Hypoplastic left heart syndrome (HLHS) is a severe form of congenital heart disease (CHD) characterized by stenosis or atresia of the mitral and aortic valves, underdevelopment of the left ventricle (LV), and aortic hypoplasia. The LV is unable to support systemic circulation, making this disease nearly uniformly fatal if left untreated. Standard treatment includes multi-stage surgical palliation that redirects blood flow such that the right ventricle (RV) provides systemic circulation. However, many patients ultimately require heart transplantation, some early in life due to unexplained single RV failure. A mouse model of HLHS revealed cardiomyocyte abnormalities in both the LV and RV despite the LV-specific hypoplasia..1 In addition, recent studies demonstrated that human induced pluripotent stem cells (iPSCs) derived from patients with HLHS have impaired cardiac differentiation compared to healthy controls, highlighting cardiomyocyte (CM)-intrinsic abnormalities.2 However, it remains unknown if iPSC-CMs derived from HLHS patients would manifest contractility deficits and could be used as a platform for understanding the pathogenesis of early RV failure in this vulnerable patient population.
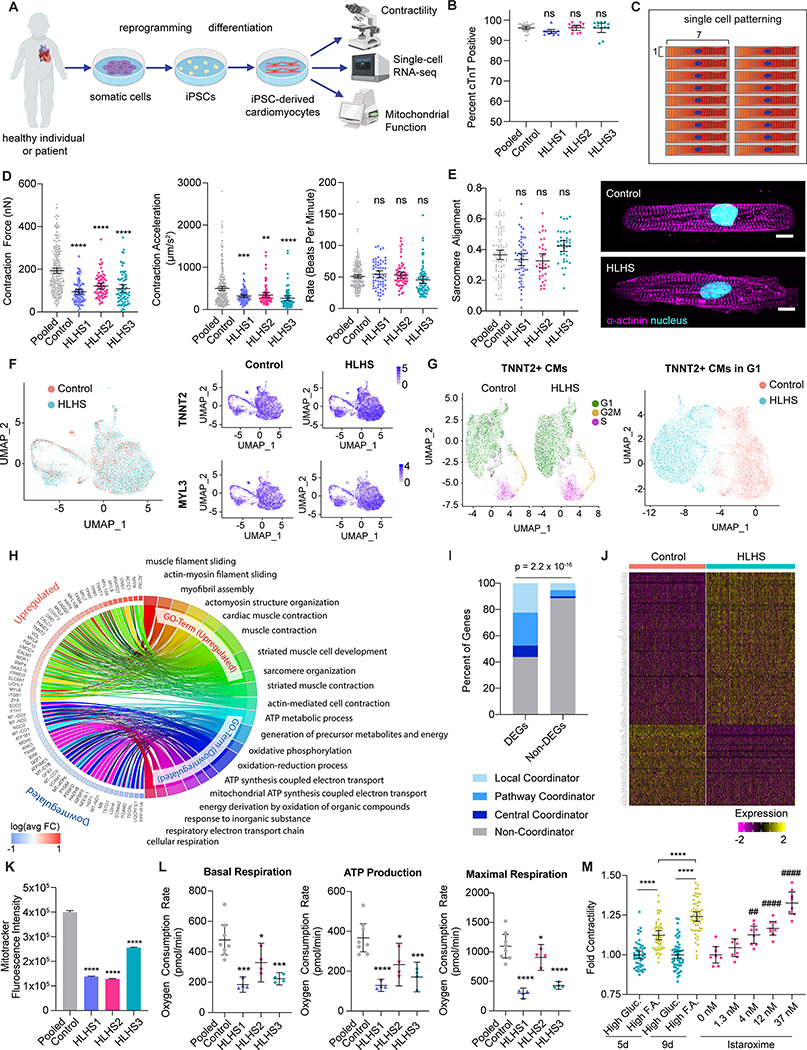
In this study, we generated iPSCs from three patients with HLHS who developed RV failure within the first decade of life (Figure A). Five healthy control lines included two unrelated individuals and one unaffected parent from each patient. Approval was obtained from the Institutional Review Boards of Mayo Clinic and Stanford University. All subjects gave consent for sample collection to derive iPSCs. Cardiac directed differentiation of iPSCs was performed utilizing small molecule modulation of the canonical Wnt/β-catenin signaling pathway. In contrast to prior reports, we did not observe a difference in cardiac differentiation efficiency between control and HLHS iPSCs based on cardiac troponin T expression (Figure B). To evaluate for functional deficits, we measured contractility in micropatterned single iPSC-CMs seeded at an optimal 7:1 aspect ratio on Day 30 of differentiation using video motion detection (Figure C).3 Contraction force and acceleration were significantly reduced in HLHS iPSC-CMs compared to controls with comparable underlying beating rates (Figure D). Sarcomere structure was preserved in HLHS iPSC-CMs based on computational alignment analysis, indicating that sarcomere disorganization was not the driver of impaired contractility (Figure E).
Figure. Impaired contractility in induced pluripotent stem cell-derived cardiomyocytes from patients with hypoplastic left heart syndrome.
A. Induced pluripotent stem cells (iPSCs) were derived from patients with hypoplastic left heart syndrome (HLHS) and controls. IPSCs were differentiated into cardiomyocytes (iPSC-CMs) using standard Wnt pathway modulation protocols. On Day 30 of differentiation, iPSC-CMs were subjected to assays of contractility, RNA sequencing, and mitochondrial function. B. Cardiac troponin T expression in control and HLHS Day 30 iPSC-CMs was measured using flow cytometry. No significant difference was identified in cardiac differentiation efficiency comparing pooled iPSC lines from control individuals (n = 5 lines, 6 biological replicates) to those from three HLHS individuals (HLHS1, HLHS2, HLHS3, n = 12 biological replicates each). C. Schematic of hydrogel micropattern for single cell plating of Day 30 iPSC-CMs D. Video motion detection of single iPSC-CMs revealed impaired contractility in HLHS iPSC-CMs (n = 72 HLHS1, 69 HLHS2, 71 HLHS3 single iPSC-CMs) relative to control (n = 276 total single iPSC-CMs from 5 iPSC lines). Both contraction force (calculated based on contraction deformation distance) and contraction acceleration were significantly reduced. Beating rate was unaffected. E. Quantification of sarcomere alignment (left) and representative images of alpha-actinin immunostaining (right) in patterned control and HLHS iPSC-CMs. Nuclei are stained with DAPI (cyan) and alpha-actinin is shown in magenta. Scale bars = 10 um. F. Single cell RNA sequencing data from Day 30 iPSC-CMs from one HLHS individual (HLHS3, n = 5310 cells) and one well-established control individual (WTC, n = 4589 cells) were analyzed and integrated using Seurat. TNNT2 and MYL3 expression revealed >98% of control and HLHS iPSC-derived cells are ventricular CMs. G. Cells identified as CMs based on normalized TNNT2 expression > 2.5 were subjected to cell cycle analysis, revealing populations of cells in G1, S, and G2/M phase in both control and HLHS iPSC-CMs (left). Re-clustering of data limited to iPSC-CMs that are in G1 phase of the cell cycle (right) shows distinct transcriptomes between iPSC-CMs from control and HLHS individuals. H. Chord plot representation of gene ontology analysis of differentially expressed genes (DEGs) between HLHS and control generated by Wilcox rank test (Bonferroni adjusted p < 0.01, logFoldChange > 0.2, expressed by >20% of cells). I. Representation of the proportion of DEGs and non-DEGs that are local coordinators, pathway coordinators, central coordinators, or non-coordinators in the heart failure network. Hypergeometric analysis showed DEGs are significantly enriched for heart failure coordinators (p = 2.2 × 10−16). J. Heatmap of all HLHS vs. control iPSC-CM DEGs that are coordinators in the heart failure network. K. Flow cytometry analysis of mitochondria labeled by MitoTracker showed reduced mitochondrial content in HLHS iPSC-CMs (n = 12,400 HLHS1, 10,556 HLHS2, and 13,000 HLHS3) compared to control iPSC-CMs (n = 16,217 cells from two unrelated controls). L. Mitochondrial function parameters, including basal respiration, ATP production, and maximal respiration, were reduced in HLHS iPSC-CMs (n = 4 replicates for each HLHS line) compared to control iPSC-CMs (n = 4 replicates for each of two unrelated controls) as measured by Seahorse analyzer. M. HLHS3 iPSC-CMs on Day 30 of differentiation were maintained as a monolayer in standard culture medium containing high glucose (High Gluc) or high fatty acid (High F.A.) content. Cells treated with high fatty acid medium showed improved contractility, as measured based on peak contraction deformation using a kinetic imaging cytometer. As a comparison, iPSC-CMs grown in high glucose medium were also treated with escalating doses of istaroxime. Absolute contractility measurements were normalized to that of cells maintained in standard high glucose medium. Data are presented as mean +/− 95% confidence interval. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 relative to pooled control (panels B, D, E, K, L) or standard high glucose medium (panel M) based on ANOVA and Dunnett’s Multiple Comparison post hoc tests. ##p<0.01, ####p<0.0001 compared to untreated cells based on ANOVA and Dunnett’s Multiple Comparison post hoc tests.
We performed single cell RNAseq on Day 30 iPSC-CMs from one control and one HLHS patient selected from our cohort based on RV failure in the first year of life. More than 98% of both control and HLHS cells expressed TNNT2 and MYL3, consistent with ventricular CM differentiation (Figure F). As cells undergoing mitosis alter contractile gene expression,4 we sought to limit our transcriptome analysis to G1 phase cells only. Visualization of the G1-phase iPSC-CMs on a UMAP plot showed distinct transcriptional profiles between HLHS and control cells (Figure G). Analysis of differentially expressed genes (DEGs) in HLHS iPSC-CMs revealed upregulation in sarcomere and cytoskeletal genes, and downregulation in genes involved in mitochondrial function and metabolism (Figure H).
We next compared the DEGs in our model of HLHS with early RV failure to a previously published analysis of weighted co-expression network connectivity in end-stage human heart failure.5 In this network, three gene sets were identified as molecular coordinators in heart failure: “local coordinators” that recruited the most neighbors in normal-to-heart failure network rewiring, “pathway coordinators” that specifically recruited neighbors from established heart failure pathways, and “central coordinators” that both recruited many neighbors, and were enriched for known heart failure pathway neighbors. The set of DEGs in HLHS was significantly enriched for these heart failure coordinators (Figure I).5 Notable examples include the canonical hypertrophy gene NPPB and pro-fibrotic gene CTGF (upregulated) and genes related to mitochondrial and metabolic function UQCRFS1 and PGAM2 (downregulated) (Figure J).
We further show that mitochondrial content in all HLHS iPSC-CMs was reduced when compared with control iPSC-CMs (Figure K). This finding was corroborated by reduced mitochondrial respiration and oxidative metabolism as evident by the significantly reduced oxygen consumption rates in HLHS iPSC-CMs (Figure L). To test for a functional consequence of decreased metabolic activity, we demonstrated that treatment of HLHS iPSC-CMs with high fatty acid content medium showed improved contractility compared to standard medium containing high glucose, demonstrating that alterations in metabolic substrate can improve contractility (Figure M). The magnitude of this increase is comparable to that seen with acute exposure to the inotrope istaroxime.
While prior studies have shown impaired cardiac differentiation in some HLHS iPSC lines, we demonstrated robust generation of ventricular CMs from our three HLHS iPSC lines. We observed significantly impaired contractility of HLHS iPSC-CMs with associated changes in gene expression that significantly overlapped prior studies of human heart failure. Moreover, our data suggest that mitochondrial dysfunction contributes to impaired contractility in HLHS iPSC-CMs, and that altering the energetic substrate of the cells can reverse the contractile phenotype. This functional impairment may contribute to susceptibility to early failure of the single RV in patients with HLHS, representing a novel therapeutic target.
Acknowledgments
We thank Sneha Venkatraman, Daniel Lee, and Boyd Rasmussen for technical assistance.
Sources of Funding
This research was supported by the Erin Hoffmann and Michael Schroepfer Foundation and Todd and Karen Wanek Family Program for Hypoplastic Left Heart Syndrome; NIH K08HL148553 (to S.L.P.); NIH F30HL149152 (to F.X.G.); NIH F32HL142205 (to S.L.); National Science Foundation Graduate Research Fellowship DGE1656518 (to E.T.C.); NIH K99HL135258 (to M.G.); NIH K08HL143185, the Sarnoff Cardiovascular Research Foundation, and the John Taylor Babbitt Foundation (to. V.N.P); NIH R01HL138539 and P01HL141084 (to M.M.); and the NIH Office of Director’s Pioneer Award (LM012179-05), the American Heart Association Established Investigator Award (17EIA33410923), the Stanford Cardiovascular Institute, and the Stanford Division of Cardiovascular Medicine, Department of Medicine (to S.M.W.); P01HL141084 and R21 HL141019 to MM; DAMF was funded by the Marie Sklodowska-Curie fellowship grant #708459 and the PLN foundation.
Footnotes
Data sharing: Raw data and complete methods can be made available upon request from the corresponding author. Single cell RNA sequencing data have been deposited in the GEO database under accession number GSE146763.
Disclosures
None
References
- 1.Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, Peterson KA, Li Y, Schwartz MC, Reynolds WT, et al. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49:1152–1159. doi: 10.1038/ng.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hrstka SC, Li X, Nelson TJ and The Wanek Program Genetics Pipeline Group. NOTCH1-dependent nitric oxide signaling deficiency in hypoplastic left heart syndrome revealed through patient-specific phenotypes detected in bioengineered cardiogenesis. Stem Cells. 2017;35:1106–1119. doi: 10.1002/stem.2582 [DOI] [PubMed] [Google Scholar]
- 3.Lee S, Yang H, Chen C, Venkatraman S, Darsha A, Wu SM, Wu JC and Seeger T. Simple Lithography-Free Single Cell Micropatterning using Laser-Cut Stencils. J Vis Exp. 2020;158:1–9. doi: 10.3791/60888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G, Tian L, Goodyer W, Kort EJ, Buikema JW, Xu A, Wu JC, Jovinge S and Wu SM. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development. 2019;146:1–11. doi: 10.1242/dev.173476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordero P, Parikh VN, Chin ET, Erbilgin A, Gloudemans MJ, Shang C, Huang Y, Chang AC, Smith KS, Dewey F, et al. Pathologic gene network rewiring implicates PPP1R3A as a central regulator in pressure overload heart failure. Nat Commun. 2019;10:2760. doi: 10.1038/s41467-019-10591-5 [DOI] [PMC free article] [PubMed] [Google Scholar]