nrf2 activation. Diabetes 65: 3171–3184, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Xie ZZ, Shi MM, Xie L, Wu ZY, Li G, Hua F, and Bian JS. Sulfhydration of p66Shc at cysteine59 mediates the antioxidant effect of hydrogen sulfide. Antioxid Redox Signal 21: 2531–2542, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yang CT, Chen L, Xu S, Day JJ, Li X, and Xian M. Recent development of hydrogen sulfide releasing/stimulating reagents and their potential applications in cancer and glycometabolic disorders. Front Pharmacol 8: 664, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yang CT, Lai ZZ, Zheng ZH, Kang JM, Xian M, Wang RY, Shi K, Meng FH, Li X, Chen L, and Zhang H. A novel pH-controlled hydrogen sulfide donor protects gastric mucosa from aspirin-induced injury. J Cell Mol Med 21: 2441–2451, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Yang G, Zhao K, Ju Y, Mani S, Cao Q, Puukila S, Khaper N, Wu L, and Wang R. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid Redox Signal 18: 1906–1919, 2013 [DOI] [PubMed] [Google Scholar]
- 124. Yu B, Zheng Y, Yuan Z, Li S, Zhu H, De La Cruz LK, Zhang J, Ji K, Wang S, and Wang B. Toward direct protein S-persulfidation: a prodrug approach that directly delivers hydrogen persulfide. J Am Chem Soc 140: 30–33, 2018 [DOI] [PubMed] [Google Scholar]
- 125. Yuan S, Shen X, and Kevil CG. Beyond a gasotransmitter: hydrogen sulfide and polysulfide in cardiovascular health and immune response. Antioxid Redox Signal 27: 634–653, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yuan Z, Zheng Y, Yu B, Wang S, Yang X, and Wang B. Esterase-sensitive glutathione persulfide donor. Org Lett 20: 6364–6367, 2018 [DOI] [PubMed] [Google Scholar]
- 127. Zhang C, Biggs TD, Devarie-Baez NO, Shuang S, Dong C, and Xian M. S-Nitrosothiols: chemistry and reactions. Chem Commun (Camb) 53: 11266–11277, 2017 [DOI] [PubMed] [Google Scholar]
- 128. Zhang D, Du J, Tang C, Huang Y, and Jin H. H2S-Induced sulfhydration: biological function and detection methodology. Front Pharmacol 8: 608, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhang D, Macinkovic I, Devarie-Baez NO, Pan J, Park CM, Carroll KS, Filipovic MR, and Xian M. Detection of protein S-sulfhydration by a Tag-switch technique. Angew Chem Int Ed Engl 53: 575–581, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhang H, Zhuang XD, Meng FH, Chen L, Dong XB, Liu GH, Li JH, Dong Q, Xu JD, and Yang CT. Calcitriol prevents peripheral RSC96 Schwann neural cells from high glucose & methylglyoxal-induced injury through restoration of CBS/H2S expression. Neurochem Int 92: 49–57, 2016 [DOI] [PubMed] [Google Scholar]
- 131. Zhang L, Zhang X, Wu YD, Xie Y, Fukuto JM, and Schaefer HF. The reaction of alkyl hydropersulfides (RSSH, R = CH3 and tBu) with H2S in the gas phase and in aqueous solution. Phys Chem Chem Phys 21: 537–545, 2019 [DOI] [PubMed] [Google Scholar]
- 132. Zhao K, Ju Y, Li S, Altaany Z, Wang R, and Yang G. S-Sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair. EMBO Rep 15: 792–800, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Zhao W, Zhang J, Lu Y, and Wang R. The vasorelaxant effect of H2S as a novel endogenous gaseous KATP channel opener. EMBO J 20: 6008–6016, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhao Y, Bhushan S, Yang C, Otsuka H, Stein JD, Pacheco A, Peng B, Devarie-Baez NO, Aguilar HC, Lefer DJ, and Xian M. Controllable hydrogen sulfide donors and their activity against myocardial ischemia-reperfusion injury. ACS Chem Biol 8: 1283–1290, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zheng L, White RH, Cash VL, and Dean DR. Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry 33: 4714–4720, 1994 [DOI] [PubMed] [Google Scholar]
- 136. Zheng Y, Liao F, Lin X, Zheng F, Fan J, Cui Q, Yang J, Geng B, and Cai J. Cystathionine gamma-lyase-hydrogen sulfide induces Runt-related transcription factor 2 sulfhydration, thereby increasing osteoblast activity to promote bone fracture healing. Antioxid Redox Signal 27: 742–753, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zheng Y, Yu B, Li Z, Yuan Z, Organ CL, Trivedi RK, Wang S, Lefer DJ, and Wang B. An esterase-sensitive prodrug approach for controllable delivery of persulfide species. Angew Chem Int Ed Engl 56: 11749–11753, 2017 [DOI] [PubMed] [Google Scholar]
- 138. Zhou H, Ding L, Wu Z, Cao X, Zhang Q, Lin L, and Bian JS. Hydrogen sulfide reduces RAGE toxicity through inhibition of its dimer formation. Free Radic Biol Med 104: 262–271, 2017 [DOI] [PubMed] [Google Scholar]