Abstract
COVID19 has changed life for people worldwide. Despite lockdowns globally, computational research has pressed on, working remotely and collaborating virtually on research questions in COVID19 and the virus it is caused by, SARS-CoV-2. Molecular simulations can help to characterize the function of viral and host proteins and have the potential to contribute to the search for vaccines and treatments. Changes in the modus operandi of research groups include broader adoption of the use of preprint servers, earlier and more open sharing of methods, models, and data, the use of social media to rapidly disseminate information, online seminars, and cloud-based virtual collaboration. Research funders and computing providers worldwide recognized the need to provide rapid and significant access to computational architectures. In this review, we discuss how the interplay of all of these factors is influencing the impact – both potential and realized – of biomolecular simulations in the fight against SARS-CoV-2.
Index Terms: HPC, molecular dynamics simulations, computational biophysics, bioRxiv deposition, open access, COVID19 HPC Consortium, COVID19, SARS-CoV-2, glycans, spike protein, protease
In January 2020, few people could have envisioned how drastically the world as we knew it would be upended, and how quickly, due to COVID19. Unusual cases of viral pneumonia were first identified in Wuhan, China, at the end of 2019. The cause was determined to be a novel coronavirus (SARS-CoV-2). The first cases subsequently appeared in the US in late January 2020. By February, cases in Europe were spreading concemingly, particularly in the Lombardy region in Italy. Lockdowns were imposed in many countries in response. By mid-March, California went into lockdown, and the UK followed suit a short time later. Halfway through 2020, lockdowns have happened all across the world. By July 1 the US hit a milestone of 50,000 new confirmed COVID19 cases in a single day. In the first half of 2020 alone, over half a million people have died from this pandemic disease and it continues to spread globally. COVID19 is unmatched in recent history in terms of the devastation it is causing, both economically and in terms of human life and health.
Scientists the world over are working to meet this challenge, in the face of added obstacles posed by lockdowns. Research funders, companies and other organizations, are making impressive efforts and commitments to share and analyze scientific and biomedical data, which have emerged rapidly in the face of the pandemic 1. Computational researchers have to some extent been less affected by lockdowns than their experimental counterparts. Computational chemists and biologists can run jobs remotely on high performance computers (HPCs), or in the cloud, regardless of whether the scientist is in their office at a university/institute or from their kitchen table in their homes. Thus, many computational scientists in the fields of biology, chemistry, medicine and allied fields realized the chance to make potentially significant near- and long- term impact. Numerous groups pivoted their efforts to address the COVID19 challenge, seizing on the opportunity to challenge their methods with targets related to the SARS-CoV-2 virus and the disease it causes, and hoping to make a contribution to tackling it. The range of activity is huge and we can mention only a few examples here.
One area of science in which computation has the potential for impact on COVID19 research is biomolecular simulation and computational biophysics. These fields use molecular models to study the structures, interactions, and dynamics of proteins and other biological macromolecules. This includes atomically detailed simulations of the components of the SARS-CoV-2 virus and its interactions with host proteins and neutralizing antibodies. Such simulations can help to reveal how viral proteins function, to explore the dynamics of its RNA genome and interactions with protein components, as well as be used to explore the effects of genetic variations (i.e., mutations that the virus adopts during spread). Molecular simulations and related techniques can also potentially contribute to the search for drugs and vaccines. These computational experiments rely on data generated from experimental biological and chemical methods, in particular X-ray crystallography and cryoelectron microscopy (cryoEM), which give highly detailed three-dimensional structural data of the viral machinery and RNA genome. Simulations can provide atomically detailed insight – in particular on protein dynamics – not readily achievable by experiment alone. A so-called force field describes the interactions between the atoms in the system, which may number several million and contain either protein, RNA, DNA, lipids, or a combination of these. The derivative of this interaction potential defines the forces on the atoms, which are numerically encoded and predict their motion, determined by integrating Newton’s equation of motion over time. The integration is performed billions or trillions of times at short (femtosecond) timesteps, to build up a trajectory over time of the protein’s atomic-level movements. Depending on the size and complexity of the system studied, these calculations can contain hundreds-of-thousands to hundreds-of-millions of atoms, and can run a simulated timescale of nanoseconds to milliseconds. Simulations of this scale are ‘compute hungry’ and with appropriate code, can scale to many hundreds of nodes, thus are often ideally suited to HPC architectures.
The first structures of SARS-CoV-2 proteins appeared in bioRxiv (a preprint server, in which researchers disclose scientific results before peer-review and publication in a scientific journal 2 ) in mid-February. The increased adoption of the preprint servers for researchers working on SARS-CoV-2 means that data are coming to light much sooner than waiting for publication in a traditional peer-reviewed journal. For molecular simulation, this is a game changer: such data are particularly important for biomolecular simulations as they generally require structural data as starting points. For example, the first cryoEM structure of the SARS-CoV-2 spike protein was published in Science on March 13, 2020, but the data it contained were available for researchers at the time of deposition into the bioRxiv on February 15, 2020. Thus, early access to data provided via bioRxiv enabled biomolecular simulation researchers to start working with the structure at least one month sooner than they otherwise would have. Similarly, crystallographers rapidly solved and shared the structure of the SARS-CoV-2 main protease, an enzyme that chops up viral polypeptide to make the proteins the virus needs to assemble in cells. The Protein Data Bank now contains many structures of several viral proteins (solved by groups across the world), including structures bound to small molecules that may be useful in the search for new drugs.
Another shift in research culture has been increased interaction via social media, such as Twitter, which together with preprint servers, webinars and video meetings, are helping to connect scientists across the globe working on this grave threat. News of manuscripts, data, and preprints quickly spreads worldwide. Virtual lab meetings and conferences held over Zoom, WebEx, Skype, BlueJeans, Google Meet, Microsoft Teams, etc. have taken hold and suddenly the global research community has been rapidly connected in new ways. This has helped to compensate for the cancelation and postponement of physical scientific meetings and conferences. The increased spread of information, data sharing and discussion through emerging communication mechanisms continues to help drive knowledge generation, links between research communities and scientific discoveries about the virus and the complex disease that it causes.
In common with other scientific fields, the biomolecular simulation community recognized that, in order to have maximum impact for COVID19, changes to standard practices would be needed. An outcome of this realization is the commitment made by over 200 biomolecular simulation groups, from many countries, to a set of shared principles to share models and data. These principles include using preprint servers to communicate models and results quickly, and sharing methods and data much more quickly than would typically happen in normal scientific publication [1], Early discussion of methods and data sharing within the simulation community led to the development of a collective site for sharing methods and data through an international joint effort by the US NSF Molecular Software Sciences Institute and European Union BioExcel project3.
Recognizing the potential of computational science – spanning domains from epidemiology to data science to aerosol modeling – governments, research funders and agencies, computer centers and companies have prioritized COVID19 applications, providing expansive access to HPC and other resources. Several initiatives have been created to support biomolecular simulation across the world (e.g. through PRACE in the EU, ARCHER via the UKRI/EPSRC and the HECBioSim and CCP-BioSim networks in the UK, RIKEN in Japan, and cloud resources specifically donated by cloud providers such as AWS, Oracle, Microsoft Azure and Google). Companies such as DE Shaw Research have carried out simulations of viral protein targets and made the results freely available. The folding@home project brought together donated resources worldwide to simulate the products of the viral genome [2], In the US, the COVID19 High Performance Computing Consortium 4 brings together the most powerful compute resources and is making them broadly available via a rapid proposal process to researchers with appropriate compute needs. What started initially as a consortium in the US quickly spread to international partnerships, including with the United Kingdom and Sweden, making available over 485 petaflops together with technical expertise in software development and other resources. A key aspect of this Consortium is that it provides a mechanism for researchers to get fast access, with application review on the order of days, to support COVID19 projects. Projects are also working to combine simulations with other types of data and modeling. An example in the EU is Fenix, which is a distributed e-infrastructure providing different types of compute and storage resources. It is being used by several different projects performing COVID19 related research. Some of them are using the HPC resources for simulations. The infrastructure is also being used for sharing data through a publicly accessible object store. Resources are being allocated through different mechanisms including the PRACE Fast Track Call for COVID19 related projects5.
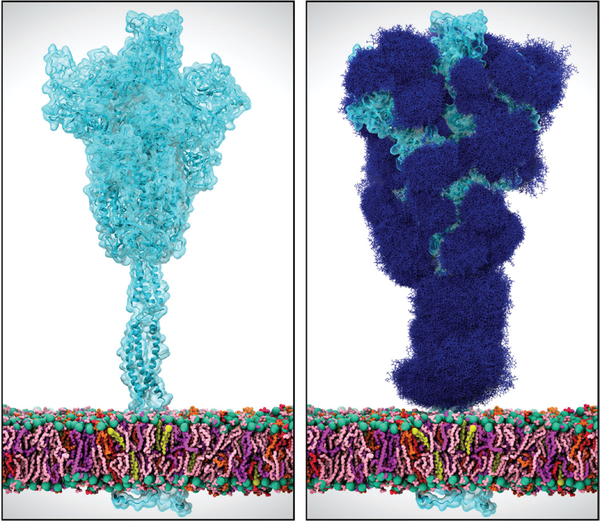
With support such as this, efforts of the biomolecular simulation community are contributing to understanding many facets of SARS-CoV-2. All of the proteins of SARS-CoV-2 are the targets of intensive modeling and simulation efforts, by many groups across the world. Similarly many groups are investigating human proteins that may be involved in the disease. Simulations are especially valuable in augmenting and extending experimental data. A particularly relevant example pertains to the sugary coating that the virus uses to mask its main infection machinery (the ‘spike’ protein) from the human immune system. This so-called ‘glycan shield’ is known to exist, with a particular make-up or composition of sugar types, but it is not possible to appreciate what the sugary shield looks like because of experimental limitations. Specifically, the glycans move too much to be captured in static images with high resolution; in other words, we know from experiments that the glycans are present, but scientists cannot ‘see’ the full structure. Molecular dynamics simulations are able to characterize the glycan shield and show how it hides the protein from the immune system (Fig. 1) [3–5]. Simulations are revealing how parts of the spike emerge from this shield to bind to human proteins to infect cells. Simulations are also being used to investigate the effects of genetic variations in the spike that have been identified by experiments. Knowing what parts of the virus surface are exposed, in what circumstances, and which parts of the virus are protected by this coating, allows researchers to understand better how neutralizing antibodies may work. Understanding the exposed portions of the spike may help in the rational design and development of vaccines. A number of efforts are directed at understanding which parts of the virus will lead to B- and T-cell epitopes and present new methods to do so [7], Simulations may also help identify parts of the spike structure, and the human proteins with which it interacts, that could be targets for binding of small molecule therapeutics.
Figure 1:
Simulations of the SARS-CoV-2 spike protein, embedded in a viral membrane (pink and purple lines), are being used to inform scientists what the spike looks like with (right panel) and without glycans (right panel, dark blue lines), in order to understand where neutralizing antibodies or drugs may bind.
Biomolecular simulations can help test and develop experimentally-testable hypotheses. They can potentially be performed rapidly and so ‘get ahead’ of experiments, e.g. delivering early predictions. Simulations can also analyze the effects of genetic variations on the structures and interactions of viral proteins. Computation can test hypotheses, in some cases more rapidly, more cheaply, and on a larger scale than experiments. Reverting to the example of the SARS-CoV-2 spike protein’s glycan shield, Casalino et al. predicted – ahead of any experimental data – that two glycans near the top of the spike head not only shielded the viral protein but also acted as a molecular trigger that would ‘lock and load’ the spike for infection. This hypothesis was developed and tested in silico, and several months later, experimentally confirmed by two independent groups. At the time of writing of this document, several other predictions are being developed and vetted by simulation groups studying SARS-CoV-2 targets as well as their interactions with host proteins. Molecular modeling is helping to analyze recently identified interaction of the spike with human neuropilin cell surface receptors [6] as well as potential interactions with the nicotinic acetylcholine receptor [7], Computing in the age of COV1D19 is providing a plethora of opportunities for simulators to work in concert with experimentalists directly, or indirectly via experimentally-testable predictions disclosed in preprints.
Molecular simulations also have the potential to contribute to the search for possible drugs. A common approach in structure-based drug development is to use in silico virtual screening methods to screen vast digitized libraries of small molecule compounds against targets of interest. ‘Docking’ codes can scan large numbers of small molecules to test whether they are likely to bind to a protein target, such as the SARS-CoV-2 main protease. Virtual libraries may contain tens-of-millions to billions of compounds, far more than can be tested experimentally. Researchers are using molecular docking codes to identify potential binders (‘hits’) among these large numbers of compounds, aiming to provide experimental labs with prospective compounds for testing. For COVID19, identification of drugs that have been approved for other conditions (drug repurposing), or compounds close to the clinic, that may have activity against SARS-CoV-2 targets (or human protein targets involved in infection or disease pathology) is an attractive approach to finding treatments that could quickly be tested in the clinic. Docking methods are highly approximate and of limited accuracy, so can be combined with more rigorous and detailed molecular simulations to include e.g. the effects of protein dynamics and to filter out false positives. Due to the large size of compound libraries and depending on the virtual screening method employed, these studies also can utilize HPC architectures [8], A remarkably early study at Oak Ridge National Lab carried out a large virtual screening campaign of FDA approved compounds to look for potential drug repurposing opportunities and the authors made the predicted results available on the bioRxiv in late February, only days after the spike structure was made available on bioRxiv. At time of writing of this article, experimental validation has not yet been disclosed, but many similar studies have made early predictions available via this route. For example, simulations are contributing to the Covid Moonshot Project, closely coordinated with experimental structural work and biochemical tests, in an effort to identify novel drug leads [9].
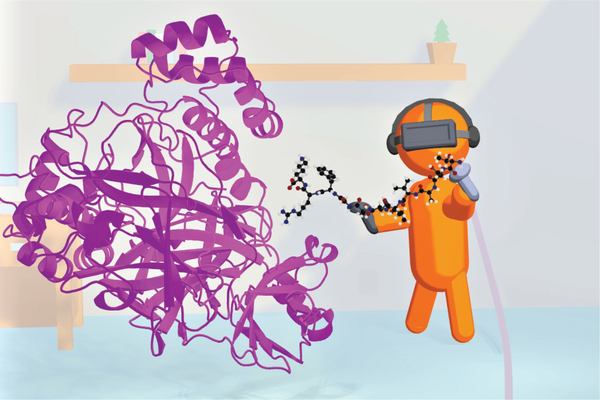
Simulations can also contribute to understanding other aspects of viral proteins and their mechanisms, which may also help in developing drugs. For example, as mentioned above, the SARS-CoV-2 main protease is a viral enzyme that breaks down long viral polypeptides into pieces that form viral proteins - essential for the manufacture of virus particles in the cell. Understanding the chemical mechanisms by which it does so, and its specificity for particular protein sequences, may help. Chemical reactions in proteins can be simulated by multiscale methods such as combined quantum mechanics/molecular mechanics (QM/MM) techniques [10,11], QM/MM methods can also be used to predict the reactivity of potential covalent binders as inhibitors and drug leads. Emerging artificial and machine learning methods will also be useful in extracting information from simulation data and connecting with experiment in the search for treatments. Interactive molecular dynamics simulations in virtual reality (iMD-VR, Figure 2) are a new way to interact with and manipulate biomolecular simulations. iMD-VR is an exciting frontier in structure-based drug design. VR offers huge potential for data sharing and distributed collaboration: when iMD-VR is cloud mounted, researchers in different physical locations can share the same virtual molecular environment, interacting together with a atomically detailed simulation and model e.g. a drug binding to its protein target. This can potentially transform how scientists collaborate, allowing researchers to work together directly even when based far from each other, to share and interrogate biomolecular models. An early example involves small molecule docking with iMD-VR to the SARS-CoV-2 main protease, where advantages to docking are realized due to the flexibility allowed by the real time protein rearrangements predicted via the MD [12].
Figure 2:
Virtual reality is emerging as a tool to interact with, and manipulate biomolecular simulations. Interactive molecular dynamics simulation in virtual reality (iMD-VR) has the potential to contribute to structure-based drug design, studies of protein structure and function, and education. This cartoon depicts a user ‘docking’ an oligopeptide substrate into the SARS-CoV-2 main protease (magenta), to model how this -viral enzyme binds the peptides that it breaks down as part of the COVID19 -viral lifecycle. The flexibility and atomically detailed interactions afforded by iMD-VR allow the user to manipulate the molecular structures to create realistic models.
The response of the biomolecular simulation community – from academic groups to computing sites, cloud providers and even chip developers such as NVIDIA – has been impressively strong and rapid, and in many cases, coordinated. Such efforts are already providing new insights and knowledge about the fundamental biology of SARS-CoV-2 as well as contributing to the discovery of novel chemical agents that could be developed into viable therapeutics. Equally striking is how COVID19 has the potential to catalyze longer-term change within the biomolecular simulation community, including the broad adoption of preprint servers for rapidly disclosing research results, and the rapid sharing of methods, models, and data, to disseminate information and knowledge, to test significance and reproducibility of models, and link to other areas of scientific investigation to tackle this global pandemic.
Acknowledgements
We thank Dr. Lorenzo Casalino (UC San Diego) and Becca Walters (University of Bristol) for creating Figure 1 and Figure 2, respectively. We also thank our research groups, as well as collaborators in the work described. We also thank especially Prof. Elisa Fadda (Maynooth University) and her research group, Prof. Michael Feig and Dr. Lim Heo (Michigan State University), Prof. Syma Khalid (University of Southampton), Prof. Carlos Simmerling and his research group (SUNY Stony Brook), Prof. Ben Neuman (Texas A&M University), Prof. Greg Voth and Dr. Viviana Monje-Galvan (University of Chicago), Prof. Julien Michel (University of Edinburgh) and his research group, Prof. Jean-Philip Piquemal (Sorbonne University), Prof. Alessio Lodola (University of Parma), Prof. Vicente Moliner (Universität Jaume I), Prof. Garrett Morris and Prof. Fernanda Duarte (Oxford University), Prof. Paolo Carloni (Jülich) and Dr. Reda Rawi (NIH Vaccine Research Center) for helpful discussions. REA thanks the Texas Advanced Computing Center for support and assistance with using Frontera. AJM thanks the Advanced Computing Research Centre, University of Bristol, the Isambard GW4 EPSRC Tier 2 HPC Centre, HECBioSim/EPSRC and Oracle for Research for computer time and technical support.
This paper was submitted for review on 6 July 2020. REA acknowledges funding by NIH GM132826, NSF RAPID MCB-2032054, an award from the RCSA Research Corp, and a UC San Diego Moore’s Cancer Center 2020 SARS-COV-2 seed grant. AJM is funded by EPSRC (CCP-BioSim, grant number EP/M022609/1), the British Society for Antimicrobial Chemotherapy (grant number BSAC-COVID-30), the Elizabeth Blackwell Institute for Health Research, University of Bristol, and for ARCHER HPC time via HECBioSim and the EPSRC/UKRI priority call for COVID19 applications.
Footnotes
Contributor Information
Rommie E. Amaro, UC San Diego.
Adrian J. Mulholland, University of Bristol.
References
- [1].Amaro RE, Mulholland AJ, A Community Letter Regarding Sharing Biomolecular Simulation Data for COVID-19, J. Chem. Inf. Model (2020) 0–6. doi: 10.1021/acs.jcim.0c00319. [DOI] [PubMed] [Google Scholar]
- [2].Zimmerman MI, Porter JR, Ward MD, Singh S, Vithani N, Meller A, Mallimadugula UL, Kuhn CE, Borowsky JH, Wiewiora RP, Hurley MFD, Harbison AM, Fogarty CA, Coifland JE, Fadda E, Voelz VA, Chodera JD, Bowman GR, Citizen Scientists Create an Exascale Computer to Combat COVID-19, BioRxiv (2020) 2020.06.27.175430. doi: 10.1101/2020.06.27.175430. [DOI] [Google Scholar]
- [3].Casalino L, Gaieb Z, Dommer AC, Harbison AM, Fogarty CA, Barros EP, Taylor BC, Fadda E, Amaro RE, Shielding and Beyond: The Roles of Glycans in SARS-CoV-2 Spike Protein, BioRxiv (2020) 2020.06.11.146522. doi: 10.1101/2020.06.11.146522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE, Aoki K, Kellman BP, Bridger R, Barouch DH, Brindley MA, Lewis NE, Tiemeyer M, Chen B, Woods RJ, Wells L, Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor, BioRxiv (2020) 2020.06.25.172403. doi: 10.1101/2020.06.25.172403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Turoñová B, Sikora M, Schürmann C, Hagen WJH, Welsch S, Blanc FEC, von Bülow S, Gecht M, Bagola K, Hömer C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Mühlebach MD, Hummer G, Krijnse Locker J, Beck M, In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges, Science (80-,). (2020) eabd5223. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Daly JL, Simonetti B, Antón-Plágaro C, Kavanagh Williamson M, Shoemark DK, Simón-Gracia L, Klein K, Bauer M, Hollandi R, Greber UF, Horvath P, Sessions RB, Helenius A, Hiscox JA, Teesalu T, Matthews DA, Davidson AD, Cullen PJ, Yamauchi Y, Neuropilin-1 is a host factor for SARS-CoV-2 infection, BioRxiv (2020) 2020.06.05.134114. doi: 10.1101/2020.06.05.134114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Oliveira ASF, Ibarra AA, Bermudez I, Casalino L, Gaieb Z, Shoemark DK, Gallagher T, Sessions RB, Amaro RE, Mulholland AJ, Simulations support the interaction of the SARS-CoV-2 spike protein with nicotinic acetylcholine receptors and suggest subtype specificity, BioRxiv (2020) 2020.07.16.206680. doi: 10.1101/2020.07.16.206680. [DOI] [Google Scholar]
- [8].Smith M, Smith JC, Repurposing Therapeutics for COVID-19: Supercomputer-Based Docking to the SARS-CoV-2 Viral Spike Protein and Viral Spike Protein-Human ACE2 Interface, (2020). doi: 10.26434/chemrxiv.H871402.v3. [DOI] [Google Scholar]
- [9].Chodera J, Lee AA, London N, von Delft F, Crowdsourcing drug discovery for pandemics, Nat. Chem 12 (2020) 581. doi: 10.1038/s41557-020-0496-2. [DOI] [PubMed] [Google Scholar]
- [10].Świderek K, Moliner V, Revealing the molecular mechanisms of proteolysis of SARS-CoV-2 Mpro by QM/MM computational methods, Chem. Sci (2020). doi: 10.1039/D0SC02823A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ramos-Guzmán CA, Ruiz-Pernía JJ, Tuñón I, Unraveling the SARS-CoV-2 Main Protease Mechanism Using Multiscale DFT/MM Methods, (2020). doi: 10.26434/chemrxiv.l2501734.v2. [DOI] [PubMed] [Google Scholar]
- [12].Deeks HM, Walters RK, Bamoud J, David R, Interactive molecular dynamics in virtual reality (iMD-VR )isan effective tool for flexible substrate and inhibitor docking to the SARS-CoV-2 main protease, (2020). doi:https://doi.org/i0.26434/chemrxiv.i2834335.v2. [DOI] [PubMed]