Abstract
Urothelial carcinoma of the upper tract (UTUC) presents specific challenges regarding accurate staging and tumor sampling. We aimed to assess the feasibility of applying next-generation sequencing to biopsy specimens and gauged the concordance of their genetic profiles with matched radical nephroureterectomy (RNU) specimens. Of the 39 biopsy specimens collected, 36 (92%) had adequate material for sequencing using a hybridization-based exon capture assay (MSK-IMPACT). The most frequently altered genes across the patient cohort were consistent with the urothelial carcinoma-associated alterations identified in a cohort of 130 RNU specimens previously sequenced at our center, including mutations in the TERT promoter (64%), hotspot activating mutations in FGFR3 (64%), and frequent mutations in chromatin remodeling genes. For 12 patients, a matching tumor sample from a subsequent RNU was sequenced. We found a high level of concordance between matched biopsy and RNU specimens, up to 92% for the likely pathogenic alterations.
Keywords: Upper tract urothelial carcinoma, Genomics, Prediction, Biomarkers, Transitional cell carcinoma
Patient summary:
We evaluated the feasibility of genomic characterization of tumor tissue collected at the time of ureteroscopic biopsy and found high concordance with subsequent radical nephroureterectomy specimens. Molecular characterization of urothelial carcinoma of the upper tract biopsies could guide treatment decision-making and identify high-risk patients who could benefit from neoadjuvant chemotherapy and low-risk patients who could benefit from conservative or organ-sparing strategies.
Urothelial carcinoma of the upper tract (UTUC) comprises 5–10% of all urothelial carcinomas and presents distinct challenges compared to bladder cancer [1]. Specifically, obtaining representative diagnostic biopsies and accurate staging are extremely difficult with current endoscopic techniques and cross-sectional imaging. Owing to these limitations, patients typically undergo radical nephroureterectomy (RNU), even though some could probably be managed conservatively if predictive information could be derived from pathological and/or molecular characterization [2]. Furthermore, there is growing evidence of the benefit of perioperative chemotherapy for patients with muscle-invasive UTUC but us use has been limited by the lack of predictive biomarkers for response and treatment benefit [3]. Patients with UTUC are often precluded from receiving adjuvant cisplatin-based chemotherapy because of poor renal function following surgery [4].
We previously identified genetic signatures in RNU specimens that are associated with adverse pathologic features [5]. Applied to diagnostic biopsy specimens patients with UTUC before treatment, such prognostic molecular data may help to identify a subset of high-risk patients most likely to derive benefit from neoadjuvant chemotherapy (NAC). Furthermore, the literature demonstrates that biomarker-based identification of biologic pathways involved in urothelial carcinogenesis can be used to predict response to systemic treatment [6].
In this study we sought to assess the feasibility of next-generation sequencing using the limited tumor material collected at the time of diagnostic ureteroscopic biopsy and to gauge the concordance of genetic profiles with subsequent primary RNU specimens from the same individuals. In the pilot phase of this study, we established that an adequate quantity of high-quality DNA could be extracted from diagnostic ureteroscopic biopsy samples. To do so, we retrospectively requested formalin-fixed and paraffin-embedded blocks of UTUC biopsies available from our institutions biorepository (n = 18). All were deemed to have adequate tumor content (>40%) on pathologic re-review and were submitted for DNA extraction and sequencing. To augment the 18 patients from this retrospective cohort, we enrolled 21 patients on a prospective tissue procurement protocol and sequenced their UTUC biopsy specimens prospectively.
All biopsies were obtained using ureteroscopic techniques. An effort was made to remove large pieces of interact tissue by performing laser tissue excision or using 3.2F cup biopsy forceps. Hematoxylin and eosin stains were reviewed to confirm the histologic diagnosis and to select the most appropriate sections for molecular characterization before DNA extraction. Matching blood was used as a source of germline DNA. Following extraction, both tumor and germline DNA was sequenced using a hybridization-based exon capture assay (MSK-IMPACT) as previously described [7]. The version of MSK-IMPACT used was capable of identifying missense mutations, small insertions and deletions, copy number alterations (CNAs), and select fusions (including FGFR3:TACC3) in 410 cancer-related genes.
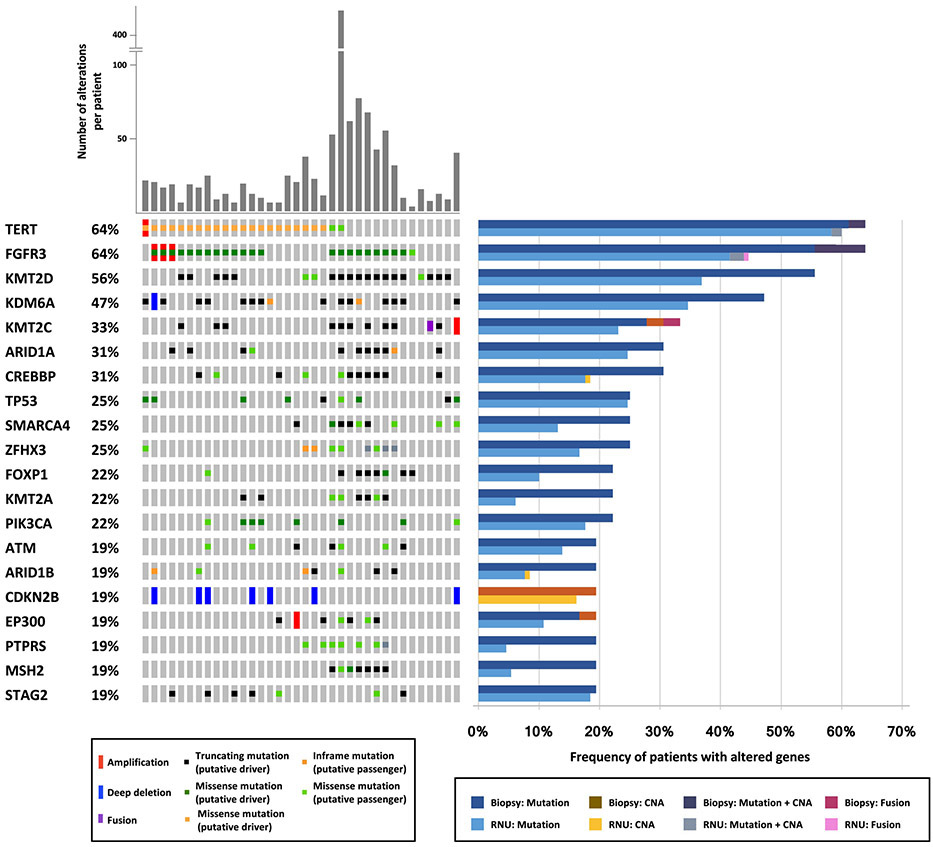
In total, 39 samples were identified and 36 (92%) had adequate material for sequencing, with a median DNA yield of 157 ng (interquartile range 31–574; Supplementary Table 1). The mean coverage for all tumors was 749×. A total of 1147 nonsynonymous mutations and 79 CNAs were identified in the cohort. The mean number of somatic alterations (mutations and copy number) per patient was 34 (range 3–416). The most frequently altered genes across the cohort are shown in Figure 1. TERT and FGFR3 were the two most frequently altered genes, with mutations observed in 64% of cases (23 of 36). Consistent with prior studies [8,9], chromatin remodeling genes including KMT2D (56%), KDM6A (47%), KMT2C (33%), ARID1A (31%), and CREBBP (31%) were commonly mutated, with most mutations in these genes predicted to result in protein inactivation. Alteration of TP53 was found in 25% of cases.
Fig. 1 –
Oncoprint of the most frequently altered genes for 36 biopsy samples successfully sequenced using MSK-IMPACT for 410 cancer-related genes. The frequency of patients with altered genes is compared to a cohort of 130 radical nephroureterectomy specimens sequenced using the same assay.
To confirm that the cohort of ureteroscopic biopsies analyzed was representative of the broader population of patients with UTUC, we compared the molecular landscape of the biopsies to a cohort of 130 RNU specimens previously sequenced at our institution using the same MSK-IMPACT assay. The two cohorts were highly similar. Of the 20 most frequently mutated genes, only five genes exhibited statistically significant differences in alteration prevalence between the ureteroscopic biopsy and RNU cohorts according to univariate analysis: FGFR3 (64% vs 45%;p = 0.041), KMT2D (56% vs 37%; p = 0.044); KMT2A (22% vs 6%; p = 0.004), PTPRS (19% vs 5%; p = 0.003), and MSH2 (19% vs 5%; p = 0.007) were all more frequently altered in the biopsy cohort. Some of these differences could be explained by a higher frequency of low-grade tumors in the biopsy cohort (34% vs 17%; p = 0.035).
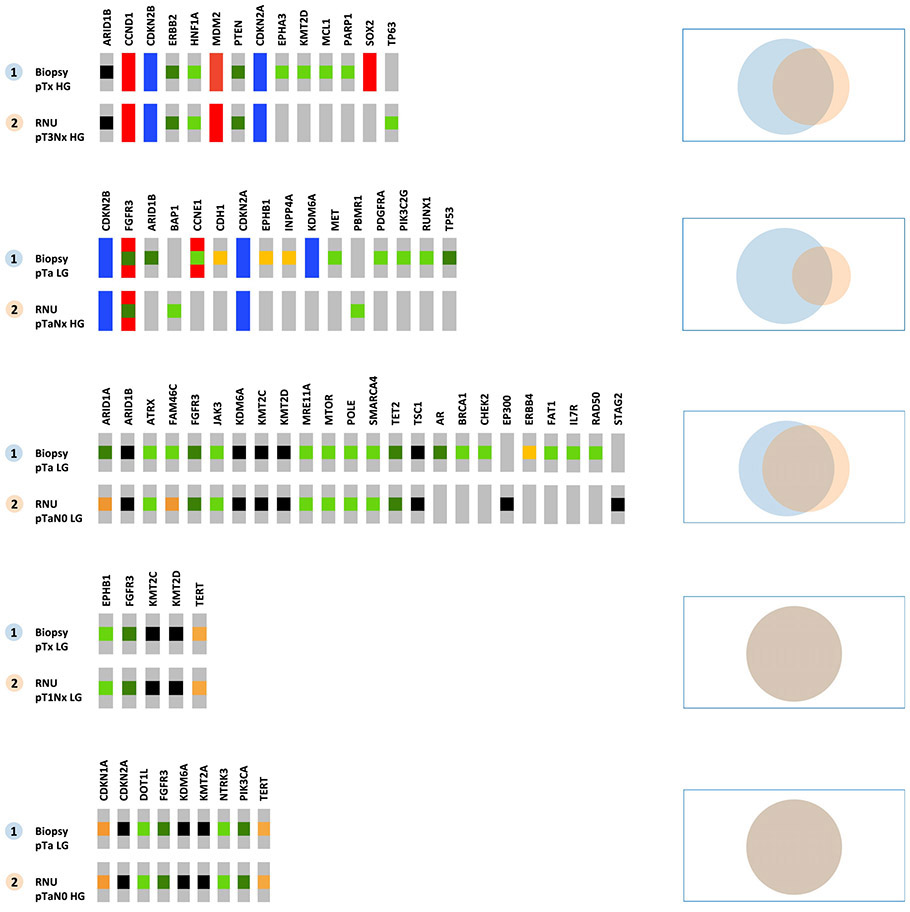
To determine whether intratumoral heterogeneity or intervening tumor evolution could be a source of molecular discordance between biopsy samples collected at diagnostic ureteroscopy and bulk tumor collected at RNU, we compared the molecular profiles of biopsy specimens and matching tumor samples from RNU for 12 patients with available material. Of these patients, 7/12 received NAC after the biopsy was performed. For the patients who did not receive NAC, 71% of all mutations and 92% of likely pathogenic mutations were present in both the biopsy and RNU specimens (Fig. 2). We found 100% concordance in 2/5 patients. For the patients who received NAC, the concordance was lower (53% and 62% for all and likely pathogenic mutations, respectively), and may have been the result of outgrowth of a drug-resistant population under the selective pressure of chemotherapy [10]. Nevertheless, 82% of the likely oncogenic mutations present in the RNU or metastasis specimen were found in the prior biopsy (Supplementary Fig 1).
Fig. 2 –
Oncoprints and Venn diagrams for matched-pair comparison of biopsy and radical nephroureterectomy specimens from patients who did not receive neoadjuvant chemotherapy.
In conclusion, genomic profiling of biopsy specimens is technically feasible via next-generation sequencing for the majority of patients (92%). This study was limited by the number of matched RNU specimen available and the absence of correlation between genomic alterations from the biopsy and the outcomes because of the small number of events. However, it is the first study to evaluate genomic characterization from UTUC biopsy and supports the validity of genomic concordance between biopsy material and primary UTUC tumors, which could be used to guide treatment decisions in the preoperative setting. Evaluation of urinary cell-free DNA could provide similar information using noninvasive collecting techniques and is under investigation.
Supplementary Material
Take Home Message.
Urothelial carcinoma of the upper tract presents specific challenges for diagnosis and treatment strategies. We demonstrated that next-generation sequencing is feasible for ureteroscopic biopsy specimens and concordant with radical nephroureterectomy specimens. This could guide treatment decisions.
Acknowledgments
Funding/Support and role of the sponsor: None.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures: Aditya Bagrodia certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
References
- 1.Green DA, Rink M, Xylinas E, et al. Urothelial carcinoma of the bladder and the upper tract: disparate twins. J Urol 2013;189:1214–21. 10.1016/j.juro.2012.05.079 [DOI] [PubMed] [Google Scholar]
- 2.Yakoubi R, Colin P, Seisen T, et al. Radical nephroureterectomy versus endoscopic procedures for the treatment of localised upper tract urothelial carcinoma: a meta-analysis and a systematic review of current evidence from comparative studies. Eur J Surg Oncol 2014;40:1629–34. 10.1016/j.ejso.2014.06.007 [DOI] [PubMed] [Google Scholar]
- 3.Leow JJ, Martin-Doyle W, Fay AP, Choueiri TK, Chang SL, Bellmunt J. A systematic review and meta-analysis of adjuvant and neoadjuvant chemotherapy for upper tract urothelial carcinoma. Eur Urol 2014;66:529–41. 10.1016/j.eururo.2014.03.003 [DOI] [PubMed] [Google Scholar]
- 4.Xylinas E, Rink M, Margulis V, et al. Impact of renal function on eligibility for chemotherapy and survival in patients who have undergone radical nephro-ureterectomy. BJU Int 2013;112:453–61. 10.1111/j.1464-410X.2012.11649.x [DOI] [PubMed] [Google Scholar]
- 5.Bagrodia A, Cha EK, Sfakianos JP, et al. Genomic biomarkers for the prediction of stage and prognosis of upper tract urothelial carcinoma. J Urol 2016;195:1684–9. 10.1016/j.juro.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plimack ER, Dunbrack RL, Brennan TA, et al. Defects in DNA repair genes predict response to neoadjuvant cisplatin-based chemotherapy in muscle-invasive bladder cancer. Eur Urol 2015;68:959–67. 10.1016/j.eururo.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sfakianos JP, Cha EK, Iyer G, et al. Genomic characterization of upper tract urothelial carcinoma. Eur Urol 2015;68:970–7. 10.1016/j.eururo.2015.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moss TJ, Qi Y, Xi L, et al. Comprehensive genomic characterization of upper tract urothelial carcinoma. Eur Urol 2017;72:641–9. 10.1016/j.eururo.2017.05.048 [DOI] [PubMed] [Google Scholar]
- 10.Faltas BM, Prandi D, Tagawa ST, et al. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat Genet 2016;48:1490–9. 10.1038/ng.3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.