Abstract

There is a demonstrated and paramount need for rapid, reliable infectious disease diagnostics, particularly those for invasive fungal infections. Current clinical determinations for an appropriate antifungal therapy can take up to 3 days using current antifungal susceptibility testing methods, a time-to-readout that can prove detrimental for immunocompromised patients and promote the spread of antifungal resistant pathogens. Herein, we demonstrate the application of intensity-based reflectometric interference spectroscopic measurements (termed iPRISM) on microstructured silicon sensors for use as a rapid, phenotypic antifungal susceptibility test. This diagnostic platform optically tracks morphological changes of fungi corresponding to conidia growth and hyphal colonization at a solid–liquid interface in real time. Using Aspergillus niger as a model fungal pathogen, we can determine the minimal inhibitory concentration of clinically relevant antifungals within 12 h. This assay allows for expedited detection of fungal growth and provides a label-free alternative to broth microdilution and agar diffusion methods, with the potential to be used for point-of-care diagnostics.
Keywords: Aspergillus niger, antifungal susceptibility testing, fungal resistance, sensor, optical sensor
Pathogenic fungi are a rising cause of disease and pose a threat to immunocompromised individuals.1 Particularly, Candida species, pathogenic Aspergilli, and Cryptococci account for invasive fungal infections in humans,2,3 resulting in ∼1.4 million deaths (annually) ascribed to fungal infection.4 Because of acquired antimicrobial resistance, species identification alone is not sufficient to target efficient therapy for fungal infections.5 Thus, antifungal susceptibility testing (AFST) is critical to direct the proper choice of treatment in a timely manner.6,7 Classical AFST methods include broth microdilution (BMD) testing, the suggested reference method (by the Clinical & Laboratory Standards Institute – CLSI or the European Committee on Antimicrobial Susceptibility Testing – EUCAST), as well as agar diffusion methods, such as the Etest (bioMérieux SA). These methods are labor-intensive and time-consuming (up to 72 h),6,8 and automated tests (e.g., Vitek2, bioMériuex SA) are only available for a limited spectrum of microorganisms.9 Thus, accelerating AFST is of great importance to improve the clinical outcome of antifungal therapy and abate the emergence of antifungal resistance.1
New approaches for molecular identification of fungal pathogens and rapid AFST include mass spectrometry (MS)10 and nucleic acid-based diagnostics.6 However, high equipment acquisition costs as well as insufficient validation and multiplexing ability limit the use of MS for AFST.10 For genomic AFST, the main disadvantage is its inability to reveal the phenotypical behavior of a pathogen.11 Therefore, research efforts are directed toward developing novel phenotypic AFST methods such as flow cytometry,12 colorimetric redox indicators,13 and isothermal microcalorimetry.14 Furthermore, microfluidic systems have recently been applied to study fungal growth and assess the activity of antimicrobials.15,16 However, some of these new techniques rely on sophisticated and expensive equipment, have a limited microorganism spectrum, or do not effectively accelerate the determination of the minimum inhibitory concentration (MIC) values.
Previously, phase-shift reflectometric interference spectroscopic measurements (PRISM) were demonstrated to monitor antibiotic susceptibility and the behavior of bacteria, colonizing within silicon microstructured arrays.17,18 In this work, we demonstrate an easy-to-perform platform that allows the growth of Aspergillus niger (A. niger) to be tracked in real time. Due to the different behavior and morphology of filamentous fungi compared to bacteria, herein, we apply intensity-based PRISM, referred to as iPRISM, as a tool for rapid and label-free, phenotypic AFST using the fungal species A. niger as a model microorganism.
Results and Discussion
iPRISM relies on the capture of fungal conidia within silicon microwells and the subsequent monitoring of fungal growth in real time by intensity-based reflectometric interference spectroscopic measurements, as depicted in Figure 1. Assays are performed in a series of temperature-controlled microfluidic channels, each containing an injection port and a waste outlet. Photonic silicon chips consisting of diffraction gratings, specifically periodic arrays of microwells with a width of ∼3 μm and depth of ∼4 μm (see scanning electron micrograph in Figure 1A-i), are individually fixed in the center of the flow channels and illuminated by a white light source positioned normal to the photonic silicon chip (Figure 1A). The resulting reflectance spectrum of the zero-order diffraction exhibits interference fringes (Figure 1B), as the incident light is partially reflected by the top and the bottom of the microwells.17 The application of frequency analysis results in a single peak where the peak position corresponds to 2nL (n represents the refractive index of the medium within the arrays, and L represents the height of the microstructures; see Figure S1) and the peak amplitude or intensity (I) corresponds to the intensity of the reflected light (Figure 1D-i,ii). The antifungal agent is introduced, and fungal growth is monitored in real time by tracking the intensity changes (ΔI) during AFST experiments.
Figure 1.
iPRISM AFST concept: Schematic representation of optical monitoring of A. niger growth and responses to antifungals by iPRISM. (A) Photonic silicon chips of microwell arrays entrap Aspergillus conidia from a conidia suspension while illuminated with a white light source. (A-i) Representative cross-sectioned HR-SEM of the photonic chip (scale bar represents 5 μm). (B) The resulting reflectance spectra are recorded and analyzed in real time, allowing for label-free monitoring of fungal growth and responses to antifungals. (C) After allowing the conidia to settle within the silicon microwells, antifungals are introduced (C-i), resulting in growth inhibition at concentrations above the MIC or (C-ii) unimpeded growth at subinhibitory antifungal concentrations. (D-i) After applying frequency analysis, growth inhibition corresponds to unchanged intensity values, while (D-ii) fungal growth on top of the microwells results in a reduction of the intensity of the reflected light.
iPRISM is performed in two steps: Initially, Aspergillus conidia suspensions at a designated concentration are introduced into the microfluidic channels and allowed to incubate on the silicon microtopologies for 15 min (Figure 1A). Subsequently, the antifungal (at varying concentrations) is introduced into the channels, and the fungal response is optically monitored by iPRISM. If the conidia germinate and hyphal growth occurs on top of the microwells (see Figure 1C-ii), the intensity of the reflected light decreases over time (Figure 1D-ii), while inhibition of growth and cell death (see Figure 1C-i) result in unchanged intensity values (Figure 1D-i).
A. niger suspensions (105 conidia mL–1) were introduced into the channels, and growth was monitored by iPRISM. The latter concentration corresponds to the suggested seeding concentration by EUCAST protocols for AFST of conidia-forming molds, including Aspergillus species.8Figure 2A presents an iPRISM curve, where the values of ΔI (%) are tracked in real time, depicting a continuous decrease in intensity over a time scale of 15 h, while for the reference channel (containing just growth medium), the signal is unchanged (see also Figure S2A, in which averaged iPRISM results and the corresponding standard deviation values are presented). The decrease in the signal is ascribed to germination and hyphal growth, which were verified by confocal laser scanning microscopy (CLSM) of A. niger stained with Calcofluor White after 15 min, 4, 5, 6, and 15 h of on-chip incubation at 30 °C (Figure 2A-i–v). Germination is visible after around 5 h (Figure 2A-iii), and further growth corresponds to a distinct reduction in signal intensity after 6 h (Figure 2A-iv) and 15 h (Figure 2A-v). Germination is a process required for vegetative growth of hyphae in which the extension of the cytoplasm and the disintegration of the outer conidial wall allows the nascent germ tube to emerge.19 In previous works, it was reported that distinct germ tube formation of A. niger conidia was visible after 6 h, which is in good agreement to our findings.20
Figure 2.
iPRISM of A. niger, at a concentration of 105 conidia mL–1, on photonic silicon chips. (A) Real-time iPRISM curve, where ΔI values were recorded over a time period of 15 h with corresponding false-colored CLSM images following Calcofluor White staining after (A-i) 15 min, (A-ii) 4 h, (A-iii) 5 h, (A-iv) 6 h, and (A-v) 15 h of incubation. Note that Figure S2 provides averaged iPRISM results, including standard deviation values, for a concentration of 105 conidia mL–1. (B) Corresponding HR-SEM images: (i) The A. niger conidia are entrapped inside the microtopologies at t = 15 min. (ii) The conidia swell and break out from the microwells at t = 4 h. (iii) A. niger spreading over the chip surface after 15 h of incubation. Fungi are false-colored green for clarity.
The confocal images of the photonic silicon chips during the first 4 h of incubation show that the conidia swell. The latter is an essential step, prior to germination, where the conidia uptake liquid and their size increases.19 To verify this behavior and rule out staining procedure artifacts, the chips were studied by high resolution scanning electron microscopy (HR-SEM). Figure 2B-i shows that, in the beginning of the assay, most of the conidia, which are spherical with a typical diameter of ∼3 μm, are entrapped within the microwells. The proportion of conidia seeded per well is typically between 2 and 5 conidia per 100 microwells at a seeding concentration of 105 conidia mL–1 (see a representative HR-SEM image in Figure S3). Over time, the confined conidia swell and break out from the microwells, as depicted in Figure 2B-ii, and after 15 h, the hyphae grow on top of the microwells and cover the chip (Figure 2B-iii). The latter further supports the observed significant decrease in intensity with time.
iPRISM at a lower initial conidia concentration of 104 conidia mL–1, which may be of potential clinical relevancy, results in unchanged intensity as depicted in Figure S2A. Optical microscopy studies reveal that the conidia germinate and form few germ tubes on the chip (Figure S2B). Yet, the iPRISM assay is not sensitive enough to detect these local morphological changes. It should be noted that there are microfluidic methods that allow for more sensitive detection of germination and fungal growth; however, these techniques require sophisticated single-cell microscopy, or the use of fluorescent labeling.21 Higher conidia concentrations, such as 106 conidia mL–1, are found to cause unstable intensity values immediately after introduction (Figure S2A). This effect is ascribed to the high number of cells that are settling onto the silicon surface. Furthermore, the germination at 106 conidia mL–1 is clearly inhibited (Figure S2B), likely due to the presence of a self-inhibitor. The latter was shown by Barrios-Gonzalez et al.22 to decrease the germination rate of A. niger at concentrations above 106 conidia mL–1. Moreover, the overall observed intensity decrease for 106 cells mL–1 is less pronounced in comparison to 105 cells mL–1 (slope values of −0.82 ΔI (%) h–1 vs −0.18 ΔI (%) h–1, respectively). Therefore, we have adhered to the EUCAST protocol, and all subsequent experiments were performed with conidial seeding concentrations of 105 conidia mL–1.
For the determination of MIC values by iPRISM, A. niger conidia were exposed to various concentrations of the clinically relevant antifungals: voriconazole and amphotericin B (Figure 3). Voriconazole is the recommended antimicrobial agent for the treatment of invasive aspergillosis,23 and it inhibits the biosynthesis of the fungal cell membrane component ergosterol.24 Amphotericin B is used as a second-line treatment, and the drug interacts with ergosterol in the cell membrane, resulting in cell death.25Figure 3 presents iPRISM curves for A. niger upon exposure to varying concentrations of voriconazole (Figure 3A) and amphotericin B (Figure 3B). For both drugs, increased concentrations result in moderate intensity changes in comparison to the untreated fungi. For example, when using 1 mg L–1 voriconazole, the intensity remains constant throughout the experiment, whereas a characteristic decrease of ∼6% is detected in the absence of the drug. This behavior is ascribed to complete growth inhibition of the fungi in the presence of increasing concentrations of the antifungal agent. Indeed, optical microscopy images (Figure 3C) reveal that, at a voriconazole concentration of 1 mg L–1, no germination is observed, while at lower drug concentrations, some germination occurs, and few germ tubes are visible on the surface of the chip. A similar behavior is observed upon exposure to amphotericin B, and at a concentration of 1 mg L–1, only minor germination is observed after 15 h (Figure 3D).
Figure 3.
A. niger iPRISM AFST. iPRISM curves, displaying ΔI (%) over time, upon exposure to varying concentrations of the antifungal drugs: (A) voriconazole and (B) amphotericin B. Corresponding optical microscope images of photonic silicon chips after 15 h, revealing the behavior of the fungi upon exposure to different concentrations of (C) voriconazole and (D) amphotericin B. Scale bar represents 50 μm.
In standard methods for AFST, such as BMD or E-test, the MIC is defined as the lowest concentration of a drug that inhibits the visible growth of a microorganism.8 In the iPRISM assay, we suggest designating the MIC as the lowest concentration of an antifungal agent at which no reduction of the intensity value ΔI (%) occurs, while at subinhibitory concentrations, a decline of intensity should be visible to some extent. As such, the iPRISM MIC values were determined to be 0.5 and 1 mg L–1 for voriconazole and amphotericin B, respectively. Figure S4 presents averaged iPRISM curves and the respective standard deviations of triplicate experiments using different chips. The statistical analysis of these results demonstrates that MIC values for amphotericin B and voriconazole can be determined within 10 and 12 h, respectively. Note that the obtained Zprime score values are negative (Table S1), indicating a low signal-to-noise ratio. This may emphasize the general challenge to define a specific MIC value for a drug–strain combination due to possible inter- and intralaboratory variation in antimicrobial susceptibility testing (AST).26 This variation is also recognized by EUCAST quality control documents stating that repeated testing of quality control strains should yield individual MIC values randomly distributed within the recommended ranges.27 Thus, the iPRISM method significantly accelerates the MIC determination in comparison to the gold-standard microbiological technique BMD, in which the MIC is determined only after 48 h.8
The iPRISM MIC values for the two antifungal drugs are compared to MIC values determined by the BMD testing, which is standardized by both EUCAST and CLSI (Table 1). The iPRISM and BMD MIC values for voriconazole are the same, while for amphotericin B, the iPRISM MIC is higher than the BMD value. Yet, the iPRISM MIC values for both voriconazole and amphotericin B comply with the EUCAST MIC distribution for these antifungals.28,29 It should be kept in mind that differences in MIC values between methods are common in both AFST and AST.7 We have previously shown that MIC values of E. coli grown on silicon gratings are slightly higher in comparison to values determined by BMD.18 The main reason for these discrepancies is likely due to the different growth environments provided for the fungi, where the BMD analyzes fungal growth in a liquid medium and the iPRISM detects changes occurring on the surface of the silicon grating. Thus, it is possible that the microtopologies provide a surface with which the A. niger can interact.
Table 1. Comparison of MIC Values as Determined by iPRISM and BMD for Voriconazole and Amphotericin B.
| iPRISM |
BMD |
|||
|---|---|---|---|---|
| antifungal agent | MIC (mg L–1) | time (h) | MIC (mg L–1) | time (h) |
| voriconazole | 0.5 | 12 | 0.5 | 48 |
| amphotericin B | 1 | 10 | 0.25 | 48 |
A proof-of-concept for a rapid and label-free assay for phenotypic AFST testing of filamentous fungi is demonstrated. The fungi are colonized on a microstructured photonic chip, which also serves as the optical transducer element. Fungal growth is monitored in real time by detecting changes in the reflectivity spectra collected from the photonic silicon chip and are correlated to characteristic morphology changes of the fungi. This approach is employed to study the behavior of A. niger upon exposure to clinically relevant antifungals, specifically voriconazole and amphotericin B. Using the iPRISM assay, distinct differentiation between growth and no growth and determination of MIC values can be achieved within <12 h for A. niger. Thus, iPRISM is significantly faster than classical methods used for filamentous fungi. Furthermore, the MIC values determined by iPRISM are in agreement with standardized BMD results for A. niger. Nevertheless, it should be noted that MIC determination for slower-growing strains may take longer compared to the model species in this work. Also, the agreement of MIC values for other fungi–drug combinations to reference methods should be validated in the future.
While this platform was demonstrated for A. niger, iPRISM can potentially be used as a tool for monitoring other microorganisms, such as Candida and Cryptococcus species due to its simple principle of detection. Thus, current and future research in our group focus on demonstrating iPRISM applicability for AFST of yeast, owing to their different morphology compared to Aspergillus species.
Methods
Materials
Glutaraldehyde, Calcofluor White, Roswell Park Memorial Institute medium (RPMI 1640), d-glucose, amphotericin B, and voriconazole were supplied by Sigma-Aldrich, Israel. Absolute ethanol, dimethyl sulfoxide (DMSO), and all buffer salts were purchased from Merck, Germany. Isopropanol and acetone were supplied by Gadot, Israel. Potato Dextrose Agar (PDA), Potato Dextrose Broth (PDB), Bacto agar, yeast extract, and casein hydrolysate were purchased from Difco, USA. 3-(N-Morpholino)propanesulfonic acid (MOPS) was supplied by Chem-Impex International, Inc., USA. Sabouraud Dextrose Agar (SDA) plates were purchased from Novamed Ltd., Israel.
Preparation of Solutions and Media
All aqueous solutions and media were prepared in Milli-Q water (18.2 MΩ cm). Phosphate buffered saline (PBS) was composed of 137 mM NaCl, 2.7 mM KCl, 1.8 mM KH2PO4, and 10 mM Na2HPO4. RPMI 1640 2% G medium (RPMI 1640 medium supplemented with 2% d-glucose; referred to as growth medium) comprised 10.4 g L–1 RPMI 1640, 34.5 g L–1 MOPS, and 18 g L–1 glucose. 1% PDA contained 10 g L–1 PDA and 15 g L–1 Bacto agar; water agar contained 20 g L–1 Bacto agar. All media and buffer were autoclaved or sterile filtered prior to use.
Aspergillus niger Isolate
The fungi were isolated from a contaminated onion into 1% PDA and incubated in darkness for 10 days at 25 °C. Conidia from the mature culture were recultured by streaking onto SDA plates supplemented with chloramphenicol to eliminate bacterial contamination and incubated as described above. For preparation of monoconidial cultures, conidia were recultured onto water agar and incubated overnight (18 h) at 25 °C. Single germinating conidia were removed by micromanipulation under a light microscope and transferred to 1% PDA. All tests were performed with a single monoconidial culture isolate, designated HCN 18, in order to limit possible genetic variability. Species identification of isolate HCN 18 was carried out by sequencing the internal transcribed spacer regions (ITS1 and ITS2; see the Supporting Information for the sequence (Table S2) and protocol), and the isolate was identified as A. niger.
Preparation of Fungal Cultures
Fungal cultures were refreshed every 2 weeks onto 1% PDA and maintained at 4 °C until used. Prior to iPRISM experiments, the cultures were transferred onto SDA plates containing chloramphenicol. Because A. niger showed the fastest growth at 30 °C, the fungal cultures were subsequently incubated at 30 °C, and after 2–5 days, sufficient sporulation for AFST experiments was reached. Conidial density was quantified by using a hemocytometer (Neubauer improved cell counting chamber).
Fabrication and Preparation of Photonic Silicon Chips
Silicon chips with microwell gratings were fabricated by standard lithography and reactive ion etching techniques at the Micro- and Nano-Fabrication and Printing Unit (Technion). The resulting wafers were coated with photoresist to protect their microstructure while dicing the wafer into 5 × 5 mm chips using an automated dicing saw (DAD3350, Disco, Japan). The photonic silicon chips were washed with acetone to remove the photoresist and oxidized for 1 h at 800 °C in a furnace (Lindberg/Blue M 1200 °C Split-Hinge, Thermo Scientific, USA).
iPRISM Assay
A custom-made, aluminum chamber with seven injection and outlet channels was used to fix and separate the photonic silicon chips during the iPRISM assays. Each injection channel was connected by tubing to a syringe injection port, and the injection of the conidia suspensions was allowed. The chamber was controlled by a motorized linear stage (Thorlabs, Inc., USA). The photonic chips were placed in a small square cavity in each channel and were separated from each other and fixed on the surface of the chamber by a rubber gasket. The system was further sealed before the experiments by an acrylic piece and by tightening the lid of the aluminum housing. Before each experiment, the system was sterilized with 70% ethanol and sterile water followed by introduction of only growth medium to allow devices, temperature, and medium to equilibrate. Subsequently, 500 μL of the conidia suspension was introduced, and after 15 min, the antifungals were introduced, while the reflectance signal was continuously recorded during the experiment.
Data Acquisition and Analysis
A bifurcated fiber optic (Ocean Optics, USA)
equipped with a collimating lens was positioned normal to the photonic
silicon chips, illuminating them via a white light source. The reflected
light was recorded by a USB4000 CCD spectrometer (Ocean Optics, USA).
The position of the chamber was controlled by a motorized stage (Thorlabs,
USA) and LabView (National Instruments, USA). Frequency analysis was
performed on acquired spectra in the range between 450 and 900 nm.
The resulting peak after fast Fourier transform (FFT) was identified
by determining the maximum peak position, where the height of the
detected peak directly corresponds to the intensity of the reflected
light. The intensity values were plotted versus time. For AFST, the
introduction time of the antifungal is referred to as time 0. The
percent changes of the intensity (ΔI) were
calculated as follows: 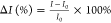 , where I is the intensity at a
given time and I0 is the intensity at
time 0.
, where I is the intensity at a
given time and I0 is the intensity at
time 0.
Characterization of Photonic Silicon Chips
The chips were examined immediately after experiments by using an optical light microscope (Axio Scope A1, Carl Zeiss, Germany) to identify fungal growth and ensure no bacterial contamination occurred.
High-resolution scanning electron microscopy (HR-SEM) was performed using a Zeiss Ultra Plus high-resolution scanning microscope. Samples were fixated using 2.5% glutaraldehyde in PBS, washed with water, and dehydrated through a dilution series in ethanol with increasing concentration from 10% to absolute ethanol.
Focused ion beam (FIB)-cross-sectional SEM images were obtained using a Dual Beam Helios NanoLab G3 instrument (FEI, USA). Platinum deposition was performed prior to observation.
Confocal laser scanning microscopy (CLSM; LSM 700, Carl Zeiss, Germany) was performed on samples stained by Calcofluor White and a drop of 10% potassium hydroxide; the excitation wavelength was 405 nm, and images were rendered by Zen software (Carl Zeiss, Germany).
Broth Microdilution
BMD was performed according to the EUCAST AFST protocol for conidia forming molds.8 Fungal growth was visually observed after 48 h and also confirmed by optical density measurements (600 nm, n = 5, Varioskan Flash, Thermo Scientific, USA). The only adjustment we made to the EUCAST protocol was decreasing the incubation temperature to 30 °C.
Acknowledgments
This work was partially supported by the Israel Innovation Authority (Kamin program). C.H. acknowledges financial aid from the Leibniz Universitätsgesellschaft and the Technion. We thank Dima Peselev and Orna Ternyak (MNFPU, Technion) for the microfabrication of the photonic chips and Omer Sabach for his artwork depicting the microfluidic channels (Figure 1A).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00234.
Figure S1, schematic of the silicon microstructure; Figure S2, iPRISM assay for different cell concentrations; Figure S3, representative seeding proportion; Figure S4, iPRISM for AFST in triplicates; Table S1, ZPrime Score for voriconazole and amphotericin B at different time points; Table S2, ITS sequencing result; protocol for DNA extraction and sequencing of the ITS regions (PDF)
Author Contributions
⊥ C.H. and H.L. contributed equally. E.S., H.L., N.N., and C.H. conceptualized the study. A.L-A., N.M-I., H.L., and C.H. performed the experiments. E.S., H.L., T.S., and C.H. wrote the manuscript. All authors commented, revised, and approved the final version of the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Srinivasan A.; Lopez-Ribot J. L.; Ramasubramanian A. K. (2014) Overcoming Antifungal Resistance. Drug Discovery Today: Technol. 11, 65–71. 10.1016/j.ddtec.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber G. (2001) An Overview of Fungal Infections. Drugs 61, 1–12. 10.2165/00003495-200161001-00001. [DOI] [PubMed] [Google Scholar]
- Montoya M. C.; Moye-Rowley W. S.; Krysan D. J. (2019) Candida auris the Canary in the Mine of Antifungal Drug Resistance. ACS Infect. Dis. 5, 1487–1492. 10.1021/acsinfecdis.9b00239. [DOI] [PubMed] [Google Scholar]
- Sanglard D. (2016) Emerging Threats in Antifungal-Resistant Fungal Pathogens. Front. Med. 3, 11–11a. 10.3389/fmed.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham C. J.; Boonstra S.; Levels S.; de Lange M.; Meis J. F.; Schneeberger P. M. (2012) Rapid Susceptibility Testing and Microcolony Analysis of Candida Spp. Cultured and Imaged on Porous Aluminum Oxide. PLoS One 7 (3), e33818 10.1371/journal.pone.0033818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M.; Posteraro B. (2017) New Approaches for Antifungal Susceptibility Testing. Clin. Microbiol. Infect. 23 (12), 931–934. 10.1016/j.cmi.2017.03.025. [DOI] [PubMed] [Google Scholar]
- Leonard H.; Colodner R.; Halachmi S.; Segal E. (2018) Recent Advances in the Race to Design a Rapid Diagnostic Test for Antimicrobial Resistance. ACS Sens 3 (11), 2202–2217. 10.1021/acssensors.8b00900. [DOI] [PubMed] [Google Scholar]
- Arendrup M. C., Meletiadis J., Mouton J. W., Lagrou K., Hamal P., and Guinea J., and the Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing. (2017) EUCAST Definitive Document E.DEF 9.3.1. [Google Scholar]
- van Belkum A.; Dunne W. M. (2013) Next-Generation Antimicrobial Susceptibility Testing. J. Clin. Microbiol. 51 (7), 2018–2024. 10.1128/JCM.00313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A.; Welker M.; Pincus D.; Charrier J. P.; Girard V. (2017) Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in Clinical Microbiology: What Are the Current Issues?. Ann. Lab. Med. 37 (6), 475–483. 10.3343/alm.2017.37.6.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes B. L.; Wiederhold N. P. (2018) Molecular Diagnostics in Medical Mycology. Nat. Commun. 9 (1), 5135. 10.1038/s41467-018-07556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichrodt R.-J.; Read N. D. (2019) Flow Cytometry and FACS Applied to Filamentous Fungi. Fungal Biol. Rev. 33 (1), 1–15. 10.1016/j.fbr.2018.06.001. [DOI] [Google Scholar]
- Leong C.; Buttafuoco A.; Glatz M.; Bosshard P. P. (2017) Antifungal Susceptibility Testing of Malassezia Spp. with an Optimized Colorimetric Broth Microdilution Method. J. Clin. Microbiol. 55 (6), 1883. 10.1128/JCM.00338-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furustrand Tafin U.; Orasch C.; Trampuz A. (2013) Activity of Antifungal Combinations against Aspergillus Species Evaluated by Isothermal Microcalorimetry. Diagn. Microbiol. Infect. Dis. 77 (1), 31–36. 10.1016/j.diagmicrobio.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Ellett F.; Jorgensen J.; Frydman G. H.; Jones C. N.; Irimia D. (2017) Neutrophil Interactions Stimulate Evasive Hyphal Branching by Aspergillus fumigatus. PLoS Pathog. 13 (1), e1006154 10.1371/journal.ppat.1006154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak N.; Olcum S.; Delgado F.; Wasserman S.; Payer K.; Murakami M.; Knudsen S.; Kimmerling R.; Stevens M.; Kikuchi Y.; Sandikci A.; Ogawa M.; Agache V.; Baléras F.; Weinstock D. M.; Manalis S. R. (2016) High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat. Biotechnol. 34, 1052–1059. 10.1038/nbt.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massad-Ivanir N.; Mirsky Y.; Nahor A.; Edrei E.; Bonanno-Young L. M.; Ben Dov N.; Sa’ar A.; Segal E. (2014) Trap and Track: Designing Self-Reporting Porous Si Photonic Crystals for Rapid Bacteria Detection. Analyst 139 (16), 3885–3894. 10.1039/C4AN00364K. [DOI] [PubMed] [Google Scholar]
- Leonard H.; Halachmi S.; Ben-Dov N.; Nativ O.; Segal E. (2017) Unraveling Antimicrobial Susceptibility of Bacterial Networks on Micropillar Architectures Using Intrinsic Phase-Shift Spectroscopy. ACS Nano 11 (6), 6167–6177. 10.1021/acsnano.7b02217. [DOI] [PubMed] [Google Scholar]
- Tsukahara T. (1968) Electron microscopy of swelling and germinating conidiospores of Aspergillus niger. Med. Mycol. 6 (3), 185–191. 10.1080/00362176885190371. [DOI] [PubMed] [Google Scholar]
- Anderson J. G.; Smith J. E. (1971) The Production of Conidiophores and Conidia by Newly Germinated Conidia of Aspergillus niger (Microcycle Conidiation). J. Gen. Microbiol. 69, 185–197. 10.1099/00221287-69-2-185. [DOI] [PubMed] [Google Scholar]
- Zhou W.; Le J.; Chen Y.; Cai Y.; Hong Z.; Chai Y. (2019) Recent Advances in Microfluidic Devices for Bacteria and Fungus Research. TrAC, Trends Anal. Chem. 112, 175–195. 10.1016/j.trac.2018.12.024. [DOI] [Google Scholar]
- Barrios-Gonzalez J.; Martinez C.; Aguilera A.; Raimbault M. (1989) Germination of concentrated suspensions of spores from Aspergillus niger. Biotechnol. Lett. 11, 551–554. 10.1007/BF01040034. [DOI] [Google Scholar]
- Maertens J. A.; Raad I. I.; Marr K. A.; Patterson T. F.; Kontoyiannis D. P.; Cornely O. A.; Bow E. J.; Rahav G.; Neofytos D.; Aoun M.; Baddley J. W.; Giladi M.; Heinz W. J.; Herbrecht R.; Hope W.; Karthaus M.; Lee D.-G.; Lortholary O.; Morrison V. A.; Oren I.; Selleslag D.; Shoham S.; Thompson G. R.; Lee M.; Maher R. M.; Schmitt-Hoffmann A.-H.; Zeiher B.; Ullmann A. J. (2016) Isavuconazole versus Voriconazole for Primary Treatment of Invasive Mould Disease Caused by Aspergillus and Other Filamentous Fungi (SECURE): A Phase 3, Randomised-Controlled, Non-Inferiority Trial. Lancet 387 (10020), 760–769. 10.1016/S0140-6736(15)01159-9. [DOI] [PubMed] [Google Scholar]
- Johnson L. B.; Kauffman C. A. (2003) Voriconazole: A New Triazole Antifungal Agent. Clin. Infect. Dis. 36 (5), 630–637. 10.1086/367933. [DOI] [PubMed] [Google Scholar]
- Gray K. C.; Palacios D. S.; Dailey I.; Endo M. M.; Uno B. E.; Wilcock B. C.; Burke M. D. (2012) Amphotericin Primarily Kills Yeast by Simply Binding Ergosterol. Proc. Natl. Acad. Sci. U. S. A. 109 (7), 2234. 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouton J. W.; Meletiadis J.; Voss A.; Turnidge J. (2018) Variation of MIC measurements: the contribution of strain and laboratory variability to measurement precision. J. Antimicrob. Chemother. 73, 2374–2379. 10.1093/jac/dky232. [DOI] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (2020) Routine and extended internal quality control for MIC determination and agar dilution for yeasts and moulds as recommended by EUCAST, version 4.0, http://www.eucast.org.
- European Committee on Antimicrobial Susceptibility Testing (2012) Voriconazole and Aspergillus spp.: Rationale for the clinical breakpoints, version 1.0, http://www.eucast.org.
- European Committee on Antimicrobial Susceptibility Testing (2012) Amphotericin B and Aspergillus spp.: Rationale for the clinical breakpoints, version 1.0, http://www.eucast.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





