Abstract
The COVID-19 pandemic is a major threat to global health1 for which there are limited medical countermeasures2,3. Moreover, we currently lack a thorough understanding of mechanisms of humoral immunity4. From a larger panel of human monoclonal antibodies (mAbs) targeting the spike (S) glycoprotein5, we identified several that exhibited potent neutralizing activity and fully blocked the receptor-binding domain of S (SRBD) from interacting with human ACE2 (hACE2). Competition-binding, structural, and functional studies allowed clustering of the mAbs into classes recognizing distinct epitopes on the SRBD as well as distinct conformational states of the S trimer. Potent neutralizing mAbs recognizing non-overlapping sites, COV2-2196 and COV2-2130, bound simultaneously to S and synergistically neutralized authentic SARS-CoV-2 virus. In two mouse models of SARS-CoV-2 infection, passive transfer of either COV2-2196 or COV2-2130 alone or a combination of both mAbs protected mice from weight loss and reduced viral burden and inflammation in the lung. In addition, passive transfer of each of two of the most potently ACE2 blocking mAbs (COV2-2196 or COV2-2381) as monotherapy protected rhesus macaques from SARS-CoV-2 infection. These results identify protective epitopes on SRBD and provide a structure-based framework for rational vaccine design and the selection of robust immunotherapeutics.
Keywords: Coronavirus; SARS-CoV-2; SARS-CoV; COVID-19; Antibodies, Monoclonal; Human; Adaptive Immunity
The S protein of SARS-CoV-2 is the molecular determinant of viral attachment, fusion, and entry into host cells6. The S protein is composed of an N-terminal subunit (S1) that mediates receptor binding and a C-terminal subunit (S2) that mediates virus–cell membrane fusion. The S1 subunit contains an N-terminal domain (NTD) and a receptor-binding domain (RBD). SARS-CoV-2 and SARS-CoV, which share approximately 78% sequence identity in their genomes1 both use human angiotensin-converting enzyme 2 (hACE2) as an entry receptor7-9. Human Abs to the (S) glycoprotein mediate protective immunity against other high-pathogenicity zoonotic betacoronaviruses including SARS-CoV10-14 and Middle East respiratory syndrome (MERS)15-24. The most potent S protein-specific mAbs appear to neutralize betacoronaviruses by blocking attachment of virus to host cells by binding to the region on SRBD that directly mediates receptor engagement. It is likely that human Abs have promise for modifying disease, when used for prophylaxis, post-exposure prophylaxis, or treatment of SARS-CoV-2 infection25. Many studies are ongoing, including randomized controlled trials evaluating convalescent plasma and one trial evaluating hyperimmune immunoglobulin, but it is not yet clear whether such treatments can reduce morbidity or mortality26.
We isolated a large panel of SARS-CoV-2 S protein-reactive mAbs from the B cells of two convalescing individuals who had been infected with SARS-CoV-2 in Wuhan China5. A subset of those antibodies bound to a recombinant RBD construct (SRBD) and exhibited neutralizing activity in a rapid screening assay with authentic SARS-CoV-25. Here, we defined the antigenic landscape of SARS-CoV-2 and determined which sites of SRBD are targets of neutralizing mAbs. We tested a panel of 40 anti-S human mAbs we previously pre-selected by a rapid neutralization screening assay in a quantitative focus reduction neutralization test (FRNT) with SARS-CoV-2 strain WA1/2020. These assays revealed the panel exhibited a range of half-maximal inhibitory concentration (IC50) values, from 15 to over 4,000 ng/mL (visualized as a heatmap in Fig. 1a, values shown in Supplemental Table 1, and full curves shown in Extended Data Fig. 1). We hypothesized that many of these SRBD-reactive mAbs neutralize virus infection by blocking SRBD binding to hACE2. Indeed, most neutralizing mAbs we tested inhibited the interaction of hACE2 with trimeric S protein directly (Fig. 1a; Extended Data Fig. 2). Consistent with these results, these mAbs also bound strongly to a trimeric S ectodomain (S2Pecto) protein or monomeric RBD SRBD (Fig. 1a; Extended Data Fig. 3). We evaluated whether S2Pecto or SRBD binding or hACE2-blocking potency predicted binding neutralization potency independently, but none of these measurements correlated with neutralization potency (Fig. 1b-d). However, each of the highest neutralizing potency tier (IC50<150 ng/mL) also had the strongest blocking activity against hACE2 (IC50< 150 ng/mL) and exceptional binding activity (EC50< 2 ng/mL) to S2Pecto trimer (Fig. 1e). Representative neutralization curves for two potently neutralizing mAbs designated COV2-2196 and COV2-2130 are shown in Fig. 1f. Potent neutralization was confirmed using pseudovirus neutralization assays, which revealed far more sensitive neutralization phenotypes than wt virus and demonstrated a requirement for the use of live virus assays for assessment of mAb potency (Fig. 1g). Both of these mAbs bound strongly to S2Pecto trimer and fully blocked hACE2 binding (Fig. 1h-i).
Figure 1. Functional characteristics of neutralizing SARS-CoV-2 mAbs.
a. Heatmap of mAb neutralization activity, hACE2 blocking activity, and binding to either trimeric S2Pecto protein or monomeric SRBD. MAbs are ordered by neutralization potency, and dashed lines indicate the 12 antibodies with a neutralization IC50 value lower than 150 ng/mL. IC50 values are visualized for viral neutralization and hACE2 blocking, while EC50 values are visualized for binding. The cross-reactive SARS-CoV SRBD mAb rCR3022 is shown as a positive control, while the anti-dengue mAb r2D22 is shown as a negative control. Data are representative of at least 2 independent experiments performed in technical duplicate. No inhibition or no binding indicates an IC50 or EC50 value of >10,000 ng/mL, respectively.
b-d. Correlation of hACE2 blocking, S2Pecto trimer binding, or SRBD binding of mAbs with their neutralization activity. e. Correlation of hACE2 blocking and S2Pecto trimer binding. R2 values are shown for linear regression analysis of log-transformed values. Purple circles indicate mAbs with a neutralization IC50 value lower than 150 ng/mL.
f. Neutralization curves for COV2-2196 and COV2-2130 against authentic SARS-CoV-2 virus. Calculated IC50 values are denoted on the graph. Error bars denote the standard deviation of each point. Data are representative of at least 2 independent experiments, each performed in technical duplicate.
g. Neutralization curves for COV2-2196 and COV2-2130 in a pseudovirus neutralization assay. Error bars denote the standard deviation of each point. Values shown are technical duplicates from a single experiment. Calculated IC50 values from a minimum of 6 experiments are denoted on the graph.
h. hACE2 blocking curves for COV2-2196, COV2-2130, and the non-blocking SARS-CoV mAb rCR3022 in the hACE2 blocking ELISA. Calculated IC50 values are denoted on the graph.. Mean ± SD of technical triplicates are shown from a representative experiment repeated twice.
i. ELISA binding of COV2-2196, COV2-2130, and rCR3022 to trimeric S2Pecto. Calculated EC50 values are denoted on the graph. Mean ± SD of technical triplicates are shown from a representative experiment repeated twice.
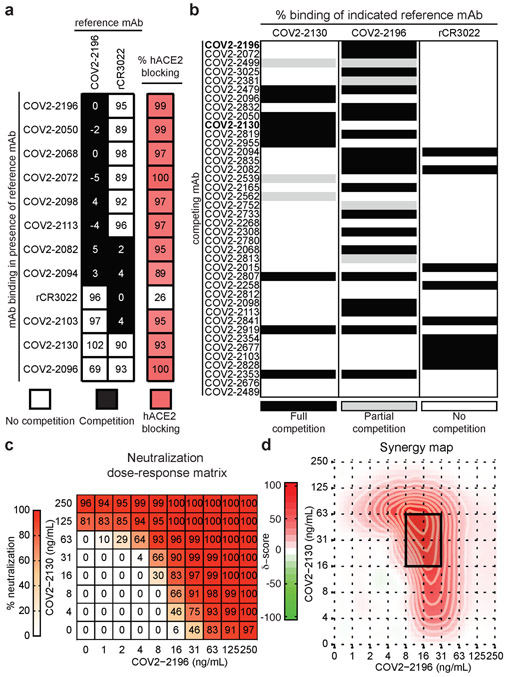
We next defined the major antigenic sites on SRBD for neutralizing mAbs by competition-binding analysis. We first used a biolayer interferometry-based competition assay with a minimal SRBD domain to screen for mAbs that competed for binding with the potently neutralizing mAb COV2-2196 or a recombinant version of the previously described SARS-CoV mAb CR3022, which recognizes a conserved cryptic epitope12,27. We identified three major groups of competing mAbs (Fig. 2a). The largest group of mAbs blocked COV2-2196 but not rCR3022, while some mAbs were blocked by rCR3022 but not COV2-2196. Two mAbs, including COV2-2130, were not blocked by either reference mAb. Most mAbs competed with hACE2 for binding, suggesting that they bound near the hACE2 binding site of the SRBD. We used COV2-2196, COV2-2130, and rCR3022 in an ELISA-based competition-binding assay with trimeric S2Pecto protein and also found that SRBD contained three major antigenic sites, with some mAbs likely making contacts in more than one site (Fig. 2b). Most of the potently neutralizing mAbs directly competed for binding with COV2-2196. The hACE2 and mAb competition binding analysis with SARS-CoV-2 convalescent patient serum or plasma from four previously described immune subjects5 showed that COV2-2196- and COV2-2130-like antibody responses are subdominant in these subjects (Extended Data Fig. 4).
Figure 2. Epitope mapping of mAbs by competition-binding analysis and synergistic neutralization by a pair of mAbs.
a. Left: mAb binding to RBD in presence of reference mAbs COV2-2196 and rCR3022. Values in squares are % binding of the mAb in the presence of the competing mAb relative to a mock-competition control. Black squares denote full competition (<33% relative binding), while white squares denote no competition (>67% relative binding). Right: biolayer interferometry-based competition binding assay measuring the ability of mAbs to prevent binding of hACE2. Values denote % blocking of hACE2 by mAb. Red color denotes high blocking activity.
b. Competition of neutralizing mAb panel with reference mAbs COV2-2130, COV2-2196, or rCR3022. Binding of reference mAbs to trimeric S2Pecto was measured in the presence of saturating competitor mAb in a competition ELISA and normalized to binding in the presence of the anti-dengue mAb r2D22. Black denotes full competition (<25% binding of reference mAb), grey denotes partial competition (25-60% binding of reference mAb), and white denotes no competition (>60% binding of reference mAb).
c. Neutralization dose-response matrix of wild-type SARS-CoV-2 by COV2-2196 and COV2-2130. Axes denote the concentration of each mAb with % neutralization shown in each square. Data is from a representative experiment that was performed in technical triplicate and repeated twice. A white-to-red heatmap denotes 0% neutralization to 100% neutralization, respectively. d. Synergy map calculated based on the SARS-CoV-2 neutralization in c. Red color denotes areas where synergistic neutralization was observed, and a black box denotes the area of maximal synergy between the two mAbs.
Since COV2-2196 and COV2-2130 did not compete for binding to SRBD, we assessed if these mAbs synergize for virus neutralization, a phenomenon previously observed for SARS-CoV mAbs12. We tested combination responses (Fig. 2c) in the FRNT using SARS-CoV-2 and compared these experimental values with the expected responses calculated by synergy scoring models28. The comparison revealed that the combination of COV2-2196 + COV2-2130 was synergistic (Fig. 2d) (with a synergy score of 17.4, where any score of >10 indicates synergy). In particular, combined mAb dose of 79 ng/mL in the cocktail (16 ng/mL of COV2-2196 and 63 ng/mL of COV2-2130) had the same activity as 250 ng/mL of each individual mAb (Fig. 2c). This finding shows that by using a cocktail the dose of each mAb can be reduced by more than three-fold to achieve the same potency of virus neutralization in vitro.
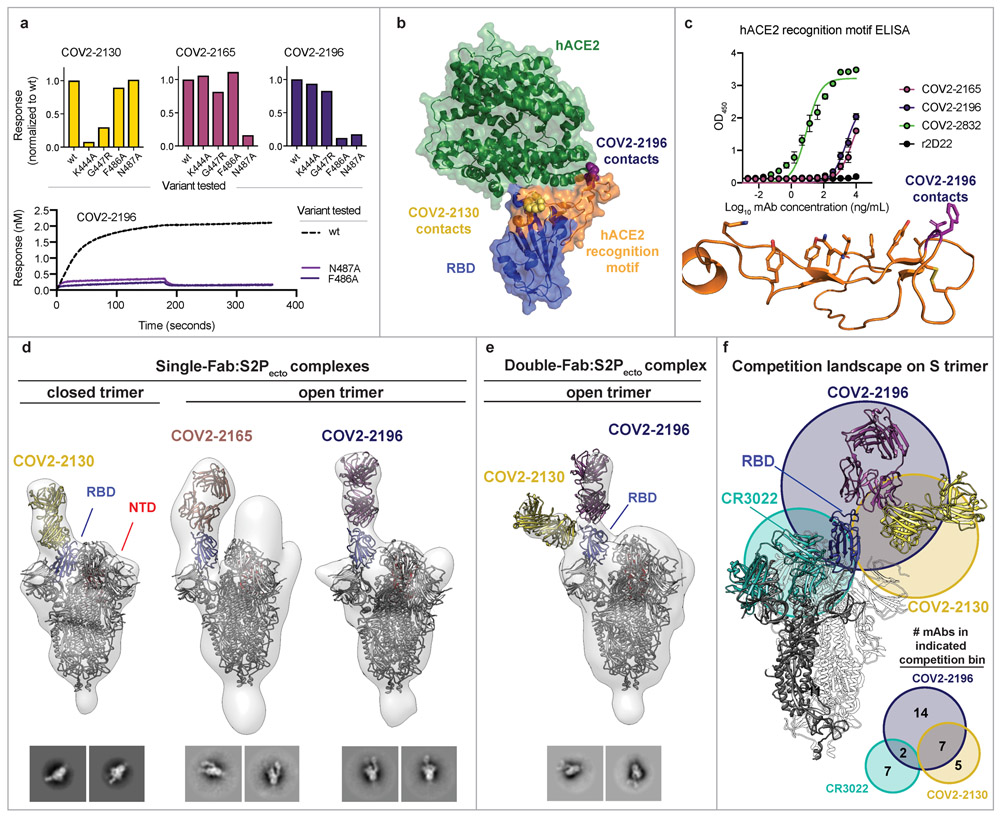
We next defined the epitopes recognized by representative mAbs in the two major competition-binding groups that synergize for neutralization. We determined critical residues for binding of neutralizing mAbs using mutagenesis. (Fig. 3a, Extended Data Fig. 5). These studies revealed F486A or N487A as critical residues for COV2-2196 and N487A as a critical residue for COV2-2165, which compete with one another for binding. Likewise, mutagenesis studies for COV2-2130 using K444A and G447R mutants defined these residues as critical for recognition (Fig. 3a). Previous structural studies have defined the interaction between the SRBD and hACE229 (schematic shown in Fig. 3b). Most of the interacting residues in the SRBD are contained within a 60-amino-acid linear peptide that defines the hACE2 recognition motif (Fig. 3c). We next tested binding of human mAbs to this minimal peptide and found that potent neutralizing members of the largest competition-binding group including COV2-2196, COV2-2165, and COV2-2832 recognized this peptide (Fig. 3c), suggesting these mAbs make critical contacts within the hACE2 recognition motif.
Figure 3. Epitope identification and structural characterization of mAbs.
a. Identification of critical contact residues by alanine and arginine mutagenesis. Top: binding of COV2-2130 (gold), COV2-2165 (maroon) or COV2-2196 (dark purple) to wild-type (wt) or mutant SRBD constructs normalized to wt. Bottom: representative binding curves for COV2-2196 to wt or SRBD constructs with mutated critical contact residues.
b. Co-crystal structure of SARS-CoV-2 RBD (blue) and hACE2 (green) (PDB 6M0J). The hACE2 recognition motif is colored orange. Critical contact residues are shown for COV2-2130 (gold spheres) and COV2-2196 (purple spheres).
c. ELISA binding of mAbs to the hACE2 recognition motif. r2D22 is shown as a negative control. Mean ± SD of technical triplicates are shown from a single experiment repeated twice. Bottom: structure of hACE2 recognition motif in orange with COV2-2196 critical contact residues shown in purple.
d. Single-Fab:S2Pecto trimer complexes visualized by negative-stain electron microscopy for COV2-2130 (gold), COV2-2165 (maroon), or COV2-2196 (dark purple). The RBD is shown in blue and the S N-terminal domain (NTD) is shown in red. Electron density is shown in grey. Trimer state (open or closed) is denoted for each complex. Representative 2D class averages for each complex are shown at the bottom (box size is 128 pixels, with 3.06 Å/pixel). Data were collected in a single experiment with detailed collection statistics in Supplemental Table 2.
e. COV2-2130 and COV2-2196 Fabs in complex with S2Pecto trimer. Colors and data collection are as in (d). Representative 2D class averages for the complexes are shown at the bottom with scales as in (d).
f. Competition-binding analysis visualized on S2Pecto trimer. The CR3022 crystal structure was docked into the double-Fab:S2Pecto trimer model. CR3022 is shown in cyan. Bottom: a quantitative Venn diagram notes the number of mAbs in each competition group.
We used negative-stain electron microscopy (NS-EM) of S2Pecto trimer/Fab complexes to determine the structural epitopes for several mAbs (Fig. 3d-e, Supplemental Table 2). The potently neutralizing antibodies COV2-2196 and COV2-2165 bound the hACE2 recognition motif of SRBD and recognized the ‘open’ conformational state of the S2Pecto trimer, in which the RBD rotates upward to expose the residues that mediate ACE2 interaction (Fig. 3d)30,31. COV2-2130, which represents a different competition-binding group, bound to the RBD in the S2Pecto trimer in the ‘closed’ position (Fig. 3d). Since COV2-2196 and COV2-2130 did not compete for binding, we attempted to make complexes of both Fabs bound at the same time to the S2Pecto trimer. We found that both Fabs bound simultaneously when the S2Pecto trimer was in the open position, indicating that COV2-2130 can recognize the SRBD in both conformations (Fig. 3e). Overlaying the two-Fab complex with the structure of the RBD:CR3022 complex27, we observed that these antibodies bind to three distinct sites on SRBD, as predicted by our competition-binding studies (Fig. 3f).
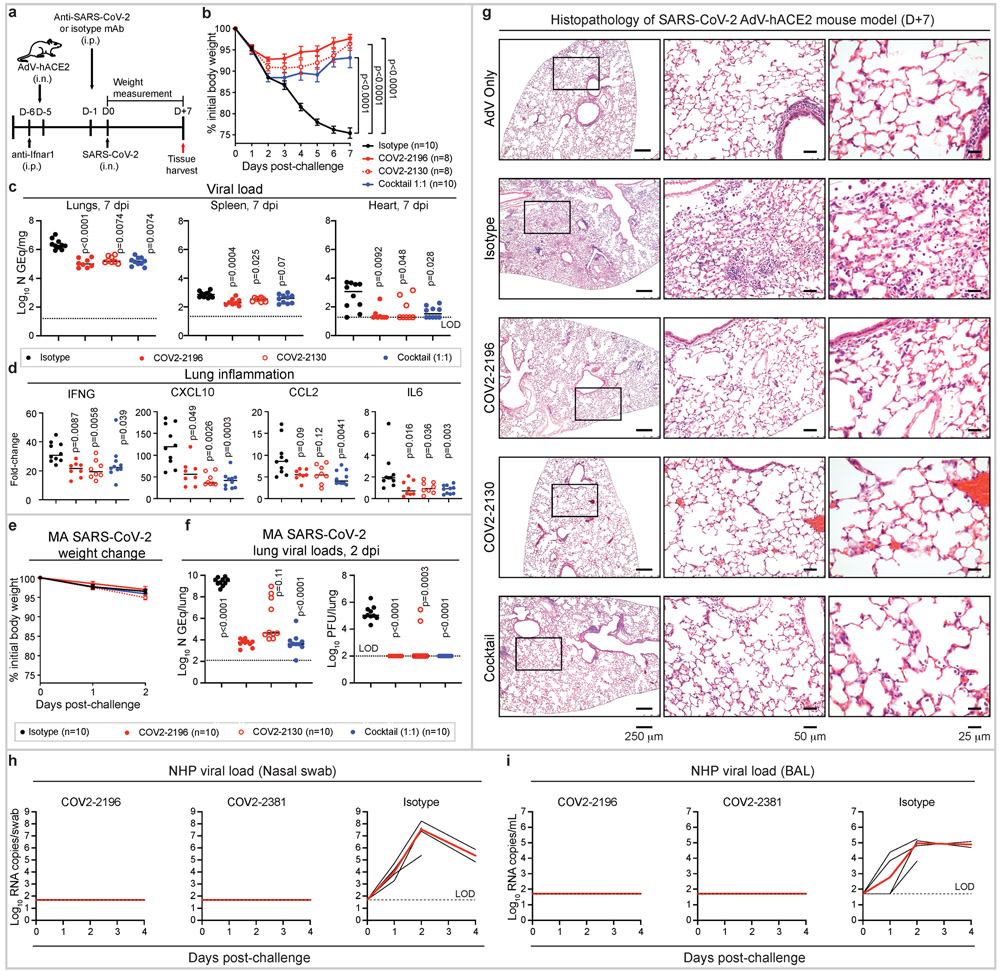
Next, we tested the prophylactic efficacy of COV2-2196 or COV2-2130 monotherapy or a combination of COV2-2196 + COV2-2130 in a newly developed SARS-CoV-2 infection model in BALB/c mice. In this model (as diagrammed in Fig. 4a), mice are first administered anti-Ifnar1 antibody and then transduced with adenovirus-expressing hACE2 (AdV-hACE2) that results in susceptibility to infection with SARS-CoV-2, viral replication, and severe bronchopneumonia32. In the present experiments, these mice were passively transferred a single dose of mAb COV2-2196, COV2-2130, a cocktail of COV2-2196 + COV2-2130, or an isotype control mAb one day before intranasal challenge with SARS-CoV-2 (4 × 105 PFU). Prophylaxis with COV2-2196 or COV2-2130 or their combination attenuated severe SARS-CoV-2-induced weight loss through the first week of infection (Fig. 4b). Viral RNA levels were reduced significantly at 7 dpi in the lung and distant sites including the heart and spleen (Fig. 4c). The expression of cytokine and chemokine genes indicative of inflammation was also reduced in the lungs of each group of COV-antibody-treated mice at 7 dpi (Fig. 4d).
Figure 4. Prophylactic efficacy of neutralizing human mAbs against SARS-CoV-2 infection in mouse and NHP models in vivo.
a. SARS-CoV-2 challenge model. Mice were treated with anti-Ifnar1 mAb, transduced with AdV-hACE2, and 200 μg of mAbs COV2-2196, −2130, or combination (1:1 ratio) or isotype control mAb were passively transferred. One day later, SARS-CoV-2 was inoculated via the i.n. route. Tissues were harvested at 7 dpi for analysis (c, d).
b. Body weight change of mice in (a) with comparison to isotype control using a repeated measurements two-way ANOVA with Tukey’s post-test. Mean ± SEM of each experimental group is shown. Numbers of animals (n) for each experimental group are shown.
Viral burden in the lung, spleen and heart (c) and cytokine and chemokine gene expression (d) were measured by RT-qPCR assay. Comparisons were performed using a Kruskal-Wallis ANOVA with Dunn’s post-test.
e. MA-SARS-CoV-2 challenge model. Mice were treated with indicated mAb and then infected intranasally with MA-SARS-CoV-2. Body weight change of mice is shown. Mean ± SEM of each experimental group is shown.
f. Viral burden in the lung was measured at 2 dpi by RT-qPCR (left) or plaque assay (right) from (e); comparisons were made using a Kruskal-Wallis ANOVA with Dunn’s post-test.
g. Hematoxylin and eosin staining of lung sections from mice that were treated and challenged as in (a). Images show low- (left), medium- (middle), and high-power magnification (right). Each image is representative of two separate experiments (n = 3 to 5 mice per group).
h-i. SARS-CoV-2 NHP challenge model. Animals received one 50 mg/kg dose of mAb COV2-2196 (n=4 NHPs per group) or mAb COV2-2381 (n = 4 NHPs per group) or isotype mAb (n = 4 NHPs per group) served as a contemporaneous control intravenously on day −3 and then challenged intranasally and intratracheally with SARS-CoV-2 in three days. Subgenomic viral RNA levels were assessed in nasal swabs (h) and the bronchioalveolar lavage (i) at multiple timepoints following challenge. Each black curve shows an individual animal, with red lines indicating the median values of animals within each treatment group. Data represent a single experiment. Dashed lines indicate assay limits of detection.
We also tested COV2-2196 or COV2-2130 or their combination for prophylactic efficacy in an immunocompetent model using a mouse-adapted (MA) SARS-CoV-2 virus33 (Fig. 4e-f). Each of the mAb treatments reduced viral RNA levels up to 105-fold at 2 dpi in the lung when compared to the isotype control group (Fig. 4f, left). All animals from COV2-2196 and COV2-2196 + COV2-2130 treatment group and 8 of 10 animals from COV2-2130 treatment no longer had infectious virus at 2 dpi in the lung as measured by plaque assay of lung tissue homogenates (Fig. 4f, right).
We evaluated the impact of mAb treatment on SARS-CoV-2-induced lung pathology. At 7 dpi, lungs from Ifnar1-antibody-treated–AdV-hACE2-transduced that were treated with isotype mAb and then infected with SARS-CoV-2- showed perivascular, peribronchial, and alveolar inflammation with immune cell infiltration and alveolar damage that are characteristic of viral pneumonia (Fig. 4g; Supplemental Table 3). In contrast, mice under the same conditions but treated instead with COV2-2196 or COV2-2130 or the combination of these mAbs developed significantly less lung disease, which was similar to that observed in AdV-hACE2-transduced control mice that were not infected with SARS-CoV-2 (Fig. 4g; Supplemental Table 3).
We next tested the protective efficacy of mAbs using a recently described SARS-CoV-2 non-human primate (NHP) challenge model34,35. For this model, we tested as monotherapy COV2-2196 and another of the most potent antibodies identified, COV2-2381, a neutralizing mAb encoded by the same variable gene segments as COV2-2196 but with a number of amino acid differences in the HCDR3 and LCDR3 (Extended Data Fig. 6a). Importantly, others have identified highly similar mAbs from multiple donors, demonstrating that these mAbs constitute a public clonotype36. Animals received one 50 mg/kg dose of mAb COV2-2196 or mAb COV2-2381 or isotype control mAb intravenously on day −3 and then were challenged intranasally and intratracheally on day 0 with a dose of 1.1 × 104 PFU SARS-CoV-2. Following challenge, we quantified subgenomic viral RNA generated by viral replication using RT-qPCR in bronchoalveolar lavage (BAL) and nasal swabs. High levels of subgenomic viral RNA were observed in the isotype control mAb-treated NHPs, with a median peak of 7.53 (range 5.37 to 8.23) log10 RNA copies/swab in nasal swab and a median peak of 4.97 (range 3.81 to 5.24) log10 RNA copies/mL in BAL (Fig. 4h-i). Subgenomic viral RNA was not detected in samples from either of the mAb treatment groups (LOD = 50 [1.7 log10] RNA copies/swab or per mL) showing protection. A pharmacokinetics analysis revealed similar concentrations of circulating human mAbs in NHPs from each treatment group (Extended Data Fig. 6b).
We next assessed the therapeutic efficacy of treatment by COV2-2196 or COV2-2130 or their combination using the MA-SARS-CoV-2 challenge model. All treatments reduced infectious virus at 2 dpi in the lung. The cocktail (1:1) delivered at a dose of 400 μg/mouse (~ 20 mg/kg) was the most efficient, as it significantly reduced lung burden up to 3 × 104-fold; 4 of 5 animals from this treatment group did not have detectable infectious virus in the lung (Fig. 5a). Similarly, treatment of AdV-hACE2-transduced mice with 400 μg/mouse of the cocktail 12 hrs after authentic SARS-CoV-2 challenge revealed full neutralization of infectious virus in the lungs in vivo (Fig. 5b). Inflammation also was reduced in the lungs of mAb cocktail-treated mice when compared to the lungs of isotype-control-treated mice (Fig. 5c). Collectively, these in vivo results suggest that either of the potently neutralizing mAbs COV2-2196 or COV2-2381 alone or COV2-2196 and COV2-2130 in combination are promising candidates for prevention or treatment of COVID-19.
Figure 5. Therapeutic efficacy of neutralizing human mAbs against SARS-CoV-2 infection.
a. Mice were inoculated via the i.n. route with MA-SARS-CoV-2 and 12 hrs later given the indicated mAb treatments by i.p. injection. Viral burden in the lungs was measured by plaque assay. Number of mice per group (n) is indicated, and data represent one experiment.
b. Mice were treated with anti-Ifnar1 mAb and transduced with AdV-hACE2. Mice were then inoculated via the i.n. route with SARS-CoV-2 and given the indicated mAb treatments by i.p. injection 12 hrs later. Two experiments were performed with 3 to 5 mice per group. Viral burden in the lungs was measured by plaque assay. Controls for plaque neutralization assay performance were included: lung homogenates from individual (n = 3) isotype-control-mAb-treated mice were mixed 1:1 (v:v) with lung homogenates from individual naïve untreated mice or mAb cocktail-treated mice. The latter mixture ensures that neutralization of infection did not occur ex vivo after tissue homogenization. For (a) and (b) measurements from individual mice and median titer are shown, and each group was compared to the isotype-control-treated group using a Kruskal-Wallis ANOVA with Dunn’s post-test.
c. Cytokine and chemokine gene expression was measured by qPCR analysis from the lungs harvested as in (b). Measurements from individual mice and median values are shown. Groups were compared using the two-sided Mann-Whitney U test. Number of mice per group (n) is indicated. Two experiments were performed with 3 to 5 mice per group.
Since the start of the SARS-CoV-2 pandemic, several groups have identified human mAbs that bind the RBD and neutralize virus36-44. Here, we defined the antigenic landscape for a number of potently neutralizing mAbs against SARS-CoV-2 derived from a larger panel of hundreds5. These studies demonstrate that although diverse human neutralizing antibodies are elicited by natural infection with SARS-CoV-2, only a small subset of those mAbs are of high potency (IC50<50 ng/mL against authentic SARS-CoV-2 virus). Biochemical and structural analysis of these potent mAbs defined three principal antigenic sites of vulnerability on the SRBD for SARS-CoV-2 neutralization. Representative mAbs from two antigenic sites were shown to synergize in vitro and protect as an in vivo cocktail in both prophylactic and therapeutic treatment. Our findings reveal critical features of effective humoral immunity to SARS-CoV-2 and suggests that the role of synergistic neutralization activity in polyclonal responses should be explored. Moreover, as SARS-CoV-2 continues to circulate, population immunity elicited by natural infection may start to select for antigenic variants that escape from the selective pressure of neutralizing antibodies. Other groups have reported selecting SARS-CoV-2 RBD escape mutations in the presence of single mAbs but not in the presence of a mixture of two mAbs45, reinforcing the need to target multiple epitopes of S in vaccines or immunotherapies. To date, the S gene has limited diversity with the exception of a D614G substitution46, far away from the amino acid positions identified in our mutational studies for the mAbs studied here. Rationally-selected therapeutic cocktails like the one described here likely offer greater resistance to SARS-CoV-2 escape. These studies set the stage for preclinical evaluation and development of the identified mAbs as candidates for use as COVID-19 immunotherapeutics in humans.
Online Methods
Antibodies.
The human antibodies studied in this paper were isolated from blood samples from two subjects in North America with previous laboratory-confirmed symptomatic SARS-CoV-2 infection that was acquired in China. The original clinical studies to obtain specimens after written informed consent were previously described5 and had been approved by the Institutional Review Board of Vanderbilt University Medical Center, the Institutional Review Board of the University of Washington, and the Research Ethics Board of the University of Toronto. The subjects (a 56-year-old male and a 56-year-old female) are a married couple and residents of Wuhan, China who traveled to Toronto, Canada, where PBMCs were obtained by leukopheresis 50 days after symptom onset. The antibodies were isolated using diverse tools for isolation and cloning of single antigen-specific B cells and the antibody variable genes encoding monoclonal antibodies5.
Cell culture
Vero E6 (American Type Culture Collection (ATCC), Cat# CRL-1586), Vero (ATCC Cat# CCL-81), HEK293 (ATCC Cat# CRL-1573), and HEK293T (ATCC Cat# CRL-3216) were maintained at 37°C in 5% CO2 in Dulbecco’s minimal essential medium (DMEM) containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 10 mM HEPES pH 7.3, 1 mM sodium pyruvate, 1× non-essential amino acids, and 100 U/mL of penicillin–streptomycin. Vero-furin cells were obtained from T. Pierson (NIH) and have been described previously47. FreeStyle 293F (ThermoFisher Scientific, R79007) were maintained at 37°C in 8% CO2. Expi293F cells (ThermoFisher Scientific, A1452) were maintained at 37°C in 8% CO2 in Expi293F Expression Medium (ThermoFisher Scientific, A1435102). ExpiCHO cells (ThermoFisher Scientific, A29127) were maintained at 37°C in 8% CO2 in ExpiCHO Expression Medium (ThermoFisher Scientific, A2910002). Mycoplasma testing of Expi293F and ExpiCHO cultures was performed on a monthly basis using a PCR-based mycoplasma detection kit (ATCC, 30-1012K).
Viruses.
SARS-CoV-2 strain 2019 n-CoV/USA_WA1/2020 was obtained from the Centers for Disease Control and Prevention (a gift from Natalie Thornburg). Virus was passaged in Vero CCL81 cells and titrated by plaque assay on Vero E6 cells. Mouse adapted SARS-CoV-2 (MA-SARS-CoV-2) virus was generated as described previously33. Virus was propagated in Vero E6 cells grown in DMEM supplemented with 10% Fetal Clone II and 1% Pen/Strep. Virus titer was determined by plaque assay. Briefly, virus was diluted serially and inoculated onto confluent monolayers of Vero E6 cells, followed by an agarose overlay. Plaques were visualized on day 2 post-infection after staining with neutral red dye. All work with infectious SARS-CoV-2 was approved by the Washington University School of Medicine or UNC-Chapel Hill Institutional Biosafety Committees and conducted in approved BSL3 facilities using appropriate powered air purifying respirators and personal protective equipment.
Recombinant antigens and proteins.
A gene encoding the ectodomain of a prefusion conformation-stabilized SARS-CoV-2 spike (S2Pecto) protein was synthesized and cloned into a DNA plasmid expression vector for mammalian cells. A similarly designed S protein antigen with two prolines and removal of the furin cleavage site for stabilization of the prefusion form of S was reported previously30. Briefly, this gene includes the ectodomain of SARS-CoV-2 (to residue 1,208), a T4 fibritin trimerization domain, an AviTag site-specific biotinylation sequence, and a C-terminal 8x-His tag. To stabilize the construct in the prefusion conformation, we included substitutions K986P and V987P and mutated the furin cleavage site at residues 682-685 from RRAR to ASVG. This recombinant spike 2P-stabilized protein (designated here as S2Pecto) was isolated by metal affinity chromatography on HisTrap Excel columns (GE Healthcare), and protein preparations were purified further by size-exclusion chromatography on a Superose 6 Increase 10/300 column (GE Healthcare). The presence of trimeric, prefusion conformation S protein was verified by negative-stain electron microscopy5. For electron microscopy with S and Fabs, we expressed a variant of S2Pecto lacking an AviTag but containing a C-terminal Twin-Strep-tag, similar to that described previously30. Expressed protein was isolated by metal affinity chromatography on HisTrap Excel columns (GE Healthcare), followed by further purification on a StrepTrap HP column (GE Healthcare) and size-exclusion chromatography on TSKgel G4000SW XL (TOSOH). To express the SRBD subdomain of SARS-CoV-2 S protein, residues 319-541 were cloned into a mammalian expression vector downstream of an IL-2 signal peptide and upstream of a thrombin cleavage site, an AviTag, and a 6x-His tag. RBD protein fused to mouse IgG1 Fc domain (designated RBD-mFc), was purchased from Sino Biological (40592-V05H). For epitope mapping by alanine scanning, SARS-CoV-2 RBD (residues 334-526) or RBD single mutation variants were cloned with an N-terminal CD33 leader sequence and C-terminal GSSG linker, AviTag, GSSG linker, and 8xHisTag. Spike proteins were expressed in FreeStyle 293 cells (Thermo Fisher) or Expi293 cells (ThermoFisher) and isolated by affinity chromatography using a HisTrap column (GE Healthcare), followed by size exclusion chromatography with a Superdex200 column (GE Healthcare). Purified proteins were analyzed by SDS-PAGE to ensure purity and appropriate molecular weights.
Electron microscopy (EM) stain grid preparation, imaging and processing of SARS-CoV-2 S2Pecto protein or S2Pecto/Fab complexes.
To perform EM imaging, Fabs were produced by digesting recombinant chromatography-purified IgGs using resin-immobilized cysteine protease enzyme (FabALACTICA, Genovis). The digestion occurred in 100 mM sodium phosphate, 150 mM NaCl pH 7.2 (PBS) for ~16 hrs at ambient temperature. In order to remove cleaved Fc and intact IgG, the digestion mix was incubated with CaptureSelect Fc resin (Genovis) for 30 min at ambient temperature in PBS buffer. If needed, the Fab was buffer exchanged into Tris buffer by centrifugation with a Zeba spin column (Thermo Scientific).
For screening and imaging of negatively-stained (NS) SARS-CoV-2 S2Pecto protein in complex with human Fabs, the proteins were incubated at a molar ratio of 4 Fab:3 spike monomer for ~1 hr and approximately 3 μL of the sample at concentrations of about 10 to 15 μg/mL was applied to a glow discharged grid with continuous carbon film on 400 square mesh copper EM grids (Electron Microscopy Sciences). The grids were stained with 0.75% uranyl formate (UF)48. Images were recorded on a Gatan US4000 4k × 4k CCD camera using an FEI TF20 (TFS) transmission electron microscope operated at 200 keV and control with SerialEM49. All images were taken at 50,000× magnification with a pixel size of 2.18 Å/pix in low-dose mode at a defocus of 1.5 to 1.8 μm. Total dose for the micrographs was ~25 to 38 e−/Å2. Image processing was performed using the cryoSPARC software package50. Images were imported, and particles were CTF estimated. The images then were denoised and picked with Topaz51. The particles were extracted with a box size of 256 pixels and binned to 128 pixels. 2D class averages were performed and good classes selected for ab-initio model and refinement without symmetry. For EM model docking of SARS-CoV-2 S2Pecto protein, the closed model (PDB: 6VXX) was used in Chimera52 for docking to the EM map (see also Supplemental Table 2 for details). For the SARS-CoV-2 S2Pecto/Fab COV2-2165, SARS-CoV-2 S2Pecto/Fab COV2-2196, and SARS-CoV-2 S2Pecto/Fab COV2-2832 complexes, the open model of SARS-CoV-2 (PDB: 6VYB) and Fab (Fab: 12E8) was used in Chimera for docking to the EM maps (see also Supplemental Table 2 for details). For the SARS-CoV-2 S2Pecto/Fab COV2-2130 complex, the closed model and Fab (PDB: 12E8) were used in Chimera for docking to the EM map (see also Supplemental Table 2 for details). For the SARS-CoV-2 S2Pecto/Fab COV2-2096 complexes, both the open (PDB: 6VYB) and closed (PDB: 6VXX) models were used. All images were made with Chimera. PyMOL (Schrödinger, Inc) was used to visualize previously solved molecular structures of the SARS-CoV-2 RBD/hACE2 complex and the 60-amino-acid hACE2 recognition motif (PDB (6M0J).
MAb production and purification.
Sequences of mAbs that had been synthesized (Twist Bioscience) and cloned into an IgG1 monocistronic expression vector (designated as pTwist-mCis_G1) were used for mammalian cell culture mAb secretion. This vector contains an enhanced 2A sequence and GSG linker that allows simultaneous expression of mAb heavy and light chain genes from a single construct upon transfection53. We previously described microscale expression of mAbs in 1 mL ExpiCHO cultures in 96-well plates5. For larger scale mAb expression, we performed transfection (1 to 300 mL per antibody) of CHO cell cultures using the Gibco™ ExpiCHO™ Expression System and protocol for 50 mL mini bioreactor tubes (Corning) as described by the vendor. Culture supernatants were purified using HiTrap MabSelect SuRe (Cytiva, formerly GE Healthcare Life Sciences) on a 24-column parallel protein chromatography system (Protein BioSolutions). Purified mAbs were buffer-exchanged into PBS, concentrated using Amicon® Ultra-4 50KDa Centrifugal Filter Units (Millipore Sigma) and stored at 4°C until use. Purified mAbs were tested routinely for endotoxin levels that found to be <30 EU/mg IgG for mouse studies and <1 EU/mg IgG for NHP studies. Endotoxin testing was performed using the PTS201F cartridge (Charles River), with sensitivity range from 10 to 0.1 EU/mL, and an Endosafe Nexgen-MCS instrument (Charles River).
ELISA binding assays.
Wells of 96-well microtiter plates were coated with purified recombinant SARS-CoV-2 S protein or SARS-CoV-2 SRBD protein at 4°C overnight. Plates were blocked with 2% non-fat dry milk and 2% normal goat serum in DPBS containing 0.05% Tween-20 (DPBS-T) for 1 hr. The bound antibodies were detected using goat anti-human IgG conjugated with HRP (horseradish peroxidase) (Southern Biotech, Cat# 2040-05, Lot B3919-XD29, 1:5,000 dilution) and TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Thermo Fisher Scientific). Color development was monitored, 1 N hydrochloric acid was added to stop the reaction, and the absorbance was measured at 450 nm using a spectrophotometer (Biotek). For dose-response assays, serial dilutions of purified mAbs were applied to the wells in triplicate, and mAb binding was detected as detailed above. Half-maximal effective concentration (EC50) values for binding were determined using Prism v8.0 software (GraphPad) after log transformation of mAb concentration using sigmoidal dose-response nonlinear regression analysis.
RBD minimal hACE2 recognition motif peptide binding ELISA.
Wells of 384-well microtiter plates were coated with 1 μg/mL streptavidin at 4°C overnight. Plates were blocked with 0.5% BSA in DPBS containing 0.05% Tween-20 (DPBS-T) for 1 hr. Plates were washed 4x with 1x PBST and 2 μg/mL biotinylated-ACE2 binding motif peptide (cat. # LT5578, from LifeTein, LLC) was added to bind streptavidin for 1 hr at ambient temperature. Purified mAbs were diluted in blocking buffer, added to the wells, and incubated for 1 hr at ambient temperature. The bound antibodies were detected using goat anti-human IgG conjugated with HRP (horseradish peroxidase) (cat. # 2014-05, Southern Biotech) and TMB (3,3′,5,5′-tetramethylbenzidine) substrate (ThermoFisher Scientific). Color development was monitored, 1N hydrochloric acid was added to stop the reaction, and the absorbance was measured at 450 nm using a spectrophotometer (Biotek). For dose-response assays, serial 3-fold dilutions starting at 10 μg/mL concentration of purified mAbs were applied to the wells in triplicate, and mAb binding was detected as detailed above.
Analysis of binding of antibodies to variant RBD proteins with alanine or arginine point mutations.
Biolayer light interferometry (BLI) was performed using an Octet RED96 instrument (ForteBio; Pall Life Sciences) and wild-type RBD protein or a mutant RBD protein with a single amino acid change at defined positions to alanine or arginine. Binding of the RBD proteins were confirmed by first capturing octa-His-tagged RBD wild-type or mutant protein from a 10 μg/mL (≈200 nM) solution onto Penta-His biosensors for 300 sec. The biosensor tips then were submerged in binding buffer (PBS/0.2% Tween 20) for a 60 sec wash, followed by immersion in a solution containing 150 nM of mAb for 180 sec (association), followed by a subsequent immersion in binding buffer for 180 sec (dissociation). Response for each RBD mutant protein was normalized to that of the wild-type RBD protein.
Focus reduction neutralization test (FRNT).
Serial dilutions of mAbs were incubated with 102 FFU of SARS-CoV-2 for 1 hr at 37°C. The mAb–virus complexes were added to Vero E6 cell culture monolayers in 96-well plates for 1 hr at 37°C. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in Minimum Essential Medium (MEM) supplemented to contain 2% heat-inactivated FBS. Plates were fixed 30 hrs later by removing overlays and fixed with 4% PFA in PBS for 20 min at room temperature. The plates were incubated sequentially with 1 μg/mL of rCR3022 anti-S antibody12 and horseradish-peroxidase (HRP)-conjugated goat anti-human IgG (Sigma-Aldrich, Cat# A6029) in PBS supplemented with 0.1% (w/v) saponin (Sigma) and 0.1% bovine serum albumin (BSA). SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot 5.0.37 Macro Analyzer (Cellular Technologies). Data were processed using Prism software version 8.0 (GraphPad). IC50 values were determined by nonlinear regression analysis using the Prism software.
Generation of S protein pseudotyped lentivirus.
Suspension cultures of 293 cells were seeded and transfected with a third-generation HIV-based lentiviral vector expressing luciferase along with packaging plasmids encoding for the following: SARS-CoV-2 spike protein with a C-terminal 19 amino acid deletion, Rev, and Gag-pol. Medium was changed 16 to 20 hrs after transfection, and the supernatant containing virus was harvested 24 hrs later. Cell debris was removed by low-speed centrifugation, and the supernatant was passed through a 0.45 μm filter unit. The pseudovirus was pelleted by ultracentrifugation and resuspended in PBS for a 100-fold concentrated stock.
Pseudovirus neutralization assay.
Serial dilutions of mAbs were prepared in a 384-well microtiter plate and pre-incubated with pseudovirus for 30 minutes at 37°C, to which 293 cells that stably express human ACE2 were added. The plate was returned to the 37°C incubator, and then 48 hrs later luciferase activity measured on an EnVision 2105 Multimode Plate Reader (Perkin Elmer) using the Bright-Glo™ Luciferase Assay System (Promega), according to manufacturer’s recommendations. Percent inhibition was calculated relative to pseudovirus-alone control. IC50 values were determined by nonlinear regression using the Prism software version 8.1.0 (GraphPad). The average IC50 value for each antibody was determined from a minimum of 3 independent experiments.
Measurement of synergistic neutralization by an antibody combination.
Synergy was defined as higher neutralizing activity mediated by a cocktail of two mAbs when compared to that mediated by individual mAbs at the same total concentration of antibodies in vitro. To assess if two mAbs synergize in a cocktail to neutralize SARS-CoV-2, we used a previously reported approach to quantitate synergy11. To evaluate the significance of the beneficial effect from combining mAbs, the observed combination responses (dose-response matrix) were compared with the expected responses calculated by means of synergy scoring models11. Virus neutralization was measured in a conventional focus reduction neutralization test (FRNT) assay using wild-type SARS-CoV-2 and Vero E6 cell culture monolayers. Individual mAbs COV2-2196 and COV2-2130 were mixed at different concentrations to assess neutralizing activity of different ratios of mAbs in the cocktail. Specifically, each of seven-fold dilutions of mAb COV2-2130 (starting from 500 ng/mL) was mixed with each of the nine dilutions of mAb COV2-2196 (starting from 500 ng/mL) in a total volume of 50 μL of per each condition and then incubated with 50 μL of live SARS-CoV-2 in cell culture medium (RPMI-1640 medium supplemented with 2% FBS) before applying to confluent Vero E6 cells grown in 96-well plates. The control values included those for determining dose-response of the neutralizing activity measured separately for the individual mAb COV2-2196 or COV2-2130, which were assessed at the same doses as in the cocktail. Each measurement was performed in duplicate. We next calculated percent virus neutralization for each condition and then calculated the synergy score value, which defined interaction between these two mAbs in the cocktail. A synergy score of less than −10 indicates antagonism, a score from −10 to 10 indicates an additive effect, and a score greater than 10 indicates a synergistic effect11.
MAb quantification.
Quantification of purified mAbs was performed by UV spectrophotometry using a NanoDrop spectrophotometer and accounting for the extinction coefficient of human IgG.
Competition-binding analysis through biolayer interferometry.
Anti-mouse IgG Fc capture biosensors (FortéBio 18-5089) on an Octet HTX biolayer interferometry instrument (FortéBio) were soaked for 10 minutes in 1x kinetics buffer (Molecular Devices 18-1105), followed by a baseline signal measurement for 60 seconds. Recombinant SARS-CoV-2 RBD fused to mouse IgG1 (RBD-mFc, Sino Biological 40592-V05H) was immobilized onto the biosensor tips for 180 seconds. After a wash step in 1x kinetics buffer for 30 seconds, the reference antibody (5 μg/mL) was incubated with the antigen-containing biosensor for 600 seconds. Reference antibodies included the SARS-CoV human mAb CR3022 and COV2-2196. After a wash step in 1x kinetics buffer for 30 seconds, the biosensor tips then were immersed into the second antibody (5 μg/mL) for 300 seconds. Maximal binding of each antibody was normalized to a buffer-only control. Self-to-self blocking was subtracted. Comparison between the maximal signal of each antibody was used to determine the percent binding of each antibody. A reduction in maximum signal to <33% of un-competed signal was considered full competition of binding for the second antibody in the presence of the reference antibody. A reduction in maximum signal to between 33 to 67% of un-competed was considered intermediate competition of binding for the second antibody in the presence of the reference antibody. Percent binding of the maximum signal >67% was considered absence of competition of binding for the second antibody in the presence of the reference antibody.
hACE2 inhibition analysis.
Wells of 384-well microtiter plates were coated with purified recombinant SARS-CoV-2 S2Pecto protein at 4°C overnight. Plates were blocked with 2% non-fat dry milk and 2% normal goat serum in DPBS-T for 1 hr. For screening assays, purified mAbs from microscale expression were diluted two-fold in blocking buffer starting from 10 μg/mL in triplicate, added to the wells (20 μL/well), and incubated for 1 hr at ambient temperature. Recombinant human ACE2 (hACE2) with a C-terminal FLAG tag peptide was added to wells at 2 μg/mL in a 5 μL/well volume (final 0.4 μg/mL concentration of hACE2) without washing of antibody and then incubated for 40 min at ambient temperature. Plates were washed, and bound hACE2 was detected using HRP-conjugated anti-FLAG antibody (Sigma-Aldrich Cat# A8592, Lot SLBV3799, 1:5,000 dilution) and TMB substrate. ACE2 binding without antibody served as a control. The signal obtained for binding of the hACE2 in the presence of each dilution of tested antibody was expressed as a percentage of the hACE2 binding without antibody after subtracting the background signal. For dose-response assays, serial dilutions of purified mAbs were applied to the wells in triplicate, and mAb binding was detected as detailed above. Half-maximal inhibitory concentration (IC50) values for inhibition by mAb of S2Pecto protein binding to hACE2 was determined after log transformation of antibody concentration using sigmoidal dose-response nonlinear regression analysis (Prism software, GraphPad Prism version 8.0).
hACE2 blocking assay using biolayer interferometry biosensor.
Anti-mouse IgG biosensors on an Octet HTX biolayer interferometry instrument (FortéBio) were soaked for 10 min in 1x kinetics buffer, followed by a baseline signal measurement for 60 seconds. Recombinant SARS-CoV-2 RBD fused to mouse IgG1 (RBD-mFc, Sino Biological 40592-V05H) was immobilized onto the biosensor tips for 180 sec. After a wash step in 1x kinetics buffer for 30 sec, the antibody (5 μg/mL) was incubated with the antigen-coated biosensor for 600 sec. After a wash step in 1x kinetics buffer for 30 sec, the biosensor tips then were immersed into the hACE2 receptor (20 μg/mL) (Sigma-Aldrich SAE0064) for 300 sec. Maximal binding of hACE2 was normalized to a buffer-only control. Percent binding of hACE2 in the presence of antibody was compared to hACE2 maximal binding. A reduction in maximal signal to <30% was considered hACE2 blocking.
High-throughput competition-binding analysis.
Wells of 384-well microtiter plates were coated with purified SARS-CoV-2 S2Pecto protein at 4°C overnight. Plates were blocked with 2% BSA in DPBS containing 0.05% Tween-20 (DPBS-T) for 1 hr. Micro-scale purified unlabeled mAbs were diluted ten-fold in blocking buffer, added to the wells (20 μL/well) in quadruplicates, and incubated for 1 hr at ambient temperature. A biotinylated preparation of a recombinant mAb based on the variable gene sequence of the previously described mAb CR302212 and also newly identified mAbs COV2-2096, −2130, and −2196 that recognized distinct antigenic regions of the SARS-CoV-2 S protein were added to each of four wells with the respective mAb at 2.5 μg/mL in a 5 μL/well volume (final 0.5 μg/mL concentration of biotinylated mAb) without washing of unlabeled antibody and then incubated for 1 hr at ambient temperature. Plates were washed, and bound antibodies were detected using HRP-conjugated avidin (Sigma) and TMB substrate. The signal obtained for binding of the biotin-labeled reference antibody in the presence of the unlabeled tested antibody was expressed as a percentage of the binding of the reference antibody alone after subtracting the background signal. Tested mAbs were considered competing if their presence reduced the reference antibody binding to less than 41% of its maximal binding and non-competing if the signal was greater than 71%. A level of 40–70% was considered intermediate competition.
Plasma or serum antibody competition-binding assays
Wells of 384-well microtiter plates were coated with purified SARS-CoV-2 S2Pecto at 4°C overnight. Plates were blocked with 2% BSA in DPBS-T for 1 hr. Plasma or serum samples were diluted in blocking buffer two-fold starting from 1:10 sample dilution, added to the wells (20 μL/well) in triplicate, and incubated for 1 hr at ambient temperature. For self-blocking controls, unlabeled mAbs COV2-2096 or COV2-2130 were added at 10 μg/mL to separate wells coated with S2Pecto. Serum from a donor without an exposure history to SARS-CoV-2 was used as a negative control for mAb binding inhibition. A biotinylated mAb COV2-2096 or COV2-2130 was added to the respective wells at 2.5 μg/mL in a 5 μL/well volume (final 0.5 μg/mL concentration of biotinylated mAb) without washing of unlabeled antibody and then incubated for 30 min at ambient temperature. Binding of biotinylated mAbs COV2-2096 or COV2-2130 alone to S2Pecto served as a control for maximal binding. Plates were washed, and bound antibodies were detected using HRP-conjugated avidin (Sigma) and TMB substrate. Inhibition of COV2-2096 or COV2-2130 binding in the presence of each dilution of tested plasma or serum was calculated as a percent of maximal COV2-2096 or COV2-2130 binding inhibition using values from COV2-2096 or COV2-2130 binding alone (maximal binding) and the corresponding self-blocking controls (maximal inhibition) after subtracting the background signal. For hACE2 inhibition assay by plasma or serum antibodies, plasma or serum samples were diluted and added to wells with S2Pecto as detailed above. Recombinant human hACE2 was added to wells at 2 μg/mL in a 5 μL/well volume (final 0.4 μg/mL concentration of hACE2) without washing of antibody and then incubated for 40 min at ambient temperature. Plates were washed, and bound hACE2 was detected using HRP-conjugated anti-FLAG antibody (Sigma) and TMB substrate. hACE2 binding without antibody served as a control. The signal obtained for binding of the hACE2 in the presence of each dilution of tested plasma or serum was expressed as a percentage of the ACE2 binding without antibody after subtracting the background signal.
Binding analysis of mAbs to alanine or arginine RBD mutants.
Biolayer light interferometry was performed using an Octet RED96 instrument (FortéBio; Pall Life Sciences). Binding was confirmed by first capturing octa-His-tagged RBD mutants 10 μg/mL (≈200 nM) onto Penta-His biosensors for 300 sec. The biosensors then were submerged in binding buffer (PBS/0.2% TWEEN 20) for a wash for 60 sec followed by immersion in a solution containing 150 nM of mAbs for 180 sec (association), followed by a subsequent immersion in binding buffer for 180 sec (dissociation). Response for each RBD mutant was normalized to that of wild-type RBD.
Protection against authentic SARS-CoV-2 in human hACE2-transduced mice.
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
Wild-type, female BALB/c mice were purchased from Jackson Laboratories (strain 000651). Animals were housed in groups of up to 5 mice/cage at 18-24°C ambient temperatures and 40-60% humidity. Mice were fed a 20% protein diet (PicoLab 5053, Purina) and maintained on a 12 hour light/dark cycle (6 am to 6 pm). Food and water were available ad libitum.
Mice (10-11-week-old) were given a single intraperitoneal injection of 2 mg of anti-Ifnar1 mAb (MAR1-5A354, Leinco) one day before intranasal administration of 2.5 × 108 PFU of AdV-hACE2. Five days after AdV transduction, mice were inoculated with 4 × 105 PFU of SARS-CoV-2 by the intranasal route. Anti-SARS-CoV-2 human mAbs or isotype control mAbs were administered 24 hrs prior (prophylaxis) or 12 hrs after (therapy) SARS-CoV-2 inoculation. Weights were monitored on a daily basis, and animals were sacrificed at 2 or 7 dpi, and tissues were harvested.
Measurement of viral burden.
For RT-qPCR tissues were weighed and homogenized with zirconia beads in a MagNA Lyser instrument (Roche Life Science) in 1 ml of DMEM media supplemented with 2% heat-inactivated FBS. Tissue homogenates were clarified by centrifugation at 10,000 rpm for 5 min and stored at −80°C. RNA was extracted using MagMax mirVana Total RNA isolation kit (Thermo Scientific) and a Kingfisher Flex 96 well extraction machine (Thermo Scientific). TaqMan primers were designed to target a conserved region of the N gene using SARS-CoV-2 (MN908947) sequence as a guide (L Primer: ATGCTGCAATCGTGCTACAA; R primer: GACTGCCGCCTCTGCTC; probe: /56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/). To establish an RNA standard curve, we generated concatenated segments of the N gene in a gBlocks fragment (IDT) and cloned this into the PCR-II topo vector (Invitrogen). The vector was linearized, and in vitro T7-DNA-dependent RNA transcription was performed to generate materials for a quantitative standard curve.
For plaque assay, homogenates were diluted serially 10-fold and applied to Vero-furin cell monolayers in 12-well plates. Plates were incubated at 37°C for 1 hr with rocking every 15 min. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were harvested 72 hrs later by removing overlays and fixed with 4% PFA in PBS for 20 min at ambient temperature. After removing the 4% PFA, plaques were visualized by adding 1 mL/well 0.05% crystal violet in 20% methanol for 20 min at ambient temperature. Excess crystal violet was washed away with PBS, and plaques were counted.
Cytokine and chemokine mRNA measurements.
RNA was isolated from lung homogenates at 7 dpi as described above. cDNA was synthesized from DNase-treated RNA using the High-Capacity cDNA Reverse Transcription kit (Thermo Scientific) with the addition of RNase inhibitor, following the manufacturer’s protocol. Cytokine and chemokine expression was determined using TaqMan Fast Universal PCR master mix (Thermo Scientific) with commercial primers/probe sets specific for IFNγ (IDT: Mm.PT.58.41769240), IL6 (Mm.PT.58.10005566), CXCL10 (Mm.PT.58.43575827), CCL2 (Mm.PT.58.42151692 and results were normalized to GAPDH (Mm.PT.39a.1) levels. Fold change was determined using the 2-ΔΔCt method comparing anti-SARS-CoV-2 specific or isotype control mAb-treated mice to naïve controls.
Histology.
Animals were euthanized, and tissues were harvested before lung inflation and fixation. The left lung lobe was tied off at the left main bronchus and collected for viral RNA analysis. The right lung lobe was inflated with ~1.2 mL of 10% neutral buffered formalin using a 3-mL syringe and catheter inserted into the trachea. For fixation after infection, inflated lungs were kept in a 40-mL suspension of neutral buffered formalin for 7 days before further processing. Tissues were embedded in paraffin, and sections were stained with hematoxylin and eosin. Tissue sections were visualized using a Leica DM6B microscope equipped with a Leica DFC7000T camera. The sections were scored by an immunopathology expert blinded to the compositions of the groups.
Viral challenge studies using MA-SARS-CoV-2 and wild-type mice.
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the UNC Chapel Hill School of Medicine (NIH/PHS Animal Welfare Assurance Number is D16-00256 (A3410-01)). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
Protection against MA-SARS-CoV-2 in wild-type mice.
12-month-old BALB/c mice from Envigo were used in experiments. Animals were housed in groups of up to 5 mice/cage at 18-24°C ambient temperatures and 40-60% humidity. Mice were fed a 20% protein diet (PicoLab 5053, Purina) and maintained on a 12 hrs light/dark cycle (8 am to 8 pm). Food and water were available ad libitum. Mice were acclimated in the BSL3 for at least 72 hrs prior to start of experiments. At 6 hrs prior to infection, mice were treated with 200 μg of human mAbs via intraperitoneal injection. The next day, mice were anesthetized with a mixture of ketamine and xylazine and intranasally inoculated with 105 PFU of MA-SARS-CoV-2 diluted in PBS. Daily weight loss was measured, and at two days post-infection mice were euthanized by isoflurane overdose prior to tissue harvest. For the post-exposure therapy study, mice were inoculated via the i.n. route with 105 PFU of MA-SARS-CoV-2 and 12 hrs later given the indicated antibody treatments by i.p. injection. The lungs were harvested at 2 dpi.
Plaque assay of lung tissue homogenates.
The lower lobe of the right lung was homogenized in 1 mL PBS using a MagnaLyser (Roche). Serial dilutions of virus were titered on Vero E6 cell culture monolayers, and virus plaques were visualized by neutral red staining at two days after inoculation. The limit of detection for the assay is 100 PFU per lung.
NHP challenge study.
The NHP research studies adhered to principles stated in the eighth edition of the Guide for the Care and Use of Laboratory Animals. The facility where this research was conducted (Bioqual Inc., Rockville, MD) is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) and approved by the Office of Laboratory Animal Welfare (NIH/PHS Assurance ID: D16-00052). NHP studies were conducted in compliance with all relevant local, state, and federal regulations and were approved by the Animal Care and Use Committee (IACUC) at Bioqual. NHP studies were conducted in compliance with all relevant local, state, and federal regulations and were approved by the relevant Institutional Animal Care and Use Committee (IACUC) at Bioqual Inc.
Twelve healthy adult rhesus macaques (Macaca mulatta) of Indian origin (5 to 15 kg body weight) were studied. Rhesus macaques were 5-7 years old and mixed male and female. Animals were allocated randomly to two anti-SARS-CoV-2 mAb treatment groups (n=4 per group) and one control (isotype-treated) group (n=4 per group). Animals received one 50 mg/kg dose of mAb COV2-2196 or mAb COV2-2381 or isotype control mAb intravenously on day −3 and then were challenged in three days with 1.1 × 104 PFU SARS-CoV-2, administered as 1 mL by the i.n route and 1 mL by the intratracheal route. Following challenge, viral RNA was assessed by RT-qPCR in bronchoalveolar lavage and nasal swabs at multiple time points as described34-35. All animals were given physical examinations. In addition, all animals were monitored daily with an internal scoring protocol approved by the Institutional Animal Care and Use Committee. These studies were not blinded.
Detection of circulating human mAbs in NHP serum.
ELISA plates were coated overnight at 4°C with 1 μg/mL of goat anti-human IgG (H+L) secondary antibody (monkey pre-adsorbed) (Novus Biologicals, Cat# NB7487) and then blocked for 2 hrs. The serum samples were assayed at 3-fold dilutions starting at a 1:3 dilution in Blocker Casein in PBS (ThermoFisher) diluent. Samples were incubated for 1 hr at ambient temperature and then removed, and plates were washed. Wells then were incubated for 1 hr with HRP-conjugated goat anti-Human IgG (monkey pre-adsorbed) (Southern Biotech Cat# 2049-05) at a 1:4,000 dilution. Wells were washed and then incubated with SureBlue Reserve TMB Microwell Peroxidase Substrate (Seracare) (100 μL/well) for 3 min followed by TMB Stop Solution (Seracare) to stop the reaction (100 μL/well). Microplates were read at 450 nm. The concentrations of the human mAbs were interpolated from the linear range of purified human IgG (Sigma) standard curves using Prism software, version 8.0 (GraphPad).
Quantification and statistical analysis.
The descriptive statistics mean ± SEM or mean ± SD were determined for continuous variables as noted. Technical and biological replicates are described in the figure legends. In the mouse studies, the comparison of weight change curves was performed using repeated measurements two-way ANOVA with Tukey’s post-test using Prism v8.0 (GraphPad). Viral burden and gene expression measurements were compared using Kruskal-Wallis ANOVA with Dunn’s post-test or two-sided Mann-Whitney U test using Prism v8.0 (GraphPad). Synergy score and dose-response matrix analysis were performed using a web application SynergyFinder28.
Data availability.
The EM maps have been deposited at the Electron Microscopy Data Bank with accession codes (Supplemental Table 2): EMD-21974, EMD-21975, EMD-21976, and EMD-21977. The EM-map EMD-21965 is publicly available. The accession numbers for cryo-EM and crystal structures used for structural analysis, including structures of the closed conformation of SARS-CoV-2 S (PDB: 6VXX), the open conformation of SARS-CoV-2 (PDB: 6VYB), Fab used for docking (PDB: 12E8), and the SARS-CoV-2 RBD/hACE2 complex and the 60-amino-acid hACE2 recognition motif (PDB (6M0J) are publicly available. Sequences of mAbs characterized here are available from GenBank under the following accession numbers: MT665032 - MT665070, MT665419 - MT665457, MT665138, and MT665525. Materials reported in this study will be made available but may require execution of a Materials Transfer Agreement.
Extended Data
Extended Data Figure 1. SARS-CoV-2 neutralization curves for mAb panel.
Neutralization of authentic SARS-CoV-2 by human mAbs. Mean ± SD of technical duplicates is shown. Data represent one of two or more independent experiments
Extended Data Figure 2. Inhibition curves for mAb inhibition of S2Pecto binding to hACE2.
Blocking of hACE2 binding to S2Pecto by anti-SARS-CoV-2 neutralizing human mAbs. Mean ± SD of triplicates of one experiment is shown. Antibodies rCR3022 and r2D22 served as controls.
Extended Data Figure 3. ELISA binding of anti-SARS-CoV-2 neutralizing human mAbs to trimeric SRBD, S2Pecto, or SARS-CoV S2Pecto antigen.
Mean ± SD of triplicates and representative of two experiments are shown. Antibodies rCR3022 and r2D22 served as controls.
Extended Data Figure 4. Serum or plasma mAbs competition binding.
a. Inhibition of hACE2 binding to S2Pecto by serum or plasma of four SARS-CoV-2 immune subjects or one non-immune control in ELISA using SARS-CoV-2 S2Pecto. mAbs were isolated from subjects 3 and 4 as described previously5. Mean ± SD of triplicates of one experiment is shown. Dotted line indicates full inhibition (100%) of hACE2 by 500 ng/mL of control mAb COV2-2196 or COV2-2130.
b. Inhibition of mAb COV2-2130 (left) or COV2-2196 (right) binding to S2Pecto by serum or plasma of four SARS-CoV-2 immune subjects or one non-immune control in ELISA using SARS-CoV-2 S2Pecto. Mean ± SD of triplicates and representative of two experiments are shown. Dotted line indicates percent of self-competition of mAb COV2-2196 and-2130 on the SARS-CoV-2 S2Pecto antigen.
Extended Data Figure 5. Mapping of mAb critical contact residues by alanine and arginine mutagenesis and biolayer interferometry.
a. Bar graphs show response values for mAb binding to wt or mutant SRBD constructs normalized to wt. Asterisks denote residues where increased dissociation of mAb was observed, likely indicating the residue is proximal to mAb epitope. Full response curves for mAb association and dissociation with wt or mutant SRBD constructs are also shown.
b. Structure of the RBD highlighting the critical contact residues for several mAbs and their location on the structure.
Extended Data Figure 6. Sequence features of human mAbs used in animal studies and mAb pharmacokinetics following infusion in NHPs.

a. Sequence features of human mAbs tested in animal models. Inferred variable genes are indicated and CDR3 amino acids are shown for heavy and light chains.
b. NHPs received one 50 mg/kg dose of mAb COV2-2196, COV2-2381, or isotype control mAb (n = 4 animals per group) intravenously on day −3 and then were challenged intranasally and intratracheally with SARS-CoV-2 at day 0. Concentration of human mAbs was determined at indicated timepoints. Each curve shows an individual animal. Data represent a single experiment.
Supplementary Material
Acknowledgements.
We thank Angela Jones and the staff of the Vanderbilt VANTAGE core laboratory for expedited sequencing, Ross Trosseth for assistance with data management and analysis, Robin Bombardi and Cinque Soto of VUMC for technical consultation on genomics approaches, Arthur Kim, Adam Bailey, Laura VanBlargan, and James Earnest of WUSTL for experimental assistance and key reagents, and Kevin M. Tuffy, Seme Diallo, Patrick M. McTamney, and Lori Clarke of AstraZeneca for generation of protein and pseudovirus reagents and related data. We thank Hanne Andersen, Mark G. Lewis, Ramya Nityanandam, Marinela Kirilova and Kaylee Verrington for research assistance with the nonhuman primate studies. This study was supported by Defense Advanced Research Projects Agency (DARPA) grants HR0011-18-2-0001 and HR00 11-18-3-0001, NIH contracts c and 75N93019C00062, NIH grants U01 AI150739, R01 AI130591 and R35 HL145242, and the Dolly Parton COVID-19 Research Fund at Vanderbilt. This work was supported by NIH grant S10 RR028106 for the Next Generation Nucleic Acid Sequencer, housed in Vanderbilt Technologies for Advanced Genomics (VANTAGE) and the Vanderbilt Institute for Clinical and Translational Research with grant support from (UL1TR002243 from NCATS/NIH). S.J.Z. was supported by NIH T32 AI095202. J.B.C. was supported by a Helen Hay Whitney Foundation postdoctoral fellowship. B.T.M was supported by NIH F32 AI138392. D.R.M. was supported by NIH T32 AI007151 and a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award. L.E.W. was supported by NIH F31 AI145189. E.C.C. was supported by NIH T32 AI138932. J.E.C. is the recipient of the 2019 Future Insight Prize from Merck KGaA, Darmstadt Germany, which supported this research with a research grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the U.S. government or the other sponsors.
Footnotes
Competing interests. R.S.B. has served as a consultant for Takeda and Sanofi Pasteur on issues related to vaccines. M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, Eli Lilly, and is on the Scientific Advisory Board of Moderna, a past recipient of unrelated research grant from Moderna and a current recipient of an unrelated research grant Emergent BioSolutions.
J.E.C. has served as a consultant for Sanofi and is on the Scientific Advisory Boards of CompuVax and Meissa Vaccines, is a recipient of previous unrelated research grants from Moderna and Sanofi and is Founder of IDBiologics, Inc. Vanderbilt University has applied for patents concerning SARS-CoV-2 antibodies that are related to this work. AstraZeneca has filed patents for materials/findings related to this work. J.J.S., K.R., Y.-M.L., and N.L.K. are employees of AstraZeneca and currently hold AstraZeneca stock or stock options. M.J.H is a member of a data safety monitoring board for AstraZeneca and founder of NuPeak Therapeutics. All other authors declared no competing interests.
REFERENCES
- 1.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tse LV, Meganck RM, Graham RL & Baric RS The Current and Future State of Vaccines, Antivirals and Gene Therapies Against Emerging Coronaviruses. Front Microbiol 11, 658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siracusano G, Pastori C & Lopalco L Humoral Immune Responses in COVID-19 Patients: A Window on the State of the Art. Front Immunol 11, 1049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zost SJ, et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. bioRxiv 10.1101/2020.05.12.091462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillay TS Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J Clin Pathol 73, 366–369 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Wan Y, Shang J, Graham R, Baric RS & Li F Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol 94(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280 e278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sui J, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A 101, 2536–2541 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ter Meulen J, et al. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet 363, 2139–2141 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ter Meulen J, et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med 3, e237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A 104, 12123–12128 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockx B, et al. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J Virol 82, 3220–3235 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, et al. Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset. J Infect Dis 215, 1807–1815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi JH, et al. Characterization of a human monoclonal antibody generated from a B-cell specific for a prefusion-stabilized spike protein of Middle East respiratory syndrome coronavirus. PLoS One 15, e0232757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu P, et al. Ultrapotent Human Neutralizing Antibody Repertoires Against Middle East Respiratory Syndrome Coronavirus From a Recovered Patient. J Infect Dis 218, 1249–1260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, et al. Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape. J Virol 92(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang N, et al. Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD. Cell Rep 28, 3395–3405 e3396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, et al. Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein. Cell Rep 24, 441–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corti D, et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci U S A 112, 10473–10478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang L, et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med 6, 234ra259 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Tang XC, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci U S A 111, E2018–2026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ying T, et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol 88, 7796–7805 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S, Hillyer C & Du L Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol 41, 355–359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valk SJ, et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: a rapid review. Cochrane Database Syst Rev 5, CD013600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan M, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ianevski A, He L, Aittokallio T & Tang J SynergyFinder: a web application for analyzing drug combination dose-response matrix data. Bioinformatics 33, 2413–2415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walls AC, et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292 e286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassan A, et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 10.1016/j.cell.2020.06.011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinnon KH, et al. A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. bioRxiv 10.1101/2020.05.06.081497 (2020). [DOI] [Google Scholar]
- 34.Chandrashekar A, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 10.1126/science.abc4776 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 10.1126/science.abc6284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robbiani DF, et al. Convergent Antibody Responses to SARS-CoV-2 Infection in Convalescent Individuals. Nature 10.1038/s41586-020-2456-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brouwer PJM, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 10.1126/science.abc5902 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 10.1016/j.cell.2020.05.025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju B, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 10.1038/s41586-020-2380-z (2020). [DOI] [PubMed] [Google Scholar]
- 40.Rogers TF, et al. Rapid isolation of potent SARS-CoV-2 neutralizing antibodies and protection in a small animal model. Science 10.1126/science.abc7520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi R, et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature 10.1038/s41586-020-2381-y (2020). [DOI] [PubMed] [Google Scholar]
- 42.Wec AZ, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 10.1126/science.abc7424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368, 1274–1278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansen J, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 10.1126/science.abd0827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baum A, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 10.1126/science.abd0831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laha S, et al. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol, 104445 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee S et al. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology 497, 33–40, 10.1016/j.virol.2016.06.022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohi M, Li Y, Cheng Y & Walz T Negative staining and image classification - Powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34, 10.1251/bpo70 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51, 10.1016/j.jsb.2005.07.007 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296, 10.1038/nmeth.4169 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Bepler T, Noble AJ, and Berger B Topaz-Denoise: general deep denoising models for cryoEM. bioRxiv 10.1101/838920 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612, 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Chng J et al. Cleavage efficient 2A peptides for high level monoclonal antibody expression in CHO cells. MAbs 7, 403–412, 10.1080/19420862.2015.1008351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheehan KC et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res 26, 804–819 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EM maps have been deposited at the Electron Microscopy Data Bank with accession codes (Supplemental Table 2): EMD-21974, EMD-21975, EMD-21976, and EMD-21977. The EM-map EMD-21965 is publicly available. The accession numbers for cryo-EM and crystal structures used for structural analysis, including structures of the closed conformation of SARS-CoV-2 S (PDB: 6VXX), the open conformation of SARS-CoV-2 (PDB: 6VYB), Fab used for docking (PDB: 12E8), and the SARS-CoV-2 RBD/hACE2 complex and the 60-amino-acid hACE2 recognition motif (PDB (6M0J) are publicly available. Sequences of mAbs characterized here are available from GenBank under the following accession numbers: MT665032 - MT665070, MT665419 - MT665457, MT665138, and MT665525. Materials reported in this study will be made available but may require execution of a Materials Transfer Agreement.