Abstract
Embryonic anterior–posterior patterning is established in Drosophila melanogaster by maternally expressed genes. The mRNAs of several of these genes accumulate at either the anterior or posterior pole of the oocyte via a number of mechanisms. Many of these mRNAs are also under elaborate translational regulation. Asymmetric RNA localization coupled with spatially restricted translation ensures that their proteins are restricted to the position necessary for the developmental process that they drive. Bicoid (Bcd), the anterior determinant, and Oskar (Osk), the determinant for primordial germ cells and posterior patterning, have been studied particularly closely. In early embryos an anterior–posterior gradient of Bcd is established, activating transcription of different sets of zygotic genes depending on local Bcd concentration. At the posterior pole, Osk seeds formation of polar granules, ribonucleoprotein complexes that accumulate further mRNAs and proteins involved in posterior patterning and germ cell specification. After fertilization, polar granules associate with posterior nuclei and mature into nuclear germ granules. Osk accumulates in these granules, and either by itself or as part of the granules, stimulates germ cell division.
This article is categorized under:
RNA Export and Localization > RNA Localization
Translation > Translation Regulation
RNA in Disease and Development > RNA in Development
Keywords: embryonic patterning, ribonucleoprotein complexes, translational control
Where the fly embryo forms its head and its abdomen is determined by maternal RNAs that are transported to the anterior or posterior poles of the developing oocyte, where they are anchored and translationally activated through complex and fascinating mechanisms.
1. INTRODUCTION
Establishment of the anterior–posterior axis of the Drosophila embryo is among the most intensively studied developmental processes. The insight of Christiane Nüsslein‐Volhard, Eric Wieschaus and their colleagues who carried out comprehensive forward genetic screens to find mutations affecting this process, and their generosity in sharing data and making their mutants available to other researchers enabled the molecular identification by the early 1990s of many of the key genes involved and basic elucidation of the relevant pathways. Since that time a wealth of new information has been obtained, leading not only to a deep understanding of this developmental process, but also providing important insights into RNA‐level mechanisms of gene regulation that are widely employed in biology. Now, 30 years later, remarkable new discoveries still emerge from studying this system, providing new knowledge about how physical processes underlie regulation of genes responsible for cellular and developmental switches. Nevertheless, important questions also still remain to be answered.
The central events of anterior–posterior patterning can be summarized as follows. bcd and oskar (osk) mRNAs are transcribed in the nurse cells and transported through cytoplasmic bridges called ring canals to the presumptive oocyte, where, during mid‐oogenesis, they later accumulate at the anterior and posterior poles respectively (Berleth et al., 1988; Ephrussi, Dickinson, & Lehmann, 1991; Kim‐Ha, Smith, & Macdonald, 1991). Upon egg deposition, bcd is translated to produce an anterior‐to‐posterior gradient of Bcd protein, a homeodomain‐containing transcription factor that regulates expression of zygotic patterning genes in a concentration‐dependent manner (Driever & Nüsslein‐Volhard, 1988a, 1988b). Osk is the key maternal determinant for germ cell specification and for establishing the posterior somatic pole of the embryo (Ephrussi et al., 1991; Kim‐Ha et al., 1991; Lehmann & Nüsslein‐Volhard, 1986). osk RNA localizes to the posterior pole of the oocyte, where it is translated, via three sequential mechanisms described in more detail below. Osk protein nucleates the assembly of germ plasm in the late oocyte and formation of polar granules in the early embryo (Ephrussi et al., 1991; Ephrussi & Lehmann, 1992; Smith, Wilson, & Macdonald, 1992). Polar granules are considered to be determinants of primordial germ cells (PGCs), as cytoplasm containing polar granules induces pole cell formation at an ectopic site (Illmensee & Mahowald, 1974). Vasa (Vas), a DEAD‐box RNA helicase, Tudor (Tud), and Aubergine (Aub), a Piwi‐family protein that interacts with piwi‐interacting RNAs (piRNAs), are key functional components of polar granules that are recruited downstream of Osk during later stages of oogenesis (Bardsley, McDonald, & Boswell, 1993; Breitwieser, Markussen, Horstmann, & Ephrussi, 1996; Hay, Jan, & Jan, 1990; Lasko & Ashburner, 1990; Wilson, Connell, & Macdonald, 1996). Many other specific mRNAs are also recruited to polar granules, including nanos (nos), germ cell‐less (gcl), and polar granule component (pgc) (Jongens, Hay, Jan, & Jan, 1992; Nakamura, Amikura, Mukai, Kobayashi, & Lasko, 1996; Wang & Lehmann, 1991; Wharton & Struhl, 1991). nos encodes the main determinant for posterior pattern, while gcl and pgc are required for development of PGCs (Gavis & Lehmann, 1992; Nakamura et al., 1996). While nos is not required for PGC specification, it is essential for their subsequent development (Asaoka‐Taguchi, Yamada, Nakamura, Hanyu, & Kobayashi, 1999; Forbes & Lehmann, 1998). After fertilization and the migration of zygotic nuclei to the posterior pole, Osk also nucleates the formation of nuclear germ granules, ribonucleoprotein particles (RNPs) that are related to polar granules but are compositionally distinct (Kistler et al., 2018). Through an unknown mechanism, nuclear germ granules, or perhaps Osk specifically, promote pole cell division.
Space constraints do not permit a comprehensive review of all relevant literature, so here I will focus on recent advances toward understanding deployment and regulation of Bicoid (Bcd), the central determinant for anterior fate, and on the assembly of RNP) complexes called polar granules, which specify posterior fate and PGCs (often called pole cells in Drosophila).
2. ESTABLISHMENT OF THE BCD GRADIENT
2.1. bcd RNA movement is microtubule‐dependent but randomly directed
bcd mRNA is transcribed in nurse cells and transferred through the ring canals into the oocyte in later stages of oogenesis. Through several intermediate steps (Figure 1) it becomes tightly localized at the anterior pole of the oocyte, and its localization depends upon the activities of three other genes, staufen (stau), swallow (swa), and exuperantia (exu), and the ESCRT‐II endosomal sorting complex (Irion & St Johnston, 2007; St Johnston, Driever, Berleth, Richstein, & Nüsslein‐Volhard, 1989). Depolymerization of microtubules with drugs blocks all steps of bcd localization (Pokrywka & Stephenson, 1991). During late oogenesis, anterior localization of bcd is achieved through continuous microtubule‐dependent active transport (Weil, Forrest, & Gavis, 2006). However, beginning at the end of oogenesis bcd is anchored at the anterior in an actin‐dependent process (Weil, Parton, Davis, & Gavis, 2008).
FIGURE 1.
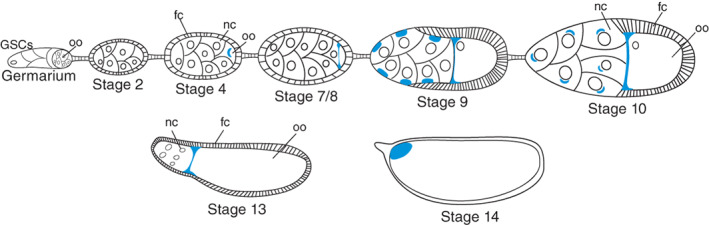
Cartoon illustrating various stages of Drosophila oogenesis, with sites of bcd RNA accumulation shown in blue. Germline stem cells (GSCs) at the anterior end of the germarium undergo four transit‐amplifying divisions and become enveloped by follicle cells (fc) to form egg chambers. The four mitotic divisions that germ cells undergo have incomplete cytokinesis, so the 16 germline cells remain interconnected by cytoplasmic bridges called ring canals. One of the germline cells that is connected to four other germline cells by ring canals becomes specified as the oocyte (oo) and is always located at the posterior end of the egg chamber. The other germline cells become nurse cells (nc). Nurse cells synthesize RNAs and proteins that can be transferred into the oocyte through ring canals. After stage 10, the cytoplasm of nurse cells is rapidly transferred into the oocyte. The nurse cells subsequently die and the mature stage 14 egg remains
A model for anterior bcd localization by which the motor protein dynein transports bcd toward the minus‐ends of microtubules emerged based primarily on observations that disruption of dynein function by overexpressing dynamitin results in dispersion of bcd RNA (Duncan & Warrior, 2002; Januschke et al., 2002). This model gained widespread acceptance, and was supported by experiments showing that injected bcd mRNA into nurse cells recruits dynein‐associated proteins required for mRNA transport (Bullock & Ish‐Horowicz, 2001; Clark, Meignin, & Davis, 2007).
However, a more recent study using improved imaging techniques to visualize bcd RNA in living oocytes confirms that bcd localization requires dynein, but paints a more complex picture than simple minus‐end directed transport (Trovisco et al., 2016). This study used a construct, bcdMS2, that expresses the bcd 3′ UTR, which is sufficient for all stages of localization (Macdonald & Struhl, 1988) fused to 11 copies of a stem‐loop element called MS2, which can be bound with very high affinity by the MS2 coat protein (MCP). Live imaging of bcd is then accomplished by coexpressing MCP conjugated to the fluorescent protein GFP (Bertrand et al., 1998; Weil et al., 2006). Using this approach, Trovisco and colleagues imaged small bcdMS2 containing particles that moved at speeds up to 2.2 μm/s. The mean velocity of these particles slowed in a hypomorphic dynein mutant background, increased in a kinesin mutant background, and the particles stopped moving altogether upon treatment with colcemid, a drug that depolymerizes microtubules. Consistent with earlier results, these experiments indicated that bcd RNA localization is dependent on dynein and microtubules and antagonized by kinesin, a plus‐end directed motor. However, assessment of the directionality of bcdMS2 particle movement showed almost no directional bias toward the anterior over the posterior, and an equal dependence on dynein for movement in either direction. These observations rule out a major contribution of directed transport to bcd RNA localization. Rather, anterior accumulation of bcd depends upon essentially random movement followed by actin‐dependent anchoring. This conclusion was supported by fluorescence recovery after photobleaching (FRAP) experiments that revealed a slow‐recovering population of bcd RNA at the anterior pole. This work also showed that the RNA‐binding protein Exu (Lazzaretti et al., 2016) is necessary to assemble bcdMS2 particles. How anterior bcd anchoring is maintained after egg activation remains to be fully determined.
2.2. bcd is translationally repressed until egg activation
bcd mRNA is translationally repressed throughout oogenesis and until egg activation (Sallés, Lieberfarb, Wreden, Gergen, & Strickland, 1994), but exactly how this is accomplished remains unclear. While early evidence suggested that translational regulation of bcd involved alterations in poly(A) tail length (Sallés et al., 1994), as occurs for many maternal mRNAs (Lim, Lee, Son, Chang, & Kim, 2016; Rouget et al., 2010), recent work using poly(A)‐tail length profiling by sequencing (PAL‐seq) (Subtelny, Eichhorn, Chen, Sive, & Bartel, 2014) showed no change in bcd poly(A) tail length upon its translational activation in early embryos (Eichhorn et al., 2016). In oocytes the average bcd poly(A) tail length was similar to those of several well‐translated mRNAs, suggesting that a repressive mechanism independent of the poly(A) tail is in operation. This mechanism remains unknown.
2.3. Bcd gradient formation occurs mostly at the protein level
Precisely how the Bcd gradient is established has been the topic of intense study. The initial model was that Bcd is translated from localized mRNA at the anterior pole, and forms a gradient by diffusion toward the posterior (Driever & Nüsslein‐Volhard, 1988a; Gregor, Wieschaus, McGregor, Bialek, & Tank, 2007; Figure 2). Localized synthesis at the anterior and a short protein half‐life ensures that a uniform Bcd concentration from the anterior to posterior is never reached. Several alternative models have also been proposed. One involves establishment of a bcd RNA gradient that extends throughout a substantial fraction of the embryo that presages the protein gradient (Spirov et al., 2009). Another postulates that nuclear trapping, rather than degradation, is important to establish the Bcd gradient (Coppey, Berezhkovskii, Kim, Boettiger, & Shvartsman, 2007). While translational control determines the time at which Bcd protein first appears, it does not seem to be a major factor in establishing its graded distribution. An early report that bcd is translationally repressed by Nanos and Pumilio in early embryos (Gamberi, Peterson, He, & Gottlieb, 2002) has recently been called into question (Wharton, Nomie, & Wharton, 2018).
FIGURE 2.
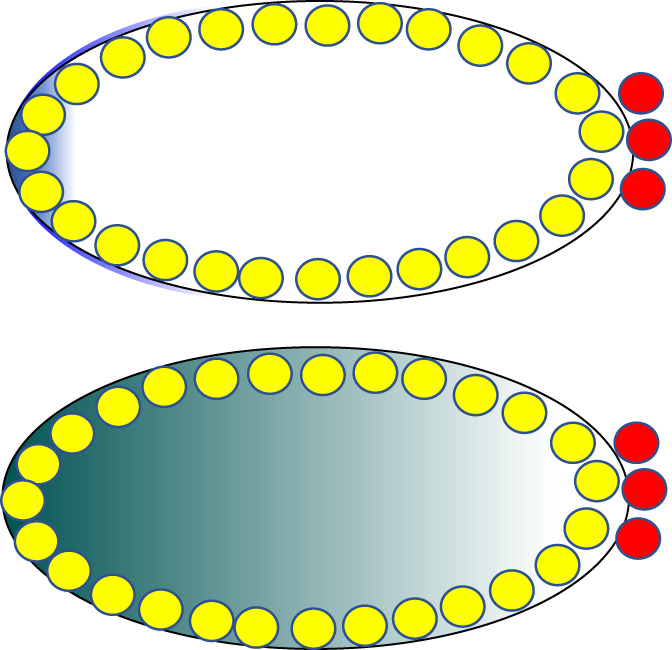
Early Drosophila embryos are syncytial, containing many nuclei that share a common cytoplasm. From a tight anterior cap of bcd RNA (blue, top panel) a gradient of Bcd protein (green, lower panel) is produced, so that nuclei are exposed to different levels of Bcd depending upon their position along the anterior–posterior gradient. Somatic nuclei are depicted in yellow while primordial germ cells (pole cells) are in red
The various models proposed make different predictions about the dynamics of synthesis, diffusion, and degradation of bcd RNA and protein along the anterior–posterior axis in early embryos. Because measuring these parameters in living embryos is technically very challenging, there have been inconsistent results among the different groups that investigate this question. However, a recent paper that used a tandem fluorescent protein timer to measure the age of Bcd protein at different positions along the anterior–posterior axis of the embryo improved upon earlier technologies and supports the original protein diffusion based model (Durrieu et al., 2018). To create such a timer, Bcd was tagged with two different fluorophores, a green one that matures quickly, and a red one that matures slowly. Newly synthesized protein is therefore mostly green, while both tags fluoresce in older protein, which consequently appears yellow. This construct was expressed in embryos and the tagged protein was quantitatively imaged with confocal multi‐view light‐sheet microscopy, with the important result that Bcd protein is older toward the posterior end of the embryo than at the anterior pole. This argues in favor of the original model of Bcd gradient formation involving protein diffusion from a localized anterior source and against the existence of an mRNA prepattern, as with an mRNA gradient newly synthesized protein would be present distant from the anterior pole.
3. LOCALIZATION AND TRANSLATIONAL REGULATION OF osk
3.1. osk is the primary determinant for germ cell specification and posterior patterning
Several lines of evidence support the primary role of osk in germ cell specification and posterior patterning. Ectopic expression of osk at the anterior can induce a second set of pole cells and a bicaudal embryonic segmentation pattern with mirror‐image posterior segments (Ephrussi & Lehmann, 1992; Smith et al., 1992). Several maternal‐effect mutants produce bicaudal embryos, in which posterior segments are duplicated at the anterior end, sometimes in a mirror‐image symmetry. A second focus of osk mRNA at the anterior pole is observed in bicaudal embryos produced by Bicaudal‐D or ik2 females (Chang et al., 2011; Ephrussi et al., 1991; Shapiro & Anderson, 2006). In contrast, embryos from hypomorphic osk females lack posterior segmentation and pole cells (Lehmann & Nüsslein‐Volhard, 1986). osk is one of a set of maternally expressed genes, called posterior‐group genes, that can mutate to give this phenotype (Nüsslein‐Volhard, Frohnhöfer, & Lehmann, 1987). The products of many of these other genes exert their effects through osk. For instance, the posterior‐group genes cappuccino, chickadee, spire, and stau are required to establish or maintain osk at the posterior pole (Manseau, Calley, & Phan, 1996; Micklem, Adams, Grünert, & St Johnston, 2000). The mRNAs and/or proteins from other posterior‐group genes, such as vas, tud, nos, and aub, accumulate in pole plasm dependent upon osk function and operate downstream (Hay, Ackerman, Barbel, Jan, & Jan, 1988; Lasko & Ashburner, 1990; Wang & Lehmann, 1991; Wilson et al., 1996).
3.2. Multiple mechanisms achieve posterior osk RNA localization
Step 1: Localization of osk to the early oocyte via dynein‐directed transport along microtubules
During oogenesis, osk mRNA accumulates at the posterior pole of the oocyte through three different sequential mechanisms (Figure 3). The first of these begins in the germarium, where osk accumulates into the presumptive oocyte, where it is readily detectable from stage 2 of oogenesis. In these early stages, the microtubule cytoskeleton is organized such that a single microtubule organizing center (MTOC) forms in the oocyte, and microtubules emanate from the MTOC through ring canals into the nurse cells (Theurkauf, Alberts, Jan, & Jongens, 1993). Thus microtubules are polarized so that their minus‐ends are concentrated in the oocyte and their plus‐ends are in the nurse cells. As will be seen, this organization is not retained in later stages of oogenesis.
FIGURE 3.
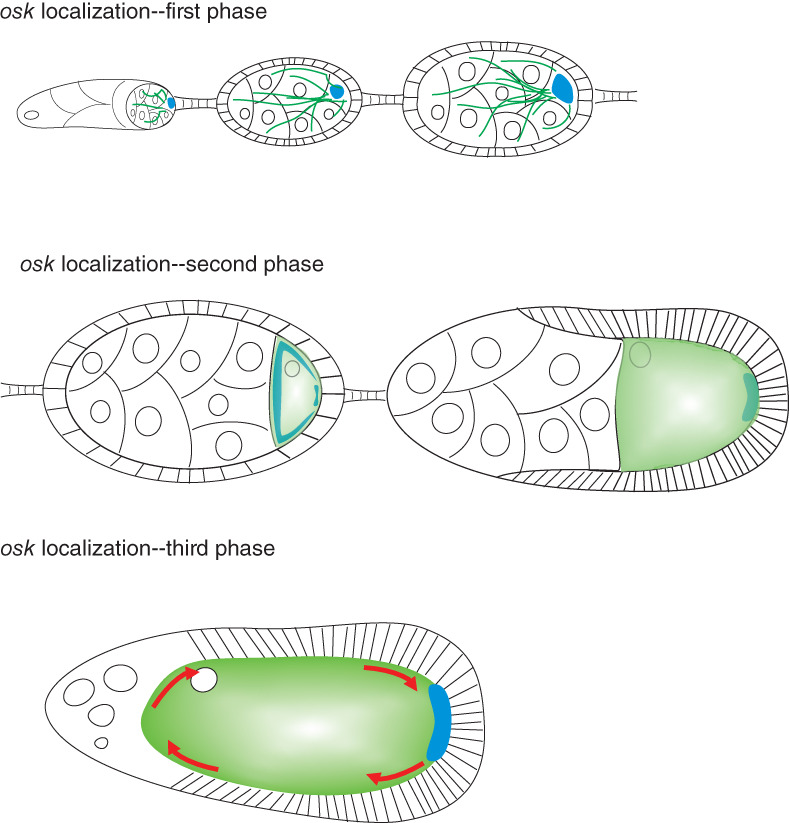
osk mRNA localization proceeds through three distinct mechanisms. In early oogenesis (top panel), a microtubule organizing center (MTOC) forms in the oocyte, and microtubules (green lines) emanate from there through the ring canals into the nurse cells. The motor protein dynein transports osk (blue) along these microtubules into the oocyte. During stages 8–9 of oogenesis (middle panel), microtubules (shaded green) reorganize so that their minus‐ends accumulate all around the oocyte cortex except at the posterior pole. The plus‐end directed motor kinesin drives osk (blue) away from the cortex and it accumulates at the posterior thanks to a weak plus‐end microtubule bias. In late oogenesis (lower panel) rapid cytoplasmic streaming (red arrows) takes place, which transports macromolecules throughout the oocyte cytoplasm. osk mRNA is carried along by these movements and is anchored at the posterior, where there is a relative absence of microtubules, by myosin V
Several reports demonstrate that osk mRNA localization into the early oocyte depends upon minus‐end directed transport by the motor protein dynein. Dynein, as assessed by immunostaining for the dynein heavy chain Dhc64C, accumulates into the early oocyte in a similar manner to osk mRNA (Li, McGrail, Serr, & Hays, 1994). In early egg chambers dynein forms part of a multiprotein complex that also contains Egalitarian (Egl), Bicaudal‐D (Bic‐D), and Lissencephaly‐1 (Lis‐1), and mutations affecting any dynein complex gene block oocyte differentiation (Gepner et al., 1996; Liu, Xie, & Steward, 1999; Mach & Lehmann, 1997; McGrail & Hays, 1997; Navarro, Puthalakath, Adams, Strasser, & Lehmann, 2004; Swan, Nguyen, & Suter, 1999). RNA injection experiments discussed in the preceding section examined both bcd and osk RNAs and suggested that this early stage of osk localization involves dynein‐directed transport (Clark et al., 2007).
Further insights into the mechanism of osk mRNA accumulation into the oocyte came from mapping of cis‐acting sequences that are required for this stage of transport. As measured by immunostaining for the proxy protein Stau, that associates with osk RNA throughout oogenesis, the osk 3′ UTR alone can localize to the oocyte (Jenny et al., 2006). The osk 3′ UTR can also rescue the oogenesis arrest observed in osk‐null alleles, indicating that it has a noncoding function. The necessary element within the 3′ UTR for oocyte entry was mapped to a 67‐nucleotide stem‐loop called the oocyte entry signal (OES) that is separable from another element, to be discussed below, that is required for posterior localization in subsequent stages of oogenesis (Jambor, Mueller, Bullock, & Ephrussi, 2014). An injection assay was established to measure dynein‐directed transport of RNAs to the apical surface of syncytial Drosophila embryos (Bullock & Ish‐Horowicz, 2001; Wilkie & Davis, 2001). A segment of the osk 3′ UTR containing the OES localizes in this assay, but localization is strongly impaired when the OES is absent (Jambor et al., 2014). However, the OES is only active in the context of larger regions of the 3′ UTR, and other partially redundant elements operate in concert with it, leading to the proposal that the presence of multiple weak 3′ UTR signals driving oocyte entry facilitates transfer of osk mRNA to the posterior localization pathway (Ryu, Kenny, Gim, Snee, & Macdonald, 2017).
As mentioned above, mutations in genes encoding dynein complex components block oocyte differentiation. While this phenotype supports a role for dynein‐mediated RNA transport in oogenesis, for many years it prevented direct analysis of the role of the dynein complex in osk localization. In the absence of an oocyte osk does not accumulate in a single cell, so this confounds cause and effect. One component of the dynein complex, Egl, is an RNA binding protein that links RNAs carrying defined localization elements to Bic‐D and dynein (Dienstbier, Boehl, Li, & Bullock, 2009). Driving short‐hairpin RNA (shRNA) targeting Egl using a maternal α‐tubulin driver that is active only after oocyte specification permits the isolation of oocytes that are depleted for it (Sanghavi, Liu, Veeranan‐Karmegam, Navarro, & Gonsalvez, 2016). In egg chambers thus depleted for Egl, osk mRNA is completely delocalized, and is present throughout the oocyte and in the cytoplasm of nurse cells. osk localization could be fully rescued by coexpression of an Egl transgene made resistant to the shRNA by introducing several point mutations into the recognition sequence. Further analysis using egl missense mutations abrogating either dynein or BicD binding supports a model for stepwise assembly of a transport complex. The first step is that Egl dimerizes through an interaction with dynein light chain, then dimerized Egl binds RNA and Bic‐D, relieving autoinhibition of Bic‐D and association with the dynein motor (Goldman, Neiswender, Veeranan‐Karmegam, & Gonsalvez, 2019). This is consistent with results from reconstitution studies using purified components (McClintock et al., 2018; Sladewski et al., 2018).
Step 2: Localization of osk to the oocyte posterior involves kinesin and microtubules
During stages 7 and 8 of oogenesis the organization of the microtubule cytoskeleton changes. Microtubules form an anterior–posterior density gradient, with a higher concentration at the anterior (Clark, Jan, & Jan, 1997). Their minus‐ends become anchored all around the cortex, a process that requires the minus‐end microtubule binding protein Patronin and Short stop (Shot), a protein that crosslinks microtubules and filamentous actin (Nashchekin, Fernandes, & St Johnston, 2016). Shot is excluded from the posterior cortex so fewer microtubules are anchored there, resulting in slightly more microtubule plus‐ends being oriented toward the posterior pole (Khoc Trong, Doerflinger, Dunkel, St Johnston, & Goldstein, 2015; Nashchekin et al., 2016; Parton et al., 2011; Zimyanin et al., 2008).
This raises the possibility that kinesin, a plus‐end directed motor protein, might conduct directed transport of osk RNA to the posterior pole of the oocyte. In fact, in kinesin heavy chain (khc) mutant oocytes posterior localization of osk fails and it accumulates around the anterior and lateral cortex (Brendza, Serbus, Duffy, & Saxton, 2000; Cha, Serbus, Koppetsch, & Theurkauf, 2002). As kinesin is however required for several cellular processes within the oocyte including cytoplasmic streaming, distinguishing direct from indirect effects remained a problem for some time. Ultimately, live imaging analysis of labeled osk in wild‐type and khc mutant oocytes produced convincing evidence that kinesin‐dependent transport toward the plus ends of microtubules results in exclusion of osk from the oocyte cortex and concentration, thanks to the weak posterior bias of plus‐ends, at the posterior pole (Parton et al., 2011; Zimyanin et al., 2008). Kinesin‐1 associates with osk‐containing ribonucleoprotein particles upon nuclear export, a process requiring the atypical RNA‐binding tropomyosin Tm1‐I/C (Gáspár, Sysoev, Komissarov, & Ephrussi, 2016).
A peculiar aspect of this second stage of osk localization is that it requires components of the exon‐junction complex (EJC) and splicing of the first intron of osk (Hachet & Ephrussi, 2001; Hachet & Ephrussi, 2004; Mohr, Dillon, & Boswell, 2001; Newmark & Boswell, 1994; Palacios, Gatfield, St Johnston, & Izaurralde, 2004). Splicing is linked to osk localization because it creates a localization element (the spliced oskar localization element, or SOLE), from the final 18 nucleotides of exon 1 of osk and the first 10 nucleotides of exon 2 (Ghosh, Marchand, Gáspár, & Ephrussi, 2012). Both the SOLE and EJC components are required for osk localization, because when SOLE is present in an RNA without splicing being required for its formation, the RNA remains unlocalized, and RNAs loaded with EJC but lacking SOLE also fail to localize (Ghosh et al., 2012). It is believed that SOLE recruits EJC, either directly or through an unknown third component (Ghosh, Obrdlik, Marchand, & Ephrussi, 2014; Simon, Masiewicz, Ephrussi, & Carlomagno, 2015). The SOLE, EJC, and 3′ UTR together are necessary for posterior transport, as interfering with any of these components results in osk mislocalization.
Step 3: Posterior osk accumulation through cytoplasmic streaming and anchorage
After stage 8 the oocyte undergoes a period of rapid growth, and during stages 10–12 the cytoplasm of the nurse cells is also rapidly transferred through the ring canals into the oocyte. Fast rotary cytoplasmic streaming occurs within the oocyte during this time, which is dependent on kinesin and the microtubule cytoskeleton (Lu, Winding, Lakonishok, Wildonger, & Gelfand, 2016; Palacios & St Johnston, 2002; Serbus, Cha, Theurkauf, & Saxton, 2005). This enables cytoplasmic content to cycle around the oocyte from the anterior to the posterior and vice versa, so in the absence of anchoring mechanisms, localized molecules would become homogeneously distributed. Conversely, cytoplasmic streaming can provide a mechanism for localizing molecules, provided that a coupled trapping mechanism also exists.
An early study showed that injected fluorescently tagged osk RNA could localize via such a mechanism (Glotzer, Saffrich, Glotzer, & Ephrussi, 1997). For some time, the importance of cytoplasmic streaming and anchoring for localization of endogenous osk remained unclear, although this mechanism became well established for RNAs that localize to the pole plasm only in late oogenesis such as nos (Forrest & Gavis, 2003). Ironically, analysis of two trans‐acting factors required for nos localization, Rumpelstiltskin (Rump) and Lost, revealed that cytoplasmic streaming and anchoring also contribute to endogenous osk localization (Jain & Gavis, 2008; Sinsimer, Jain, Chatterjee, & Gavis, 2011). Oocytes doubly mutant for lost and rump support osk localization through stage 10 of oogenesis, but the resulting embryos have clearly reduced posterior osk localization and form fewer pole cells. A defect in osk localization in late oogenesis in lost rump mutants was also directly shown by live imaging of GFP‐tagged osk RNA.
Kinesin‐1 is required both for cytoplasmic streaming (Lu et al., 2016; Palacios & St Johnston, 2002; Serbus et al., 2005) and for the initial posterior localization of osk. This complicated the unraveling of the relative importance of the two mechanisms. To resolve this, a mutant was produced that blocked microtubule sliding required for cytoplasmic streaming (KhcmutA, carrying four missense mutations in the C‐terminal microtubule binding site) (Lu et al., 2018). Simultaneous RNA interference targeting klc and btz was used to specifically block directed transport. Using these strains it was found that cytoplasmic streaming can partially rescue posterior localization of osk and fluorescently labeled Stau when the initial microtubule‐dependent transport phase is blocked. Supporting redundancy between these two localization mechanisms, over 80% of embryos produced by Khc mutA or klc btz double knockdown females produced pole cells (Lu et al., 2018).
How is osk anchored at the posterior pole during cytoplasmic streaming? Several lines of evidence suggest that anchoring involves an actomyosin‐based mechanism. From a forward‐genetic screen aimed at identifying new maternal‐effect lethal loci, several alleles of didum, which encodes myosin V, were obtained (Krauss, López de Quinto, Nüsslein‐Volhard, & Ephrussi, 2009; Luschnig et al., 2004). These mutants presented a posterior‐group phenotype related to that of osk mutants, and didum oocytes showed defects in osk mRNA localization with some osk being retained in the central region of the oocyte. This work led to the suggestion that transport or entrapment of osk through a process dependent on F‐actin and myosin V is a necessary final step in its localization. However, as myosin V is present throughout all the oocyte cortex, not just the posterior pole (Krauss et al., 2009) it remained unclear how its activity becomes spatially restricted.
A very recent elegant study resolves this issue, showing an antagonistic relationship between cortical myosin V and microtubules with respect to osk accumulation (Lu et al., 2020). Consistent with previous work (Clark et al., 1997; Nashchekin et al., 2016; Zimyanin et al., 2008), an anterior‐to‐posterior gradient of microtubule density with a relative absence at the posterior pole was demonstrated. Depolymerization of microtubules by germline overexpression of Klp10A, a depolymerizing kinesin, resulted in uniform accumulation of the osk proxy Stau around the entire oocyte cortex, while overexpression of Patronin, which prevents depolymerization and increases microtubule density, resulted in accumulation of Stau in the center of the oocyte cytoplasm (Lu et al., 2020). Next, optogenetic tools were used to recruit Klp10A to particular spatial locations on the oocyte cortex, depleting microtubules at the site of recruitment. When this was done, Stau accumulated in the same positions, and this was reversible when the light stimulus leading to Klp10A recruitment was removed. These experiments confirm a model whereby osk/Stau localization and anchoring critically depends upon local microtubule density, with low density favoring cortical trapping of osk/Stau by myosin V, and high density favoring cortical exclusion of osk/Stau by kinesin‐1.
3.3. osk is under complex translational control
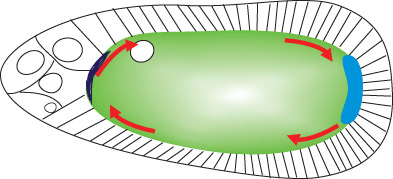
osk mRNA generates two protein isoforms from different translational start sites (Markussen, Michon, Breitwieser, & Ephrussi, 1995). The long isoform (Long Osk) anchors mRNAs and mitochondria in the pole plasm (Hurd et al., 2016; Tanaka, Kato, Matsuda, Hanyu‐Nakamura, & Nakamura, 2011; Tanaka & Nakamura, 2008; Vanzo & Ephrussi, 2002; Vanzo, Oprins, Xanthakis, Ephrussi, & Rabouille, 2007). Short Osk, meanwhile, supports all Osk functions in assembly of polar granules and in posterior patterning and germ cell specification (Markussen et al., 1995). It is unknown how initiation from these two start sites, which are in the same open reading frame, is controlled. Complex temporal and spatial regulation of translation ensures that Osk accumulates abundantly only at the posterior pole, not while the mRNA is in transit, and not before stage 9 of oogenesis (Figure 4). Failure of these mechanisms results in lethality. Bruno1 (Bru1) represses osk translation in early oogenesis and outside the pole plasm, through interactions with binding sites in the osk 3′ UTR called Bru1 response elements (BREs). Deleting or mutating the BREs generally results in ectopic Osk expression (Kim‐Ha, Kerr, & Macdonald, 1995; Reveal et al., 2010; Webster, Liang, Berg, Lasko, & Macdonald, 1997). Bru1 represses osk through two distinct, but coupled mechanisms. The first such mechanism to be revealed involves Bru1‐mediated recruitment of Cup to osk mRNA. Cup is a competitive inhibitor of eIF4G for binding to eIF4E, therefore it inhibits assembly of an active cap‐binding complex (Nakamura, Sato, & Hanyu‐Nakamura, 2004) (see Box for further details). osk translation is also inhibited by oligomerization of osk mRNA into large RNPs that are not readily accessed by ribosomes and the rest of the translational machinery (Besse, López de Quinto, Marchand, Trucco, & Ephrussi, 2009; Chekulaeva, Hentze, & Ephrussi, 2006; Kim et al., 2015). The two mechanisms are linked. Concentrating osk mRNA molecules into RNPs brings them into close juxtaposition and facilitates regulation in trans; Bru1 bound to the 3′ end of one osk mRNA molecule can recruit Cup to eIF4E bound to the 5′ cap structure of an adjacent osk mRNA molecule (Macdonald, Kanke, & Kenny, 2016; Reveal et al., 2010). osk translation is tightly linked to maintenance of posterior localization of osk mRNA localization. In nonsense osk alleles or when translational activation elements are mutated, osk mRNA localizes only transiently to the pole plasm (Ephrussi et al., 1991; Kim‐Ha et al., 1991; Munro, Kwon, Schnapp, & St Johnston, 2006).
FIGURE 4.
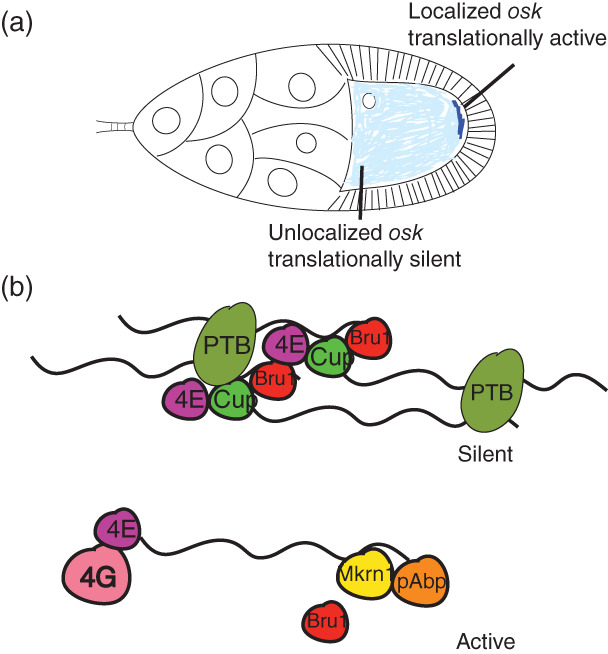
osk is under complex translational control. (a). Unlocalized osk is translationally repressed by oligomerization and by the translational repressor Bru1. osk is oligomerized by PTB, which can bind to multiple sites on the osk 3’ UTR. Bru1 binds to specific sites in the osk 3’ UTR and recruits Cup, a competitive inhibitor for eukaryotic initiation factor (eIF) 4E (4E) binding to eIF4G. (b). In the pole plasm osk translation is activated. Makorin1 (Mkrn1) displaces Bru1 and recruits poly(A) binding protein (pAbp), which stimulates translation
BOX 1. REGULATION OF CAP‐DEPENDENT TRANSLATION.
The step of translation initiation that is regulated by some of the proteins discussed here is the assembly of the 48S initiation complex. This complex includes the mRNA, the initiator tRNA, the 40S (small) ribosomal subunit, and various translation initiation factors (eIFs) (Eliseev et al., 2018). eIF4F plays a central role in assembling this complex. It is composed of three subunits: eIF4E, which binds the cap structure present at the 5′ end of mRNA; eIF4A, an RNA helicase, and eIF4G, which binds both eIF4E and eIF3. eIF4G binding to eIF4E is necessary to assemble eIF4F at the cap, and eIF4G binding to eIF3 enables recruitment of the small ribosomal subunit, which is also bound by eIF3. Translational repressors discussed here disrupt eIF4F formation in different ways. Cup competes with eIF4G for binding to eIF4E, thus prohibiting eIF4F assembly on mRNAs to which it is bound. Conversely, 4EHP binds the cap structure but does not bind eIF4G, sequestering mRNA to which it is bound away from eIF4F and ribosomes.
Less is known about how osk translation is activated in the pole plasm, although several proteins have been implicated in this process (Chang, Tan, & Schedl, 1999; Micklem et al., 2000; Wilson et al., 1996). One aspect of osk translational activation is inhibition of Bru1 (Kim et al., 2015), and one BRE‐containing region in the distal part of the osk 3′ UTR (BRE‐C) functions in activation as well as in repression (Reveal et al., 2010). A recent study implicated the RNA binding protein Makorin‐1 (Mkrn1) as a key activator of osk translation (Dold et al., 2020). Mkrn1 binds the osk 3′ UTR at an A‐rich region immediately adjacent to BRE‐C, where it antagonizes Bru1 binding and promotes recruitment of poly(A) binding protein.
3.4. Osk associates with Vas and binds RNA
Despite extensive analysis of the developmental role of Osk, its precise molecular function has long remained unclear. Structural analysis of two domains of Osk, its N‐terminal LOTUS domain (Osk‐N), and its C‐terminal hydrolase‐like domain (Osk‐C), indicates that it can directly associate with RNA (Yang et al., 2015), which could explain, at least in part, its role in polar granule assembly. The LOTUS domain is an established RNA binding motif (Anantharaman, Zhang, & Aravind, 2010; Callebaut & Mornon, 2010), but in Osk‐N it homodimerizes in a manner that masks the nucleic acid binding surface. Osk‐C forms a structure resembling a lipase‐like fold, but with nonconservative changes in several key amino acids that would rule out any catalytic activity. However, Osk‐C was found to avidly bind some RNAs, including the osk and nos 3′ UTRs, in EMSA assays. Thus sequence similarity analysis in this case is misleading, and Osk‐C, rather than the predicted Osk‐N, possesses a direct RNA binding activity. Point mutation analysis implicated three specific Arg residues within Osk‐C in RNA binding, which all reside on a positively charged ridge that can accommodate RNA. Several other residues present in this ridge and/or spatially proximate to these Arg residues have been implicated by mutation in Osk‐dependent developmental functions (Ephrussi et al., 1991; Kim‐Ha et al., 1991; Lehmann & Nüsslein‐Volhard, 1986; Rongo, 1996; Yang et al., 2015).
The DEAD‐box helicase Vas appears to be a cofactor for many functions of Osk, and as described above the two proteins are often present in close association within RNPs. Osk interacts directly with Vas through an interaction between the LOTUS domain and an alpha‐helical region of Vas, with two phenylalanine residues of Vas (F504 and F508) essential for Osk binding (Jeske et al., 2015; Jeske, Müller, & Ephrussi, 2017). Mutation of one of these residues (F504E) blocks recruitment of Vas to the pole plasm. Vas also functions in piRNA biogenesis and localizes to the perinuclear nuage in nurse cells, the structure where piRNA processing takes place (ElMaghraby et al., 2019; Zhang et al., 2012). Two other LOTUS domain proteins, Tejas and Tapas, also interact with Vas and play roles in piRNA‐mediated processes (Jeske et al., 2017; Patil, Anand, Chakrabarti, & Kai, 2014; Patil & Kai, 2010).
4. POLAR GRANULE FORMATION INVOLVES SPECIFIC MOLECULAR ASSOCIATIONS AND NONSPECIFIC PHYSICAL PROCESSES
Key proteins and mRNAs involved in germ cell specification associate in polar granules, RNA‐protein complexes that are linked to germ cell development throughout the animal kingdom. Polar granules are one class of related particles that are collectively called germ granules. Polar granules in early Drosophila melanogaster embryos are nonmembrane bound, electron‐dense, approximately round particles with diameters ranging from 200 to 500 nm (Mahowald, 1962). They are first seeded by localization of single RNA molecules, which subsequently recruit additional transcripts from the same gene through homotypic interactions (Niepielko, Eagle, & Gavis, 2018). They later become associated with the embryonic nuclei that are destined to become PGCs (Lerit & Gavis, 2011), and further grow in size and number through the cellular blastoderm stage of development. During this period they are referred to as nuclear germ granules (Kistler et al., 2018). Beginning in the 1980s, molecular genetic studies revealed numerous protein and RNA components of polar granules, many of which have been described above. However, the mechanisms by which these particles assemble only began to come to light in recent years, and important questions about this remain to be answered.
A critical insight was provided by work on P granules, structures analogous to polar granules in the nematode Caenorhabditis elegans. This work showed that P granules exhibit physical properties similar to those of a liquid with a viscosity comparable to that of glycerol (Brangwynne et al., 2009). Furthermore, it provided compelling evidence that accumulation of P granules in the germ plasm is not dependent on active transport or on cytoplasmic flows. Rather, accumulation comes about through establishment, by polarity proteins including MEX‐5 and PAR‐1, of a greater likelihood of condensation versus dissolution of P granule material into droplets at the posterior pole where the germ plasm forms.
4.1. Homotypic RNA–RNA interactions underlie germ granule assembly
In contrast, evidence began to emerge in Drosophila that higher‐order supramolecular assemblies arise in the pole plasm through specific RNA–RNA and RNA–protein interactions. Much earlier work had already shown that Stau, which is required for a late stage of bcd localization, binds best to RNA when a base‐paired structure involving two bcd RNA molecules is present (Ferrandon, Koch, Westhof, & Nüsslein‐Volhard, 1997). In a study of osk RNA in mid‐oogenesis, Ephrussi and colleagues showed that polypyrimidine tract binding (PTB) protein can bind to several sites in its osk 3′ UTR, contributing to its translational repression prior to its posterior localization (Besse et al., 2009). PTB has four RNA recognition motifs (RRMs), and the third and fourth of these domains are oriented in such a way that they can bind to PTB binding sites on different RNA molecules, thus serving as a bridge and potentially serving as a basis for assembly of higher‐order structures. Interestingly, osk‐containing RNPs in stage‐9 oocytes that are precursors to polar granules are smaller and more numerous in hephasteus mutants (hephasteus encodes PTB).
Subsequent advances in microscopy have enabled more detailed study of the molecular topography of polar granules and related RNPs, providing evidence for internal organization built around homotypic clustering of RNA molecules. Super‐resolution, single‐molecule level imaging of polar granules in early embryos showed that while Osk, Vas, Tud, and Aub proteins all occupy the same space within the granules, different RNA components of polar granules have different distributions, suggesting that polar granules have internal structure (Trcek et al., 2015; Figure 5). cycB and nos RNAs largely colocalized with the four proteins, while pgc and gcl RNA occupy more peripheral regions of the polar granule. Despite its importance in seeding pole plasm assembly, osk RNA is ultimately excluded from polar granules. However, it is present in other particles, one type including only Vas and another type including only pgc among the molecules examined. Exclusion of osk from embryonic polar granules was also reported in other studies (Herpers, Xanthakis, & Rabouille, 2010; Little et al., 2015).
FIGURE 5.
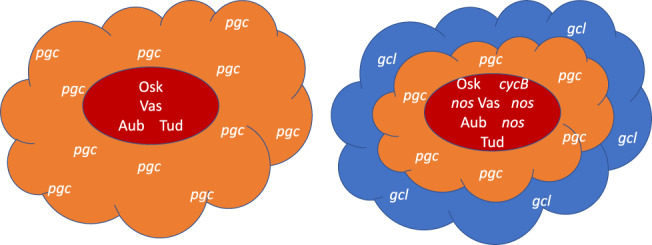
Cartoon illustrating the structured and heterogeneous nature of polar granules. Osk, Vas, Aub, and Tud proteins accumulate in the core region of polar granules along with cycB and nos mRNAs. pgc and gcl mRNAs accumulate in more peripheral regions (Trcek et al., 2015). Some polar granules contain only one species of mRNA while others accumulate several (Little, Sinsimer, Lee, Wieschaus, & Gavis, 2015)
4.2. Pole plasm RNPs fall into several classes that assemble by different mechanisms
Such a highly organized internal structure argues against a model of Drosophila polar granule formation proceeding simply by liquid droplet condensation, although concentration‐dependent biophysical processes surely have an important role. Importantly, internal structure has also been reported in C. elegans P granules using a similar approach (Wang et al., 2014).
Another study used high‐resolution fluorescent in situ hybridization (FISH) to follow the assembly of germ plasm RNPs during oogenesis and early embryogenesis (Little et al., 2015). From this work it was concluded that germ granules share a common set of proteins including Osk, Vas, and Tud, but are heterogeneous in terms of their RNA composition. Germ granules with different RNAs assemble by different mechanisms. RNPs containing single copies of nos RNA assemble into granules containing an average of 10–20 nos RNA molecules at the posterior oocyte cortex. Conversely, osk RNA already resides within multi‐copy RNPs during its transport, and assembles into large granules often containing >100 RNA molecules that are compositionally distinct from nos particles. This work was further extended (Niepielko et al., 2018) to show that single‐copy nos and pgc RNPs can first populate the same granule, and then granules grow by self‐recruitment of additional RNA molecules into homotypic clusters, a process that requires simultaneous acquisition of Osk protein. Polar granule targeting and assembly into homotypic clusters appears to involve different 3′ UTR elements, at least for pgc RNA (Eagle, Yeboah‐Kordieh, Niepielko, & Gavis, 2018). A more recent study indicates that two polar granule proteins, Aub and Tud, also form distinct clusters within the granules (Vo et al., 2019).
4.3. Many RNAs associate with germ granules by base‐pairing with Aub‐associated piRNAs
All of the studies discussed above looked at a few canonical polar granule RNAs (osk, nos, pgc, cycB) but well over 100 other mRNAs accumulate in pole plasm (Frise, Hammonds, & Celniker, 2010; Jambor et al., 2015; Lécuyer et al., 2007; Rangan et al., 2009). These do not include extra‐mitochondrial mtlrRNA, which was reported based on electron microscopy evidence to be present in the pole plasm (Akiyama & Okada, 1992; Amikura, Kashikawa, Nakamura, & Kobayashi, 2001; Iida & Kobayashi, 1998; Kobayashi, Amikura, & Okada, 1993). Colocalization analysis using quantitative single‐molecule RNA FISH showed essentially complete overlap between mtlrRNA and EYFP‐labeled mitochondria in the pole plasm (Hurd et al., 2016). Localization of other pole plasm RNAs is abrogated in osk mutants, suggesting a mechanism in common with their better characterized counterparts. How then are all these different RNAs targeted? Vourekas, Alexiou, Vrettos, Maragkakis, and Mourelatos (2016) demonstrated a central role for Aub and piwi‐interacting RNAs (piRNAs) in this process. Aub interacts with piRNAs, many of which have complementary sequences to transposon‐encoded mRNAs, to form piRNPs, which interact with target mRNAs through base pairing and silence their expression (Siomi, Sato, Pezic, & Aravin, 2011). However, Aub‐loaded piRNAs are highly diverse and more than half are derived from mRNA, not transposon, sequences (Barckmann et al., 2015; Vourekas et al., 2016). Consistent with the high concentration of Aub in polar granules, Aub‐associated mRNAs from wild‐type embryos, but not from embryos produced by tud mutant females, are highly enriched for those that are localized to the posterior of the embryo and/or to germ cells (Barckmann et al., 2015; Vourekas et al., 2016). This suggests that Aub‐associated piRNAs recognize and play a role in posterior accumulation of germ plasm mRNAs. Transcriptome‐wide prediction of targets for Aub‐associated piRNAs shows that sites with modest complementarity are present essentially at random. However, posterior‐localized mRNAs are generally longer than average, therefore they tend to contain more such sites, and thus a random process becomes biased toward specific mRNAs (Vourekas et al., 2016). Stabilization of posterior‐localized mRNAs is further accomplished through Aub‐mediated recruitment of the poly(A) polymerase Wispy (Dufourt et al., 2017).
4.4. Short Osk nucleates germ granules in pole cell nuclei, which promote their proliferation
After fertilization, particles containing Vas and nos are recruited by dynein‐dependent transport along centrosome‐associated astral microtubules to become associated with posterior nuclei (Lerit & Gavis, 2011). These particles are similar in size and shape to polar granules. However, as soon as pole cells form, nuclear Osk‐ and Vas‐containing particles increase in size and number, becoming prominent in germ cells by the cellular blastoderm stage of development (Kistler et al., 2018). Nuclear germ granules behave as phase‐transitioned condensates, with both liquid‐like and hydrogel‐like properties (Kistler et al., 2018). They are seeded by Short Osk and, like polar granules, are nonmembrane bound RNPs. While they contain Vas, they do not accumulate several other polar granule components including Aub and Tud proteins nor nos, pgc, or gcl RNAs. Nuclear germ granules are not completely liquid‐like and labile, because they do not fuse with one another; however, they are dynamic in that they can exchange Osk and Vas from particle to particle. When formation of nuclear germ granules is prevented by deleting the Osk nuclear localization signal, the number of pole cells, but not of pole buds, is reduced, indicating a role for nuclear Osk and potentially these granules in promoting germ cell division (Kistler et al., 2018). Each pole bud normally forms two pole cells (Pae, Cinalli, Marzio, Pagano, & Lehmann, 2017) but in OskΔNLS embryos the division that accomplishes this often fails to occur, and subsequent divisions occur even less frequently (Kistler et al., 2018).
5. CONCLUSION
Despite all that has been learned important questions about the structure and function of polar granules remain unresolved. Some of these are specific, for instance more needs to be learned about how bcd RNA anchoring is maintained at the anterior cortex, and about how bcd translation is repressed until egg activation. We also lack understanding about how translation from the two initiation codons that produce the long and short isoforms of Osk is properly balanced.
Other open questions are wider in scope. For instance, is there a functional importance of the restricted structural domains within polar granules? Since the position of an RNA within a granule does not specify the onset of its translation, and granules do not reorganize as some RNAs become translationally active (Trcek et al., 2015), perhaps there is none. Related to this, it is unclear how the translational machinery can gain access to RNAs embedded within polar granules, as association into large RNPs like polar granules more typically masks RNAs from translation. Finally, it has long been known that polar granules in different Drosophilid species have different characteristic shapes and sizes (Counce, 1963; Mahowald, 1968). In some manner Osk controls these species‐specific differences, as D. immigrans Osk expressed in D. melanogaster oocytes lacking endogenous osk produces D. immigrans‐type polar granules, which perhaps surprisingly are functional, rescuing the osk mutant phenotype (Jones & Macdonald, 2007). How any of the mechanisms described above can influence higher‐order structure at the scale of nearly a micron remains to be determined.
CONFLICT OF INTEREST
The author has declared no conflicts of interest for this article.
RELATED WIREs ARTICLE
Posttranscriptional regulation in Drosophila oocytes and early embryos
ACKNOWLEDGMENTS
I am very grateful to Sanjay Ghosh for critical reading of the manuscript and helpful comments. I am also grateful to NSERC for support from Discovery Grant RGPIN‐2014‐06340 and from a Visiting Professorship from the Radboud University Excellence Initiative.
Lasko P. Patterning the Drosophila embryo: A paradigm for RNA‐based developmental genetic regulation. WIREs RNA. 2020;11:e1610 10.1002/wrna.1610
Funding information Natural Sciences and Engineering Research Council of Canada, Grant/Award Number: RGPIN‐2014‐06340; Radboud Universiteit, Grant/Award Number: Visiting Professorship
References
REFERENCES
- Akiyama, T. , & Okada, M. (1992). Spatial and developmental changes in the respiratory activity of mitochondria in early Drosophila embryos. Development, 115(4), 1175–1182. [DOI] [PubMed] [Google Scholar]
- Amikura, R. , Kashikawa, M. , Nakamura, A. , & Kobayashi, S. (2001). Presence of mitochondria‐type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proceedings of the National Academy of Sciences of the United States of America, 98(16), 9133–9138. 10.1073/pnas.171286998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman, V. , Zhang, D. , & Aravind, L. (2010). OST‐HTH: A novel predicted RNA binding domain. Biology Direct, 5, 13 10.1186/1745-6150-5-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka‐Taguchi, M. , Yamada, M. , Nakamura, A. , Hanyu, K. , & Kobayashi, S. (1999). Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nature Cell Biology, 1(7), 431–437. 10.1038/15666 [DOI] [PubMed] [Google Scholar]
- Barckmann, B. , Pierson, S. , Dufourt, J. , Papin, C. , Armenise, C. , Port, F. , … Simonelig, M. (2015). Aubergine iCLIP reveals piRNA‐dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Reports, 12(7), 1205–1216. 10.1016/j.celrep.2015.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardsley, A. , McDonald, K. , & Boswell, R. E. (1993). Distribution of Tudor protein in the Drosophila embryo suggests separation of functions based on site of localization. Development, 119(1), 207–219. [DOI] [PubMed] [Google Scholar]
- Berleth, T. , Burri, M. , Thoma, G. , Bopp, D. , Richstein, S. , Frigerio, G. , … Nüsslein‐Volhard, C. (1988). The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. The EMBO Journal, 7(6), 1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand, E. , Chartrand, P. , Schaefer, M. , Shenoy, S. M. , Singer, R. H. , & Long, R. M. (1998). Localization of ASH1 mRNA particles in living yeast. Molecular Cell, 2(4), 437–445. 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Besse, F. , López de Quinto, S. , Marchand, V. , Trucco, A. , & Ephrussi, A. (2009). Drosophila PTB promotes formation of high‐order RNP particles and represses oskar translation. Genes & Development, 23(2), 195–207. 10.1101/gad.505709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne, C. P. , Eckmann, C. R. , Courson, D. S. , Rybarska, A. , Hoege, C. , Gharakhani, J. , … Hyman, A. A. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science, 324(5935), 1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Breitwieser, W. , Markussen, F.‐H. , Horstmann, H. , & Ephrussi, A. (1996). Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes & Development, 10(17), 2179–2188. 10.1101/gad.10.17.2179 [DOI] [PubMed] [Google Scholar]
- Brendza, R. P. , Serbus, L. R. , Duffy, J. B. , & Saxton, W. M. (2000). A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science, 289(5487), 2120–2122. 10.1126/science.289.5487.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock, S. L. , & Ish‐Horowicz, D. (2001). Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature, 414(6864), 611–616. 10.1038/414611a [DOI] [PubMed] [Google Scholar]
- Callebaut, I. , & Mornon, J. P. (2010). LOTUS, a new domain associated with small RNAs in the germline. Bioinformatics, 26(9), 1140–1144. 10.1093/bioinformatics/btq122 [DOI] [PubMed] [Google Scholar]
- Cha, B. J. , Serbus, L. R. , Koppetsch, B. S. , & Theurkauf, W. E. (2002). Kinesin I‐dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nature Cell Biology, 4(8), 592–598. 10.1038/ncb832 [DOI] [PubMed] [Google Scholar]
- Chang, C. W. , Nashchekin, D. , Wheatley, L. , Irion, U. , Dahlgaard, K. , Montague, T. G. , … St Johnston, D. (2011). Anterior‐posterior axis specification in Drosophila oocytes: Identification of novel bicoid and oskar mRNA localization factors. Genetics, 188(4), 883–896. 10.1534/genetics.111.129312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J. S. , Tan, L. , & Schedl, P. (1999). The Drosophila CPEB homolog, Orb, is required for Oskar protein expression in oocytes. Developmental Biology, 215(1), 91–106. 10.1006/dbio.1999.9444 [DOI] [PubMed] [Google Scholar]
- Chekulaeva, M. , Hentze, M. W. , & Ephrussi, A. (2006). Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell, 124(3), 521–533. 10.1016/j.cell.2006.01.031 [DOI] [PubMed] [Google Scholar]
- Clark, A. , Meignin, C. , & Davis, I. (2007). A dynein‐dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development, 134(10), 1955–1965. 10.1242/dev.02832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, I. , Jan, L. Y. , & Jan, Y. N. (1997). Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron, and muscle. Development, 124(2), 461–470. [DOI] [PubMed] [Google Scholar]
- Coppey, M. , Berezhkovskii, A. M. , Kim, Y. , Boettiger, A. N. , & Shvartsman, S. Y. (2007). Modeling the Bicoid gradient: diffusion and reversible nuclear trapping of a stable protein. Developmental Biology, 312(2), 623–630. 10.1016/j.ydbio.2007.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counce, S. J. (1963). Developmental morphology of polar granules in Drosophila including observations on pole cell behavior and distribution during embryogenesis. Journal of Morphology, 112, 129–145. [Google Scholar]
- Dienstbier, M. , Boehl, F. , Li, X. , & Bullock, S. L. (2009). Egalitarian is a selective RNA‐binding protein linking mRNA localization signals to the dynein motor. Genes & Development, 23(13), 1546–1558. 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dold, A. , Han, H. , Liu, N. , Hildebrandt, A. , Brüggemann, M. , Rücklé, C. , … Lasko, P. (2020). Makorin 1 controls embryonic patterning by alleviating Bruno1‐mediated repression of oskar translation. PLoS Genetics, 16(1), e1008581 10.1371/journal.pgen.1008581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever, W. , & Nüsslein‐Volhard, C. (1988a). A gradient of Bicoid protein in Drosophila embryos. Cell, 54(1), 83–93. 10.1016/0092-8674(88)90182-1 [DOI] [PubMed] [Google Scholar]
- Driever, W. , & Nüsslein‐Volhard, C. (1988b). The Bicoid protein determines position in the Drosophila embryo in a concentration‐dependent manner. Cell, 54(1), 95–104. 10.1016/0092-8674(88)90183-3 [DOI] [PubMed] [Google Scholar]
- Dufourt, J. , Bontonou, G. , Chartier, A. , Jahan, C. , Meunier, A.‐C. , Pierson, S. , … Simonelig, M. (2017). piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nature Communications, 8, 1305 10.1038/s41467-017-01431-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, J. E. , & Warrior, R. (2002). The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Current Biology, 12(23), 1982–1991. 10.1016/S0960-9822(02)01303-9 [DOI] [PubMed] [Google Scholar]
- Durrieu, L. , Kirrmaier, D. , Schniedt, T. , Kats, I. , Raghavan, S. , Hufnagel, L. , … Knop, M. (2018). Bicoid gradient formation mechanism and dynamics revealed by protein lifetime analysis. Molecular Systems Biology, 14(9), e8355 10.15252/msb.20188355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagle, W. V. I. , Yeboah‐Kordieh, D. K. , Niepielko, M. G. , & Gavis, E. R. (2018). Distinct cis‐acting elements mediate targeting and clustering of Drosophila polar granule RNAs. Development, 145(22), dev164657 10.1242/dev164657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn, S. W. , Subtelny, A. O. , Kronja, I. , Kwasnieski, J. C. , Orr‐Weaver, T. L. , & Bartel, D. P. (2016). mRNA poly(A)‐tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and embryos. eLife, 5, e16955 10.7554/eLife.16955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseev, B. , Yeramala, L. , Leitner, A. , Karuppasamy, M. , Raimondeau, E. , Huard, K. , … Schaffitzel, C. (2018). Structure of a human cap‐dependent 48S translation pre‐initiation complex. Nucleic Acids Research, 46(5), 2678–2689. 10.1093/nar/gky054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElMaghraby, M. F. , Andersen, P. R. , Pühringer, F. , Hohmann, U. , Meixner, K. , Lendl, T. , … Brennecke, J. (2019). A heterochromatin‐specific RNA export pathway facilitates piRNA production. Cell, 178(4), 964–979. 10.1016/j.cell.2019.07.007 [DOI] [PubMed] [Google Scholar]
- Ephrussi, A. , Dickinson, L. K. , & Lehmann, R. (1991). Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell, 66(1), 37–50. 10.1016/0092-8674(91)90137-n [DOI] [PubMed] [Google Scholar]
- Ephrussi, A. , & Lehmann, R. (1992). Induction of germ cell formation by oskar . Nature, 358(6385), 387–392. 10.1038/358387a0 [DOI] [PubMed] [Google Scholar]
- Ferrandon, D. , Koch, I. , Westhof, E. , & Nüsslein‐Volhard, C. (1997). RNA‐RNA interaction is required for the formation of specific bicoid mRNA 3’ UTR‐STAUFEN ribonucleoprotein particles. The EMBO Journal, 16(7), 1751–1758. 10.1093/emboj/16.7.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes, A. , & Lehmann, R. (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development, 125(4), 679–690. [DOI] [PubMed] [Google Scholar]
- Forrest, K. M. , & Gavis, E. R. (2003). Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA in Drosophila . Current Biology, 13(14), 1159–1168. 10.1016/s0960-9822(03)00451-2 [DOI] [PubMed] [Google Scholar]
- Frise, E. , Hammonds, A. S. , & Celniker, S. E. (2010). Systematic image‐driven analysis of the spatial Drosophila embryonic expression landscape. Molecular Systems Biology, 6, 345 10.1038/msb.2009.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberi, C. , Peterson, D. S. , He, L. , & Gottlieb, E. (2002). An anterior function for the Drosophila posterior determinant Pumilio. Development, 129(11), 2699–2710. [DOI] [PubMed] [Google Scholar]
- Gáspár, I. , Sysoev, V. , Komissarov, A. , & Ephrussi, A. (2016). An RNA‐binding atypical tropomyosin recruits kinesin‐1 dynamically to oskar mRNPs. The EMBO Journal, 36(3), 319–333. 10.15252/embj.201696038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis, E. R. , & Lehmann, R. (1992). Localization of nanos RNA controls embryonic polarity. Cell, 71(2), 301–313. 10.1016/0092-8674(92)90358-j [DOI] [PubMed] [Google Scholar]
- Gepner, J. , Li, M. , Ludmann, S. , Kortas, C. , Boylan, K. , Iyadurai, S. J. , … Hays, T. S. (1996). Cytoplasmic dynein function is essential in Drosophila melanogaster . Genetics, 142(3), 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S. , Marchand, V. , Gáspár, I. , & Ephrussi, A. (2012). Control of RNP motility and localization by a splicing‐dependent structure in oskar mRNA. Nature Structural & Molecular Biology, 19(4), 441–449. 10.1038/nsmb.2257 [DOI] [PubMed] [Google Scholar]
- Ghosh, S. , Obrdlik, A. , Marchand, V. , & Ephrussi, A. (2014). The EJC binding and dissociating activity of PYM is regulated in Drosophila . PLoS Genetics, 10(6), e1004455 10.1371/journal.pgen.1004455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, J. B. , Saffrich, R. , Glotzer, M. , & Ephrussi, A. (1997). Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Current Biology, 7(5), 326–337. 10.1016/s0960-9822(06)00156-4 [DOI] [PubMed] [Google Scholar]
- Goldman, C. H. , & Gonsalvez, G. B. (2017). The role of microtubule motors in mRNA localization and patterning within the Drosophila oocyte. Results and Problems in Cell Differentiation, 63, 149–168. 10.1007/978-3-319-60855-6_7 [DOI] [PubMed] [Google Scholar]
- Goldman, C. H. , Neiswender, H. , Veeranan‐Karmegam, R. , & Gonsalvez, G. B. (2019). The Egalitarian binding partners Dynein light chain and Bicaudal‐D act sequentially to link mRNA to the dynein motor. Development, 146, dev176529 10.1242/dev176529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor, T. , Wieschaus, E. F. , McGregor, A. P. , Bialek, W. , & Tank, D. W. (2007). Stability and nuclear dynamics of the Bicoid morphogen gradient. Cell, 130(1), 141–152. 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet, O. , & Ephrussi, A. (2001). Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Current Biology, 11(21), 1666–1674. 10.1016/s0960-9822(01)00508-5 [DOI] [PubMed] [Google Scholar]
- Hachet, O. , & Ephrussi, A. (2004). Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature, 428(6986), 959–963. 10.1038/nature02521 [DOI] [PubMed] [Google Scholar]
- Hay, B. , Ackerman, L. , Barbel, S. , Jan, L. Y. , & Jan, Y. N. (1988). Identification of a component of Drosophila polar granules. Development, 103(4), 625–640. [DOI] [PubMed] [Google Scholar]
- Hay, B. , Jan, L. Y. , & Jan, Y. N. (1990). Localization of Vasa, a component of Drosophila polar granules, in maternal‐effect mutants that alter embryonic anteroposterior polarity. Development, 109(2), 425–433. [DOI] [PubMed] [Google Scholar]
- Herpers, B. , Xanthakis, D. , & Rabouille, C. (2010). ISH‐IEM: A sensitive method to detect endogenous mRNAs at the ultrastructural level. Nature Protocols, 5(4), 678–687. 10.1038/nprot.2010.12 [DOI] [PubMed] [Google Scholar]
- Hurd, T. R. , Herrmann, B. , Sauerwald, J. , Sanny, J. , Grosch, M. , & Lehmann, R. (2016). Long Oskar controls mitochondrial inheritance in Drosophila melanogaster . Developmental Cell, 39(5), 560–571. 10.1016/j.devcel.2016/11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida, T. , & Kobayashi, S. (1998). Essential role of mitochondrially encoded large rRNA for germ‐line formation in Drosophila embryos. Proceedings of the National Academy of Sciences of the United States of America, 95(19), 11274–11278. 10.1073/pnas.95.19.11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illmensee, K. , & Mahowald, A. (1974). Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proceedings of the National Academy of Sciences of the United States of America, 71(4), 1016–1020. 10.1073/pnas.71.4.1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion, U. , & St Johnston, D. (2007). bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature, 445(7127), 554–558. 10.1038/nature05503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R. A. , & Gavis, E. R. (2008). The Drosophila hnRNP M homolog Rumpelstiltskin regulates nanos mRNA localization. Development, 135(5), 973–982. 10.1242/dev.015438 [DOI] [PubMed] [Google Scholar]
- Jambor, H. , Mueller, S. , Bullock, S. L. , & Ephrussi, A. (2014). A stem‐loop structure directs oskar mRNA to microtubule minus ends. RNA, 20(4), 429–439. 10.1281/rna.041566.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor, H. , Surendranath, V. , Kalinka, A. T. , Mejstrik, P. , Saalfeld, S. , & Tomancak, P. (2015). Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. eLife, 4, e05003 10.7554/eLife.05003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke, J. , Gervais, L. , Dass, S. , Kaltschmidt, J. A. , Lopez‐Schier, A. , St Johnston, D. , … Guichet, A. (2002). Polar transport in the Drosophila oocyte requires Dynein and kinesin I cooperation. Current Biology, 12(23), 1971–1981. 10.1016/S0960-9822(02)01302-7 [DOI] [PubMed] [Google Scholar]
- Jenny, A. , Hachet, O. , Závorszky, P. , Cyrklaff, A. , Weston, M. D. J. , St Johnston, D. , … Ephrussi, A. (2006). A translation‐independent role of oskar RNA in early Drosophila oogenesis. Development, 133(15), 2827–2833. 10.1242/dev02456 [DOI] [PubMed] [Google Scholar]
- Jeske, M. , Bordi, M. , Glatt, S. , Müller, S. , Rybin, V. , Müller, C. W. , & Ephrussi, A. (2015). The crystal structure of the Drosophila germline inducer Oskar identifies two domains with distinct Vasa helicase‐ and RNA‐binding activities. Cell Reports, 12(4), 587–598. 10.1016/jcelrep.2015.06.055 [DOI] [PubMed] [Google Scholar]
- Jeske, M. , Müller, C. W. , & Ephrussi, A. (2017). The LOTUS domain is a conserved DEAD‐box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage. Genes & Development, 31(9), 939–952. 10.1101/gad.297051.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J. R. , & Macdonald, P. M. (2007). Oskar controls morphology of polar granules and nuclear bodies in Drosophila . Development, 134(2), 233–236. 10.1242/dev02729 [DOI] [PubMed] [Google Scholar]
- Jongens, T. A. , Hay, B. , Jan, L. Y. , & Jan, Y. N. (1992). The germ cell‐less gene product: A posteriorly localized component necessary for germ cell development in Drosophila . Cell, 70(4), 569–584. 10.1016/0092-8674(92)90427-e [DOI] [PubMed] [Google Scholar]
- Khoc Trong, P. , Doerflinger, H. , Dunkel, J. , St Johnston, D. , & Goldstein, R. E. (2015). Cortical microtubule nucleation can organise the cytoskeleton of Drosophila oocytes to define the anteroposterior axis. eLife, 4, e06088 10.7554/eLife.06088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G. , Pai, C. I. , Sato, K. , Person, M. D. , Nakamura, A. , & Macdonald, P. M. (2015). Region‐specific activation of oskar mRNA translation by inhibition of Bruno‐mediated repression. PLoS Genetics, 11(2), e1004992 10.1371/journal.pgen.1004992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim‐Ha, J. , Kerr, K. , & Macdonald, P. M. (1995). Translational regulation of oskar mRNA by Bruno, an ovarian RNA‐binding protein, is essential. Cell, 81(3), 403–412. 10.1016/0092-8674(95)90393-3 [DOI] [PubMed] [Google Scholar]
- Kim‐Ha, J. , Smith, J. L. , & Macdonald, P. M. (1991). oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell, 66(1), 23–35. 10.1016/0092-8674(91)90136-m [DOI] [PubMed] [Google Scholar]
- Kistler, K. E. , Trcek, T. , Hurd, T. R. , Chen, R. , Liang, F. X. , Sall, J. , … Lehmann, R. (2018). Phase‐transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. eLife, 7, e37949 10.7554/eLife.37949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, S. , Amikura, R. , & Okada, M. (1993). Presence of mitochondrial large ribosomal RNA outside mitochondria in germ plasm of Drosophila melanogaster . Science, 260(5113), 1521–1524. 10.1126/science.7684857 [DOI] [PubMed] [Google Scholar]
- Krauss, J. , López de Quinto, S. , Nüsslein‐Volhard, C. , & Ephrussi, A. (2009). Myosin‐V regulates oskar mRNA localization in the Drosophila oocyte. Current Biology, 19(23), 1058–1063. 10.1016/j.cub.2009.04.062 [DOI] [PubMed] [Google Scholar]
- Lasko, P. F. , & Ashburner, M. (1990). Posterior localization of Vasa protein correlates with, but is not sufficient for, pole cell formation. Genes & Development, 4(6), 905–921. 10.1101/gad.4.6.905 [DOI] [PubMed] [Google Scholar]
- Lazzaretti, D. , Veith, K. , Kramer, K. , Basquin, C. , Urlaub, H. , Irion, U. , & Bono, F. (2016). The bicoid mRNA localization factor Exuperantia is an RNA‐binding pseudonuclease. Nature Structural and Molecular Biology, 23(8), 705–713. 10.1038/nsmb.3254 [DOI] [PubMed] [Google Scholar]
- Lécuyer, E. , Yoshida, H. , Parthasarathy, N. , Alm, C. , Babak, T. , Cerovina, T. , … Krause, H. M. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell, 131(1), 174–187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Lehmann, R. (2016). Germ plasm biogenesis—An Oskar‐centric perspective. Current Topics in Developmental Biology, 116, 679–707. 10.1016/bs.ctdb.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, R. , & Nüsslein‐Volhard, C. (1986). Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila . Cell, 47(1), 141–152. 10.1016/0092-8674(86)90375-2 [DOI] [PubMed] [Google Scholar]
- Lerit, D. A. , & Gavis, E. R. (2011). Transport of germ plasm on astral microtubules directs germ cell development in Drosophila . Current Biology, 21(6), 439–448. 10.1016/j.cub.2011.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. G. , McGrail, M. , Serr, M. , & Hays, T. S. (1994). Drosophila cytoplasmic dynein, a microtubule motor that is asymmetrically localized in the oocyte. The Journal of Cell Biology, 126(6), 1475–1494. 10.1083/jcb.126.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. , Lee, M. , Son, A. , Chang, H. , & Kim, V. N. (2016). mTAIL‐seq reveals dynamic poly(A) tail regulation in oocyte‐to‐embryo development. Genes & Development, 30(14), 1671–1682. 10.1101/gad.284802.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, S. C. , Sinsimer, K. S. , Lee, J. J. , Wieschaus, E. , & Gavis, E. R. (2015). Independent and coordinate trafficking of single Drosophila germ plasm RNAs. Nature Cell Biology, 17(5), 558–568. 10.1038/ncb3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Xie, T. , & Steward, R. (1999). Lis1, the Drosophila homolog of a human lissencephaly gene, is required for germline cell division and oocyte differentiation. Development, 126, 4477–4488. [DOI] [PubMed] [Google Scholar]
- Lu, W. , Lakonishok, M. , Liu, R. , Billington, N. , Rich, A. , Glotzer, M. , … Gelfand, V. I. (2020). Competition between kinesin‐1 and myosin‐V defines Drosophila posterior determination. eLife, 14(9), e54216 10.7554/eLife.54216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W. , Lakonishok, M. , Serpinskaya, A. S. , Kirchenbüchler, D. , Ling, S. C. , & Gelfand, V. I. (2018). Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination. The Journal of Cell Biology, 217(10), 3497–3511. 10.1083/jcb/201709174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W. , Winding, M. , Lakonishok, M. , Wildonger, J. , & Gelfand, V. I. (2016). Microtubule‐microtubule sliding by kinesin‐1 is essential for normal cytoplasmic streaming in Drosophila oocytes. Proceedings of the National Academy of Sciences of the United States of America, 113(34), E4995–E5004. http://doi.org/10/1073/pnas.1522424113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig, S. , Moussian, B. , Krauss, J. , Desjeux, I. , Perkovic, C. , & Nüsslein‐Volhard, C. (2004). An F1 genetic screen for maternal‐effect mutations affecting embryonic pattern formation in Drosophila melanogaster . Genetics, 167(1), 325–342. 10.1534/genetics.167.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. , He, F. , Xie, G. , & Deng, W. M. (2016). Maternal AP determinants in the Drosophila oocyte and embryo. Wiley Interdisciplinary Reviews: Developmental Biology, 5(5), 562–581. 10.1002/wdev.235 [DOI] [PubMed] [Google Scholar]
- Macdonald, P. M. , Kanke, M. , & Kenny, A. (2016). Community effects in regulation of translation. eLife, 5, e10965 10.7554/eLife.10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald, P. M. , & Struhl, G. (1988). cis‐acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature, 336(6199), 595–598. 10.1038/336595a0 [DOI] [PubMed] [Google Scholar]
- Mach, J. M. , & Lehmann, R. (1997). An Egalitarian‐Bicaudal‐D complex is essential for oocyte specification and axis determination in Drosophila . Genes & Development, 11(4), 423–435. 10.1101/gad.11.4.423 [DOI] [PubMed] [Google Scholar]
- Mahowald, A. P. (1962). Fine structure of pole cells and polar granules in Drosophila melanogaster . The Journal of Experimental Zoology, 151(3), 201–215. 10.1002/jez.1401510302 [DOI] [Google Scholar]
- Mahowald, A. P. (1968). Polar granules in Drosophila. II. Ultrastructural changes during early embryogenesis. The Journal of Experimental Zoology, 167(2), 237–262. 10.1002/jez.1401670211 [DOI] [PubMed] [Google Scholar]
- Manseau, L. , Calley, J. , & Phan, H. (1996). Profilin is required for posterior patterning of the Drosophila oocyte. Development, 122(7), 2109–2116. [DOI] [PubMed] [Google Scholar]
- Markussen, F.‐H. , Michon, A. M. , Breitwieser, W. , & Ephrussi, A. (1995). Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development, 121(11), 3723–3732. [DOI] [PubMed] [Google Scholar]
- McClintock, M. A. , Dix, C. I. , Johnson, C. M. , McLaughlin, S. H. , Maizels, R. J. , Hoang, H. T. , & Bullock, S. L. (2018). RNA‐directed activation of cytoplasmic dynein‐1 in reconstituted transport RNPs. eLife, 7, e36312 10.7554/eLife.36312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail, M. , & Hays, T. S. (1997). The microtubule motor cytoplasmic dynein is required for spindle orientation during germline cell divisions and oocyte differentiation in Drosophila . Development, 124(12), 2409–2419. [DOI] [PubMed] [Google Scholar]
- Micklem, D. R. , Adams, J. , Grünert, S. , & St Johnston, D. (2000). Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. The EMBO Journal, 19(6), 1366–1377. 10.1093/emboj/19.6.1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, S. , Dillon, S. , & Boswell, R. (2001). The RNA‐binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes & Development, 15(21), 2886–2899. 10.1101/gad.927001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, T. P. , Kwon, S. , Schnapp, B. J. , & St Johnston, D. (2006). A repeated IMP‐binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. The Journal of Cell Biology, 172(4), 577–588. 10.1083/jcb.200510044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A. , Amikura, R. , Mukai, M. , Kobayashi, S. , & Lasko, P. F. (1996). Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science, 274(5295), 2075–2079. 10.1126/science.274.5295.2075 [DOI] [PubMed] [Google Scholar]
- Nakamura, A. , Sato, K. , & Hanyu‐Nakamura, K. (2004). Drosophila Cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Developmental Cell, 6(1), 69–78. 10.1016/s1534-5807(03)00400-3 [DOI] [PubMed] [Google Scholar]
- Nashchekin, D. , Fernandes, A. R. , & St Johnston, D. (2016). Patronin/Shot cortical foci assemble the noncentrosomal microtubule array that specifies the Drosophila anterior‐posterior axis. Developmental Cell, 38(1), 61–72. 10.1016/j.devcel.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, C. , Puthalakath, H. , Adams, J. M. , Strasser, A. , & Lehmann, R. (2004). Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nature Cell Biology, 6(5), 427–435. 10.1038/ncb1122 [DOI] [PubMed] [Google Scholar]
- Newmark, P. , & Boswell, R. (1994). The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila . Development, 120(5), 1303–1313. [DOI] [PubMed] [Google Scholar]
- Niepielko, M. G. , Eagle, W. V. I. , & Gavis, E. R. (2018). Stochastic seeding coupled with mRNA self‐recruitment generates heterogeneous Drosophila germ granules. Current Biology, 28(12), 1872–1881. 10.1016/j.cub.2018.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nüsslein‐Volhard, C. , Frohnhöfer, H.‐G. , & Lehmann, R. (1987). Determination of anteroposterior polarity in Drosophila . Science, 238(4834), 1675–1681. 10.1126/science.3686007 [DOI] [PubMed] [Google Scholar]
- Pae, J. , Cinalli, R. M. , Marzio, A. , Pagano, M. , & Lehmann, R. (2017). GCL and CUL3 control the switch between cell lineages by mediating localized degradation of an RTK. Developmental Cell, 42(2), 130–142. 10.1016/j.devcel.2017.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, I. , Gatfield, D. , St Johnston, D. , & Izaurralde, E. (2004). An eIF4AIII‐containing complex required for mRNA localization and nonsense‐mediated mRNA decay. Nature, 427(6976), 753–757. 10.1038/nature02351 [DOI] [PubMed] [Google Scholar]
- Palacios, I. M. , & St Johnston, D. (2002). Kinesin light chain‐independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localization in the Drosophila oocyte. Development, 129(23), 5473–5485. 10.1242/dev.00119 [DOI] [PubMed] [Google Scholar]
- Parton, R. M. , Hamilton, R. S. , Ball, G. , Yang, L. , Cullen, C. F. , Lu, W. , … Davis, I. (2011). A PAR‐1‐dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. The Journal of Cell Biology, 194(1), 121–135. 10.1083/jcb.201103160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, V. S. , Anand, A. , Chakrabarti, A. , & Kai, T. (2014). The Tudor domain protein Tapas, a homolog of the vertebrate Tdrd7, functions in piRNA pathway to regulate retrotransposons in germline of Drosophila melanogaster . BMC Biology, 12, 61 10.1186/s12915-014-0061-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, V. S. , & Kai, T. (2010). Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Current Biology, 20(8), 724–730. 10.1016/j.cub.2010.02.046 [DOI] [PubMed] [Google Scholar]
- Pokrywka, N. J. , & Stephenson, E. C. (1991). Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development, 113(1), 55–66. [DOI] [PubMed] [Google Scholar]
- Quinlan, M. E. (2016). Cytoplasmic streaming in the Drosophila oocyte. Annual Review of Cell and Developmental Biology, 32, 173–195. 10.1146/annurev-cellbio-111315-125416 [DOI] [PubMed] [Google Scholar]
- Rangan, P. , DeGennaro, M. , Jaime‐Bustamente, K. , Coux, R.‐X. , Martinho, R. G. , & Lehmann, R. (2009). Temporal and spatial control of germ‐plasm RNAs. Current Biology, 19(1), 72–77. 10.1016/j.cub.2008.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveal, B. , Yan, N. , Snee, M. J. , Pai, C. I. , Gim, Y. , & Macdonald, P. M. (2010). BREs mediate both repression and activation of oskar mRNA translation and act in trans . Developmental Cell, 18(3), 496–502. 10.1016/j.devcel.2009.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas‐Rios, P. , & Simonelig, M. (2018). piRNAs and PIWI proteins: Regulators of gene expression in development and stem cells. Development, 145(17), dev161786 10.1242/dev.161786 [DOI] [PubMed] [Google Scholar]
- Rongo, C. G. (1996). The role of RNA localization and translational regulation in Drosophila germ cell determination (PhD thesis). Massachusetts Institute of Technology. Available at http://hdl.handle.net/1721.1/10562
- Rouget, C. , Papin, C. , Boureux, A. , Meunier, A. C. , Franco, B. , Robine, N. , … Simonelig, M. (2010). Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature, 467(7319), 1128–1132. 10.1038/nature09465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, P. V. , & Lerit, D. A. (2018). RNA localization regulates diverse and dynamic cellular processes. Traffic, 19(7), 496–502. 10.1111/tra.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, Y. H. , Kenny, A. , Gim, Y. , Snee, M. , & Macdonald, P. M. (2017). Multiple cis‐acting signals, some weak by necessity, collectively direct robust transport of oskar mRNA to the oocyte. Journal of Cell Science, 130(18), 3060–3071. 10.1242/jcs.202069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallés, F. J. , Lieberfarb, M. E. , Wreden, C. , Gergen, J. P. , & Strickland, S. (1994). Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science, 266(5193), 1996–1999. 10.1126/science.7801127 [DOI] [PubMed] [Google Scholar]
- Sanghavi, P. , Liu, G. , Veeranan‐Karmegam, R. , Navarro, C. , & Gonsalvez, G. B. (2016). Multiple roles for Egalitarian in polarization of the Drosophila egg chamber. Genetics, 203(1), 415–432. 10.1534/genetics.115.184622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus, L. R. , Cha, B. J. , Theurkauf, W. E. , & Saxton, W. M. (2005). Dynein and the actin cytoskeleton control kinesin‐driven cytoplasmic streaming in Drosophila oocytes. Development, 132(16), 3743–3752. 10.1242/dev.01956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, R. S. , & Anderson, K. V. (2006). Drosophila Ik2, a member of the I kappa B kinase family, is required for mRNA localization during oogenesis. Development, 133(8), 1467–1475. 10.1242/dev.02318 [DOI] [PubMed] [Google Scholar]
- Simon, B. , Masiewicz, P. , Ephrussi, A. , & Carlomagno, T. (2015). The structure of the SOLE element of oskar mRNA. RNA, 21(8), 1444–1453. 10.1261/rna.049601.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsimer, K. S. , Jain, R. A. , Chatterjee, S. , & Gavis, E. R. (2011). A late phase of germ plasm accumulation during Drosophila oogenesis requires lost and rumpelstiltskin . Development, 138(16), 3431–3440. 10.1242/dev.065029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, M. C. , Sato, K. , Pezic, D. , & Aravin, A. A. (2011). PIWI‐interacting small RNAs: The vanguard of genome defense. Nature Reviews Molecular Cell Biology, 12(4), 246–258. 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Sladewski, T. E. , Billington, N. , Ali, M. Y. , Bookwalter, C. S. , Lu, H. , Krementsova, E. B. , … Trybus, K. M. (2018). Recruitment of two dyneins to an mRNA‐dependent Bicaudal D transport complex. eLife, 7, e36306 10.7554/eLife.36306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. L. , Wilson, J. E. , & Macdonald, P. M. (1992). Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell, 70(5), 849–859. 10.1016/0092-8674(92)90318-7 [DOI] [PubMed] [Google Scholar]
- Spirov, A. , Fahmy, K. , Schneider, M. , Frei, E. , Noll, M. , & Baumgartner, S. (2009). Formation of the Bicoid morphogen gradient: an mRNA gradient dictates the protein gradient. Development, 136(4), 605–614. 10.1242/dev.031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston, D. , Driever, W. , Berleth, T. , Richstein, S. , & Nüsslein‐Volhard, C. (1989). Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development, 107(Suppl.), 13–19. [DOI] [PubMed] [Google Scholar]
- Subtelny, A. O. , Eichhorn, S. W. , Chen, G. R. , Sive, H. , & Bartel, D. P. (2014). Poly(A)‐tail profiling reveals an embryonic switch in translational control. Nature, 508(7494), 66–71. 10.1038/nature13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter, B. (2018). RNA localization and transport. Biochimica et Biophysica Acta – Gene Regulatory Mechanisms, 1861, 938–951. 10.1016/j.bbagrm.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Swan, A. , Nguyen, T. , & Suter, B. (1999). Drosophila Lissencephaly‐1 functions with Bic‐D and Dynein in oocyte determination and nuclear positioning. Nature Cell Biology, 1(7), 444–449. 10.1038/15680 [DOI] [PubMed] [Google Scholar]
- Tanaka, T. , Kato, Y. , Matsuda, K. , Hanyu‐Nakamura, K. , & Nakamura, A. (2011). Drosophila Mon2 couples Oskar‐induced endocytosis with actin remodeling for cortical anchorage of the germ plasm. Development, 138(12), 2523–2532. 10.1242/dev.062208 [DOI] [PubMed] [Google Scholar]
- Tanaka, T. , & Nakamura, A. (2008). The endocytic pathway acts downstream of Oskar in Drosophila germ plasm assembly. Development, 135(6), 1107–1117. 10.1242/dev.017293 [DOI] [PubMed] [Google Scholar]
- Theurkauf, W. E. , Alberts, B. , Jan, Y. N. , & Jongens, T. A. (1993). A central role for microtubules in the differentiation of Drosophila oocytes. Development, 118(4), 1169–1180. [DOI] [PubMed] [Google Scholar]
- Trcek, T. , Grosch, M. , York, A. , Shroff, H. , Lionnet, T. L. , & Lehmann, R. (2015). Drosophila germ granules are structured and contain homotypic RNA clusters. Nature Communications, 6, 7962 10.1038/ncomms8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek, T. , & Lehmann, R. (2019). Germ granules in Drosophila . Traffic, 20(9), 650–660. 10.1111/tra.12674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovisco, V. , Belaya, K. , Nashchekin, D. , Irion, U. , Sirinakis, G. , Butler, R. , … St Johnston, D. (2016). bicoid mRNA localises to the Drosophila oocyte anterior by random Dynein‐mediated transport and anchoring. eLife, 5, e17537 10.7554/eLife.17537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo, N. , Oprins, A. , Xanthakis, D. , Ephrussi, A. , & Rabouille, C. (2007). Stimulation of endocytosis and actin dynamics by Oskar polarizes the Drosophila oocyte. Developmental Cell, 12(4), 543–555. 10.1016/j.devcel.2007.03.002 [DOI] [PubMed] [Google Scholar]
- Vanzo, N. F. , & Ephrussi, A. (2002). Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development, 129(15), 3705–3714. [DOI] [PubMed] [Google Scholar]
- Vo, H. D. L. , Wahlduzzaman, Tindell, S. J. , Zheng, J. , Gao, M. , & Arkov, A. (2019). Protein components of ribonucleoprotein granules from Drosophila germ cells oligomerize and show distinct spatial organization during germline development. Scientific Reports, 9(1), 19190 10.1038/s41598-019-55747-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas, A. , Alexiou, P. , Vrettos, N. , Maragkakis, M. , & Mourelatos, Z. (2016). Sequence‐dependent but not sequence‐specific piRNA adhesion traps mRNAs to the germ plasm. Nature, 531(7594), 390–394. 10.1038/nature17150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , & Lehmann, R. (1991). Nanos is the localized posterior determinant in Drosophila . Cell, 66(4), 637–647. 10.1016/0092-8674(91)90110-k [DOI] [PubMed] [Google Scholar]
- Wang, J. T. , Smith, J. , Chen, B. C. , Schmidt, H. , Rasoloson, D. , Paix, A. , … Seydoux, G. (2014). Regulation of RNA granule dynamics by phosphorylation of serine‐rich intrinsically disordered proteins in C. elegans . eLife, 3, e04591 10.7554/eLife.04591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, P. J. , Liang, L. , Berg, C. A. , Lasko, P. , & Macdonald, P. M. (1997). Translational repressor Bruno plays multiple roles in development and is widely conserved. Genes & Development, 11(19), 2510–2521. 10.1101/gad.11.19.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil, T. T. , Forrest, K. M. , & Gavis, E. R. (2006). Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Developmental Cell, 11(2), 251–262. 10.1016/j.devcel.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Weil, T. T. , Parton, R. , Davis, I. , & Gavis, E. R. (2008). Changes in bicoid mRNA anchoring highlight conserved mechanisms during the oocyte‐to‐embryo transition. Current Biology, 18(14), P1055–P1061. 10.1016/j.cub.2008.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton, R. P. , & Struhl, G. (1991). RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos . Cell, 67(5), 955–967. 10.1016/0092-8674(91)90368-9 [DOI] [PubMed] [Google Scholar]
- Wharton, T. H. , Nomie, K. J. , & Wharton, R. P. (2018). No significant regulation of bicoid mRNA by Pumilio or Nanos in the early Drosophila embryo. PLoS One, 13(3), e0194865 10.1371/journal.pone.0194865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus, E. (2016). Positional information and cell fate determination in the early Drosophila embryo. Current Topics in Developmental Biology, 117, 567–579. 10.1016/bs.ctdb.2015.11.020 [DOI] [PubMed] [Google Scholar]
- Wilkie, G. S. , & Davis, I. (2001). Drosophila wingless and pair‐rule transcripts localize apically by dynein‐mediated transport of RNA particles. Cell, 105(2), 209–219. 10.1016/s0092-8674(01)00312-9 [DOI] [PubMed] [Google Scholar]
- Wilson, J. E. , Connell, J. E. , & Macdonald, P. M. (1996). aubergine enhances oskar translation in the Drosophila ovary. Development, 122(5), 1631–1639. [DOI] [PubMed] [Google Scholar]
- Yang, N. , Yu, Z. , Hu, M. , Wang, M. , Lehmann, R. , & Xu, R.‐M. (2015). Structure of Drosophila Oskar reveals a novel RNA binding protein. Proceedings of the National Academy of Sciences of the United States of America, 112(37), 11541–11546. 10.1073/pnas.1515568112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Wang, J. , Xu, J. , Zhang, Z. , Koppetsch, B. S. , Schultz, N. , … Theurkauf, W. E. (2012). UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell, 151(4), 871–884. 10.1016/j.cell.2012.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin, V. L. , Belaya, K. , Pecreaux, J. , Gilchrist, M. J. , Clark, A. , Davis, I. , … St Johnston, D. (2008). In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell, 134(5), 843–853. 10.1016/j.cell.2008.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
FURTHER READING
- Others have recently reviewed this general topic, each with their own different emphasis. The interested reader should also consult the following references (Goldman & Gonsalvez, 2017; Lehmann, 2016; Ma, He, Xie, & Deng, 2016; Quinlan, 2016; Rojas‐Rios & Simonelig, 2018; Ryder & Lerit, 2018; Suter, 2018; Trcek & Lehmann, 2019; Wieschaus, 2016).
- Briscoe, J. , & Small, S. (2015). Morphogen rules: Design principles of gradient‐mediated embryo patterning. Development, 142(23), 3996–4009. 10.1242/dev.129452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, P. F. , Poulin, F. , Cho‐Park, Y. A. , Cho‐Park, I. B. , Chicoine, J. D. , Lasko, P. , & Sonenberg, N. (2005). A new paradigm for translational control: Inhibition via 5′‐3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell, 121(3), 411–423. 10.1016/j.cell.2005.02.024 [DOI] [PubMed] [Google Scholar]
- Rivera‐Pomar, R. , Niessing, D. , Schmitt‐Ott, U. , Gehring, W. J. , & Jäckle, H. (1996). RNA binding and translational suppression by Bicoid. Nature, 379(6567), 746–749. 10.1038/379746a0 [DOI] [PubMed] [Google Scholar]
- Rödel, C. J. , Gilles, A. F. , & Averof, M. (2013). MicroRNAs act as cofactors in Bicoid‐mediated translational repression. Current Biology, 23(16), 1579–1584. 10.1016/j.cub.2013.06.041 [DOI] [PubMed] [Google Scholar]
- Sinsimer, K. S. , Lee, J. J. , Thiberge, S. Y. , & Gavis, E. R. (2013). Germ plasm anchoring is a dynamic state that requires persistent trafficking. Cell Reports, 12(5), 1169–1177. 10.1016/jcelrep.2013.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winding, M. , Kelliher, M. T. , Lu, W. , Wildonger, J. , & Gelfand, V. I. (2016). Role of kinesin‐1‐based microtubule sliding in Drosophila nervous system development. Proceedings of the National Academy of Sciences of the United States of America, 113(34), E4985–E4994. 10.1073/pnas.1522416113 [DOI] [PMC free article] [PubMed] [Google Scholar]


