Abstract
Molecular heterogeneity determines the differences in the pathological features, prognosis and survival after relapse when comparing left-sided colon cancer (LCC) and right-sided colon cancer (RCC). At present, the discrepancy in the underlying molecular events between the two types of colon cancer has not been thoroughly investigated. The present study aimed to explore novel targets to predict the disease stage and prognosis of LCC and RCC. Expression analysis of guanine nucleotide binding-protein γ subunit 4 (GNG4) was performed using the Gene Expression Profiling Interactive Analysis (GEPIA) and Oncomine databases. Survival and association analyses were performed using GEPIA and the colon adenocarcinoma dataset from The Cancer Genome Atlas database. GNG4-positive cells in a tissue microarray were examined using immunohistochemistry. According to the GNG4 expression data from Caucasian patients included in the TCGA dataset, GNG4 was highly expressed and positively associated with pathological stage and overall survival (OS) rates in colon cancer. GNG4 expression was higher in LCC than in RCC. Patients with LCC with high GNG4 expression exhibited higher pathological stage and lower survival rates, whereas this was not observed in patients with RCC. In addition, the clinical tissues used in the microarray showed that GNG4 expression was increased in Chinese patients with LCC compared with that in patients with RCC. Consistently, GNG4 expression was negatively associated with OS in patients with LCC, but not in patients with RCC. However, no association was observed between GNG4 expression and the disease stage of colon cancer in both patients with LCC and RCC. Overall, the molecular heterogeneity of GNG4 in LCC and RCC suggests that GNG4 may be used as a diagnostic and prognostic biomarker in patients with LCC.
Keywords: GNG4, gene heterogeneity, LCC, RCC, overall survival, disease stage
Introduction
Colon adenocarcinoma (COAD) is the third leading cause of morbidity (46.9 per 100,000 men and 35.6 per 100,000 women) and mortality (17.7 per 100,000 men and 12.4 per 100,000 women) among malignant neoplasms worldwide, according to statistics from 2013 (1). Alcohol and processed meat consumption, advanced age, family history and tumor metastasis are closely associated with the incidence of COAD (2). Although surgical resection, chemotherapy and radiotherapy have been widely used to treat colon cancer, most patients with advanced disease exhibit drug resistance, leading to a particularly poor prognosis (3,4). Increasing evidence has suggested that left-sided colon cancer (LCC), which arises from the embryonic midgut, and right-sided colon cancer (RCC), which originates from the hindgut, exhibit distinct differences in embryonic origin, biology, anatomy, genetic mutations and alterations, and clinical outcomes, and this may be the major cause of the poor prognosis following surgery and medical procedures in COAD (5). Therefore, identification of tumor-driven genes underlying the progression of LCC and RCC is of great importance for monitoring the progression and improving the prognosis of patients with COAD.
The cecum, ascending colon, hepatic flexure and transverse colon are anatomically referred to as the right colon, while the descending colon and sigmoid colon are classified as the left colon (6). Cancer located in the right and left colon is defined as RCC or LCC, respectively. The incidence of RCC is associated with sex, age, cancer history and insulin resistance, whereas LCC is closely associated with a low fiber diet, heavy smoking and alcoholism (7). Cancer located in the right colon has a prevalence toward being of a more advanced tumor stage and having a large tumor size, and often metastasizes to the lymph nodes or peritoneal region (8). Poor survival rate is observed in patients with RCC compared with LCC (9). LCC is associated with a reduced risk of death, which may be due to the observation that patients with LCC more commonly present with early-stage disease, and this results in the disparity in prognosis according to tumor site (10). Numerous differences in molecular signaling pathways have been reported between RCC and LCC (11). Notably, using molecular biology techniques, numerous molecular events that cause distinct efficacy of molecular targeted agents, such as TP53, KRAS, CpG Island Methylator Phenotype (CIMP) and microsatellite instability (MSI), in patients with LCC and RCC have been revealed in recent years (12). For example, deregulated genes for CIMP high and MSI tumors are often downregulated. In contrast, genes that are deregulated in TP53 are likely to be upregulated compared to normal paired samples. However, to the best of our knowledge, the specific molecules that underlie the difference between LCC and RCC have not yet been determined.
Guanine nucleotide binding-protein γ subunit 4 (GNG4) is a member of the G-protein trimer complex, and was first identified as the brain-specific subunit (13). In the human brain, GNG4 is more highly expressed in the hippocampus compared with in other brain regions, and GNG4 expression is reduced with advanced age, which suggests that GNG4 is associated with cognitive decline (14). A previous study has suggested that GNG4 is hypermethylated and notably decreased in glioblastoma (GBM) (15). In addition to its roles in the brain, GNG4 is involved in pathological processes, it activates signaling pathways in acute myocardial infarction, and it may contribute to the prevention and treatment of recurrent cardiovascular events (16). Additionally, it has been demonstrated that GNG4 is a hub gene that has a high degree of connectivity in colon cancer (17). According to bioinformatics analysis, the hub gene GNG4 has a clinical diagnostic value in patients with colorectal cancer (18). Therefore, GNG4 is an important regulator during tumor progression, particularly in colon cancer. However, whether there is molecular heterogeneity of GNG4 in LCC and RCC remains unclear.
The present study was aimed to investigate the molecular heterogeneity of GNG4 in LCC and RCC. Additionally, the association between GNG4 expression and clinical characteristics including disease stage and overall survival (OS) rate was also assessed in LCC and RCC of Caucasian and Chinese patients. These findings demonstrated that GNG4 may be used as an effective prognostic factor for LCC and RCC.
Materials and methods
Tissue microarray
A tissue and cDNA microarray containing 78 colon carcinoma and paired para-carcinoma tissues was purchased from Shanghai Outdo Biotech Co., Ltd. Among the 78 cases, 32 were tumors located in the left colon and the other 46 were tumors located in the right colon. All experiments were approved by the Ethics Committee of the Affiliated Cancer Hospital of Fujian Medical University (Fuzhou, China).
Immunohistochemistry (IHC)
For IHC staining, 5 µm thick paraffin embedded sections were dewaxed in an oven for 30 min at 60°C. After deparaffinization in xylene and rehydration in gradient ethanol, antigens were retrieved with 0.01 M citrate salt solution (pH 6.0) using high pressure method (100°C) for 15 min. Following washing with PBS plus Tween-20 and blocking with 5% BSA (cat. no. A8010; Beijing Solarbio Science & Technology Co., Ltd.) for 1 h at room temperature, the slides were incubated with primary antibody against GNG4 (cat. no. ab238868; 1:250; Abcam) at 4°C overnight. Following incubation with the secondary antibody (cat. no. ab6721; 1:2,000; Abcam) for 15 min at room temperature, antigen-antibody complexes were visualized using 3,3-diaminobenzidine reagent (cat. no. ZL1-9081; OriGene Technologies, Inc.). The results were observed under a biological inverted light microscope (×5 and ×20 magnifications; IX51; Olympus Corporation). Comprehensive analysis included measuring staining intensity and the number of positive cells as follows: Negative (−), 0%; weakly positive (+), <20%; moderately positive (++), 20–50%; and strongly positive (+++), >50%.
Reverse transcription-quantitative PCR (RT-qPCR)
cDNA extracted from 78 cases of colon carcinoma, corresponding to the same samples as the tissue microarray, was purchased from Shanghai Outdo Biotech Co., Ltd. Samples were plated in 96-well plates and subsequently, RT-qPCR was performed using the SYBR Green kit (Roche Diagnostics GmbH) according to manufacturers protocols. GNG4 expression was normalized to GAPDH expression. The primer sequences were shown in Table SI. Relative gene expression levels were calculated using the 2−∆∆Cq method (19).
Western blotting
A total of 15 clinical samples from patients aged between 38 and 62 years (13 men and 2 women; mean age, 50.47±2.035), including 5 normal colon tissues, 5 left-sided colon tumor tissues and 5 right-sided colon tumor tissues obtained from resection were collected at The Affiliated Cancer Hospital of Fujian Medical University between October 2019 and February 2020. All these patients had been diagnosed with colon cancer previously. Written informed consent was obtained from all subjects, and the present study was approved by the Ethics Committee of The Affiliated Cancer Hospital of Fujian Medical University. Total protein was extracted from normal colon and tumor tissues using RIPA protein lysate (cat. no. P0013B; Beyotime Institute of Biotechnology). Protein concentration was determined using BCA Protein Assay Kit (23225; Thermo Fisher Scientific, Inc.). Equal amounts of protein (30 µg) were loaded and separated on 12% gels by SDS-PAGE. Subsequently, protein was transferred to an activated PVDF membrane (IPVH00010; EMD Millipore). The membrane was blocked with 5% skimmed milk dissolved in PBS-Tween-20 (0.1%) at room temperature (~25°C) for 1 h. Next, primary antibody against GNG4 (cat. no. PA5-103877; 1:800; Thermo Fisher Scientific, Inc.) was added for an incubation at 4°C overnight. β-actin (cat. no. BM0627; 1:5,000; Boster Biological Technology) served as the internal control. Following incubation with goat anti-rabbit secondary antibody conjugated to HRP (cat. no. BA1054; 1:5,000; Boster Biological Technology) at room temperature (~25°C) for an additional 1 h, the membrane was incubated with ECL visualization reagent (34065; Thermo Fisher Scientific, Inc.) and the bands were visualized using the Imagequant LAS 4000 mini machine (Cytiva).
Data collection and analysis
The samples collected from TCGA were divided into LCC (n=181) and RCC (n=258). No information in anatomic neoplasm was excluded from the dataset. The mRNA expression levels of GNG4 in normal colon tissues and colon cancer tissues were collected from the Oncomine database (https://www.oncomine.org/resource/login.html) and the Gene Expression Profiling Interactive Analysis (GEPIA) version 2 database (20). GNG4 expression in LCC (n=99) and RCC (n=167) excluding the normal samples was determined using the raw data from The Cancer Genome Atlas (TCGA, n=524; http://portal.gdc.cancer.gov/) and analyzed using GraphPad v6.0 software (GraphPad Software, Inc.). The samples collected from the Oncomine database included 12 normal colon samples and 70 COAD samples. The association between GNG4 expression and disease stage in patients with COAD was analyzed with one-way ANOVA (followed by Tukeys post hoc test) using raw data downloaded from the GEPIA database. In the tissue microarray, the association between GNG4 and disease stage was determined based on the IHC staining results. The disease stage information was mentioned in the tissue microarray.
Survival analysis
OS rate was analyzed using GEPIA on the basis of GNG4 expression status. The difference in OS rate between patients with LCC and RCC was compared using the raw data from TCGA and analyzed with GraphPad v6.0 software. The data from the tissue microarray were used to evaluate the OS rate in Chinese patients with tumors in the left and right colon.
Statistical analysis
The association between pathological stage and GNG4 expression in tissue microarray was analyzed with GraphPad Prism 6.0 (GraphPad Software, Inc.) using χ2 and Fishers exact tests. The association analysis between tumor stage and GNG4 status in patients with COAD was performed using one-way ANOVA, followed by Tukeys post hoc test. Histological staining was evaluated using Image Pro Plus 6.0 software (Media Cybernetics). GNG4 gene expression and relative protein expression for tissue microarray were compared with GraphPad Prism using two-tailed t-tests (unpaired) and one-way ANOVA followed by Tukeys post hoc test, respectively. Kaplan-Meier plotter was used to generate the survival curves using GraphPad Prism software (version 6.0; GraphPad Software, Inc.), and data were compared between groups using the log-rank (Mantel-Cox) test (univariate Cox regression analysis). P<0.05 was considered to indicate a statistically significant difference.
Results
GNG4 is highly expressed in White patients with COAD
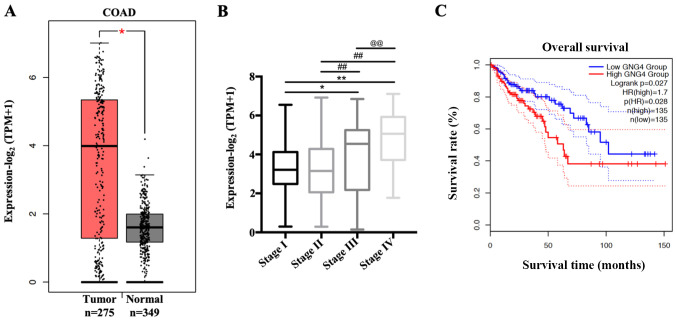
To determine GNG4 expression in White patients with COAD included in TCGA database, the present study analyzed the transcriptional levels of GNG4 in 275 patients with COAD and 349 normal controls. The mean expression level of GNG4 was increased 2.2-fold in the tumor group compared with that in the normal group (Fig. 1A). Subsequently, the association between GNG4 and disease stage of COAD was assessed. The results indicated that GNG4 status exhibited no difference when comparing patients with COAD in stages I and II. However, GNG4 expression significantly increased in disease stages III and IV compared with that in low disease stages (I and II) (stage I vs. III, P=0.0428; stage I vs. IV, P<0.01; stage II vs. III, P<0.01; and stage II vs. IV, P<0.01), with the highest levels observed in stage IV (stage III vs. IV, P=0.0427) (Fig. 1B). GNG4 expression was divided into two groups, using as a cut-off the median method which was obtained from GEPIA 2.0. Additionally, patients with COAD and high GNG4 expression (n=135) had a decreased survival rate compared with patients with low GNG4 expression (n=135) (Fig. 1C). Therefore, high expression levels of GNG4 were positively associated with pathological grade and prognosis in White patients with COAD.
Figure 1.
Association of GNG4 status with disease stage and overall survival in patients with COAD. (A) Different transcriptional expression levels of GNG4 in COAD samples (n=275) and normal controls (n=349). Gene expression data were collected from the GEPIA 2.0 database and analyzed with GraphPad Prism using two-tailed t-tests (unpaired). (B) Patients with COAD were divided into four groups based on disease stage, including stage I, II, III and IV. Subsequently, the association between GNG4 expression and disease stage was analyzed by one-way ANOVA followed by Tukeys post hoc test. (C) Survival analysis was performed using GEPIA 2.0 in patients with COAD with low and high GNG4 expression. *P<0.05; **, ##, @@P<0.01. COAD, colon adenocarcinoma; GEPIA, Gene Expression Profiling Interactive Analysis; GNG4, guanine nucleotide binding-protein γ subunit 4; TPM, transcripts per million.
Molecular heterogeneity of GNG4 is observed in White patients with LCC and RCC
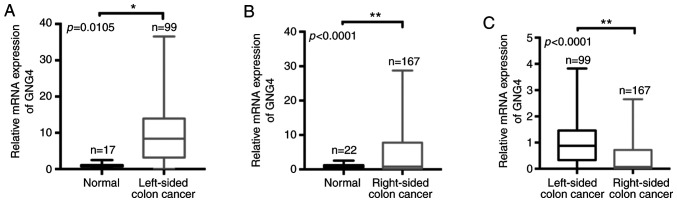
Given that GNG4 is an important hub gene during the tumorigenesis of COAD, the present study aimed to investigate the differences in GNG4 expression between White patients with LCC and those with RCC. Firstly, the raw data of patients with COAD was collected from TCGA and screened according to grouping criteria. Subsequently, all samples (ethnicity, White) were divided into two groups according to the tumor site in the colon, i.e. LCC and RCC. Based on the sample information, only 15 normal samples in LCC and 22 normal samples in RCC were included. In LCC cohorts, there were 99 tumor samples and the corresponding normal samples (n=15), and there were 167 RCC tumor samples and corresponding normal tissues (n=22). When comparing normal control tissues (n=15) with LCC tissues (n=99), it was revealed that GNG4 expression was significantly elevated in the LCC tissues (P=0.0105; Fig. 2A). Consistent with this, there was a significant increase in GNG4 expression in RCC tissues (n=167) compared with the corresponding normal tissues (n=22) (P<0.0001; Fig. 2B). However, compared with the level in patients with RCC (n=167), the GNG4 expression level was increased in patients with LCC (n=99) (P<0.0001; Fig. 2C). Therefore, molecular heterogeneity of GNG4 was present in LCC and RCC.
Figure 2.
GNG4 expression in LCC and RCC. Comparison of GNG4 mRNA expression (A) between normal controls and patients with LCC, (B) between normal controls and patients with RCC, and (C) between patients with LCC and RCC based on raw data from the colon adenocarcinoma dataset in The Cancer Genome Atlas database. Data were analyzed with GraphPad Prism using two-tailed t-tests (unpaired). *P<0.05; **P<0.01. GNG4, guanine nucleotide binding-protein γ subunit 4; LCC, left-sided colon cancer; RCC, right-sided colon cancer.
GNG4 is associated with disease stage and prognosis in White patients with LCC
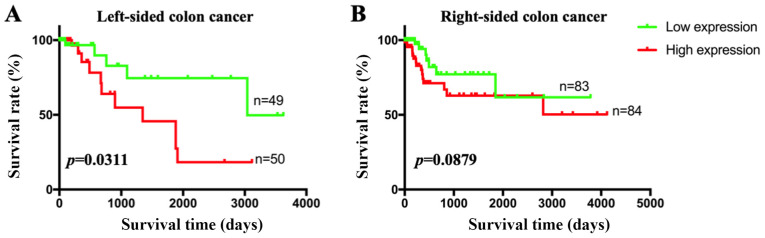
Since genetic heterogeneity of GNG4 was observed between LCC and RCC, it was speculated that there may be a difference in the clinical diagnostic value of GNG4 in the two cancer types. First, patients were divided into two groups based on GNG4 expression (cut-off-high, 50%; cut-off-low, 50%). High GNG4 expression was associated with high disease stage in patients with LCC (P=0.0356; Table I). However, no association between GNG4 status and disease stage was identified in patients with RCC (P=0.6260; Table I). Additionally, patients with LCC in the high GNG4 expression group had a lower survival rate than those in the low GNG4 expression group (P=0.0311; Fig. 3A). In patients with RCC, no association between GNG4 expression and OS was observed (P=0.0879; Fig. 3B). Therefore, GNG4 status was associated with pathological stage and OS in White patients with LCC, but not in those with RCC.
Table I.
Association between GNG4 expression and pathological stage in LCC and RCC.
| LCC | RCC | |||
|---|---|---|---|---|
| Stage | Low GNG4, n | High GNG4, n | Low GNG4, n | High GNG4, n |
| I | 8 | 5 | 18 | 15 |
| II | 24 | 18 | 35 | 29 |
| III | 9 | 18 | 22 | 28 |
| IV | 4 | 13 | 9 | 11 |
| P-value | 0.0356a | 0.6260 | ||
P<0.05. GNG4, guanine nucleotide binding-protein γ subunit 4; LCC, left-sided colon cancer; RCC, right-sided colon cancer.
Figure 3.
Association between GNG4 status and overall survival in patients with LCC and RCC. Clinical data of patients with LCC and RCC were separated from among the raw data in the colon adenocarcinoma dataset in The Cancer Genome Atlas database. Subsequently, the patients with LCC and RCC were grouped based on GNG4 status. The association between GNG4 status and (A) LCC or (B) RCC was analyzed using univariate Cox regression analysis. GNG4, guanine nucleotide binding-protein γ subunit 4; LCC, left-sided colon cancer; RCC, right-sided colon cancer.
GNG4 is highly expressed in Chinese patients with LCC
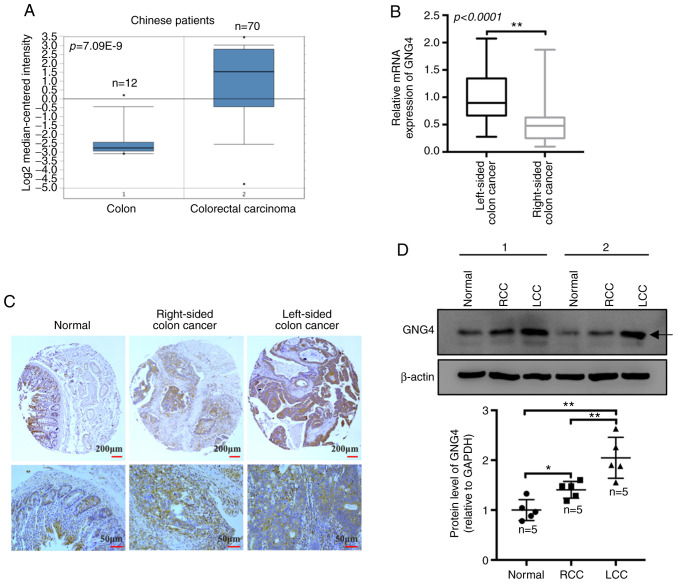
To further ascertain the expression pattern of GNG4 in Chinese patients with colorectal cancer, the present study collected raw data, including that of 70 cases of colorectal cancer and 12 normal colon samples, from the Oncomine database. All 82 samples were from a Chinese population. The results revealed that GNG4 expression was significantly increased in patients with colorectal cancer compared with that in normal controls (P<0.0001; Fig. 4A). Subsequently, tissue and cDNA microarrays containing 78 pairs of COAD samples and paired adjacent colon tissues were used to evaluate the expression levels of GNG4 in human specimens by IHC staining and RT-qPCR. The microarray comprised 32 cases of LCC and 46 cases of RCC. The data indicated that the relative GNG4 mRNA expression in patients with LCC was markedly increased compared with that in patients with RCC (P<0.0001; Fig. 4B). Additionally, the IHC results demonstrated that GNG4 expression was notably increased in both LCC and RCC samples compared with that in the normal colonic mucosal tissues, with higher GNG4 expression observed in LCC compared with that in RCC (Fig. 4C). Furthermore, the increase in GNG4 protein expression was also observed in the fresh clinical samples. GNG4 protein expression was markedly increased in LCC (P=0.0002) and RCC tissues (P=0.0491) compared with that in normal colon tissues. Consistently, higher GNG4 expression was observed in LCC compared with that in RCC (P=0.0099; Fig. 4D). Based on the IHC results, high GNG4 expression (++ and +++) was identified in 16 LCC tumor tissues, with a higher percentage (14/32) in the strong positive (+++) category. However, only 7/46 RCC samples exhibited strong positive staining (Table II). Based on multiple factor analysis, it was observed that there was a significant difference in GNG4 status between LCC and RCC in the Chinese patients (P=0.0019; Table II). These data implied that GNG4 expression was increased in Chinese patients with colorectal cancer, particularly in patients with LCC, and that it may be involved in the progression of COAD.
Figure 4.
Differential analysis of GNG4 expression in Chinese patients with COAD. (A) Transcriptional expression levels of GNG4 in normal controls and patients with COAD. The raw data were obtained from the Oncomine database. Data were analyzed with GraphPad Prism using two-tailed t-tests (unpaired). (B) Differential GNG4 mRNA expression between LCC and RCC was determined using reverse transcription-quantitative PCR. (C) Immunohistochemical staining of GNG4 in normal, LCC and RCC tissues in the tissue microarray. (D) Protein expression levels of GNG4 in normal, LCC and RCC tissues, as measured by western blotting. The graph presents the relative protein expression levels of GNG4 in the aforementioned three tissues (one-way ANOVA, followed by Tukeys post hoc test). *P<0.05; **P<0.01. COAD, colon adenocarcinoma; GNG4, guanine nucleotide binding-protein γ subunit 4; LCC, left-sided colon cancer; RCC, right-sided colon cancer.
Table II.
Immunohistochemical staining of guanine nucleotide binding-protein γ subunit 4 in samples from Chinese patients with LCC and RCC.
| High expression | Low expression | ||||
|---|---|---|---|---|---|
| Type | Strong (+++), n | Elevated (++), n | Moderate (+), n | Absent (−), n | P-value |
| LCC | 14 | 2 | 13 | 3 | 0.0019a |
| RCC | 7 | 16 | 13 | 10 | |
P<0.05. (−), (+), (++) and (+++) represent different degrees of staining. The data were analyzed with GraphPad Prism 6.0 using χ2 and Fisher's exact tests. LCC, left-sided colon cancer; RCC, right-sided colon cancer.
GNG4 is negatively associated with OS in Chinese patients with LCC
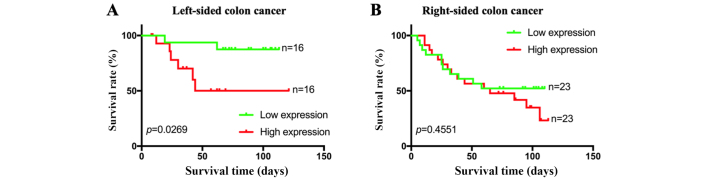
When combining the mRNA expression levels of GNG4 and the clinical data of patients in the microarray, it was observed that both high GNG4 expression and low GNG4 expression were more frequently present in patients with stage II colorectal cancer, in both LCC and RCC (Table III). Additionally, no associations between GNG4 status and disease stage were identified in patients with LCC (P=0.4324) or in patients with RCC (P=0.2717; Table III). Survival analysis of the 32 patients with LCC indicated that low expression levels of GNG4 were associated with a good prognosis and high survival rate compared with high expression levels of GNG4 (P=0.0269; Fig. 5A). However, GNG4 expression status did not affect the prognosis of patients with colorectal cancer with tumors located in the right-sided colon (P=0.4551; Fig. 5B). Overall, GNG4 expression was only associated with the prognosis of Chinese patients with LCC.
Table III.
Association between GNG4 expression and pathological stage of LCC and RCC in Chinese patients.
| LCC | RCC | |||
|---|---|---|---|---|
| Stage | Low GNG4, n | High GNG4, n | Low GNG4, n | High GNG4, n |
| I | 1 | 1 | 2 | 3 |
| II | 13 | 10 | 18 | 13 |
| III | 2 | 5 | 3 | 7 |
| P-value | 0.4324 | 0.2717 | ||
GNG4, guanine nucleotide binding-protein γ subunit 4.
Figure 5.
Association between GNG4 status and overall survival in Chinese patients with LCC and RCC. Clinical data of patients with LCC and RCC were obtained from the product documentation of the tissue microarray. Based on the status of GNG4, patients with LCC or RCC were divided into two groups each. Subsequently, the association between GNG4 status and (A) LCC or (B) RCC was analyzed using univariate Cox regression analysis. GNG4, guanine nucleotide binding-protein γ subunit 4; LCC, left-sided colon cancer; RCC, right-sided colon cancer.
Discussion
Genetic and immunological differences between the proximal (right-sided) colon and the distal (left-sided) colorectum have a strong impact on surgical and oncological outcomes, which increases the difficulty of the management of patients with COAD (21,22). Investigation of the diagnostic markers for different anatomical sites (left or right) facilitates personalized medicine and the treatment of colon cancer (23). The present study revealed the potential diagnostic and prognostic values of GNG4 in LCC. To the best of our knowledge, the present study is the first to illuminate the genetic heterogeneity of GNG4 between LCC and RCC.
Based on sequencing and the Search Tool for the Retrieval of Interacting Genes/Proteins database, GNG4 has been demonstrated to be involved in the development of gastric cancer in patients with H. pylori infection (24). In comparison with normal samples, GNG4 is highly expressed in rectal adenocarcinoma samples; however, there is no association between GNG4 expression and the survival rate of patients with rectal cancer (25). In the present study, GNG4 was upregulated in COAD samples, including both LCC and RCC samples. The data indicated that the increase in GNG4 expression in COAD samples was positively associated with tumors on both sides of the colon. However, GNG4 expression was higher in patients with LCC compared with that in patients with RCC. To the best of our knowledge, the present study was the first to reveal the genetic heterogeneity of GNG4 in colorectal cancer. Hypermethylated GNG4 has been identified in thymic carcinoma in relation to thymoma and thymus, and is positively associated with poor relapse-free survival rate in patients with all types of thymic epithelial tumor (26). Aberrant DNA methylation of GNG4, which is involved in cellular regulatory events, including cell adhesion and signal transduction, can serve as a diagnostic and therapeutic biomarker for bladder cancer (27). In the present study, GNG4 status was only associated with disease stage and prognosis in patients with LCC, but not in patients with RCC. These data suggested that GNG4 may be a biomarker for diagnosis and prognosis in patients with LCC, which is partly consistent with the results from a previous study (17). Due to the limited sample size, this result should be further confirmed in large-scale cohorts. In summary, GNG4 might be a good indicator for the treatment of patients with LCC.
There are clear differences in genetic heterogeneity and prognosis of colon cancer among different ethnicities. African American (AA) patients with COAD have a poor prognosis compared with Caucasian (CA) patients with COAD (28). Compared with that in Iranian patients (a White population), hypermethylation of the glycoprotein nmb, intercellular adhesion molecule 5 and chromodomain helicase DNA binding protein 5 genes is identified in AA patients, and these are the candidate cancer genes specifically involved in the progression of COAD in the AA population (29). The present study revealed that GNG4 was notably upregulated in Chinese patients with COAD, and there was genetic heterogeneity of GNG4 between LCC and RCC. In this study, most of the cohorts downloaded from datasets from the TCGA dataset were of Caucasian ethnicity. This finding indicated that abnormal GNG4 expression did not differ between the Chinese study population and other CA patients. Despite the limited sample size, the present study also demonstrated that GNG4 was associated with the OS of patients with LCC, whereas there was no association between disease stage and GNG4 status in Chinese patients with COAD, including LCC and RCC. The data in the microarray results were different from the database results of Caucasian patients, implying that the clinical application value of GNG4 differs depending on geographical location. More subjects of different ethnicities are required to investigate the diagnostic and prognostic role of GNG4 in COAD, particularly in patients with LCC.
GNG4, c-Myc, DNA polymerase α1, catalytic subunit and ribonucleotide reductase catalytic subunit M1 could serve as prognostic factors for the response to treatment in patients with locally advanced rectal cancer (30). Whether there is a association between GNG4 and drug-sensitivity during the treatment of patients with LCC remains unclear and requires more comprehensive analysis.
Mechanistically, forced expression of GNG4 inhibits GBM cell migration by decreasing the activation of ERK and JNK via stromal cell-derived factor 1α/C-X-C motif chemokine receptor 4-dependent chemokine signaling (31). Depletion of PSMC3-interacting protein represses cell viability and xenograft tumorigenesis of hepatocellular carcinoma (HCC) cells via upregulation of GNG4 (32). Exogenous GNG4 decreases cell proliferation in renal cell carcinoma by affecting hypoxic response signaling pathways (33). These studies suggest the inhibitory effect of GNG4 on tumorigenesis in GBM, HCC and renal cell carcinoma. Protein-protein interaction analysis of different genes in LCC and RCC has demonstrated that GNG4 may be a hub gene at the core of the interaction network (34). This implies that GNG4 is an important molecular switch in the development of LCC.
However, further in vitro and in vivo experiments are required to investigate the biological function of GNG4 in colorectal cancer. Additionally, the sample volume of Chinese patients in the present study was small and limited the analysis on the clinical significance of GNG4 in patients with LCC and RCC. Future studies with larger cohorts are needed for verification of the findings in the present study.
In summary, high expression levels of GNG4 and genetic heterogeneity of GNG4 between LCC and RCC were present in both White and Chinese patients. However, GNG4 was only associated with the OS of Chinese patients with LCC, while GNG4 status was associated with the disease stage and prognosis of LCC in White patients. GNG4 may be a good prognostic factor for patients with LCC worldwide.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- LCC
left-sided colon cancer
- RCC
right-sided colon cancer
- GNG4
guanine nucleotide binding-protein γ subunit 4
- GEPIA
Gene Expression Profiling Interactive Analysis
- TCGA
The Cancer Genome Atlas
- COAD
colon adenocarcinoma
- GBM
glioblastoma
- HCC
hepatocellular carcinoma
Funding
The present study was supported by a grant from the Startup Fund for Scientific Research, Fujian Medical University (no. 2017XQ1211).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the [GEPIA2] repository, [http://gepia2.cancer-pku.cn/#general].
Authors contributions
YC and JS conceived the idea and drafted the manuscript. JS and JY analyzed the expression pattern of GNG4 in LCC and RCC. RL, XC and LZ performed the statistical analysis. All authors discussed the results, edited this manuscript, read and approved the final version.
Ethics approval and consent to participate
All experiments were approved by the Ethics Committee of the Affiliated Cancer Hospital of Fujian Medical University (Fuzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G, II, Samuel S, Kim MP, Lim SJ, Ellis LM. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–1957. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoyagi T, Terracina KP, Raza A, Takabe K. Current treatment options for colon cancer peritoneal carcinomatosis. World J Gastroentero. 2014;20:12493–12500. doi: 10.3748/wjg.v20.i35.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brulé SY, Jonker DJ, Karapetis CS, OCallaghan CJ, Moore MJ, Wong R, Tebbutt NC, Underhill C, Yip D, Zalcberg JR, et al. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in NCIC CO.17. Eur J Cancer. 2015;51:1405–1414. doi: 10.1016/j.ejca.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Nitsche U, Stögbauer F, Späth C, Haller B, Wilhelm D, Friess H, Bader FG. Right sided colon cancer as a distinct histopathological subtype with reduced prognosis. Digest Surg. 2016;33:157–163. doi: 10.1159/000443644. [DOI] [PubMed] [Google Scholar]
- 7.Xiang L, Zhan Q, Zhao XH, Wang YD, An SL, Xu YZ, Li AM, Gong W, Bai Y, Zhi FC, Liu SD. Risk factors associated with missed colorectal flat adenoma: A multicenter retrospective tandem colonoscopy study. World J Gastroentero. 2014;20:10927–10937. doi: 10.3748/wjg.v20.i31.10927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss JM, Pfau PR, OConnor ES, King J, LoConte N, Kennedy G, Smith MA. Mortality by stage for right- versus left-sided colon cancer: Analysis of surveillance, epidemiology, and end results-medicare data. J Clin Oncol. 2011;29:4401–4409. doi: 10.1200/JCO.2011.36.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yahagi M, Okabayashi K, Hasegawa H, Tsuruta M, Kitagawa Y. The worse prognosis of right-sided compared with left-sided colon cancers: A systematic review and meta-analysis. J Gastrointest Surg. 2016;20:648–655. doi: 10.1007/s11605-015-3026-6. [DOI] [PubMed] [Google Scholar]
- 10.Petrelli F, Tomasello G, Borgonovo K, Ghidini M, Turati L, Dallera P, Passalacqua R, Sgroi G, Barni S. Prognostic survival associated with left-sided vs. right-sided colon cancer: A systematic review and meta-analysis. JAMA Oncol. 2017;3:211–219. doi: 10.1001/jamaoncol.2016.4227. [DOI] [PubMed] [Google Scholar]
- 11.Lee GH, Malietzis G, Askari A, Bernardo D, Al-Hassi HO, Clark SK. Is right-sided colon cancer different to left-sided colorectal cancer?-A systematic review. Eur J Surg Oncol. 2015;41:300–308. doi: 10.1016/j.ejso.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Slattery ML, Pellatt DF, Mullany LE, Wolff RK, Herrick JS. Gene expression in colon cancer: A focus on tumor site and molecular phenotype. Genes Chromosomes Cancer. 2015;54:527–541. doi: 10.1002/gcc.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalyanaraman S, Copeland NG, Gilbert DG, Jenkins NA, Gautam N. Structure and chromosomal localization of mouse G protein subunit gamma 4 gene. Genomics. 1998;49:147–151. doi: 10.1006/geno.1998.5223. [DOI] [PubMed] [Google Scholar]
- 14.Bonham LW, Evans DS, Liu Y, Cummings SR, Yaffe K, Yokoyama JS. Neurotransmitter pathway genes in cognitive decline during aging: Evidence for GNG4 and KCNQ2 Genes. Am J Alzheimers Dis Other Demen. 2018;33:153–165. doi: 10.1177/1533317517739384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shukla S, Pia Patric IR, Thinagararjan S, Srinivasan S, Mondal B, Hegde AS, Chandramouli BA, Santosh V, Arivazhagan A, Somasundaram K. A DNA methylation prognostic signature of glioblastoma: Identification of NPTX2-PTEN-NF-κB nexus. Cancer Res. 2013;73:6563–6573. doi: 10.1158/0008-5472.CAN-13-0298. [DOI] [PubMed] [Google Scholar]
- 16.Liao JQ, Chen Z, He QH, Liu YM, Wang J. Differential gene expression analysis and network construction of recurrent cardiovascular events. Mol Med Rep. 2016;13:1746–1764. doi: 10.3892/mmr.2015.4707. [DOI] [PubMed] [Google Scholar]
- 17.Yang W, Ma J, Zhou W, Li Z, Zhou X, Cao B, Zhang Y, Liu J, Yang Z, Zhang H, et al. Identification of hub genes and outcome in colon cancer based on bioinformatics analysis. Cancer Manag Res. 2019;11:323–338. doi: 10.2147/CMAR.S173240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Lu D, Sun K, Xu Y, Hu P, Li X, Xu F. Identification of biomarkers associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Gene. 2019;692:119–125. doi: 10.1016/j.gene.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Tang ZF, Li CW, Kang BX, Gao G, Li C, Zhang ZM. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bufill JA. Colorectal cancer: Evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779–788. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 22.Hu W, Yang Y, Li X, Huang M, Xu F, Ge W, Zhang S, Zheng S. Multi-omics approach reveals distinct differences in left-and right-sided colon cancer. Mol Cancer Res. 2018;16:476–485. doi: 10.1158/1541-7786.MCR-17-0483. [DOI] [PubMed] [Google Scholar]
- 23.Gervaz P, Bucher P, Morel P. Two colons-two cancers: Paradigm shift and clinical implications. J Surg Oncol. 2004;88:261–266. doi: 10.1002/jso.20156. [DOI] [PubMed] [Google Scholar]
- 24.Chu AN, Liu JW, Yuan Y, Gong YH. Comprehensive Analysis of Aberrantly Expressed ceRNA network in gastric cancer with and without H. pylori infection. J Cancer. 2019;10:853–863. doi: 10.7150/jca.27803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu BX, Huang GJ, Cheng HB. Comprehensive analysis of core genes and potential mechanisms in rectal cancer. J Comput Biol. 2019;26:1262–1277. doi: 10.1089/cmb.2019.0073. [DOI] [PubMed] [Google Scholar]
- 26.Kishibuchi R, Kondo K, Soejima S, Tsuboi M, Kajiura K, Kawakami Y, Kawakita N, Sawada T, Toba H, Yoshida M, et al. DNA methylation of GHSR, GNG4, HOXD9 and SALL3 is a common epigenetic alteration in thymic carcinoma. Int J Oncol. 2020;56:315–326. doi: 10.3892/ijo.2019.4915. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Fang L, Zang Y, Xu Z. Identification of core genes and key pathways via integrated analysis of gene expression and DNA methylation profiles in bladder cancer. Med Sci Monit. 2018;24:3024–3033. doi: 10.12659/MSM.909514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Govindarajan R, Posey J, Chao CY, Lu R, Jadhav T, Javed AY, Javed A, Mahmoud FA, Osarogiagbon RU, Manne U. A comparison of 12-gene colon cancer assay gene expression in African American and Caucasian patients with stage II colon cancer. BMC Cancer. 2016;16:368. doi: 10.1186/s12885-016-2365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, Nouraie M, Green R, Lee E, Smoot DT, Ashktorab H. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4:e7012. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palma P, Cano C, Conde-Muiño R, Comino A, Bueno P, Ferrón JA, Cuadros M. Expression profiling of rectal tumors defines response to neoadjuvant treatment related genes. PLoS One. 2014;9:e112189. doi: 10.1371/journal.pone.0112189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal J, Patil V, Mondal B, Shukla S, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Epigenetically silenced GNG4 inhibits SDF1α/CXCR4 signaling in mesenchymal glioblastoma. Genes Cancer. 2016;7:136–147. doi: 10.18632/genesandcancer.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding JL, Li Y, Fan HX, Xu W, Gao R, Bai S, Zhu Z, Yang W, Gong Y, Yang J, Zhou J. Knockdown of PSMC3IP suppresses the proliferation and xenografted tumorigenesis of hepatocellular carcinoma cell. J Cell Biochem. 2019;120:5449–5458. doi: 10.1002/jcb.27824. [DOI] [PubMed] [Google Scholar]
- 33.Maina EN, Morris MR, Zatyka M, Raval RR, Banks RE, Richards FM, Johnson CM, Maher ER. Identification of novel VHL target genes and relationship to hypoxic response pathways. Oncogene. 2005;24:4549–4558. doi: 10.1038/sj.onc.1208649. [DOI] [PubMed] [Google Scholar]
- 34.Liang L, Zeng JH, Qin XG, Chen JQ, Luo DZ, Chen G. Distinguishable prognostic signatures of left- and right-sided colon cancer: A study based on sequencing data. Cell Physiol Biochem. 2018;48:475–490. doi: 10.1159/000491778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the [GEPIA2] repository, [http://gepia2.cancer-pku.cn/#general].