Abstract
Cleft lip with or without cleft palate (CL/P) is considered a multifactorial genetic disorder. Folic acid metabolism has been suggested to underlie the development of CL/P. The gene for the enzyme 5,10-methylentetrahydrofolate reductase (MTHFR) contributes to folic acid metabolism, and polymorphisms of this gene at C677T (rs1801133) and A1298C (rs1801131) are reported to alter its enzyme activity and are suggested to be involved in CL/P development. We investigated C677T and A1298C polymorphisms of the MTHFR gene in Japanese patients with nonsyndromic CL/P and cleft palate only (CPO). We examined 240 patients with CL/P, 103 fathers and 153 mothers of the patients, and 68 healthy controls. Restriction fragment length polymorphisms (RFLPs) of C677T and A1298C of MTHFR were analyzed. We determined the frequencies of the polymorphisms in the patients and controls and performed a transmission equilibrium test and haplotype analysis of both MTHFR C677T and A1298C. There were no significant differences in the frequencies of MTHFR C677T and A1298C polymorphisms between the patients and controls. We did not observe transmission equilibrium or linkage equilibrium among the cases. In this experimental condition, we did not detect an association of MTHFR C677T and/or A1298C polymorphisms with the development of CL/P in this Japanese cohort.
Keywords: MTHFR gene, cleft lip with or without palate, folic acid
Introduction
Cleft lip with or without cleft palate (CL/P) is the most frequent congenital abnormality in the maxillofacial region. The prevalence of cleft lip with or without cleft palate in Japan is 1.73 per 1,000 individuals (1). As a disease model for CL/P, a multi-factorial threshold model involving several genetic and environmental factors has been proposed to explain the development of CL/P, but it is difficult to identify key factors (2). Modifications in various genes have been proposed to be related to the development of CL/P, including IRF6, VAX1, 8q24 locus, ABCA4, MAFB, and 17q22 locus (which were identified mainly by genome-wide association studies), and BMP4, FGFR2, FOXE1, MSX1, MYH9, CHIRSPLD2 FGF8, GSTT1, MTHFR, PDGFC, PVRL1, SUMO1, TGFα, and TGFβ3 (which were revealed mainly by candidate gene association or mutation detection studies) (3,4). Smoking, alcohol consumption, drug use, and viral infection during pregnancy have been proposed to be associated with the development of CL/P as environmental factors (2).
It has been known for decades that vitamin deficiencies during pregnancy can cause congenital abnormalities. Folic acid deficiency is well known to affect neural tube closure and the development of orofacial clefts, and supplementation with folic acid was demonstrated to reduce the frequencies of congenital abnormalities (2,5-7). In A/J mice, folic acid administered before and during pregnancy reduced the onset of naturally developed CL/P (8,9). Tolarova and Harris and Shaw et al reported a decrease in the prevalence of CL/P when mothers took vitamin supplements including folic acid before and during pregnancy until the organogenesis stage (10,11).
Regarding the metabolism of folic acid, the enzyme activity of 5,10-methylenetetrahydrofolate reductase (MTHFR) was proposed to be a rate-determining step influencing serum folic acid levels (12). Polymorphisms of the MTHFR gene at base pair 677 and 1298 (C677T; rs1801133, A1298C; rs1801131) were demonstrated to decrease the enzyme activity (13). These polymorphisms were also suggested to be correlated with CL/P (11,14). We conducted the present study to investigate the relationship between MTHFR gene polymorphisms and the development of CL/P in a Japanese population.
Patients and methods
Patients
The patient series included Japanese individuals with non-syndromic CL/P treated at the Department of Oral and Maxillofacial Surgery, Dokkyo Medical University Hospital or the Cleft Lip and Palate Center, Aichi-Gakuin University Dental Hospital. The patients, parents and guardians of the patients and control subjects provided informed consent for the investigation of gene polymorphisms for MTHFR by polymerase chain reaction (PCR) amplification of their DNA fragment. The study was approved by the Clinical Research Ethics Committee of Dokkyo University Hospital (approval no. R-33-22J), and the Research Ethics Committee of Aichi-Gakuin University School of Dentistry (approval no. 55). The samples were collected from April 2004 to March 2007 at Aichi-Gakuin University Dental Hospital and from June 2007 to May 2008 at Dokkyo Medical University Hospital. We were able to analyze 240 patients as well as 103 fathers and 153 mothers of these patients. As controls, 68 volunteers with no congenital abnormalities (including cleft lip and palate) and no family history of congenital abnormality were also enrolled (Table I).
Table I.
Demographic data of the study cohort (n=240), parents and controls.
| Sex | Total no. of cases | ||
|---|---|---|---|
| Male | 129 | ||
| Female | 111 | ||
| Demographic data labeled by type of abnormality | |||
| Type of abnormality | Sex | No. of cases | |
| CLA | 73 | ||
| Right cleft | Male | 16 | |
| Incomplete | 12 | ||
| Complete | 3 | ||
| Unknown | 1 | ||
| Female | 5 | ||
| Incomplete | 3 | ||
| Complete | 2 | ||
| Unknown | 0 | ||
| Left cleft | Male | 18 | |
| Incomplete | 13 | ||
| Complete | 4 | ||
| Unknown | 1 | ||
| Female | 21 | ||
| Incomplete | 10 | ||
| Complete | 10 | ||
| Unknown | 1 | ||
| Bilateral cleft | Male | 8 | |
| Incomplete | 3 | ||
| Complete | 4 | ||
| Unknown | 1 | ||
| Female | 4 | ||
| Incomplete | 3 | ||
| Complete | 1 | ||
| Unknown | 0 | ||
| Unidentified | 1 | ||
| CLP | 124 | ||
| Right cleft | Male | 23 | |
| Incomplete | 6 | ||
| Complete | 15 | ||
| Unknown | 2 | ||
| Female | 22 | ||
| Incomplete | 3 | ||
| Complete | 17 | ||
| Unknown | 2 | ||
| Left cleft | Male | 32 | |
| Incomplete | 9 | ||
| Complete | 21 | ||
| Unknown | 2 | ||
| Female | 22 | ||
| Incomplete | 3 | ||
| Complete | 18 | ||
| Unknown | 1 | ||
| Bilateral cleft | Male | 20 | |
| Incomplete | 2 | ||
| Complete | 17 | ||
| Unknown | 1 | ||
| Female | 5 | ||
| Incomplete | 1 | ||
| Complete | 2 | ||
| Unknown | 2 | ||
| CP | 43 | ||
| Demographic data labeled by family members, and controls | |||
| Family members | Type of abnormality | No of cases | |
| Fathers: | 103 | ||
| of CLA | 32 | ||
| of CP | 17 | ||
| of CLP | 54 | ||
| Mothers: | 153 | ||
| of CLA | 51 | ||
| of CP | 34 | ||
| of CLP | 68 | ||
| Controls: | 68 | ||
| Male | 29 | ||
| Female | 39 | ||
CLA, cleft lip; CLP, cleft lip palate; CP, cleft palate.
PCR amplification and detection of the restriction fragment length polymorphisms (RFLPs)
DNA was extracted from all of the subject peripheral blood samples with the use of the DNA Blood Mini kit (Qiagen GmbH) following the manufacturer's instructions. The extracted DNA was dissolved in Tris-EDTA buffer and stored at -80˚C until analysis. For the detection of the C677T polymorphism, the following primers were used: Forward primer, 5'-TGAAGGAGAAGGTGTCTGCGGGA-3' and reverse primer, 5'-AGGACGGTGCGGTGAGAGTG-3' (15). DNA (200 ng) was used as a template, and each primer set (0.2 µM final concentration) was treated in 10 µl of reaction solution containing 0.25 U Takara Ex Taq® (Takara Bio), dNTPs (0.4 mM each), and reaction buffer containing 4 mM Mg2+. After preheating for 5 min at 94˚C, the samples were denatured for 1 min at 94˚C followed by amplification at 64˚C for 1 min and extension at 72˚C for 1 min; the reaction was operated for 30 cycles following extension at 72˚C for 5 min.
For the detection of the A1298C polymorphism, the following primers were used: Forward primer, 5'-CTTTGGGGAGCTGAAGGACTACTAC-3' and reverse primer: 5'-CACTTTGTGACCATTCCGGTTTG-3' (16). We used 200 ng of DNA as a template, and each primer set (0.2 µM final concentration) was treated in 10 µl of reaction solution containing 0.25 U Takara Ex Taq®, dNTPs (0.4 mM each), and reaction buffer containing 4 mM Mg2+. After preheating at 94˚C for 5 min, the samples were denatured at 94˚C for 1 min followed by amplification at 57˚C for 1 min and extension at 72˚C for 1 min; the reaction was repeated for 30 cycles following extension at 72˚C.
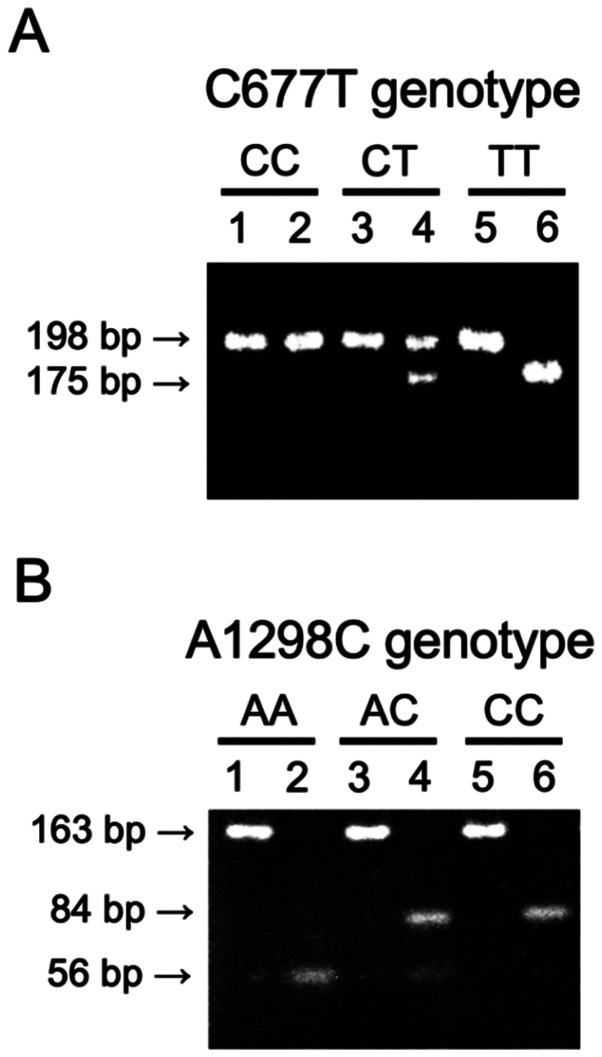
The PCR products amplified by the two primer sets were digested by restriction enzymes for the detection of polymorphisms. For C677T detection, the PCR products were a 198-bp fragment. Conversion of C to T creates an HinfI restriction site, the 198 bp fragment was digested into 175-bp and 23-bp fragments (Fig. 1A). For the A1298C detection, the PCR products were 163 bp and MboII digested the fragment into 56-bp, 31-bp, 30-bp, 28-bp and 18-bp fragments. Conversion of A to C abolishes the MboII restriction site, it digests into 84, 31, 30, 18 bp fragments (Fig. 1B). All the subject genotypes were determined by PCR-RFLP and the allele and genotype frequencies were determined. With regard to the C677T polymorphism, the wild-type 677th base of the MTHFR gene is referred to as CC (677CC), the heterozygous type as CT (677CT), and the recessive type as TT (677TT). With regard to the A1298C polymorphism, the wild-type is referred to as AA (1298AA), the heterozygous type as AC (1298AC), and the recessive type as CC (1298CC).
Figure 1.
Representative data of PCR-RFLP. (A) MTHFR C677T polymorphism. The 198 fragment represents the C allele and the 175 bp fragment represents the T allele. Lane 1, PCR product of genotype CC; lane 2, PCR products of genotype CC digested with HinfI; lane 3, PCR product of genotype CT; lane 4, PCR products of genotype CT digested with HinfI; lane 5, PCR products of genotype TT; lane 6, PCR products of genotype TT digested with HinfI. (B) MTHFR A1298C polymorphism. The fragment of 163 bp is the total length of PCR products, the 56 bp fragment represents the A allele, the fragment of 84 bp represents the C allele, respectively. Lane 1, PCR product of genotype AA; lane 2, PCR products of genotype AA digested with MboII; lane 3, PCR product of genotype AC; lane 4, PCR products of genotype AC digested with MboII; lane 5, PCR products of genotype CC; lane 6, PCR products of genotype CC digested with MboII. PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphisms; MTHFR, 5,10-methylentetrahydrofolate reductase.
Transmission disequilibrium test (TDT)
We examined the over-transmission of C677T and A1298C from heterozygous parents to their children with CL/P (patients) by conducting a transmission disequilibrium test (TDT). The family-based association test (FBAT) developed by Laird and Lange was conducted as previously described (17).
Haplotype analysis
Since the transmission of an allele from parents to their children is known to differ from family to family, we investigated the linkage disequilibrium of the subjects' haplotypes. In linkage disequilibrium test, D, D' and r, coefficient of linkage disequilibrium, were calculated. D indicates the coefficient of linkage disequilibrium, D' is the normalized coefficient of linkage disequilibrium, r is the coefficient of correlation. SNPstats, a web tool for SNP analysis, (https://snpstats.net) was used to estimate the haplotypes from genotype frequencies and to further analyze the linkage disequilibrium and haplotype association with CL/P (18).
Statistical analysis
We tested the Hardy-Weinberg equilibrium (HWE) for each patient, the patient's father and mother, and the controls with the use of the R package ‘HardyWeinberg’ (19,20). Fisher's exact test was used for the statistical analyses. The results of the TDT were analyzed using McNemar's test, and haplotypes were analyzed with the use of SNPstats software. We calculated the odds ratios (ORs) with confidence intervals (CIs) to assess the associations of genotypes. The strength of the association was tested under genetic models comparing the allele, genotype, and dominant (TT+CT vs. CC for C677T; CC+AC vs. AA for A1298C) or recessive (CC+CT vs. TT for C677T; AA+AC vs. CC for A1298C) models, respectively. A P-value <0.05 was indicative of a statistically significant difference.
Results
Polymorphism at base pair (bp) 677 of the MTHFR gene
The genotype frequency of the MTHFR C677T polymorphism of the patients, patients' fathers and mothers, and controls were in agreement with the HWE: P=0.89, P=0.55, P=0.17, and P=0.60, respectively. There were no significant differences in the allele frequencies of C and T at bp 677 of the MTHFR gene between the patients and controls. The genotype frequencies also showed no significant differences between the patients and controls (Table II). The dominant or recessive genetic model for C677T was tested to investigate the association of genotypes and CL/P, although the results revealed no association (Table II).
Table II.
Polymorphisms of MTHFR C677T.
| Odds ratio | ||||||
|---|---|---|---|---|---|---|
| Group | Polymorphism | Cases (n=240) n (%) | Controls (n=68) n (%) | OR | 95% CI | P-value |
| Patient | Genotype | |||||
| CC | 89 (37.1) | 27 (39.7) | 1.000 | |||
| CT | 116 (48.3) | 34 (50.0) | 1.035 | 0.582-1.841 | 0.907 | |
| TT | 35 (14.6) | 7 (10.3) | 1.517 | 0.605-3.801 | 0.372 | |
| Allele | ||||||
| C | 294 (61.3) | 88 (64.7) | 1.000 | |||
| T | 186 (38.8) | 48 (35.3) | 1.160 | 0.780-1.725 | 0.464 | |
| Genetic model | ||||||
| Dominant | 151 (62.9) | 41 (60.3) | 1.117 | 0.643-1.940 | 0.694 | |
| Recessive | 205 (85.4) | 61 (89.7) | 0.672 | 0.284-1.589 | 0.363 | |
| Type of cleft | ||||||
| CLA | Genotype | |||||
| CC | 32 (43.8) | 27 (39.7) | 1.000 | |||
| CT | 32 (43.8) | 34 (50.0) | 0.794 | 0.393-1.605 | 0.521 | |
| TT | 9 (12.4) | 7 (10.3) | 1.085 | 0.357-3.300 | 0.886 | |
| Allele | ||||||
| C | 95 (66.0) | 88 (64.7) | 1.000 | |||
| T | 49 (34.0) | 48 (35.3) | 0.946 | 0.578-1.547 | 0.824 | |
| Genetic model | ||||||
| Dominant | 41 (56.2) | 41 (60.3) | 0.844 | 0.431-1.650 | 0.619 | |
| Recessive | 64 (87.7) | 25 (78.1) | 1.991 | 0.669-5.925 | 0.210 | |
| CLP | Genotype | |||||
| CC | 41 (33.1) | 27 (39.7) | 1.000 | |||
| CT | 61 (49.2) | 34 (50.0) | 1.181 | 0.622-2.245 | 0.610 | |
| TT | 22 (17.7) | 7 (10.3) | 2.070 | 0.777-5.512 | 0.141 | |
| Allele | ||||||
| C | 143 (57.7) | 88 (64.7) | 1.000 | |||
| T | 105 (42.3) | 48 (35.3) | 1.346 | 0.873-2.075 | 0.177 | |
| Genetic model | ||||||
| Dominant | 83 (66.9) | 41 (60.3) | 1.333 | 0.722-2.461 | 0.357 | |
| Recessive | 102 (82.3) | 61 (89.7) | 0.532 | 0.215-1.319 | 0.168 | |
| CLA+CLP | Genotype | |||||
| CC | 73 (37.1) | 27 (39.7) | 1.000 | |||
| CT | 93 (47.2) | 34 (50.0) | 1.012 | 0.560-1.827 | 0.969 | |
| TT | 31 (15.7) | 7 (10.3) | 1.638 | 0.645-4.158 | 0.296 | |
| Allele | ||||||
| C | 239 (60.7) | 88 (64.7) | 1.000 | |||
| T | 155 (39.3) | 48 (35.3) | 1.189 | 0.793-1.784 | 0.403 | |
| Genetic model | ||||||
| Dominant | 124 (62.9) | 41 (60.3) | 1.119 | 0.636-1.969 | 0.697 | |
| Recessive | 166 (84.3) | 61 (89.7) | 0.614 | 0.257-1.468 | 0.270 | |
| CP | Genotype | |||||
| CC | 16 (37.2) | 27 (39.7) | 1.000 | |||
| CT | 23 (53.5) | 34 (50.0) | 1.142 | 0.506-2.576 | 0.750 | |
| TT | 4 (9.30) | 7 (10.3) | 0.964 | 0.244-3.815 | 0.959 | |
| Allele | ||||||
| C | 55 (64.0) | 88 (64.7) | 1.000 | |||
| T | 31 (36.0) | 48 (35.3) | 1.033 | 0.588-1.815 | 0.909 | |
| Genetic model | ||||||
| Dominant | 27 (62.8) | 41 (60.3) | 1.111 | 0.506-2.440 | 0.793 | |
| Recessive | 39 (90.7) | 61 (89.7) | 1.119 | 0.307-4.075 | 0.865 | |
| Parents | ||||||
| Father | Genotype | |||||
| CC | 31 (30.1) | 10 (34.5) | 1.000 | |||
| CT | 54 (52.4) | 15 (51.7) | 1.161 | 0.466-2.896 | 0.748 | |
| TT | 18 (17.5) | 4 (13.8) | 1.452 | 0.397-5.31 | 0.572 | |
| Allele | ||||||
| C | 116 (56.3) | 35 (60.3) | 1.000 | |||
| T | 90 (43.7) | 23 (39.7) | 1.181 | 0.652-2.138 | 0.583 | |
| Genetic model | ||||||
| Dominant | 72 (69.9) | 19 (65.5) | 1.222 | 0.51-2.929 | 0.652 | |
| Recessive | 85 (82.5) | 25 (86.2) | 0.756 | 0.234-2.438 | 0.638 | |
| Mother | Genotype | |||||
| CC | 56 (36.6) | 17 (43.6) | 1.000 | |||
| CT | 80 (52.3) | 19 (48.7) | 1.278 | 0.611-2.674 | 0.514 | |
| TT | 17 (11.1) | 3 (7.70) | 1.720 | 0.45-6.583 | 0.424 | |
| Allele | ||||||
| C | 192 (62.7) | 53 (67.9) | 1.000 | |||
| T | 114 (37.3) | 25 (32.1) | 1.259 | 0.742-2.136 | 0.393 | |
| Genetic model | ||||||
| Dominant | 97 (63.4) | 22 (56.4) | 1.338 | 0.656-2.731 | 0.422 | |
| Recessive | 136 (88.9) | 36 (92.3) | 0.667 | 0.185-2.401 | 0.533 | |
MTHFR, 5,10-methylentetrahydrofolate reductase; CLA, cleft lip; CLP, cleft lip palate; CP, cleft palate.
Polymorphism at base pair (bp) 1298 of the MTHFR gene
The genotype frequency of MTHFR A1298C polymorphism of the patients, patients' fathers and mothers, and controls were in agreement with the HWE: P=1.00, P=0.40, P=0.79, and P=1.00, respectively. There were no significant differences in the allele frequencies of A and C at bp 1298 of the MTHFR gene between the patients and controls. The genotype frequency also showed no significant difference between the patients and controls (Table III). The dominant or recessive genetic model for A1298C was tested to investigate the association of genotypes and CL/P, although the results revealed no association (Table III).
Table III.
Polymorphism of the MTHFR A1298C.
| Odds ratio | ||||||
|---|---|---|---|---|---|---|
| Group | Polymorphism | Cases (n=240) n (%) | Controls (n=68) n (%) | OR | 95% CI | P-value |
| Patient | Genotype | |||||
| AA | 155 (64.6) | 43 (63.2) | 1.000 | |||
| AC | 77 (32.1) | 22 (32.4) | 0.971 | 0.552-1.382 | 0.921 | |
| CC | 8 (3.30) | 3 (4.40) | 0.740 | 0.223-2.050 | 0.665 | |
| Allele | ||||||
| A | 232 (73.2) | 65 (72.2) | 1.000 | |||
| C | 85 (26.8) | 25 (27.8) | 0.927 | 0.603-1.288 | 0.753 | |
| Genetic model | ||||||
| Dominant | 85 (35.4) | 25 (36.8) | 0.943 | 0.557-1.357 | 0.838 | |
| Recessive | 232 (96.7) | 65 (95.6) | 1.338 | 0.293-2.274 | 0.672 | |
| Type of cleft | ||||||
| CLA | Genotype | |||||
| AA | 51 (69.9) | 43 (63.2) | 1.000 | |||
| AC | 21 (28.8) | 22 (32.4) | 0.766 | 0.430-1.437 | 0.474 | |
| CC | 1 (1.40) | 3 (4.40) | 0.562 | 0.124-2.533 | 0.533 | |
| Allele | ||||||
| A | 72 (76.6) | 65 (72.2) | 1.000 | |||
| C | 22 (23.4) | 25 (27.8) | 0.759 | 0.485-1.336 | 0.369 | |
| Genetic model | ||||||
| Dominant | 22 (30.1) | 25 (36.8) | 0.742 | 0.435-1.406 | 0.404 | |
| Recessive | 71 (97.3) | 65 (95.6) | 1.638 | 0.201-3.074 | 0.592 | |
| CLP | Genotype | |||||
| AA | 73 (58.9) | 43 (63.2) | 1.000 | |||
| AC | 45 (36.3) | 22 (32.4) | 1.205 | 0.575-1.483 | 0.564 | |
| CC | 6 (4.80) | 3 (4.40) | 0.356 | 0.142-1.930 | 0.165 | |
| Allele | ||||||
| A | 118 (69.8) | 65 (72.2) | 1.000 | |||
| C | 51 (30.2) | 25 (27.8) | 1.151 | 0.638-1.370 | 0.589 | |
| Genetic model | ||||||
| Dominant | 51 (41.1) | 25 (36.8) | 1.202 | 0.589-1.461 | 0.554 | |
| Recessive | 118 (95.2) | 65 (95.6) | 0.908 | 0.232-2.191 | 0.894 | |
| CLA+CLP | Genotype | |||||
| AA | 124 (62.9) | 43 (63.2) | 1.000 | |||
| AC | 65 (33.0) | 22 (32.4) | 1.025 | 0.557-1.407 | 0.936 | |
| CC | 8 (4.10) | 3 (4.40) | 0.925 | 0.245-2.141 | 0.911 | |
| Allele | ||||||
| A | 189 (72.1) | 65 (72.2) | 1.000 | |||
| C | 73 (27.9) | 25 (27.8) | 0.998 | 0.617-1.313 | 0.994 | |
| Genetic model | ||||||
| Dominant | 73 (37.1) | 25 (36.8) | 1.013 | 0.568-1.385 | 0.966 | |
| Recessive | 189 (95.9) | 65 (95.6) | 1.090 | 0.267-2.190 | 0.900 | |
| CP | Genotype | |||||
| AA | 31 (72.1) | 43 (63.2) | 1.000 | |||
| AC | 11 (25.6) | 22 (32.4) | 0.694 | 0.362-1.517 | 0.402 | |
| CC | 1 (2.30) | 3 (4.40) | 0.462 | 0.071-3.194 | 0.504 | |
| Allele | ||||||
| A | 42 (77.8) | 65 (72.2) | 1.000 | |||
| C | 12 (22.2) | 25 (27.8) | 1.369 | 0.513-1.672 | 0.443 | |
| Genetic model | ||||||
| Dominant | 12 (27.9) | 25 (36.8) | 0.666 | 0.366-1.480 | 0.335 | |
| Recessive | 42 (97.7) | 65 (95.6) | 1.938 | 0.134-4.153 | 0.566 | |
| Parents | ||||||
| Father | Genotype | |||||
| AA | 63 (61.2) | 18 (62.1) | 1.000 | |||
| AC | 33 (32.0) | 10 (34.5) | 0.943 | 0.404-1.627 | 0.896 | |
| CC | 7 (6.80) | 1 (3.40) | 2.000 | 0.156-3.867 | 0.522 | |
| Allele | ||||||
| A | 158 (76.7) | 46 (79.3) | 1.000 | |||
| C | 48 (23.3) | 12 (20.7) | 1.133 | 0.517-1.533 | 0.731 | |
| Genetic model | ||||||
| Dominant | 40 (38.8) | 11 (37.9) | 1.039 | 0.435-1.628 | 0.930 | |
| Recessive | 96 (93.2) | 28 (96.6) | 0.490 | 0.087-2.928 | 0.504 | |
| Mother | Genotype | |||||
| AA | 104(68) | 25 (64.1) | 1.000 | |||
| AC | 44 (28.8) | 12 (30.8) | 0.881 | 0.437-1.512 | 0.749 | |
| CC | 5 (3.30) | 2 (5.10) | 0.601 | 0.147-2.372 | 0.553 | |
| Allele | ||||||
| A | 252 (82.4) | 62 (79.5) | 1.000 | |||
| C | 54 (17.6) | 16 (20.5) | 0.830 | 0.495-1.374 | 0.558 | |
| Genetic model | ||||||
| Dominant | 49 (32.0) | 14 (35.9) | 0.841 | 0.444-1.469 | 0.646 | |
| Recessive | 148 (96.7) | 37 (94.9) | 1.600 | 0.229-2.825 | 0.580 | |
MTHFR, 5,10-methylentetrahydrofolate reductase; CLA, cleft lip; CLP, cleft lip palate; CP, cleft palate.
Polymorphism of the MTHFR gene in each cleft type
Since the onset mechanism of each cleft type differs, we compared the polymorphisms of the MTHFR gene in the patients with cleft lip, those with cleft lip with palate, and those with cleft palate only. Our analyses revealed that there were no significant differences in the allele frequency or genotype frequencies at either bp 677 or bp 1,298 of the MTHFR gene (Tables II and III). The dominant or recessive genetic model also did not reveal an association to each cleft type.
TDT
We investigated 28 families in which both parents had heterozygous C677T polymorphism (677CT) and nine families with heterozygous A1298C polymorphism (1298AC). No significant transmission disequilibrium was revealed in 677CT or 1298AC by the TDT results (Table IV).
Table IV.
Results of the transmission disequilibrium test (TDT) of MTHFR C677T and A1298C polymorphisms.
| Allele | Transmitted | Not transmitted | P-value |
|---|---|---|---|
| 677C | 33 | 23 | 0.181 |
| 677T | 23 | 33 | |
| 1298A | 8 | 10 | 0.637 |
| 1298C | 10 | 8 |
MTHFR, 5,10-methylentetrahydrofolate reductase.
Haplotype analysis
The haplotype analysis showed no linkage disequilibrium between C677T and A1298C (Table V). The haplotype estimation determined three types: C_A, T_A, and C_C. The frequency of haplotypes was similar to those reported in the Japanese population from the 1000 Genome Project: C_A, 44.2%; T_A, 38.0%; C_C, 17.8%; T_C, not detected in the Japanese population (21). We detected a rare haplotype (T_C) in one patient. No significant between-group differences were observed except for the rare case of T_C (Table V).
Table V.
Haplotype analysis results for MTHFR 677 and 1,298 polymorphisms.
| Linkage disequilibrium analysis | |||||
|---|---|---|---|---|---|
| C677T | A1298C | ||||
| C677T | - | D=-0.0663 | |||
| D'=0.8814 | |||||
| r=-0.3428 | |||||
| A1298C | - | - | |||
| Haplotype frequency estimation (n=308) and association analysis | |||||
| Haplotype | Total | Controls | Patients | OR (95% CI) | P-value |
| C_A | 0.4316 | 0.4412 | 0.4287 | 1.00 | |
| T_A | 0.3709 | 0.3529 | 0.3754 | 1.09 (0.71-1.070) | 0.69 |
| C_C | 0.1891 | 0.2059 | 0.1838 | 0.93 (0.55-1.57) | 0.79 |
| T_C | 0.0089 | 0 | 0.0121 | 38636069.94 | <0.0001 (38636069.93-38636069.95) |
D, coefficient of linkage disequilibrium; D', normalized coefficient of linkage disequilibrium; r, coefficient of correlation. In comparison of the odds ratio, the most frequent haplotype C_A was used as reference. MTHFR, 5,10-methylentetrahydrofolate reductase.
Discussion
As one of the most common congenital abnormalities, cleft lip with or without cleft palate (CL/P) is a multifactorial genetic disorder that involves multiple genetic and environmental factors. Folic acid deficiency in mothers causes developmental defects in human fetuses and increases the risk of a malformed fetus, and folic acid supplementation has been reported to prevent the onset of cleft palate (6,7). Folic acid is a member of the vitamin B family that is involved in the synthesis of DNA, RNA, and amino acids. Folic acid is also involved in the reaction in which homocysteine is methylated and converted to methionine (8). In addition, folic acid is thought to play important roles in organogenesis (22).
Single-nucleotide polymorphisms of C677T and A1298C in MTHFR were shown to change the serum folic acid concentration (13). Genotype 677TT or 1298CC would decrease the enzyme activity of MTHFR and lower the production of 5-methyl tetrahydrofolic acid (5-methyl THF). The 5-methyl THF regulates the methylation of homocysteine to methionine as a coenzyme, and a decrease in 5-methyl THF disturbs this process and increases the serum level of homocysteine. Hyperhomocysteinemia is known to be involved in neural tube defects by affecting neural crest cells. It is believed that in such a scenario, mesenchymal cells derived from neural crest cells would also be affected, thus ultimately resulting in CL/P (9,11,22). To date, there have been no studies investigating MTHFR C677T polymorphism in relation to cleft types in Japanese subjects. The present study is the first to analyze MTHFR A1298C polymorphism together with MTHFR C677T polymorphism in Japanese patients with CL/P.
Contrary to our expectations, there was no significant difference in A1298C or in C677T of the MTHFR gene among the patients with any cleft type and the healthy control subjects. In addition, there was no significant transmission disequilibrium in either 677CT or 1298AC. The haplotype analysis also revealed no significant difference. Tolarova and Harris reported that MTHFR C677T polymorphism is involved in the onset of CL/P, and Martinelli et al noted that they observed no notable differences of MTHFR C677T polymorphism between patient and parent groups, but they did observe marked differences in the allele frequency between the mothers and the control group, and the frequency of T allele was high (10,23). In their study of mothers with low folic acid intake during pregnancy, van Rooij et al reported that the onset of CL/P was correlated with MTHFR C677T and A1298C polymorphisms (24). However, Shaw et al observed no correlations between C677T polymorphism and the development of cleft lip or cleft lip and palate (11), and Kanno et al documented no correlation between MTHFR C677T polymorphism and the development of CL/P in a Japanese series (25). According to a study by Pezzetti et al, MTHFR C677T polymorphism was correlated with the development of cleft lip with or without cleft palate, while A1298C polymorphism was not (4). With these conflicting results, no conclusions can be drawn regarding the relationship between the development of CL/P and MTHFR C677T or A1298C polymorphisms.
During the years 2007 to 2020, there were 15 meta-analyses concerning MTHFR C677T and A1298C polymorphisms and CL/P. Among them, 12 studies concerned C677T; nine studies suggested an increased risk for CL/P (26-34), whereas the other three studies denied the association (35-37). Most studies have thus suggested that the C677T polymorphism is likely to increase the risk for CL/P, which disagrees with our findings. Regarding the A11298C polymorphism, there are seven studies and six of them suggested no association with CL/P (27,29,34,36,38,39), which is consistent with our findings. There is a single study that suggested an association between A1298C and CL/P (30). Most of the studies mentioned the heterogeneity of the association due to the different ethnicities of the subjects. They also noted that environmental factors such as alcohol consumption, smoking habits, and maternal folic acid supplementation are important (37,38,40). According to the meta-analyses conducted to date, in Asians, C677T T allele or recessive genotype TT tends to be a risk for CL/P.
Ethnic differences in the distribution of MTHFR C677T and A1298C polymorphisms have been reported (13,41). Yoshida reported that the distribution of polymorphisms in Vietnamese and Mongolians differed from that in other Asians (41), and there are differences in distribution between our findings in Japanese and the frequency of MTHFR C677T polymorphism in Vietnamese. We also collected other cohorts by searching PubMed. We identified 21 matches reporting the prevalence of MTHFR C677T and A1298C in various ethnic populations, and there were some differences in allele frequencies. For example, genotype CC at bp 677 is likely to be dominant in Moroccan patients (42) whereas genotype TT in 677 is dominant in Chinese patients and Iranian patients (43,44). Some ethnicities even showed higher frequencies of a functionally superior genotype of MTHFR C677T and A1298C in the patient groups, which is the opposite of our hypothesis. This discrepancy may be due in part to the multiplicity of the enzymes, including MTHFR, that regulate the serum level of folic acid. In addition, it is known that folic acid nutritional supplementation can overcome the decreased activity of, at least, MTHFR. There is also a possibility that if the mother's folic acid serum level is sufficient, the child may overcome the recessive genotype.
Concerning the nutritional status of Japanese women, it has been reported that although their daily intake of folic acid is not sufficient, their serum concentrations are higher compared to women in other countries due to the tendency of Japanese women's dietary behavior during pregnancy (45). They tend to adopt healthy behaviors such as eating more colored vegetables instead of meat, which results in their consumption of folic acid as a highly bioavailable natural food (46). In the earlier study (45), even without a folic acid-fortified diet, most of the subjects exhibited a folic acid concentration that was sufficient to prevent neural tube defects.
A few studies have investigated gene-environment interactions; one study conducted in Caucasians demonstrated a reduced effect of maternal folic acid supplementation on the development of CL/P, although there was no association with C677T genetic status (37). Although, there have been no investigations of gene-environment interactions, especially maternal folic acid supplementation, in Asian populations including Japanese. The gene-environment interactions in these populations remain to be elucidated in future studies.
It is possible that our present sample size was not large enough to observe the association, and we therefore combined the patients examined by Tohoku University in a previous Japanese cohort with our present results in order to increase the sample number (26). We observed no significant difference in the allele frequencies or dominant genetic model frequency (C vs. T, case n=411, control n=317, odds ratio=0.954, 95% CI: 0.845-1.076, P-value=0.694; dominant model, cases n=205, control n=162, odds ratio=0.984, 95% CI: 0.830-1.168, P-value=0.926).
Concerning the meta-analysis data, C677T mutation might be a risk for CL/P development in Asian population. However, we could not observe such an association neither C677T nor A1298C in the Japanese population. It is likely that MTHFR C677T and A1298C polymorphisms have less impact on CL/P development in Japanese populations. Part of the reason for this may be because of the typical diet consumed by pregnant Japanese women, which may be the reason that we did not observe significant differences in the present case-control study. In this context, it is necessary to investigate both serum levels of vitamins, other major environmental factors including folic acid supplementation, smoking, alcohol consumption, and genetic factors at the same time points in order to determine whether decreased folic acid metabolism and/or gene polymorphisms of metabolic enzymes are risk factors for the development of CL/P.
Acknowledgements
We express our deepest gratitude to Dr Yutaka Imai for encouraging us to conduct this study, and to Dr Kazumoto Kimura of the Medical Information Center for his guidance concerning the statistical analysis.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
CK and WY performed and analyzed the RFLP-PCR. YK and CK interpreted the data and were the major contributors to the manuscript's writing, and they contributed equally to this work. NN and HK contributed to the presentation of the data and the manuscript's revision. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by both the Clinical Research Ethics Committee of Dokkyo University Hospital (approval no. R-33-22J), and the Research Ethics Committee of Aichi-Gakuin University School of Dentistry (approval no. 55). The consent of the patients and controls for publication was obtained before their participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cooper ME, Ratay JS, Marazita ML. Asian oral-facial cleft birth prevalence. Cleft Palate Craniofac J. 2006;43:580–589. doi: 10.1597/05-167. [DOI] [PubMed] [Google Scholar]
- 2.Vieira AR. Genetic and environmental factors in human cleft lip and palate. Front Oral Biol. 2012;16:19–31. doi: 10.1159/000337521. [DOI] [PubMed] [Google Scholar]
- 3.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: Understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pezzetti F, Martineli M, Scapolo L, Carinci F, Palmieri A, Marchesini J, Carinci P, Caramelli E, Rullo R, Gombos F, Tognon M. Maternal MTHFR variant forms increase the risk in offspring of isolated nonsyndromic cleft lip with or without cleft palate. Hum Mutat. 2004;24:104–105. doi: 10.1002/humu.9257. [DOI] [PubMed] [Google Scholar]
- 5.Sato F. Candidate gene analysis of non-syndromic cleft lip with or without cleft palate and isolated cleft palate in Japanese families. Nihon Koku Geka Gakkai Zasshi. 2000;46:1–8. (In Japanese) [Google Scholar]
- 6.Milunsky A, Jick H, Jick SS, Bruell CL, MacLaughlin DS, Rothman KJ, Willett W. Multivitamin/folic acid supplementation in early pregnancy reduces the prevalence of neural tube defects. JAMA. 1989;262:2847–2852. doi: 10.1001/jama.262.20.2847. [DOI] [PubMed] [Google Scholar]
- 7.Werler MM, Shapiro S, Mitchell AA. Periconceptional folic acid exposure and risk of occurrent neural tube defects. JAMA. 1993;269:1257–1261. [PubMed] [Google Scholar]
- 8.Bailey LB, Gregory JF III. Folate metabolism and requirements. J Nutr. 1999;129:779–782. doi: 10.1093/jn/129.4.779. [DOI] [PubMed] [Google Scholar]
- 9.Niimi T. Experimental study on effects of folic acid on cleft lip and/or palate, thymic anomalies and cardiovascular anomalies. Aichi Gakuin Shigakkai Shi. 1998;36:663–669. (In Japanese) [Google Scholar]
- 10.Tolarova M, Harris J. Reduced recurrence of orofacial clefts after periconceptional supplementation with high-dose folic acid and multivitamins. Teratology. 1995;51:71–78. doi: 10.1002/tera.1420510205. [DOI] [PubMed] [Google Scholar]
- 11.Shaw GM, Rozen R, Finnell RH, Todoroff K, Lammer EJ. Infant C677T mutation in MTHFR, maternal periconceptional vitamin use, and cleft lip. Am J Med Genet. 1998;80:196–198. doi: 10.1002/(sici)1096-8628(19981116)80:3<196::aid-ajmg2>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 12.Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, Rozen R. Human methylenetetrahydrofolate reductase: Isolation of cDNA mapping and mutation identification. Nat Genet. 1994;7:195–200. doi: 10.1038/ng0694-195. [DOI] [PubMed] [Google Scholar]
- 13.Hiraoka M, Kato K, Saito Y, Yasuda K, Kagawa Y. Gene-nutrient and gene-gene interactions of controlled folate intake by Japanese women. Biochem Biophys Res Commun. 2004;316:1210–1216. doi: 10.1016/j.bbrc.2004.02.174. [DOI] [PubMed] [Google Scholar]
- 14.Tolarova M, Van Rooij I, Pastor M, van der Put NMJ, Goldberg AC, Hol F, Capozzi A, Thomas CMG, Pastor L, Mosby T, et al. A common mutation in the MTHFR gene is a risk factor for nonsyndromic cleft lip and palate anomalies. Am J Hum Genet. 1998;63(A27) [Google Scholar]
- 15.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP, et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 16.van der Put NM, Gabreëls F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel LP, Blom HJ. A second common mutation in the methylenetetrahydrofolate reductase gene: An additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- 18.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: A web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 19.Graffelman J. Exploring diallelic genetic markers: The hardyweinberg package. J Stat Softw. 2015;64:1–23. [Google Scholar]
- 20. R Core Team: R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, 2016. https://www.R-project.org/. [Google Scholar]
- 21.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. The 1000 Genomes Project Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selhub J, Rosenberg IH. Folic acid. In: International Life Sciences Institute-Nutrition Foundation. Present Knowledge in Nutrition. Ziegler EE and Filer LJ Jr (eds). 7th edition. ILSI Press, International Life Sciences Institute Nutrition Foundation, Washington, DC, pp206-219, 1996. [Google Scholar]
- 23.Martinelli M, Scapoli L, Pezzetti F, Carinci F, Stabellini G, Bisceglia L, Gombos F, Tognon M. C677T variant form at the MTHFR gene and CL/P: A risk factor for mothers? Am J Med Genet. 2001;98:357–360. doi: 10.1002/1096-8628(20010201)98:4<357::aid-ajmg1108>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 24.van Rooij IA, Vermeij-Keers C, Kluijtmans LA, Ocké MC, Zielhuis GA, Goorhuis-Brouwer SM, van der Biezen JJ, Kuijpers-Jagtman AM, Steegers-Theunissen R. Does the interaction between maternal folate intake and the methylenetetrahydrofolate reductase polymorphisms affect the risk of cleft lip with or without cleft palate? Am J Epidemiol. 2003;157:583–591. doi: 10.1093/aje/kwg005. [DOI] [PubMed] [Google Scholar]
- 25.Kanno K, Yamada A, Suzuki Y, Matsubara Y. C677T Polymorphism at the MTHFR gene and cleft lip with/ without cleft palate. J Jpn Soc Plast Reconst Surg. 2001;21:690–694. [Google Scholar]
- 26.Amooee A, Dastgheib SA, Niktabar SM, Noorishadkam M, Lookzadeh MH, Mirjalili SR, Heiranizadeh N, Neamatzadeh H. doi: 10.1080/15513815.2019.1707918. Association of Fetal MTHFR 677C > T polymorphism with non-syndromic cleft lip with or without palate risk: A systematic review and meta-analysis. Fetal Pediatr Pathol: 1-17, 2019. [DOI] [PubMed] [Google Scholar]
- 27.Pan X, Wang P, Yin X, Liu X, Li D, Li X, Wang Y, Li H, Yu Z. Association between maternal MTHFR polymorphisms and nonsyndromic cleft lip with or without cleft palate in offspring, a meta-analysis based on 15 case-control studies. Int J Fertil Steril. 2015;8:463–480. doi: 10.22074/ijfs.2015.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo YL, Cheng YL, Ye P, Wang W, Gao XH, Chen Q. Association between MTHFR polymorphisms and orofacial clefts risk: A meta-analysis. Birth Defects Res A Clin Mol Teratol. 2012;94:237–244. doi: 10.1002/bdra.23005. [DOI] [PubMed] [Google Scholar]
- 29.Zhao M, Ren Y, Shen L, Zhang Y, Zhou B. Association between MTHFR C677T and A1298C polymorphisms and NSCL/P risk in Asians: A meta-analysis. PLoS One. 2014;9(e88242) doi: 10.1371/journal.pone.0088242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan Y, Zhang W, Ma J, Du Y, Li D, Cai Q, Jiang H, Wang M, Zhang Z, Wang L. Infants'MTHFR polymorphisms and nonsyndromic orofacial clefts susceptibility: A meta-analysis based on 17 case-control studies. Am J Med Genet A. 2012;58A:2162–2169. doi: 10.1002/ajmg.a.35503. [DOI] [PubMed] [Google Scholar]
- 31.Jiang C, Yin N, Zhao Z, Wu D, Wang Y, Li H, Song T. Lack of association between MTHFR, MTR, MTRR, and TCN2 genes and nonsyndromic CL±P in a Chinese population: Case-control study and meta-analysis. Cleft Palate Craniofac J. 2015;52:579–587. doi: 10.1597/14.067. [DOI] [PubMed] [Google Scholar]
- 32.Verkleij-Hagoort A, Bliek J, Sayed-Tabatabaei F, Ursem N, Steegers E, Steegers-Theunissen R. Hyperhomocysteinemia and MTHFR polymorphisms in association with orofacial clefts and congenital heart defects: A meta-analysis. Am J Med Genet A. 2007;143A:952–960. doi: 10.1002/ajmg.a.31684. [DOI] [PubMed] [Google Scholar]
- 33.Li Q, Xu L, Jia X, Saleem K, Zaib T, Sun W, Fu S. SNPs in folate pathway are associated with the risk of nonsyndromic cleft lip with or without cleft palate, a meta-analysis. Biosci Rep. 2020;40(BSR20194261) doi: 10.1042/BSR20194261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rai V. Strong association of C677T polymorphism of methylenetetrahydrofolate reductase gene with nonsyndromic cleft lip/palate (nsCL/P) Indian J Clin Biochem. 2018;33:5–15. doi: 10.1007/s12291-017-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imani MM, Golchin N, Safaei M, Rezaei F, Abbasi H, Sadeghi M, Lopez-Jornet P, Mozaffari HR, Sharifi R. Methylenetetrahydrofolate reductase C677T polymorphism is not associated with the risk of nonsyndromic cleft lip/palate: An updated meta-analysis. Sci Rep. 2020;10(1531) doi: 10.1038/s41598-020-58357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Assis Machado R, de Toledo IP, Martelli-Júnior H, Reis SR, Neves Silva Guerra E, Coletta RD. Potential genetic markers for nonsyndromic oral clefts in the Brazilian population: A systematic review and meta-analysis. Birth Defects Res. 2018;110:827–839. doi: 10.1002/bdr2.1208. [DOI] [PubMed] [Google Scholar]
- 37.Johnson CY, Little J. Folate intake, markers of folate status and oral clefts: Is the evidence converging? Int J Epidemiol. 2008;37:1041–1058. doi: 10.1093/ije/dyn098. [DOI] [PubMed] [Google Scholar]
- 38.Niktabar SM, Aarafi H, Dastgheib SA, Noorishadkam M, Mirjalili SR, Lookzadeh MH, Akbarian-Bafghi MJ, Morovati-Sharifabad M, Neamatzadeh H. doi: 10.1080/15513815.2019.1683918. Association of MTHFR 1298A > C polymorphism with susceptibility to non-syndromic cleft lip with or without palate: A case-control study and meta-analysis. Fetal Pediatr Pathol: Nov 4, 2019 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 39.Imani MM, Rezaei F, Mire H, Delavarian M, Sadeghi M, Safaei M, Mozaffari HR. A meta-analysis and meta-regression of association between MTHFR A1298C polymorphism and nonsyndromic cleft lip/palate risk: An evaluation based on five genetic models. Int Orthod. 2020;18:191–202. doi: 10.1016/j.ortho.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Butali A, Little J, Chevrier C, Cordier S, Steegers-Theunissen R, Jugessur A, Oladugba B, Mossey PA. Folic acid supplementation use and the MTHFR C677T polymorphism in orofacial clefts etiology: An individual participant data pooled-analysis. Birth Defects Res A Clin Mol Teratol. 2013;97:509–514. doi: 10.1002/bdra.23133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida W. A study of 5,10-methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism on cleft lip and/or palate in the Vietnamese. Aichi Gakuin J Dent Sci. 2005;43:421–437. (In Japanese) [Google Scholar]
- 42.Rafik A, Rachad L, Kone AS, Nadifi S. MTHFR C677T polymorphism and risk of nonsyndromic cleft lip with or without cleft palate in the Moroccan population. Appl Clin Genet. 2019;12:51–54. doi: 10.2147/TACG.S194166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Jiao XH, Wang XP, Sun XY, Dong C. MTR, MTRR, MTHFR gene polymorphisms and susceptibility to nonsyndromic cleft lip with or without cleft palate. Genet test Mol Biomarkers. 2016;20:297–303. doi: 10.1089/gtmb.2015.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebadifar A, Ameli N, Khorramkhorshid HR, Salehi Zeinabadi4 M, Kamali K, Khoshbakht T. Incidence assessment of MTHFR C677T and A1298C polymorphisms in Iranian nonsyndromic cleft lip and/or palate patients. J Dent Res Dent Clin Dent Prospects. 2015;9:101–104. doi: 10.15171/joddd.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mito N, Takimoto H, Umegaki K, Ishiwaki A, Kusama K, Fukuoka H, Ohta S, Abe S, Yamawaki M, Ishida H, Yoshiike N. Folate intakes and folate biomarker profiles of pregnant Japanese women in the first trimester. Eur J Clin Nutr. 2007;61:83–90. doi: 10.1038/sj.ejcn.1602497. [DOI] [PubMed] [Google Scholar]
- 46.Takimoto H, Yoshiike N, Katagiri A, Ishida H, Abe S. Nutritional status of pregnant and lactating women in Japan: A comparison with non-pregnant/non-lactating controls in the national nutrition survey. J Obstet Gynaecol Res. 2003;29:96–103. doi: 10.1046/j.1341-8076.2002.00078.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.