Abstract
Primary ovarian insufficiency (POI) is a heterogeneous disorder associated with several genes. The majority of cases are still unsolved. Our aim was to identify the molecular diagnosis of a Brazilian cohort with POI. Genetic analysis was performed using a customized panel of targeted massively parallel sequencing (TMPS) and the candidate variants were confirmed by Sanger sequencing. Additional copy number variation (CNV) analysis of TMPS samples was performed by CONTRA. Fifty women with POI (29 primary amenorrhea and 21 secondary amenorrhea) of unknown molecular diagnosis were included in this study, which was conducted in a tertiary referral center of clinical endocrinology. A genetic defect was obtained in 70% women with POI using the customized TMPS panel. Twenty-four pathogenic variants and two CNVs were found in 48% of POI women. Of these variants, 16 genes were identified as BMP8B, CPEB1, INSL3, MCM9, GDF9, UBR2, ATM, STAG3, BMP15, BMPR2, DAZL, PRDM1, FSHR, EIF4ENIF1, NOBOX, and GATA4. Moreover, a microdeletion and microduplication in the CPEB1 and SYCE1 genes, respectively, were also identified in two distinct patients. The genetic analysis of eleven patients was classified as variants of uncertain clinical significance whereas this group of patients harbored at least two variants in different genes. Thirteen patients had benign or no rare variants, and therefore the genetic etiology remained unclear. In conclusion, next-generation sequencing (NGS) is a highly effective approach to identify the genetic diagnoses of heterogenous disorders, such as POI. A molecular etiology allowed us to improve the disease knowledge, guide decisions about prevention or treatment, and allow familial counseling avoiding future comorbidities.
Introduction
Primary ovarian insufficiency (POI), also known as premature ovarian failure (POF), results in primary or secondary amenorrhea, hypoestrogenism, infertility, and elevated gonadotrophin levels (FSH>LH) [1]. POI patients have shown widely varying clinical phenotypes starting with women at puberty up to 40-year-old women. These patients can present primary amenorrhea, usually diagnosed at a younger age with delay of puberty and absence of breast development, whereas secondary amenorrhea is diagnosed at ages from <20 up to 40 years and is characterized by an irregular menstrual cycle and most often normal pubertal development and is the most frequent POI phenotype [2].
POI appears to have a genetic component presenting as sporadic or familial. However, the inheritance pattern is known to be autosomal dominant or autosomal recessive and monogenic, and recently, oligogenic inheritance has been proposed [3]. The heterogenous genetic basis of POI can rely on over 75 genes [2]. These genes are involved in several pathways such as gonadal development, meiosis (DNA replication and repair), hormonal signaling, immune function, and metabolism, although the majority of POI cases are yet to be elucidated [2,4]. This limitation may be narrowed with advancements in massively parallel sequencing, also known as next-generation sequencing (NGS). This method has been used as an effective tool to elucidate the genetic origin of heterogeneous diseases, such as POI. The aim of this study was to identify the genetic diagnosis of 50 patients with POI by using targeted massively parallel sequencing (TMPS).
Patients and methods
POI cohort
Approval from the institutional review board and written informed consent were obtained from all subjects or the parents or guardians of the minors before blood collection for DNA analysis. This study was approved by the Ethics Committee of Hospital das Clínicas, University of São Paulo Medical School, Brazil (protocol number 2015/12837/1.015.223). A cohort of 50 women presenting with POI was selected for this study between 2014 and 2018 at the Hospital das Clínicas University of São Paulo Medical School (Table 1). All patients had high levels of gonadotropins (FSH>20 U/L), hypoestrogenism, absence of FMR1 premutation, and positive anti-ovary or 21-hydroxylase antibodies. In primary amenorrhea cases, the patients also presented delay of puberty and absence of spontaneous breast development. The patients were treated with conjugated estrogens daily followed by progesterone replacement in the first 12 days of the month, resulting in menstrual bleeding and complete breast development in patients with primary amenorrhea.
Table 1. Clinical features of 50 Brazilian women with primary ovarian insufficiency.
| Patient ID | Amenorrhea | Age at first appointment (yr) | Previous treatment | FSH (IU/L) | LH (IU/L) | Height (cm) | Tanner stage at diagnosis | Syndromic features | Associate phenotype |
|---|---|---|---|---|---|---|---|---|---|
| POI-1 | Secondary | 27 | No | 76 | 43 | 163 | B5/P5 | ||
| POI-2 | Primary | 23 | Yes (at 17yr) | 99 | 45 | 166 | B5/P4 | High arched palate; Cubitus valgus | |
| POI-3 | Primary | 19 | 29 | 9 | 170 | B1/P2 | Cubitus valgus | Hearing loss, sensorineural | |
| POI-4 | Primary | 17 | 128 | 51 | 147 | B1/P1 | |||
| POI-5 | Primary | 32 | Yes | 48 | 25 | 161 | B4/P4 | Type 2 Diabetes mellitus | |
| POI-6 | Primary | 17 | Yes (at 16yr) | 65 | 25 | 166 | B5/P4 | ||
| POI-7 | Primary | 17 | 21 | 12 | NA | B1/P2 | |||
| POI-8 | Secondary | 28 | Yes (at 16yr) | 71 | 21 | 168 | B4/P4 | ||
| POI-9 | Primary | 18 | 118 | 64 | 145 | B1/P2 | High arched palate; Cubitus valgus; wide-spaced nipples, growth deficit | Macrocytic anemia | |
| POI-10 | Secondary | 32 | 89 | 19 | NA | NA | |||
| POI-11 | Primary | 13 | 100 | 44 | 141 | B3/P3 | Short stature | ||
| POI-12 | Secondary | 37 | 38 | 32 | 147 | B5/P5 | |||
| POI-13 | Primary | 23 | 83 | 28 | NA | B2/P4 | |||
| POI-14 | Secondary | 21 | Yes (at 19yr) | 63 | 39 | 162 | B2/P2 | ||
| POI-15 | Secondary | NA | 64 | 20 | NA | NA | |||
| POI-16 | Primary | 19 | 87 | 51 | 151 | B2/P3 | |||
| POI-17 | Secondary | 25 | 76 | 36 | NA | NA | |||
| POI-18 | Primary | 19 | Yes (at 16yr) | 46 | 28 | 163 | B3/P4 | High arched palate | Dyslipidemia |
| POI-19 | Primary | 30 | Yes (at 12yr) | 58 | 24 | 156 | B2/P5 | High arched palate | |
| POI-20 | Primary | 26 | Yes (at 25yr) | 138 | 47 | 167 | B4/P4 | Cubitus valgus | Tremor on right/dominant hand; no muscle atrophy |
| POI-21 | Primary | 21 | 89 | 37 | 161 | B3/P3 | |||
| POI-22* | Primary | 21 | Yes (at 17yr) | 94 | 25 | 164 | B5/P5 | Migraine | |
| POI-23* | Primary | 17 | Yes (at 15yr) | 45 | 21 | 176 | B4/P5 | Wilson's disease and keratoconus | |
| POI-24 | Primary | 16 | No | 106 | 42 | 157 | B1/P3 | Cubitus valgus; Late psychomotor development | |
| POI-25 | Secondary | 21 | 96 | 61 | NA | NA | |||
| POI-26 | Primary | 14 | No | 87 | 51 | 164 | B1/P1 | High arched palate | Chronic telogen effluvium, hypothryroidism, type 2 diabetes mellitus |
| POI-27 | Secondary | 17 | Yes (at 15yr) | 98 | 27 | 164 | NA | Precocious puberty (no treated) | |
| POI-28 | Primary | 18 | 48 | 10 | 167 | B1/P3 | |||
| POI-29 | Primary | 14 | 114 | 34 | 150 | B1/P1 | |||
| POI-30 | Primary | 31 | Yes | 86 | 32 | 170 | NA | ||
| POI-31 | Secondary | 37 | 103 | 10 | 160 | B5/P5 | |||
| POI-32 | Secondary | 34 | Yes (at 18yr) | 47 | 15 | 164 | B5/P5 | ||
| POI-33 | Secondary | 32 | Yes (at 28yr) | 18 | 14 | 148 | B5/P5 | ||
| POI-34 | Primary | 34 | Yes (at 18yr) | 65 | 30 | 156 | NA | ||
| POI-35 | Primary | 22 | Yes (at 15yr) | 42 | 19 | 157 | B4/NA | High arched palate; sindactilia | |
| POI-36 | Secondary | 38 | Yes (at 27yr) | 63 | 36 | 155 | B5/P5 | ||
| POI-37 | Primary | 14 | No | 96 | 32 | 157 | B1/P4 | Cafe au lait spots; high arched palate; cubitus valgus; hyperdontia | Congenital heart murmur |
| POI-38 | Primary | 23 | Yes (at 19yr) | 69 | NA | 154 | B4/P5 | ||
| POI-39 | Secondary | 19 | Yes (at 16yr) | 72 | 44 | 166 | B5/P4 | High arched palate | |
| POI-40 | Secondary | 38 | Yes (at 27yr) | 52 | 35 | 155 | B5/P5 | Type 2 Diabetes mellitus | |
| POI-41 | Secondary | 34 | 38 | 13 | 168 | B5/P5 | |||
| POI-42 | Secondary | 35 | 50 | 21 | 172 | B5/P5 | |||
| POI-43 | Secondary | 35 | 88 | 35 | 151 | B4/P3 | |||
| POI-44 | Secondary | 30 | 94 | 25 | 160 | B5/P5 | |||
| POI-45 | Primary | 43 | Yes (at 33yr) | 75 | 28 | 153 | B4/P5 | Hearing loss, sensorineural; Kidney transplant, congenital heart murmur | |
| POI-46 | Secondary | 40 | Yes (at 39yr) | 32 | 7 | 162 | B5/P5 | ||
| POI-47 | Secondary | 27 | Yes | 75 | 56 | 172 | B5/P5 | High arched palate, low-set posteriorly rotated ears | |
| POI-48 | Primary | 17 | No | 119 | 36 | 175 | B1/P4 | Familial Ectrodactyly | |
| POI-49# | Primary | 18 | 63 | 32 | 155 | B1/P2 | |||
| POI-50# | Primary | 16 | 66 | 28 | 151 | B2/P4 |
NA: not available.
* Siblings of Family 1.
# Sibling of Family 2.
Molecular analyses
Targeted Massively Parallel Sequencing (TMPS)
Genomic DNA was extracted from peripheral blood leukocytes using standard procedures. A custom SureSelectXT DNA target enrichment panel was designed using SureDesign tools (Agilent Technologies Santa Clara, CA, USA). Based on human and animal gonadal development and function, known and candidate genes were selected as follows:
1) Gonadal formation: CBX2, CTNNB1, DHH, EMX2, FGF9, FST, GATA4, LHX9, NR0B1, NR5A1, RSPO1, SOX9, SOX8, SRY, WNT4, WT1, and ZFPM2;
2) Ovarian development: AMH, AMHR2, BCL2, BCL2L2, BMP15, BMP4, BMP8B, BMPR1B, BMPR2, CDKN1B, CYP11A1, CYP17A1, CYP19A1, DAZL, DIAPH2, DND1, EIF2B2, EIF2B5, eIF4ENIF1, FIGLA, FMN2, FOXL2, FOXO1, FOXO3, FOXO4, FSHR, GDF9, GJA4, INHA, INHBA, INHBB, INSL3, KIT, KITLG, LHCGR, LHX8, NANOS1, NANOS2, NANOS3, NBN, NOBOX, PGRMC1, POF1B, POLR3H, POR, POU5F1, PDGFRA, PRDM1, PTEN, SMAD1, SMAD4, SMAD5, SOHLH1, SOHLH2, SOX3, STAR, STRA8, TCF21, TGFBR3, TIAL1, UBE3A, and ZFX;
3) Meiosis and DNA repair genes: ATM, BRWD1, CDC25B, CDK2, CKS2, CPEB1, CYP26B1, DMC1, ERCC1, ERCC2, CBS-PGBD3, FANCA, FANCC, FANCG, FANCL, GJA4, GPR3, HFM1, HSF2, MCM8, MCM9, MEI1, MLH1, MLH3, MOS, MSH4, MSH5, NOS3, NUP107, PMS2, PSMC3IP, RAD51B, REC8, SGOL2, SMC1B, SPO11, STAG3, SYCE1, SYCP1, SYCP2, SYCP3, TOP3B, TRIP13, UBB, and UBR2;
4) Putative variants, detected by GWAS, and causative genes without a clear mechanism in ovary function: ADAMTS16, ADAMTS19, BRSK1, CHM, COL4A6, DACH2, DMRT1, ERS1, ESR2, FMR1, HARS2, HK3, HSD17B4, LARS2, NCOA1, NXF5, PAPPA, RSPO2, TSHB, and XPNPEP2;
5) Candidate and known genes of disorders/differences in sex development: AKR1C2, AKR1C4, AR, ARX, ATRX, CDH7, CYP21A2, DHCR7, DMRT1, DMRT2, HNF1B, HSD11B1, HSD17B3, HSD3B2, FGFR2, LHX1, MAMLD1, MAP3K1, SRD5A2, SOX3, and WWOX.
Exonic regions and 25 base pairs of intronic flanking sequence of all genes were included. The proband’s libraries were prepared according to the SureSelectXT Target Enrichment Protocol (Agilent Technologies Santa Clara, CA, USA). Deep sequencing of these amplicon libraries was performed on a NextSeq 500 next-generation sequencer (Illumina San Diego, CA, USA). Alignment of raw data and variant calling were performed following the steps described by França and collaborators [5]. The first criterion used to distinguish new variants from polymorphisms was filtered variants with a MAF<0.01 in 1000 Genomes, Exome Variant Server NHLBI GO Exome Sequencing Project (ESP), and Exome Aggregation Consortium (ExAC) databases. The variants were evaluated in the gnomAD database and in the results section; only gnomAD is shown since all public databases described above are included in it. Moreover, only missense, nonsense, and frameshift variants in coding regions and splice sites were included. The pathogenicity predictions for new point mutations leading to aminoacid changes were evaluated on SIFT, Polyphen2, Mutation Taster, CADD, and GERP. The allelic variants were classified based on the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines [6] combined with InterVar analysis [7], and additional information obtained from animal model findings already reported in the literature. For recessive inheritance (homozygous or compound heterozygous variants), a PM3 criterion was used. Three out of five computational in silico tools were used as evidence to support a deleterious effect on the gene or gene product (S1 Table). New mutations reported in this study were not validated at RNA and/or protein levels.
Sanger sequencing
The mutations detected by TMPS were confirmed in the patients and their available families by Sanger sequencing. Primers flanking the variants were used for PCR amplification. Primer sequences are available on request. All PCR products were sequenced using the BigDye terminator v3.1 cycle sequencing kit followed by automated sequencing using the ABI PRISM 3130xl (Applied Biosystems, Foster City, CA). Moreover, Sanger sequencing was performed to evaluate 200 fertile Brazilian women controls for the putative damaging mutation found in the patients with GATA4, GDF9, and STAG3.
Copy number variation
For analyzing the copy number variation (CNV) in TMPS samples, CONTRA (COpy Number Targeted Resequencing Analysis) software was utilized. This method for the detection of CNV using NGS data is based on empirical relationships between log-ratio and coverage and therefore is capable of identifying copy numbers of gains and losses for each target region based on normalized depth of coverage [8]. We evaluated log-ratio +1 for duplication and log-ratio -1 for deletion, adjusting the P-value below 0.05. Integrative Genomics Viewer (IGV) software was used to confirm the decreased or increased depth coverage [9,10]. Some CNVs were confirmed by Multiplex Ligation-dependent Probe Amplification (MLPA) (MRC Holland, Amsterdam, The Netherlands), when commercial probes were available. The MLPA reaction was performed according to the manufacturer’s recommendations.
Results
The mean coverage depth of the targeted regions data was x173.6 (SD ± ×79), with at least 98% of the sequenced bases covering more than 10‐fold. We recruited 50 patients from different Brazilian endocrinology institutes, including 29 patients with primary amenorrhea and 21 with secondary amenorrhea (Table 1), all of them with the 46,XX karyotype. As listed in Table 1, 37 patients presented with isolated POI, and 13 syndromic POI, which were characterized by the presence of high arched palate, cubitus valgus, late psychomotor development, short stature, and other skeletal abnormalities.
A genetic defect was identified in 70% (35 of 50) of women with POI using the customized TMPS panel. A total of 24 pathogenic variants and 2 CNVs were identified in 48% (24 of 50) of POI patients and considered a molecular genetic diagnosis of POI. These twenty-four pathogenic variants are related to 16 genes: BMP8B (POI-1 and POI-36), CPEB1 (POI-4), INSL3 (POI-5), MCM9 (POI-11 and POI-25), GDF9 (POI-16, POI-49, and POI-50), UBR2 (POI-17), ATM (POI-20), STAG3 (POI-21), BMP15 (POI-22 and POI-23), BMPR2 (POI-29), DAZL (POI-31), PRDM1 (POI-37), FSHR (POI-38), EIF4ENIF1 (POI-40), NOBOX (POI-42), and GATA4 (POI-12 and POI-45) (Table 2). These changes included 18 missense variants, 3 nonsense variants, and 3 frameshift variants. A total of 13 patients carried a single heterozygous pathogenic variant (13 of 22, 59%), 4 patients presented compound heterozygous variants (4 of 22, 18%), and 5 homozygous variants were found (5 of 22, 23%). Furthermore, two pathogenic CNVs were detected in 2 patients (2 of 50, 4%). Among the identified CNVs, POI-14 had a microdeletion in the CPEB1 gene (83.8-Kb) and POI-7 carried a microduplication of the SYCE1 gene (11.4-Kb) (Table 2).
Table 2. Pathogenic variants detected in a Brazilian cohort of 50 women with primary ovarian insufficiency.
| Patient ID | Gene | Accession number | Genotype | Variant annotation | gnomAD1 | dbSNP | Novel or previously reported variant in POI patient | ACMG classification2 | Supporting evidence related to infertility/POI in animal models and humans |
|---|---|---|---|---|---|---|---|---|---|
| POI-1 | BMP8B | NM_001720.5 | Heterozygous | c.1024A>G:p.M342V | 0.00006 | rs149276444 | Novel | P: PS3+PM1+PM2+PP2+PP3 | Ref. [11] |
| POI-4 | CPEB1 | NM_030594.5 | Heterozygous | c.259C>T:p.R87C | 0.0004 | rs200188266 | Novel | LP: PS3+PM1+PM2+PP3+BP1 | Ref. [12–15] |
| POI-5 | INSL3 | NM_001265587.2 | Homozygous | c.52G>A:p.V18M | 0.001 | rs200056709 | Novel | LP: PS3+PM1+PM3+PP2+BP4 | Ref. [16] |
| POI-7 | Chr10: SYCE1 | NM_130784 | Heterozygous | 11.4Kb duplication | NA | NA | Novel | NA | Ref. [12–15,17] |
| POI-11 | MCM9 | NM_017696.2 | Compound Heterozygous | c.2059T>C:p.F687L | Absent | rs1046135510 | Novel | LP: PS3+PM2+PM3+BP4 | Ref. [2,4,18–22] |
| MCM9 | NM_017696.2 | Compound Heterozygous | c.3223C>T:p.P1075S | 0.003 | rs61744508 | Novel | LP: PS3+PM3+BP4 | Ref. [2,4,18–22] | |
| POI-12 | GATA4 | NM_002052.5 | Heterozygous | c.280G>A:p.A94T | 0.0001 | rs780764610 | Novel | LP: PS3+PM1+PM2+PP2+BP4 | Ref. [23] |
| POI-14 | Chr15: CPEB1 | NM_030594 | Heterozygous | 83.8Kb deletion | NA | NA | Novel | NA | Ref. [12–15] |
| POI-16 | GDF93 | NM_005260.5 | Homozygous | c.783delC:p.S262Hfs*2 | Absent | rs1216260561 | Novel | P: PVS1+PS3+PM1+PM2PM3+PP3 | Ref. [2,4,19,22,24] |
| POI-17 | UBR2 | NM_015255.2 | Heterozygous | c.4843T>A:p.S1615T | Absent | rs1017000245 | Novel | LP: PS3+PM2+PP2+PP3 | Ref. [25] |
| POI-20 | ATM | NM_000051.3 | Heterozygous | c.334G>A:p.A112T | 0.0002 | rs146382972 | Novel | LP: PM1+PM2+PM3+PP3+PP5+BP1 | Ref. [4] |
| ATM | NM_000051.3 | Heterozygous | c.7313C>T:p.T2438I | 0.0001 | rs147604227 | Novel | LP: PM1+PM2+PM3+PP3+PP5+BP1 | Ref. [4] | |
| POI-21 | STAG34 | NM_001282716.1 | Heterozygous | c.290dupC:p.N98Qfs*2 | Absent | Absent | Novel | P: PVS1+PS3+PM2+PM3+PP3+PP5 | Ref. [2,4,19,22,26,27] |
| STAG34 | NM_001282716.1 | Heterozygous | c.1950C>A:p.Y650* | Absent | Absent | Novel | P: PVS1+PS3+PM2+PM3+PP3 | Ref. [2,4,19,22,26,27] | |
| POI-22 | BMP15* | NM_005448.2 | Homozygous | c.343C>T:p.Q115* | 0.00001 | rs782799707 | Novel | P: PVS1+PS3+PM1+PM2+PP3 | Ref. [4,19,22,28,29] |
| POI-23 | BMP15* | NM_005448.2 | Homozygous | c.343C>T:p.Q115* | 0.00001 | rs782799707 | Novel | P: PVS1+PS3+PM1+PM2+PP3 | Ref. [4,19,22,28,29] |
| POI-25 | MCM9 | NM_017696.2 | Heterozygous | c.1163C>A:p.T388N | 0.0005 | rs545524695 | Novel | LP: PS3+PM1+PM2+PP3+BP1 | Ref. [18,20,21] |
| POI-29 | BMPR2 | NM_001204.7 | Heterozygous | c.1357G>A:p.V453M | Absent | Absent | Novel | P: PS3+PM1+PM2+PP2+PP3 | Ref. [30] |
| POI-31 | DAZL | NM_001190811.1 | Heterozygous | c.640C>T:p.Q214* | Absent | Absent | Novel | P: PVS1+PM2+PP3 | Ref. [31,32] |
| POI-36 | BMP8B | NM_001720.5 | Heterozygous | c.778C>T:p.R260C | 0.002 | rs199806017 | Novel | LP: PS3+PM1+PP2+PP3 | Ref. [11] |
| POI-37 | PRDM1 | NM_001198.4 | Heterozygous | c.1250C>G:p.P417R | Absent | rs200035233 | Novel | LP: PS3+PM2+PP2+PP3 | Ref. [33,34] |
| POI-38 | FSHR | NM_000145.4 | Compound Heterozygous | c.1298C>A:p.A433D | 0.000008 | rs763676828 | Reported (Ref. [5]) | P: PS3+PM1+PM2+PM3+PP2+PP3+PP5 | Ref. [2,4,5,22] |
| FSHR | NM_000145.4 | Compound Heterozygous | c.507delC:p.F170Lfs*4 | 0.000004 | rs746673169 | Novel | P: PVS1+PS3+PM2+PM3+PP3 | Ref. [2,4,22] | |
| POI-40 | EIF4ENIF1 | NM_001164501.2 | Heterozygous | c.2006A>G:p.K669R | 0.00002 | rs374538489 | Novel | LP: PS3+PM2+PP3 | Ref. [35–37] |
| POI-42 | NOBOX | NM_001080413.3 | Heterozygous | c.479C>T:p.P160L | 0.00003 | rs372037920 | Novel | LP: PS3+PM1+PP2+BP4 | Ref. [2,4,38,39] |
| POI-45 | GATA4 | NM_002052.5 | Homozygous | c.1220C>G:p.P407R | 0.00006 | rs115099192 | Novel | P: PS3+PM2+PM3+PM5+PP2+PP3+BP1 | Ref. [23] |
| POI-49 | GDF9# | NM_005260.5 | Heterozygous | c.389A>G:p.Q130R | Absent | Absent | Novel | LP: PS3+PM2+PP1+PP2+BP4 | Ref. [2,4,19,22,24] |
| POI-50 | GDF9# | NM_005260.5 | Heterozygous | c.389A>G:p.Q130R | Absent | Absent | Novel | LP: PS3+PM2+PP1+PP2+BP4 | Ref. [2,4,19,22,24] |
* Siblings of Family 1
# Siblings of Family 2.
1The variant frequency was assessed in the gnomAD database (https://gnomad.broadinstitute.org/). Accessed in July 2019.
2ACMG/AMP classification was done according to Richards et al. (Ref. [6]) combined with InterVar evaluation (Ref. [7]). Accessed in June 2020.
3Published as a case report in Franca et al., 2018 (Ref. [24]).
4Published as a case report in Franca et al., 2019 (Ref. [26]).
P: Pathogenic variant; LP: Likely Pathogenic variant; VUS: Variant of uncertain significance; LB: Likely Benign variant; B: Benign variant. NA: not available.
In this current study, eleven patients (11 of 50, 22%) harbored more than one variant in different genes (Table 3), and most of these variants were classified as variants of uncertain clinical significance (VUS). A total of 24 variants were identified in 16 unrelated genes (POU5F1, HK3, NXF5, GATA4, NBN, ATM, COL4A6, XPNPEP2, SYCP1, FANCL, ERCC2 NOBOX, HARS2, SMC1B, GDF9, HELQ).
Table 3. Variants detected in multiple genes in a Brazilian cohort of 50 women with primary ovarian insufficiency.
| Patient ID | Gene | Accession number | Genotype | Variant annotation | gnomAD1 | dbSNP | Novel or previously reported variant in POI patient | ACMG classification2 | Supporting evidence related to infertility/POI in animal models and humans |
|---|---|---|---|---|---|---|---|---|---|
| POI-3 | POU5F1 | NM_002701.6 | Heterozygous | c.133C>T:p.P45S | Absent | Absent | Novel | VUS: PM2+BP4 | Ref. [19] |
| HK3 | NM_002115.3 | Heterozygous | c.1945C>T:p.R649C | 0.00005 | rs376092049 | Novel | VUS: PM1+PM2+PP3+BP1 | Ref. [19] | |
| POI-9 | NXF5 | NM_032946.2 | Homozygous | c.959G>A:p.R320Q | 0.0004 | rs113591248 | Novel | LP: PM1+PM2+PM3+BP4 | Ref. [19,40] |
| NXF5 | NM_032946.2 | Homozygous | c.145A>G:p.I49V | 0.0003 | rs113468014 | Novel | LP: PM1+PM2+PM3+BP4 | Ref. [19,40] | |
| GATA4 | NM_002052.5 | Heterozygous | c.280G>A:p.A94T | 0.0001 | rs780764610 | Novel | LP: PS3+PM1+PM2+PP2+BP4 | Ref. [23] | |
| NBN | NM_002485.5 | Heterozygous | c.456G>A:p.M152I | 0.0001 | rs201816949 | Novel | VUS: PM1+PM2+PP3+BP1 | Ref. [4] | |
| POI-9 | ATM | NM_000051.3 | Heterozygous | c.5879T>A:p.I1960N | 0.000004 | rs587782503 | Novel | VUS: PM1+PM2+PP3+BP1 | Ref. [4] |
| POI-10 | NXF5 | NM_032946.2 | Heterozygous | c.958C>T:p.R320* | 0.001 | rs140252282 | Novel | VUS: PVS1+PP3+BP6 | Ref. [19,40] |
| COL4A6 | NM_033641.4 | Heterozygous | c.2371G>A:p.G791S | 0.001 | rs143895379 | Novel | VUS: PP3+BP6 | Ref. [19] | |
| XPNPEP2 | NM_003399.6 | Heterozygous | c.644C>T:p.T215I | 0.002 | rs138365897 | Novel | VUS: PP3 | Ref. [19] | |
| POI-13 | NXF5 | NM_032946.2 | Heterozygous | c.526G>A:p.G176S | 0.000006 | Absent | Novel | VUS: PM1+PM2+PP3 | Ref. [19,40] |
| SYCP1 | NM_003176.4 | Heterozygous | c.433C>G:p.R145G | 0.000004 | Absent | Novel | VUS: PM2+PP3 | Ref. [41] | |
| POI-15 | POU5F1 | NM_002701.6 | Heterozygous | c.87G>T:p.W29C | 0.0001 | rs200769740 | Novel | VUS: PM2+PP3 | Ref. [19] |
| HK3 | NM_002115.3 | Heterozygous | c.2026C>T:p.P676S | 0.0003 | rs199684264 | Novel | VUS: PM2+PP3+BP1 | Ref. [19] | |
| POI-24 | FANCL | NM_001114636.1 | Homozygous | c.1111_1114dup:p.T372Nfs*13 | 0.003 | rs759217526 | Novel | P: PVS1+PM3+PP3+BS2 | Ref. [42] |
| ATM | NM_000051.3 | Homozygous | c.1273G>T:p.A425S | 0.000004 | rs769214234 | Novel | VUS: PM1+PM2+PM3+PM5+BP1+BP4 | Ref. [4] | |
| POI-30 | ERCC2 | NM_000400.4 | Heterozygous | c.1606G>A:p.V536M | 0.0002 | rs142568756 | Novel | VUS: PM1+PM2+PP3 | Ref. [43] |
| POI-35 | NOBOX | NM_001080413.3 | Heterozygous | c.271G>T:p.G91W | 0.003 | rs77587352 | Reported (Ref. [39]) | LB: PS3+PP5+BS2+BP4 | Ref. [2,4,38,39] |
| HK3 | NM_002115.3 | Heterozygous | c.521C>T:p.T174M | 0.002 | rs141123858 | Novel | VUS: PM1+PP3+BP1 | Ref. [19] | |
| POI-41 | HARS2 | NM_012208.4 | Heterozygous | c.1105G>C:p.G369R | 0.0007 | rs61736946 | Novel | VUS: PM1+PP3+PP5 | Ref. [22] |
| SMC1B | NM_148674.5 | Heterozygous | c.2683C>T:p.R895W | 0.0001 | rs199797179 | Novel | VUS: PM1+PP3+BP1 | Ref. [22,44] | |
| POI-43 | SYCP1 | NM_003176.4 | Heterozygous | c.1747C>G:p.L583V | 0.0001 | rs147626229 | Novel | VUS: PM2+PP3 | Ref. [41] |
| POI-44 | GDF9 | NM_005260.5 | Heterozygous | c.191C>T:p.A64V | 0.00004 | rs751002918 | Novel | P: PS3+PM2+PP3 | Ref. [2,4,19,22,24] |
| FANCL | NM_001114636.1 | Heterozygous | c.1111_1114dup:p.T372Nfs*13 | 0.003 | rs759217526 | Novel | VUS: PVS1+PP3+BS2 | Ref. [42] | |
| HELQ | NM_133636.4 | Heterozygous | c.3095delA:p.Y1032Sfs*4 | 0.00002 | rs761786816 | Novel | P: PVS1+PM2+PP3 | Ref. [45] |
* Siblings of Family 1
# Siblings of Family 2.
1The variant frequency was assessed in the gnomAD database (https://gnomad.broadinstitute.org/). Accessed in July 2019.
2ACMG/AMP classification was done according to Richards et al. (Ref. [6]) combined with InterVar evaluation (Ref. [7]). Accessed in June 2020.
P: Pathogenic variant; LP: Likely Pathogenic variant; VUS: Variant of uncertain significance; LB: Likely Benign variant; B: Benign variant.
Among the identified VUS, 19 variants were heterozygous and 5 were homozygous. Most of the identified VUS variants were missense (21 of 24, 88%), three were frameshifts (2 of 24, 8%), and one was a nonsense variant (1 of 24, 4%). Therefore, the first reported variants in novel genes and/or mode of inheritance in POI patients supported by animal findings are listed in Table 4.
Table 4. List of novel genes or mode of inheritance in primary ovarian insufficiency patients supported by animal model findings.
| Patient ID | Gene | Genotype | Variant annotation | Supported by animal model findings | Inheritance or genotype previously identified | References |
|---|---|---|---|---|---|---|
| POI-1 | BMP8B | Heterozygous | c.1024A>G:p.M342V | Reduced number of PGCs | - | [11] |
| POI-36 | BMP8B | Heterozygous | c.778C>T:p.R260C | Reduced number of PGCs | - | [11] |
| POI-5 | INSL3 | Homozygous | c.52G>A:p.V18M | Disruption of female cycle and reduced number of litters | - | [16] |
| POI-12 | GATA4 | Heterozygous | c.280G>A:p.A94T | Regulation of ovarian steroidogenesis | - | [23] |
| POI-45 | GATA4 | Heterozygous | c.1220C>G:p.P407R | Regulation of ovarian steroidogenesis | - | [23] |
| POI-16 | GDF93 | Homozygous | c.783delC:p.S262Hfs*2 | Block in follicular development leading to complete infertility | Heterozygous/Missense | [2,4,46] |
| POI-17 | UBR2 | Heterozygous | c.4843T>A:p.S1615T | Reduced fertility | - | [25] |
| POI-30 | ERCC2 | Heterozygous | c.1606G>A:p.V536M | No signs of estrus cycle, small ovaries, and no preovulatory follicles | - | [43] |
| POI-31 | DAZL | Heterozygous | c.640C>T:p.Q214* | Subfertility | Missense | [31,32] |
| POI-37 | PRDM1 | Heterozygous | c.1250C>G:p.P417R | Arterial pole defects, reduced and failed migration and proliferation of PGCs | - | [33,34] |
| POI-44 | FANCL | Homozygous and Heterozygous | c.1111_1114dup:p.T372Nfs*13 | Reduced fertility and defective of germ cells | - | [42] |
| POI-44 | HELQ | Heterozygous | c.3095delA:p.Y1032Sfs*4 | Subfertility and germ cell attrition | - | [45] |
In addition, 12 of 50 patients (24%) had no rare variants of these screened genes. Two potential pathogenic missense variants in the BMP8B and ATM genes and one 14.4 Kb microdeletion in the TOP3B gene were identified in 3 patients (POI-8, POI-32, and POI-48). However, these variants/CNVs were not considered to be causative since the fertile mothers also carried these deletion/variants, and thus they were classified as benign. Thereby, fifteen POI patients have remained without a genetic diagnosis.
Discussion
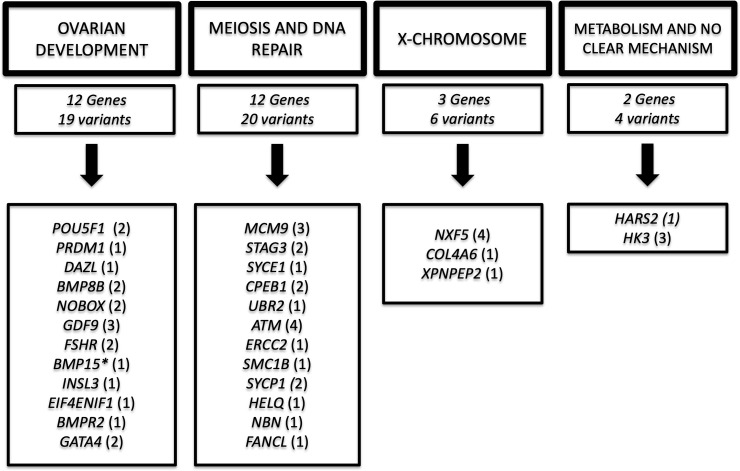
POI is a heterogeneous disorder characterized by a strong genetic basis that comprises at least 75 genes [2]. Defects in genes involved in gonadal development (oogenesis and folliculogenesis), meiosis and DNA repair, hormonal signaling, immune function, and metabolism are related to the POI phenotype [2,4]. Herein, a genetic defect of POI patients was obtained in 70% of affected women using a customized TMPS panel (Tables 2 and 3) and is discussed below. The majority of these defects were found in autosomal genes related to oogenesis and folliculogenesis and meiosis/DNA repair genes summarized in Fig 1.
Fig 1. Diagram showing the number of variants classified as pathogenic, likely pathogenic, and variant of uncertain significance in each gene group identified in 50 primary ovarian insufficiency patients.
*BMP15 is located on the X-chromosome; however, it is shown in ovarian development due to its well-known role in this category.
A) Ovarian development: Oogenesis and foliculogenesis
Genes related to primordial germ cells (POU5F1, PRMD1, DAZL, BMP8B)
Primordial germ cells (PGCs) emerge from the extraembryonic mesoderm and migrate to the genital ridge, giving rise to oocytes [22]. Several proteins are involved in migration, proliferation, and survival of PGCs, such as TGFB factors and Wnt/B-catenin pathways. PRDM1 is able to drive these pathways by obtaining pluripotent cells [22]. Moreover, POU5F1 (OCT4), DAZL, and SYCP1 are also implicated in oocyte development.
POU5F1
POU class 5 homeobox 1 is a pluripotent gene downregulated in Nobox KO mice [19]. One heterozygous missense variant (p.Pro13Thr) in POU5F1 was reported in one Chinese POI woman [19], and herein, one novel heterozygous variant in POU5F1 (c.133C>T;p.Pro45Ser), combined with a second novel heterozygous variant in the HK3 gene (Table 3), was found in a 19-year-old woman presenting with secondary amenorrhea, cubitus valgus, and hearing loss (POI-3) (Table 1). We also identified a second combination of POU5F1 (c.87G>T;p.Trp29Cys) and HK3 (c.2026C>T;p.Pro676Ser) (Table 3) in POI-15 presenting with secondary amenorrhea (Table 1). Interestingly, HK3 was revealed as a potential candidate leading to POI previously identified by GWAS associated with susceptibility between POI and early menopause [19].
PRMD1
One novel heterozygous missense variant in PRDM1 (Table 2) was found in POI-37, who was diagnosed at 14 years of age presenting with primary amenorrhea, delay of puberty, cafe au lait spots, high arched palate, cubitus valgus, hyperdontia, and congenital heart defect (Table 1). Consistent with this associated phenotype, Prdm1 mutation in mouse embryos promoted arterial pole defects characterized by misalignment or reduction of the aorta and pulmonary trunk and abnormalities in the arterial tree [33]. Moreover, heterozygous and null Prdm1-deficient mutant embryos form a tight cluster of PGCs that fail to show the migration, proliferation and repression of homeobox genes (Hox) [34].
DAZL
DAZL expression (Deleted in azoospermia-like), specifically expressed in germ cells, is essential in the beginning of meiosis, as it induces STRA8 and, consequently, the activation of SYCP1, 2 and 3, which are members of the synaptonemal complex [22]. Jung and collaborators [31] have shown by expressing DAZL with recombinant human GDF9 and BMP15 that embryonic stem cells can be induced to differentiate into ovarian follicle-like cells, such as oocytes and granulosa cells, underlying DAZL as a key player in ovarian development. In addition, the homozygous female and male Dazl-/- mice are infertile, while heterozygous mice exhibit a subfertile phenotype. The novel nonsense heterozygous variant (c.640C>T;p.Gln214*) in DAZL (Table 2) was identified in POI-31, who was diagnosed with secondary amenorrhea at 37 years of age (Table 1). Heterozygous and homozygous missense variants in DAZL were reported in infertile men and woman associated with secondary amenorrhea [32]. Although DAZL plays a key role in human and animal germ cells, it seems that pathogenic variants are a rare cause of male and female infertility.
SYCP1
Although SYCP1 is an essential member of the synaptonemal complex in labeling the axes of the chromosome during meiotic prophase I [22], no pathogenic variants associated with female infertility have been identified in this gene. Sycp1-/- male and female mice were infertile, whereas Sycp1+/- mice were fertile [41]. Herein, POI-43, who presented secondary amenorrhea at 35 years of age (Table 1), has a novel heterozygous missense variant in SYCP1 (c.1747C>G;p.Leu583Val), which is rare and deleterious according to in silico analysis (Table 3). Further investigation of heterozygous variants in SYCP1 should be done in order to evaluate a dominant negative or haploinsufficiency effect in this patient.
BMP8B
BMP8B, a member of the BMP superfamily, is also implicated in the primordial germ cell (PGC) process through the events of the development stages to generate mature sperm and oocytes. Bmp8b-/- male mice have shown small testes and infertility [47]. In addition, Bmp8b is required for PGC generation in female rodent physiology, whereas null and heterozygous Bmp8b mice have shown a lack of PGCs and reduced number of PGCs, respectively. In this study, two patients had novel and deleterious heterozygous BMP8B variants (POI-1:c.1024A>G;p.Met342Val; POI-36:c.778C>T;p.Arg260Cys) (Tables 2 and 4). POI-1 and POI-36 were diagnosed with secondary amenorrhea, and no additional phenotype or syndromic features were found (Table 1). Although no mutations in the BMP8B gene have been reported in POI patients, the decreased number of PGCs described in rodents could explain the secondary amenorrhea phenotype, and these mutations could lead to POI in these patients [11].
Genes related to folliculogenesis (NOBOX, GDF9, FSHR, BMP15, INSL3, EIF4ENIF1, BMPR2, GATA4)
NOBOX
NOBOX, GDF9, and FSHR are well-known genes associated with POI, and a summary of these genes was described in França et al [5,24,38]. Here, POI-42, who was diagnosed at 35 years of age presenting with secondary amenorrhea (Table 1), harbored a novel and pathogenic heterozygous variant in NOBOX (c.479C>T:p.Pro160Leu) (Table 2). Moreover, POI-35 has a novel heterozygous missense variant in HK3 (Table 3), which is associated with an early age of menopause [19], and she also has a reported NOBOX variant (c.271G>T:p.Gly91Trp) (Table 3) previously described by Bouilly and collaborators [39]. A cosegregation was evaluated in the unaffected mother of POI-35, and both variants were also found in her. This NOBOX variant has been classified as a likely benign variant according to ACMG guidelines due to its frequency in the population database (0.003), absence of segregation, and no impact on the gene according to in silico analysis. However, Bouilly and collaborators [39] demonstrated impaired transcriptional activity of NOBOX_p.Gly91Trp for binding to the GDF9 promoter in vitro. Hence, we classified this variant as VUS. These authors described two patients harboring this variant: one woman presenting with primary amenorrhea and absence of puberty without segregation data and another woman presenting with secondary amenorrhea and cosegregation of this variant in her family. Indeed, they were unable to rule out additional factors to explain the different phenotypes in those patients, and we were also unable to determine the genetic etiology of POI-35 since the fertile mother carried both variants. Further investigation should be performed on this patient.
FSHR
A pathogenic homozygous missense variant in FSHR was identified in two siblings presenting with hypergonadotropic hypogonadism using whole-exome sequencing [5] and references therein. In this study, one novel heterozygous frameshift deletion (c.507delC:p.Phe170Leufs*4) and one reported heterozygous missense (c.1298C>A:p.Ala433Asp) in the FSHR (Table 2) were identified in POI-38, a 23-year-old girl presenting with primary amenorrhea and delayed puberty (Tables 1 and 2). Interestingly, the c.1298C>A:p.Ala433Asp homozygous missense variant in the FSHR gene was previously reported by our group as causative of POI [5].
GDF9. The first homozygous 1-bp deletion in the GDF9 gene was identified in POI-16 with primary amenorrhea published as a case report [24]. Furthermore, a novel pathogenic heterozygous missense (c.389A>G:p.Gln130Arg) was found in POI-49 and her affected sister (POI-50) (Table 2). Both siblings were diagnosed with primary amenorrhea, puberty delay, and no other associate features were found (Table 1). Interestingly, POI-44 carried a rare and novel heterozygous missense variant in GDF9 (c.191C>T;p.Ala64Val), but two other variants in distinct genes were also found to cause a frameshift insertion in FANCL and a novel frameshift deletion in HELQ (c.3095delA;p.Tyr1032Serfs*4) (Table 3). Although the FANCL gene has been associated with Fanconi anemia, no clinical features of Fanconi were found in this patient, and therefore it is unlikely that the FANCL gene might cause ovarian failure. HELQ is a member of the DNA repair process related to tumor predisposition. In addition, Helq-deficient female mice showed subfertility and germ cell attrition. Moreover, heterozygous female mice exhibited a similar but less severe phenotype, indicative of haploinsufficiency [45] correlating to secondary amenorrhea found in POI-44 (Table 1). A cohort of Chinese women with POI was evaluated by Sanger and no mutations were found in the HELQ gene [48]. Unfortunately, the parental DNAs were unavailable for segregation analysis. Therefore, the molecular diagnosis of POI-44 needs further effort and remains uncertain, as we were unable to rule out a combined effect of these variants.
BMP15
Bone morphogenetic protein 15 has already been reported as an X-linked POI cause in patients with primary and secondary amenorrhea (MIM 300510), and recently, a homozygous missense variant in this gene was described in a patient with secondary amenorrhea [28]. In addition, homozygous ablation of BMP15 in sheep caused impaired follicular development beyond the primary stage [29]. In contrast with these models, Bmp15-/- mice exhibited minimal ovarian histopathological defects but showed reduced ovulation and fertilization rates and no phenotype was observed in Bmp15+/- mice. In this study, a nonsense homozygous variant (c.343C>T;p.Gln115*) in the BMP15 gene was identified in two siblings (POI-22 and POI-23) with primary amenorrhea (Table 2). Both affected sisters presented delayed puberty with hypergonadotropic hypogonadism (Table 1). Although heterozygous pathogenic variants in BMP15 have been reported in POI patients, the mechanism of haploinsufficiency or dominant negative effect was barely demonstrated [22]. Indeed, this is the first familial case showing a fertile mother carrier of a pathogenic heterozygous variant, arguing in favor of no heterozygous phenotype being found in animal models.
INSL3
INSL3 is a member of the insulin-like group of peptide hormones. It was first identified as a testis-specific gene transcript sequence in Leydig cells. Nevertheless, INSL3 is also produced in steroidogenic theca internal cells of antral follicles, which are equivalent cells to Leydig cells in the female physiology in bovines, rodents, monkeys, and humans [16]. INSL3 plays a key role in androstenedione synthesis, a major steroid precursor for granulosa cells to produce estrogens. In fact, the loss of INSL3 was shown to be a marker for follicle atresia, earlier than any steroidogenic expression depletion. Furthermore, the role of INSL3 in POI pathology is poorly understood since no patients have been described yet. In this study, POI-5, born from a consanguineous marriage, was evaluated at 32 years of age with a history of primary amenorrhea (Table 1). She harbored a novel homozygous variant in the exon 1 of the INSL3 gene (c.52G>A:p.Val18Met) (Table 2). In addition, knockout female mice have shown an impairment in fertility with disruption of the female cycle and reduced number of litters [16].
EIF4ENIF1
Eukaryotic translation initiation factor 4E nuclear import factor 1 (EIF4ENIF1) has been implicated in female germ cell development as a translational repressor in fruit fly, rodents, and human [35,36]. Functional studies in Drosophila and mice demonstrated that haploinsufficiency of EIF4ENIF1 promotes abnormal oocyte growth and impaired meiotic maturation [35,36]. In addition, a novel heterozygous nonsense variant (p.Ser429*) in EIF4ENIF1 was found in one woman with isolated POI and secondary amenorrhea [37]. A rare and deleterious heterozygous missense variant (c.2006A>G:p.Lys669Arg) in this gene (Table 2) was identified in POI-40, who was also diagnosed with secondary amenorrhea and born from a nonconsanguineous marriage (Table 1). This second report reinforces the EIF4ENIF1 gene as a dominant inheritance of POI.
BMPR2
Bone morphogenetic protein receptor type II is a transmembrane serine/threonine kinase that belongs to the TGF-beta superfamily, of which GDF9 and BMP15 are also members. In addition, BMPR2, the main genetic cause of pulmonary hypertension, is involved in embryonal development and in bone formation [49]. A POI patient presenting with an isolated secondary amenorrhea phenotype was reported to be harboring one heterozygous variant (p.Ser987Phe) in BMPR2 in association with a second heterozygous variant in LHCGR (p.Ans99Ser) [50]. No functional studies were exhibited regarding the oligogenic hypothesis, although in vitro results of the p.Ser987Phe in BMPR2 showed a potential implication of this gene in POI [30]. In this study, POI-29 was diagnosed at 14 years of age with primary amenorrhea, puberty delay, and normal height (Table 1). She has a novel heterozygous variant in BMPR2 (c.1357G>A;p.Val453Met) (Table 2). Consistent with the first report [50], no clinical features of pulmonary hypertension have been found in POI-29, but we cannot exclude future development of this disorder.
GATA4
GATA-binding protein 4, a member of the GATA transcription factor family, is associated with organ development in mesodermal and endodermal tissues, such as heart, gut, and gonads. The bipotential gonads emerge as genital ridges that originate from the proliferation of the coelomic epithelium, a process in which GATA4 is present. GATA4 is observed in ovarian somatic cells being expressed during the entire fetal period. In addition, GATA4 and GATA6 play a pivotal role in follicle assembly, granulosa cell differentiation, postnatal follicle growth, and luteinization. Conditional knockdown of these factors has led to female infertility at any stage of ovarian development [23]. Moreover, GATA factors have been associated with ovarian development in different species including mammals, fish, birds, and fruit fly [23]. Some in vitro studies have shown that steroidogenic enzymes implicated in the synthesis of progesterone, androgens, and estrogens may be regulated by GATA4 and GATA6. In the absence of GATA4 and GATA6 expression, estradiol synthesis is impaired, and its inactivation is also stimulated by high expression of CYP1B1 [23]. Furthermore, GATA4 interacts with SF1/NR5A1 and LRH1/NR5A2 to activate the promoter of 3βHSD2, AMH, inhibin-α, and CYP19A1, indicating GATA4 as a partner of these proteins in the regulation of ovarian steroidogeneses [23]. Interestingly, no pathogenic variants in GATA4 have been reported in the POI phenotype, although heterozygous missense variants in this gene are associated with 46, XY DSD with or without congenital heart defects [51,52]. Herein, POI-45 carries a pathogenic homozygous missense variant in GATA4 (c.1220C>G:p.Pro407Arg) (Table 2). She was diagnosed with primary amenorrhea, hearing loss, and additional kidney failure. Moreover, a congenital murmur was diagnosed at early age, and a surgery was done. Unfortunately, no clinical history of POI-45 congenital heart failure is available. In addition, a heterozygous missense variant in GATA4 (c.280G>A:p.Ala94Thr) (Table 3) was also found in POI-12, who was diagnosed with secondary amenorrhea (Table 1). These new findings could expand our knowledge of GATA4 in POI etiology.
B) Meiosis and DNA repair genes (MCM9, STAG3, SYCE1, CPEB1, UBR2, ATM, ERCC2, SYCP1, SMC1B)
MCM9
Minichromosome maintenance complex component 9 is involved in homologous recombination (HR) during meiosis process. Null MCM9 mice were sterile for showing defects in HR and gametogenesis [21]. Some homozygous and compound heterozygous variants were identified in different POI cohorts by using an NGS approach [21]. In our cohort, compound heterozygous missense variants in the MCM9 [(c.2059T>C) and (c.3223C>T)] (Table 2) were found in POI-11, a patient presenting short stature and primary amenorrhea (Table 1). Indeed, the same phenotype was observed in the first report of this gene [18]. Unfortunately, chromosomal instability and segregation analyses in POI-11 could not be assessed. Moreover, twelve women with POI carried heterozygous variants in the MCM9 gene, and most of these women were diagnosed as having secondary amenorrhea [20]. A rare and deleterious heterozygous variant in this gene (c.1163C>A;p.Thr388Asn) (Table 2) was also identified in POI-25 presenting with a mild phenotype characterized by secondary amenorrhea without short stature (Table 1) as previously described by Desai and collaborators [20]. It seems that MCM9 may be causing POI in the autosomal dominant and recessive mode of inheritance.
STAG3
Two pathogenic heterozygous loss-of-function variants in STAG3 were identified in one woman presenting with primary amenorrhea using whole-exome sequencing (Table 2) [26].
SYCE1 and CPEB1
These genes play a key role in meiosis by maintaining synaptonemal complex stability. In mouse studies, the impaired function of these genes has shown a reproductive phenotype of infertility due to the absence of synaptonemal complexes and meiotic arrest in meiosis I [53,54]. Pathogenic copy number variations (CNVs) and point variants in SYCE1 and CPEB1 genes are already implicated in the POI phenotype in distinct cohorts [12–15,17]. Jaillard and collarators reported a 123-kb duplication in SYCE1 in one patient presenting with isolated POI [13]. Herein, POI-7 has a microduplication (11.4-kb) in this gene (Table 2). Furthermore, an 83.8-kb deletion in the CPBE1 gene was found in POI-14 (Table 2), the same region previously identified in some POI cohorts [12,14,15]. In addition, the rare and deleterious heterozygous missense variant in CPEB1 (c.259C>T;p.Arg87Cys) was also identified in POI-4 (Table 2). The 10q and 15q loci appear to be important POI loci, and these findings corroborate the role of CPEB1 and SYCE1 in POI etiology.
UBR2
UBR2 encodes E3 ubiquitin ligase of the N-end rule proteolytic pathway for ubiquitin-mediated protein degradation. Null Ubr2 mice showed chromosome fragility and impaired HR repair [55]. Moreover, these null mice were not viable, and Ubr2+/- showed reduced fertility [25]. POI-17, who was diagnosed with isolated POI presenting with secondary amenorrhea, carries a novel and pathogenic heterozygous missense variant in the UBR2 gene (c.4843T>A;p.Ser1615Thr) (Tables 2 and 4). This is the first POI patient harboring a defect in the UBR2 gene.
ATM
ATM, the first DNA repair gene associated with POI, is required for cell-cycle checkpoint [4]. Mutations in ATM led to syndromic POI, characterized by primary amenorrhea and associated with autosomal recessive ataxia telangiectasia [4]. POI-20 was diagnosed with primary amenorrhea, cubitus valgus, and tremor in her dominant hand without muscle atrophy (Table 1). She was identified with a compound heterozygous missense variant in the ATM gene, suggesting a recessive inheritance as previously described. Although severe cerebellar degeneration was not found in this patient, ATM variant features could lead to impaired DNA repair, reducing the germ cell pool of this patient. Interestingly, a rare homozygous missense (Table 3) was also identified in POI-24, a primary amenorrhea case with syndromic features, such as cubitus valgus and late psychomotor development (Table 1). However, POI-24 has either a homozygous frameshift insertion in the FANCL gene, which is also associated with chromosomal instability and Fanconi anemia. POI-24 has no Fanconi anemia features, however, although we were unable to rule out FANCL contribution in the POI phenotype of this patient.
ERCC2
The ERCC2 DNA repair factor is associated with complex phenotypes such as cerebrooculofacioskeletal syndrome 2 (MIM 610756), trichothiodystrophy 1 (MIM 601615), xeroderma pigmentosum D (MIM 278730). Female knock-in mice showed no signs of estrus cycle, small ovaries, and no preovulatory follicles. These mice also exhibited osteoporosis, kyphosis, osteosclerosis, early graying, cachexia, and reduced life span [43]. Herein, a rare heterozygous variant (c.1606G>A;p.Val536Met) in ERCC2 (Table 3) was identified in POI-30, who was diagnosed with isolated primary amenorrhea (Table 1). In addition, this variant is predicted to be deleterious in all available in silico tools. Interestingly, three heterozygous pathogenic variants in the ERCC6, another member of the DNA repair cascade such as ERCC2, was found in one Chinese familial and sporadic case with POI [56]. Further studies may be needed to understand the contribution of ERCC2 gene in POI; however, DNA repair genes have been strongly associated with POI etiology.
SYCP1
Although SYCP1 is an essential member of synaptonemal complex in labeling the axes of the chromosome during meiotic prophase I [22], no pathogenic variants associated with female infertility have been identified in these genes. Sycp1-/- male and female mice were infertile whereas Sycp1+/- mice were fertile [41]. Herein, POI-43, who presented secondary amenorrhea at 35 years of age (Table 1), has a heterozygous missense variant in SYCP1 (c.1747C>G;p.Leu583Val), which is rare and deleterious in silico analysis (Table 3).
SMC1B
Structural Maintenance of Chromosomes 1B encodes a protein which belongs to the cohesin family, which is specific to the meiosis process [22]. Bouilly and collaborators [44] have reported two POI patients with different phenotypes harboring heterozygous variants in SMC1B in association with a second gene defect. One woman presenting primary amenorrhea had the p.Gln1177Leu heterozygous missense variant in SMC1B and the p.Ser5Arg heterozygous missense variant in the BMP15 gene [44]. Moreover, a secondary amenorrhea case had one heterozygous missense variant in SMC1B (p.Ile221Thr) and one heterozygous variant in NOBOX (p.Gly91Trp, discussed above) [44]. In this current study, POI-41, who was diagnosed with secondary amenorrhea and isolated POI, carries two rare heterozygous missense variants in distinct genes, SMC1B and HARS2 (Table 3). HARS2 has been associated with Perrault syndrome, which is an autosomal recessive disorder characterized by ovarian dysgenesis and deafness [22]. No deafness onset was found in POI-41 at 38 years of age. Interestingly, an affected sibling of POI-41, who was also diagnosed with secondary amenorrhea, carried both variants. DNA of the parents was unavailable for cosegregation analysis. Based on genetic features, these combined variants were classified as VUS (Table 3).
C) X-chromosome genes: NXF5, XPNPEP2, COL4A6
The association of the X chromosome, region from Xq13.3 to Xq27, has been shown as a critical region for normal ovarian development [19]. Several genes disrupted by breakpoints in balanced X-autosome translocations have been associated with POI etiology, including DIAPH2, XPNPEP2, DACH2, POF1B, CHM, PGRMC1, COL4A6, and NXF5 [19].
A cytogenetic analysis in a patient presenting with delay of puberty and primary amenorrhea, and no other clinical features showed a de novo translocation 46,XX, t(X;15)(Xq22;p11) with a breakpoint interval containing the NXF5 gene [40]. Although the NXF5 function is not well known in ovary development, functional data demonstrated that the NXF5 protein is implicated in the posttranscriptional regulation of mRNA, and thus its heterozygous deficiency results in altered mRNA metabolism, similar to the proposed mechanism for the fragile-X-associated protein FMR1 [40]. Herein, three patients (POI-9, POI-10, POI-13) had NXF5 variants combined with another variant in distinct genes (Table 3). POI-9 was diagnosed at 18 years of age with primary amenorrhea, puberty delay, and syndromic features, such as high arched palate, cubitus valgus, wide-spaced nipples, and short stature (Table 1). She carried two rare homozygous variants in the NXF5 gene, one rare heterozygous variant in the GATA4, a rare heterozygous variant in the NBN gene, and one rare heterozygous variant in the ATM (Table 3). All variants were classified as VUS. The cytogenic analysis confirmed 46, XX karyotype and no deletions or duplications were found in this patient. A homozygous pathogenic variant in NBN was recently reported in an isolated POI case [4]; however, no heterozygous NBN and ATM variants were reported as POI cause, although they have been associated with cancer predisposition. It seems that POI is likely caused by the homozygous variants in the NXF5; however, we cannot eliminate an additional effect by other rare and deleterious heterozygous variants found in this patient. Moreover, a rare heterozygous nonsense variant in NXF5 was found in POI-10, a secondary amenorrhea case. In addition, three missense variants in COL4A6, in XPNPEP2, and in INHBB were identified in this patient (Table 3). Inhibin, which belongs to the superfamily of TGF-β, acts as a negative regulator of FSH secretion, and impaired inhibin B bioactivity has shown increased susceptibility to POI [19]. POI-13 was diagnosed at 23 years of age presenting with primary amenorrhea and delay of puberty (Table 1). She carried two undescribed heterozygous missense variants, one in the NXF5 and another one in the SYCP1 (Table 3), a DNA repair gene discussed above. The mechanism of how these genes may contribute to POI in an oligogenic manner needed to be elucidated, and therefore, these variants were classified as VUS.
Conclusion
The majority of the genetic etiology of POI remains uncharacterized in the literature. In this study, this gap has been narrowed with the massively parallel sequencing technique, which allowed us to expand genotype-phenotype correlations and to improve familial counseling, i.e., the identification of additional affected members at a younger age improves the quality of patients’ lives, such as self-esteem in young women who had no breast development or planning for crypreservation of eggs for future intervention. Ultimately, a genetic diagnosis could better characterize the risk of developing underlying conditions, such as osteoporosis, cardiovascular disease, allowing physicians to provide an efficient and appropriate hormone replacement therapy.
Supporting information
(DOCX)
Acknowledgments
The authors thank the patients and their families for participating in this study. They are grateful to Ana Caroline Afonso, LIM42, and SELA teams for providing technical assistance.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received the following funding in support of this study: Fundação de Amparo à Pesquisa do Estado de São Paulo, 2014/14231-0 to Dr. Monica M França; Fundação de Amparo à Pesquisa do Estado de São Paulo, 2013/02162-8 to Berenice B. Mendonca; Conselho Nacional de Desenvolvimento Científico e Tecnológico, 303002/2016-6 to Berenice B. Mendonca; and Fundação de Amparo à Pesquisa do Estado de São Paulo, 2014/50137-5.
References
- 1.Webber L, Davies M, Anderson R, Bartlett J, Braat D, Cartwright B, et al. ESHRE Guideline: management of women with premature ovarian insufficiency. Hum Reprod. 2016;31(5):926–37. Epub 2016/03/22. 10.1093/humrep/dew027 . [DOI] [PubMed] [Google Scholar]
- 2.França MM, Mendonca BB. Genetics of Primary Ovarian Insufficiency in the Next-Generation Sequencing Era. J Endocr Soc. 2020;4(2):bvz037 Epub 2019/02/19. 10.1210/jendso/bvz037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laissue P. The molecular complexity of primary ovarian insufficiency aetiology and the use of massively parallel sequencing. Mol Cell Endocrinol. 2018;460:170–80. Epub 2017/07/23. 10.1016/j.mce.2017.07.021 . [DOI] [PubMed] [Google Scholar]
- 4.Tucker EJ, Grover SR, Bachelot A, Touraine P, Sinclair AH. Premature Ovarian Insufficiency: New Perspectives on Genetic Cause and Phenotypic Spectrum. Endocr Rev. 2016;37(6):609–35. Epub 2016/10/03. 10.1210/er.2016-1047 . [DOI] [PubMed] [Google Scholar]
- 5.França MM, Lerario AM, Funari MFA, Nishi MY, Narcizo AM, de Mello MP, et al. A Novel Homozygous Missense FSHR Variant Associated with Hypergonadotropic Hypogonadism in Two Siblings from a Brazilian Family. Sex Dev. 2017;11(3):137–42. Epub 2017/06/08. 10.1159/000477193 . [DOI] [PubMed] [Google Scholar]
- 6.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. Epub 2015/03/05. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am J Hum Genet. 2017;100(2):267–80. Epub 2017/01/26. 10.1016/j.ajhg.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28(10):1307–13. Epub 2012/04/02. 10.1093/bioinformatics/bts146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92. Epub 2012/04/19. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying Y, Liu XM, Marble A, Lawson KA, Zhao GQ. Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol. 2000;14(7):1053–63. 10.1210/mend.14.7.0479 . [DOI] [PubMed] [Google Scholar]
- 12.McGuire MM, Bowden W, Engel NJ, Ahn HW, Kovanci E, Rajkovic A. Genomic analysis using high-resolution single-nucleotide polymorphism arrays reveals novel microdeletions associated with premature ovarian failure. Fertil Steril. 2011;95(5):1595–600. Epub 2011/01/22. 10.1016/j.fertnstert.2010.12.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaillard S, Akloul L, Beaumont M, Hamdi-Roze H, Dubourg C, Odent S, et al. Array-CGH diagnosis in ovarian failure: identification of new molecular actors for ovarian physiology. J Ovarian Res. 2016;9(1):63 Epub 2016/10/03. 10.1186/s13048-016-0272-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tšuiko O, Nõukas M, Žilina O, Hensen K, Tapanainen JS, Mägi R, et al. Copy number variation analysis detects novel candidate genes involved in follicular growth and oocyte maturation in a cohort of premature ovarian failure cases. Hum Reprod. 2016;31(8):1913–25. Epub 2016/06/14. 10.1093/humrep/dew142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bestetti I, Castronovo C, Sironi A, Caslini C, Sala C, Rossetti R, et al. High-resolution array-CGH analysis on 46,XX patients affected by early onset primary ovarian insufficiency discloses new genes involved in ovarian function. Hum Reprod. 2019;34(3):574–83. 10.1093/humrep/dey389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivell R, Anand-Ivell R. Insulin-like peptide 3 (INSL3) is a major regulator of female reproductive physiology. Hum Reprod Update. 2018;24(6):639–51. 10.1093/humupd/dmy029 . [DOI] [PubMed] [Google Scholar]
- 17.de Vries L, Behar DM, Smirin-Yosef P, Lagovsky I, Tzur S, Basel-Vanagaite L. Exome sequencing reveals SYCE1 mutation associated with autosomal recessive primary ovarian insufficiency. J Clin Endocrinol Metab. 2014;99(10):E2129–32. Epub 2014/07/25. 10.1210/jc.2014-1268 . [DOI] [PubMed] [Google Scholar]
- 18.Wood-Trageser MA, Gurbuz F, Yatsenko SA, Jeffries EP, Kotan LD, Surti U, et al. MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am J Hum Genet. 2014;95(6):754–62. 10.1016/j.ajhg.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin Y, Jiao X, Simpson JL, Chen ZJ. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum Reprod Update. 2015;21(6):787–808. Epub 2015/08/04. 10.1093/humupd/dmv036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai S, Wood-Trageser M, Matic J, Chipkin J, Jiang H, Bachelot A, et al. MCM8 and MCM9 Nucleotide Variants in Women With Primary Ovarian Insufficiency. J Clin Endocrinol Metab. 2017;102(2):576–82. 10.1210/jc.2016-2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiao X, Ke H, Qin Y, Chen ZJ. Molecular Genetics of Premature Ovarian Insufficiency. Trends Endocrinol Metab. 2018;29(11):795–807. Epub 2018/08/02. 10.1016/j.tem.2018.07.002 . [DOI] [PubMed] [Google Scholar]
- 22.Huhtaniemi I, Hovatta O, La Marca A, Livera G, Monniaux D, Persani L, et al. Advances in the Molecular Pathophysiology, Genetics, and Treatment of Primary Ovarian Insufficiency. Trends Endocrinol Metab. 2018;29(6):400–19. Epub 2018/04/26. 10.1016/j.tem.2018.03.010 . [DOI] [PubMed] [Google Scholar]
- 23.Bennett-Toomey J, Stocco C. GATA Regulation and Function During the Ovarian Life Cycle. Vitam Horm. 2018;107:193–225. Epub 2018/02/13. 10.1016/bs.vh.2018.01.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.França MM, Funari MFA, Nishi MY, Narcizo AM, Domenice S, Costa EMF, et al. Identification of the first homozygous 1-bp deletion in GDF9 gene leading to primary ovarian insufficiency by using targeted massively parallel sequencing. Clin Genet. 2018;93(2):408–11. Epub 2017/12/26. 10.1111/cge.13156 . [DOI] [PubMed] [Google Scholar]
- 25.Kwon YT, Xia Z, An JY, Tasaki T, Davydov IV, Seo JW, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23(22):8255–71. 10.1128/mcb.23.22.8255-8271.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.França MM, Nishi MY, Funari MFA, Lerario AM, Baracat EC, Hayashida SAY, et al. Two rare loss-of-function variants in the STAG3 gene leading to primary ovarian insufficiency. Eur J Med Genet. 2019;62(3):186–9. Epub 2018/07/10. 10.1016/j.ejmg.2018.07.008 . [DOI] [PubMed] [Google Scholar]
- 27.Caburet S, Arboleda VA, Llano E, Overbeek PA, Barbero JL, Oka K, et al. Mutant cohesin in premature ovarian failure. N Engl J Med. 2014;370(10):943–9. 10.1056/NEJMoa1309635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Wang J, Wang X, Li L, Pan H, Chen B, et al. A novel homozygous mutation of bone morphogenetic protein 15 identified in a consanguineous marriage family with primary ovarian insufficiency. Reprod Biomed Online. 2018;36(2):206–9. Epub 2017/10/26. 10.1016/j.rbmo.2017.10.104 . [DOI] [PubMed] [Google Scholar]
- 29.Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta TS, et al. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet. 2000;25(3):279–83. 10.1038/77033 . [DOI] [PubMed] [Google Scholar]
- 30.Patiño LC, Silgado D, Laissue P. A potential functional association between mutant BMPR2 and primary ovarian insufficiency. Syst Biol Reprod Med. 2017;63(3):145–9. Epub 2017/03/17. 10.1080/19396368.2017.1291767 . [DOI] [PubMed] [Google Scholar]
- 31.Jung D, Xiong J, Ye M, Qin X, Li L, Cheng S, et al. In vitro differentiation of human embryonic stem cells into ovarian follicle-like cells. Nat Commun. 2017;8:15680 Epub 2017/06/12. 10.1038/ncomms15680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tung JY, Rosen MP, Nelson LM, Turek PJ, Witte JS, Cramer DW, et al. Novel missense mutations of the Deleted-in-AZoospermia-Like (DAZL) gene in infertile women and men. Reprod Biol Endocrinol. 2006;4:40 Epub 2006/08/02. 10.1186/1477-7827-4-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent SD, Mayeuf-Louchart A, Watanabe Y, Brzezinski JA, Miyagawa-Tomita S, Kelly RG, et al. Prdm1 functions in the mesoderm of the second heart field, where it interacts genetically with Tbx1, during outflow tract morphogenesis in the mouse embryo. Hum Mol Genet. 2014;23(19):5087–101. Epub 2014/05/12. 10.1093/hmg/ddu232 . [DOI] [PubMed] [Google Scholar]
- 34.Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207–13. Epub 2005/06/05. 10.1038/nature03813 . [DOI] [PubMed] [Google Scholar]
- 35.Zappavigna V, Piccioni F, Villaescusa JC, Verrotti AC. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc Natl Acad Sci U S A. 2004;101(41):14800–5. Epub 2004/10/01. 10.1073/pnas.0406451101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villaescusa JC, Allard P, Carminati E, Kontogiannea M, Talarico D, Blasi F, et al. Clast4, the murine homologue of human eIF4E-Transporter, is highly expressed in developing oocytes and post-translationally modified at meiotic maturation. Gene. 2006;367:101–9. Epub 2005/12/15. 10.1016/j.gene.2005.09.026 . [DOI] [PubMed] [Google Scholar]
- 37.Kasippillai T, MacArthur DG, Kirby A, Thomas B, Lambalk CB, Daly MJ, et al. Mutations in eIF4ENIF1 are associated with primary ovarian insufficiency. J Clin Endocrinol Metab. 2013;98(9):E1534–9. Epub 2013/07/31. 10.1210/jc.2013-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.França MM, Funari MFA, Lerario AM, Nishi MY, Pita CC, Fontenele EGP, et al. A novel homozygous 1-bp deletion in the NOBOX gene in two Brazilian sisters with primary ovarian failure. Endocrine. 2017;58(3):442–7. Epub 2017/10/24. 10.1007/s12020-017-1459-2 . [DOI] [PubMed] [Google Scholar]
- 39.Bouilly J, Bachelot A, Broutin I, Touraine P, Binart N. Novel NOBOX loss-of-function mutations account for 6.2% of cases in a large primary ovarian insufficiency cohort. Hum Mutat. 2011;32(10):1108–13. Epub 2011/09/09. 10.1002/humu.21543 . [DOI] [PubMed] [Google Scholar]
- 40.Bertini V, Ghirri P, Bicocchi MP, Simi P, Valetto A. Molecular cytogenetic definition of a translocation t(X;15) associated with premature ovarian failure. Fertil Steril. 2010;94(3):1097.e5–8. Epub 2010/03/24. 10.1016/j.fertnstert.2010.02.013 . [DOI] [PubMed] [Google Scholar]
- 41.de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19(11):1376–89. 10.1101/gad.329705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agoulnik AI, Lu B, Zhu Q, Truong C, Ty MT, Arango N, et al. A novel gene, Pog, is necessary for primordial germ cell proliferation in the mouse and underlies the germ cell deficient mutation, gcd. Hum Mol Genet. 2002;11(24):3047–53. 10.1093/hmg/11.24.3047 . [DOI] [PubMed] [Google Scholar]
- 43.de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296(5571):1276–9. Epub 2002/04/11. 10.1126/science.1070174 . [DOI] [PubMed] [Google Scholar]
- 44.Bouilly J, Beau I, Barraud S, Bernard V, Azibi K, Fagart J, et al. Identification of Multiple Gene Mutations Accounts for a new Genetic Architecture of Primary Ovarian Insufficiency. J Clin Endocrinol Metab. 2016;101(12):4541–50. Epub 2016/09/07. 10.1210/jc.2016-2152 . [DOI] [PubMed] [Google Scholar]
- 45.Adelman CA, Lolo RL, Birkbak NJ, Murina O, Matsuzaki K, Horejsi Z, et al. HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis. Nature. 2013;502(7471):381–4. Epub 2013/09/04. 10.1038/nature12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383(6600):531–5. 10.1038/383531a0 . [DOI] [PubMed] [Google Scholar]
- 47.Zhao GQ, Deng K, Labosky PA, Liaw L, Hogan BL. The gene encoding bone morphogenetic protein 8B is required for the initiation and maintenance of spermatogenesis in the mouse. Genes Dev. 1996;10(13):1657–69. 10.1101/gad.10.13.1657 . [DOI] [PubMed] [Google Scholar]
- 48.Wang W, Zhao S, Zhuang L, Li W, Qin Y, Chen ZJ. The screening of HELQ gene in Chinese patients with premature ovarian failure. Reprod Biomed Online. 2015;31(4):573–6. Epub 2015/06/29. 10.1016/j.rbmo.2015.06.012 . [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Rivas G, Jerjes-Sánchez C, Rodriguez D, Garcia-Pelaez J, Trevino V. A systematic review of genetic mutations in pulmonary arterial hypertension. BMC Med Genet. 2017;18(1):82 Epub 2017/08/02. 10.1186/s12881-017-0440-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fonseca DJ, Patiño LC, Suárez YC, de Jesús Rodríguez A, Mateus HE, Jiménez KM, et al. Next generation sequencing in women affected by nonsyndromic premature ovarian failure displays new potential causative genes and mutations. Fertil Steril. 2015;104(1):154–62.e2. Epub 2015/05/16. 10.1016/j.fertnstert.2015.04.016 . [DOI] [PubMed] [Google Scholar]
- 51.Eggers S, Sadedin S, van den Bergen JA, Robevska G, Ohnesorg T, Hewitt J, et al. Disorders of sex development: insights from targeted gene sequencing of a large international patient cohort. Genome Biol. 2016;17(1):243 Epub 2016/11/29. 10.1186/s13059-016-1105-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez de LaPiscina I, de Mingo C, Riedl S, Rodriguez A, Pandey AV, Fernández-Cancio M, et al. GATA4 Variants in Individuals With a 46,XY Disorder of Sex Development (DSD) May or May Not Be Associated With Cardiac Defects Depending on Second Hits in Other DSD Genes. Front Endocrinol (Lausanne). 2018;9:142 Epub 2018/04/04. 10.3389/fendo.2018.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1(2):201–13. 10.1016/s1534-5807(01)00025-9 . [DOI] [PubMed] [Google Scholar]
- 54.Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, et al. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 2009;5(2):e1000393 Epub 2009/02/27. 10.1371/journal.pgen.1000393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ouyang Y, Kwon YT, An JY, Eller D, Tsai SC, Diaz-Perez S, et al. Loss of Ubr2, an E3 ubiquitin ligase, leads to chromosome fragility and impaired homologous recombinational repair. Mutat Res. 2006;596(1–2):64–75. Epub 2006/02/20. 10.1016/j.mrfmmm.2005.12.016 . [DOI] [PubMed] [Google Scholar]
- 56.Qin Y, Guo T, Li G, Tang TS, Zhao S, Jiao X, et al. CSB-PGBD3 Mutations Cause Premature Ovarian Failure. PLoS Genet. 2015;11(7):e1005419 Epub 2015/07/28. 10.1371/journal.pgen.1005419 [DOI] [PMC free article] [PubMed] [Google Scholar]