Abstract
Coronaviruses (CoVs) classified in the Coronaviridae family infect a very large spectrum of vertebrate group. Seven CoVs that cause human disease consist of Alpha-CoVs, which are HCoV-229E, and NL63 and beta-CoVs, which are MERS-CoV, SARS-CoV, HCoV-OC43, HCoV-HKU1, and SARS-CoV-2. SARS-CoV-2 is an enveloped, positive-polarity, single-stranded RNA virus responsible for a new Coronavirus disease 2019 (COVID-19). The mutagenic ability of the SARS-CoV-2 directs its evolution and genome variability, thus allowing viruses to escape from host immunity and develop drug resistance. Tracing viral mutations is also important for the development of new vaccines, antiviral drugs, and diagnostic systems. During replication in the host cell, genomic mutations occur in the virus and these mutations are transferred to new generations. For this reason, systematic monitoring of mutations in the SARS-CoV-2 genome allows observation of the national and international molecular epidemiology of the virus. SARS-CoV-2 spike (S) glycoprotein is vital in the binding of the virus to the host cell receptor that is angiotensin converting-enzyme 2 (ACE2), membrane fusion, vaccine studies and immune response to the virus. Therefore, mutations in the gene encoding the S glycoprotein and especially the possible variations in the receptor binding domain (RBD) in S gene are important issues to be emphasized. In this article, information about the mutations observed in the SARS-CoV-2 S glycoprotein and their possible effects are presented.
Keywords: COVID-19, ACE-2, spike (S) protein, mutations
ÖZ
Coronaviridae familyası içerisinde sınıflandırılan koronavirüsler (CoVs) oldukça geniş bir omurgalı grubu enfekte etmektedir. İnsanlarda hastalık oluşturan yedi tip koronavirüsten HCoV-229E ve NL63 alfa, MERS-CoV, SARS-CoV, HCoV-OC43, HCoV-HKU1, SARS-CoV-2 ise beta cinsinde yer almaktadır. SARS-CoV-2, yeni bir koronavirüs hastalığına (Coronavirus disease 2019; COVID-19) yol açan zarflı, pozitif-polariteli, tek sarmallı bir RNA virüsüdür. SARS-CoV-2’nin mutasyon kabiliyeti viral evrimi ve genom değişkenliğini yönlendirir. Böylece virüslerin konakçı bağışıklığından kaçmasına ve ilaç direnci geliştirmesine olanak tanır. Ayrıca viral mutasyonların izlenmesi yeni aşılar, anti-viral ilaçlar ve tanı sistemlerinin geliştirilmesi için de önemlidir. Konak hücrede replikasyon esnasında virüste mutasyon oluşmakta ve bu mutasyonlar yeni soylara aktarılmaktadır. Bu nedenle SARS-CoV-2 genomundaki mutasyonların sistematik bir biçimde izlenmesi virüsün ulusal ve uluslararası moleküler epidemiyolojisinin gözlenmesine olanak sağlamaktadır. SARS-CoV-2’nin spike (S) proteini virüsün konak hücre reseptörü olan anjiyotensin dönüştürücü enzim 2’ye (ACE2) bağlanması, membrana füzyonu, aşı çalışmaları ve oluşacak bağışık yanıtta hayati öneme sahiptir. Bu nedenle S glikoproteinini kodlayan gendeki mutasyonlar ve özellikle virüsün ACE-2 reseptörüne bağlanma bölgesindeki (RBD olası varyasyonların irdelenmesi, üzerinde durulması gereken önemli konulardandır. Bu yazıda SARS-CoV-2 S glikoproteininde saptanmış olan mutasyonlar ve olası etkileri hakkında bilgi sunulmuştur.
Keywords: COVID-19, ACE-2, spike (S) protein, mutasyonlar
Introduction
Coronaviruses (CoVs) belonging to the Coronaviridae family infect a variety of vertebrates. They are commonly seen in bats, but can also be found in many birds and mammals, including humans. Coronaviridae consist of four genera as alfa, beta, gamma and delta. Among the seven types of Coronaviruses causing diseases in humans, HCoV-229E and NL63 are in Alpha-CoVs; MERS-CoV, SARS-CoV, HCoV-OC43, HCoV-HKU1, and SARS-CoV-2 are in the Beta-CoV genus1.
Coronaviruses can lead to a wide range of health problems in humans from a common cold to severe diseases that may result in death. In the past few decades, there have been outbreaks of serious and sometimes fatal respiratory diseases caused by CoVs, the new human pathogens. These CoV strains, which have phylogenetically differed from common human CoVs, are detected in bats and are typically transmitted to humans through an intermediate host2. These new strains have stronger virulence and can cause fatal infections in some individuals. In severe cases, death occurs due to gradual respiratory failure as a result of alveolar damage3-5.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the new Coronavirus disease 2019 (COVID-19), which emerged in Wuhan, China in December 20196. COVID-19 has caused a pandemic affecting 215 countries around the world. The total number of 7.765 626 cases was reported on June 13, 2020, and the total number of deaths has reached 428.7457. SARS-CoV-2 mainly spreads by the large respiratory droplets, which infected people expel when sneezing or coughing. Its propagation speed is quite high, it is possible for one person to infect 2-4 people8. Incubation period is very long (five to seven days, but can sometimes extend to 14 days)9. Infected people can spread the virus 1-2 days before clinical symptoms appear until disappearance of symptoms10. Fever, cough, shortness of breath, and diarrhea are the most common clinical symptoms. The mean mortality rate is 3%9,11. Approximately 25% of infected people are asymptomatic and they contribute to the transmission of the virus12.
General Characteristics of Sars-CoV-2
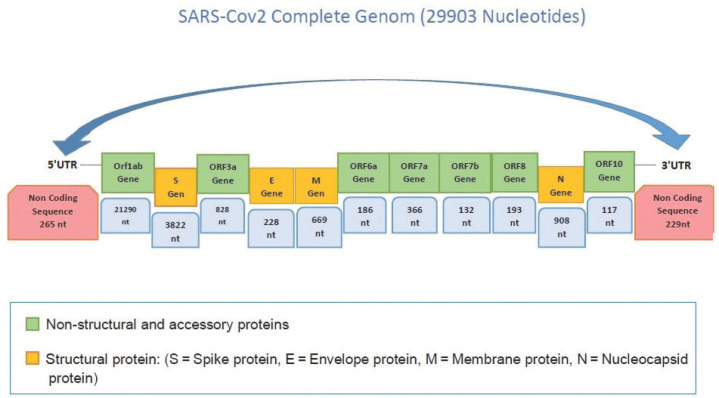
SARS-CoV-2, belonging to the subgenus of Sarbecovirus, is a spherical enveloped virus with virion size of 70-90 nm. It is a single-stranded positive sense RNA virus. The genome consists of 29,903 nucleotides and 11 open reading frame (ORF) regions and encodes structural proteins, non-structural proteins (nsp1-16) and accessory proteins (ORF3a, ORF6, ORF7a and b, ORF8, ORF10) (Figure 1). There are four structural proteins; spike (S), envelope (E), membrane (M), and nucleocapsid (N). Together, S, E and M proteins form the viral envelope. M and E proteins are essential for virus morphology, packaging, and cell detachment. S glycoprotein consists of 3.822 nucleotides responsible for the attachment and fusion of the virus to the host cell membrane. Non-structural proteins are involved in the replication-transcription and the vesicles formation, while accessory proteins counteract the natural defense mechanisms of the host13-16.
Figure 1.
Genome structure of SARS-CoV-2. ORF3a, ORF6, ORF7a and b, ORF8, ORF10 encode accessory proteins; Orf1ab encodes non-structural proteins. non-stractural and accessory proteins.
Structure and Function of S Glycoprotein
Recognition and entry of virus into the host cell are the most important step in determining the infectivity and pathogenesis of the disease17. Using S glycoproteins, SARS-CoV-2 interacts with human respiratory and epithelial cells that express angiotensin-converting enzyme-2 (ACE2) receptors18. The sequence analysis of the SARS-CoV-2 S gene has shown that it has only 75% homology with the SARS-CoV S protein13,19. However, the analysis of the receptor-binding motif (RBM) in S protein revealed that most of the amino acid residues required for binding to the ACE-2 receptor are commonly protected between SARS-CoV and SARS-CoV-2. This data confirms that both CoV viruses use the same ACE-2 receptor for entry into host cell20,21.
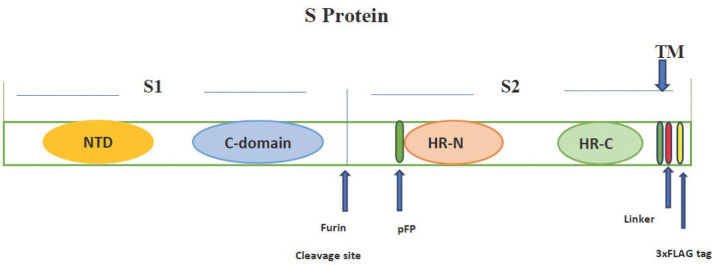
S glycoprotein consists of a short intracellular tail, a transmembrane anchor, and a receptor-binding ectodomain. The ectodomain of S protein is a trimetric class 1 fusion protein of approximately 1.200 amino acids in length. This ectodomain contains a S1 subunit that provides adhesion to the target cell and a S2 subunit that ensures membrane-fusion22. Each of these subunits consists of subdomains with different functions (Figure 2). In the S1 subunit, there are approximately 200 amino acid-length receptor binding domains (RBDs) localized between glutamine 321 (Q312) and asparagine 536 (N536) amino acids. RBD has a receptor-binding determining region (RBDR) that recognizes ACE-223, 24. The conformation of RBDR is controlled by the hinge-like conformational movement of RBD. Based on the hinge-like movement of RBD, the trimeric S protein is temporarily located in symmetrical (RBD down) or asymmetrical (RBD standing) conformation. RBD is located in the C-terminal domain (CTD) in the S1 subunit in MERS-CoV, SARS-CoV, and SARS-CoV-2. The CDT of SARS-CoV-2 contains two structural subdomains: one is the preserved core and the other is the external subdomain. The CTD of the SARS-CoV-2 recognizes subdomain I in the N-terminal domain (NTD) of ACE-2 using its external subdomain. Interaction of SARS-CoV-2-CTD with hACE-2 receptor is similar overall to that observed for SARS-CoV-RBD. However, key residue substitutions in SARS-CoV-2-CTD slightly strengthen the interaction and lead to higher affinity for receptor binding than SARS-CoV-RBD. It has been shown that 24 residues in the ACE-2 receptor have function on its contact with SARS-CoV-2 CTD and SARS-CoV-RBD. Among these 24 residues in hACE-2, 15 amino acids are in greater contact with the SARS-CoV-2-CTD than SARS-CoV-RBD (Table 1)18.
Figure 2.
Schematic parts of the S protein. NTD, N-terminal domain; CTD, C-terminal domain; pFP, potential fusion protein; HR-N, heptad repeat N; HR-C, heptad repeat C; TM, a transmembrane domain. Modified from reference (22).
Table 1.
Amino acids having a role in the interaction of hACE-2 with SARS-CoV-2-CTD and SARS-CoV-RBD18.
| hACE-2 | SARS-CoV-2-CTD | SARS-CoV-RBD |
|---|---|---|
| S19 | A475, G476 | P462 |
| Q24 | A475, G476, N478 | N473 |
| T27 | F456, Y473, A475, Y489 | L443, Y475 |
| F28 | Y489 | Y475 |
| D30 | K417, L455, F456 | Y442 |
| K31 | L455, F456, E484, Y489, F490, Q493 | Y442, Y475 |
| H34 | Y453, L455, Q493 | Y440, Y442, N479 |
| E35 | Q493 | - |
| E37 | Y505 | Y491 |
| D38 | Y449, G496, Q498 | Y436, G482, Y484 |
| Y41 | Q498, T500, N501 | Y484, T486, T487 |
| Q42 | G446, Y449, Q498 | Y436, Y484 |
| L45 | Q498, T500 | Y484, T486 |
| L79 | F486 | L472 |
| M82 | F486 | L472 |
| Y83 | F486, N487, Y489 | N473, Y475 |
| Q325 | - | R426 |
| E329 | - | R426 |
| N330 | T500 | T486 |
| K353 | G496, N501, G502, Y505 | Y481, G482, Y484, T487, G488, Y491 |
| G354 | Y502, Y505 | G488, Y491 |
| D355 | T500, G502 | T486, T487, G488 |
| R357 | T500 | T486 |
| R393 | Y505 | Y491 |
Sars-CoV-2 Mutations
SARS-CoV-2-dependent mortality rates show significant differences between countries ranging from 0.9% to15.44%. For example, mortality rate is 0.9% in Singapore, 2.56% in Turkey, 5.6% in China, and 15.44% in Belgium25. Undoubtedly, the infrastructure facilities of the countries, quarantine measures, compliance of society with personal protective measures, immune status and genetics of people are factors playing a role in these differences. However, in order to understand the difference between the mortality rates, it is also necessary to know the mutation rate of the virus and its ability to evolve over time. The biological characterization of mutations is useful to monitor viral drug resistance, elucidate how the virus escapes immune response, and understand mechanisms associated with pathogenesis. In addition, viral mutation studies are important for the development of new vaccines, antiviral drugs, and diagnostic systems26,27.
Analyzing mutations in 95 SARS-CoV-2 strains with full genome sequence data available in GISAID from different countries showed that there was a strong correlation between the detected mutations and sampling time and geographic location. A total of 116 mutations were identified, with the most common mutations being 8782C>T in the ORF1ab gene, 28144T>C in the ORF8 gene, and 29095C>T in the N gene. In addition, mutations were also detected in S and M genes. The authors emphasized the importance of the continuity of studies on viral genome variations, stating that these mutations might have an impact on the transmission of SARS-CoV-2 and the severity of the disease28.
A second research group analyzed the whole genome sequence data of 95 SARS-CoV-2 strains and they found 99.99% similarity in both nucleotides (nt) (99.91-100%) and amino acid levels (99.91-100%) among these strains. While the overall variation in the ORF regions of these strains was at a low frequency, 13 mutation points were detected in the ORF1a, 1b, S, 3a, M, 8 and N regions. Among these, nt28144 in ORF8 and nt8782 in ORF1a had the highest mutation rates of 30.53% and 29.47%, respectively. Based on these findings, the authors suggested that there might be selective mutations in SARS-CoV-2 and emphasized that further studies on mutation analysis would be useful in terms of diagnosis, clinical follow-up, and control, as well as monitoring the biology of the virus29.
In a whole genome sequence study conducted with two samples in Italy, mutations were observed in the nucleotide and amino acid levels in the ORF1a, 1b, S and N genes30. In another study, mutations were analyzed among 56 SARS-CoV-2 strains from China, USA and Italy, and it was reported that the homology between the strains was over 99%. Two hot mutation zones were identified at positions 8789 and 28151. At position 8789, either T (U) or C mutation fell within the polyprotein gene. On the other hand, the mutations characterized by the presence of C or U formed at position 28151 in ORF8 could lead to the amino acid substitution of Ser/Leu, which can affect the conformation of the peptide14.
Mutations in S gene
SARS-CoV-2 having higher infectivity than SARS-CoV has led to questions concerning the binding affinity of SARS-CoV-2 S protein to ACE-2 and its neutralization with anti-SARS-CoV monoclonal antibodies (mAbs). In a previous study, Val-Lys417 alteration was recorded in the RBD of SARS-CoV-2. The Lys417 amino acid strongly interacts with the Asp30 amino acid of ACE-2. Therefore, RBD-ACE-2 interaction in SARS-CoV-2 is stronger than the SARS-CoV-ACE-2 interaction. It is suggested that this situation may be effective in the higher transmission rate of SARS-CoV-2 than SARS-CoV. In addition, the transformation of Pro-Ala475 and insertion of Gly482 into the AGSTPCNGV ring of RBD has prevented the neutralization of SARS-CoV-2 by anti-SARS-CoV mAbs31.
Analysis of 2.058 high-quality SARS-CoV-2 genomes uploaded to GISAID up to March 31, 2020 revealed 1.221 single-nucleotide polymorphisms with 753 missense, 452 silent, 12 nonsense, and four intergenic substitutions. The authors reported that mutations mostly concentrated on the S, N, and ORF3a genes, in which real/expected rates of mutations were 1.21, 1.99 and 1.82, respectively. These mutations may indicate that the virus has originated from an animal reservoir and adapted to humans. In fact, structural proteins in Coronaviruses undergo higher levels of antigenic variation, which increases the permanence of the virus by mediating its escape from immune response and adapting it to the host. This study showed mutations at 15 different points in the RBD region of SARS-CoV-2 strains32 (Table 2).
Table 2.
Localization of the 15 amino acid variations observed in RBD of mutant SARS-CoV-232.
| Codon | SARS-CoV-2 reference strain | SARS-CoV-2 mutant strain |
|---|---|---|
| 321 | Glutamine | Leucine |
| 341 | Valine | Isoleucine |
| 344 | Alanine | Serine |
| 348 | Alanine | Threonine |
| 354 | Alanine | Threonine |
| 364 | Aspartate | Tyrosine |
| 378 | Lysine | Arginine |
| 408 | Arginine | Isoleucine |
| 409 | Glutamine | Glutamate |
| 435 | Alanine | Serine |
| 458 | Lysine | Arginine |
| 508 | Tyrosine | Histidine |
| 516 | Glutamate | Glutamine |
| 519 | Histidine | Proline |
| 520 | Alanine | Serine |
In a study published on the April 30, 2020, a group of scientists from the United States and the UK examined mutations in the SARS-CoV-2 S protein and detected 14 mutations. It was emphasized that the mutations were distributed over a wide geographic area and time interval (Table 3). It was also suggested that these mutations provide advantages to virus for transmission and evasion form treatment. In particular, the D614G mutation in S gene was to be a major concern. It was noted that this mutation, which started to spread in Europe at the beginning of February 2020, rapidly became dominant in every region it entered. In addition, the study provided the evidence of recombination between locally circulating strains33. In a current study carried on the whole genome sequence data of 30 SARS-CoV-2 strains submitted from Turkey to GISAID, 29 of the genomes were classified as type L, which is considered to be derived from ancestral clade S. Turkish SARS-CoV-2 strains are distributed across three of four major clusters (2, 3, and 4) identified around the world. In the mutation analysis related to the S gene region, D614G mutation [substitution of aspartic acid (D) with glycine (G) in codon 614], which is associated with the facilitation of the transmission of the virus and virulence, was detected in 23 genomes. Other mutations observed in the S gene region were N679K, V772I, and T1238I, with the latter two being detected in the samples sequenced from Turkey34. None of these mutations observed in S gene of the Turkish strains was in the RBD.
Table 3.
Mutations detected in the S gene region and their geographical distribution33.
| S mutation* | Location and possible effect | Geographical distribution |
|---|---|---|
| D614G | SARS-CoV epitope/inter-protomer stabilization | Global |
| L5F | Single peptide | 13 countries |
| L8V/W | Single peptide | Hong Kong |
| H49Y | S1 NTD domain | China |
| Y145H/del | S1 NTD domain | Six countries |
| Q239K | S1 NTD domain | Europe |
| V367F | Up/Down conformation | Europe /Hong Kong |
| G476S | RBD | Washington, USA |
| V615I/F | SARS-CoV ADE epitope | Wales |
| A831V | Potential fusion protein in S2 | Iceland |
| D839Y/N/E | S2 subunit | Europe |
| S943P | HR1 fusion core | Belgium |
| P1263L | Cytoplasmic tail | United Kingdom, Iceland, Australia |
NTD: N-terminal domain, RBD: receptor binding domain, HR1: N-terminal domain, ADE: antibody-dependent enhancement. *D: aspartic acid, G: glycine, L: leucin, F: phenyl alanine, V: valine, W: tryptophan, H: histidine, Y: tyrosine, Q: glutamine, K: lysine, S: serin, I: isolysin, A: alanine, N: asparagine, E: glutamic acid, P: proline.
Conclusion
In vaccine studies S glycoprotein has been chosen as an ideal target molecule. Antibodies against this protein block the binding of the virus to ACE-2 and prevent entry of the virus into the cell. Therefore, monitoring variations in this protein is very important. To date, there are more than 42000 whole-genome sequences of SARS-CoV-2 strains from all over the world. Although some mutations have been recognized in the S gene as well as other genes in the SARS-CoV-2 genome, currently any mutation has not resulted in major variation in the viral virulence and antigenic structure of the S proteins. Gathering and sharing information on S variants will provide important data on the emergence of possible new variants and subvariants. Using these data, it will be possible to evaluate the effectiveness of vaccine studies to be developed and to monitor the changes related to the pathogenesis of the disease. It is also useful to determine the rate and speed of mutations since they play an important role in the virus escaping host immune response and thus developing resistance to drugs.
References
- 1.Center for disease control and prevention. Human Coronavirus types. Available from: https://www.cdc.gov/coronavirus/types.html. Accessed date: June 15, 2020.
- 2.Zhou P, Yang XL, Wang XG et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Peng F, Wang R et al. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;109:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, Lu L, Cao W, Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect. 2020;9:727–32. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ho W, Huang Y, Jin D-Y et al. SARS-CoV-2 is an appropriate name for the new coronavirus. The Lancet. 2020;395:949–50. doi: 10.1016/S0140-6736(20)30557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worldmeters. COVID-19 Coronavirus pandemic. 2020. Available from: https://www.worldometers.info/coronavirus/. Accessed date: June 15, 2020.
- 8.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)-China. Chin J Epidemiol. 2020;41:145–51. [PMC free article] [PubMed] [Google Scholar]
- 9.Guan WJ, Ni ZY, Hu Y et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382:1708–20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.T.C. Ministry of Health. COVID-19 (SARS-CoV-2 Infection) General Information, Epidemiology and Diagnosis. 2020. Accessed date: June 15 2020.
- 11.Chen N, Zhou M, Dong X et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai Y, Yao L, Wei T et al. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020;323:1406–7. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu R, Zhao X, Li J et al. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceraolo C, Giorgi FM. Genomic variance of the 2019-n-CoV coronavirus. J Med Virol. 2020;92:522–8. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C, Liu Y, Yang Y Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020. [DOI] [PMC free article] [PubMed]
- 17.Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020. [DOI] [PMC free article] [PubMed]
- 18.Wang Q, Zhang Y, Wu L Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020. [DOI] [PMC free article] [PubMed]
- 19.Zhou P, Yang XL, Wang XG et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Moore MJ, Vasilieva N et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–4. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou X, Liu Y, Lei X et al. Characterization of spike glycoprotein of SARS CoV2 on virus entry and its immune crossreactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F. Structure, function, and evolution of Coronavirus spike proteins. Annu Rev Virol. 2016;3:237–61. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zablon FM, Aryal S. The novel Coronavirus, COVID-19: An overview. The Biology Notes. 2020. Accessed from: https://thebiologynotes.com/the-novel-coronavirus-2019-ncov-an-overview/
- 25.CEBM. Global COVID-19 case fatality rates. https://www.cebm.net/covid19/global-covid-19-case-fatality-rates/). 2020. Accessed date June 15, 2020.
- 26.Coppée F, Lechien J, Declèves A-E, Tafforeau L, Saussez S. SARSCoV-2: virus mutations in specific European populations. New Microbes New Infect. 2020;36:100696. doi: 10.1016/j.nmni.2020.100696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pachetti M, Marini B, Benedetti F et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Liu Z, Chen Z The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol. 2020. [DOI] [PMC free article] [PubMed]
- 30.Stefanelli P, Faggioni G, Presti AL et al. Whole genome and phylogenetic analaysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020;25:2000305. doi: 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah M, Rather BA, Choi S, Woo HG. Sequence variation in COVID-19 is predicted to facilitate stronger interaction with ACE2 promoting high infectivity. Structural Biology Immunology. 2020.
- 32.Guan Q, Sadykov M, Nugmanova R, J. Carr M, Arold ST, Arnab P. The genomic variation landscape of globally-circulating clades of SARS-CoV-2 defines a genetic barcoding scheme. bioRxiv. 2020.
- 33.Korber B, Fischer WM, Gnanakaran S Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020.
- 34.Adebali O, Bircan A, Cirici D Phylogenetic analysis of SARS-CoV-2 genomes in Turkey. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]