Abstract
Context
Metabolic flexibility is the physiologic acclimatization to differing energy availability and requirement states. Effectively maintaining metabolic flexibility remains challenging, particularly since metabolic dysregulations in meal consumption during cardiometabolic disease (CMD) pathophysiology are incompletely understood.
Objective
We compared metabolic flexibility following consumption of a standardized meal challenge among adults with or without CMDs.
Design, Setting, and Participants
Study participants (n = 349; age 37-54 years, 55% female) received a standardized meal challenge (520 kcal, 67.4 g carbohydrates, 24.3 g fat, 8.0 g protein; 259 mL). Blood samples were collected at baseline and 2 hours postchallenge. Plasma samples were assayed by high-resolution, nontargeted metabolomics with dual-column liquid chromatography and ultrahigh-resolution mass spectrometry. Metabolome-wide associations between features and meal challenge timepoint were assessed in multivariable linear regression models.
Results
Sixty-five percent of participants had ≥1 of 4 CMDs: 33% were obese, 6% had diabetes, 39% had hypertension, and 50% had metabolic syndrome. Log2-normalized ratios of feature peak areas (postprandial:fasting) clustered separately among participants with versus without any CMDs. Among participants with CMDs, the meal challenge altered 1756 feature peak areas (1063 reversed-phase [C18], 693 hydrophilic interaction liquid chromatography [HILIC]; all q < 0.05). In individuals without CMDs, the meal challenge changed 1383 feature peak areas (875 C18; 508 HILIC; all q < 0.05). There were 108 features (60 C18; 48 HILIC) that differed by the meal challenge and CMD status, including dipeptides, carnitines, glycerophospholipids, and a bile acid metabolite (all P < 0.05).
Conclusions
Among adults with CMDs, more metabolomic features differed after a meal challenge, which reflected lower metabolic flexibility relative to individuals without CMDs.
Keywords: cardiometabolic disease, metabolomics, metabolic health, meal challenge, postprandial state
Cardiometabolic diseases (CMDs) and other noncommunicable diseases account for over 70% of deaths globally [1]. More effectively reducing the disease burden from CMDs remains a major challenge [1]. One critical research gap is how to more effectively maintain cardiometabolic health, including metabolic flexibility in daily physiological processes, across the life course [2, 3]. Metabolic flexibility is broadly defined as the dynamic acclimatization that occurs to maintain energy homeostasis throughout heterogeneous physiologic needs and conditions [2, 3]. Healthy systems are considered those with metabolic resilience—the ability to effectively regulate—in response to differing energy states [4].
Impaired metabolic flexibility has been hypothesized to influence CMD pathophysiology [2, 3, 5]; however, specific contributions remain incompletely understood. One challenge is the lack of a single, standard definition of metabolic flexibility, particularly in light of numerous interrelated physiological processes involved in maintaining energy homeostasis [2, 3, 6]. Previous studies have begun characterizing different aspects of reduced metabolic flexibility, including the decreased ability to sense nutrients, maintain energy homeostasis, and shift between macronutrient fuels [6]. Few studies have evaluated activity metabolomics in response to meal consumption among people with versus without CMDs.
In this study, we defined metabolic flexibility as the changes in metabolomic profiles following a physiologic meal challenge, relative to while fasting. High-resolution, nontargeted metabolomic profiling provides greater granularity and metabolite coverage relative to standard diagnostic biomarkers [5, 7-10]. Our study objective was to compare the metabolomic profile response following a standardized meal challenge of adults with CMDs, relative to metabolically healthy participants. We hypothesized that: (i) there would be increased changes in metabolomic features after the meal challenge among people with CMDs, compared to those without CMDs; and (ii) metabolomic features in major energy, macronutrient, and bile acid pathways would be responsive in the meal challenge and differ by CMD status.
1. Materials and Methods
Study participants
We studied adult participants of the Institute of Nutrition of Central America and Panama (INCAP) Nutritional Supplementation Trial Longitudinal Study [11, 12]. The present analysis focuses on participants enrolled in the follow-up wave conducted from 2015 to 2017 [13].
Meal challenge
After an overnight fast, participants visited the research clinic in the morning. Baseline venous blood samples were drawn after confirming fasting. Individuals were excluded from the meal challenge if their fasting blood glucose was ≥180 mg/dL or they self-reported having diabetes. Study participants then received a standardized beverage, which was comprised of Incaparina (a vegetable protein mixture developed by INCAP), skim milk (lactose-free), safflower oil, and sugar. Each portion (259 mL) contained 520.0 kcal, 8.0 g protein, 24.3 g fat, and 67.4 g carbohydrate (Supplemental Table 1) [14]. This composition was intended to reflect a physiologic meal consisting of relatively higher fat and higher sugar content. Many geographic settings undergoing the nutrition transition have greater availability of higher fat and higher sugar-containing diets, which have been hypothesized to contribute to CMDs [15]. Hyperglycemia and hyperlipidemia are components of diabetes and metabolic syndrome, and often co-occur with obesity and hypertension [16-19].
Data collection
Trained phlebotomists collected venous blood samples at baseline (fasting) and 120 minutes after the meal challenge (postprandial). Plasma samples for metabolomic analysis were collected in heparin tubes. Samples were maintained on ice prior to centrifugation at 3000 revolutions per minute for 10 minutes, and subsequently at −80 °C until assay. Blood samples were assayed for plasma glucose, insulin, and lipids (further details regarding assay methodology and instruments in Supplemental Table 2) [14].
Trained study staff interviewed study participants regarding sociodemographic and clinical information, including use of medications [13]. Standard protocols were used to obtain anthropometric measurements [13].
Random selection of analysis subset
From the 1139 individuals who participated in the 2015-2017 data collection wave, we sampled a random subset (n = 401; Supplemental Fig. 1) [14]. Exclusion criteria included: (i) pregnancy or lactation (n = 4) or missing key clinical variables (n = 4); (ii) incomplete metabolomic data (<2 timepoints of blood samples; n = 44 for reversed-phase [C18], n = 47 for hydrophilic interaction liquid chromatography [HILIC]); and (iii) failed quality check between technical replicates (n = 0 for C18; n = 3 for HILIC). After these exclusions, 349 participants with C18 data and 343 participants with HILIC data were included in the analysis.
High-resolution metabolomics
Pairs of plasma samples (fasting, postprandial state) were randomly ordered for nontargeted metabolomic assay. Each plasma sample (fasting or postprandial state) was assayed in triplicate for high-resolution metabolomic data. After frozen samples were thawed, acetonitrile (2:1, v/v; HPLC grade; Millipore, MA, USA; Sigma Aldrich, MO, USA) was added to precipitate protein [20]. Samples were maintained on ice for 30 minutes, centrifuged (14 000g for 10 minutes at 4 °C), and the supernatant was stored at 4 °C in a refrigerated autosampler prior to assay [20, 21].
The following commercially available, stable isotope-labeled internal standards were included: [13C6]-d-glucose, [15N]-indole, [2-15N]-l-lysine dihydrochloride, [13C5]-l-glutamic acid, [13C7]-benzoic acid, [3,4-13C2]-cholesterol, [15N]-l-tyrosine, [trimethyl-13C3]-caffeine, [15N2]-uracil, [3,3-13C2]-cystine, [1,2-13C2]-palmitic acid, [15N,13C5]-l-methionine, [15N]-choline chloride, and 2’-deoxyguanosine-15N2,13C10-5’-monophosphate [20, 21]. Human reference plasma from the National Institute of Standards and Technology (standard reference material 1950) and a pooled reference plasma (Q-std3) prepared from commercial human plasma samples (Equitech Bio, TX, USA) were included for quality control [22]. Q-std3 was included before and after every 20 samples; specifically, triplicates of 2 Q-Std3 samples were included as the beginning, middle, or end of every batch of 40 samples.
Liquid chromatography mass spectrometry
Plasma samples were assayed by liquid chromatography–Fourier transform mass spectrometry (LC-FT-MS) with 2 chromatographic columns: C18 (Higgins Analytical, Targa, 2.1 × 50 mm) with negative electrospray ionization, and HILIC (Waters BEH Amide 2.1 × 50 mm) with positive electrospray ionization [21, 23]. A Switchos control valve (LC Packings) allowed for alternation between the 2 columns [21]. Data acquisition occurred by mass spectrometer (Orbitrap Fusion mass spectrometer; Thermo Fisher Scientific, MA, USA) with specifications as previously described [21]. The scan range for the detection of mass-to-charge ratio (m/z) scan was between 85 and 1250, and mass resolution was 120 000.
Data extraction
Raw data (.raw files) were collected throughout the chromatographic separation and converted to .cdf files (Xcalibur software; Thermo Fisher Scientific, CA, US). apLCMS [24] and xMSanalyzer [25] were used for the extraction and initial preprocessing of chromatographic data. Preprocessing included noise reduction, peak identification, retention time correction, peak alignment, feature quantification, weak signal detection, and batch effect adjustment with ComBat [24-26]. A feature was defined as a unique combination of m/z and retention time. The ion intensity (peak area) of each feature was integrated.
Standard operating procedures and quality control were based on prior studies, including via xMSanalyzer [25]. For each sample, pairwise Pearson correlations between replicates were assessed and averaged. Prior to exclusions of participants and data filtering of features (Supplemental Fig. 1 [14]), the median averaged pairwise Pearson correlation between replicates was 98.7% (interquartile range [IQR], 97.0%, 99.5%) across samples from C18 data, and 99.5% (IQR, 98.6%, 99.8%) across all samples from HILIC data. Samples (n = 0 for C18; n = 3 for HILIC) with mean Pearson correlation coefficients <0.75 across technical replicates were excluded.
Features observed in <80% of study participant samples in each column (C18, HILIC) were filtered [27]. Any feature peak area values that were missing or zero were subsequently assigned as half of the lowest observed value (limit of detection) of the feature peak area across all samples for each column.
Definitions
CMD definitions were based on guidelines of the World Health Organization, American Diabetes Association, American College of Cardiology, American Heart Association, and National Cholesterol Education Program (Adult Treatment Panel III report; Supplemental Table 2) [14, 16-19]. Participants were characterized as being metabolically unhealthy if they had any of 4 CMDs (obesity, hypertension, diabetes, metabolic syndrome).
Statistical analysis
Statistical analysis was conducted utilizing R (version 3.5.1; R Foundation for Statistical Computing; Vienna, Austria) and SAS (version 9.4; SAS Institute Inc.; Cary, NC, US). We used a complete case approach (Supplemental Fig. 1) [14]. Prior to analysis, we converted feature intensities (peak areas) to log2-normalized values. For each feature, we calculated the peak area ratio as the postprandial timepoint divided by the fasting timepoint. Statistical significance was based on 2-sided hypothesis tests, and α value <0.05. After feature-by-feature multivariable regressions with metabolomic data, the P values were collectively adjusted for a false discovery rate (FDR), which controls the proportion of selected features that incorrectly reject the null hypothesis. FDR-adjusted q < 0.05 was considered significant for Stages 1 and 2A. In regressions with interaction terms (Stages 2B), P < 0.05 was considered significant.
Descriptive analysis and visualizations
Continuous variables were reported as mean (standard deviation) or median (IQR), and categorical variables were reported as n (%). Subgroups were compared by Kruskal-Wallis, Wilcoxon rank-sum, or Mantel-Haenszel chi-square test statistics. Correlations between log2-normalized feature peak areas and their ratios (postprandial/fasting), and CMD biomarkers were evaluated by Spearman rank correlation coefficients. We compared differences of log2-normalized feature peak area ratios between participants with versus without any CMDs with MetaboAnalystR [28]. As examples, we visualized the clustering of feature peak area ratios with unsupervised dimensionality reduction (principal components analysis) and supervised discriminant analysis approaches (eg, partial least squares–discriminant analysis [PLS-DA], orthogonal partial least squares–discriminant analysis [OPLS-DA]) [28].
Feature selection approach via regressions
We used a 2-stage feature selection approach based on multivariable linear mixed regressions with repeated measurements (Supplemental Fig. 2) [14]. In Stage 1, we assessed whether the peak areas of each feature differed by the meal challenge timepoint, adjusting for age and sex. The model equation was:
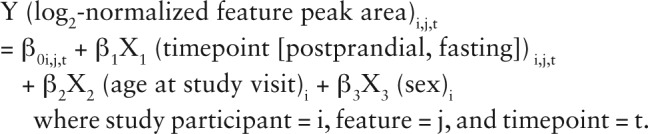
Features with beta coefficients of the meal challenge (β 1) with FDR-adjusted q < 0.05 were annotated, considered in functional pathway analysis, and eligible for Stage 2. In Stage 2A, we assessed whether feature peak areas responded to the meal challenge, accounting for age and sex, among participants either with or without CMDs. The same Stage 1 regression equation was used among these 2 stratified groups of participants. In Stage 2B, we additionally included CMD status and the 2-way multiplicative interaction term between the meal challenge timepoint (postprandial, fasting) as independent variables in the Stage 1 regressions. The Stage 2B regression equation was:

Features with beta coefficients of the interaction term (β 5) with P < 0.05 were annotated and considered in functional pathways.
Feature annotations
We used annotations from xMSannotator, which considers multiple criteria in its algorithm [29] and incorporates Human Metabolome Database reference database information [30] for putative identification of features (Supplemental Fig. 3) [14]. We considered feature annotation confidence and assigned identification confidence scores to some features, based on the 5-level system proposed by the Metabolomics Standards Initiative (MSI; Supplemental Fig. 3) [14, 31]. Annotations of select metabolites in Stage 2B regressions were compared to identities by co-elution and MS/MS fragmentation patterns relative to authentic standards.
Ethical conduct of research
The Institutional Review Boards at Emory University (Atlanta, Georgia, US) and INCAP (Guatemala City, Guatemala) approved the study protocol. All participants provided their written informed consent.
2. Results
Sociodemographic and cardiometabolic health characteristics of the 349 study participants are in Table 1. Briefly, among study participants (37-54 years of age), 116 (33%) had body mass index (BMI) considered obese, 22 (6%) had diabetes, 135 (39%) had hypertension, and 173 (50%) had metabolic syndrome (Table 2). One hundred twenty-three adults were considered metabolically healthy insofar as they had none of these 4 CMDs (35%; Table 2). A lower proportion of men (51%) had at least 1 CMD compared with women (76%; P < 0.01; Table 2). Relative to men, greater proportions of women had obesity (43% vs 21%), metabolic syndrome (65% vs 31 %), and any CMD (76% vs 51%; all P < 0.01; Table 2). Several CMD indicators (postprandial glucose, glycated hemoglobin, cholesterol [total, HDL, non-HDL]) differed between men and women (all P < 0.01; Table 2).
Table 1.
Sociodemographic and Clinical Characteristics of Adult Study Participants (n = 349)
| Overall | Males | Females | P a | ||||
|---|---|---|---|---|---|---|---|
| n | 349 | n | 157 | n | 192 | ||
| Median (IQR) | Median (IQR) | Median (IQR) | |||||
| Sociodemographics | |||||||
| Age at follow-up (years)b | 349 | 44.0 (40.0, 47.0) | 157 | 44.0 (40.0, 47.0) | 192 | 44.0 (41.0, 47.5) | 0.79 |
| Anthropometry | |||||||
| BMI (kg/m2) | 349 | 27.8 (24.8, 31.1) | 157 | 26.5 (24.3, 29.4) | 192 | 28.9 (25.9, 32.4) | <0.01 |
| Biochemical indicators | |||||||
| Glucose profile | |||||||
| Fasting blood glucose (mg/dL) | 349 | 98.6 (93.1, 105.2) | 157 | 97.5 (93.2, 104.0) | 192 | 99.3 (93.0, 105.6) | 0.65 |
| Postprandial glucose (mg/dL) | 349 | 109.3 (95.6, 123.6) | 157 | 100.7 (90.8, 116.4) | 192 | 116.6 (102.9, 128.4) | <0.01 |
| Glycated hemoglobin (%) | 348 | 5.8 (5.5, 6.0) | 157 | 5.7 (5.5, 5.9) | 191 | 5.8 (5.6, 6.0) | <0.01 |
| Lipid profile | |||||||
| Triglycerides (mg/dL) | 349 | 144.0 (100.0, 207.0) | 157 | 142.0 (98.0, 213.0) | 192 | 144.5 (101.5, 197.0) | 0.89 |
| Total cholesterol (mg/dL) | 349 | 175.0 (151.0, 198.0) | 157 | 167.0 (143.0, 189.0) | 192 | 182.0 (158.0, 208.0) | <0.01 |
| HDL-cholesterol (mg/dL) | 349 | 40.7 (36.3, 46.8) | 157 | 37.8 (33.7, 44.6) | 192 | 42.5 (37.9, 48.9) | <0.01 |
| Non-HDL-cholesterol (mg/dL)c | 349 | 132.1 (110.7, 154.3) | 157 | 124.4 (104.0, 149.6) | 192 | 136.9 (116.7, 159.1) | <0.01 |
| Clinical | |||||||
| Systolic blood pressure (mm Hg) | 349 | 122.0 (113.5, 132.0) | 157 | 123.0 (114.5, 132.0) | 192 | 120.5 (112.0, 131.8) | 0.24 |
| Diastolic blood pressure (mm Hg) | 349 | 73.5 (67.5, 80.5) | 157 | 73.5 (67.5, 79.5) | 192 | 73.3 (67.5, 80.5) | 0.60 |
Data values are either median (IQR) or n (%). Among study participants with available metabolomic data at both timepoints and key variables of interest (CMDs, Atole exposure).
Abbreviations: BMI, body mass index; CMDs, cardiometabolic diseases; HDL, high-density lipoprotein; IQR, interquartile range.
a P values based on Wilcoxon rank-sum tests.
bAt study visit date (of biological sample collection) in 2015–2017 data collection.
cNon-HDL-cholesterol (mg/dL) calculated as the difference between total (mg/dL) and HDL-cholesterol (mg/dL) plasma concentrations.
Table 2.
CMDs Among Guatemalan Adults (n = 349)
| Overall | Males | Females | P a | |
|---|---|---|---|---|
| N = 349 | n = 157 | n = 192 | ||
| n (%) | n (%) | n (%) | ||
| CMDs | ||||
| Obesityb | 116 (33%) | 33 (21%) | 83 (43%) | <0.01 |
| Diabetesc | 22 (6%) | 8 (5%) | 14 (7%) | 0.40 |
| Pre-diabetesc | 130 (37%) | 52 (33%) | 78 (41%) | 0.15 |
| Hypertensiond | 135 (39%) | 59 (38%) | 76 (40%) | 0.70 |
| Pre-hypertensiond | 67 (19%) | 36 (23%) | 31 (16%) | 0.11 |
| Metabolic syndromee | 173 (50%) | 48 (31%) | 125 (65%) | <0.01 |
| Central obesitye | 210 (60%) | 35 (22%) | 175 (91%) | <0.01 |
| High fasting blood glucose or medication usee | 151 (43%) | 60 (38%) | 91 (47%) | 0.09 |
| High triglycerides or statin usee | 167 (48%) | 76 (48%) | 91 (47%) | 0.85 |
| Low HDL- cholesterole | 241 (69%) | 91 (58%) | 150 (78%) | <0.01 |
| High blood pressure or medication usee | 113 (32%) | 48 (31%) | 65 (34%) | 0.52 |
| Any CMDf | 226 (65%) | 80 (51%) | 146 (76%) | <0.01 |
Data values are either median (IQR) or n (%). Among study participants with available metabolomic data at both timepoints and key variables of interest (CMDs, Atole exposure).
Abbreviations: BMI, body mass index; CMDs, cardiometabolic diseases; HDL, high-density lipoprotein; IQR, interquartile range.
a P values based on Mantel-Haenszel chi-square tests.
b According to World Health Organization categorization [16].
c Defined by recommendations of the American Diabetes Association [17] (See Supplemental Table 2 [14]).
d Per hypertension diagnosis cutoff values from the 2017 American College of Cardiology/American Heart Association Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults [18] (See Supplemental Table 2 [14]).
e Metabolic syndrome was defined as having 3 or more of the 5 criteria, based on the National Cholesterol Education Program guidelines [19] (See Supplemental Table 2 [14]).
f Metabolically unhealthy defined as having any of the 4 assessed CMDs. In other words, presence of obesity, hypertension, diabetes, and/or metabolic syndrome, including comorbidities.
We observed 9849 C18 and 13 908 HILIC metabolomic features in total (Supplemental Fig. 1) [14]. After data filtering, 5085 C18 and 7444 HILIC features remained eligible for the feature selection process, (Supplemental Fig. 1) [14]. Among all participants, 2090 features (C18: 1288; HILIC: 802) had peak areas that were altered following the meal challenge, compared to fasting (all q < 0.05; Table 3). Among these features, 1180 (56.5%) had putative annotations (49.2% C18; 68.1% HILIC, Table 3).
Table 3.
Summary of Metabolomic Features Differing by Meal Challenge and CMD Statusa
| Differing features (#) | Meal Challenge | Any CMD | ||
|---|---|---|---|---|
| LC-FT-MS columns | C18 | HILIC | C18 | HILIC |
| nb,c | 349 | 343 | 349 | 343 |
| Feature selection | ||||
| Stage 1: Overall linear regressiond | ||||
| q < 0.05 e | 1288 | 802 | --- | --- |
| Stage 2A: Stratified linear regressionf,g | ||||
| CMDs subgroupg | ||||
| q < 0.05e | --- | --- | 1063 | 693 |
| No CMDs subgroupg | ||||
| q < 0.05e | --- | --- | 875 | 508 |
| Stage 2B: Overall linear regressiong,h | ||||
| Interaction term (subgroup × time)g | ||||
| P < 0.05e | --- | --- | 60 | 48 |
| q < 0.05 | --- | --- | 0 | 0 |
| Annotations | Stage 1 | Stage 2B | ||
| Annotated features | 634 | 546 | 27 | 32 |
| Total annotations | 3406 | 5238 | 153 | 281 |
Abbreviations: CMD, cardiometabolic disease; FDR, false discovery rate; HILIC, hydrophilic interaction liquid chromatography; LC-FT-MS, liquid chromatography–Fourier transform mass spectrometry.
aValues in this table indicate the number of features with log2-normalized peak areas, which differed by the key subgroups of interest (meal challenge, any CMDs).
bThe total features observed were 9849 (C18) and 13 908 (HILIC). After data filtering, 5085 (C18) and 7444 (HILIC) features remained eligible for the feature selection approach. A complete case approach was utilized in multivariable regressions.
cAll regressions with C18 data were available among 349 participants with 2 samples (fasting, postprandial) of metabolomic data and key variables of interest (utilized to define CMDs). HILIC regressions were among 343 participants, based on data availability of key variables.
d Stage 1 feature selection was based on multivariable regressions. For each feature, a linear model with repeated measurements was utilized (Proc Mixed in SAS); the model equation was: Y (log2-normalized feature peak area)i,j,t = β 0i,j,t + β 1X1 (timepoint [postprandial, baseline]) i,j,t + β 2X2 (age at study visit)i + β 3X3 (sex)i, where each study participant was denoted as i, feature was j, and timepoint was t. Features remained eligible for Stage 2 feature selection if the beta-coefficient of the meal challenge timepoint (β 1) had FDR-adjusted P-value (q) < 0.05.
e These features were subsequently eligible for visualizations, annotations, and pathway analysis.
f Stage 2A feature selection utilized the same regression equation as in Stage 1, except these sets of regressions were stratified by CMD status (any versus none). In each subgroup, the total number of features with q < 0.05 of the beta-coefficient (β 1) of the meal challenge timepoint are included in this table.
g Among 1288 C18 features, or among 802 HILIC features
h Among features eligible from Stage 1 selection, these were also considered with multivariable regressions that additionally considered subgroups of interest and their interaction terms with meal challenge as independent variables (Stage 2B). A linear model with repeated measurements was utilized (Proc Mixed in SAS); the model equation was: Y (log2-normalized feature peak area)i,j,t = β 0i,j,t + β 1X1 (timepoint [postprandial challenge, baseline]) i,j,t + β 2X2 (any CMDs)i + β 3X3 (age at study visit)i + β 4X4 (sex)i, + β 5X5 (anyCMDs*timepoint)i where each study participant was denoted as i, feature was j, and timepoint was t. The number of features with P < 0.05 and q < 0.05 the beta-coefficient (β 5) of the interaction term were included in this table.
Comparing metabolomic response to meal challenge among participants with versus without CMDs
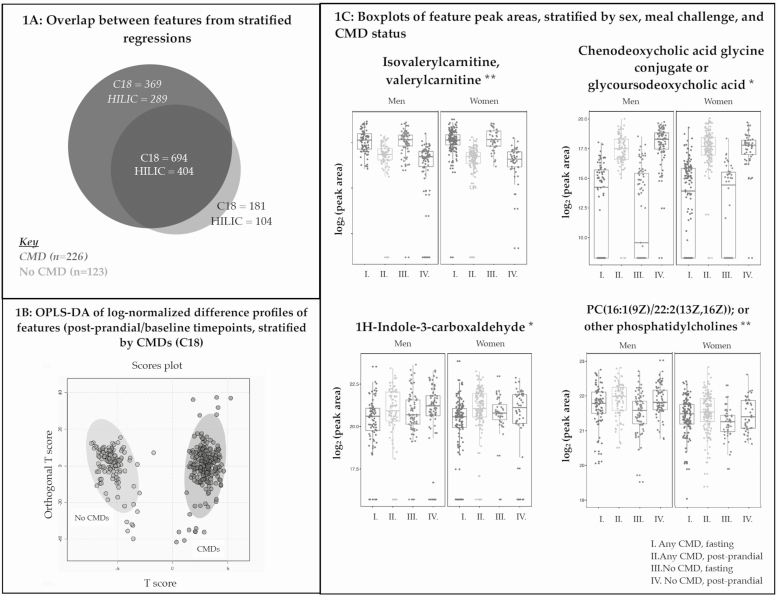
Among participants with CMDs (n = 226), 1063 C18 and 693 HILIC features had differential peak areas pre– versus post–meal challenge (all q < 0.05; Fig. 1A; Table 3). Of these C18 features, 747 increased and 316 decreased after the meal challenge. Among the HILIC features, 400 increased and 293 decreased following the meal challenge.
Figure 1.
Metabolomic feature peak area changes following standardized meal challenge differed between metabolically healthy versus unhealthy individuals. 1A: In this Venn diagram, each circle represents the numbers of features with beta coefficients with FDR-adjusted P values (q) < 0.05 in stratified regressions (Stage 2A in feature selection approach) in each subgroup (metabolically healthy or unhealthy). Circle diameters were proportionally scaled by the number of features (C18, HILIC) represented. 1B: OPLS-DA was used to compare whether feature peak area ratios (postprandial/fasting) clustered in distinct patterns among metabolically healthy versus unhealthy participants. Each circle represents an individual (metabolically healthy—light gray, metabolic unhealthy—dark gray). Data are from LC-FT-MS (C18, negative electrospray ionization). 1C: Examples of C18 (*) and HILIC (**) feature peak areas with putative annotations (from xMSannotator results among features selected in Stage 2B regressions with P < 0.05) are represented in boxplots, stratified by sex, meal challenge, and CMD status (Supplemental Table 4 [14]). Abbreviations: CMD, cardiometabolic disease; FDR, false discovery rate; HILIC, hydrophilic interaction liquid chromatography; LC-FT-MS, liquid chromatography Fourier transform mass spectrometry; OPLS-DA, orthogonal partial least squares–discriminant analysis.
In individuals without the 4 CMDs (n = 123), 875 C18 and 508 HILIC features had peak areas that differed after the meal challenge (all q < 0.05; Fig. 1A; Table 3). Of these C18 features, 613 increased and 262 decreased after the meal challenge. Among these HILIC features that were responsive to the meal challenge, 325 features increased and 183 features decreased.
Subtracting the overlapping differential features observed in both groups, 658 feature peak areas (369 C18; 289 HILIC) were altered following the meal challenge only among individuals with CMDs (Fig. 1A). In participants without CMDs, 285 unique features (181 C18; 104 HILIC) changed in the meal challenge and were not observed in the CMD subgroup (Fig. 1A). Log2-normalized postprandial:baseline peak area ratios clustered separately among metabolically healthy versus unhealthy participants, based on OPLS-DA (Fig. 1B) and PLS-DA (Supplemental Fig. 4) [14].
We also assessed whether the response of each metabolomic feature peak area to the meal challenge varied by CMD status by examining the 2-way multiplicative interaction between CMD status and the meal challenge timepoint. A total of 108 feature peak areas (60 C18; 48 HILIC) were associated with this interaction term (all P < 0.05; Table 3). Among these features, 59 (54.6%) had putative annotations (45.0% C18; 66.7% HILIC; Table 3). Excluding features with low xMSannotator confidence scores (MSI Level 4), delta parts per million (ppm) ≤5, there were 13 features from C18 data and 9 features from HILIC data with annotations (Supplemental Table 3) [14]. The interaction term was respectively associated with increased carnitines (trans-2-dodecanoylcarnitine) and dipeptides (histidinyl-tryptophan or tryptophyl-histidine) peak areas (all P < 0.05; MSI Levels 2 and 3 [31]; Supplemental Table 3 [14], Table 4). The interaction term was also associated with phospholipids, specifically increased phosphatidic acid (PA) 34:2 and decreased phosphatidylcholine (PC) 38:3 peak areas (all P < 0.05; Supplemental Table 3 [14], Table 4). Decreased peak area of a bile acid metabolite (chenodeoxycholic acid glycine conjugate or glycoursodeoxycholic acid) was associated with the interaction term (P < 0.05; Table 4). Increased 3b-17-b-dihydroxyetiocholane (a steroid hormone) and 1H-indole-3-carboxaldehyde (a microbiome-derived product) peak areas were respectively associated with the interaction term (both P < 0.05; MSI Levels 2 and 3 [31]; Table 4).
Table 4.
Associations Between Metabolomic Response to Meal Challenge and CMD Status
| Featurea | Monoisotopic mass | Adduct | Adjusted association with interaction term (meal challenge × any CMDs)b | Technical column | |||||
|---|---|---|---|---|---|---|---|---|---|
| β | SE | P | |||||||
| M/Z | RT | Annotation(s) | HMDB | ||||||
| 671.4670 | 275 | PA (34:2) | HMDB07860 | 672.4730 | M-H | 0.54 | 0.23 | 0.02 | C18 |
| 834.5970 | 27 | PC (38:3) c | HMDB08020 c | 811.6091 c | M+Na c | -0.10 | 0.04 | 0.01 | HILIC |
| 246.1697 | 28 | Isovalerylcarnitine, valerylcarnitine | HMDB00688, HMDB13128 | 245.1627 | M+H | -0.19 | 0.06 | <0.01 | HILIC |
| 343.2669 | 23 | Trans-2-dodecenoylcarnitine | HMDB13326 | --- | M+H [+1] | 0.60 | 0.29 | 0.03 | HILIC |
| 364.1367 | 60 | Histidinyl-tryptophan, tryptophyl-histidine | HMDB28896, HMDB29085 | 341.1488 | M+Na | 1.80 | 0.79 | 0.02 | HILIC |
| 386.1198 | 57 | Histidinyl-tryptophan, tryptophyl-histidine | HMDB28896, HMDB29085 | 341.1488 | M+2Na-H | 0.86 | 0.40 | 0.03 | HILIC |
| 159.0652 | 282 | Succinylacetone | HMDB00635 | 158.0579 | M+H | -0.19 | 0.09 | 0.04 | HILIC |
| 292.2369 | 232 | 3b,17b-dihydroxyetiocholane | HMDB00369 | --- | M-H [-1] | 0.73 | 0.35 | 0.04 | C18 |
| 450.3143 | 26 | Chenodeoxycholic acid glycine conjugate, glycoursodeoxycholic acid | HMDB00637, HMDB00708 | --- | M-H [+2] | -0.87 | 0.42 | 0.04 | C18 |
| 144.0454 | 240 | 1H-indole-3-carboxaldehyde | HMDB29737 | 145.0528 | M-H | 0.72 | 0.27 | 0.01 | C18 |
Abbreviations: CMD, cardiometabolic disease; HILIC, hydrophilic interaction liquid chromatography; LC-FT-MS, liquid chromatography–Fourier transform mass spectrometry; m/z, mass-to-charge ratio; OPLS-DA, orthogonal partial least squares–discriminant analysis; PA, phosphatidic acid; PC, phosphatidylcholine; RT, retention time.
a In this table, features were selected in Stage 2B regressions among all participants (n = 349), based on associations with interaction terms between any CMDs and meal challenge (P < 0.05). Among annotated features, those with energy, macronutrient metabolism, excretory or bile acids, and microbiome-derived metabolites included in this table.
b For each feature peak area, a linear model with repeated measurements was utilized (Proc Mixed in SAS; Stage 2B regressions). The model equation was: Y (log2-normalized feature peak area)i,j,t = β 0i,j,t + β 1X1 (timepoint [postprandial, baseline]) i,j,t + β 2X2 (any CMDs)i + β 3X3 (age at study visit)i + β 4X4 (sex)i, + β 5X5 (any CMDs*timepoint)i where each study participant was denoted as i, feature was j, and timepoint was t. In this table, the beta-coefficient, SE, and P value of the interaction term in each regression were included.
c Details regarding other phosphatidylcholines in Supplemental Table 3 [14].
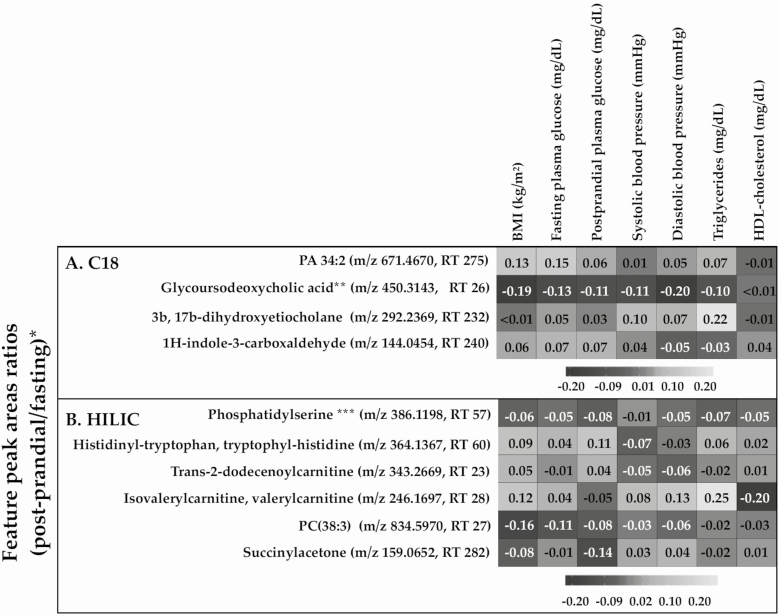
Correlations between log2 feature peak area ratios (postprandial/fasting) and a panel of CMD biomarkers are shown in Fig. 2. The peak area ratio of isovalerylcarnitine or valerylcarnitine (m/z 246.1697, retention time [RT] 28) was positively correlated with BMI, diastolic blood pressure, and triglycerides (all P < 0.05), and negatively correlated with HDL-cholesterol (P < 0.01). The peak area ratio of phosphatidylcholine (38:3; m/z 834.5970, RT 27) was negatively correlated with BMI and fasting plasma glucose (both P < 0.05). The peak area ratio of histidinyl-tryptophan or tryptophyl-histidine (m/z 346.1367, RT 60) was positively correlated with postprandial glucose (P = 0.03).
Figure 2.
Correlations between CMD biomarkers and metabolomic feature peak area ratios (postprandial/fasting), stratified by column. *All values are Spearman rank correlations of each bivariate associations between a log2-normalized feature peak area ratio (postprandial/fasting) and CMD biomarker. ** Chenodeoxycholic acid glucine conjugate was another potential annotation. *** Other potential annotations included histidinyl-tryptophan and tryptophyl-histidine. Abbreviations: m/z, mass-to-charge ratio; RT, retention time.
3. Discussion
Our results showed that a greater number of metabolomic feature peak areas differed following a standardized meal challenge among adults with CMDs (1756 features), compared with those without CMDs (1383 features). This finding reflected reduced metabolic flexibility, or worsened ability to acclimatize to the standardized meal challenge, among adults with CMDs. Metabolomic features that differed by the meal challenge and CMD status included dipeptides, glycerophospholipids, carnitines, and a bile acid metabolite.
Metabolic flexibility in CMDs
Our finding that more features changed after a meal challenge among adults with CMDs was consistent with the increased metabolic dysregulation in CMDs that has been previously described [2-4]. Healthy metabolism has been described as having metabolic resilience, or the ability to maintain stability in energy homeostasis during dynamic changes, such as caloric availability in a postabsorptive state or deficit in a fasting state [2-4]. One hypothesis is that during CMD pathophysiology, physiologic challenges result in short-term compensatory mechanisms of the body that gradually create an “allostatic load” or an inability to adequately respond, such as abnormal accumulation of lipids [32]. Different facets of metabolic inflexibility, including dysregulation of substrate availability (“push”) and requirement (“pull”), are associated with elevated or reduced biomarkers in obesity, insulin resistance, and deficiency [2, 3]. Increased triglycerides in skeletal muscle are often observed with insulin resistance and are hypothesized to be explained by abnormal muscle glucose oxidation (increased in a basal state, decreased with insulin administration) [33]. Other evidence has also shown metabolomic profiles associated with CMD status although many prior studies have been cross-sectional [5, 7-10, 34] and difficult to directly compare to our findings.
Protein metabolism in CMDs
We found that the meal challenge response of dipeptides and carnitines differed among participants with versus without CMDs. Prior evidence has corroborated that protein catabolites differed in CMDs [5, 7-10]. Murine studies have provided potential mechanisms of how catabolic protein metabolites affect systemic insulin resistance and obesity [35, 36]. Decreased relative concentrations of histidine and tryptophan were found in urine of rats with diabetes, compared with healthy controls [37]. One explanation is that large neutral amino acids, including tryptophan, phenylalanine, leucine, isoleucine, and valine, compete for cellular transport in mammalian cells [10]. In a dysregulated state, an imbalance of amino acids could result in decreased activities involving the other amino acids [10]. For example, it has been hypothesized that excess branched-chain amino acids (BCAAs) could result in decreased neurotransmitters such as serotonin, which is derived from tryptophan [10]. Other issues arising from an imbalance of amino acids include: (i) a BCAA catabolite increases trans-endothelial fatty acid transport; (ii) greater BCAAs activate the mammalian target of rapamycin (mTOR), which can cause the uncoupling of insulin signaling; and (iii) mitotoxic metabolites result in dysregulated beta-cell mitochondrial function [35, 36]. In vivo human studies have found that plasma tryptophan and histidine concentrations differed by type 2 diabetes [8, 37] and obesity [38] status.
We found that trans-2-dodecanoylcarnitine peak areas differed by CMD status and the meal challenge. There is limited previous evidence that can be directly compared, however prior studies with other study designs have highlighted the plausibility of our result. Dodecanoylcarnitine (C12) and other acylcarnitine peak intensities were increased among overweight participants after being randomly assigned a low-calorie diet for 12 weeks [39]. Several previous studies have reported the association between acylcarnitines and diabetes indicators, however the directionality of findings have been inconsistent [40-42]. One study found that lower concentrations of acylcarnitines were associated with improved diabetes indicators such as insulin sensitivity [40]. A separate study found that arachidonoyl-carnitine was associated with greater plasma glucose concentration [41]. A supplementation study among participants with impaired glucose tolerance showed that 2 g/day of L-carnitine increased acylcarnitine concentrations in skeletal muscle but had no effects on whole-body insulin sensitivity [42].
Putative mechanisms have been hypothesized to explain the observed links between acylcarnitines, obesity, and diabetes [43]. Bariatric surgery has been associated with improved type 2 diabetes and major metabolic alterations, including fatty acid metabolism [43, 44]. Medium- and long-chain fatty acids are transported as acylcarnitines across mitochondrial membranes for β-oxidation [6, 45]. Dodecanoylcarnitine and other acylcarnitines have a key role in fatty acid β-oxidation and have been hypothesized to indicate metabolic flexibility [43, 44].
Three dietary supplementation studies have shown that L-carnitine, a natural form found in red meats, improved metabolic flexibility [42, 46, 47]. A randomized, placebo-controlled, double-blind crossover trial showed that L-carnitine supplementation restored metabolic flexibility (evaluated by hyperinsulinemic-euglycemic clamp and high-energy meal challenge) among individuals with impaired glucose tolerance [42]. Murine studies have also confirmed the links between carnitines and metabolic flexibility [48, 49]. One study found that mice with a gene deletion of carnitine acetyltransferase had lower whole-body carbohydrate oxidation after a meal challenge [49]. L-carnitine supplementation resulted in increased average daily respiratory exchange ratio, which reflected systemic carbohydrate oxidation, among mice with obesity and insulin resistance [46].
Phospholipids in CMDs
Our findings showed that 2 glycerophospholipids (phosphatidylcholine and phosphatidic acid) were associated with the interaction between CMD status and meal challenge timepoint. Prior literature has established links between phospholipids with mitochondrial function [50], which is linked with metabolic flexibility [6]. Mitochondria have a central role in energy homeostasis at the cellular level, and the ability to alternate between converting different carbon sources (fatty acids, glucose, amino acids) to acetyl-CoA, which is substrate for the tricarboxyclic acid cycle [6]. Phosphatidylcholines and phosphatidylethanolamines account for the majority of phospholipids in mitochondrial membranes [51]. Fasting plasma insulin was positively associated with membrane lipids (phosphatidylcholine, phosphatidylethanolamine, sphingomyelin; all P < 0.05) among women with obesity and no diabetes [52].
Bile acids in dysregulated metabolism
In our study, the response of bile acid metabolite (chenodeoxycholic acid glycine conjugate or glycoursodeoxycholic acid) in a meal challenge differed by CMD status. Prior studies have found associations between glycoursodeoxycholic acid and diabetes treatment (metformin) [53] as well as outcomes (gestational diabetes) [54]. Metformin upregulates conjugated bile acids such as glycoursodeoxycholic acid, which is hypothesized to explain the lower circulating cholesterol concentrations observed with metformin treatment [53].
Bile acids have nonclassical functions in regulating energy homeostasis, including glucose and lipid metabolism [55-57]. Bile acid sequestration reduces plasma glucose and glycated hemoglobin among people with type 2 diabetes mellitus [56]. Putative mechanisms include the ability of bile acids to activate a cell surface receptor (G-protein–coupled receptor [GPCR] TGR5 [GPR131]) to improve glucose tolerance [58], and nuclear receptors (pregnane X receptor, constitutive androstane receptor, vitamin D receptor) that can subsequently affect transcriptional activities of bile acid, lipid, and glucose metabolism [59-61].
Limitations and strengths
We were unable to consider each of the 4 CMDs separately, given the limited power to detect differences across the respective subsets of people with each individual CMDs (eg, diabetes). Causal inferences could not be made based on our findings, given that CMD status was assessed during the same study visit as the meal challenge response. Our interpretation that a greater number of feature changes could reflect lower metabolic flexibility was based on the assumption that most metabolomic features have homeostatic regulation between the study timepoints among healthy individuals. There are still unresolved challenges in nontargeted metabolomics, particularly in resolving the major bottleneck of accurately identifying large numbers of known and unknown features [62]. Interpretations of our findings should account for the methodological limitations of nontargeted metabolomics, including that feature peak areas are relative quantifications. Future studies with orthogonal methods, including MS/MS, are needed for validation and confirmation. Features could not be converted to concentrations by reference standardization [22] as known concentrations of the calibrated reference were not available for annotated features selected by Stage 2B regressions. We did not assess the potential influences of genetics, the gut microbiome, or adipose or skeletal tissues on our associations of interest [3, 63].
Strengths of our study included the standardized meal challenge, which addresses the heterogeneity of meal challenge (eg, macronutrient composition, intake frequency, follow-up duration) in prior literature considering metabolomics. To our knowledge, our sample size was larger than other metabolomic studies with meal challenges among individuals with metabolic diseases [64, 65]. Nontargeted high-resolution metabolomic data have greater coverage of metabolites compared with other approaches [5]. The utilization of dual columns (C18, HILIC) and different electrospray ionization modes (positive, negative) additionally provided increased feature coverage [21]. Stable isotope-labeled internal standards and 2 sets of quality controls were included in the metabolomic assays, in order to monitor and correct potential sources of bias (eg, instrument drift).
4. Summary
Following a standardized meal challenge, a greater number of metabolomic feature peak areas differed among participants with CMDs, relative to those without CMDs, which could reflect increased metabolic dysregulation and decreased flexibility. Features that were altered by the meal challenge and CMD status included dipeptides, glycerophospholipids, carnitines, and a bile acid metabolite.
Acknowledgments
We thank ViLinh Tran, Ken Liu, and Kristine Dennis for their technical assistance.
Financial Support:. Research reported in this publication was supported by the National Institutes of Health: HD075784 (A.D.S., Eunice Kennedy Shriver National Institute of Child Health and Human Development); HL007745 (E.A.Y., National Heart, Lung and Blood Institute); S10 OD018006 (D.P.J., Office of the Director). D.P.J. was supported in part by R01 ES023485, U2C ES030163 and P30 ES019776 (National Institute of Environmental Health Sciences). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health; Eunice Kennedy Shriver National Institute of Child Health and Human Development; National Heart, Lung and Blood Institute; or National Institute of Environmental Health Sciences.
Author Contributions: A.D.S. conceived and designed this study. D.P.J. developed the high-resolution metabolomic assay protocol and workflow. M.R.Z. directed all field activities and supervised the glucose assays. T.Y. designed the bioinformatic workflow for the initial processing of mass spectrometry data. E.A.Y. and A.D.S. analyzed data, and E.A.Y. wrote the initial manuscript draft. All authors (E.A.Y., T.Y., D.P.J., M.R.Z., and A.D.S.) contributed to critically revising the paper. A.D.S. had primary responsibility for final content. All authors read and approved the final manuscript.
Glossary
Abbreviations
- BCAA
branched-chain amino acid
- BMI
body mass index
- CMD
cardiometabolic disease
- FDR
false discovery rate
- HILIC
hydrophilic interaction liquid chromatography
- INCAP
Institute of Nutrition of Central America and Panama
- IQR
interquartile range
- LC-FT-MS
liquid chromatography–Fourier transform mass spectrometry
- m/z
mass-to-charge ratio
- MSI
Metabolomics Standards Initiative
- OPLS-DA
orthogonal partial least squares–discriminant analysis
- PLS-DA
partial least squares–discriminant analysis
- RT
retention time
Additional Information
Disclosure Summary: E.A.Y., T.Y., D.P.J., M.R.Z., and A.D.S. have no conflicts of interest.
Data Availability
The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
- 1. World Health Organization. Noncommunicable Diseases Progress Monitor 2020. Geneva: World Health Organization; 2020. https://www.who.int/publications/i/item/ncd-progress-monitor-2020 [Google Scholar]
- 2. Smith RL, Soeters MR, Wüst RCI, Houtkooper RH. Metabolic flexibility as an adaptation to energy resources and requirements in health and disease. Endocr Rev. 2018;39(4):489-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodpaster BH, Sparks LM. Metabolic flexibility in health and disease. Cell Metab. 2017;25(5):1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van der Greef J, van Wietmarschen H, van Ommen B, Verheij E. Looking back into the future: 30 years of metabolomics at TNO. Mass Spectrom Rev. 2013;32(5):399-415. [DOI] [PubMed] [Google Scholar]
- 5. Newgard CB. Metabolomics and metabolic diseases: where do we stand? Cell Metab. 2017;25(1):43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muoio Deborah M. Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell. 2014;159(6):1253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15(5):606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roberts LD, Koulman A, Griffin JL. Towards metabolic biomarkers of insulin resistance and type 2 diabetes: progress from the metabolome. Lancet Diabetes Endocrinol. 2014;2(1):65-75. [DOI] [PubMed] [Google Scholar]
- 10. Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stein AD, Melgar P, Hoddinott J, Martorell R. Cohort Profile: the Institute of Nutrition of Central America and Panama (INCAP) Nutrition Trial Cohort Study. Int J Epidemiol. 2008;37(4):716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martorell R, Schroeder DG, Rivera JA. History and design of the INCAP longitudinal study (1969-77) and its follow-up (1988-89). J Nutr. 1995;125(4 Suppl):1027s-1041s [DOI] [PubMed] [Google Scholar]
- 13. Ford ND, Behrman JR, Hoddinott JF, et al. Exposure to improved nutrition from conception to age 2 years and adult cardiometabolic disease risk: a modelling study. Lancet Glob Health. 2018;6(8):e875-e884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu EA, Yu T, Jones DP, Ramirez-Zea M, Stein AD. Supplemental Materials - Metabolomic profiling after a meal shows greater changes and lower metabolic flexibility in cardiometabolic diseases. UNC Dataverse; 2020. 10.15139/S3/XSZGXR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Popkin BM, Adair LS, Ng SW. Global nutrition transition and the pandemic of obesity in developing countries. Nutr Rev. 2012;70(1):3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization. Obesity: preventing and managing the global epidemic. Report of a WHO Consultation. WHO Technical Report Series 894. Geneva: World Health Organization; 2000. [PubMed] [Google Scholar]
- 17. American Diabetes Association. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2019. Diabetes Care. 2019;42(1):S13-S28. [DOI] [PubMed] [Google Scholar]
- 18. Whelton PK, Carey RM, Aronow WS, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension. 2018;71(6):e13-e115. [DOI] [PubMed] [Google Scholar]
- 19. National Cholesterol Education Program. Third report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143-3421. [PubMed] [Google Scholar]
- 20. Go YM, Uppal K, Walker DI, et al. Mitochondrial metabolomics using high-resolution Fourier-transform mass spectrometry. Methods Mol Biol. 2014;1198:43-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013;9(1 Suppl):S132-S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Go YM, Walker DI, Liang Y, et al. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol Sci. 2015;148(2):531-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fernandes J, Chandler JD, Liu KH, Uppal K, Go YM, Jones DP. Putrescine as indicator of manganese neurotoxicity: dose-response study in human SH-SY5Y cells. Food Chem Toxicol. 2018;116(Pt B):272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu T, Park Y, Johnson JM, Jones DP. apLCMS–adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25(15):1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uppal K, Soltow QA, Strobel FH, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118-127. [DOI] [PubMed] [Google Scholar]
- 27. Smilde AK, van der Werf MJ, Bijlsma S, van der Werff-van der Vat BJ, Jellema RH. Fusion of mass spectrometry-based metabolomics data. Anal Chem. 2005;77(20):6729-6736. [DOI] [PubMed] [Google Scholar]
- 28. Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486-W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uppal K, Walker DI, Jones DP. xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Anal Chem. 2017;89(2):1063-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0–the human metabolome database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801-D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 2007;3(3):211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Orešič M. Systems Biology in Human Health and Disease. In: Orešič M, Vidal-Puig A, eds. A Systems Biology Approach to Study Metabolic Syndrome. Springer; 2014:17-23. [Google Scholar]
- 33. Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000;49(5):677-683. [DOI] [PubMed] [Google Scholar]
- 34. McGarrah RW, Crown SB, Zhang GF, Shah SH, Newgard CB. Cardiovascular metabolomics. Circ Res. 2018;122(9):1238-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jang C, Oh SF, Wada S, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med. 2016;22(4):421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10(12):723-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salek RM, Maguire ML, Bentley E, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics. 2007;29(2):99-108. [DOI] [PubMed] [Google Scholar]
- 38. Breum L, Rasmussen MH, Hilsted J, Fernstrom JD. Twenty-four-hour plasma tryptophan concentrations and ratios are below normal in obese subjects and are not normalized by substantial weight reduction. Am J Clin Nutr. 2003;77(5):1112-1118. [DOI] [PubMed] [Google Scholar]
- 39. Kang M, Yoo HJ, Kim M, Kim M, Lee JH. Metabolomics identifies increases in the acylcarnitine profiles in the plasma of overweight subjects in response to mild weight loss: a randomized, controlled design study. Lipids Health Dis. 2018;17(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liepinsh E, Makrecka-Kuka M, Makarova E, et al. Decreased acylcarnitine content improves insulin sensitivity in experimental mice models of insulin resistance. Pharmacol Res. 2016;113(Pt B):788-795. [DOI] [PubMed] [Google Scholar]
- 41. Weng L, Gong Y, Culver J, et al. Presence of arachidonoyl-carnitine is associated with adverse cardiometabolic responses in hypertensive patients treated with atenolol. Metabolomics. 2016;12(10):160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bruls YM, de Ligt M, Lindeboom L, et al. Carnitine supplementation improves metabolic flexibility and skeletal muscle acetylcarnitine formation in volunteers with impaired glucose tolerance: a randomised controlled trial. Ebiomedicine. 2019;49:318-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Samczuk P, Luba M, Godzien J, et al. “Gear mechanism” of bariatric interventions revealed by untargeted metabolomics. J Pharm Biomed Anal. 2018;151:219-226. [DOI] [PubMed] [Google Scholar]
- 44. Grenier-Larouche T, Carreau AM, Geloën A, et al. Fatty acid metabolic remodeling during type 2 diabetes remission after bariatric surgery. Diabetes. 2017;66(11):2743-2755. [DOI] [PubMed] [Google Scholar]
- 45. Bremer J. Carnitine–metabolism and functions. Physiol Rev. 1983;63(4):1420-1480. [DOI] [PubMed] [Google Scholar]
- 46. Power RA, Hulver MW, Zhang JY, et al. Carnitine revisited: potential use as adjunctive treatment in diabetes. Diabetologia. 2007;50(4):824-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Noland RC, Koves TR, Seiler SE, et al. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J Biol Chem. 2009;284(34):22840-22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koves TR, Ussher JR, Noland RC, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7(1):45-56. [DOI] [PubMed] [Google Scholar]
- 49. Muoio DM, Noland RC, Kovalik JP, et al. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 2012;15(5):764-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tian R, Colucci WS, Arany Z, et al. Unlocking the secrets of mitochondria in the cardiovascular system: path to a cure in heart failure—a report from the 2018 National Heart, Lung, and Blood Institute Workshop. Circulation. 2019;140(14):1205-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013;52(4):590-614. [DOI] [PubMed] [Google Scholar]
- 52. Zeghari N, Younsi M, Meyer L, Donner M, Drouin P, Ziegler O. Adipocyte and erythrocyte plasma membrane phospholipid composition and hyperinsulinemia: a study in nondiabetic and diabetic obese women. Int J Obes Relat Metab Disord. 2000;24(12):1600-1607. [DOI] [PubMed] [Google Scholar]
- 53. Pascale A, Marchesi N, Govoni S, Coppola A, Gazzaruso C. The role of gut microbiota in obesity, diabetes mellitus, and effect of metformin: new insights into old diseases. Curr Opin Pharmacol. 2019;49:1-5. [DOI] [PubMed] [Google Scholar]
- 54. Li J, Huo X, Cao YF, et al. Bile acid metabolites in early pregnancy and risk of gestational diabetes in Chinese women: a nested case-control study. Ebiomedicine. 2018;35: 317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Nierop FS, Scheltema MJ, Eggink HM, et al. Clinical relevance of the bile acid receptor TGR5 in metabolism. Lancet Diabetes Endocrinol. 2017;5(3):224-233. [DOI] [PubMed] [Google Scholar]
- 56. Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap–bile acids in metabolic control. Nat Rev Endocrinol. 2014;10(8):488-498. [DOI] [PubMed] [Google Scholar]
- 57. Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7(8):678-693. [DOI] [PubMed] [Google Scholar]
- 58. Thomas C, Gioiello A, Noriega L, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vallim TQdA, Edwards PA. Bile acids have the gall to function as hormones. Cell Metab. 2009;10(3):162-164. [DOI] [PubMed] [Google Scholar]
- 60. Zhang Y, Edwards PA. FXR signaling in metabolic disease. FEBS Lett. 2008;582(1):10-18. [DOI] [PubMed] [Google Scholar]
- 61. Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89(1):147-191. [DOI] [PubMed] [Google Scholar]
- 62. Viant MR, Kurland IJ, Jones MR, Dunn WB. How close are we to complete annotation of metabolomes? Curr Opin Chem Biol. 2017;36:64-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sayin SI, Wahlström A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225-235. [DOI] [PubMed] [Google Scholar]
- 64. Ferslew BC, Xie G, Johnston CK, et al. Altered bile acid metabolome in patients with nonalcoholic steatohepatitis. Dig Dis Sci. 2015;60(11):3318-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu P, Zhang J, Chen Y, et al. Analysis of human C24 bile acids metabolome in serum and urine based on enzyme digestion of conjugated bile acids and LC-MS determination of unconjugated bile acids. Anal Bioanal Chem. 2018;410(21): 5287-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.