Abstract
Introduction
Patients with community-acquired pneumonia (CAP) undergo a dysregulated host response that is related to mortality. MicroRNAs (miRNAs) participate in this response, but their expression pattern and their role as biomarkers in CAP have not been fully characterized.
Methods
A prospective observational study was performed in a cohort of 153 consecutive patients admitted to hospital with CAP. Clinical and analytical variables were collected, and the main outcome variable was 30-day mortality. Small RNA was purified from plasma of these patients obtained on the first day of admission, and miRNA expression was analyzed by RT-PCR. Univariate and multivariate analyses were carried out through the construction of a logistic regression model. The proposed model was compared with established prognostic clinical scales using ROC curve analysis.
Results
The mean age of the patients included was 74.7 years [SD 15.9]. Their mean PSI was 100.9 [SD 34.6] and the mean modified Charlson index was 2.9 [SD 3.0]. Both miR-146a and miR-16-5p showed statistically significant association with 30-day mortality after admission due to CAP (1.10 vs. 0.23 and 51.74 vs. 35.23, respectively), and this association remained for miR-16-5p in the multivariate analysis adjusted for age, gender and history of bronchoaspiration (OR 0.95, p = 0.021). The area-under-the-curve (AUC) of our adjusted multivariate model (AUC = 0.954 95%CI [0.91–0.99]), was better than those of prognostic scales such as PSI (AUC = 0.799 [0.69–0.91]) and CURB-65 (AUC = 0.722 [0.58–0.86]).
Conclusions
High levels of miR-146a-5p and miR-16-5p upon admission due to CAP are associated with lower mortality at 30 days of follow-up. Both miRNAs could be used as biomarkers of good prognosis in subjects hospitalized with CAP.
Introduction
Community-acquired pneumonia (CAP) is a frequent and severe infection. Low tract respiratory infections are the fifth cause of overall mortality and the first infectious cause of mortality worldwide [1, 2]. In addition to its impact on survival, suffering from CAP affects post-episode quality of life and functionality [3], which represents a considerable burden on the health system [4].
Numerous strategies have been studied to improve the prediction of CAP prognosis, and thus help in decision-making regarding the management of these patients [5]. Various widely validated clinical scores have been developed, such as the Pneumonia Severity Index (PSI) [6] or CURB-65 [7], capable of evaluating the clinical situation at the time of diagnosis and predicting its evolution. In parallel, several factors of the inflammatory response associated with CAP have been studied as potential prognostic markers in these patients [8], such as procalcitonin, C-reactive protein (CRP) or leukocyte count, showing prognostic utility that was not better than common clinical scales [9].
MicroRNAs (miRNAs) are small non-coding RNA molecules that have a complementary antiparallel sequence to messenger RNAs (mRNAs). Their binding to specific mRNAs allows post-transcriptional regulation of gene expression, blocking protein synthesis [10]. They can be secreted to the extracellular milieu included in small extracellular vesicles called exosomes. Through these exosomes or bound to transport proteins, miRNAs can travel in the bloodstream and can be incorporated into other cells, thereby regulating their gene expression [11].
These molecules are very abundant, widely present in multiple tissues and biological fluids, and have evolutionarily conserved sequences [12]. They play a role in processes such as embryonic development, cell death and proliferation, hematopoiesis, neurodevelopment, and metabolic regulation [13, 14]. But perhaps one of their most important functions is their role in regulating immunological processes, including the innate and adaptive immune response, the development and differentiation of immune cells, and the prevention of autoimmune disorders [15].
Regarding CAP pathophysiology, miRNAs can influence the development and function of immune cells by blocking the translation of key proteins such as transcription factors or intermediate molecules in cell receptor signaling cascades [16]. In addition, some miRNAs have been identified as key players in modulating the immune response to severe bacterial infection, by controlling neutrophil activation and recruitment and the chemotactic signal that initiates the inflammatory process [17]. MiRNA determination in peripheral blood has been used for the diagnosis of malignancies, cardiovascular diseases or autoimmune disorders [18]. Moreover, several studies have established the utility of circulating miRNAs in the diagnosis of sepsis [19] and in various specific infections (e.g. HIV, viral hepatitis or tuberculosis) [20].
However, there is limited scientific literature on the usefulness of these molecules as prognosis markers in CAP.
In an attempt to find more accurate prognostic predictive tools for CAP, our research group set out to analyze the use of circulating microRNAs as prognostic biomarkers for mortality in this disease.
Materials and methods
Prospective observational study in a cohort of 153 consecutive patients admitted for CAP in 2015 at a university hospital in Spain. Patients older than 18 years diagnosed with CAP in the Emergency Room were included in the study. CAP was considered when patients presented symptoms of lower respiratory tract infection together with the appearance of a new infiltrate on a chest radiograph and the absence of an alternative diagnosis during follow-up, according to the usual definition [21]. Sociodemographic and clinical variables, presence of comorbidities (individual and grouped, such as the modified Charlson index [22]), characteristics of the infectious process (including the CAP severity indices CURB65 and PSI) and analytical and radiological parameters at admission were collected. These patients underwent a blood test on the first day of admission, and were treated according to the clinical practice guidelines in force at that time [5]. The main outcome variable was 30-day mortality.
This cohort has been previously used in other studies, in the context of a larger research project on prognostic biomarkers in CAP [23, 24]. All the data generated during this research are openly available in the public repository of Zenodo.org [https://doi.org/10.5281/zenodo.3930832]. Furthermore, the methodology followed can be found in the previously published protocol [25].
Laboratory procedures
Small RNA was purified from patients´ 250 μl plasma samples by column-based protocol, and retrotranscribed to cDNA (Exiqon’s miRCURY™ series kits, 4 μl of RNA in-put); synthetic RNA controls were added in this process (spike-ins UniSP2, UniSp4 and UniSp5 before RNA extraction, and UniSP6 before retrotranscription to cDNA). After cDNA was diluted 1:40, the quality of the process was evaluated (QC control Panel) and only 117 samples passed the test (A detailed explanation of the technical criteria used for the exclusion of samples in the quality control process can be found in S1 Fig in S1 Appendix). Eight samples paired by age and gender were selected (4 patients who had suffered a cardiovascular event or death during follow-up and 4 who had not) and a panel of 752 human miRNAs was tested (miRCURY LNA™ Universal—Ready-to-Use Human Panel, Exiqon), in order to determine a preliminary pattern of differential miRNA expression between patients with different CAP progression.
According to the preliminary data obtained, 25 candidate miRNAs were selected: 4 intended to be used as normalizers, 5 selected by statistical criteria (univariate association with mortality) and 16 selected from an exhaustive bibliographic search on miRNAs, sepsis, inflammation and / or cardiovascular disease, prioritizing those that appeared in a greater number of publications and those related to respiratory diseases. RT-PCR was carried out in triplicate by hybridization with double-stranded flurochrome (ExiLENT SYBR® Green Master Mix) using the C1000 Touch CFX384 thermocycler (Bio-Rad). A PCR efficiency of 2 was assumed.
The relative amount of each miRNA was calculated with ΔCt = CtmiRNA—CtUniSp2, and it was later normalized using the GeNorm algorithm and the geometric mean of the most stable miRNAs. The final data was calculated with the formula 2-ΔCt and the values were expressed as the fold change (FC) of each miRNA with respect to UniSP2, as described by Marabita et al. [26].
Only miRNAs whose Cts were less than 2 standard deviation (SD) above the average of the least abundant spike-in, UniSp5, were taken into consideration for the analysis.
Statistical analysis
For the descriptive analysis of the cohort, mean and SD were calculated for quantitative variables with equal variances, and median and interquartile range for quantitative variables with unequal variances. Normality of data was assessed with the Kolmogorov-Smirnov test and homoscedasticity with the Levene’s test. Qualitative variables were expressed as proportion and total cases. The relationship of the different independent variables with the cumulative incidence of the main dependent variable was analyzed using the Student’s t-test for quantitative variables with equal or unequal variances, or the χ2 test or the Fisher’s exact test for qualitative variables, as appropriate. For the correlation analysis of the candidate miRNAs, the Pearson´s test (represented as a heat-map) was used, followed by the Spearman´s correlation test. Subsequently, a multivariate analysis was carried out by constructing a logistic regression model (for 30-day mortality), in order to study possible confounding and intermediate variables. All p values ≤ 0.05 were considered statistically significant, although another threshold (p ≤ 0.10) was used in the processes of variable selection, following the principle of parsimony. Selection of the most parsimonious model was made with the Likelihood-Ratio test (LR test). The predictive capacity of the estimated model as well as the comparison with established scales was made using Receiver operator characteristic (ROC) curves and subsequent comparison between areas under the curve (AUC). In addition, net reclassification index (NRI) and integrated discrimination index (IDI) were calculated. Statistical analysis was carried out using Stata v15 and R v3.5.2.
Ethical principles
This study was previously approved by the Research Ethics Committee (REC) of Hospital Universitario de La Princesa and it was carried out following the ethical principles established in the Declaration of Helsinki, recommendations related to Good Clinical Practice, and the legislation in force regarding confidentiality. All the included patients were informed about the study and signed the informed consent, which was an inclusion criterion in this study.
Results
A total of 153 patients were included in the study. Mean age was 75.7 years [SD 16.1], with a greater proportion of men (58.2%, n = 86). Most had a previous history of smoking (65.1%, n = 99), with a chronic obstructive pulmonary disease (COPD) prevalence of 31.4% (n = 48). The most frequent cardiovascular risk factor was high blood pressure (58.2%, n = 89), and the most frequent cardiovascular comorbidity was chronic heart failure (18.9%, n = 29). The modified Charlson index was 3.12 points [SD 2.9]. The severity of pneumonia was quantified using the usual scales: average PSI index was 103 points [SD 35.2] and average CURB-65 index was 2.78 points [SD 1.1].
Analytical and radiological variables, as well as all the prognostic scales measured were compared between surviving and deceased patients 30 days after admission. Results are shown in Table 1 and S1 and S2 Tables in S1 Appendix. Eighteen patients died in the first 30 days after admission (11.8%).
Table 1. Sociodemographic variables and comorbidities.
| TOTAL n = 153 | 30-day mortality | p* | |||||
|---|---|---|---|---|---|---|---|
| ALIVE n = 135 | DECEASED n = 18 | ||||||
| % | n | % | n | % | n | ||
| Age—mean / SD | 75.68 | 16.13 | 73.82 | 16.18 | 89.69 | 5.15 | <0.001 |
| Gender (male) | 58.17 | 89 | 58.52 | 79 | 55.56 | 10 | 0.811 |
| Ethnicity (caucasian) | 97.39 | 149 | 97.04 | 131 | 100 | 18 | 1.000 |
| Life habits and vaccination | |||||||
| Alcoholism (active o former) | 8.50 | 13 | 8.89 | 12 | 5.56 | 1 | 1.000 |
| Tobacco use (active o former) | 65.10 | 99 | 65.93 | 89 | 58.82 | 10 | 0.563 |
| Pack-years—mean / SD | 27.24 | 29.66 | 27.17 | 29.37 | 27.86 | 33.38 | 0.935 |
| Pneumococcal vaccination | 41.33 | 62 | 40.60 | 54 | 47.06 | 8 | 0.611 |
| Flu vaccination (previous year) | 62.00 | 93 | 62.41 | 83 | 58.82 | 10 | 0.774 |
| Comorbidities | |||||||
| HBP | 58.17 | 89 | 59.26 | 80 | 50.00 | 9 | 0.454 |
| DM | 16.34 | 25 | 17.04 | 23 | 11.11 | 2 | 0.739 |
| Hypercholesterolemia | 32.68 | 50 | 30.37 | 41 | 50.00 | 9 | 0.095 |
| Obesity | 42.48 | 65 | 42.96 | 58 | 38.89 | 7 | 0.743 |
| TIA | 4.58 | 7 | 3.7 | 5 | 11.11 | 2 | 0.192 |
| Stroke | 11.76 | 18 | 9.63 | 13 | 27.78 | 5 | 0.041 |
| Ischemic cardiomyopathy | 8.5 | 13 | 7.41 | 10 | 16.67 | 3 | 0.183 |
| Chronic heart failure | 18.95 | 29 | 18.52 | 25 | 22.22 | 4 | 0.750 |
| VTE | 1.96 | 3 | 1.48 | 2 | 5.56 | 1 | 0.315 |
| CKD | 14.38 | 22 | 14.81 | 20 | 11.11 | 2 | 1.000 |
| Chronic hapatopathy | 3.27 | 5 | 3.7 | 5 | 0.00 | 0 | 0.406 |
| COPD | 31.37 | 48 | 31.85 | 43 | 27.78 | 5 | 0.794 |
| Asthma | 5.23 | 8 | 5.93 | 8 | 0.00 | 0 | 0.597 |
| HIV infection | 5.88 | 9 | 6.67 | 9 | 0.00 | 0 | 0.600 |
| Solid neoplasm | 9.15 | 14 | 8.15 | 11 | 16.67 | 3 | 0.216 |
| Modified Charlson Index—mean / SD | 3.12 | 2.94 | 2.98 | 2.98 | 4.22 | 2.39 | 0.092 |
| Chronic treatment | |||||||
| Bronchodilators (any type) | 33.33 | 51 | 34.07 | 46 | 27.78 | 5 | 0.791 |
| Oral corticosteroids | 3.97 | 6 | 3.73 | 5 | 5.88 | 1 | 0.518 |
| Statins | 29.14 | 44 | 28.36 | 38 | 35.29 | 6 | 0.553 |
| Antiplatelets | 26.14 | 40 | 25.19 | 34 | 33.33 | 6 | 0.460 |
| Functional status | |||||||
| Institutionalized | 7.84 | 12 | 5.93 | 8 | 22.22 | 4 | 0.037 |
| Cognitive impairment | 17.65 | 27 | 11.85 | 16 | 61.11 | 11 | <0.001 |
| Malnutrition | 11.11 | 17 | 9.63 | 13 | 22.22 | 4 | 0.119 |
| History of bronchoaspiration | 8.5 | 13 | 4.44 | 6 | 38.89 | 7 | <0.001 |
CKD: Chronic kidney disease; COPD: Chronic obstructive pulmonary disease; DM: Diabetes mellitus; HBP: High blood pressure; HIV: Human immunodeficiency virus; TIA: Transient ischemic attack; VTE: Venous thromboembolism.
Quantitative variables: Mean and SD; Qualitative variables: % and n;
* Qualitative variables: Chi-square test or Fisher´s exact test. Quantitative variables: t-test for equal or unequal variances as appropriate.
A blood sample was taken from all included patients upon admission. Small RNA was extracted from plasma samples and after quality control evaluation, only 117 samples were considered valid for miRNA analysis. A flowchart of the detailed technical criteria for exclusion of samples can be found in S1 Fig in S1 Appendix.
To assess whether sample exclusion was random, the main sociodemographic and clinical variables were compared between the group of 117 patients with valid samples and the group of 36 patients excluded. No statistically significant differences were found (p≤0.05) (S3 Table in S1 Appendix).
Analyzing the main outcome variable among those 117 patients with a valid sample, 11 patients (9.4%) died during the 30 days after admission for CAP.
Not all the microRNAs selected as candidates for analysis were measurable with guarantees in the set of 117 patients; of the 25 candidate miRNAs, 11 were excluded from the final analysis as they were not abundant enough in one or more patients (S4 Table in S1 Appendix). Four miRNAs were used as normalizers (miR-103a-3p, miR-23b-3p, miR-23a-3p and miR -25-3p).
Association of normalized expression of each miRNA (FCs) with to 30-days mortality was analyzed (Table 2).
Table 2. miRNA relative levels according to 30-day mortality.
| microRNAs | Total (n = 117) | 30-day mortality | p* | ||||
|---|---|---|---|---|---|---|---|
| No (n = 106) | Yes (n = 11) | ||||||
| mean | SD | mean | SD | mean | SD | ||
| hsa-miR-107 | 0.17 | 0.08 | 0.17 | 0.08 | 0.19 | 0.09 | 0.343 |
| hsa-miR-17-5p | 0.67 | 0.23 | 0.69 | 0.22 | 0.56 | 0.27 | 0.074 |
| hsa-miR-21 | 3.00 | 1.42 | 2.97 | 1.45 | 3.26 | 1.05 | 0.529 |
| hsa-miR-144-3p | 4.08 | 3.98 | 4.04 | 4.06 | 4.46 | 3.17 | 0.740 |
| hsa-miR-16-5p | 50.19 | 40.26 | 51.74 | 41.76 | 35.23 | 14.91 | 0.010 |
| hsa-miR- 486 | 1.29 | 1.24 | 1.32 | 1.29 | 0.99 | 0.63 | 0.159 |
| hsa-miR-20a | 0.65 | 0.29 | 0.67 | 0.28 | 0.52 | 0.38 | 0.099 |
| hsa-miR-34a-3p | 0.02 | 0.03 | 0.01 | 0.03 | 0.03 | 0.05 | 0.223 |
| hsa-miR-106b-5p | 2.01 | 1.19 | 2.02 | 1.23 | 1.88 | 0.70 | 0.709 |
| hsa-miR-146a | 1.02 | 1.78 | 1.10 | 1.85 | 0.23 | 0.14 | <0.001 |
| hsa-miR-483-5p | 0.02 | 0.04 | 0.01 | 0.04 | 0.04 | 0.08 | 0.285 |
| hsa-miR-125b | 0.03 | 0.06 | 0.03 | 0.04 | 0.07 | 0.17 | 0.453 |
* t-test for equal or unequal variances, as appropriate.
MiR-16-5p and miR-146a levels were both significantly higher in patients who survived compared to those who died after 30 days of follow-up (p = 0.010 and p <0.001, respectively). Distribution of these miRNAs according to mortality is shown in Fig 1.
Fig 1. Relative expression levels (FCs) at admission of miR-16-5p (A) and miR-146a (B) according to 30-day mortality after hospitalization for CAP.
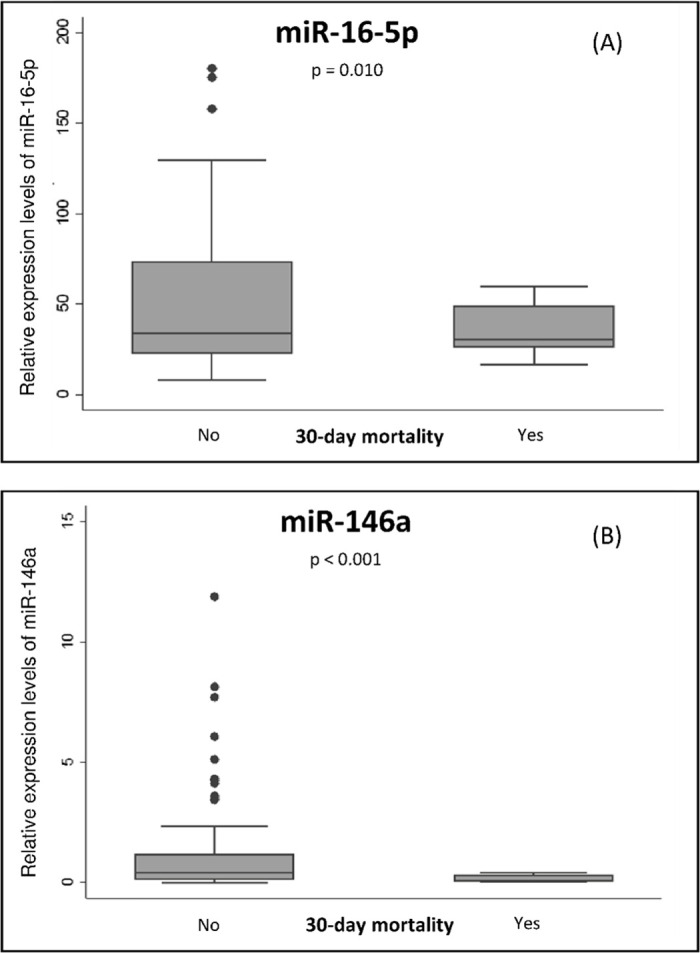
Data are represented as box plots. Dots represent outliers. Differences were analyzed by Student´s t test for unequal variances p values ≤ 0.05 are considered statistically significant.
Next, we analyzed whether the expression of both candidate miRNAs showed correlation. Heatmap representation of correlations between normalized relative quantities of the candidate miRNAs showed that miR-16-5p expression did not show a strong correlation with miR-146a, although it correlated with three other miRNAs (miR-106-5p, miR-486p and miR-144-3p; Fig 2). Moreover, a direct analysis of the correlation between miR-146a and miR-16-5p showed a weak correlation (rho = -0.57, p <0.001).
Fig 2. Heatmap representation of correlations between miRNAs in plasma of CAP patients.
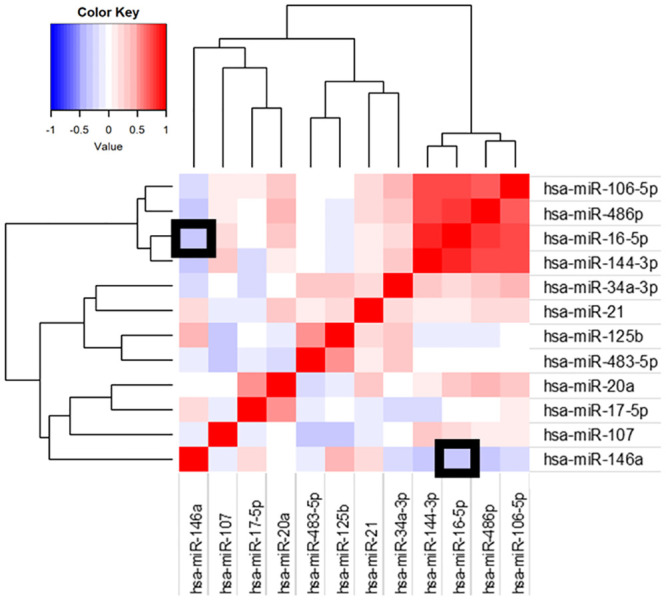
Graphic represents correlation between plasma levels of different miRNAs in CAP patients at admission. Correlation coefficients are represented by a color scale. Red colors represent positive correlations and blue colors negative correlations. Darker hues present higher coefficients. Squares outlined in black show the correlation between miR-146a and miR-16-5p.
Subsequently, to assess the prognostic power of both selected miRNAs, a multivariate model was constructed through a logistic regression. For this analysis, all variables with p≤0.10 in the univariate analysis were included, as well as variables of clinical significance such as sex or comorbidity assessed by modified Charlson index. The PSI and CURB65 prognostic scales were excluded from the model, since they were constructed from variables already included in the multivariate analysis, and also to be able to later compare them with the fitted model.
Finally, after comparing the models using the LR test, and always keeping in the model the two significant miRNAs from the univariate analysis, the most parsimonious model was constructed with the variables age, sex, history of bronchoaspiration, miR-146a and miR-16-5p (Table 3).
Table 3. Multivariate models of selected miRNAs, non-adjusted or adjusted for confounding variables.
| NON-ADJUSTED MULTIVARIATE MODEL | MULTIVARIATE MODEL ADJUSTED FOR CONFOUNDING VARIABLES | |||||
|---|---|---|---|---|---|---|
| OR | 95% CI OR | p | OR | 95% CI OR | p | |
| miR-16-5p | 0.97 | 0.94-0.99 | 0.025 | 0.95 | 0.91-0.99 | 0.021 |
| miR-146a | 0.04 | 0.00-0.81 | 0.036 | 0.05 | 0.00-2.00 | 0.109 |
| Age | 1.36 | 1.05-1.77 | 0.022 | |||
| Sex | 0.09 | 0.001-1.50 | 0.093 | |||
| History of bronchoaspiraton | 36.49 | 1.48-899.17 | 0.028 | |||
Sex (female vs male), History of bronchoaspiration (yes vs no).
MiR-16-5p maintained statistical significance as a prognostic marker; based on the estimated OR, it could be interpreted that for each decrease of one unit of miR-16-5p the probability of survival would increase 1.05 folds. This model presents a high LR Test (χ2 = 41.231; p <0.001), and good predictive capacity (R2Cox & Snell = 0.297; R2Nagalkerke = 0.640). Moreover, it is superior to the unadjusted multivariate model (χ2 = 14.887; p = 0.001), with lower generalized coefficients of determination (R2Cox & Snell = 0.119 and R2Nagalkerke = 0.258).
Lastly, the predictive capacity for 30-day mortality of both models and the validated prognosis clinical scales was assessed by ROC curve analysis (Fig 3).
Fig 3. ROC curve analysis of non-adjusted and adjusted multivariate models and common CAP severity scales PSI y CURB-65 for 30-day mortality.
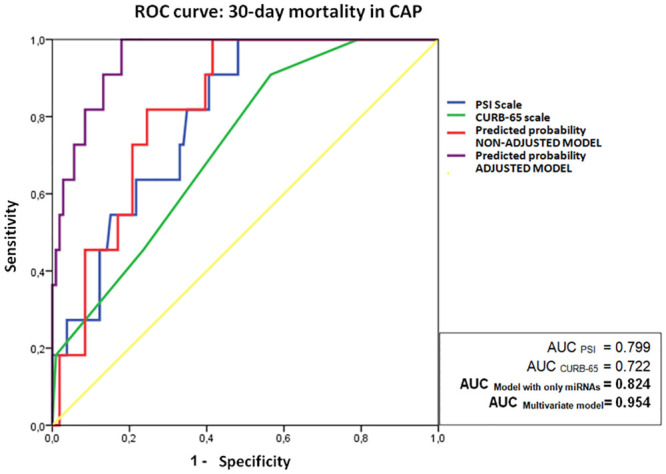
The prognostic capacity of the adjusted model (AUC = 0.954 95%CI [0.91–0.99]), was better than the unadjusted model (AUC = 0.824 [0.73–0.92]), and likewise better than the prognostic scales PSI (AUC = 0.799 [0.69–0.91]) and CURB-65 (AUC = 0.722 [0.58–0.86]).
In addition, we sought to test how the model proposed could classify patients according to 30-day mortality compared to classic prognosis scales such as CURB-65 and PSI. For that purpose, NRI and IDI were assessed. The NRI estimated for our multivariate model vs CURB-65 was 59.61% (SE = 1391.69; p = 1) and vs PSI was 39.54% (SE = 1029.78; p = 1); IDI estimation for our model vs. CURB-65 was -0.40 (SE = 0.12; p = 0.007) and vs. PSI was -0.41 (SE = 0.11; p = 0.004).
Discussion
The present study tries to assess the utility of circulating microRNA levels as prognostic biomarkers in patients admitted to hospital for CAP. For the selection of candidates, a two-step study was performed, with a first global approach using microarrays, together with a selection based on previous literature, followed by a second confirmation stage using semi-quantitative RT-PCR. After analysis of twenty-five candidate miRNAs, only two of them, miR-146a and miR-16-5p, showed a statistically significant association with mortality 30 days after admission for CAP. High levels of both miRNAs were associated with greater survival. This association remained for miR-16-5p in the multivariate analysis after adjusting for age, gender, and history of bronchoaspiration. In our sample of patients admitted with CAP, this adjusted model was at least as good at predicting mortality at 30 days as the classic CURB-65 and PSI prognostic scales, after comparison of AUCs and evaluation of reclassification indices NRI and IDI. Therefore, and waiting to standardize the method and replicate it in other cohorts, our results show that the measurement of miR-146a and miR-16-5p could be useful for predicting short-term mortality after admission for CAP.
The use of circulating miRNAs as biomarkers is not new, and although is not yet widespread as routine clinical practice, it has been successfully applied in the field of respiratory diseases [27].
Regarding diagnosis, some authors have studied in depth the use of miRNAs as biomarkers for pneumonia with respect to other respiratory diseases. For this purpose miRNA levels have been determined in various biological fluids: in serum, allowing patients with pulmonary tuberculosis to be distinguished from healthy controls and patients with CAP [28]; in exosomes from pleural fluid, distinguishing between CAP and lung cancer [29]; or in sputum, discriminating active pulmonary tuberculosis from other diseases [30]. Within pneumonias, the determination of circulating miRNAs has allowed differentiating viral pneumonia from bacterial pneumonia in pediatric population [31]. It has also been used in the adult population, differentiating bacterial etiology (Streptococcus pneumoniae) from viral etiology (Influenza H3N2 virus) [32]. Interestingly, apart from studies in respiratory diseases, miR-146a determination in plasma has been successfully used as a diagnostic biomarker of sepsis in patients with clinical criteria for systemic inflammatory response syndrome (SIRS) [33].
Regarding prognosis, few studies have evaluated the ability of these molecules to predict disease progression. Wu et al. found that elevated miR-146a, miR-27a, miR-126, and miR-155 in serum exosomes were associated with increased occurrence of acute respiratory distress syndrome in patients with CAP; they even concluded that miR-126 could be used as a prognostic marker, as it was statistically associated with 28-day mortality [34]. In another recent article, Zhang et al., using a sepsis-specific preloaded microarray concluded that miR-223-3p could be used to predict the development of sepsis in CAP [35]. As far as we know, there are no other studies–in the literature that have investigated the use of the determination of circulating miRNAs levels to evaluate CAP prognosis.
Our finding of lower 30-day mortality in patients with elevated levels of miR-16-5p and miR-146a at admission for CAP could reflect a better inflammatory response against the invading pathogen.
MiR-16-5p has been linked to mechanisms of protection from lung damage after infection. In cell models subjected to lipopolysaccharide (LPS)-induced damage, overexpression of miR-16-5p reduced acute lung damage through inhibition of the systemic inflammatory response via inhibition of TNF-α and interleukin-6 [36]. These results have subsequently been replicated in an animal model of chronic lung infection with Mycoplasma gallisepticum; overexpression of miR-16-5p was able to stop the inflammatory response, exerting its inhibitory effect directly on PI3K kinase, which is a key component in the NF-κB activation cascade, and therefore, for TNF-α production [37].
Likewise, elevated miR-146a levels have been associated with reduction of LPS induced lung inflammation: exogenous addition of miR-146a significantly suppress LPS-induced inflammatory response (TNF-α, IL-6, and IL-1β expression) in alveolar macrophages, through inhibition of IRAK-1 and TRAF-6 expression, both key components of the NF-κB activation cascade [38].
Interestingly, in murine models of pneumococcal pneumonia, exogenous mimetic miRNAs that inhibit this pathway—such as miR 124 3p [39] and miR-302 [40]—promote the regeneration of alveolar epithelial cells and improve the recovery of mice affected by bacterial pneumonia.
Thus, a physiopathogenic explanation of the protective effect of circulating miR-16-5p and miR-146a observed in our patients could be related to their inhibitory effect on the inflammatory response. High levels of both miRNAs detected in CAP patients upon admission could be involved in reducing activation of the inflammatory cascades secondary to lung infection, thereby decreasing systemic inflammatory burden, and allowing a better clinical evolution in the medium term.
Furthermore, these results would be in line with those already published by our research group showing that uncontrolled inflammation in CAP and its quantification by means of blood markers allows predicting adverse prognosis in short and medium term follow-up [23, 24, 41].
We consider that the main weaknesses of our study are the difficult standardization of miRNA quantification, a common problem in this type of studies, and the exclusive recruitment of hospitalized patients, which makes it difficult to compare our miRNA data with widely used prognostic scales.
The main strengths are the sample size reached, which was sufficient to achieve statistically significant results, the use of strict quality criteria in the selection of valid samples, the use of various endogenous and exogenous miRNAs in the standardization process, and above all, the selection process of miRNAs in two steps, which ensured a good initial selection of candidates.
Nevertheless, the prognostic value of miR-16-5p and miR-146a described in this work needs to be further confirmed in routine clinical practice.
Conclusions
In CAP patients requiring hospitalization, elevated plasma levels of miR-146a-5p and miR-16-5p measured at admission are associated with lower mortality at 30 days of follow-up. These two miRNAs could be used in the future as biomarkers of good prognosis in patients hospitalized for CAP.
Supporting information
(DOCX)
Acknowledgments
We would like to thank Mrs. Gloria Mateos and Emilia Roy, MD, of Biomedical Research Institute La Princesa for their contribution to the revision of the Methodology section of this manuscript. We are also grateful to Fernando Moldenhauer, MD PhD, for his constant support and reviews and to Ana Gómez Berrocal, MD PhD, for her sincere opinions and the style corrections introduced in the final writing of this manuscript.
Abbreviations
- AUC
Area Under the Curve
- CAP
Community-acquired pneumonia
- CRP
C-reactive protein
- FC
Fold changes
- IL
Interleukin
- miRNAs
Micro RNAs
- PSI
Pneumonia Severity Index
- ROC
Receiver operator characteristic
- RT-PCR
Real-time Polymerase chain reaction
- SIRS
Systemic inflammatory response syndrome
- TNF
Tumor necrosis factor
Data Availability
All data generated during this research are openly available from Zenodo.org (https://doi.org/10.5281/zenodo.3930832 or https://zenodo.org/record/3930832).
Funding Statement
This work has been funded by the Carlos III Health Institute (ERDF, European Regional Development Fund), by the Spanish Society of Pneumology and Thoracic Surgery and by the Ministry of Science, Innovation and Universities of Spain. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Solomon CG, Wunderink RG, Waterer GW. Community-Acquired Pneumonia. N Engl J Med. 2014;370: 543–551. 10.1056/NEJMcp1214869 [DOI] [PubMed] [Google Scholar]
- 2.Troeger C, Forouzanfar M, Rao PC, Khalil I, Brown A, Swartz S, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17: 1133–1161. 10.1016/S1473-3099(17)30396-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangen M-JJ, Huijts SM, Bonten MJM, de Wit GA. The impact of community-acquired pneumonia on the health-related quality-of-life in elderly. BMC Infect Dis. 2017;17: 208 10.1186/s12879-017-2302-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Miguel-Díez J, Jiménez-García R, Hernández-Barrera V, Jiménez-Trujillo I, de Miguel-Yanes JM, Méndez-Bailón M, et al. Trends in hospitalizations for community-acquired pneumonia in Spain: 2004 to 2013. Eur J Intern Med. 2017;40: 64–71. 10.1016/j.ejim.2016.12.010 [DOI] [PubMed] [Google Scholar]
- 5.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis Off Publ Infect Dis Soc Am. 2007;44 Suppl 2: S27–72. 10.1086/511159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fine MJ, Auble TE, Yealy DM, Hanusa BH, Weissfeld LA, Singer DE, et al. A Prediction Rule to Identify Low-Risk Patients with Community-Acquired Pneumonia. N Engl J Med. 1997;336: 243–250. 10.1056/NEJM199701233360402 [DOI] [PubMed] [Google Scholar]
- 7.Lim WS, Lewis S, Macfarlane JT. Severity prediction rules in community acquired pneumonia: a validation study. Thorax. 2000;55: 219–223. 10.1136/thorax.55.3.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sungurlu S, Balk RA. The Role of Biomarkers in the Diagnosis and Management of Pneumonia. Clin Chest Med. 2018;39: 691–701. 10.1016/j.ccm.2018.07.004 [DOI] [PubMed] [Google Scholar]
- 9.Sibila O, Restrepo MI. Biomarkers in community-acquired pneumonia: still searching for the one. Eur Respir J. 2019;53: 1802469 10.1183/13993003.02469-2018 [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75: 843–854. 10.1016/0092-8674(93)90529-y [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Liang H, Zhang J, Zen K, Zhang C-Y. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22: 125–132. 10.1016/j.tcb.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107: 823–826. 10.1016/s0092-8674(01)00616-x [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431: 350–355. 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 14.Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci CMLS. 2008;65: 545–562. 10.1007/s00018-007-7355-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauley KM, Chan EKL. MicroRNAs and Their Emerging Roles in Immunology. Ann N Y Acad Sci. 2008;1143: 226–239. 10.1196/annals.1443.009 [DOI] [PubMed] [Google Scholar]
- 16.Fernández-Messina L, Gutiérrez-Vázquez C, Rivas-García E, Sánchez-Madrid F, de la Fuente H. Immunomodulatory role of microRNAs transferred by extracellular vesicles. Biol Cell Auspices Eur Cell Biol Organ. 2015;107: 61–77. 10.1111/boc.201400081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar Kingsley SM, Vishnu Bhat B. Role of MicroRNAs in the development and function of innate immune cells. Int Rev Immunol. 2017;36: 154–175. 10.1080/08830185.2017.1284212 [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Chen J, Sen S. MicroRNA as Biomarkers and Diagnostics. J Cell Physiol. 2016;231: 25–30. 10.1002/jcp.25056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benz F, Roy S, Trautwein C, Roderburg C, Luedde T. Circulating MicroRNAs as Biomarkers for Sepsis. Int J Mol Sci. 2016;17 10.3390/ijms17010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma P, Pandey RK, Prajapati P, Prajapati VK. Circulating MicroRNAs: Potential and Emerging Biomarkers for Diagnosis of Human Infectious Diseases. Front Microbiol. 2016;7: 1274 10.3389/fmicb.2016.01274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim WS, Baudouin SV, George RC, Hill AT, Jamieson C, Jeune IL, et al. BTS guidelines for the management of community acquired pneumonia in adults: update 2009. Thorax. 2009;64: iii1–iii55. 10.1136/thx.2009.121434 [DOI] [PubMed] [Google Scholar]
- 22.Bordon J, Wiemken T, Peyrani P, Paz ML, Gnoni M, Cabral P, et al. Decrease in Long-term Survival for Hospitalized Patients With Community-Acquired Pneumonia. Chest. 2010;138: 279–283. 10.1378/chest.09-2702 [DOI] [PubMed] [Google Scholar]
- 23.Curbelo J, Luquero Bueno S, Galván-Román JM, Ortega-Gómez M, Rajas O, Fernández-Jiménez G, et al. Inflammation biomarkers in blood as mortality predictors in community-acquired pneumonia admitted patients: Importance of comparison with neutrophil count percentage or neutrophil-lymphocyte ratio. PLOS ONE. 2017;12: e0173947 10.1371/journal.pone.0173947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luquero-Bueno S, Galván-Román JM, Curbelo J, Ortega-Gómez M, Lancho-Sánchez Á, Vega-Piris L, et al. Interleukin-6 levels as an evolution marker of late mortality and cardiovascular events after an episode of community-acquired pneumonia. Eur Respir J. 2018;52: PA2606 10.1183/13993003.congress-2018.PA2606 [DOI] [Google Scholar]
- 25.Rajas O, Ortega-Gómez M, Román JMG, Curbelo J, Jiménez GF, Piris LV, et al. The incidence of cardiovascular events after hospitalization due to CAP and their association with different inflammatory markers. BMC Pulm Med. 2014;14: 197 10.1186/1471-2466-14-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marabita F, de Candia P, Torri A, Tegnér J, Abrignani S, Rossi RL. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. 2016;17: 204–212. 10.1093/bib/bbv056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abd-El-Fattah AA, Sadik NAH, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys. 2013;67: 875–884. 10.1007/s12013-013-9575-y [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Guo J, Fan S, Li Y, Wei L, Yang X, et al. Screening and Identification of Six Serum microRNAs as Novel Potential Combination Biomarkers for Pulmonary Tuberculosis Diagnosis. PLOS ONE. 2013;8: e81076 10.1371/journal.pone.0081076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin J, Wang Y, Zou Y-Q, Chen X, Huang B, Liu J, et al. Differential miRNA expression in pleural effusions derived from extracellular vesicles of patients with lung cancer, pulmonary tuberculosis, or pneumonia. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2016. 10.1007/s13277-016-5410-6 [DOI] [PubMed] [Google Scholar]
- 30.Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PloS One. 2012;7: e43184 10.1371/journal.pone.0043184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang F, Bai J, Zhang J, Yang D, Fan H, Huang L, et al. Identification of potential diagnostic biomarkers for pneumonia caused by adenovirus infection in children by screening serum exosomal microRNAs. Mol Med Rep. 2019;19: 4306–4314. 10.3892/mmr.2019.10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poore GD, Ko ER, Valente A, Henao R, Sumner K, Hong C, et al. A miRNA Host Response Signature Accurately Discriminates Acute Respiratory Infection Etiologies. Front Microbiol. 2018;9 10.3389/fmicb.2018.02957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Wang H-C, Chen C, Zeng J, Wang Q, Zheng L, et al. Differential expression of plasma miR-146a in sepsis patients compared with non-sepsis-SIRS patients. Exp Ther Med. 2013;5: 1101–1104. 10.3892/etm.2013.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Wu C, Gu W, Ji H, Zhu L. Serum Exosomal MicroRNAs Predict Acute Respiratory Distress Syndrome Events in Patients with Severe Community-Acquired Pneumonia. BioMed Res Int. 2019;2019: 3612020 10.1155/2019/3612020 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Zhang W, Jia J, Liu Z, Si D, Ma L, Zhang G. Circulating microRNAs as biomarkers for Sepsis secondary to pneumonia diagnosed via Sepsis 3.0. BMC Pulm Med. 2019;19: 93 10.1186/s12890-019-0836-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai Z-G, Zhang S-M, Zhang Y, Zhou Y-Y, Wu H-B, Xu X-P. MicroRNAs are dynamically regulated and play an important role in LPS-induced lung injury. Can J Physiol Pharmacol. 2012;90: 37–43. 10.1139/y11-095 [DOI] [PubMed] [Google Scholar]
- 37.Zhang K, Han Y, Zhao Y, Sun Y, Zou M, Fu Y, et al. Upregulated gga-miR-16-5p Inhibits the Proliferation Cycle and Promotes the Apoptosis of MG-Infected DF-1 Cells by Repressing PIK3R1-Mediated the PI3K/Akt/NF-κB Pathway to Exert Anti-Inflammatory Effect. Int J Mol Sci. 2019;20 10.3390/ijms20051036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng Z, Gong H, Li Y, Jie K, Ding C, Shao Q, et al. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp Lung Res. 2013;39: 275–282. 10.3109/01902148.2013.808285 [DOI] [PubMed] [Google Scholar]
- 39.Gao W, Yang H. MicroRNA‑124‑3p attenuates severe community‑acquired pneumonia progression in macrophages by targeting tumor necrosis factor receptor‑associated factor 6. Int J Mol Med. 2019;43: 1003–1010. 10.3892/ijmm.2018.4011 [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Li Y, Zhang P, Baker ST, Wolfson MR, Weiser JN, et al. Regenerative therapy based on miRNA-302 mimics for enhancing host recovery from pneumonia caused by Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 2019;116: 8493–8498. 10.1073/pnas.1818522116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curbelo J, Rajas O, Arnalich B, Galván-Román JM, Luquero-Bueno S, Ortega-Gómez M, et al. Estudio del porcentaje de neutrófilos y el cociente de neutrófilos-linfocitos como marcadores pronósticos en pacientes hospitalizados por neumonía adquirida en la comunidad. Arch Bronconeumol. 2019;55: 472–477. 10.1016/j.arbres.2019.02.005 [DOI] [PubMed] [Google Scholar]


