Abstract
Background
The classical functions of the skeleton encompass locomotion, protection and mineral homeostasis. However, cell-specific gene deletions in the mouse and human genetic studies have identified the skeleton as a key endocrine regulator of metabolism. The bone-specific phosphatase, Phosphatase, Orphan 1 (PHOSPHO1), which is indispensable for bone mineralisation, has been recently implicated in the regulation of energy metabolism in humans, but its role in systemic metabolism remains unclear. Here, we probe the mechanism underlying metabolic regulation by analysing Phospho1 mutant mice.
Results
Phospho1−/− mice exhibited improved basal glucose homeostasis and resisted high-fat-diet-induced weight gain and diabetes. The metabolic protection in Phospho1−/− mice was manifested in the absence of altered levels of osteocalcin. Osteoblasts isolated from Phospho1−/− mice were enriched for genes associated with energy metabolism and diabetes; Phospho1 both directly and indirectly interacted with genes associated with glucose transport and insulin receptor signalling. Canonical thermogenesis via brown adipose tissue did not underlie the metabolic protection observed in adult Phospho1−/− mice. However, the decreased serum choline levels in Phospho1−/− mice were normalised by feeding a 2% choline rich diet resulting in a normalisation in insulin sensitivity and fat mass.
Conclusion
We show that mice lacking the bone mineralisation enzyme PHOSPHO1 exhibit improved basal glucose homeostasis and resist high-fat-diet-induced weight gain and diabetes. This study identifies PHOSPHO1 as a potential bone-derived therapeutic target for the treatment of obesity and diabetes.
Keywords: PHOSPHO1, Osteocalcin, Choline, Bone, Energy metabolism, Insulin, Endocrine organ, Skeleton, Obesity
Background
In addition to its classical structural functions, the skeleton is a site of significant glucose uptake and is involved in the regulation of whole-body glucose metabolism [1–9]. Osteocalcin (OCN) is the most abundant osteoblast-specific non-collagenous protein derived from bone and is thought to maintain the mechanical properties of the bone matrix by regulating calcium binding when fully carboxylated (GLA13-OCN) [10]. However, when OCN is not γ-carboxylated (uncarboxylated (GLU-OCN) or undercarboxylated (GLU13-OCN)), it is released from bone into the circulation where it is able to regulate whole-body glucose metabolism in an endocrine manner [7, 11–14]. Mice deficient in OCN have increased fat mass and are hyperglycemic, hypoinsulinemic and insulin-resistant in muscle. Furthermore, serum GLU17-OCN (human form of GLU13) levels and β-cell function show an inverse correlation with glycated haemoglobin (HbA1c), fat mass and plasma glucose levels [15–19]. Osteoblasts regulate glucose metabolism through OCN-dependent and independent mechanisms [20, 21]. An alternative candidate is the bone-specific cytosolic phosphatase, Phosphatase, Orphan 1 (PHOSPHO1) [22–29]. PHOSPHO1 initiates bone matrix mineralisation, and PHOSPHO1 deficiency causes significant skeletal pathology, bowed long bones, osteomalacia and scoliosis in early life [30–33]. Within the osteoblast, PHOSPHO1 forms choline and inorganic phosphate (Pi) from the hydrolyses of phosphocholine (PCho). The liberated Pi is incorporated into hydroxyapatite during the mineralisation process whereas the choline may alter glucose homeostasis. Elevated levels of choline result in insulin resistance in mice, and choline supplementation induces hyperglycaemia and insulin intolerance in mice via the modulation of plasma glucagon [34].
A number of recent studies have implicated PHOSPHO1 in the regulation of energy metabolism in humans [35–38]. Within the PHOSPHO1 gene, differential methylation sites have been identified as potentially useful biomarkers for clinical application in the early detection of type 2 diabetes [35] and significant associations between methylation at loci within the PHOSPHO1 gene and the future risk of type 2 diabetes exist [36, 37]. Differential methylation in PHOSPHO1 was associated with three lipid traits (total cholesterol, high-density lipoprotein cholesterol, and triglycerides) [39, 40]. Most recently, genetic variants of PHOSPHO1 in a bivariate twin study were found to be associated with body mass index and waist-hip ratio [38]. Taken together, these findings suggest that in addition to the established role of PHOSPHO1 in biomineralisation of the skeleton and dentition, Phospho1 ablation may result in improved glucose homeostasis and a reduction in metabolic disease susceptibility. We hypothesised that bone-derived choline may be an important regulator of global metabolism. To address this, we examined the metabolic phenotype of juvenile and adult Phospho1−/− mice.
Results
Phospho1 inactivation improves glucose tolerance and insulin sensitivity in juvenile mice
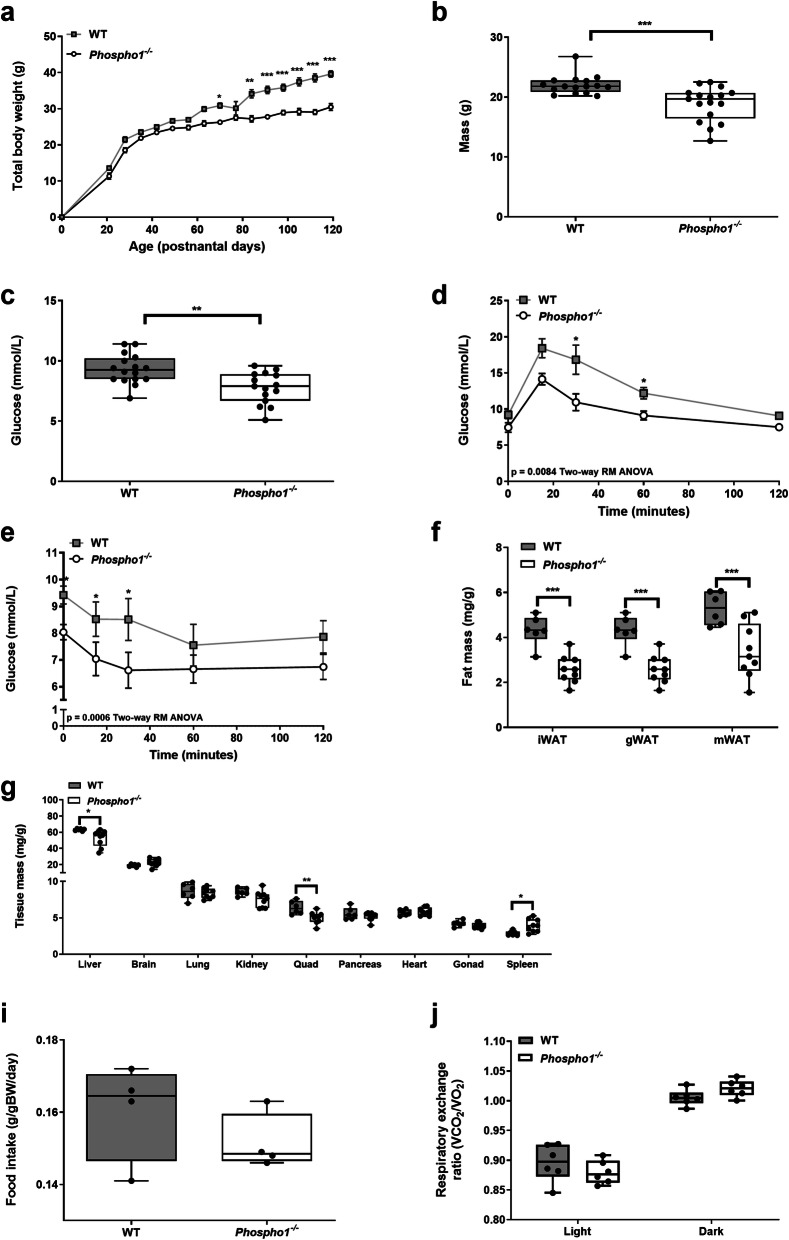
Growth of Phospho1−/− mice was decreased compared to wild-type (WT) mice (Fig. 1a). Juvenile Phospho1−/− mice (35-day-old) had reduced body weight and blood glucose levels (WT, 9.31 ± 0.31 mmol/L; Phospho1−/−, 7.76 ± 0.32 mmol/L; p < 0.01) (Fig. 1b, c), improved glucose tolerance (Fig. 1d) and whole body insulin sensitivity compared to WT counterparts (Fig. 1e). Consistent with this, adipose depots were smaller in Phospho1−/− mice: inguinal (iWAT; WT, 4.31 ± 0.27 mg/g; Phospho1−/−, 2.58 ± 0.20 mg/g; p < 0.001), mesenteric (mWAT; WT, 5.30 ± 0.30 mg/g; Phospho1−/−, 3.39 ± 0.40 mg/g; p < 0.01) and gonadal (gWAT; WT, 4.31 ± 0.27 mg/g; Phospho1−/−, 2.58 ± 0.20 mg/g; p < 0.001) adipose tissue (Fig. 1f). Phospho1−/− mice had significantly smaller livers (WT, 64.36 ± 0.49 mg/g; Phospho1−/−, 52.71 ± 3.37 mg/g; p < 0.05), and quadriceps (WT, 6.39 ± 0.36 mg/g; Phospho1−/−, 5.01 ± 0.26 mg/g; p < 0.01) (Fig. 1g). Food intake (WT, 0.13 ± 0.01 g/gBW/day; Phospho1−/−, 0.12 ± 0.01 g/gBW/day) (Fig. 1i), activity (Additional file 1: Fig. S1) and energy expenditure (day and night respiratory exchange rate (RER)) (Fig. 1j) were comparable between genotypes.
Fig. 1.
Juvenile Phospho1−/− mice display increased insulin sensitivity and decreased fat mass. Phospho1−/− mice showed decreased (a, b) growth and live weight at 35 days of age and (c) fasting glucose, (d, e) improved glucose and insulin tolerance and (f) decreased adipose tissue. Notable differences in tissue mass were also observed in the liver, quadriceps and spleen. These changes were not a consequence of (i) altered food intake or (j) energy expenditure. Data in (a) and are shown as mean ± SEM of 16–17 mice per group, (b) mean ± SEM 6–8 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001 and the data were assessed as follows: (a, d, e) repeated measures two-way ANOVA with Sidak’s multiple comparisons test; (b, c) two-tailed t test; (f, g) two-tailed unpaired t test; (i) two-tailed Mann–Whitney test; (j) two-tailed unpaired t test
Phospho1 deficiency protects from diet-induced diabetes in adult mice
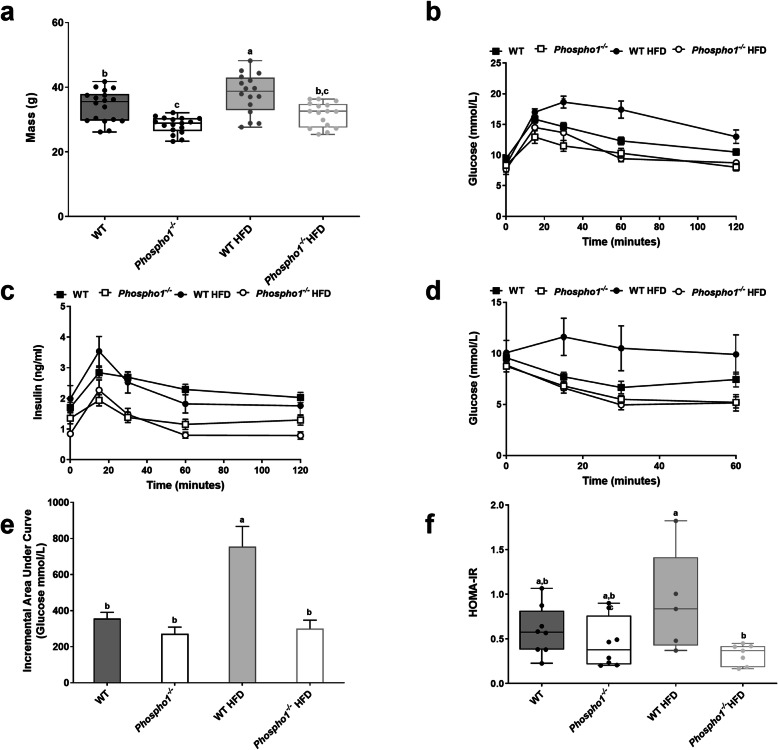
We next fed WT and Phospho1−/− mice a chronic high fat diet (HFD) from weaning until adulthood (120 days of age). Adult Phospho1−/− mice maintained a lower body weight when fed the HFD compared to WT HFD mice (control diet (CD)—WT, 34.20 ± 1.12 g; Phospho1−/−, 28.30 ± 0.59 g; HFD—WT: 38.0 ± 1.54 g, Phospho1−/−: 32.4 ± 1.26 g; p < 0.05; Fig. 2a). Fasting glucose levels were raised in WT mice fed a HFD but not in Phospho1−/− mice (CD—WT, 9.50 ± 0.37 mmol/l; Phospho1−/−, 8.59 ± 0.27 mmol/l; HFD—WT, 10.3 ± 0.53 mmol/l; Phospho1−/−, 9.27 ± 0.77) (Additional file 2: Fig. S2).
Fig. 2.
Phospho1−/− mice are protected from glucose intolerance. a Body mass. b Glucose tolerance test (GTT). c Glucose stimulate insulin secretion (GSIS). d Insulin tolerance test (ITT). e Incremental area under the curve for GTT. f HOMA-IR. All data are shown as mean ± SEM. b–e 5–8 mice per group. Different letters above the error bar show significant difference at p < 0.05 and the data were assessed as follows: a, e, f Two-way ANOVA with multiple comparisons; b–d repeated measures two-way ANOVA with Sidak’s multiple comparisons test
Glucose tolerance was improved in Phospho1−/− mice after chronic HFD (Fig. 2b). Insulin secretion across the glucose tolerance test (GTT) was also lower in Phospho1−/− mice on both CD and HFD, suggestive of insulin sensitisation rather than exaggerated β-cell insulin secretion as the major basis of the phenotype (Fig. 2c). This was confirmed with insulin tolerance tests (ITT) after chronic HFD, which revealed greater glucose disposal in Phospho1−/− mice (Fig. 2d–f).
Phospho1 deficiency protects from diet-induced obesity in adult mice
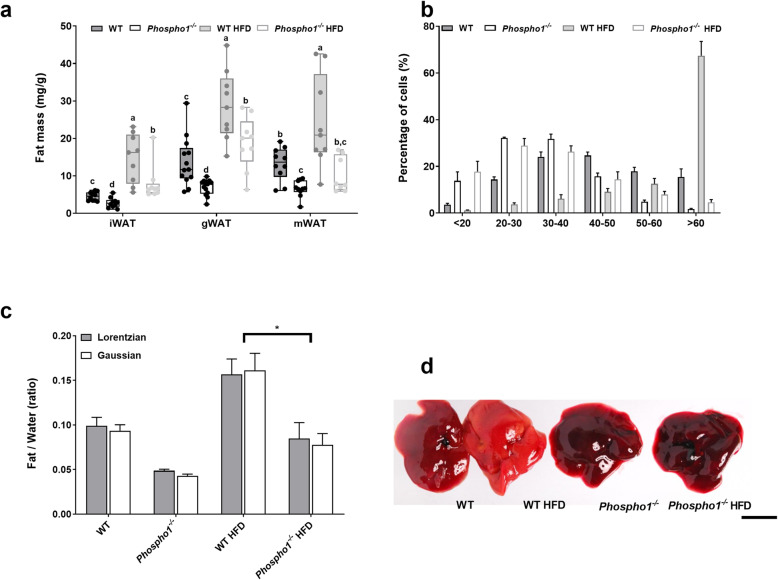
The insulin sensitivity observed in Phospho1−/− mice was consistent with the finding of smaller inguinal (CD—WT, 4.51 ± 0.37 mg/g BW; Phospho1−/−, 2.79 ± 0.42 mg/g BW; HFD—WT, 14.67 ± 2.12 mg/g BW, Phospho1−/−, 7.95 ± 1.56 mg/g BW; p < 0.01), mesenteric (CD—WT, 13.2 ± 1.34 mg/g BW; Phospho1−/−, 5.56 ± 1.61 mg/g BW; HFD—WT, 24.14 ± 4.05 mg/g BW; Phospho1−/−, 10.22 ± 1.57 mg/g BW; p < 0.01) and gonadal (CD—WT, 13.7 ± 1.81 mg/g BW; Phospho1−/−, 6.96 ± 0.58 mg/g BW; HFD—WT, 28.77 ± 3.12 mg/g BW; Phospho1−/−, 18.78 ± 2.37 mg/g BW; p < 0.01) fat depots noted in CD and HFD Phospho1−/− mice at necropsy (Fig. 3a). Moreover, confirmation that Phospho1−/− mice did not become obese when fed a HFD was shown by μMRI (Additional file 3: Fig. S3). These observations were also not explained by altered activity or increased food intake in 120-day-old adult male mice (data not shown).
Fig. 3.
Phospho1−/− are protected from NAFLD. a Fat analysis of 120-day-old WT and Phospho1−/− mice on both a control and HFD. b Quantification of gonadal fat adipocyte diameter. c Quantitative assessment of liver fat utilising spectroscopy. d Gross livers of representative mice left to right (WT, WT HFD, Phospho1−/−, Phospho1−/− HFD; scale bar = 10 mm). All data are represented as mean ± S.E.M. b, c n = 3–4 mice per group. *p < 0.05. Different letters above the error bar show significant difference at p < 0.05 and the data were assessed as follows: a–c Two-way ANOVA with multiple comparisons
Histological analysis revealed smaller gonadal adipocytes in Phospho1−/− mice fed both a CD and a HFD. High fat feeding had no significant effect on gonadal adipocyte size in Phospho1−/− mice; however, it significantly increased the number of large adipocytes (> 60 μm in diameter) in WT mice (p < 0.0001) (Fig. 3b). Phospho1−/− mice were also protected from the pronounced hepatic fat accumulation that was noted in WT mice following HFD feeding (Fig. 3c, d).
Insulin sensitivity in Phospho1−/− mice is independent of elevated adiponectin serum levels
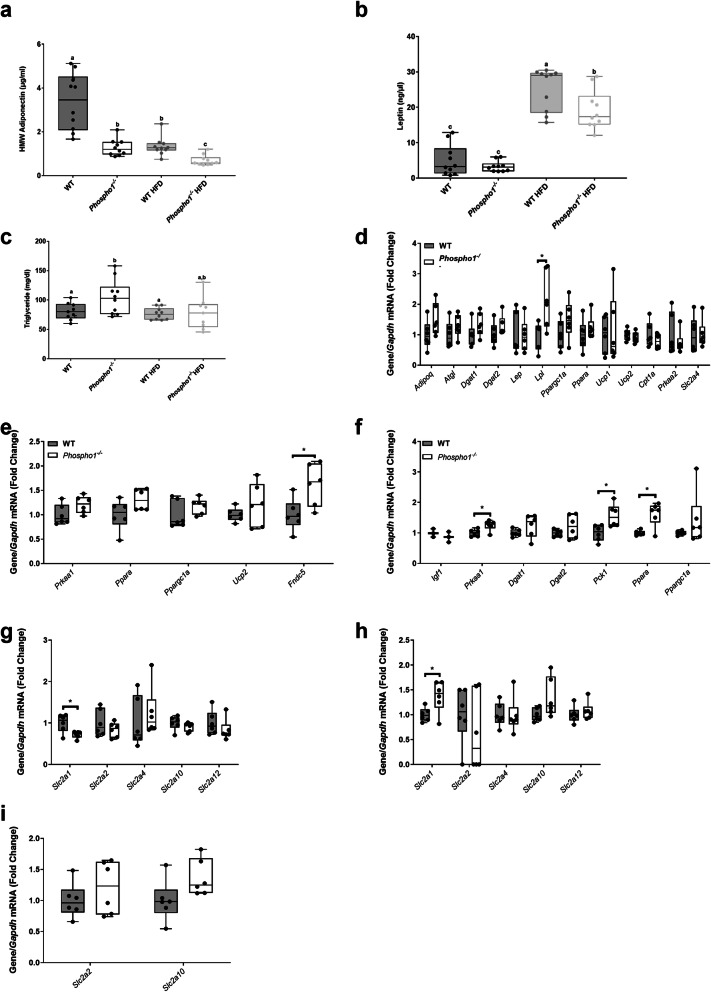
In an attempt to uncover the mechanism(s) responsible for the increased insulin sensitivity in Phospho1−/− mice, serum levels of adiponectin and leptin were measured. Levels of high molecular weight adiponectin, a hormone linked to insulin-sensitisation [41], were decreased in Phospho1−/− mice fed either a CD (2.62-fold) or a HFD (1.92-fold) (both p < 0.001) suggesting that insulin sensitivity and protection from obesity are independent of adiponectin (Fig. 4a). The observed decrease in circulating adiponectin in Phospho1−/− mice was not due to decreased bone marrow adipose tissue, an endocrine organ that contributes significantly to serum adiponectin (Additional file 4: Fig. S4) [42].
Fig. 4.
Phospho1−/− mice are insulin sensitive despite decreased adiponectin. a Adiponectin, b leptin and c triglyceride serum quantification. RT-qPCR analysis of tissue extracted from 120-day-old WT and Phospho1−/− mice, d adipose tissue, e quadriceps femoris and f liver. RT-qPCR analysis of GLUT receptors from g adipose tissue, h quadriceps femoris and i liver. Data are represented as mean ± S.E.M. * p < 0.05. Different letters above the error bar show significant difference at p < 0.05 and the data were assessed as follows: a–c Two-way ANOVA with multiple comparisons, d–i two-tailed Mann–Whitney test for non-normally distributed data and two-tailed unpaired t test for normally distributed data
Serum leptin levels in CD-fed mice were unaffected by Phospho1 deficiency, whereas in comparison to WT mice fed a HFD serum leptin levels were significantly decreased 1.31-fold (p < 0.05) in Phospho1−/− mice fed a HFD (Fig. 4b), accordant with reduced fat mass [43]. Phospho1−/− CD mice had increased circulating serum triglycerides compared to WT CD mice, but no change was observed in WT and Phospho1−/− mice fed a HFD (Fig. 4c). Consistent with increased oxidative metabolism of carbohydrate and lipids in other peripheral tissues, mRNA levels of genes encoding key metabolic proteins were increased in adipose tissue (Lpl) muscle (Fndc5) and liver (Prkaa1, Pepck1 and Ppara) (Fig. 4d-f). The mRNA levels of genes encoding GLUT receptors (Slc2a1, 2, 4, 10 and 12) were largely unchanged (Fig. 4g-i).
Canonical thermogenesis does not underlie the metabolic protection observed in adult Phospho1 deficient mice
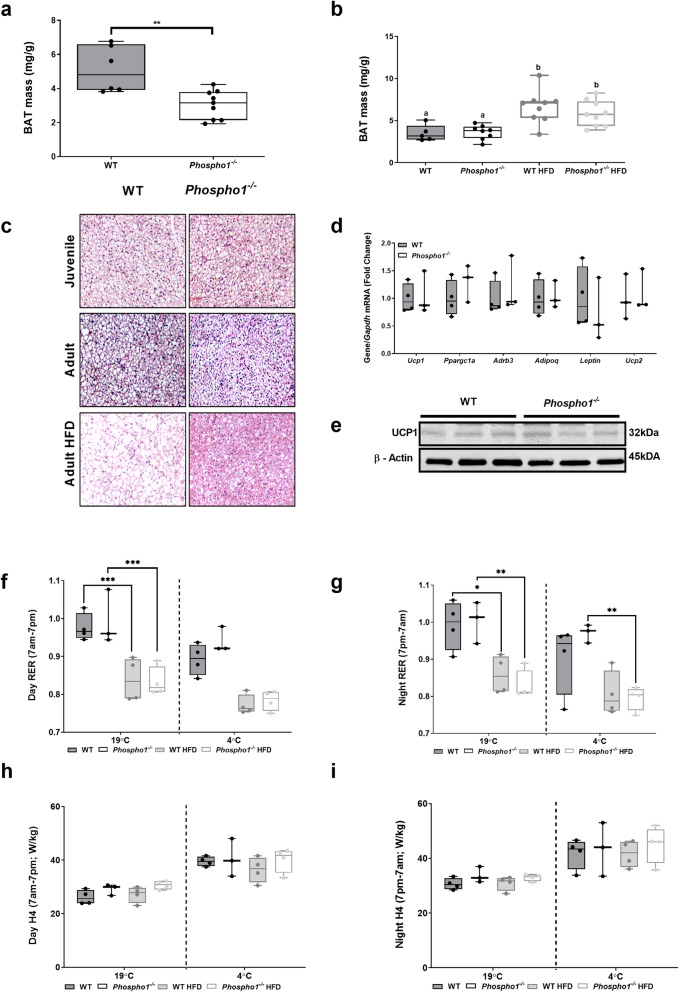
Thirty-five-day-old Phospho1−/− mice had decreased interscapular brown adipose tissue (BAT) mass compared to WT counterparts (WT, 5.11 ± 0.57 mg/g; Phospho1−/−, 3.05 ± 0.40 mg/g; p < 0.01) (Fig. 5a). However this reduction in BAT mass did not persist to adulthood nor during high-fat feeding (Fig. 5b). Strikingly, adult and high-fat-fed Phospho1−/− mice had smaller brown adipocytes compared to WT controls (Fig. 5c). In order to see if BAT activation and thermogenesis might be responsible for the observed phenotype, we measured key brown fat genes including uncoupling protein 1 (Ucp1) (Fig. 5d, e); no differences were observed in the mRNA and protein levels. Furthermore, there were no significant differences in respiratory exchange ratio (RER, indicative of metabolic substrate preference) or energy expenditure between WT or Phospho1−/− mice fed either a chow or HFD housed at either room temperature or during cold exposure (4 °C) (Fig. 5f-i). These in vivo data show that increased canonical thermogenesis does not underlie the metabolic protection observed in the Phospho1-deficient mice so this line of investigation was not pursued further.
Fig. 5.
Canonical thermogenesis does not underlie the metabolic protection observed in the adult Phospho1-deficient mice. Brown adipose tissue (BAT) mass in a juvenile (35-day-old) and b adult (120-day-old) WT and Phospho1−/− mice. c Representative micrographs of BAT from WT and Phospho1−/− mice. d Brown fat gene expression and e UCP1 protein analysis. f–i Insulin sensitivity and protection from diet induced obesity in Phospho1−/− mice was not a consequence of altered energy expenditure (RER—respiratory exchange ratio; H4 = H3 (W)/lean mass (kg)). Data are represented as mean ± S.E.M. *p < 0.05, **p < 0.01, ***p < 0.001. Different letters above the error bar show significant difference at p < 0.05 and the data were assessed as follows: a, d two-tailed Mann–Whitney test. b, d, f–i Two-way ANOVA with multiple comparisons
Diabetes mellitus-associated genes are enriched in Phospho1−/− primary osteoblasts
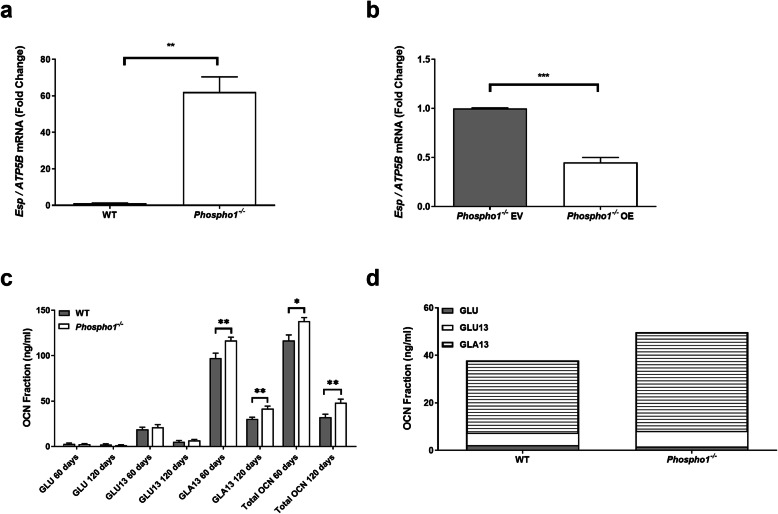
We next sought to unravel the genetic circuitry responsible for the improved glucose tolerance in Phospho1−/− mice. To address this, a transcriptomic analysis of Phospho1-deficient osteoblasts was completed. There was a striking 20-fold upregulation of embryonic stem cell phosphatase (Esp) mRNA, the gene encoding the protein osteotesticular protein tyrosine phosphatase (OST-PTP) in Phospho1-deficient mice [7]. These data were validated in primary WT and Phospho1−/− calvarial osteoblasts and Phospho1-deficient osteoblast overexpressing Phospho1 (Fig. 6a, b). Protein tyrosine phosphatases are recognised master regulators of insulin receptor signalling (INSR), negatively modifying osteoblast-insulin signalling and thereby controlling GLU13-OCN release [7, 44–48]. The identification of elevated Esp expression in Phospho1−/− mice was strongly suggestive of a reciprocal regulation between OST-PTP and PHOSPHO1 in the control of glucose homeostasis. This increased Esp expression was however inconsistent with the improved glucose tolerance in the Phospho1−/− mice. Therefore in an attempt to reconcile this anomaly, we measured circulating GLU–OCN and GLU13-OCN, which were found to be unchanged in juvenile and adult Phospho1−/− mice (Fig. 6c). Only GLU–OCN and GLU13-OCN present in the serum have been shown to act as a hormone [5, 49]. Therefore, these data implied that elevated serum levels of uncarboxylated or undercarboxylated OCN did not mediate the improved metabolic phenotype (Fig. 6c, d). The elevated levels of carboxylated and total OCN in Phospho1 deficiency was consistent with increased bone turnover in these mice as previously reported [30].
Fig. 6.
Osteocalcin-independent mechanism of PHOSPHO1-regulated energy metabolism. To assess the relative change in Esp mRNA expression in primary calvarial osteoblasts, RT-qPCR was conducted to compare a primary WT and Phospho1−/− osteoblasts and b Phospho1−/− osteoblast transfected with empty (EV) or overexpressing (OE) vectors. c, d Osteocalcin content of serum from WT and Phospho1−/− mice at 60 and 120 days of age. Data are represented as mean ± S.E.M n = 5–6 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001 and the data were assessed as follows: a–c Two-tailed Mann–Whitney test or two-tailed unpaired t test
Phospho1−/− calvarial osteoblasts show a greater metabolic capacity in utilising exogenous substrates compared to WT osteoblasts
Primary calvarial osteoblast metabolism analysis revealed that differentiated Phospho1−/− osteoblasts had elevated basal and FCCP (protonophoric uncoupler) induced oxygen consumption rates (indicative of oxidative phosphorylation) compared to WT osteoblasts when supplied exogenously with glucose, pyruvate and glutamine (Additional file 5: Fig. S5). There was a significant increase in the glycolytic rate of both the non-differentiated and differentiated Phospho1−/− osteoblasts compared to WT osteoblasts suggesting that they have increased glucose metabolism. Based on these observations, we conclude that differentiating Phospho1−/− calvarial osteoblasts show a greater metabolic capacity in utilising exogenous substrates compared to WT osteoblasts.
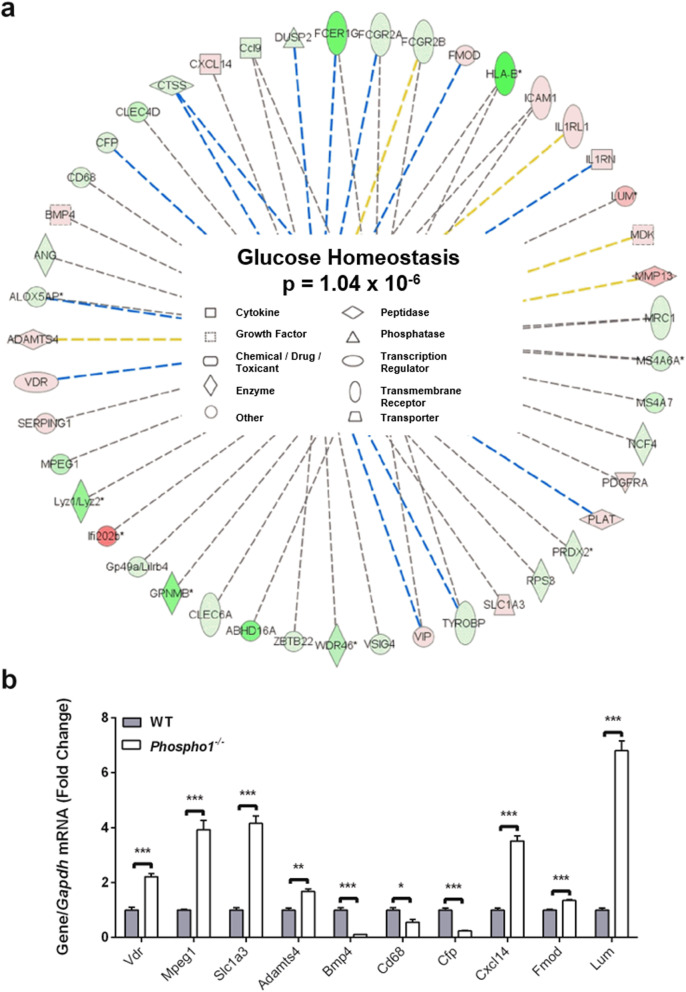
To further clarify the genetic pathways underpinning these observations, 22 differentially expressed genes identified from the microarray analysis were found by Ingenuity Pathway Analysis (IPA) to be associated with glucose homeostasis (Additional file 6: Table. S6). Further predictive analysis of the differentially expressed genes of the original microarray data set identified > 40 genes to be associated with energy metabolism (p = 1.04 × 10−6) (Fig. 7a) of which 10 were found to be associated with both diabetes and bone, following an NCBI (PubMed) in silico search. Validation by RT-qPCR confirmed that 70% of the genes predicted by IPA (Vdr, Slc1a3, Adamts4, Cd68, Cfp, Fmod and Lum) were differentially regulated in Phospho1-deficient osteoblasts (p < 0.05; Fig. 7b and Additional file 7: Table. S7). Furthermore, GeneMANIA network analysis predicted that Phospho1 both directly and indirectly interacts with 36 genes associated with glucose transport and metabolic processes and insulin receptor signalling (Additional file 8: Fig. S8). Of the output genes, Atf4, Foxo1 and Insr are recognised skeletal modulators of energy metabolism, suggesting crosstalk between Phospho1 and other metabolic regulatory genes [5, 50, 51].
Fig. 7.
Ingenuity Pathways Analysis network summary predictions. a Ingenuity Pathways Analysis was used to predict further genes associated with glucose homeostasis based upon the 22 genes found to be differentially expressed in the microarray. b Predicated genes were analysed by RT-qPCR in WT and Phospho1−/− primary calvarial osteoblast’s. Results were normalised to the Atp5b housekeeping gene. Data are represented as mean ± S.E.M (n = 3 replicates). *P < 0.05, **P < 0.01, ***P < 0.001. Red = Upregulated. Green = Downregulated (the darker the shade of green and red colour indicates a more extreme up/down regulation, conversely the paler the shade indicates a more subtle up/down regulation. Dashed line = indirect interaction (blue = inhibition, yellow = findings underlying the relationship are inconsistent with the state of the downstream node, grey = Ingenuity Pathways Analysis prediction) and the data were assessed in (b) using a two-tailed unpaired t test
Identification of differentially expressed serum proteins in Phospho1−/− mice
To explore the secretome profile, quantitative SWATH (sequential window acquisition of all theoretical spectra) MS (mass spectrometry) proteomics [52] was conducted on serum from WT and Phospho1−/− mice fed CD and HFD. Differentially expressed proteins (> 100) were identified in HFD Phospho1−/− serum compared to HFD-fed WT mice. These proteins were highly associated with glycolysis, gluconeogenesis and ‘metabolic pathways’ (Additional file 9: Table. S9). Pathway and network analysis predicted that the identified proteins interacted with miR-34a, a microRNA that is known to affect diverse parts of insulin signalling in the pancreas, liver, muscle and adipose tissue [53].
Bone-derived choline is involved in global energy regulation
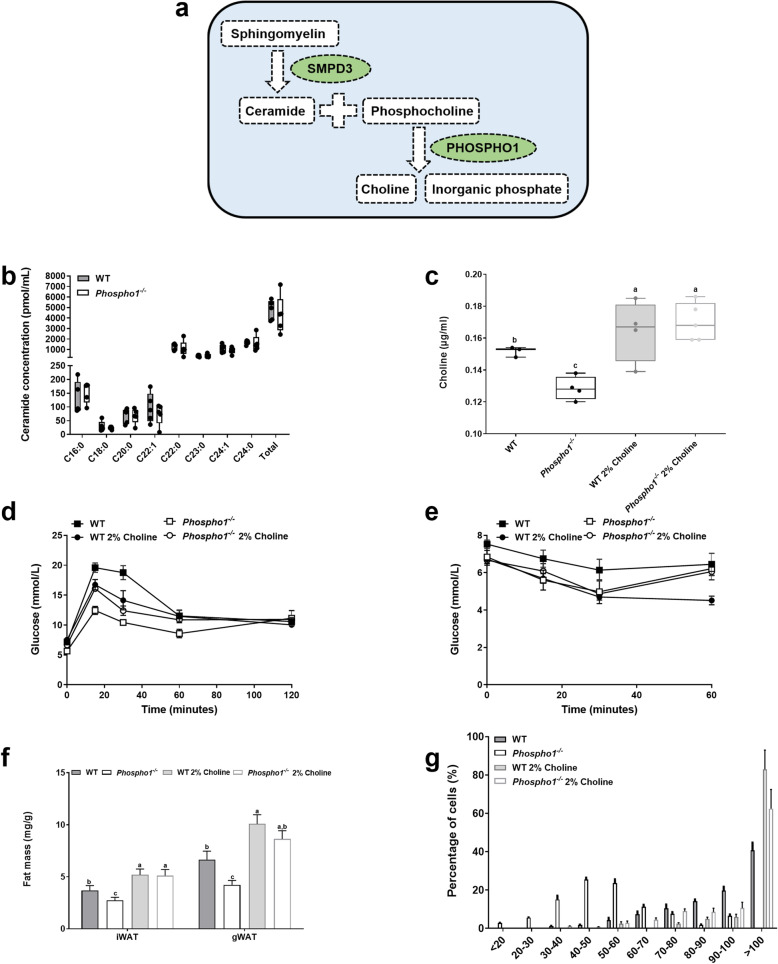
Neutral sphingomyelinase 2 (nSMase2) catalyses the hydrolysis of sphingomyelin to form ceramide and PCho [54]. Furthermore, PCho is the preferred substrate for PHOSPHO1 yielding choline and Pi (Fig. 8a) [55]. As elevated levels of both ceramide and choline result in insulin resistance in mice [34, 56] we targeted these intermediates to establish if alterations in their serum levels could explain the insulin sensitive phenotype in Phospho1−/− mice [34, 56]. The levels of various ceramide species were unchanged (Fig. 8b); however, Phospho1−/− mice had a significant decrease in serum choline levels (WT, 0.152 ± 0.001 μg/ml; Phospho1−/−, 0.128 ± 0.003 μg/ml; p < 0.01) (Fig. 8c) which were normalised upon choline supplementation. Supplementation of WT and Phospho1−/− mice with a 2% choline diet (a well-tolerated, palatable diet [57]), also normalised the insulin sensitivity measured in Phospho1−/−, measured by GTT (Fig. 8d). However, unlike WT mice which when fed a 2% choline diet took longer to recover from the insulin challenge, Phospho1−/− showed no metabolic change in response to insulin between the diets (Fig. 8e). Furthermore, choline supplementation normalised the lean phenotype observed in Phospho1−/− mice (Fig. 8f, g). These results support the notion that Phospho1 deficiency improves the metabolic profile of mice in vivo and confers resistance to obesity and diabetes in part via the alteration of serum choline levels.
Fig. 8.
Bone derived choline regulates insulin sensitivity. a Schematic diagram outlining the mechanisms by which ceramide and choline and linked. b Mouse serum ceramide and choline (c) analysis by LC-MS/MS (ceramide) and assay (choline). d GTT. e ITT. f Dissected fat depot weights. g Quantification of epididymal fat adipocyte diameter and representative histology. Data are represented as mean ± S.E.M (c–g) n = 3–5 per group. Different letters above the error bar show significant difference at p < 0.05 and the data were assessed as follows: c, f, g Two-way ANOVA with multiple comparisons. b One-way ANOVA with Dunnet’s or Tukey’s tests for multiple comparisons. d, e repeated measures two-way ANOVA with Sidak’s multiple comparisons test
Discussion
The fundamental observations presented in this study further strengthen the concept that the skeleton acts as an endocrine organ and provides empirical evidence for a critical role for PHOSPHO1 in energy metabolism. Phospho1 deficiency results in decreased blood glucose levels, improved insulin sensitivity, glucose tolerance and conferred protection from diet-induced obesity and diabetes in mice despite a 60-fold upregulation of Esp expression by Phospho1−/− osteoblasts. Mice lacking Esp in osteoblasts present with severe hypoglycaemia and hyperinsulinemia resulting in postnatal lethality in the first 2 weeks of life [7]. Conversely, mice overexpressing Esp exclusively in osteoblasts were glucose intolerant and insulin resistant [7, 44–48]. Intriguingly, the increased insulin sensitivity in Phospho1−/− mice was not associated with the expected rise in serum GLU13-OCN levels suggesting that PHOSPHO1-regulated energy metabolism is via OCN-independent mechanisms. This notion has previously been observed when partial genetic ablation of osteoblasts profoundly affected energy expenditure, gonadal fat weight and insulin sensitivity which were not restored by the administration of OCN [20, 21]. Nevertheless, it is possible that the increased insulin sensitivity noted in Phospho1−/− mice may be primed by an initial rise in GLU13-OCN levels, which is eventually normalised in a compensatory manner by the observed increase in Esp expression. This being the case we would predict that the loss of Esp on a Phospho1−/− background would exacerbate the insulin sensitivity due to increased GLU13-OCN serum levels. These data strengthen the concept that a novel pathway exists between osteoblasts and glucose homeostasis; however, it does highlight the potential cross-talk between OCN-dependent and OCN-independent mechanisms of glucose metabolism.
The role of PHOSPHO1 in controlling bone mineralisation has been extensively investigated through the use of both in vitro and in vivo mouse models. Crucial for the initiation of mineralisation within matrix vesicles, PHOSPHO1 hydrolyses membrane lipid derivatives, primarily PCho to produce Pi (utilised in hydroxyapatite formation) and choline [55]. Phosphocholine can be formed from choline via choline kinase activity or phosphatidylcholine via PLA2 and ENPP6 as well as from the hydrolysis of sphingomyelin, via nSMase2 to form PCho and ceramide [58]. Mindful of this, it has been reported that elevated levels of both ceramide and choline result in insulin resistance in mice [34, 56]. We saw no change in ceramide species in Phospho1−/− mice; however, there was a significant decrease in serum choline levels in Phospho1−/− mice, which was normalised in Phospho1−/− mice fed a 2% choline rich diet resulting in a normalisation in insulin sensitivity and fat mass. This study highlights for the first time the importance of bone-derived choline in the regulation of energy metabolism; however, it remains clear that choline supplementation alone does not fully correct all metabolic defects in Phospho1−/− mice.
The regulation of global energy metabolism by the skeleton is a complex, multifactorial process, and it is therefore likely that presently undefined bone secreted factors may have a significant role in PHOSPHO1’s ability to regulate global energy. We identified > 100 secreted proteins that may contribute to the energy regulation via the skeleton. These unique proteins were highly associated with glycolysis, gluconeogenesis and ‘metabolic pathways’ and association with miR-34a, a microRNA that affects diverse parts of insulin signalling in the pancreas, liver, muscle and adipose tissue [53, 59]. Furthermore, lumican, a proteoglycan secreted by differentiating osteoblasts and a constituent of the bone matrix was found to be enriched in serum of Phospho1−/− mice by both proteomic and microarray analysis. Interestingly, lumican has also been observed in the decidua of diabetic patients [60, 61]. Further investigations of these and other candidates may uncover new skeletal regulators of energy metabolism that act via osteocalcin independent mechanisms.
The metabolic phenotype observed in the Phospho1-deficient mice was not due to increased BAT activation despite striking histological differences between WT and Phospho1−/− tissue. BAT is a thermogenic organ that increases energy expenditure to generate heat, maintaining body temperature in a cold environment [62]. When activated by cold exposure or through activation of β3-adrenergic receptors, BAT improves insulin sensitivity and lipid clearance, highlighting its key role in metabolic health [63]. PHOSPHO1 has been previously suggested to have a role in murine BAT function. Increased expression of Phospho1 in both isolated brown and white adipocytes was observed following treatment with the β-adrenergic agonist (CL316,243) compared to placebo-treated white adipocytes [64]. Mice with an adipose-specific defect in fatty acid oxidation (Cpt2A−/−) showed a loss of β3-adrenergic induced Phospho1 expression in BAT [64] and expression of Phospho1 was shown to be elevated in Ucp1-deficient animals; however, WT mice had very low Phospho1 expression in BAT and no detectable PHOSPHO1 was observed in WT iWAT and BAT [65]. No difference in Phospho1 gene expression was observed between human supraclavicular and subcutaneous adipose progenitor cells (GEO Dataset: GDS5171/8016540) and murine WAT had elevated Phospho1 gene expression compared to BAT (GEO Dataset: GDS2813/1452485_at). In our hands, we have noted high expression of Phospho1 in murine BAT but no protein expression was detected (Additional file 10: Fig. S10). Taken together, these data suggest that Phospho1 is likely to play a role in BAT function; however, the BAT phenotype observed in Phospho1 deficient mice does not appear to underlay the metabolic protection we see in these animals. Further studies are necessary to unravel the role of PHOSPHO1 in BAT using a BAT conditional knock-out model of Phospho1.
Conclusions
Collectively, the results of this study add further credibility to the concept that GLU13-OCN is not the sole mediator of the endocrine function of the skeleton [20]. We suggest, as others have, that further undefined bone-derived proteins/lipids work in partnership with OCN to regulate the metabolic function of the skeleton and affect other metabolic organs such as muscle and liver [20]. Indeed, this study has identified other potential protein mediators and raised the possibility that bone-derived choline may contribute to the regulation of the development of the metabolic syndrome. Furthermore, our results suggest that Esp may act as a fine controller of insulin sensitivity in mice, offering protection from severe hypoglycaemia and dyslipidaemia.
Several previous reports have suggested an association between PHOSPHO1 expression in disorders of altered energy metabolism such as obesity and diabetes [35–39]. The data from this present study is both supportive of such an association but also provides insight into the mechanisms by which PHOSPHO1 may contribute to the regulation of energy metabolism, inclusive of insulin sensitivity, glucose tolerance and fat metabolism. Also, inhibitors of PHOSPHO1 activity such as the proton pump inhibitor lansoprazole, commonly prescribed to control and prevent symptoms of gastroesophageal reflux disease and dyspepsia, have been associated with improved glycaemic control in diabetic patients [28, 66–68]. Taken together, the identification of PHOSPHO1 in the role of energy metabolism in both the human and mouse offers the potential to manipulate key targets of the PHOSPHO1 pathway to improve metabolic health [36, 37].
Methods
Aim
Examine glucose metabolism and the energy status of Phospho1−/− mice.
Reagents
All chemicals, tissue culture medium and buffers were from Sigma-Aldrich (Dorset, UK) and Invitrogen (Paisley, UK) unless otherwise stated. PCR oligonucleotides were purchased from MWG Eurofins (Ebersberg, Germany) and Primer Design (Southampton, UK). PHOSPHO1 HuCAL Fab bivalent antibody was purchased from AbD Serotech (Kidlington, UK). All antibodies were diluted 1:1000 unless otherwise noted.
Animals
Phospho1 null mice were generated as previously described [31]. Offspring carrying the mutant Phospho1 gene were identified by genotyping (F: 5′-TCCTCCTCACCTTCGACTTC-3′, R: 5′-TCCTCCTCACCTTCGACTTC-3′). All in vivo studies were conducted at 120 days of age unless otherwise stated. Male mice were fed a high fat diet consisting of 58% of calories from fat (DBM Scotland, Broxburn, UK) or control diet (6.2% calories from fat; Harlan Laboratories, Indianapolis, IN, USA) starting at 4 weeks of age. Male mice were fed a 2% supplemented choline diet (Harlan Laboratories) or control diet (Harlan Laboratories) for 5 weeks prior to cull at 120 days. Ad libitum food consumption was monitored for 6 days and basal nocturnal activity was quantified using an AM524 Single Layer X, Y IR activity monitor and associated Amonlite software (Linton Instrumentations, Norfolk, UK). Juvenile metabolic activity was measured using indirect calorimetry (Oxymax Lab Animal Monitoring System: CLAMS (Columbus Instruments, OH USA). Adult metabolic rate was measured using indirect calorimetry (TSE PhenoMaster 1.0, with software version 6.1.9). Cold exposed mice were first housed in these cages for 3 days at room temperature (RT) for acclimation and baseline measurements. Mice were then house for 72 h at 4 °C. All experiments were conducted blind to the operator. Animals were maintained under conventional housing conditions with a 12-h light/dark cycle with free access to food and water (except when food was restricted during fasting). All animal experiments were approved by The Roslin Institute’s Animal Users Committee, and the animals were maintained in accordance with UK Home Office guidelines for the care and use of laboratory animals.
Metabolic studies
Male juvenile and adult (35 and 120 days old respectively) were weighed and fasted for 4 h between 9 am and 1 pm. Prior to the start of the tests, a basal blood sample was collected by venesection into EDTA powder-coated capillary tubes (Starstedt, Leicester, UK). Basal glucose levels were measured using a glucose monitoring system (Accu-Chek® Aviva, Roche, Leicester, UK). Two milligrammes of D-glucose (Sigma, Poole, UK) per g of body weight was administered by gavage or 0.5 mU of insulin (Actrapid, NovoNordisk, Bagsvaerd, Denmark) per g/body weight was administered intraperitoneally (i.p.). At precisely 15, 30, 60 and 120 min following administration, blood glucose was measured with an Accu-Chek® Aviva glucose meter (Roche Diagnostics Ltd., Lewes, UK) and insulin was measured by ELISA (ChrystalChem, Chicago, IL, USA). Animals were allowed to recover for 2 weeks prior to euthanasia. Tissues were collected for protein, gene and histological analysis.
Serum measurements
Serum samples were prepared from blood collected by heart puncture of CO2 culled mice in the fasted state. Total, carboxylated (GLA13-OCN), undercarboxylated (GLU13-OCN) and uncarboxylated (GLU-OCN) osteocalcin [49], adiponectin and leptin (CrystalChem) were quantified by ELISA.
Primary osteoblast isolation and culture
Under sterile conditions, calvaria were isolated from 2- to 4-day-old new-born WT and Phospho1−/− mice as previously described [69]. Osteoblasts were expanded in flasks in growth medium consisting of α-MEM supplemented with 10% FBS and 1% gentamicin in a humidified atmosphere of 95% air/5% CO2 and maintained at 37 °C. When the cells reached 80–90% confluency, they were seeded at a density of 2.5 × 104/cm2 in multi-well plates. Conditioned medium was collected upon plate confluency, centrifuged to remove particulates and frozen at − 80 °C until required. For overexpression studies, primary osteoblasts were transfected with empty (EV) or overexpressing (OE) vectors as previously described [70].
Mito stress test
An XF24 Analyser (Agilent Technologies, Santa Clara, CA, USA) was used to measure the respiratory function of primary osteoblasts. Osteoblasts were plated at a density of 50,000 cells per well and transferred to a 37 °C CO2 incubator until and calvarial osteoblasts were differentiated using osteogenic differentiation media containing 8 mM β-glycerophosphate and 50 μg/ml ascorbic acid for 3 days. On the day of the assay, cells were washed in XF Assay Media supplemented with 25 mM glucose and 10 mM pyruvate and placed in a non-CO2 incubator at 37 °C for 1 h prior to start of assay. Reagents were prepared for the assay (injection volume of 75 μL for each reagent per well) from 2.5 mM Seahorse stock solutions, Oligomycin (1.2 μM). Following equilibration, the Seahorse plate was placed in the Seahorse XF24 Analyser for sample analysis. The raw data was normalised to protein content in each well at the end of the assay [71].
Gene expression analyses and immunoblotting
RNA extractions from tissues and cells were performed using the RNeasy Lipid Tissue Kit (Qiagen). The SuperScript First Strand Synthesis System (Invitrogen) was used for reverse transcription. Real-time PCR amplification with the 2 x precision master mix (Primer design, Southampton, UK) using the Stratagene Mx3000P real-time QPCR system (Agilent Technologies, Santa Clara, CA, USA). Each sample was tested in triplicate and compared to a housekeeping gene (Atp5B in osteoblasts and bone tissue, Lrp10 in adipose tissue and Gapdh in all other tissues) using MxPro software (Cheshire, UK), and the relative expression of the analysed genes was calculated using the ΔΔCT method [72]. All primer sequences are described in Table 1. Primers sequences are available in Table 1. For protein extraction, cells were scraped and tissues homogenised in an appropriate volume of radio-immunoprecipitation assay (RIPA) buffer containing 15% of complete mini protease inhibitor cocktail (Roche, Burgess Hill, West Sussex, UK). Protein concentration was determined by the Bio-Rad DC protein assay (Bio-Rad, Hertfordshire, UK). Immunoblotting was conducted with specific antibodies and protein bands were visualised using the enhanced chemiluminescence (ECL) Western Blotting Detection System (GE Healthcare, Chalfont St Giles, UK) or the Odyssey infrared detection system (LICOR). All antibodies are described in Table 2.
Table 1.
Sequences of primers used for qPCR
| Species | Transcript | Primer sequences 5′ to 3′ |
|---|---|---|
| M. musculus | Adamts4 | F: ATGGCCTCAATCCATCCCAG |
| R: GCAAGCAGGGTTGGAATCTTTG | ||
| M. musculus | Adipoq | F: GGCCGTTCTCTTCACCTACG |
| R: TGGAGGAGCACAGAGCCAG | ||
| M. musculus | Adrb3 | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Atgl | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Atp5b | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Bmp4 | F: ATTCCTGGTAACCGAATGCTG |
| R: CCGGTCTCAGGTATCAAACTAGC | ||
| M. musculus | Cd68 | F: CCATCCTTCACGATGACACCT |
| R: GGCAGGGTTATGAGTGACAGTT | ||
| M. musculus | Cfp | F: CTGCTACTGGTTATCCTGCCA |
| R: TCTACCCTGATGTCTCTCCCA | ||
| M. musculus | Cpt1a | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Cxcl14 | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Dgat1 | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Dgat2 | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Esp | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Fmod | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Fndc5 | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Igf1 | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Lep | F: ATTTCACACACGCAGTCGGTAT |
| R: GGTGGAGCCCAGGAATGAAG | ||
| M. musculus | Lpl | F: TCTGTACGGCACAGTGG |
| R: CCTCTCGATGACGAAGC | ||
| M. musculus | Lum | F: CTCTTGCCTTGGCATTAGTCG |
| R: GGTCATCACAGTACATGGCAGT | ||
| M. musculus | Mpeg1 | F: CTGGATGATAATAGCGTGTGCT |
| R: AAGACAGGTAGTTTCAGGGCA | ||
| M. musculus | Pck1 | F: CATATGCTGATCCTGGGCATAAC |
| R: CAAACTTCATCCAGGCAATGTC | ||
| M. musculus | Phospho1 | F: TTCTCATTTCGGATGCCAACA |
| R: TGAGGATGCGGCGGAATAA | ||
| M. musculus | Ppara | F: CCCTGAACATCGAGTGTCGA |
| R: AATAGTTCGCCGAAAGAAGCC | ||
| M. musculus | Ppargc1a | F: CCCTGCCATTGTTAAGACC |
| R: TGCTGCTGTTCCTGTTTTC | ||
| M. musculus | Prkaa1 | Primer Design, Southampton, UK; sequence not disclosed |
| M. musculus | Prkaa2 | F: GTCAAAGCCGACCCAATGATA |
| R: CGTACACGCAAATAATAGGGGTT | ||
| M. musculus | Slc1a3 | F: ACCAAAAGCAACGGAGAAGAG |
| R: GGCATTCCGAAACAGGTAACTC | ||
| M. musculus | Slc2a1 | F: TCAACACGGCCTTCACTG |
| R: CACGATGCTCAGATAGGACATC | ||
| M. musculus | Slc2a10 | F: ACCAAAGGACAGTCTTTAGCTG |
| R: ATCTTCCAAGCAGACGGATG | ||
| M. musculus | Slc2a12 | F: GGGTGTCAACCTTCTCATCTC |
| R: CCAAAGAGCATCCCTTAGTCTC | ||
| M. musculus | Slc2a2 | F: TGTGCTGCTGGATAAATTCGCCTG |
| R: AACCATGAACCAAGGGATTGGACC | ||
| M. musculus | Slc2a4 | F: CCAGTATGTTGCGGATGCTAT |
| R: TTTTAGGAAGGTGAAGATGAAGAAG | ||
| M. musculus | Ucp1 | F: GGATGGTGAACCCGACAACT |
| R: AACTCCGGCTGAGAAGATCTTG | ||
| M. musculus | Vdr | F: GAATGTGCCTCGGATCTGTGG |
| R: ATGCGGCAATCTCCATTGAAG |
Table 2.
Antibodies used for Western blotting
| Protein | Species | Source | Catalogue Number | Dilution | Band size (kDa) |
|---|---|---|---|---|---|
| PHOSPHO1 | Human | AbD Serotec | HCA093 | 1:1000 | 30 |
| UCP1 | Rabbit | Cell Signaling | #14670 | 1:1000 | 32 |
| β-actin (HRP-linked) | Mouse | Sigma | A3854 | 1:50,000 | 45 |
Tissue histology
Tissue was fixed in 4% PFA and embedded in paraffin wax. Five-micrometre sections were stained with haematoxylin and eosin (H&E) using the Leica Autostainer and mounted in DePeX (VWR, Lutterworth, UK). Adipocyte diameter and pancreatic β-cell islet number and size were quantified using ImageJ software as previously described [7, 73].
Osmium staining
Mouse tibiae were fixed in 10% neutral-buffered formalin and decalcified in 14% EDTA, pH 7.4. Mouse bones were stained according to [74]. Briefly, bones were stained with a 1% osmium tetroxide solution for 48 h at room temperature. Bones were washed in Sorensen’s buffer and embedded in 1% agarose prior to μCT scanning (μCT100 Scanco Medical, Bassersdorf, Switzerland) − 12 μm, medium resolution, 70 kVp, 114 μA, 0.5 mm AL filter and integration time 500 ms. Analysis was performed using the manufacturer’s software.
Micro-magnetic resonance, computed tomography and liver spectroscopy
CD and HFD mice were sacrificed immediately before imaging. For micro-magnetic resonance, mice were imaged on a Varian 7 Tesla magnet using VnmrJ Pre-Clinical MRI Software. T2-weighted images were acquired both in the axial (1 mm thickness, 192 × 192 pixels, TR – 3000 ms, TE 24 ms, 1 average, FOV – 38.4 × 38.4) and coronal planes (0.5 mm thickness, 512 × 256 pixels, TR – 3000 ms, TE 24 ms, 4 averages, FOV – 102.4 × 51.2). Liver spectroscopy was conducted on user defined areas (TR - 1800 ms, TE – 11.5 ms, 16 averages, Vauxhall 3x3x3). Lorentzian and Gaussian lineshape were used to fit peaks to MR data (jMRUI http://www.mrui.uab.es/mrui/mrui).
Microarray and pathway analysis
Labelled cRNA was prepared from 500 ng of WT and Phospho1−/− primary calvarial osteoblast RNA using the Illumina® RNA Amplification Kit from Ambion (Austin, TX, USA). The labelled cRNA (1500 ng for mouse and 750 ng for human) was hybridised overnight at 58 °C to the SentrixMouseWG-6 Expression BeadChip or humanHT-12 Expression BeadChip (> 46,000 gene transcripts; Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. BeadChips were subsequently washed and developed with fluorolink streptavidin-Cy3 (GE Healthcare). BeadChips were scanned with an Illumina BeadArray Reader. Data was generated from Imagedata using Illumina software, GenomeStudio. Normalised data was generated using 3Cubic Spline2 Model in software. Pathway analysis was performed with Ingenuity Pathway Analysis (IPA, Ingenuity® Systems, www.ingenuity.com) and GeneMANIA (http://www.genemania.org).
Proteomic analysis
Proteins from serum was extracted and prepared as previously described [75–77], and the extracted peptides was analysed using a RSLC 3000 nanoscale capillary LC followed by qTOF mass spectrometry (5600 Triple-TOF, Sciex). Sequential window acquisition of all theoretical spectra (SWATH) was used to profile all proteins in each sample using a data-independent acquisition method [78]. ProteinPilot™ was used for protein identification and quantitation, as well as visualising peptide-protein associations and relationships.
Choline extraction
Serum samples were analysed using tandem mass spectrometry (LC-MS/MS) and a multiple-reaction monitoring (MRM) methodology. Five microliters of serum was extracted with 90 μL of an organic solution (10% methanol and 90% acetonitrile), containing the deuterium-labelled internal standard (IS, D9-Cho at 10 μg/mL). This resulted in precipitation of proteins, which were removed by filtration with a Millex 0.45-μm filter followed by centrifugation for 2 min at 6000g. The mass transitions used to measure the analytes are choline (mass transition m/z 104 → 60) and D9-choline (mass transition m/z 113 → 69). A QTRAP 5500 triple-quadrupole mass spectrometer (AB Sciex, Warrington, Cheshire, UK) with ESI ion source was used for data acquisition. Separation of analytes was performed in an Acquity UPLC-MS/MS (Waters, Hertfordshire, UK), with a binary pump system at a flow rate of 0.3 mL/min, connected to the mass spectrometer. The injection volume was 10 μL. Samples were separated using a Cogent 100 mm × 2.1 mm, 4 μm Diamond Hydride silica column (Microsolv Technologies, NJ, USA) and a linear gradient from 65% buffer B (0.1% formic acid in Acetronitrile) and 35% buffer A (0.1% formic acid in water) to 35% buffer B over 7 min. Analyst software (AB SCIEX) was used for HPLC system control, data acquisition and data processing.
Statistics
The data were analysed using various statistical models. All data were analysed for normal distribution within each experimental group using the Shapiro–Wilk normality test. Linear regression and correlation analysis based on Excel (Microsoft Office 10) built-in functions with interval of confidence and testing of the correlation coefficients were performed according to standard procedures. The SAS software was used to fit the generalised linear model (Microsoft Office 10). Normally distributed data were analysed by ANOVA or t tests, as appropriate. Where data were not normally distributed, non-parametric tests were used. When appropriate, P values were adjusted for multiple comparisons. Data are presented as histograms (means ± standard error) or box and whisker plots (boxes indicate the 25th and 75th percentiles; whiskers display the range; and horizontal lines in each box represent the median). Regression and correlation coefficients are given with the intervals of confidence (p = 0.05). Statistical analyses were performed using Sigma Plot software (v 11.0) (Systat Software Inc., London, UK) and Prism software (GraphPad, USA). P value < 0.05 was considered statistically significant.
Supplementary information
Additional file 1: Fig. S1. Ambulatory activity of WT and Phospho1−/− mice. (a) Total activity, (b) fast activity, (c) slow activity, (d) total static counts, (e) fast static counts, (f) slow static counts, (g) total mobile counts, (h) fast mobile counts, (i) slow mobile counts, (j) active time, (k) static time (l) mobile time, (m) front to back, (n) inactive time, (o) distance travelled. Data are represented as mean ± S.E.M (n = 6 replicates). *p < 0.05.
Additional file 2: Fig. S2. Fasted glucose levels of 120 day old WT and Phospho1−/− mice on the control and HFD. Different letters above the error bar show significant difference at p < 0.05.
Additional file 3: Fig. S3. μMRI adipose quantification from WT and Phospho1−/− mice on both a control and HFD. (a) Representative reconstructed μMRI scan. Green = subcutaneous adipose tissue, Red = mesenteric adipose tissue, Blue = brown adipose tissue. (b) Inguinal WAT (iWAT), mesenteric WAT (mWAT) and brown adipose tissue (BAT) mass determined by μMRI. Results were normalised to body weight (mg/g). Data are represented as mean ± S.E.M (n = 3 replicates). Different letters above the error bar show significant difference at p < 0.05.
Additional file 4: Fig. S4. Marrow adipose tissue μCT osmium quantification. (a) Region-specific quantification of tibial marrow adipose tissue (MAT) volume. Regions include the proximal epiphysis (Prox Epi), the growth plate to the tibia/fibula (Tib/Fib) junction (GP to T/F J) and the tibia/fibula junction to the end of the bone (T/F J to end). (b) Representative images of osmium-stained tibiae scanned by μCT. Marrow fat is dark grey and bone is light grey. Data are represented as mean ± S.E.M (n = 3 replicates). * p < 0.05, ** p < 0.01.
Additional file 5: Fig. S5. Seahorse analysis of WT and Phospho1−/− osteoblasts. (a - b) Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) using the Seahorse X-24 analyser in WT and Phospho1−/− primary calvarial osteoblasts cultured in growth media and growth media supplemented with osteogenic differentiation media for 3 days following a Mito Stress Test. Data represented as means ±S.E.M from average of two independent seahorse runs, with n = 5 wells per group. **p < 0.01, ***p < 0.001.
Additional file 6: Table S6. Osteoblast microarray candidates involved associated with glucose homeostasis. 21 genes from the WT and Phospho1−/− osteoblast microarray were identified by Ingenuity Pathway Analysis to be associated with glucose homeostasis p = 1.04 × 10− 6.
Additional file 7: Table S7. In Silico analysis of Ingenuity pathways predictions In Silico analysis of genes predicated to be associated with bone and diabetes mellitus.
Additional file 8: Fig. S8 GeneMANIA network summary predictions GeneMANIA network generated using Ingenuity Pathways Analysis gene predictions. The network highlights potential interactions between Phospho1 and related osteoblast genes involved in the glucose metabolic process, encompassing; glucose transport, insulin receptor signalling, response to insulin and cellular response to insulin stimulus. Query genes (black) with the exception of Spic and Runx2 which were inputted manually, other genes (grey) were generated by the programme using a large set of inbuilt functional association data. Node size are based on GO terms. Network line colour corresponds to interaction: purple = co-expression, pink = physical interactions, blue = co-localisation, green = shared protein domains orange = predicted, grey = other.
Additional file 9: Table S9. In Silico analysis top diseases associated with unique WT HFD and Phospho1−/− HFD proteins. (CSV 3 kb)
Additional file 10: Fig. S10. Gene and protein expression of Phospho1 mRNA and PHOSPHO1 protein in murine tissue (a) RT-qPCR of Phospho1 in murine tissues, high expression was seen in the gonad and brown adipose tissue (BAT) (b) Protein expression of PHOSPHO1 was detectable by western blot in the calvaria and bone. Non-specific binding of the PHOSPHO1 antibody was observed in the pancreas, seen in both WT and Phospho1−/− pancreatic tissue.
Acknowledgements
We thank Elaine Seawright for technical assistance; Darren Smith for providing animal support; Dr. Calum Gray, Prof. Maurits Jansen and Ross Lennen for MRI assistance (Edinburgh Imaging, University of Edinburgh); Prof. Roland. Stimson (Centre for Cardiovascular Science, University Edinburgh) for critical feedback on this manuscript.
Abbreviations
- PHOSPHO1
Phosphatase Orphan 1
- ANOVA
Analysis of variance
- BAT
Brown adipose tissue
- CD
Control diet
- Esp
Embryonic stem cell phosphatase
- EV
Empty vector
- FCCP
Protonophoric uncoupler
- GLA13-OCN
Carboxylated osteocalcin
- GLU13-OCN
Undercarboxylated osteocalcin
- GLU-OCN
Uncarboxylated osteocalcin
- GSIS
Glucose stimulate insulin secretion
- GTT
Glucose tolerance test
- gWAT
Gonadal white adipose tissue
- HbA1c
Glycated haemoglobin
- HFD
High fat diet
- HOMA-IR
Homeostatic Model Assessment of Insulin Resistance
- IPA
Ingenuity Pathway Analysis
- ITT
Insulin tolerance test
- iWAT
Inguinal white adipose tissue
- LC-MS/MS
Liquid chromatography with tandem mass spectrometry
- mWAT
Mesenteric white adipose tissue
- nSMase2
Neutral sphingomyelinase 2
- OCN
Osteocalcin
- OE
Overexpressing vector
- OST-PTP
Osteotesticular protein tyrosine phosphatase
- pCho
Phosphocholine
- Pi
Inorganic phosphate
- RER
Respiratory exchange rate
- SWATH
Sequential window acquisition of all theoretical spectra
- Ucp1
Uncoupling protein 1
- WAT
White adipose tissue
- WT
Wild-type
Authors’ contributions
Conceptualisation: K.J.S. and C.F. Methodology: K.J.S., N.M.M., C.V., C.H., S.R., L.B., D.B., M.B-L., A.R.G., Z. K and M.F. Investigation: K.J.S., B.J.T., N.M.M., C.V., C.H., S.R., L.B., M.B-L., A.R.G., and M.F. Formal analysis: K.J.S., C.H., M.C.Y., M.F. and A.R.G. Resources: N.M.M., C.V., L.B., G.K., M.M., C.J.R, V.E.M., J.L.R. and C.F. Writing—original draft: K.J.S. and C.F. Writing—review and editing: all authors. Visualisation: K.J.S. and A.G. Supervision: C.J.R., G.K., M.M., J.L.M., W.P.C., V.E.M and C.F. Funding acquisition: K.J.S., V.E.M. and C.F. All authors read and approved the final manuscript.
Funding
This project was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) UK through a studentship award (KJS), and Institute Strategic Programme Grant Funding (BB/J004316/1 and BB/P013732/1) (CF, VEM), BBSRC Institute Career Path Fellowship funding from the BBSRC (VEM) and grant AR53102 from the National Institute of Arthritis and Musculoskeletal Diseases (NIAMS) from the National Institutes of Health (NIH), USA. Small project grants and lab exchange funds were provided by the Roslin Institute and The Society for Endocrinology.
Availability of data and materials
Data generated or analysed during this study are included in this published article and its supplementary information files or available from the corresponding author on request. All analysed datasets were sourced from the authors. Raw data can be found in Additional file 11.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12915-020-00880-7.
References
- 1.Clemens TL, Karsenty G. The osteoblast: an insulin target cell controlling glucose homeostasis. J Bone Miner Res. 2011;26(4):677–680. doi: 10.1002/jbmr.321. [DOI] [PubMed] [Google Scholar]
- 2.Confavreux CB, Levine RL, Karsenty G. A paradigm of integrative physiology, the crosstalk between bone and energy metabolisms. Mol Cell Endocrinol. 2009;310(1–2):21–29. doi: 10.1016/j.mce.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ducy P, Amling M, Takeda S, Priemel M, Schilling AF, Beil FT, et al. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell. 2000;100(2):197–207. doi: 10.1016/S0092-8674(00)81558-5. [DOI] [PubMed] [Google Scholar]
- 4.Ferron M, Hinoi E, Karsenty G, Ducy P. Osteocalcin differentially regulates beta cell and adipocyte gene expression and affects the development of metabolic diseases in wild-type mice. Proc Natl Acad Sci U S A. 2008;105(13):5266–5270. doi: 10.1073/pnas.0711119105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferron M, Wei J, Yoshizawa T, Del Fattore A, DePinho RA, Teti A, et al. Insulin signaling in osteoblasts integrates bone remodeling and energy metabolism. Cell. 2010;142(2):296–308. doi: 10.1016/j.cell.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karsenty G. The mutual dependence between bone and gonads. J Endocrinol. 2012;213(2):107–114. doi: 10.1530/JOE-11-0452. [DOI] [PubMed] [Google Scholar]
- 7.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130(3):456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zoch ML, Abou DS, Clemens TL, Thorek DL, Riddle RC. In vivo radiometric analysis of glucose uptake and distribution in mouse bone. Bone Res. 2016;4:16004. doi: 10.1038/boneres.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suchacki KJ, Tavares AAS, Mattiucci D, Scheller EL, Papanastasiou G, Gray C, et al. Bone marrow adipose tissue is a unique adipose subtype with distinct roles in glucose homeostasis. Nat Commun. 2020;11(1):3097. doi: 10.1038/s41467-020-16878-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauschka PV, Lian JB, Cole DE, Gundberg CM. Osteocalcin and matrix Gla protein: vitamin K-dependent proteins in bone. Physiol Rev. 1989;69(3):990–1047. doi: 10.1152/physrev.1989.69.3.990. [DOI] [PubMed] [Google Scholar]
- 11.Pandey A, Khan HR, Alex NS, Puttaraju M, Chandrasekaran TT, Rudraiah M. Under-carboxylated osteocalcin regulates glucose and lipid metabolism during pregnancy and lactation in rats. J Endocrinol Invest. 2020;43(8):1081–95. [DOI] [PubMed]
- 12.Pepene CE. Serum under-carboxylated osteocalcin levels in women with polycystic ovary syndrome: weight-dependent relationships with endocrine and metabolic traits. J Ovarian Res. 2013;6(1):4. doi: 10.1186/1757-2215-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JJ, Toy WC, Wong MD, Tan CS, Tavintharan S, Wong MS, et al. Elevated undercarboxylated and reduced carboxylated osteocalcin are associated with metabolic syndrome in middle age Asian females. Exp Clin Endocrinol Diabetes. 2013;121(6):329–333. doi: 10.1055/s-0033-1334883. [DOI] [PubMed] [Google Scholar]
- 14.Al-Suhaimi EA, Al-Jafary MA. Endocrine roles of vitamin K-dependent- osteocalcin in the relation between bone metabolism and metabolic disorders. Rev Endocr Metab Disord. 2020;21(1):117–125. doi: 10.1007/s11154-019-09517-9. [DOI] [PubMed] [Google Scholar]
- 15.Prats-Puig A, Mas-Parareda M, Riera-Perez E, Gonzalez-Forcadell D, Mier C, Mallol-Guisset M, et al. Carboxylation of osteocalcin affects its association with metabolic parameters in healthy children. Diabetes Care. 2010;33(3):661–663. doi: 10.2337/dc09-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanazawa I, Tanaka K, Ogawa N, Yamauchi M, Yamaguchi T, Sugimoto T. Undercarboxylated osteocalcin is positively associated with free testosterone in male patients with type 2 diabetes mellitus. Osteoporos Int. 2013;24(3):1115–1119. doi: 10.1007/s00198-012-2017-7. [DOI] [PubMed] [Google Scholar]
- 17.Hwang YC, Jeong IK, Ahn KJ, Chung HY. The uncarboxylated form of osteocalcin is associated with improved glucose tolerance and enhanced beta-cell function in middle-aged male subjects. Diabetes Metab Res Rev. 2009;25(8):768–772. doi: 10.1002/dmrr.1045. [DOI] [PubMed] [Google Scholar]
- 18.Pollock NK, Bernard PJ, Gower BA, Gundberg CM, Wenger K, Misra S, et al. Lower uncarboxylated osteocalcin concentrations in children with prediabetes is associated with beta-cell function. J Clin Endocrinol Metab. 2011;96(7):E1092–E1099. doi: 10.1210/jc.2010-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonneau J, Ferland G, Karelis AD, Doucet E, Faraj M, Rabasa-Lhoret R, et al. Association between osteocalcin gamma-carboxylation and insulin resistance in overweight and obese postmenopausal women. J Diabetes Complicat. 2017;31(6):1027–1034. doi: 10.1016/j.jdiacomp.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Yoshikawa Y, Kode A, Xu L, Mosialou I, Silva BC, Ferron M, et al. Genetic evidence points to an osteocalcin-independent influence of osteoblasts on energy metabolism. J Bone Miner Res. 2011;26(9):2012–2025. doi: 10.1002/jbmr.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al Rifai O, Chow J, Lacombe J, Julien C, Faubert D, Susan-Resiga D, et al. Proprotein convertase furin regulates osteocalcin and bone endocrine function. J Clin Invest. 2017;127(11):4104–4117. doi: 10.1172/JCI93437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart AJRS, Seawright E, Davey MG, Fleming RH, Farquharson C. The presence of PHOSPHO1 in matrix vesicles and its developmental expression prior to skeletal mineralization. Bone. 2006;39(5):1000–1007. doi: 10.1016/j.bone.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Houston BSA, Farquharson C. PHOSPHO1-a novel phosphatase specifically expressed at sites of mineralisation in bone and cartilage. Bone. 2004;34(4):629–637. doi: 10.1016/j.bone.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 24.Ciancaglini P, Yadav MC, Simao AM, Narisawa S, Pizauro JM, Farquharson C, et al. Kinetic analysis of substrate utilization by native and TNAP-, NPP1-, or PHOSPHO1-deficient matrix vesicles. J Bone Miner Res. 2010;25(4):716–723. doi: 10.1359/jbmr.091023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houston BSE, Jefferies D, Hoogland E, Lester D, Whitehead C, Farquharson C. Identification and cloning of a novel phosphatase expressed at high levels in differentiating growth plate chondrocytes. Biochim Biophys Acta. 1999;1448(3):500–506. doi: 10.1016/S0167-4889(98)00153-0. [DOI] [PubMed] [Google Scholar]
- 26.Stewart AJSR, Blindauer CA, Paisey SJ, Farquharson C. Comparative modelling of human PHOSPHO1 reveals a new group of phosphatases within the haloacid dehalogenase superfamily. Protein Eng. 2003;16(12):889–895. doi: 10.1093/protein/gzg126. [DOI] [PubMed] [Google Scholar]
- 27.Roberts S, Narisawa S, Harmey D, Millan J, Farquharson C. Functional involvement of PHOSPHO1 in matrix vesicle-mediated skeletal mineralization. J Bone Miner Res. 2007;22(4):617–627. doi: 10.1359/jbmr.070108. [DOI] [PubMed] [Google Scholar]
- 28.Roberts SJ, Stewart AJ, Schmid R, Blindauer CA, Bond SR, Sadler PJ, et al. Probing the substrate specificities of human PHOSPHO1 and PHOSPHO2. Biochim Biophys Acta. 2005;1752(1):73–82. doi: 10.1016/j.bbapap.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Macrae VEDM, McTeir L, Narisawa S, Yadav MC, Millan JL, Farquharson C. Inhibition of PHOSPHO1 activity results in impaired skeletal mineralization during limb development of the chick. Bone. 2010;46(4):1146–1155. doi: 10.1016/j.bone.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huesa C, Yadav MC, Finnila MA, Goodyear SR, Robins SP, Tanner KE, et al. PHOSPHO1 is essential for mechanically competent mineralization and the avoidance of spontaneous fractures. Bone. 2011;48(5):1066–1074. doi: 10.1016/j.bone.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yadav MC, Simao AM, Narisawa S, Huesa C, McKee MD, Farquharson C, et al. Loss of skeletal mineralization by the simultaneous ablation of PHOSPHO1 and alkaline phosphatase function: a unified model of the mechanisms of initiation of skeletal calcification. J Bone Miner Res. 2011;26(2):286–297. doi: 10.1002/jbmr.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Javaheri B, Carriero A, Staines KA, Chang YM, Houston DA, Oldknow KJ, et al. Phospho1 deficiency transiently modifies bone architecture yet produces consistent modification in osteocyte differentiation and vascular porosity with ageing. Bone. 2015;81:277–291. doi: 10.1016/j.bone.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyde A, Staines KA, Javaheri B, Millan JL, Pitsillides AA, Farquharson C. A distinctive patchy osteomalacia characterises Phospho1-deficient mice. J Anat. 2017;231(2):298–308. doi: 10.1111/joa.12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G, Zhang L, Li T, Zuniga A, Lopaschuk GD, Li L, et al. Choline supplementation promotes hepatic insulin resistance in phosphatidylethanolamine N-methyltransferase-deficient mice via increased glucagon action. J Biol Chem. 2013;288(2):837–847. doi: 10.1074/jbc.M112.415117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willmer T, Johnson R, Louw J, Pheiffer C. Blood-based DNA methylation biomarkers for type 2 diabetes: potential for clinical applications. Front Endocrinol (Lausanne). 2018;9:744. [DOI] [PMC free article] [PubMed]
- 36.Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3(7):526–34. [DOI] [PMC free article] [PubMed]
- 37.Dayeh T, Tuomi T, Almgren P, Perfilyev A, Jansson PA, de Mello VD, et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics. 2016;11(7):482–488. doi: 10.1080/15592294.2016.1178418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Duan H, Tian X, Xu C, Wang W, Jiang W, et al. Genetics of Obesity Traits: A Bivariate Genome-Wide Association Analysis. Front Genet. 2018;9:179. doi: 10.3389/fgene.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sayols-Baixeras S, Subirana I, Lluis-Ganella C, Civeira F, Roquer J, Do AN, et al. Identification and validation of seven new loci showing differential DNA methylation related to serum lipid profile: an epigenome-wide approach. The REGICOR study. Human Mol Genet. 2016;25(20):4556–65. [DOI] [PMC free article] [PubMed]
- 40.Fan J, Cai Q, Zhang D, Weinstock J, Qu X, Jiang S. PHOSPHO1 gene DNA methylations are associated with a change in HDL-C response to simvastatin treatment. Curr Pharm Des. 2020. 10.2174/1381612826666200720234604. Online ahead of print. [DOI] [PubMed]
- 41.Whitehead JP, Richards AA, Hickman IJ, Macdonald GA, Prins JB. Adiponectin--a key adipokine in the metabolic syndrome. Diabetes Obes Metab. 2006;8(3):264–280. doi: 10.1111/j.1463-1326.2005.00510.x. [DOI] [PubMed] [Google Scholar]
- 42.Cawthorn WP, Scheller EL, Learman BS, Parlee SD, Simon BR, Mori H, et al. Bone marrow adipose tissue is an endocrine organ that contributes to increased circulating adiponectin during caloric restriction. Cell Metab. 2014;20(2):368–375. doi: 10.1016/j.cmet.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldberg IJ, Eckel RH, Abumrad NA. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J Lipid Res. 2009;50(Suppl):S86–S90. doi: 10.1194/jlr.R800085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80(2):225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 45.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103(2):211–225. doi: 10.1016/S0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 46.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 47.Dacquin R, Mee PJ, Kawaguchi J, Olmsted-Davis EA, Gallagher JA, Nichols J, et al. Knock-in of nuclear localised beta-galactosidase reveals that the tyrosine phosphatase Ptprv is specifically expressed in cells of the bone collar. Dev Dyn. 2004;229(4):826–834. doi: 10.1002/dvdy.20003. [DOI] [PubMed] [Google Scholar]
- 48.Mauro LJ, Olmsted EA, Skrobacz BM, Mourey RJ, Davis AR, Dixon JE. Identification of a hormonally regulated protein tyrosine phosphatase associated with bone and testicular differentiation. J Biol Chem. 1994;269(48):30659–30667. [PubMed] [Google Scholar]
- 49.Ferron M, Wei J, Yoshizawa T, Ducy P, Karsenty G. An ELISA-based method to quantify osteocalcin carboxylation in mice. Biochem Biophys Res Commun. 2010;397(4):691–696. doi: 10.1016/j.bbrc.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshizawa T, Hinoi E, Jung DY, Kajimura D, Ferron M, Seo J, et al. The transcription factor ATF4 regulates glucose metabolism in mice through its expression in osteoblasts. J Clin Invest. 2009;119(9):2807–2817. doi: 10.1172/JCI39366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rached MT, Kode A, Silva BC, Jung DY, Gray S, Ong H, et al. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J Clin Invest. 2010;120(1):357–368. doi: 10.1172/JCI39901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vowinckel J, Capuano F, Campbell K, Deery MJ, Lilley KS, Ralser M. The beauty of being (label)-free: sample preparation methods for SWATH-MS and next-generation targeted proteomics. F1000Res. 2013;2:272. doi: 10.12688/f1000research.2-272.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoffel W, Jenke B, Block B, Zumbansen M, Koebke J. Neutral sphingomyelinase 2 (smpd3) in the control of postnatal growth and development. Proc Natl Acad Sci U S A. 2005;102(12):4554–4559. doi: 10.1073/pnas.0406380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts SJSA, Sadler PJ, Farquharson C. Human PHOSPHO1 exhibits high specific phosphoethanolamine and phosphocholine phosphatase activities. Biochem J. 2004;382(Pt 1):59–65. doi: 10.1042/BJ20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang G, Badeanlou L, Bielawski J, Roberts AJ, Hannun YA, Samad F. Central role of ceramide biosynthesis in body weight regulation, energy metabolism, and the metabolic syndrome. Am J Physiol Endocrinol Metab. 2009;297(1):E211–E224. doi: 10.1152/ajpendo.91014.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guseva MV, Hopkins DM, Scheff SW, Pauly JR. Dietary choline supplementation improves behavioral, histological, and neurochemical outcomes in a rat model of traumatic brain injury. J Neurotrauma. 2008;25(8):975–983. doi: 10.1089/neu.2008.0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart AJ, Leong DTK, Farquharson C. PLA2 and ENPP6 may act in concert to generate phosphocholine from the matrix vesicle membrane during skeletal mineralization. FASEB J. 2018;32(1):20–25. doi: 10.1096/fj.201700521r. [DOI] [PubMed] [Google Scholar]
- 59.Kim HR, Roe JS, Lee JE, Cho EJ, Youn HD. p53 regulates glucose metabolism by miR-34a. Biochem Biophys Res Commun. 2013;437(2):225–231. doi: 10.1016/j.bbrc.2013.06.043. [DOI] [PubMed] [Google Scholar]
- 60.Favaro RR, Salgado RM, Covarrubias AC, Bruni F, Lima C, Fortes ZB, et al. Long-term type 1 diabetes impairs decidualization and extracellular matrix remodeling during early embryonic development in mice. Placenta. 2013;34(12):1128–1135. doi: 10.1016/j.placenta.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 61.Raouf A, Ganss B, McMahon C, Vary C, Roughley PJ, Seth A. Lumican is a major proteoglycan component of the bone matrix. Matrix Biol. 2002;21(4):361–367. doi: 10.1016/S0945-053X(02)00027-6. [DOI] [PubMed] [Google Scholar]
- 62.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 63.Lee P, Smith S, Linderman J, Courville AB, Brychta RJ, Dieckmann W, et al. Temperature-acclimated brown adipose tissue modulates insulin sensitivity in humans. Diabetes. 2014;63(11):3686–3698. doi: 10.2337/db14-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee J, Choi J, Aja S, Scafidi S, Wolfgang MJ. Loss of adipose fatty acid oxidation does not potentiate obesity at thermoneutrality. Cell Rep. 2016;14(6):1308–1316. doi: 10.1016/j.celrep.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell. 2015;163(3):643–655. doi: 10.1016/j.cell.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mefford IN, Wade EU. Proton pump inhibitors as a treatment method for type II diabetes. Med Hypotheses. 2009;73(1):29–32. doi: 10.1016/j.mehy.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 67.Boj-Carceller D, Bocos-Terraz P, Moreno-Vernis M, Sanz-Paris A, Trincado-Aznar P, Albero-Gamboa R. Are proton pump inhibitors a new antidiabetic drug? A cross sectional study. World J Diabetes. 2011;2(12):217–220. doi: 10.4239/wjd.v2.i12.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barchetta I, Guglielmi C, Bertoccini L, Calella D, Manfrini S, Secchi C, et al. Therapy with proton pump inhibitors in patients with type 2 diabetes is independently associated with improved glycometabolic control. Acta Diabetol. 2015;52(5):873–880. doi: 10.1007/s00592-015-0721-4. [DOI] [PubMed] [Google Scholar]
- 69.Staines KA, Zhu D, Farquharson C, MacRae VE. Identification of novel regulators of osteoblast matrix mineralization by time series transcriptional profiling. J Bone Miner Metab. 2014;32(3):240–251. doi: 10.1007/s00774-013-0493-2. [DOI] [PubMed] [Google Scholar]
- 70.Huesa C, Houston D, Kiffer-Moreira T, Yadav MM, Millan JL, Farquharson C. The functional co-operativity of tissue-nonspecific alkaline phosphatase (TNAP) and PHOSPHO1 during initiation of skeletal mineralization. Biochem Biophys Rep. 2015;4:196–201. doi: 10.1016/j.bbrep.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guntur AR, Gerencser AA, Le PT, DeMambro VE, Bornstein SA, Mookerjee SA, et al. Osteoblast-like MC3T3-E1 cells prefer glycolysis for ATP production but adipocyte-like 3T3-L1 cells prefer oxidative phosphorylation. J Bone Miner Res. 2018;33(6):1052–1065. doi: 10.1002/jbmr.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 73.Huesa C, Zhu D, Glover JD, Ferron M, Karsenty G, Milne EM, et al. Deficiency of the bone mineralization inhibitor NPP1 protects mice against obesity and diabetes. Dis Model Mech. 2014;7(12):1341–1350. doi: 10.1242/dmm.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scheller EL, Troiano N, Vanhoutan JN, Bouxsein MA, Fretz JA, Xi Y, et al. Use of osmium tetroxide staining with microcomputerized tomography to visualize and quantify bone marrow adipose tissue in vivo. Methods Enzymol. 2014;537:123–139. doi: 10.1016/B978-0-12-411619-1.00007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Young K, Conley B, Romero D, Tweedie E, O'Neill C, Pinz I, et al. BMP9 regulates endoglin-dependent chemokine responses in endothelial cells. Blood. 2012;120(20):4263–4273. doi: 10.1182/blood-2012-07-440784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Romero D, O’Neill C, Terzic A, Contois L, Young K, Conley BA, et al. Endoglin regulates cancer-stromal cell interactions in prostate tumors. Cancer Res. 2011;71(10):3482–3493. doi: 10.1158/0008-5472.CAN-10-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Young K, Tweedie E, Conley B, Ames J, FitzSimons M, Brooks P, et al. BMP9 crosstalk with the hippo pathway regulates endothelial cell Matricellular and chemokine responses. PLoS One. 2015;10(4):e0122892. doi: 10.1371/journal.pone.0122892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11(6):O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Ambulatory activity of WT and Phospho1−/− mice. (a) Total activity, (b) fast activity, (c) slow activity, (d) total static counts, (e) fast static counts, (f) slow static counts, (g) total mobile counts, (h) fast mobile counts, (i) slow mobile counts, (j) active time, (k) static time (l) mobile time, (m) front to back, (n) inactive time, (o) distance travelled. Data are represented as mean ± S.E.M (n = 6 replicates). *p < 0.05.
Additional file 2: Fig. S2. Fasted glucose levels of 120 day old WT and Phospho1−/− mice on the control and HFD. Different letters above the error bar show significant difference at p < 0.05.
Additional file 3: Fig. S3. μMRI adipose quantification from WT and Phospho1−/− mice on both a control and HFD. (a) Representative reconstructed μMRI scan. Green = subcutaneous adipose tissue, Red = mesenteric adipose tissue, Blue = brown adipose tissue. (b) Inguinal WAT (iWAT), mesenteric WAT (mWAT) and brown adipose tissue (BAT) mass determined by μMRI. Results were normalised to body weight (mg/g). Data are represented as mean ± S.E.M (n = 3 replicates). Different letters above the error bar show significant difference at p < 0.05.
Additional file 4: Fig. S4. Marrow adipose tissue μCT osmium quantification. (a) Region-specific quantification of tibial marrow adipose tissue (MAT) volume. Regions include the proximal epiphysis (Prox Epi), the growth plate to the tibia/fibula (Tib/Fib) junction (GP to T/F J) and the tibia/fibula junction to the end of the bone (T/F J to end). (b) Representative images of osmium-stained tibiae scanned by μCT. Marrow fat is dark grey and bone is light grey. Data are represented as mean ± S.E.M (n = 3 replicates). * p < 0.05, ** p < 0.01.
Additional file 5: Fig. S5. Seahorse analysis of WT and Phospho1−/− osteoblasts. (a - b) Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) using the Seahorse X-24 analyser in WT and Phospho1−/− primary calvarial osteoblasts cultured in growth media and growth media supplemented with osteogenic differentiation media for 3 days following a Mito Stress Test. Data represented as means ±S.E.M from average of two independent seahorse runs, with n = 5 wells per group. **p < 0.01, ***p < 0.001.
Additional file 6: Table S6. Osteoblast microarray candidates involved associated with glucose homeostasis. 21 genes from the WT and Phospho1−/− osteoblast microarray were identified by Ingenuity Pathway Analysis to be associated with glucose homeostasis p = 1.04 × 10− 6.
Additional file 7: Table S7. In Silico analysis of Ingenuity pathways predictions In Silico analysis of genes predicated to be associated with bone and diabetes mellitus.
Additional file 8: Fig. S8 GeneMANIA network summary predictions GeneMANIA network generated using Ingenuity Pathways Analysis gene predictions. The network highlights potential interactions between Phospho1 and related osteoblast genes involved in the glucose metabolic process, encompassing; glucose transport, insulin receptor signalling, response to insulin and cellular response to insulin stimulus. Query genes (black) with the exception of Spic and Runx2 which were inputted manually, other genes (grey) were generated by the programme using a large set of inbuilt functional association data. Node size are based on GO terms. Network line colour corresponds to interaction: purple = co-expression, pink = physical interactions, blue = co-localisation, green = shared protein domains orange = predicted, grey = other.
Additional file 9: Table S9. In Silico analysis top diseases associated with unique WT HFD and Phospho1−/− HFD proteins. (CSV 3 kb)
Additional file 10: Fig. S10. Gene and protein expression of Phospho1 mRNA and PHOSPHO1 protein in murine tissue (a) RT-qPCR of Phospho1 in murine tissues, high expression was seen in the gonad and brown adipose tissue (BAT) (b) Protein expression of PHOSPHO1 was detectable by western blot in the calvaria and bone. Non-specific binding of the PHOSPHO1 antibody was observed in the pancreas, seen in both WT and Phospho1−/− pancreatic tissue.
Data Availability Statement
Data generated or analysed during this study are included in this published article and its supplementary information files or available from the corresponding author on request. All analysed datasets were sourced from the authors. Raw data can be found in Additional file 11.