Abstract
It is common practice for data providers to include text descriptions for each column when publishing datasets in the form of data dictionaries. While these documents are useful in helping an end-user properly interpret the meaning of a column in a dataset, existing data dictionaries typically are not machine-readable and do not follow a common specification standard. We introduce the Semantic Data Dictionary, a specification that formalizes the assignment of a semantic representation of data, enabling standardization and harmonization across diverse datasets. In this paper, we present our Semantic Data Dictionary work in the context of our work with biomedical data; however, the approach can and has been used in a wide range of domains. The rendition of data in this form helps promote improved discovery, interoperability, reuse, traceability, and reproducibility. We present the associated research and describe how the Semantic Data Dictionary can help address existing limitations in the related literature. We discuss our approach, present an example by annotating portions of the publicly available National Health and Nutrition Examination Survey dataset, present modeling challenges, and describe the use of this approach in sponsored research, including our work on a large NIH-funded exposure and health data portal and in the RPI-IBM collaborative Health Empowerment by Analytics, Learning, and Semantics project. We evaluate this work in comparison with traditional data dictionaries, mapping languages, and data integration tools.
Keywords: Semantic Data Dictionary, Dictionary Mapping, Codebook, Knowledge Modeling, Data Integration, Data Dictionary, Mapping Language, Metadata Standard, Semantic Web, Semantic ETL, FAIR, Data
1. Introduction
With the rapid expansion of data-driven applications and the expansion of data science research over the past decade, data providers and users alike have relied on datasets as a means for recording and accessing information from a variety of distinct domains. Datasets are composed of distinct structures that require additional information to help users understand the meaning of the data. A common approach used by data providers involves providing descriptive information for a dataset in the form of a data dictionary, defined as a “centralized repository of information about data such as meaning, relationships to other data, origin, usage, and format” [1]. Data dictionaries are useful for many data management tasks, including aiding users in data conversion processes, testing data generation, validating data, and storing data usage criteria [2].
When storing data into a system that adheres to the structure of a particular data dictionary, that document can be used to aid in validation both when inputting new data into the system or updating existing data. By including additional information about a dataset itself, data dictionaries can be used to store data usage criteria. Additionally, data conversion is aided by the inclusion of the format and units of the data points, which allows users to use conversion formulae to convert the data into another format or unit. When considering these benefits, we see that the use of data dictionaries has had a significant impact on data use and reuse. Nevertheless, we argue that data dictionaries can be improved by leveraging emerging Semantic Web technologies.
The use of data dictionaries to record descriptions about datasets and their elements has become widely adopted by data providers, often with the intent of aiding reusability. These data dictionaries are useful to data users in reducing ambiguity when interpreting dataset content. Considering the structure and annotations that traditional data dictionaries are comprised of, we find that for each column header in a dataset, these documents often contain a label that is more informative than the column name, as well as a comment describing the column header. Such annotations in themselves are essential for an end-user to understand the data, as column names are often arbitrary or encoded. Existing data dictionaries often contain structural information about a dataset column, such as the format of the data, the data type, or the associated units of measurement. As this information is required for the proper analysis of data, we commend data providers for including it in their data dictionaries. For datasets that contain categorical codes, data providers have done well to document the possible values and include descriptive labels for each category.
While many publicly available datasets include documents resembling data dictionaries, we find that, across institutions, these documents do not adhere to a common metadata standard. Metadata, defined as “structured data about data” [3], should be able to be processed using software. Existing data dictionary standards typically are aimed at human consumption and do not subscribe to models that are machine-understandable, and thus lack support for formal semantics. Consequently, tasks involving the combination of data from multiple datasets that are described using data dictionaries are not easily automated.
1.1. A need for semantics
From the dataset production perspective, datasets can convey much more information than the data itself. Dataset entries often correspond to physical observations, such as the weight of a sample, an event duration, or a person’s gender. Traditional data dictionaries do well in describing these measurements but cannot represent the measured objects. There is a need to annotate these implicit concepts (representing the measured objects) that are indispensable to a complete understanding of the data but do not correspond to columns in the dataset. Annotations of both explicit and implicit concepts allow for the conversion of a tabular format of data into a semantically richer graphical representation.
There may be a variety of ways that a data user can benefit from a semantic representation of data, such as enhanced provenance attributions, query capabilities, and the ability to infer new knowledge. We argue for the applicability of the Semantic Data Dictionary (SDD) as a standard model for representing machine-readable metadata for datasets. The SDD comprises a set of specifications formalizing the assignment of a semantic representation to data by annotating dataset columns and their values using concepts from best practice vocabularies and ontologies. It is a collection of individual documents, where each plays a role in creating a concise and consistent knowledge representation. Each of these components, described in Section 3, is implemented using tables. In Appendix B, we provide the specifications for each of the SDD tables. Throughout the remainder of this article, we describe modeling methods, include informative examples from projects employing this approach, discuss modeling challenges, and evaluate our approach against traditional data dictionaries, mapping languages, and data integration tools.
As science moves towards a more open approach, priority has been given to publishing scientific data in a way that is Findable, Accessible, Interoperable, and Reusable (FAIR) [4]. The FAIR principles are used to evaluate the quality of published datasets or the workflow that is used to produce data. As part of our approach to evaluating our methodology, we examine adherence to the FAIR guiding principles. While we have considered these guidelines in designing our approach, and they have been adopted for many projects, the FAIR principles are not without limitations. For example, methods for the facilitation of data sharing are not specified, which may result in error perpetuation from differing interpretations of design choices, and more vigorous privacy concerns need to be addressed [5]. The use of the FAIR guidelines and traditional data integration approaches alone do not guarantee enough granularity of representation to support the pooling of data across studies, thereby limiting the potential impact for more significant statistical analyses. However, this capability has been demonstrated using the SDD approach for the Children’s Health Exposure Analysis Resource (CHEAR) project [6].
1.2. Supporting biomedical research
While the SDD approach can and has been used for the semantic annotation of data in multiple domains, we will limit our examples in this paper to the field of biomedicine. The application of semantic technologies in areas like healthcare or the life sciences has the potential to facilitate scientific research in these fields. Many vocabularies and ontologies that define concepts and relationships in a formal graphical structure have been created to describe critical terms related to anatomy, genetics, diseases, and pharmaceuticals [7, 8]. Best practice ontologies should be leveraged for the annotation of biomedical and clinical data to create knowledge representations that align with existing semantic technologies, services, and workflows. Ideally, the desired representation model would allow for improved data discovery, interoperability, and reuse, while supporting provenance, trust, traceability, and reproducibility.
Challenges arise for biomedical researchers who are unfamiliar with approaches for performing semantic annotation. Existing methods to provide machine-understandable interpretations of data are difficult for most researchers to learn [9]. The biomedical community has traditionally used data dictionaries to provide information regarding the use of a dataset. While such documents are useful for a human interpreter, they generally cannot be used by themselves to automate the creation of a structured knowledge representation of the corresponding data. We recognize the need for an approach for annotating biomedical data that feels familiar to domain scientists while adhering to Semantic Web standards and machine-understandability. Since SDDs consist of tabular documents that resemble traditional data dictionaries, they can be used by biomedical scientists to annotate data naturally. In order to aid researchers who do not have a computer science background, we leverage the traits of SDDs, being both machine-readable and unambiguous, to provide interpretation software1 that can be used to create a knowledge model that meets the desired semantic representation characteristics mentioned above.
1.3. Motivation
In Section 2.1, we consider institutions that provide guidelines for the use of data dictionaries to record descriptive content for a dataset. While existing guidelines have helped create human-understandable documents, we believe that there is room for improvement by introducing a formalization that is machine-readable. With the current advances in Artificial Intelligence technologies, there is an increased need for data users to have annotated data that adhere to Semantic Web standards [10, 11]. We consider the benefits of combining data from disparate sources in such a way that it can be used in a unified manner. Harmonization across datasets allows for the comparison between similar columns, using a controlled vocabulary. The ability to combine data from various sources and formats into a single cohesive knowledge base allows for the implementation of innovative applications, such as faceted browsers or data visualizers.
Data and provenance understanding refer respectively to data interpretability and the ability to discern provenance attributions, both by humans and machines. This level of knowledge is necessary for the reuse of data and the reproduction of scientific experiments. Annotation of data improves query and integration capabilities [12], and the use of Semantic Web standards enhances the ability to find the data through a web search [13]. Unfortunately, it is difficult for data users, who have a second-hand understanding of the data compared to data providers, to create these annotations themselves. As an example, a study related to data dissemination revealed that three researchers, independently analyzing a single dataset and using similar approaches, arrived at noticeably dissimilar interpretive conclusions [14].
Additionally, difficulties arise for someone without a technology background to develop competence in technical approaches, due to challenges associated with technological semantics, such as research problems being defined, clarified, and communicated in a way that is perceptable by a general audience [15]. Therefore, the desire to create a standard for people from a wide variety of domains, including those who are untrained in Computer Science and semantic technologies, is an additional motivation. Easing the semantic annotation process for these users is a significant challenge. A machine-readable standard for dataset metadata can improve data harmonization, integration, reuse, and reproducibility.
1.4. Claims
We claim that the formalism of the Semantic Data Dictionary addresses some of the limitations of existing data dictionary approaches. Traditional data dictionaries provide descriptions about the columns of a dataset, which typically represent physical measurements or characteristics, but omit details about the described entities. Existing data dictionaries do not acknowledge the notion that the data values are instances of concepts that may have relationships with other instances of concepts, such as entity-entity, attribute-attribute, or entity-attribute relations.
In contrast, the SDD approach allows for the direct annotation of concepts implicitly referenced in a dataset. Existing data dictionaries focus on the structure of the data rather than the inherent meaning, including value ranges, formats, and data types. Further information about the data, including the units, meaning, and associated objects, is provided in text descriptions that are not machine-interpretable. The SDD, on the other hand, focuses on the semantics of the data and includes the above information in a way that is readily able to be processed. The SDD consists of an intrinsic model with relationships that can be further customized, allowing the annotator to describe relationships between both explicit and implicit concepts inherent in the dataset. By considering these characteristics of SDDs, we argue that a standardized machine-readable representation for recording dataset metadata and column information is achieved.
We also claim that the SDD approach presents a level of abstraction over methodologies that use mapping languages. This is achieved by simplifying the programming knowledge requirements by separating the annotation portion of the approach from the software component. As a result, the SDD approach improves the ease of use for a domain scientist over other semantic tools. Additionally, by presenting the annotation component in a form that resembles traditional data dictionaries, this approach provides a bridge between the conventional data dictionary approaches, used by domain scientists, and the formal techniques used by Semantic Web researchers.
2. Related Work
The SDD approach leverages state-of-the-art advancements in many data and knowledge related areas: traditional data dictionaries, data integration, mapping languages, semantic extract, transform, and load (ETL) methods, and metadata standards. In this section, we present related work in each of those extensive areas by highlighting their accomplishments and discussing their limitations.
2.1. Data Dictionaries
There are several patents relating to the use of dictionaries to organize metadata [16, 17, 18]. However, published articles mentioning data dictionaries tend to refrain from including the associated formalism. Thus, we expanded our scope to search for data dictionaries that included standards published on the web, several of which are discussed below.
The Stony Brook Data Governance Council recommendations list required elements and present principles associated with data dictionaries.2 However, the ability to semantically represent the data is not permitted. Additionally, while data columns can be explicitly described, this approach does not allow the description of implicit concepts that are being described by the dataset, which we refer to as object elicitation. The ability to annotate implicit concepts (described in Section 3.2) is one of the distinguishing features of our work. The Open Science Framework3 and the United States Government (USG) Statistical Community of Practice and Engagement (SCOPE)4 also guide the creation of a data dictionary that includes required, recommended, and optional entries. These data dictionaries support the specification of data types and categorical values, but minimally allow for the encorporation of semantics and do not leverage existing ontologies or vocabularies. The data dictionary specifications for the Biosystematic Database of World Diptera include both general and domain-specific elements [19]. Nevertheless, use of this data dictionary outside of the biological domain appears improbable. Based on the Data Catalog Vocabulary (DCAT [20]), the Project Open Data Metadata Schema provides a data dictionary specification.5 Of the data dictionaries’ recommendations examined, the Project Open Data Metadata Schema was the most general and the only one to use Semantic Web standards.
There are many recommendations for constructing data dictionaries; however, we found that most are project- or domain-specific, and we find no clear evidence that they are consistently applied by users outside of these individual groups. The exploration of these data dictionaries reveals the need for a standard formalization that can be used across institutions and projects.
2.2. Data Integration Approaches
Data integration is a technique that utilizes data from multiple sources to construct a unified view of the combined data [21]. Here we consider existing approaches that have been employed to address data integration challenges.
The Semantic Web Integration Tool (SWIT) can be used to perform transformation and integration of heterogeneous data through a web interface in a manner that adheres to the Linked Open Data (LOD) principles [22]. While the writing of mapping rules is simplified through the use of a web interface, the use of this approach may still prove difficult for users without a Semantic Web background. Neo4j is designed as a graph database (GDB) system that supports data integration based on the labeled property graph (LPG) model, which consists of attributed nodes with directed and labeled edges [23]. Despite being implemented using an LPG model rather than RDF, Neo4j can read and write RDF, and by using GraphScale [24], it can further employ reasoning capabilities [25]. Nevertheless, data integration capabilities, such as using ontologies to semantically annotate data schema concepts and the associated objects, are limited.
To provide an integrated view of data collected on moving entities in geographical locations, RDF-Gen was developed as a means of SPARQL-based knowledge graph generation from heterogeneous streaming and archival data sources [26]. While this approach is promising and does support the representation of implicit objects, we find, due to the requirement of creating SPARQL-based graph transformation mappings, that it would likely be difficult for domain scientists to use. DataOps is an integration toolkit that supports the combination of data in varying, different formats, including relational databases, CSV, Excel, and others, that can be accessed via R [27]. While existing user interface components can be used to ease the annotation process and the use of DataOps in industry is expanding, the expertise required to use this approach presents a steep learning curve. OpenRefine is a standalone, open-source tool capable of cleaning and transforming large datasets [28]. Some limitations of this approach pertain to difficulties in performing subset selection, cell-based operations, and dataset merging.
It is important to note that most data integration approaches fall short when eliciting objects and relations to comprehensively characterize the semantics of the data. We continue this discussion on data integration by considering mapping languages and semantic extract, transform, and load applications.
2.2.1. Mapping Languages
In this section, we introduce mapping languages that can be used to convert a relational database (RDB), tabular file, or hierarchical structure to an RDF format and their related tool support.
The RDB to RDF Mapping Language (R2RML) is a W3C standard language for expressing mappings from relational databases to RDF datasets [29]. R2RML mappings contain properties to define the components of the mapping, including the source table, columns retrieved using SQL queries, relationships between columns, and a template for the desired output URI structure. The R2RML limitations stem from the requirement of writing the mapping using RDF format, the need to be familiar with the R2RML vocabulary to write mappings, and the support for only relational databases. R2RML extensions exist to address these limitations. The RDF Mapping Language (RML) extends the R2RML vocabulary to support a broader set of possible input data formats, including CSV, XML, and JSON [30]. In this regard, RML extends the R2RML logical table class to be instead defined as a logical source, which allows the user to specify the source URI, reference, reference formulation, and iterator. RML is supported by a tool to define mappings called the RMLEditor, which allows users to make edits to heterogeneous data source mappings using a graphical user interface (GUI) [31]. Both R2RML and RML are robust and provide a solid cornerstone for general RDF generation from tabular data. Still, they fall short when dealing with some particularities of our problem scenario, including the creation of implicit relationships for elicited objects and the annotation of categorical data values. The xR2RML language leverages RML to expand the R2RML vocabulary to support the increase of several RDF data formats as well as the mapping of non-relational databases [32]. With the use of R2RML mappings, the OpenLink Virtuoso Universal Server has an extension to import relational databases or CSV files that can then transform into RDF [33]. Due to the usage requirement of a mapping language to specify graph transformations, a domain scientist may be reluctant to employ the above approaches.
KR2RML is an extension to R2RML addressing several of its limitations, including support for multiple input and output data formats, new serialization formats, transformations and modeling that do not rely on knowledge about domain-specific languages, and scalability when handling large amounts of data [34]. KR2RML is implemented in an open-source application called Karma. Karma is a system that uses semantics to integrate data by allowing users to import data from a variety of sources, clean and normalize the data, and create semantic descriptions for each of the data sources used [35]. Karma includes a visual interface that helps automate parts of the modeling process by suggesting proposed mappings based on semantic type assignments, and hence reduces some of the usage barriers associated with other mapping language methodologies. Nevertheless, some distinguishing factors between this and our approach include the following: when using the SDD approach, there is no need to write mapping transformation rules, and through the use of the Codebook (described in Section 3.3), the SDD approach supports cell value annotation.
CSV2RDF is a W3C standard for converting tabular data into RDF [36]. Introduced to address the limitation of R2RML that only relational data could be mapped, CSV2RDF extends R2RML to allow the mapping of additional structured data formats, such as CSV, TSV, XML and JSON [37]. The applicability of CSV2RDF for converting large amounts of data has been demonstrated using publicly available resources from a data portal [38]. CSV2RDF has also been used in an approach to automatically convert tabular data to RDF [39].
The Sparqlification Mapping Language (SML) progresses towards a formal model for RDB2RDF mappings, maintaining the same expressiveness as R2RML while simplifying usage by providing a more concise syntax, achieved by combining traditional SQL CREATE VIEW statements with SPARQL CONSTRUCT queries [40]. SML is intended to be a more human-readable mapping language than R2RML. The R2R Mapping Language, also based on SPARQL, is designed for writing dataset mappings represented as RDF using dereferenceable URIs [41]. While it is possible for the user to specify metadata about each mapping, the possible mappings that can be specified correspond to direct translations between the data and the vocabulary being used, rather than allowing for detailed object elicitation.
Another mapping language based on SPARQL is Tarql, where databases are referenced in FROM clauses, mappings can be specified using a SELECT or ASK clause, and RDF can be generated using a CONSTRUCT clause [42]. One limitation of this approach is that it uses SPARQL notation for tasks that were not originally intended by the grammar, rather than extending SPARQL with additional keywords. The D2RQ mapping language, which allows for querying on mapped databases using SPARQL, is a declarative language that allows for querying through the use of the RDF Data Query Language (RDQL), publication of a database on the Semantic Web with the RDF Net API, reasoning over database content using the Jena ontology API, and accessing database information through the Jena model API [43]. Some limitations of D2RQ include integration capabilities over multiple databases, write operations such as CREATE, DELETE, or UPDATE, and support for Named Graphs [44].
While many of the mapping languages above focus on the conversion of RDBs to knowledge graphs, RDB2OWL is a high-level declarative RDB-to-RDF/OWL mapping language used to generate ontologies from RDBs [45]. It is achieved by mapping the target ontology to the database structure. RDB2OWL supports the reuse of RDB table column and key information, includes an intuitive human-readable syntax for mapping expressions, allows for both built-in and user-defined functions, incorporates advanced mapping definition primitives, and allows for the utilization of auxiliary structures defined at the SQL level [45].
In addition to the difficulties associated with writing mapping transformations, we find that mapping-language-based methodologies have limited object and relation elicitation capabilities, and cell value annotation is typically not permitted. These limitations are addressed in the SDD approach.
2.2.2. Semantic Extract, Transform, & Load
Extract, transform, and load (ETL) operations refer to processes that read data from a source database, convert the data into another format, and write the data into a target database. In this section, we examine several ETL approaches that leverage semantic technologies. LinkedPipes ETL (LP-ETL) is a lightweight, linked data preparation tool supporting SPARQL queries, including debug capabilities, and can be integrated into external platforms [46]. LP-ETL contains both back-end software for performing data transformations, as well as a front-end web application that includes a pipeline editor and an execution monitor. A pipeline is defined as “a repeatable data transformation process consisting of configurable components, each responsible for an atomic data transformation task” [46]. As transformations in this approach are typically written as SPARQL construct statements, this methodology would be difficult to employ for someone who is unfamiliar with SPARQL. Semantic Extract, Transform, and Load-er (SETLr) is a scalable tool that uses the JSON-LD Template (JSLDT) language6 for the creation of RDF from a variety of data formats [47]. This approach permits the inclusion of conditionals and loops (written in JSLDT) within the mapping file, allowing for the transformation process to iterate through the input data in interesting ways. Nevertheless, there may be a steep learning curve for researchers without a programming background to adopt this approach.
Eureka! Clinical Analytics is a web application that performs ETL on Excel spreadsheets containing phenotype data [48]. Since this application was designed for use on clinical projects, it cannot easily be generalized for use in domains outside of biomedicine. The open-source Linked Data Integration Framework (LDIF) leverages Linked Data to provide both data translation and identity resolution capabilities [49]. LDIF uses runtime environments to manage data flow between a set of pluggable modules that correspond to data access, transformation, and output components. Improvements in the framework resulted in the extension of the importer capabilities to allow for input in the form of RDF/XML, N-Triples, and Turtle, import data by crawling RDF links through the use of LDspider, and replicate data through SPARQL construct queries [50]. One limitation of LDIF is that the runtime environment that supports RDF is slower than the in-memory and cluster environment implementations that do not support RDF. Other approaches use existing semantic technologies to perform ETL [51, 52, 53]. These approaches, however, have a similar hurdle for adoption, in that they are often perceived as challenging by those unfamiliar with Semantic Web vocabularies and standards. SDDs provide a means of performing Semantic ETL without the requiring the writing of complex transformation scripts.
2.3. Metadata Standards
The collection of SDD specifications that we discuss in Section 3 serve to provide a standard guideline for semantically recording the metadata associated with the dataset being annotated. In this section, we examine existing metadata standards for describing data that incorporate semantics. The ISO/IEC 11179 standard includes several components, including the (1) framework, (2) conceptual model for managing classification schemes, (3) registry metamodel and basic attributes, (4) formulation of data definitions, (5) naming and identification principles, (6) registration instructions, and (7) registry specification for datasets.7 This standard is intended to address the semantics, representation, and registration of data. Nevertheless, a limitation of ISO/IEC 11179 is that it mainly focuses on the lifestyle management of the metadata describing data elements rather than of events associated with the data values [54]. The Cancer Data Standards Repository (caDSR) implements the ISO/IEC 111791 standard to organize a set of common data elements (CDEs) used in cancer research [55]. The Clinical Data Interchange Standards Consortium (CDISC) has produced several Unified Modeling Language (UML) models that provide schemas for expressing clinical data for research purposes [56]. However, as these schemas are based on the Health Level 7 (HL7) reference implementation model (RIM), which focuses on representing information records instead of things in the world, semantic concepts are used as codes that tag records rather than to provide types for entities.
3. The Semantic Data Dictionary
The Semantic Data Dictionary approach provides a way to create semantic annotations for the columns in a dataset, as well as for categorical or coded cell values. This is achieved by encoding mappings to terms in an appropriate ontology or set of ontologies, resulting in an aggregation of knowledge formed into a graphical representation. A well-formed SDD contains information about the objects and attributes represented or referred to by each column in a dataset, utilizing the relevant ontology URIs to convey this information in a manner that is both machine-readable and unambiguous.
The main output of interpreting SDDs are RDF graphs that we refer to as knowledge graph fragments, since they can be included as part of a larger knowledge graph. Knowledge graphs, or structured graph-based representations that encode information, are variably defined but often contain a common set of characteristics: (i) real world entities and their interrelations are described, (ii) classes and relations of entities are defined, (iii) interrelating of entities is allowed, and (iv) diverse domains are able to be covered [57]. We have published a number of SDD resources, such as tutorials, documentation, complete examples, and the resulting knowledge graph fragments.8 Full sets of annotated SDDs for several public datasets are also available here.
To support the modularization and ease of adoption of the annotation process, we implement the SDD as a collection of tabular data that can be written as Excel spreadsheets or as Comma Separated Value (CSV) files. The SDD is organized into several components to help modularize the annotation process. We introduce the components here and go into further detail on each throughout the remainder of this section. A document called the Infosheet is used to specify the location of each of the SDD component tables. Furthermore, the user can record descriptive metadata about the dataset or SDD in this document. The Dictionary Mapping (DM) is used to specify mappings for the columns in the dataset that is being annotated. If only this component is included with the SDD, an interpreter can still be used to convert the data into an RDF representation. Therefore, we focus the majority of our discussion in this section on the DM table. We also briefly describe the remaining SDD components that allow for richer annotation capabilities and ease the annotation process. The Codebook is used to interpret categorical cell values, allowing the user to assign mappings for data points in addition to just the column headers. The Code Mapping table is used to specify shorthand notations to help streamline the annotation process. For example, the user can specify ‘mm’ to be the shorthand notation for uo:0000016,9 the class in the Units of Measurement Ontology (UO [58]) for millimeter. The Timeline table is used to include detailed annotations for events or time intervals. Finally, the Properties table allows the user to specify custom predicates employed during the mapping process. We use SmallCaps font when referring to columns in an SDD table and italics when referring to properties from ontologies. Further information on the SDD modeling process is available on the SDD documentation website.10
3.1. Infosheet
To organize the collection of tables in the SDD, we use the Infosheet (Appendix Table B.1), which contains location references for the Dictionary Mapping, Code Mapping, Timeline, Codebook, and Properties tables. The Infosheet allows for the use of absolute, relative, or web resource locations. In addition to location references, the Infosheet is used to include supplemental metadata (Appendix Table B.2) associated with the SDD, such as a title, version information, description, or keywords. In this regard, the Infosheet serves as a configuration document, weaving together each of the individual pieces of the Semantic Data Dictionary and storing the associated dataset-level metadata.
The properties that are included support distribution level dataset descriptions based on the Health Care and the Life Sciences (HCLS) standards,11 as well as the Data on the Web Best Practices (DWBP).12 The HCLS standards contain a set of metadata concepts that should be used to describe dataset attributes. While the resulting document was developed by stakeholders working in health related domains, the properties included are general enough to be used for datasets in any domain. The DWBP were developed by a working group to better foster communications between data publishers and users, improve data management consistency, and promote data trust and reuse. The associated document lists 35 best practices that should be followed when publishing data on the web, each of which includes an explanation for why the practice is relevant, the intended outcome, possible implementation and testing strategies, and potential benefits of applying the practice.
In Section 4, we provide an example of using the SDD approach to annotate the National Health and Nutrition Examination Survey (NHANES). An example Infosheet for the Demographics table of this dataset is provided in Appendix Table C.1.
3.2. Dictionary Mapping
The Dictionary Mapping (DM) table includes a row for each column in the dataset being annotated (referred to as explicit entries), and columns corresponding to specific annotation elements, such as the type of the data (Attribute, Entity)13, label (Label), unit (Unit), format (Format), time point (Time), relations to other data columns (inRelationTo, Relation), and provenance information (wasDerivedFrom, wasGeneraTEDBy). Figure 1 shows the conceptual diagram of the DM. Such a representation is similar to the structure of general science ontologies, such as the Semanticscience Integrated Ontology (SIO) [59] or the Human-Aware Science Ontology (HAScO) [60]. We use SIO properties for the mapping of many of the DM columns, as shown in the Dictionary Mapping specification in Appendix Table B.3, while also leveraging the PROV-O ontology [61] to capture provenance information. Despite specifying this default set of mappings, we note that the Properties table of the SDD can be used to determine the set of predicates used in the mapping process, allowing the user to customize the foundational representation model.
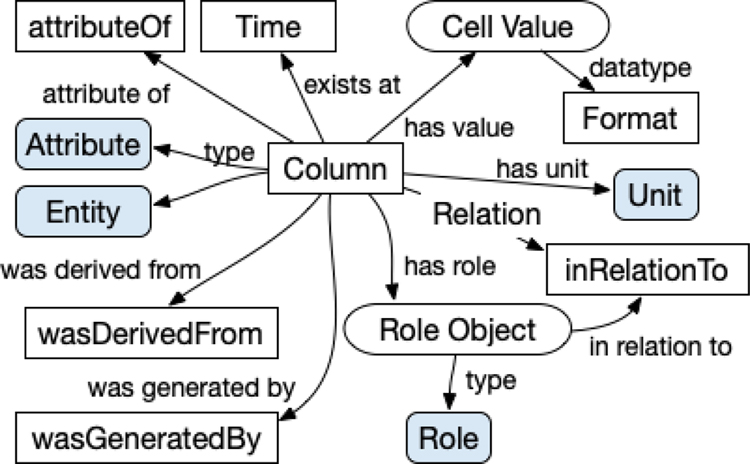
Figure 1:
A conceptual diagram of the Dictionary Mapping that allows for a representation model that aligns with existing scientific ontologies. The Dictionary Mapping is used to create a semantic representation of data columns. Each box, along with the “Relation” label, corresponds to a column in the Dictionary Mapping table. Blue rounded boxes correspond to columns that contain resource URIs, while white boxes refer to entities that are generated on a per-row/column basis. The actual cell value in concrete columns is, if there is no Codebook for the column, mapped to the has value object of the column object, which is generally either an attribute or an entity.
In addition to allowing for the semantic annotation of dataset columns, unlike traditional mapping approaches, the SDD supports the annotation of implicit concepts referenced by the data. These concepts, referred to as implicit entries, are typically used to represent the measured entity or the time of measurement. For example, for a column in a dataset for a subject’s age, the concept of age is explicitly included, while the idea that the age belongs to a human subject is implicit. These implicit entries can then be described to have a type, a role, relationships, and provenance information in the same manner as the explicit entries. For example, to represent the subject that had their age measured, we could create an implicit entry, ??subject.14
3.2.1. Attributes and Entities
Attribute and Entity are included in the DM to allow for the type assignment of an entry. While both of these columns map to the property rdf:type,15 they are both included as it may be semantically significant to distinguish between characteristics and objects. If an entry describes a characteristic, Attribute should be populated with an appropriate ontology class. The entity that contains the characteristic described, which can be either explicit or implicit, should be referenced in attributeOf. While columns in a dataset typically describe an observed characteristic, this is not always the case. If an entry describes an object, such as a person, place, thing, or event, Entity should be populated with an appropriate ontology class.
3.2.2. Annotation Properties and Provenance
A set of annotation properties, including comments, labels, or definitions, allows for the description of an explicit or implicit entry in further detail. While Label is the only column included in the DM Specification for an annotation property, if support for comments and definitions is included in an SDD interpreter, we recommend the use of the rdfs:comment and skos:definition predicates, respectively. In terms of including provenance, wasDerivedFrom can be used to reference pre-existing entities that are relevant in the construction of the entry, and wasGeneraTEDBy can be used to describe the generation activity associated with the entry.
3.2.3. Additional Dictionary Mapping Columns
The Role, Relation, and inRelationTo columns of the DM are used to specify roles and relationships associated with entries. A reference to objects or attributes an entry is related to should be populated in inRelationTo. By populating Role, the sio:hasRole property is used to assign the specified role to the entry. Custom relationships using properties that are not included in the SDD can be specified using Relation. Events in the form of time instances or intervals associated with an entry should be referenced in Time. The unit of measurement of the data value can be specified in Unit. In general, we recommend the use of concepts in the Units of Measurement Ontology (UO) for the annotation of units, as many existing vocabularies in various domains leverage this ontology. A W3C XML Schema Definition Language (XSD) primitive data type16 can be included in Format to specify the data type associated with the data value.
3.2.4. Dictionary Mapping Formalism
We define a formalism for the mapping of DM columns to an RDF serialization. The notation we use for formalizing the SDD tables is based on an approach for translating constraints into first-order predicate logic [62]. While most of the DM columns have one-to-one mappings, we can see the interrelation of the mapping of Role, Relation, and inRelationTo. In the formalism included below, ‘Value’ represents the cell value of the data point that is being mapped.
3.3. Codebook
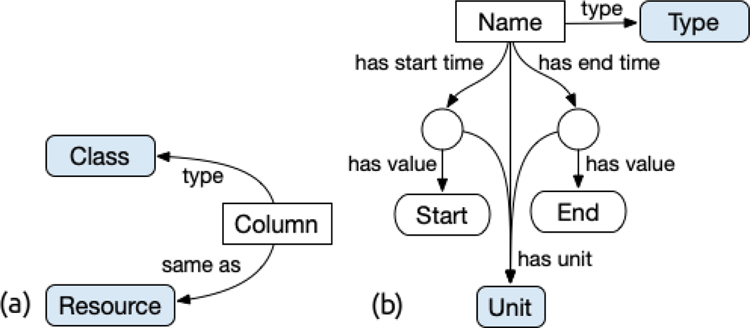
The Codebook table of the SDD allows for the annotation of individual data values that correspond to categorical codes. The Codebook table contains the possible values of the codes in Code, their associated labels in Label, and a corresponding ontology concept assignment in Class. If the user wishes to map a Codebook value to an existing web resource or instance of an ontology class, rather than a reference to a concept in an ontology, Resource can be populated with the corresponding URI. We recommend that the class assigned to each code for a given column be a subclass of the attribute or entity assigned to that column. A conceptual diagram of the Codebook is shown in Figure 2 (a). The Codebook Specification is provided in Appendix Table B.4. The formalism for mapping the Codebook is included below.
Figure 2:
(a) A conceptual diagram of the Codebook, which can be used to assign ontology classes to categorical concepts. Unlike other mapping approaches, the use of the Codebook allows for the annotation of cell values, rather than just columns. (b) A conceptual diagram of the Timeline, which can be used to represent complex time associated concepts, such as time intervals.
3.4. Code Mapping
The Code Mapping table contains mappings of abbreviated terms or units to their corresponding ontology concepts. This aids the human annotator by allowing the use of shorthand notations instead of repeating a search for the URI of the ontology class. The set of mappings used in the CHEAR project is useful for a variety of domains and is available online.17
3.5. Timeline
If an implicit entry for an event included in the DM corresponds to a time interval, the implicit entry can be specified with greater detail in the Timeline table. Timeline annotations include the corresponding class of the time associated entry, the units of the entry, start and end times associated with an event entry, and a connection to other entries that the Timeline entry may be related to. Shown in Figure 2(b) is a conceptual diagram of the Timeline. The Timeline Specification is provided in Appendix Table B.5. The formalism for mapping the Timeline is included below.
3.6. Property Customization
The Semantic Data Dictionary approach creates a linked representation of the class or collection of datasets it describes. The default model provided is based on SIO, which can be used to express a wide variety of objects using a fixed set of terms, incorporates annotation properties from RDFS and SKOS, and uses provenance predicates from PROV-O. Shown in Appendix Table B.6 are the default set of properties that we recommend.
By specifying the associated properties with specific columns of the Dictionary Mapping Table, the properties used in generating the knowledge graph can be customized. This means that it is possible to use an alternate knowledge representation model, thus making this approach ontology-agnostic. Nevertheless, we urge the user to practice caution when customizing the properties used to ensure that the resulting graph is semantically consistent (for example, not to replace an object property with a datatype property).
In the formalism presented above and the DM, CB, and TL specifications of Appendix Tables B.3, B.4, & B.5, fourteen distinct predicates are used.18 Fourteen of the sixteen rows of the Properties Table are included to allow the alteration of any of these predicates. The two additional rows pertain to Attribute and Entity, which, like Type, by default map to rdf:type, but can be customized to use an alternate predicate if the user wishes. In this way, by allowing for the complete customization of the predicates that are used to write the formalism, the SDD approach is ontology-agnostic. Note that the predicates used in the Infosheet Metadata Supplement of Table B.2, which are based on the best practices described in Section 3.1, are not included in the Properties Specification.
4. Example – The National Health and Nutrition Examination Survey
The National Health and Nutrition Examination Survey (NHANES) contains publicly available demographic and biomedical information. A challenge in creating a knowledge representation from this dataset is determining how to represent the implicit entities referenced by the data, such as a participant of the study or the household that they live in. Additionally, information about a participant may be dispersed throughout multiple tables that consequently need to be integrated, resulting in difficulties when following traditional mapping approaches.
NHANES data dictionaries include a variable list that contains names and descriptions for the columns in a given dataset component, as well as a documentation page that consists of a component description, data processing and editing information, analytic notes, and a codebook. Unfortunately, the dataset description provided is textual and is therefore not readily processed.
We find that neither the data documentation nor the codebooks included in NHANES incorporate mappings to ontology concepts. Thus, we provide a simple example of how several columns from the NHANES Demographics dataset would be represented using the SDD approach. The terms in this example are annotated using the CHEAR, SIO, and National Cancer Institute Thesaurus (NCIT) ontologies. Shown in Tables 1, 2, and 3 are a portion of the SDD we encoded for the NHANES Demographics dataset, in which we respectively present a subset of the explicit DM entries, implicit DM entries, and the Codebook entries. An example Infosheet for the NHANES Demographic dataset is provided in Appendix Table C.1. The complete set of explicit and implicit entries are provided in Appendix Table C.3 and Appendix Table C.2, respectively. An expanded codebook is included in Appendix Table C.4. Additional NHANES tables not included in this article were also encoded as part of this annotation effort.19
Table 1:
Subset of Explicit Entries identified in NHANES Demographics Data
| Column | Label | Attribute | attributeOF | Unit | Time |
|---|---|---|---|---|---|
| SEQN | Respondent number | sio:Identifier | ??part | ||
| RIDEXAGM | Age in months at exam | sio:Age | ??part | mth | ??exam |
| DMDEDUC3 | Education level | chear:EducationLevel | ??part | ||
| DMDHRAGE | HH age in years | sio:Age | ??HHRef | yr | |
Table 2:
Subset of Implicit Entries identified in NHANES Demographics Data
| Column | Label | Entity | Role | inRelationTo |
|---|---|---|---|---|
| ??part | Participant | ncit:C29867, sio:Human | sio:SubjectRole | ??exam |
| ??exam | Examination | ncit:C131902 | ||
| ??HHRef | Household head | sio:Human | chear:HeadOfHousehold | ??hh |
| ??hh | Household | chear:Household | ??part | |
Table 3:
Subset of NHANES Demographic Codebook Entries
In Table 1, we provide the explicit entries that would be included in the DM. The data column SEQN corresponds to the identifier of the participant. The resource created from this column can be used to align any number of NHANES tables, helping address the data integration problem. Another column included is the categorical variable that corresponds to education level. Also included are two variables that correspond to the age of the participant taking the survey and the age of the specified reference person of the household, defined as the person who owns or pays rent for the house. We see how the use of implicit entries, as well as the use of specified Code Mapping units, helps differentiate the two ages. The corresponding implicit entries referenced by the explicit entries are annotated in Table 2.
In Table 3, we include a subset of the Codebook for this example. The SDD Codebook here is similar to the original NHANES codebook, with the addition of Column, so that multiple codebooks do not have to be created to correspond to each categorical variable, and Class, used to specify a concept from an ontology to which the coded value maps.
5. Current Use
In this section, we provide a case study on projects that have leveraged the SDD for health-related use cases. We focus on work done for the Health Empowerment by Analytics, Learning, and Semantics (HEALS) project, while also briefly discussing efforts in other programs. In our funded research, our sponsors often desire the representation their data in a semantically consistent way that supports their intended applications. They wish to play a role in the annotation process by contributing their subject matter expertise. We find that the SDD approach is more accessible to domain scientists than other programming intensive approaches. Additionally, they appreciate that the ability to reuse SDDs limits the amount of necessary future updates when, for example, a data schema changes.
5.1. Health Empowerment by Analytics, Learning, and Semantics
As part of the RPI and IBM collaborative Health Empowerment by Analytics, Learning, and Semantics (HEALS) project,20 SDDs have been used to aid in semantic representation tasks for use cases involving breast cancer and electronic health record (EHR) data.
5.1.1. Breast Cancer Use Case
For the creation of an application used for the automatic re-staging breast cancer patients, the SDD approach was used to create a knowledge representation of patient data from the Surveillance, Epidemiology, and End Results (SEER) program [63]. In order to integrate treatment recommendations associated with a given biomarker into the application, an SDD for the Clinical Interpretation of Variants in Cancer (CIViC) database was also created. By applying the SDD approach to help solve this problem, seamless data integration between these two distinct sources was demonstrated, which would have been more difficult to achieve using some of the methods described in Section 2.2. For example, if any of the mapping language or Semantic ETL approaches were applied, the writing of a script that requires an intrinsic understanding of the dataset would be necessary, rather than needing to just fill out the SDD tables. While this approach still requires an understanding of the dataset, if the SDD approach was used for describing the datasets mentioned above, the data apprehension requirement on the user would greatly reduced. Another advantage demonstrated by using this approach was that, since a limited set of properties are leveraged in the semantic model that was created, the cost of implementing the application, in terms of programming resources and overhead, was reduced. A subset of the explicit entries from the SEER DM are shown in Table 4.
Table 4:
Subset of Explicit Entries identified in SEER
| COLUMN | ATTRIBUTE | ATTRIBUTEOF | UNIT | TIME |
|---|---|---|---|---|
| T | ncit:C120284 | ??tumor | mm | |
| N | sio:Count | ??lymph node | ||
| M | sio:StatusDescriptor | ??metastasis | ||
| Age at diagnosis | sio:Age | ??subject | ??diagnosis | |
| Vital status recode | sio:LifeStatus | ??subject | ||
| Year of diagnosis | sio:TimeInstant | ??diagnosis | xsd:gYear | |
| HER2 | sio:StatusDescriptor | ??her2 gene | ||
| ER | sio:StatusDescriptor | ??er gene | ||
| PR | sio:StatusDescriptor | ??pr gene | ||
Additional cancer-related work for the HEALS project involves the annotation of a subset of The Cancer Genome Atlas (TCGA) through the NCI Genomic Data Commons (GDC) portal. While these SDDs are not included here, they are openly available on our SDD resources web-page. The clinical subset of the TCGA data that was annotated contains patient demographic and tumor information, and the methylation portion contains genetic information. By using the same ontology classes that were used for the SEER dataset to annotate these concepts, we are able to leverage TCGA data to further enrich the cancer staging application described above.
5.1.2. Electronic Health Record Data
To create a knowledge representation from electronic health record (EHR) data, we annotated the Medical Information Mart for Intensive Care III (MIMIC-III) dataset using SDDs. While this effort involved annotating 26 relational tables, we only include a subset of the Dictionary Mapping of the admission table in Table 5. Using this approach, we can represent implicit concepts associated with the data. The inclusion of implicit concepts provides connection points for linking the various EHR data tables into a single coherent knowledge representation model that reflects the reality recorded by the data. This would be difficult to accomplish using many of the alternate approaches we examined that do not support object elicitation.
Table 5:
Subset of Dictionary Mapping for the MIMIC-III Admission table
| Column | Attribute | attributeOF | Entity | Role | inRelationTo |
|---|---|---|---|---|---|
| SUBJECT_ID | sio:Identifier | ??subject | |||
| ADMITTIME | sio:TimeInstant | ??admission | |||
| DISCHTIME | sio:TimeInstant | ??discharge | |||
| DEATHTIME | sio:TimeInstant | ??death | |||
| INSURANCE | chear:InsuranceType | ??subject | |||
| RELIGION | chear:Religion | ??subject | |||
| MARITAL_STATUS | chear:MaritalStatus | ??subject | |||
| ETHNICITY | sio:Ethnicity | ??subject | |||
| DIAGNOSIS | ogms:0000073 | ??subject | |||
| ??subject | sio:Human | sio:SubjectRole | |||
| ??admission | ncit:C25385 | ??subject | |||
| ??discharge | genepio:0001849 | ??subject | |||
| ??death | ncit:C28554 | ??subject | |||
5.2. Additional Use Cases
Several institutions are employing the Semantic Data Dictionary approach for a variety of projects. The Icahn School of Medicine at Mount Sinai uses SDDs for the NIH CHEAR and the follow on HHEAR projects to annotate data related to demographics, anthropometry, birth outcomes, pregnancy characteristics, and biological responses. The Lighting Enabled Systems & Applications (LESA) Center is using SDDs to annotate sensor data. SDDs are being used in Brazil for the Big Data Ceara project, through Universidade de Fortaleza, and the Global Burden of Disease project, through Universidade Federal de Minas Gerais.
5.3. Remarks
In this section, we discussed how SDDs help represent knowledge for a variety of other projects that involve collaborative efforts with domain scientists, exhibiting the applicability of this approach for researchers in a variety of specializations. For the HEALS project, we have shown DMs for use cases that involve breast cancer and EHR records. As well as patient demographic characteristics from the SEER data, we encode the size of the patient’s tumor, the number of lymph nodes affected, whether or not the cancer metastasized, and several genetic biomarkers. Using this data, the successful automation of re-staging breast cancer patients was accomplished. While we only show a single DM for the MIMIC-III dataset, this use case involves the annotation of multiple relational data tables and demonstrates how data integration can be performed using SDDs.
6. Modeling Challenges for Domain Scientists
An initial strategy of training that was followed by qualitative evaluation was used to examine the difficulty experienced by researchers who do not have a Semantic Web background when first using the Semantic Data Dictionary. Domain scientists, including epidemiologists and bio-statisticians, were presented with initial training by a Semantic Web expert. Supporting materials were developed in collaboration with a domain expert and then were made available to provide guidance and examples to facilitate domain scientists’ use of the Semantic Data Dictionary.
First, a template for completing the Semantic Data Dictionary that included pre-populated fields for common demographic concepts, such as age, race, and gender, was provided to domain scientists to use for each study. Second, a help document was created that included instructions and representations of more complex concepts, including measurements of environmental samples, measurements of biological samples, and measurements taken at specific time-points. Third, a practical workshop was held where a semantic scientist provided training in semantic representation to the domain scientists. Following the workshop and distribution of supporting materials, domain scientists completed at least one Semantic Data Dictionary for an epidemiologic study and were then asked about the challenges they faced. Despite this training and workshop being conducted in a context related to epidemiology and health, the key takeaways resulted in general lessons learned.
The first identified challenge was the representation of implicit objects implied by the features in the dataset. This is an uncommon representation in the public health domain. While the modeling of simple concepts may be intuitive (e.g. maternal age has a clear implicit reference to mother), the representation of complex ideas, such as fasting blood glucose levels, proves to be more difficult as the implicit object, and relationships between concepts, is not as intuitive for domain scientists. A second modeling challenge involved discussions on how to represent time-associated concepts that power the ontology-enabled tools and allow domain scientists to harmonize data across studies. Additionally, when a concept was not found in a supporting ontology, there were questions of how to best represent the concept in a semantically-appropriate way. In many cases, these challenges resulted in a need to go back to a Semantic Web expert for clarification.
To alleviate these challenges, we have refined and expanded the number of publicly-available resources that include documentation, step-by-step modeling methods, tutorials, demonstrations, and informative examples. We increased the complexity of examples and incorporated time-associated concepts to initial templates and help documents. To facilitate further communication, a web-based Q&A document has been shared between the Semantic Web experts and the domain scientists to enable timely feedback and answers to specific questions on the representation of concepts and the need to generate new concepts.
In addition to the solutions presented above, we plan for future training events to explicitly demonstrate the use of the Semantic Data Dictionary. We will provide an overview on the semantic representation, as well as guidelines for using the corresponding documentation and training materials.
7. Evaluation
To evaluate the Semantic Data Dictionary approach, we categorize metrics from earlier evaluations on mapping languages [64, 65] and requirements of data integration frameworks. In addition to evaluating the SDD for adherence to these metrics, we survey similar work to determine the extent to which they meet the metrics in comparison. We include a set of evaluation metrics that we organized into four categories. These categories are respectively related to data, semantics, the FAIR principles, and generality.
To measure the degree to which an approach meets each metric, we provide a value of 0, 0.5, or 1, depending on the extent to which an approach responds to an evaluation parameter. In general, if an approach does not meet a metric, it is given a score of 0. If it meets a metric partially, we assign a score of 0.5. We also assign this score to approaches that meet a metric by omission, such as being ontology-agnostic by not supporting the use of ontologies at all. If an approach completely meets the metric, it is given a score of 1. We list the criteria used for the assignment of numerical values below (refer to Table 6 for the complete list of categorized metrics).
Table 6:
High-level comparison of Semantic Data Dictionaries, Traditional Data Dictionaries, approaches involving Mapping Languages, and general Data Integration Tools
| Metric | SDD | Traditional DD | Mapping Language | Data Integration Tool |
|---|---|---|---|---|
| Data | 1 | 0.25 | 1 | 1 |
| Harmonizable | 1 | 0 | 1 | 1 |
| Ingestible | 1 | 0 | 1 | 1 |
| Subset selection | 1 | 0.5 | 1 | 1 |
| Data type assignment | 1 | 0.5 | 1 | 1 |
| Semantics | 0.89 | 0.11 | 0.5 | 0.56 |
| Object elicitation | 1 | 0 | 0 | 0.5 |
| Relation elicitation | 1 | 0 | 0 | 0 |
| Queryable | 1 | 0 | 1 | 1 |
| Value annotation | 1 | 1 | 0 | 0 |
| Time annotation | 1 | 0 | 0.5 | 0 |
| Space annotation | 0.5 | 0 | 0 | 1 |
| Domain knowledge support | 0.5 | 0 | 1 | 1 |
| Top-level ontology foundation | 1 | 0 | 1 | 0.5 |
| Graph materialization | 1 | 0 | 1 | 1 |
| FAIR | 1 | 0.33 | 1 | 0.83 |
| Accessible | 1 | 0.5 | 1 | 0.5 |
| Findable | 1 | 0 | 1 | 1 |
| Interoperable | 1 | 0 | 1 | 1 |
| Reusable | 1 | 0.5 | 1 | 1 |
| Reproducible | 1 | 0.5 | 1 | 1 |
| Transparent | 1 | 0.5 | 1 | 0.5 |
| Generality | 0.92 | 0.33 | 0.92 | 0.92 |
| Domain-agnostic | 1 | 1 | 1 | 1 |
| Ontology-agnostic | 1 | 0.5 | 0.5 | 1 |
| Leverages best practices | 0.5 | 0.5 | 1 | 0.5 |
| Provenance | 1 | 0 | 1 | 1 |
| Documentation | 1 | 0 | 1 | 1 |
| Machine-readable | 1 | 0 | 1 | 1 |
7.1. Data integration capabilities
In this category, we consider how the approach can harmonize and ingest data, allows for subset data selection, and permits a data type assignment. We evaluate whether the approach is harmonizable in the sense that it has the capability of creating a cohesive representation for similar concepts across columns or datasets in general. We check that knowledge generated across datasets can be compared using similar terms from a controlled set of vocabularies. For this metric, we respectively assign a score of 0, 0.5, or 1 if data integration capabilities are not supported, somewhat supported, or wholly supported.
Next, we consider whether the approach is ingestible, outputting data in a standard format that can be uploaded and stored (ingested) and supports inputs of varying formats. We assign a score of 1 if the resulting data representation can be stored in a database or triplestore, and if it can input data of varying formats. If one of the two features are supported, we assign a score of 0.5. If neither are supported, we assign a score of 0.
Furthermore, we consider a subset selection metric, where we check if the approach allows the user to select a subset of the data, either in terms of columns and rows, on which to perform the annotation. For this metric, a score of 0 is assigned if this capability is not included in the approach. We assign a score of 0.5 if either a subset of the rows or the columns can be specified for annotation, but not both. If the approach allows for the selection of both a subset of rows or of columns to be annotated, we assign a score of 1.
Finally, we include the data type assignment metric, measuring the extent to which XML data types can be assigned to attributes when mapping data. We assign a score of 0 for this metric if the approach does not allow for the assignment of data types when mapping data. If the assignment of a limited set of data types that are not based on XML standards is incorporated, a score of 0.5 is assigned. If the approach allows the assignment of XML data types, a score of 1 is given.
7.2. Formal semantics capabilities
In this category, we consider if the approach allows for object or relation elicitation, as well as value, time, or space annotation. We also check if the resulting data representation is queryable and if the approach supports both domain-specific and general ontology foundations. Finally, graph materialization is the last assessment metric we apply. Data usually consists of attributing value to observations, measurements, or survey results. Dataset descriptions contain metadata, but often omit details on the objects that the values describe. For a complete semantic representation, one must also consider the ability to represent implicit objects that are associated with the data points, which we measure using the object elicitation metric. If the approach does not include the ability to represent implicit objects, a score of 0 is assigned. If implicit objects are considered but not annotated in detail, we assign a score of 0.5. We assign a score of 1 if implicit objects can be represented and richly annotated.
In addition to being able to represent implicit concepts, we consider relation elicitation, where relationships between implicitly elicited objects can be represented. A score of 0 is assigned if an approach does not allow for the representation of relationships between elicited objects. If relationships between elicited objects can be represented, but not annotated in detail, a score of 0.5 is assigned. We assign a score of 1 if relationships between elicited objects can be represented and richly annotated.
Next, we consider if the resulting representation is queryable, so that specific data points can be easily retrieved using a query language. A score of 0 is assigned for this metric if specific content from the knowledge representation cannot be queried. If it can be queried using a relational querying method, such as SQL, but not a graph querying method, a score of 0.5 is assigned. If content can be queried using a graph querying method, such as SPARQL, we assign a score of 1.
We further consider the annotation of cell values, rather than just column headers, using the value annotation metric. This covers the ability to annotate categorical cell values, assign units to annotate non-categorical cell values, and specify attribute mappings of object properties related to cell values. If the approach does not allow for the annotation of cell values at all, or allows for a limited set of annotations for cell values, we assign scores of 0 and 0.5, respectively. We assign a score of 1 if an approach includes the ability to annotate categorical cell values, assigns units to annotate non-categorical cell values, and specifies attribute mappings of object properties related to cell values.
We consider the ability to represent specific scientific concepts, including time and space. Using the time annotation metric, we check for the ability to use timestamps to annotate time-series values, as well as named time instances to annotate cell values. A score of 0 is assigned for this metric if an approach does not allow for the representation of time. If the approach allows for the representation of time, but does not permit detailed annotations, we assign a score of 0.5. We assign a score of 1 if the approach allows for detailed annotation of time, such as the use of timestamps to annotate time-series values and named time instances to annotate cell values.
The space annotation metric is added to check for the use of semantic coordinate systems to annotate the acquisition location of measurements. We assign a score of 0 if an approach does not allow for the representation of space. If it allows for the representation of space, but does not permit detailed annotations, we assign a score of 0.5. A score of 1 is assigned if the use of semantic coordinate systems to annotate the acquisition location of measurements is supported. We examine domain knowledge support by checking if the approach permits the design of mappings using pre-existing domain-specific ontologies or controlled vocabularies. A score of 0 is assigned for this metric if the approach does not permit the design of reusable mappings driven by domain knowledge. We assign a score of 0.5 if it permits the design of reusable mappings using either pre-existing ontologies or controlled vocabularies, but not both. If annotations from both pre-existing ontologies or controlled vocabularies are allowed, we assign a score of 1.
Using the top-level ontology foundation metric, we consider the ability to use general upper ontologies as a foundation for the resulting model. If an approach cannot specify mapping rules based on foundation ontologies, a score of 0 is assigned for this metric. If a subset of mapping rules based on general foundation ontologies can be specified, we assign a score of 0.5. A score of 1 is assigned if the approach allows for the specifiation of all mapping rules based on general foundation ontologies. Essentially, we are checking if the semantic model that results from the annotation approach is structured based on a given ontology. While we recommend the use of well known upper ontologies such as SIO or Basic Formal Ontology (BFO [66]), in evaluating this metric we allow the approach the leverage any ontology.
Finally, with the graph materialization metric, we assess the persistence of the generated knowledge graph into an accessible endpoint or file. If the approach does not allow for the materialization of the generated graph, a score of 0 is assigned. If the generated graph is reified into an accessible endpoint or downloadable file, but not both, a score of 0.5 is assigned. If both materializations into an accessible endpoint and a downloadable file are supported, we assign a score of 1.
7.3. FAIR
In the FAIR category, we consider the metrics associated with the FAIR guiding principles, including if the approach and resulting artifacts are findable, accessible, interoperable, and reusable. Furthermore, we also consider the related metrics of reproducibility and transparency, which are not included in the FAIR acronym. While several of the metrics we measure in the other categories of our evaluation aid with the creation of FAIR data, such as the incorporation of provenance or the inclusion of documention as discussed in Section 7.3.1, we include these six metrics in the FAIR category since they are directly associated with intent of the principles in enhancing data reuse and are explicitly discussed in the introductory article on the FAIR principles [4].
For the findable metric, we consider the use of unique persistent identifiers, such as URLs, as well as the inclusion of web searchable metadata so that the knowledge is discoverable on the web. If the knowledge representation is neither persistent nor discoverable, we assign a score of 0 for this metric. If the knowledge representation is one of the two, we assign a score of 0.5. A score of 1 is assigned if the knowledge representation is both persistent and discoverable.
We consider a knowledge representation to be accessible if resources are openly available using standardized communication protocols, with the consideration that data that cannot be made publicly available is accessible through authentication. Accessibility also includes the persistence of metadata, that even if data is retired or made unavailable, its description still exists on the Web. As additional consideration for evaluating accessibility, we examine whether or not the associated software for an approach is free and publicly available. If resources and metadata are not published openly, a score of 0 is assigned for this metric. If some resources and metadata are persistent and openly available, we assign a score of 0.5. A score of 1 is assigned if all of the resources and metadata from a given approach are both persistent and openly available using standardized communication protocols.
For the interoperable metric, we consider the use of structured vocabularies, such as best practice ontologies, that are RDF compliant. Mainly, we are checking to see if the knowledge representation is published using an RDF serialization. If the knowledge representation does not use a structured vocabulary, a score of 0 is assigned. If it uses structured vocabularies that are not RDF compliant, we assign a score of 0.5. A score of 1 is assigned if the knowledge representation uses formal vocabularies or ontologies that are RDF compliant.
To test if an approach or the resulting knowledge representation is reusable, we consider the inclusion of a royalty-free license that permits unrestricted reuse, and that consent or terms of agreement documents are available when applicable. We also discuss if included metadata about the resource is detailed enough for a new user to understand. A score of 0 is assigned for this metric if an approach does not include a royalty-free license. If a royalty-free license that permits unrestricted use of some portions of the tool is included, a score of 0.5 is assigned. We assign a score of 1 if the approach includes a royalty-free license that permits unrestricted use of all portions of the tool.
We examine if an approach is reproducible in terms of scientific activities introduced within a given methodology, such that experiments can be independently conducted and verified by an outside party. If the approach creates a knowledge representation that cannot be reproduced, a score of 0 is assigned. If the knowledge representation that can be produced by an outside party with the help of the involved party, rather than entirely independently, we assign a score of 0.5. A score of 1 is assigned if the approach for creating a knowledge representation can be independently produced.
Finally, we consider if data and software are transparent, such that there are no “black boxes” used in the process of creating a knowledge representation. Transparency is readily achieved by making sure that software is made openly available. If the associated code for a given approach is not openly accessible, we assign a score of 0. We assign a score of 0.5 if some of the associated code is open, while other portions are not openly available. This generally applies to approaches that are both free and paid versions of software. If all of the associated code for an approach is open source, a score of 1 is given.
7.3.1. Generality assessment
To evaluate the generality of an approach, we investigate whether or not the method is domain-agnostic, is ontology-agnostic, and adheres to existing best practices. We weigh whether the method incorporates provenance attributions, is machine-understandable, and contains documents to aid the user, such as documentation, tutorials, or demonstrations.
We analyze whether an approach is domain-agnostic, in that its applicability does not restrict usage to a particular domain. A score of 0 is assigned for this metric if the approach only applies to a single field of study. If the approach applies to multiple fields of study but does not work for specific domains, a score of 0.5 is assigned. We assign a score of 1 if the approach can be generalized to any areas of study.
On a similar vein, we judge if the method is ontology-agnostic, where usage is not limited to a particular ontology or set of ontologies. If the approach depends on a particular ontology or set of ontologies, a score of 0 is assigned. If the dependence on particular ontologies is unclear from the examined literature and documentation, we assign a score of 0.5. A score of 1 is assigned for this metric if the approach is independent of any particular ontology.
We examine the literature and documentation associated with a given approach or knowledge representation to see if it leverages best practices. In particular, we consider the applicable best practices related to the HCLS and DWBP guidelines. Among the practices we test for include the ability of the approach to incorporate descriptive metadata, license and provenance information, version indicators, standardized vocabularies, and use locale-neutral data representations. A score of 0 is assigned if the literature associated with an approach does not acknowledge or adhere to existing best practice standards. If existing standards are acknowledged but are not adhered to or are partially adhered to, we assign a score of 0.5. If the literature acknowledges and adheres to existing best practices, a score of 1 is assigned.
We consider the inclusion of provenance, involving the capture of existential source information, such as attribution information for how a data point was measured or derived. A score of 0 is assigned for this metric if the approach does not include attributions to source or derivation information. If attribution information that does not use Semantic Web standards is included, we assign a score of 0.5. If the approach covers attributions recorded using a Semantic Web vocabulary, such as the PROV-O ontology, a score of 1 is assigned. In terms of documentation, we further search for the inclusion of assistive documents, tutorials, and demonstrations. We assign a score of 0 for this metric if just one of either documentation, tutorials, or demonstrations is included. If two or all of the above are involved, we assign scores of 0.5 or 1, respectively.
Finally, we consider the machine-readable metric, determining whether the resulting knowledge representation from an approach is discernable by software. In addition to the consideration of the machine-readability of output artifacts such as produced knowledge graphs, we also examine input artifacts, such as the document that contains the set of semantic mappings. If neither input nor output artifacts can be parsed using software, a score of 0 is assigned for this metric. If either input or output artifacts can be parsed, but not both, a score of 0.5 is assigned. We assign a score of 1 if both input and output artifacts are machine-readable.
8. Results
In Table 6, we provide a high-level comparison between the Semantic Data Dictionary, traditional data dictionaries, mapping languages and semantic approaches that leverage them, and data integration tools. Of the conventional data dictionaries examined in Section 2.1, we use the Project Open Metadata Schema data dictionary for comparison since it was the only reviewed guideline that used a standard linked data vocabulary. Of the mapping languages, we use R2RML for comparison, as it is a standard that is well adopted by the Semantic Web community. Of the data integration tools we surveyed, we use Karma for this evaluation, as it is an example of a data integration approach that was designed with both the FAIR principles and ease of use for the end-user in mind. Rather than only using these approaches in conducting the evaluation, we think of these examples as guidelines and consider traditional data dictionaries, mapping languages, and data integration tools in general when assigning numerical scores.
We have demonstrated the benefits of using a standardized machine-readable representation for recording dataset metadata and column information, which is achieved through SDDs, over earlier data dictionary formats. Furthermore, we demonstrate that the SDD approach presents a level of abstraction over methodologies that use mapping languages, allowing improved ease of use for a domain scientist over other semantic tools. In this regard, SDDs tend to provide a bridge between conventional data dictionary approaches used by domain scientists and formal semantic approaches used by Semantic Web researchers, thereby accommodating both user groups. We recognize that the RDF mapping tools that exist are intended to provide a bridge by reducing manual mapping or KG creation work that would otherwise be necessary, but also acknowledge that they may be unusable to domain scientists.
9. Discussion
In presenting this work, we consider two general types of users. We consider those using SDDs to semantically annotate data as well as those using SDDs in place of traditional data dictionaries in order to understand the data being described. For the first group of users, benefits of using SDDs include that the annotation process is accessible for users outside of the Semantic Web domain and that existing SDDs can be reused to ease the creation of new annotations. Some benefits for the second group include that (i) traditionally humans alone can understand data descriptions in existing data dictionaries but SDDs can be interpreted by machines as well, (ii) SDDs are written using fixed vocabularies which reduces ambiguity, and (iii) the SDD provides a standard specification that can be used to interpret existing annotations.
By including a fixed set of tables for the annotator to fill out that are interpreted and converted using a standard set of rules, the SDD framework provides consistency by creating a formal semantic representation using direct RDF mappings, resulting in an increased likelihood of diverse annotators creating similar representations. This is in contrast with other mapping approaches, where multiple annotators are much less likely to produce similar results when addressing the same dataset. The SDD approach reduces such representational biases as it abstracts away structural modeling decisions from the user, both cultivating scalability of production and simultaneously lowering the barrier of entry since not all of the authors have to be computer scientists. Moreover, the vocabulary used in an SDD can be easily updated by replacing terms from any of the tables, where similar updates are much less amenable when using standard mapping methods. An advantage of these features of the SDD is that users can focus on their topic of specialization rather than on the RDF, reducing the need for domain scientists to also become ontology experts. Given a recommended set of ontologies to use, any user should be able to create their own SDD for a given dataset.
From the evaluation of Section 7, we find that in the data category, SDDs perform much better than traditional data dictionaries, and equally well as mapping languages and data integration tools. SDDs outperform the three other approaches in the semantics category. In terms of semantics, a notable impact of this work is our approach to object and relation elicitation, where detailed annotations for objects implicitly referenced by the data can be included. SDDs and mapping languages perform equally well in the FAIR category, surpassing the scores of data integration tools and traditional data dictionaries. SDDs, mapping languages, and data integration tools tied for the best performance in the generality category, greatly outperforming traditional data dictionaries. While traditional DDs performed the worst over all four categories, they do outperform mapping languages and data integration tools in the value annotation metric.
10. Conclusion
While the use of SDDs addresses many of the shortcomings associated with the prior art, we do acknowledge several limitations of this approach. In Section 6, we mention several challenges faced by epidemiologists in creating SDDs. We found that the domain scientists had difficulties representing complex ideas, implicit concepts, and time associations. Additionally, determining the best ontology term to use when creating annotations was not always clear. These challenges relate to the limitation that this approach has some reliance on the annotator containing knowledge about relevant ontologies in the domain of discourse. Several steps to help alleviate these challenges are discussed in Section 6.
Another limitation of this approach is that it currently only supports the annotation of tabular data. Adopting techniques from some of the methods discussed in Section 2.2.2 can help with a future extension to support XML data. Additions to support the annotation of unstructured text data is beyond the scope of this work. Finally, we acknowledge that the annotation process discussed in this article is mostly done manually. This limitation decreases the likelihood of the adoption of this approach by those wishing to streamline the annotation process or incorporate the approach as part of a larger workflow. While automated annotation is not yet supported, existing research on an SDD editor is being conducted by members of the Tetherless World Constellation (TWC) involves the incorporation of Natural Language Processing (NLP) techniques to suggest concepts from ontologies based on text descriptions.
Our approach was outperformed in a few of the evaluation metrics, including space annotation, domain knowledge support, and the leveraging of best practices. Space annotation, to some degree, is supported through the use of implicit entries and property customization. Nevertheless, the SDD approach received a 0.5 rather than a 1 for this metric since, unlike Karma, which supports the annotation of geospatial data, and contains tutorials for how to annotate such data and tools developed specifically for geospatial data integration [67, 68, 69], it does not readily allow for the incorporation of the longitudinal and latitudinal coordinates. While the SDD approach allows the use of domain ontologies during the annotation process, a score of 0.5 was assigned to the domain knowledge support metric since we have not developed tools that suggest to the user the most appropriate domain concept to use. Nevertheless, as mentioned above, on-going work on an SDD editor will leverage NLP techniques to allow for this capability. Finally, while many of the DWBP and HCLS recommendations are incorporated into our approach, a score of 0.5 was received in terms of leveraging best practices because additional standards for these guidelines have yet to be incorporated. Additionally, further alignment with that standards mentioned in Section 2.3 should be achieved. The relevant best practices associated with our approach has been a subject of much discussion; further incorporation of these recommendations will be included in future revisions.
An ideal knowledge model promotes improved discovery, interoperability, reuse, traceability, and reproducibility. The knowledge model resulting from the SDD approach adheres to Semantic Web standards, resulting in improved discovery on the web, as well as interoperability with systems that also use RDF data serializations. These artifacts are reusable, as SDD tables created for one dataset can be reused to annotate another similar dataset. Scientific studies involving SDDs are traceable and reproducible by design, as the artifacts designed during the modeling process can be published and shared, helping to ensure consistency for other researchers attempting to examine the studies.
In this work, we advance the state of the art of metadata capture of datasets by improving on existing standards with the formalization of the Semantic Data Dictionary specification, which produces machine-readable knowledge representations by leveraging Semantic Web technologies. This is achieved by formalizing the assignment of a semantic representation of data and annotating dataset columns and their values using concepts from best practice ontologies. We provide resources such as documentation, examples, tutorials, and modeling guidelines to aid those who wish to create their own Semantic Data Dictionaries. We claim that this approach and the resulting artifacts are FAIR, help address limitations of traditional data dictionaries, and provides a bridge between representation methods used by domain scientists and semantic mapping approaches. We evaluate this work by defining metrics over several relevant categorizations, and scoring the Semantic Data Dictionary, traditional data dictionaries, mapping languages, and data integration tools for each metric. As we provide a methodology to aid in scientific workflows, this work eases the semantic annotation process for data providers and users alike.
Acknowledgements
This work is supported by the National Institute of Environmental Health Sciences (NIEHS) Award 0255-0236-4609 / 1U2CES026555-01, IBM Research AI through the AI Horizons Network, and the CAPES Foundation Senior Internship Program Award 88881.120772 / 2016-01. We acknowledge the members of the Tetherless World Constellation (TWC) and the Institute for Data Exploration and Applications (IDEA) at Rensellaer Polytechnic Institute (RPI) for their contributions, including Rebecca Cowan, John Erickson, and Oshani Seneviratne.
Author Biography
 Sabbir M. Rashid is a Ph.D. student at Rensselaer Polytechnic Institute
working with Professor Deborah McGuinness on research related to data annotation and
harmonization, ontology engineering, knowledge representation, and various forms of
reasoning. Prior to attending RPI, Mr. Rashid completed a double major at Worcester
Polytechnic Institute, where he received B.S. degrees in both Physics and Electrical
& Computer Engineering. Much of his graduate studies at RPI have involved the
research discussed in this article. His current work includes the application of
deductive and abductive inference techniques over linked health data, such as in the
context of chronic diseases like diabetes.
Sabbir M. Rashid is a Ph.D. student at Rensselaer Polytechnic Institute
working with Professor Deborah McGuinness on research related to data annotation and
harmonization, ontology engineering, knowledge representation, and various forms of
reasoning. Prior to attending RPI, Mr. Rashid completed a double major at Worcester
Polytechnic Institute, where he received B.S. degrees in both Physics and Electrical
& Computer Engineering. Much of his graduate studies at RPI have involved the
research discussed in this article. His current work includes the application of
deductive and abductive inference techniques over linked health data, such as in the
context of chronic diseases like diabetes.

James P. McCusker is the Director of Data Operations at the Tetherless World Constellation at Rensselaer Polytechnic Institute. He works with Deborah McGuinness on using knowledge graphs to further scientific research, especially in biomedical domains. He has worked on applying semantics to numerous projects, including drug repurposing using systems biology, cancer genome resequencing, childhood health and environmental exposure, analysis of sea ice conditions, and materials science. He is the architect of the open source Whyis knowledge graph development and management framework, which has been used across many of these domains.
 Paulo Pinheiro is a data scientist and software engineer managing projects
at the frontier between artificial intelligence and databases. His areas of
expertise includes the following: data policies and information assurance, such as
security and privacy; data operation including curation, quality monitoring,
semantic integration, provenance management, and uncertainty assessment; data
visualization; and data analytics including automated reasoning. Paulo holds a Ph.D.
in Computer Science from the University of Manchester, UK.
Paulo Pinheiro is a data scientist and software engineer managing projects
at the frontier between artificial intelligence and databases. His areas of
expertise includes the following: data policies and information assurance, such as
security and privacy; data operation including curation, quality monitoring,
semantic integration, provenance management, and uncertainty assessment; data
visualization; and data analytics including automated reasoning. Paulo holds a Ph.D.
in Computer Science from the University of Manchester, UK.

Marcello P. Bax is a professor and researcher in the Postgraduate Program in Knowledge Management and Organization (PPG-GOC) at the School of Information Science at Federal University of Minas Gerais, Brazil. Prior to joining the School of Information Science, Dr. Bax was a postdoctoral fellow in the Computer Science Department at UFMG, a leading Computer Science research group in Latin America. Dr. Bax spent a year on sabbatical with Professor McGuinness’ group and the Tetherless World Constellation at RPI, during which he worked with the coauthors on the research described in this article. His research seeks to develop methods for the curating of scientific data, with a focus on semantic annotation, the goal of building curatorial repositories for data reuse and reproduction of scientific research results.

Henrique Santos is a Research Scientist in the Tetherless World Constellation at Rensselaer Polytechnic Institute, where he researches and applies Semantic Web technologies in multidisciplinary domains for supporting more flexible, more efficient, and improved solutions in comparison with traditional approaches. His research interests include data integration, knowledge representation, domain-specific reasoning, and explainable artificial intelligence. He has over 10 years of experience working with Semantic Web technologies and holds a Ph.D. in Applied Informatics from Universidade de Fortaleza.
 Jeanette A. Stingone is an Assistant Professor in the Department of
Epidemiology at Columbia University’s Mailman School of Public Health. She
couples data science techniques with epidemiologic methods to address research
questions in children’s environmental health. She currently leads the Data
Science Translation and Engagement Group of the Human Health and Exposure Analysis
Resource Data Center. In this role, she supports the use of metadata standards and
ontologies for data harmonization efforts across disparate studies of environmental
health. Dr. Stingone’s interests also include the use of collective science
initiatives to advance public health research.
Jeanette A. Stingone is an Assistant Professor in the Department of
Epidemiology at Columbia University’s Mailman School of Public Health. She
couples data science techniques with epidemiologic methods to address research
questions in children’s environmental health. She currently leads the Data
Science Translation and Engagement Group of the Human Health and Exposure Analysis
Resource Data Center. In this role, she supports the use of metadata standards and
ontologies for data harmonization efforts across disparate studies of environmental
health. Dr. Stingone’s interests also include the use of collective science
initiatives to advance public health research.

Amar K. Das is the Program Director of Integrated Care Research at IBM Research and an Adjunct Associate Professor of Biomedical Data Science at Dartmouth College. His research activities include the development of biomedical ontologies and Semantic Web technologies for clinical decision support, information retrieval, and machine learning. In his role in the RPI-IBM HEALS initiative, Dr. Das is the IBM technical lead for advancing knowledge representation and reasoning in healthcare. Dr. Das holds an M.D. and Ph.D. in Biomedical Informatics from Stanford University, and has completed a residency in Psychiatry and a postdoctoral fellowship in Clinical Epidemiology at Columbia University/New York State Psychiatric Institute.
 Deborah L. McGuinness is the Tetherless World Senior Constellation Chair
and Professor of Computer and Cognitive Science. She is also the founding director
of the Web Science Research Center at Rensselaer Polytechnic Institute. Dr.
McGuinness has been recognized with awards as a fellow of the American Association
for the Advancement of Science (AAAS) for contributions to the Semantic Web,
knowledge representation, and reasoning environments and as the recipient of the
Robert Engelmore award from Association for the Advancement of Artificial
Intelligence (AAAI) for leadership in Semantic Web research and in bridging
Artificial Intelligence (AI) and eScience, significant contributions to deployed AI
applications, and extensive service to the AI community. Deborah is a leading
authority on the Semantic Web and has been working in knowledge representation and
reasoning environments for over 30 years and leads the research group that designed
and implemented the research presented in this paper.
Deborah L. McGuinness is the Tetherless World Senior Constellation Chair
and Professor of Computer and Cognitive Science. She is also the founding director
of the Web Science Research Center at Rensselaer Polytechnic Institute. Dr.
McGuinness has been recognized with awards as a fellow of the American Association
for the Advancement of Science (AAAS) for contributions to the Semantic Web,
knowledge representation, and reasoning environments and as the recipient of the
Robert Engelmore award from Association for the Advancement of Artificial
Intelligence (AAAI) for leadership in Semantic Web research and in bridging
Artificial Intelligence (AI) and eScience, significant contributions to deployed AI
applications, and extensive service to the AI community. Deborah is a leading
authority on the Semantic Web and has been working in knowledge representation and
reasoning environments for over 30 years and leads the research group that designed
and implemented the research presented in this paper.
Appendix A. Namespace Prefixes
Table A.1:
Namespace Prefixes and IRIs for Relevant Ontologies
| Ontology | Prefix | IRI |
|---|---|---|
| Children’s Health Exposure Analysis Resource | chear | http://hadatac.org/ont/chear# |
| Dublin Core Terms | dct | http://purl.org/dc/terms/ |
| Exposure Ontology | exo | http://purl.obolibrary.org/obo/ExO_ |
| National Cancer Institute Thesaurus | ncit | http://purl.obolibrary.org/obo/NCIT_ |
| Provenance, Authoring and Versioning | pav | http://purl.org/pav/ |
| Provenance Ontology | prov | http://www.w3.org/ns/prov# |
| RDF Schema | rdfs | http://www.w3.org/2000/01/rdf-schema# |
| Resource Description Framework | rdf | http://www.w3.org/1999/02/22-rdf-syntax-ns# |
| Schema.org | schema | https://schema.org/ |
| Semanticscience Integrated Ontology | sio | http://semanticscience.org/resource/ |
| Simple Knowledge Organization System | skos | http://www.w3.org/2004/02/skos/core# |
| Units of Measurement Ontology | uo | http://purl.obolibrary.org/obo/UO_ |
| Web Ontology Language | owl | http://www.w3.org/2002/07/owl# |
| XML Schema Data types | xsd | http://www.w3.org/2001/XMLSchema# |
Appendix B. Specifications
Due to the subjective nature of deciding the importance of each component, the rows in each of the specifications are shown in alphabetical order rather than in a meaningful sequence.
Appendix B.1. Infosheet Specification
Table B.1:
Infosheet Specification
| Infosheet Row | Description |
|---|---|
| Code Mapping | Reference to Code Mapping table location |
| Codebook | Reference to Codebook table location |
| Dictionary Mapping | Reference to Dictionary Mapping table location |
| Properties | Reference to Properties table location |
| Timeline | Reference to Timeline table location |
Table B.2:
Infosheet Metadata Supplement
| Infosheet Row | Related Property | Description |
|---|---|---|
| Contributors | dct:contributor | Contributors to the SDD |
| Creators | dct:creator | Creators of the SDD |
| Date Created | dct:created | Date the SDD was created |
| Description | dct:description | Description of the KG fragment |
| ImporTS | owl:imports | Ontologies that the SDD references |
| Keywords | schema:keywords | Keywords to be associated with the KG fragment |
| License | dct:license | License URL |
| Previous Version | pav:previousVersion | Previous version URL |
| Publisher | dct:publisher | Publisher of the SDD |
| Title | dct:title | Title of KG fragment |
| Version | owl:versionInfo | Current version URL |
| Version OF | dct:isVersionOf | Resource URL for primary version |
Appendix B.2. Dictionary Mapping Specification
Table B.3:
Dictionary Mapping Specification
| DM Column | Related Property | Description |
|---|---|---|
| Attribute | rdf:type | Class of attribute entry |
| attributeOF | sio:isAttributeOf | Entity having the attribute |
| Column | Entry column header in dataset | |
| Entity | rdf:type | Class of entity entry |
| Format | Specifies the structure of the cell value | |
| inRelationTo | sio:inRelationTo | Entity that the role is linked to |
| Label | rdfs:label | Label for the entry |
| Relation | Custom property used in inRelationTo | |
| Role | sio:hasRole | Type of the role of the entry |
| Time | sio:existsAt | Time point of measurement |
| Unit | sio:hasUnit | Unit of measure for entry |
| wasDerivedFrom | prov:wasDerivedFrom | Entity from which the entry was derived |
| wasGeneraTEDBy | prov:wasGeneratedBy | Activity from which the entry was produced |
Appendix B.3. Codebook Specification
Table B.4:
Codebook Specification
| Codebook Column | Related Property | Description |
|---|---|---|
| Class | rdf:type | Class the Code refers to |
| Code | sio:hasValue | Value of the dataset entry |
| Column | Entry column header in dataset | |
| Label | rdfs:label | Label for the codebook entry |
| Resource | rdf:type | Web Resource URI the Code refers to |
Appendix B.4. Timeline Specification
Table B.5:
Timeline Specification
| Timeline Column | Related Property | Description |
|---|---|---|
| End | sio:hasEndTime | The starting time point associated with the Timeline entry |
| inRelationTo | sio:inRelationTo | Entity that the Timeline entry is associated with |
| Name | Implicit entry reference for the Timeline entry | |
| Start | sio:hasStartTime | The starting time point associated with the Timeline entry |
| Type | rdf:type | Class the Timeline entry refers to |
| Unit | sio:hasUnit | Unit of measure for Timeline entry |
Appendix B.5. Properties Specification
Table B.6:
Properties Specification
| Row | Property |
|---|---|
| Attribute | rdf:type |
| attributeOF | sio:isAttributeOf |
| Comment | rdfs:comment |
| Definition | skos:definition |
| End | sio:hasEndTime |
| Entity | rdf:type |
| inRelationTo | sio:inRelationTo |
| Label | rdfs:label |
| Role | sio:hasRole |
| Start | sio:hasStartTime |
| Time | sio:existsAt |
| Type | rdf:type |
| Unit | sio:hasUnit |
| Value | sio:hasValue |
| wasDerivedFrom | prov:wasDerivedFrom |
| wasGeneraTEDBy | prov:wasGeneratedBy |
Appendix C. National Health and Nutrition Examination Survey Annotations
Table C.1:
NHANES Demographics Infosheet
| Attribute | Value |
|---|---|
| Creators | Sabbir M. Rashid |
| Code Mapping | NHANES/config/code mappings.csv |
| Codebook | NHANES/input/CB/DEMO H Doc-CB.csv |
| Contributors | “James P. McCusker, Paulo Pinheiro, Marcello P. Bax, Henrique O. Santos, Alexander New, Shruthi Chari, Mathew Johnson,John S. Erickson, Kristin P. Bennett, Jeanette A. Stingone, Deborah L. McGuinness” |
| Date Created | 2018-10-14 |
| Description | KG fragment from manually annotated NHANES Demographics SDD. |
| Dictionary Mapping | NHANES/input/DM/DEMO H Doc-DM.csv |
| ImporTS | “http://semanticscience.org/ontology/sio-subset-labels.owl, http://hadatac.org/ont/chear/, http://purl.obolibrary.org/obo/ncit.owl” |
| Keywords | “demographics, gender, age, race, citizenship, marital status, household” |
| License | https://opensource.org/licenses/MIT |
| Previous Version | http://tw.rpi.edu/heals/kb/nhanes/1.1 |
| Properties | NHANES/config/Properties.csv |
| Publisher | Tetherless World Constellation |
| Timeline | NHANES/input/TL/DEMO H Doc-TL.csv |
| Title | The National Health and Nutrition Examination (NHANES) SDD KG |
| Version | http://tw.rpi.edu/heals/kb/nhanes/1.2 |
| Version OF | http://tw.rpi.edu/heals/kb/nhanes/ |
Table C.2:
NHANES Demographic Implicit Entries
| Column | Label | Entity | Role | inRelationTo |
|---|---|---|---|---|
| ??participant | Participant | ncit:C29867, sio:Human | sio:SubjectRole | |
| ??screening | Screening | chear:Screening | ||
| ??exam | Examination | ncit:C131902 | ||
| ??birth | Birth | sio:Birthing | ||
| ??pregnancy | Pregnancy | chear:Pregnancy | ||
| ??interview | Interview | ncit:C16751 | ||
| ??instrument | Instrumentation | ncit:C16742 | ||
| ??household | Household | chear:Household | ??participant | |
| ??HHRef | Household reference | sio:Human | chear:HeadOfHousehold | ??household |
Table C.3:
NHANES Demographic Explicit Entries
| Column | Label | Attribute | attributeOF | Unit | Time | Entity | Relation | inRelationTo |
|---|---|---|---|---|---|---|---|---|
| SEQN | Respondent sequence number | sio:Identifier | ??participant | |||||
| RIAGENDR | Gender | sio:BiologicalSex | ??participant | |||||
| RIDAGEYR | Age in years at screening | sio:Age | ??participant | yr | ??screening | |||
| RIDAGEMN | Age in months at screening | sio:Age | ??participant | mth | ??screening | |||
| RIDRETH1 | Race/Hispanic origin | sio:Race | ??participant | |||||
| RIDEXAGM | Age in months at exam | sio:Age | ??participant | mth | ??exam | |||
| DMDBORN4 | Country of birth | ??birth | sio:Country | sio:isLocationOf | ??participant | |||
| DMDCITZN | Citizenship status | sio:StatusDescriptor | ??participant | |||||
| DMDYRSUS | Length of time in US | sio:TimeInterval | ??participant | |||||
| DMDEDUC3 | Education level - Children/Youth | chear:EducationLevel | ??participant | |||||
| DMDEDUC2 | Education level - Adults 20+ | chear:EducationLevel | ??participant | |||||
| DMDMARTL | Marital status | chear:MaritalStatus | ??participant | |||||
| RIDEXPRG | Pregnancy status at exam | sio:StatusDescriptor | ??pregnancy | ??exam | ??participant | |||
| SIALANG | Language of SP Interview | chear:Language | ??instrument | ??interview | ??participant | |||
| DMDHRGND | HH ref person’s gender | sio:BiologicalSex | ??HHRef | |||||
| DMDHRAGE | HH ref person’s age in years | sio:Age | ??HHRef | yr | ||||
| DMDHRBR4 | HH ref person’s country of birth | ??birth | sio:Country | sio:isLocationOf | ??HHRef | |||
| DMDHREDU | HH ref person’s education level | chear:EducationLevel | ??HHRef | |||||
| DMDHRMAR | HH ref person’s marital status | chear:MaritalStatus | ??HHRef | |||||
| WTINT2YR | Full sample 2 year interview wt | chear:Weight | ??participant | ??interview | ||||
| WTMEC2YR | Full sample 2 year MEC exam wt | chear:Weight | ??participant | ??exam | ||||
| INDHHIN2 | Annual household income | chear:Income | ??household |
Table C.4:
Expanded NHANES Demographic Codebook Entries
| Column | Code | Label | Class |
|---|---|---|---|
| RIAGENDR | 1 | Male | sio:Male |
| RIAGENDR | 2 | Female | sio:Female |
| RIAGENDR | . | Missing | ncit:C142610 |
| RIDRETH1 | 1 | Mexican American | exo:0000151 |
| RIDRETH1 | 2 | Other Hispanic | exo:0000145 |
| RIDRETH1 | 3 | Non-Hispanic White | exo:0000158 |
| RIDRETH1 | 4 | Non-Hispanic Black | exo:0000132 |
| RIDRETH1 | 5 | Other Race - Including Multi-Racial | exo:0000153 |
| RIDRETH1 | . | Missing | ncit:C142610 |
| DMDEDUC3 | 0 | Never attended / kindergarten only | chear:NoFormalEducation |
| DMDEDUC3 | 1 | 1st grade | chear:EducationGrade |
| DMDEDUC3 | 2 | 2nd grade | chear:EducationGrade |
| DMDEDUC3 | 3 | 3rd grade | chear:EducationGrade |
| DMDEDUC3 | 4 | 4th grade | chear:EducationGrade |
| DMDEDUC3 | 5 | 5th grade | chear:EducationGrade |
| DMDEDUC3 | 6 | 6th grade | chear:EducationGrade |
| DMDEDUC3 | 7 | 7th grade | chear:EducationGrade |
| DMDEDUC3 | 8 | 8th grade | chear:EducationGrade |
| DMDEDUC3 | 9 | 9th grade | chear:EducationGrade |
| DMDEDUC3 | 10 | 10th grade | chear:EducationGrade |
| DMDEDUC3 | 11 | 11th grade | chear:EducationGrade |
| DMDEDUC3 | 12 | “12th grade, no diploma” | chear:SomeHighSchool |
| DMDEDUC3 | 13 | High school graduate | chear:HighSchoolGraduate |
| DMDEDUC3 | 14 | GED or equivalent | ncit:C67135 |
| DMDEDUC3 | 15 | More than high school | chear:HigherEducation |
| DMDEDUC3 | 55 | Less than 5th grade | chear:SomeElementarySchool |
| DMDEDUC3 | 66 | Less than 9th grade | chear:SomeMiddleSchool |
| DMDEDUC3 | 77 | Refused | ncit:C49161 |
| DMDEDUC3 | 99 | Don’t Know | ncit:C67142 |
| DMDEDUC3 | . | Missing | ncit:C142610 |
| DMDEDUC2 | 1 | Less than 9th grade | chear:SomeMiddleSchool |
| DMDEDUC2 | 2 | 9–11th grade | chear:SomeHighSchool |
| DMDEDUC2 | 3 | High school graduate/GED or equivalent | chear:HighSchoolGraduate |
| DMDEDUC2 | 4 | Some college or AA degree | chear:SomeCollege |
| DMDEDUC2 | 5 | College graduate or above | chear:CollegeGraduate |
| DMDEDUC2 | 7 | Refused | ncit:C49161 |
| DMDEDUC2 | 9 | Don’t Know | ncit:C67142 |
| DMDEDUC2 | . | Missing | ncit:C142610 |
| DMDMARTL | 1 | Married | ncit:C51773 |
| DMDMARTL | 2 | Widowed | ncit:C51775 |
| DMDMARTL | 3 | Divorced | ncit:C51776 |
| DMDMARTL | 4 | Separated | ncit:C51777 |
| DMDMARTL | 5 | Never married | ncit:C51774 |
| DMDMARTL | 6 | Living with partner | ncit:C53262 |
| DMDMARTL | 77 | Refused | ncit:C49161 |
| DMDMARTL | 99 | Don’t Know | ncit:C67142 |
| DMDMARTL | . | Missing | ncit:C142610 |
Footnotes
https://github.com/USG-SCOPE/data-dictionary/blob/gh-pages/Metadata-Scheme-for-Data-Dictionaries.md
A listing of ontology prefixes used in this article is provided in Appendix Table A.1.
When referencing columns from any of the SDD tables, the Small Caps typeface is used.
When including implicit entries in an SDD table, the prefix “??” is used as a distinguishing labeling feature. The typewriter typeface is used in this article when referring to instances of implicit entries.
The italics typeface is used when a property from an ontology is mentioned.
rdf:type, sio:isAttributeOf, rdfs:comment, skos:definition, sio:hasStartTime, sio:existsAt, sio:hasEndTime, sio:inRelationTo, rdfs:label, sio:hasRole, sio:hasUnit, sio:hasValue, prov:wasDerivedFrom, & prov:wasGeneratedBy
References
- [1].IBM, IBM Dictionary of Computing, 10th ed., McGraw-Hill, Inc., New York, NY, USA, 1993. https://dl.acm.org/doi/book/10.5555/541721. [Google Scholar]
- [2].Uhrowczik PP, Data dictionary/directories, IBM Systems Journal 12 (1973) 332–350. 10.1147/sj.124.0332. [DOI] [Google Scholar]
- [3].Duval E, Hodgins W, Sutton S, Weibel SL, Metadata principles and practicalities, D-lib Magazine 8 (2002) 1082–9873. 10.1045/april2002-weibel. [DOI] [Google Scholar]
- [4].Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, et al. , The fair guiding principles for scientific data management and stewardship, Scientific data 3 (2016). 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boeckhout M, Zielhuis GA, Bredenoord AL, The fair guiding principles for data stewardship: fair enough?, European journal of human genetics 26 (2018) 931 10.1038/s41431-018-0160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McCusker JP, Rashid SM, Liang Z, Liu Y, Chastain K, Pinheiro P, Stingone JA, McGuinness DL, Broad, interdisciplinary science in tela: An exposure and child health ontology, in: WebSci ‘17: Proceedings of the 2017 ACM on Web Science Conference, 2017, pp. 349–357. 10.1145/3091478.3091497. [DOI] [Google Scholar]
- [7].Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. , The obo foundry: coordinated evolution of ontologies to support biomedical data integration, Nature biotechnology 25 (2007) 1251 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, Jonquet C, Rubin DL, Storey M-A, Chute CG, et al. , Bioportal: ontologies and integrated data resources at the click of a mouse, Nucleic acids research 37 (2009) W170–W173. 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Joo J, Adoption of semantic web from the perspective of technology innovation: A grounded theory approach, International journal of human-computer studies 69 (2011) 139–154. 10.1016/j.ijhcs.2010.11.002. [DOI] [Google Scholar]
- [10].Staab S, Maedche A, Handschuh S, An annotation framework for the semantic web, Inst. AIFB, Univ, 2001. http://citeseerx.ist.psu.edu/viewdoc/citations?doi=10.1.1.25.910.
- [11].Handschuh S, Staab S, Annotation for the semantic web, volume 96, IOS Press, 2003. https://doi.org/10.10072F978-3-642-20323-7_2. [Google Scholar]
- [12].Chen B, Ding Y, Wild DJ, Improving integrative searching of systems chemical biology data using semantic annotation, Journal of cheminformatics 4 (2012) 6 10.1186/1758-2946-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wei W, Barnaghi PM, Bargiela A, Semantic-enhanced information search and retrieval, in: Sixth International Conference on Advanced Language Processing and Web Information Technology (ALPIT 2007), IEEE, 2007, pp. 218–223. 10.1109/ALPIT.2007.59. [DOI] [Google Scholar]
- [14].Atici L, Kansa SW, Lev-Tov J, Kansa EC, Other people’s data: a demonstration of the imperative of publishing primary data, Journal of Archaeological Method and Theory 20 (2013) 663–681. 10.1007/s10816-012-9132-9. [DOI] [Google Scholar]
- [15].Willoughby KW, Technological semantics and technological practice: Lessons from an enigmatic episode in twentieth-century technology studies, Knowledge, Technology & Policy 17 (2004) 11–43. 10.1007/s12130-004-1002-7. [DOI] [Google Scholar]
- [16].Haskell RE, Heil JA, Cassidy J, Dynamic dictionary and term repository system, 2009. US: Patent 7,580,831 https://patents.google.com/patent/US7580831B2/en.
- [17].Lau L, Endo J, Karren S, Willis M, Harada S, Beeney S, Larsen B, Cassin E, Gerard M, Mapping clinical data with a health data dictionary, 2002. US: Patent App. 09/755,966 https://patents.google.com/patent/US20020128861A1/en.
- [18].Apacible JT, Nolan SP, Kalmady GD, Varadan V, Extensible and localizable health-related dictionary, 2013. US: Patent 8,417,537 https://patents.google.com/patent/US8417537B2/en.
- [19].Thompson FC, Data dictionary and standards for fruit fly information database, Myia (1999). https://repository.si.edu/handle/10088/18512.
- [20].W. W. W. Consortium, et al. , Data catalog vocabulary (dcat) (2014). https://www.w3.org/TR/2020/SPSD-vocab-dcat-20200204/.
- [21].Lenzerini M, Data integration: A theoretical perspective, in: Proceedings of the twenty-first ACM SIGMOD-SIGACT-SIGART symposium on Principles of database systems, ACM, 2002, pp. 233–246. 10.1145/543613.543644. [DOI] [Google Scholar]
- [22].del Carmen Legaz-García M, Miñarro-Giménez JA, Menárguez-Tortosa M, Fernández-Breis JT, Generation of open biomedical datasets through ontology-driven transformation and integration processes, Journal of biomedical semantics 7 (2016) 32 10.1186/s13326-016-0075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Miller JJ, Graph database applications and concepts with neo4j, in: Proceedings of the Southern Association for Information Systems Conference, Atlanta, GA, USA, volume 2324, 2013, pp. 141–147. https://pdfs.semanticscholar.org/322a/6e1f464330751dea2eb6beecac24466322ad.pdf. [Google Scholar]
- [24].Liebig T, Vialard V, Opitz M, Metzl S, Graphscale: Adding expressive reasoning to semantic data stores., in: International Semantic Web Conference (Posters & Demos), 2015. https://www.semanticscholar.org/paper/GraphScale%3A-Adding-Expressive-Reasoning-to-Semantic-Liebig-Vialard/1f3652b98b825f2ec4fd4a5c2bd2416377eef8b0. [Google Scholar]
- [25].Liebig T, Neo4j: A reasonable rdf graph database & reasoning engine, 2018. Accessed: 2019-05-21 https://neo4j.com/blog/neo4j-rdf-graph-database-reasoning-engine/.
- [26].Santipantakis GM, Kotis KI, Vouros GA, Doulkeridis C, Rdf-gen: Generating rdf from streaming and archival data, in: Proceedings of the 8th International Conference on Web Intelligence, Mining and Semantics, ACM, 2018, p. 28 10.1145/3227609.3227658. [DOI] [Google Scholar]
- [27].Pinkel C, Schwarte A, Trame J, Nikolov A, Bastinos AS, Zeuch T, Dataops: seamless end-to-end anything-to-rdf data integration, in: European Semantic Web Conference, Springer, 2015, pp. 123–127. 10.1007/978-3-319-25639-9_24. [DOI] [Google Scholar]
- [28].Ham K, Free open-source tool for cleaning and transforming data, Journal of the Medical Library Association: JMLA 2013 101 (2013) 233–234. http://openrefine.org. [Google Scholar]
- [29].Arenas M, Bertails A, Prud’hommeaux E, Sequeda J, A direct mapping of relational data to rdf, W3C recommendation 27 (2012) 1–11. https://www.w3.org/TR/rdb-direct-mapping/. [Google Scholar]
- [30].Dimou A, Vander Sande M, Colpaert P, De Vocht L, Verborgh R, Mannens E, Van de Walle R, Extraction and semantic annotation of workshop proceedings in html using rml, in: Semantic Web Evaluation Challenge, Springer, 2014, pp. 114–119. 10.1007/978-3-319-12024-9_15. [DOI] [Google Scholar]
- [31].Heyvaert P, Dimou A, Herregodts A-L, Verborgh R, Schuurman D, Mannens E, Van de Walle R, Rmleditor: a graph-based mapping editor for linked data mappings, in: European Semantic Web Conference, Springer, 2016, pp. 709–723. 10.1007/978-3-319-34129-3_43. [DOI] [Google Scholar]
- [32].Michel F, Djimenou L, Zucker CF, Montagnat J, Translation of relational and non-relational databases into rdf with xr2rml, in: 11th International Confenrence on Web Information Systems and Technologies (WEBIST’15), 2015, pp. 443–454. 10.5220/0005448304430454. [DOI] [Google Scholar]
- [33].V. J. Provider, Openlink virtuoso universal server: Documentation, OpenLink Software (2009). http://docs.openlinksw.com/virtuoso/.
- [34].Slepicka J, Yin C, Szekely PA, Knoblock CA, Kr2rml: An alternative interpretation of r2rml for heterogenous sources., in: Cold, 2015. http://usc-isi-i2.github.io/papers/slepicka15-cold.pdf.
- [35].Knoblock CA, Szekely P, Exploiting semantics for big data integration., AI Magazine 36 (2015). 10.1609/aimag.v36i1.2565. [DOI] [Google Scholar]
- [36].Tennison J, Kellogg G, Herman I, Generating rdf from tabular data on the web. recommendation, World Wide Web Consortium (W3C) (2015). https://www.w3.org/TR/csv2rdf.
- [37].Dimou A, Vander Sande M, Colpaert P, Mannens E, Van de Walle R, Extending r2rml to a source-independent mapping language for rdf., in: International Semantic Web Conference (Posters & Demos), volume 1035, 2013, pp. 237–240. https://dl.acm.org/doi/10.5555/2874399.2874459. [Google Scholar]
- [38].Ermilov I, Auer S, Stadler C, Csv2rdf: User-driven csv to rdf mass conversion framework, in: Proceedings of the ISEM, volume 13, 2013, pp. 04–06. https://www.bibsonomy.org/bibtex/23bc97cec6ee1214c47991bf4f70f479c/soeren. [Google Scholar]
- [39].HAIDER N, HOSSAIN F, Csv2rdf: generating rdf data from csv file using semantic web technologies, Journal of Theoretical and Applied Information Technology 96 (2018). http://www.jatit.org/volumes/Vol96No20/19Vol96No20.pdf. [Google Scholar]
- [40].Stadler C, Unbehauen J, Westphal P, Sherif MA, Lehmann J, Simplified rdb2rdf mapping, in: LDOW@WWW, volume 1409, 2015. http://publica.fraunhofer.de/documents/N-481451.html. [Google Scholar]
- [41].Bizer C, Schultz A, The r2r framework: Publishing and discovering mappings on the web., COLD 665 (2010) 97–108. https://dl.acm.org/doi/10.5555/2878947.2878956. [Google Scholar]
- [42].Cyganiak R, Tarql (sparql for tables): Turn csv into rdf using sparql syntax, Technical Report, 2015. http://tarql.github.io.
- [43].Bizer C, Seaborne A, D2rq-treating non-rdf databases as virtual rdf graphs, in: Proceedings of the 3rd international semantic web conference (ISWC2004), volume 2004, Proceedings of ISWC2004, 2004. 10.1038/npre.2011.5660.1. [DOI] [Google Scholar]
- [44].Bizer C, Cyganiak R, D2rq-lessons learned, in: W3C Workshop on RDF Access to Relational Databases, 2007, p. 35 https://www.w3.org/2007/03/RdfRDB/papers/d2rq-positionpaper/. [Google Scholar]
- [45].Čerāns K, Būmans G, Rdb2owl: a rdb-to-rdf/owl mapping specification language, Information Systems (2011) 139–152. http://rdb2owl.lumii.lv/pubs/rdb2owl_main_2011.pdf. [Google Scholar]
- [46].Kl J, Škoda P, Nečasky M, Linkedpipes etl: Evolved linked data preparation, in: European Semantic Web Conference, Springer, 2016, pp. 95–100. 10.1007/978-3-319-47602-5_20. [DOI] [Google Scholar]
- [47].McCusker JP, Chastain K, Rashid S, Norris S, McGuinness DL , Setlr: the semantic extract, transform, and load-r, PeerJ Preprints 6 (2018) e26476v1 10.7287/peerj.preprints.26476v1. [DOI] [Google Scholar]
- [48].Post AR, Krc T, Rathod H, Agravat S, Mansour M, Torian W, Saltz JH, Semantic etl into i2b2 with eureka!, AMIA Summits on Translational Science Proceedings 2013 (2013) 203 https://www.ncbi.nlm.nih.gov/pubmed/24303265. [PMC free article] [PubMed] [Google Scholar]
- [49].Schultz A, Matteini A, Isele R, Bizer C, Becker C, Ldif-linked data integration framework, in: Proceedings of the Second International Conference on Consuming Linked Data-Volume 782, CEUR-WS. org, 2011, pp. 125–130. http://ldif.wbsg.de/. [Google Scholar]
- [50].Schultz A, Matteini A, Isele R, Mendes PN, Bizer C, Becker C, Ldif-a framework for large-scale linked data integration, in: 21st International World Wide Web Conference (WWW 2012), Developers Track, Lyon, France, 2012. 10.17169/refubium-18883. [DOI] [Google Scholar]
- [51].Skoutas D, Simitsis A, Designing etl processes using semantic web technologies, in: Proceedings of the 9th ACM international workshop on Data warehousing and OLAP, ACM, 2006, pp. 67–74. 10.1145/1183512.1183526. [DOI] [Google Scholar]
- [52].Bansal SK, Towards a semantic extract-transform-load (etl) framework for big data integration, in: 2014 IEEE International Congress on Big Data, IEEE, 2014, pp. 522–529. 10.1109/BigData.Congress.2014.82. [DOI] [Google Scholar]
- [53].Bansal SK, Kagemann S, Integrating big data: A semantic extract-transform-load framework, Computer 48 (2015) 42–50. 10.1109/MC.2015.76. [DOI] [Google Scholar]
- [54].Zozus MN, Bonner J, Rock L, Towards data value-level metadata for clinical studies., in: ITCH, 2017, pp. 418–423. 10.3233/978-1-61499-742-9-418. [DOI] [PubMed]
- [55].Warzel DB, Andonyadis C, McCurry B, Chilukuri R, Ishmukhamedov S, Covitz P, Common data element (cde) management and deployment in clinical trials, in: AMIA annual symposium proceedings, volume 2003, American Medical Informatics Association, 2003, p. 1048 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1480162/. [PMC free article] [PubMed] [Google Scholar]
- [56].Kuchinke W, Wiegelmann S, Verplancke P, Ohmann C, Extended cooperation in clinical studies through exchange of cdisc metadata between different study software solutions, Methods of information in medicine 45 (2006) 441–446. 10.1055/s-0038-1634102. [DOI] [PubMed] [Google Scholar]
- [57].Paulheim H, Knowledge graph refinement: A survey of approaches and evaluation methods, Semantic web 8 (2017) 489–508. 10.3233/SW-160218. [DOI] [Google Scholar]
- [58].Gkoutos GV, Schofield PN, Hoehndorf R, The units ontology: a tool for integrating units of measurement in science, Database 2012 (2012). 10.1093/database/bas033. [DOI] [PMC free article] [PubMed]
- [59].Dumontier M, Baker CJ, Baran J, Callahan A, Chepelev L, Cruz-Toledo J, Del Rio NR, Duck G, Furlong LI, Keath N, Klassen D, McCusker JP, Queralt-Rosinach N, Samwald M, Villanueva-Rosales N, Wilkinson MD, Hoehndorf R, The semanticscience integrated ontology (sio) for biomedical research and knowledge discovery, Journal of Biomedical Semantics 5 (2014) 14 10.1186/2041-1480-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Pinheiro P, Bax MP, Santos H, Rashid SM, Liang Z, Liu Y, Ne’eman Y, McCusker JP, McGuinness DL, Annotating diverse scientific data with hasco., in: ONTOBRAS, 2018, pp. 80–91. http://ceur-ws.org/Vol-2228/paper4.pdf.
- [61].Lebo T, Sahoo S, McGuinness D, Belhajjame K, Cheney J, Corsar D, Garijo D, Soiland-Reyes S, Zednik S, Zhao J, Prov-o: The prov ontology, W3C recommendation (2013). https://www.w3.org/TR/prov-o/.
- [62].Beckert B, Keller U, Schmitt PH, Translating the object constraint language into first-order predicate logic, in: Proc. of the VERIFY Workshop at Federated Logic Conferences (FLoC), 2002, pp. 113–123. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.18.5551. [Google Scholar]
- [63].Seneviratne O, Rashid SM, Chari S, McCusker JP, Bennett KP, Hendler JA, McGuinness DL, Knowledge integration for disease characterization: A breast cancer example, in: International Semantic Web Conference, Springer, 2018, pp. 223–238. 10.1007/978-3-030-00668-6_14. [DOI] [Google Scholar]
- [64].Crotti Junior A, Debruyne C, Brennan R, O’Sullivan D, An evaluation of uplift mapping languages, International Journal of Web Information Systems 13 (2017) 405–424. 10.1108/IJWIS-04-2017-0036. [DOI] [Google Scholar]
- [65].Hert M, Reif G, Gall HC, A comparison of rdb-to-rdf mapping languages, in: Proceedings of the 7th International Conference on Semantic Systems, ACM, 2011, pp. 25–32. 10.1145/2063518.2063522. [DOI] [Google Scholar]
- [66].Smith B, Kumar A, Bittner T, Basic Formal Ontology for Bioinformatics, IFOMIS Reports, 2005. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.89.3787.
- [67].Butenuth M, Gösseln G. v., Tiedge M, Heipke C, Lipeck U, Sester M, Integration of heterogeneous geospatial data in a federated database, ISPRS Journal of Photogrammetry and Remote Sensing 62 (2007) 328–346. 10.1016/j.isprsjprs.2007.04.003. [DOI] [Google Scholar]
- [68].Janowicz K, Scheider S, Pehle T, Hart G, Geospatial semantics and linked spatiotemporal data–past, present, and future, Semantic Web 3 (2012) 321–332. 10.3233/SW-2012-0077. [DOI] [Google Scholar]
- [69].Huang W, Mansourian A, Harrie L, Geospatial data integration and visualisation using linked data., in: AGILE PhD School, 2017. http://ceur-ws.org/Vol-2088/paper6.pdf.