ABSTRACT
Folate-mediated one-carbon metabolism (FOCM) is compartmentalized within human cells to the cytosol, nucleus, and mitochondria. The recent identifications of mitochondria-specific, folate-dependent thymidylate [deoxythymidine monophosphate (dTMP)] synthesis together with discoveries indicating the critical role of mitochondrial FOCM in cancer progression have renewed interest in understanding this metabolic pathway. The goal of this narrative review is to summarize recent advances in the field of one-carbon metabolism, with an emphasis on the biological importance of mitochondrial FOCM in maintaining mitochondrial DNA integrity and mitochondrial function, as well as the reprogramming of mitochondrial FOCM in cancer. Elucidation of the roles and regulation of mitochondrial FOCM will contribute to a better understanding of the mechanisms underlying folate-associated pathologies.
Keywords: folate, thymidylate, mitochondrial DNA, mitochondrial metabolism, cancer metabolism
Introduction to Folate-Mediated One-Carbon Metabolism—Associations with Pathology and Biosynthetic Outputs
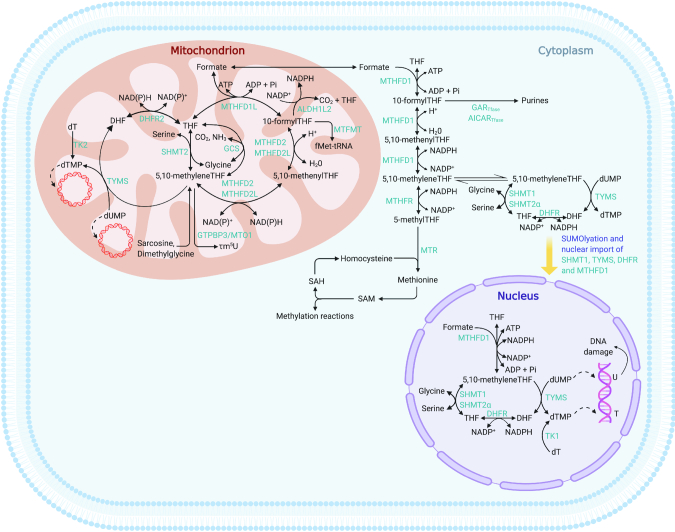
Folate-mediated one-carbon metabolism (FOCM) is an interconnected metabolic network that serves to activate and transfer one-carbon units for many biochemical processes, including purine and deoxythymidine (dT) monophosphate (dTMP) biosynthesis, mitochondrial protein translation, and methionine regeneration, all of which support diverse cellular functions such as cell proliferation, protein synthesis, and mitochondrial respiration (Figure 1). As described below, FOCM is compartmentalized within the cell. The expression of enzymes in the mitochondrial FOCM pathway has been reviewed extensively (1). The objective of this review is to summarize recent findings linking impairments in mitochondrial FOCM to alterations in mitochondrial genome stability and ultimately mitochondrial function.
FIGURE 1.
Folate-mediated one-carbon metabolism. Cellular compartmentalization of FOCM. dTMP synthesis occurs in the cytosol, nucleus, and mitochondria, whereas purine synthesis and methionine synthesis take place within the cytosol. Mitochondrial FOCM generates formate for cytosolic and nuclear FOCM and biosynthetic precursors for mtDNA synthesis and mitochondrial protein translation. AICARTfase, aminoimidazolecarboxamide ribonucleotide transformylase; ALDH1L2, aldehyde dehydrogenase 1 family member L2; DHF, dihydrofolate; DHFR, dihydrofolate reductase; dT, deoxythymidine; dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; fMet, N-formylmethionine; FOCM, folate-mediated one-carbon metabolism; GARTfase, glycinamide ribonucleotide tranformylase; GCS, glycine cleavage system; GTPBP3, GTP-binding protein 3, mitochondrial; mtDNA, mitochondrial DNA; MTFMT, mitochondrial methionyl-tRNA formyltransferase; MTHFD, methylenetetrahydrofolate dehydrogenase; MTHFD1L, methylenetetrahydrofolate dehydrogenase 1–like; MTHFR, methylene-tetrahydrofolate reductase; MTO1, mitochondrial tRNA translation optimization 1; MTR, methionine synthase; Pi, phosphate; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SHMT, serine hydroxymethyltransferase; SUMO, small ubiquitin-like modifier; T, thymidine; THF, tetrahydrofolate; TK, thymidine kinase; TYMS, thymidylate synthase; U, uracil; τm5U, modified nucleoside 5-taurinomethyluridine.
Disruptions of folate metabolism are associated with several human pathologies including neural tube defects (NTDs) and cancer. NTDs are a class of birth defects that result from failure of neurulation during early human embryonic development. NTDs are among the most common human birth defects and even the least severe NTDs can result in lifelong disability. Numerous genetic and environmental factors are involved in the etiology of NTDs; however, among all these risk factors, the strongest predictor is low blood folate concentrations (2). Observational studies in the 1960s led to the recognition that reduced maternal folate concentrations were associated with an elevated risk for NTDs (3). Although further studies indicated that folic acid supplementation decreased NTD incidence and widely implemented folic acid fortification programs have been successfully used for this purpose across the globe, the biochemical mechanisms underlying the folic acid–responsive NTD pathogenesis remain largely undefined (2, 4).
Similarly, folate has been studied extensively with respect to both cancer development and treatment. Epidemiological and animal studies have demonstrated that folate deficiency increases the risk for several cancers, including colorectal cancer, pancreatic cancer, and breast cancer (5–10). However, given the role of folate in nucleotide biosynthesis for rapid cell growth and the use of antifolates to inhibit cell proliferation, elevated folate status has been hypothesized to promote the progression of established tumors (11). Following the initial successful use of antifolates including aminopterin for the treatment of leukemia, FOCM emerged as a therapeutic target for cancers, including mesothelioma, lung cancer, and gastrointestinal cancer (12–14). Understanding of the FOCM pathway in the context of cancer pathogenesis has led to discoveries of chemotherapeutic agents such as methotrexate, fluorouracil, and trifluridine (12). As described in more detail below, the recently appreciated importance of mitochondrial folate metabolism in supporting cancer cell proliferation may also lead to the development of novel therapeutic targets.
De novo purine synthesis is a 10-reaction process that requires 2 folate-dependent enzymes—5′-phosphoribosyl-glycinamide (GAR) transformylase and 5′-phosphoribosyl-5-aminoimidazole-4-carboxamide (AICAR) transformylase—to add 2 one-carbon units donated from 10-formyl-tetrahydrofolate (-THF) into the purine backbone (Figure 1) (15). Purines provide essential components for the synthesis of DNA and RNA as well as energy to promote cell proliferation (16). When there is an increased demand for purines, such as in proliferating cells and tumor cells, de novo purine synthesis plays a critical role in maintaining the cellular pool of purines and has the largest demand for folate one-carbon units (15). Therefore, the de novo purine synthesis pathway has been a frequent target for drugs used for cancer chemotherapy and for the treatment of autoimmune disease (17). The de novo dTMP synthesis involves the folate-dependent enzymes serine hydroxymethyltransferase (SHMT), thymidylate synthase (TYMS), and dihydrofolate reductase (DHFR) for the conversion of deoxyuridine monophosphate (dUMP) to dTMP (Figure 1). In this pathway, serine is used by SHMT to generate 5,10-methylene-THF, which provides one-carbon group for the methylation of dUMP to dTMP in a reaction catalyzed by TYMS. Dihydrofolate (DHF) generated by this reaction is converted to THF by the NAD(P)H-dependent enzyme DHFR, which is a target for chemotherapeutic agents including methotrexate and trimetrexate (18). The regenerated THF can then be used for another round of de novo dTMP biosynthesis. Distinct from other nucleotides, the de novo synthesis of dTMP takes place in the cytosol, the nucleus, and the mitochondria (19, 20). Perturbed de novo dTMP synthesis results in an accumulation of its precursor, dUMP, leading to uracil misincorporation into DNA (21). In the event that uracil is incorporated into the DNA, the uracil base can be removed and replaced with a thymine base by base excision repair, which is initiated by the enzyme uracil-DNA N-glycosylase (UNG). However, when the dUTP to dT triphosphate (dTTP) ratio is too high, DNA polymerases continue misincorporating uracil during both DNA replication and repair. As uracil accumulates to a critical level in DNA, double-strand breaks (DSBs) can be generated as a result of the repetitive base excision repair of the uracil residues by UNG. In addition, uracil accumulation near replication origins can stall DNA replication fork progression and lead to fork collapse, contributing to DSB formation, DNA instability, and ultimately cell death (22, 23).
The metabolic relation between folate nutrition and uracil in DNA is not fully understood. Although uracil misincorporation in DNA has been suggested to be a biomarker of folate and/or vitamin B-12 status (21), it is not well established whether uracil misincorporation is causally related to the development of FOCM-associated pathologies (2). Further research is needed to characterize the role of uracil misincorporation in FOCM-associated pathologies along causal pathways.
The most reduced form of folate, 5-methyl-THF, is used for the remethylation of homocysteine to methionine. 5-Methyl-THF is generated by a cytosolic NAD(P)H-dependent reaction catalyzed by methylene tetrahydrofolate reductase (MTHFR) and it serves as a methyl donor for the cobalamin (vitamin B-12)-dependent conversion of homocysteine to methionine by methionine synthase (MTR) (17). Methionine is an essential amino acid that is required for protein synthesis and can also be adenosylated to S-adenosylmethionine (SAM). SAM, a reactive methyl group carrier, plays a crucial role in numerous methylation reactions such as the methylation of proteins, DNA, and histones as well as biosynthetic processes including phosphatidylcholine, creatine, and polyamine synthesis, as well as for sulfur metabolism (17, 24–26). Changes in methionine concentrations alter the ratio of SAM to S-adenosylhomocysteine (SAH), which then impacts methylation reactions (27). Methionine metabolism that supplies SAM is therefore critical for the maintenance and adaptation of the epigenome.
Intracellular Compartmentalization of One-Carbon Metabolism
Nuclear folate-mediated one-carbon metabolism
The folate metabolic network is compartmentalized within mammalian cells in the nucleus, the cytosol, and the mitochondria (Figure 1). In the nucleus, there are 2 pathways for dTMP synthesis: dTMP salvage pathway and de novo dTMP synthesis pathway. The salvage pathway involves phosphorylation of thymidine to dTMP by the enzyme, thymidine kinase (TK) 1 (TK1), while the de novo synthesis of dTMP is catalyzed by the enzymes SHMT1 and SHMT2α (encoded by SHMT1 and SHMT2, respectively), methylenetetrahydrofolate dehydrogenase (MTHFD) 1, TYMS, and DHFR. Both serine (catalyzed by SHMT1/2α) and formate (catalyzed by MTHFD1) act as one-carbon sources for the production of 5,10-methylene-THF, which is required to methylate dUMP to dTMP. However, formate, produced through mitochondrial FOCM, provides most of the one-carbon units required for nuclear de novo dTMP synthesis (2). Nuclear de novo dTMP synthesis enzymes localize to the cytosol during the G1 phase of the cell cycle and are modified by the small ubiquitin-like modifier (SUMO) protein during S and G2/M phase and then translocated from the cytosol into the nucleus. Following SUMO-dependent translocation to the nucleus, de novo dTMP biosynthesis enzymes form a multienzyme complex associated with the nuclear lamin proteins and many DNA replication and repair proteins (28). In animal models and cultured cells, SHMT1 and/or SHMT2α act as essential scaffold proteins in tethering this multienzyme nuclear de novo dTMP synthesis complex to the nuclear lamina, which effectively increases rates of de novo dTMP synthesis (28, 29). Interestingly, expression of a catalytically inactive SHMT1 mutant also increased de novo dTMP biosynthesis in cultured cells (30), emphasizing this contribution of SHMT1 as a scaffold protein in accelerating de novo dTMP synthesis is independent of its catalytic activity (31). This finding is consistent with observations demonstrating that MTHFD1 also translocates to the nucleus and provides most of one-carbon units, originating from formate, for nuclear de novo dTMP synthesis (1, 32).
Nuclear localization of enzymes responsible for nuclear de novo dTMP biosynthesis is essential to prevent uracil accumulation in DNA, as has been demonstrated in an SHMT1 overexpression mouse model (33). In this mouse model, overexpression of SHMT1 unexpectedly impaired the localization of SHMT1 and TYMS, and increased uracil content in hepatic nuclear DNA. Similarly, Shmt1+/−and Shmt1−/− mice exhibit elevated uracil in genomic DNA and develop folic acid–responsive NTDs (29, 34). Taken together, these data suggest that impaired SHMT1 nuclear localization or impaired nuclear de novo dTMP complex formation may underlie the folic acid–responsive NTDs in mice (2).
Mitochondrial folate-mediated one-carbon metabolic pathway
Mitochondrial FOCM exists in parallel to the cytosolic FOCM pathway (Figure 1) (35). These 2 pathways are connected by transport of one-carbon sources that can readily traverse the mitochondrial inner membrane, including serine, glycine, and formate. Mitochondrial FOCM produces formate, which, in turn, is used in the cytosolic/nuclear pathway for purine, dTMP, and methionine biosynthesis and also generates dTMP and N-formylmethionine tRNA for mitochondrial DNA and protein synthesis, respectively (1, 17). Mitochondrial FOCM can be categorized into 3 phases: integration of one-carbon units derived from one-carbon sources (such as serine and glycine) to the mitochondrial pool of THF, interconversion of the activated one-carbon units transported by THF between different oxidation states, and donation of one-carbon units carried by THF by their export to cytoplasm as formate (1).
At least 4 one-carbon donors (serine, glycine, dimethylglycine, and sarcosine) have been identified in the mitochondria, with serine being the major one-carbon donor in most organisms, including humans (1, 36–38). Serine donates the one-carbon unit from its side chain to THF in a reaction catalyzed by SHMT2, thereby converting serine to glycine and THF to 5,10-methylene-THF. The product of this reaction, glycine, can also serve as a source of one-carbon units (39). Glycine, the second main donor of one-carbon units in mitochondria, can be broken down by a mitochondrial multienzyme system, glycine cleavage system (GCS), to generate 5,10-methylene-THF (40, 41). GCS is highly tissue-specific, being fully expressed in the liver, kidney, and brain while inactive in the heart (42). Whole-body glycine flux studies reveal that mitochondrial GCS produces 5,10-methylene-THF at a very high rate, suggesting that GCS may support a high degree of purine and thymidylate synthesis; however, it appears that almost all GCS-derived one-carbon units end up in serine synthesis, with the remainder entering nucleotide synthesis and homocysteine remethylation (43). Furthermore, although serine and glycine can be interconverted, it has been reported that exogenous glycine cannot substitute for serine to support nucleotide synthesis (44). In some cell types, dimethylglycine and sarcosine, which are produced from choline oxidation, also contribute to the generation of 5,10-methylene-THF through dimethylglycine dehydrogenase and sarcosine dehydrogenase, respectively (36, 37). In addition to choline metabolism, sarcosine is also synthesized from glycine in a reaction catalyzed by an abundant enzyme glycine N-methyltransferase (GNMT) (45).
The SHMT2 gene produces a 56-kDa “full length” SHMT2 protein containing a mitochondrial leader sequence that localizes to the cytosol and mitochondria. Cleavage of the mitochondrial leader sequence results in a 50-kDa processed SHMT2 that presents exclusively in the mitochondria. The 53-kDa SHMT2α isoform results from a second transcript with an additional translational start site, which results in a protein that lacks the mitochondrial leader sequence. Therefore, this SHMT2α isoform localizes to the cytosol and the nucleus (19). These SHMT2 isoforms exhibit tissue-specific expression patterns. The SHMT2 isoform is the primary form in liver, while SHMT2α and the unprocessed SHMT2 protein precursor are found at higher concentrations in kidney (19). Mitochondria provides the major source of one-carbon units for cytosolic FOCM through serine catabolism by SHMT2 (46). Similar to the nuclear FOCM, in the mitochondria, SHMT2 catalyzes the reversible conversion of serine and THF to glycine and 5,10-methylene-THF (Figure 1). Using 5,10-methylene-THF as the substrate, TYMS transfers a one-carbon unit to dUMP to generate dTMP and DHF. DHFR2 then regenerates THF from DHF to allow for another round of dTMP biosynthesis (20). 10-Formyl-THF in mitochondrial FOCM acts as a branch point where the formyl group can be converted into formate or carbon dioxide (1). The formate branch is catalyzed by the mitochondrial C1-tetrahydrofolate synthase 1-like (MTHFD1L) to generate formate for the cytosolic FOCM (47, 48), whereas the carbon dioxide branch is catalyzed by the NADP-dependent mitochondrial 10-formyl-THF dehydrogenase [aldehyde dehydrogenase 1 family member L2 (ALDH1L2)] to produce carbon dioxide and THF (49). 10-Formyl-THF also contributes to mitochondrial protein synthesis by serving as the formyl donor for the formylation of the initiator methionyl-tRNAMet catalyzed by methionyl-tRNA formyltransferase (MTFMT) (50).
Mitochondrial FOCM and mitochondrial DNA integrity
Maintenance of an adequate and balanced cellular dTMP pool is essential to preserve DNA integrity for both the nuclear and mitochondrial genomes. Both depletion and expansion of dTMP pools impact genomic (both nuclear and mitochondrial) DNA integrity (51). Disruption of de novo dTMP synthesis results in uracil misincorporation into both nuclear DNA and mitochondrial DNA (mtDNA), which subsequently contributes to genome instability (21). Depletion of mitochondrial dTMP pools due to mitochondrial TK2 mutation, which reduces mitochondrial dTMP concentrations by blocking dTMP salvage synthesis (conversion of dT to dTMP by mitochondrial TK2), has been suggested to impede mtDNA replication, leading to mtDNA-depletion syndromes (MDS) (52). Elevations in dTMP pools also lead to mtDNA depletion (53), mtDNA deletions, and site-specific point mutations (54), as observed in mitochondrial neurogastrointestinal encephalomyopathy, an autosomal recessive disorder caused by cytosolic thymidine phosphorylase (TP) deficiency (55).
dTMP synthesis from the salvage pathway is not sufficient to sustain a continuous supply of dTMP to mtDNA replication and thus the de novo dTMP synthesis pathway is also required (52). As described above, the de novo dTMP synthesis pathway requires the folate-dependent enzymes SHMT2, TYMS, and DHFR2 to catalyze the conversion of dUMP to dTMP. mtDNA from HeLa cells grown in folate-deplete media exhibited 84% more uracil than cells grown in folate-replete media (56). In fact, in HeLa cells, mtDNA is more sensitive to uracil incorporation as a result of folate deficiency than is nuclear DNA (56, 57). In mtDNA from glyA Chinese hamster ovary (CHO) cells, which are glycine autotrophs due to the lack of mitochondrial SHMT2, 40% more uracil was observed than in wild-type CHO cells (20). Taken together, these data suggest than mtDNA is highly sensitive to uracil misincorporation when FOCM is disturbed.
Several studies in model systems have also demonstrated that folate deficiency can lead to accumulation of mtDNA deletions (58–63), which may cause reduced expression of genes within mtDNA that are essential to mitochondrial function and energy production. In rats, the 4834-kb mtDNA common deletion was increased by 3.5-fold in lymphocytes after 4 wk consuming a folate-deficient diet. Lymphocyte mtDNA common deletion frequency was inversely associated with blood folate concentrations and positively correlated with the mtDNA deletions in the lungs, muscles, heart, liver, kidneys, pancreas, and brain. This suggests that the accumulated mtDNA common deletions observed in lymphocyte mtDNA reflect mtDNA damage in other body tissues and organs (61). In another study in rats, the effect of folate deficiency on both mtDNA common deletions and mitochondria-related gene expression was investigated (60). Dietary folate deprivation significantly increased the frequency of the mtDNA common deletion and reduced mtDNA content in several tissues, including the brain, the heart, and the liver. The increased frequency of the common deletion was associated with enhanced mitochondrial biogenesis, possibly in an attempt to compensate for mitochondrial dysfunction resulting from the accumulation of large-scale deletions (60). In addition, the frequency of the mtDNA common deletion was reduced by folate supplementation in the liver of old (12 mo) rats (62). In agreement with the above studies, mice lacking uracil DNA glycosylase (Ung−/−) exhibited an increase in mtDNA mutagenesis in aged brain, together with an apparently compensatory increase in mtDNA content in response to low folate status (64).
In addition to salvage synthesis and de novo synthesis pathways, the mitochondrial inner membrane protein MPV17 (MPV17) also appears to play a role in mitochondrial dTMP accumulation (56). MPV17 has been proposed to be a channel in the mitochondrial inner membrane that imports deoxynucleotide triphosphate (dNTP) into the mitochondria (65). Depressed MPV17 expression depletes mitochondrial folate concentrations and increases uracil content in mtDNA in HeLa cells, which indicates that mitochondrial dTMP synthesis capacity is not sufficient to maintain mitochondrial dTMP levels to prevent uracil misincorporation and that import of cytosolic dTMP may be required to maintain mitochondrial dTMP pools (56).
Inborn errors of metabolism in genes involved in mtDNA maintenance, including mitochondrial nucleotide pool maintenance, mtDNA replication, and mitochondrial fusion, are associated with MDS (66). As described above, defects in dTMP salvage pathway including TK2 result in an imbalanced mitochondrial nucleotide pool leading to MDS (52). Interestingly, a transcriptomic study revealed an induction of SHMT2 and TYMS in patients with TK2-related myopathic MDS (67). In the context of mtDNA homeostasis, SHMT2 and TYMS are 2 key enzymes involved in the de novo dTMP biosynthesis pathway in mitochondria. Thus, upregulation of the mitochondrial de novo dTMP synthesis pathway is likely to reflect a biological mechanism attempting to compensate for the lack of dTMP as a result of the severely reduced TK2 activity in these patients. Similarly, mtDNA replication defects caused by dominant mutations in TWINKLE helicase remodel mitochondrial FOCM through inducing MTHFD2 and MTHFD1L and upregulating de novo serine biosynthesis. Replication stalling as a result of TWINKLE defects promotes dNTP synthesis, leading to an increased and imbalanced cellular dNTP pool, which accelerates mtDNA deletion mutagenesis (68). Future studies into how mitochondrial FOCM is modified in response to defects in mtDNA maintenance and the consequences of this remodeling will increase our understanding of the pathogenesis of mtDNA maintenance disorders.
Mitochondrial FOCM and mitochondrial function
Recent studies have shown that mitochondrial FOCM enzymes are particularly strongly upregulated in proliferating lymphocytes and human cancers (17, 69–71), which, in part, reflects the role of mitochondrial FOCM in supporting critical cellular functions such as cell proliferation and mitochondrial respiration. Some mitochondrial compartment–specific uses of mitochondrial folate one-carbon units include the local biosynthesis of dTTP (20) and of N-formylmethionine (fMet) (72), tRNA modification (73), oxidative phosphorylation (OXPHOS) complex assembly (74) and redox state regulation (75, 76).
One study identified the mitochondrial FOCM enzyme SHMT2 as being required for robust mitochondrial oxygen consumption and cell proliferation in a low-glucose environment (77). More specifically, SHMT2 deletion in Jurkat cells caused defects in mitochondrial respiration and impaired translation of mitochondria-encoded proteins, without affecting cytoplasmic protein translation. In addition, SHMT2-null Jurkat cells exhibited a loss of the fMet-tRNAMet, which is generated by a tRNA formylation reaction catalyzed by the mitochondrial MTFMT, using 10-formyl-THF as the formyl donor. This modified tRNA is required to initiate translation specifically in mitochondria, and its loss is observed in patients with the mitochondrial disease Leigh syndrome (72, 77). These results suggest that proper translation of mitochondria-encoded proteins supported by Met-tRNAMet formylation is an additional key function of mitochondrial FOCM. However, maintenance of mitochondria-encoded proteins appeared to require only limited amounts of mitochondrial one-carbon units, as minimal expression of SHMT2 was sufficient to maintain the expression of mitochondrially translated proteins (77). As such, the use of mitochondrial one-carbon units for the biosynthesis of fMet is unlikely to underlie the upregulation of mitochondrial FOCM enzymes observed in cancers (69, 71, 77).
Another study also demonstrates that loss of SHMT2, but not of other folate-dependent enzymes, leads to defective OXPHOS in human HCT116 colon cancer cells due to impaired mitochondrial protein translation (73). This study demonstrated that mitochondrial 5,10-methylene-THF is essential to maintain mitochondrial respiratory capacity by providing the methyl group for tRNA taurinomethylation, a relatively novel tRNA base modification and a unique use of mitochondrial 5,10-methylene-THF. Mitochondrial ribosome profiling in SHMT2-knockout cells revealed that the lack of this tRNA modification caused defective mitochondrial translation, with ribosome stalling at certain lysine and leucine codons, which can lead to defective translation for a subset of the mitochondria-encoded OXPHOS proteins. On the other hand, as described above, the decrease in fMet-tRNAMet synthesis observed in SHMT2 knockout Jurkat cells resulted in impaired mitochondrial translation initiation (77), which can cause defective global OXPHOS protein synthesis in mitochondria. The differential translational responses to SHMT2 deletion observed from the different cell types may indicate that SHMT2 modulates mitochondrial protein translation in a cell type–specific manner (73, 77). Ribosome stalling at the specific lysine and leucine codons (73) was also observed in patients with mitochondrial disorders, including myoclonic epilepsy with ragged red fibers (MERRF) and mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) (78, 79). Taken together, these results reveal a biochemical mechanism whereby mitochondrial FOCM contributes to translation of mitochondrial proteins, linking mitochondrial FOCM with certain inborn errors of mitochondrial metabolism (77). In addition to SHMT2 deficiency, mutations in mitochondrial GCS are associated with glycine encephalopathy, an inborn error that results in glycine accumulation to pathologic concentrations in cerebrospinal fluid and plasma, which can cause severe neuronal dysfunction (78).
In contrast to the above observations (73, 77) that loss of SHMT2 leads to defective mitochondrial translation in cells, another study argued that the ability of an SHMT2-knockout 293A cell line to synthesize mitochondria-encoded proteins was not impaired when compared with the wild-type cells as determined using stable-isotope labeling (74). Instead, this study demonstrated that the regulation of the OXPHOS system by SHMT2 may take place after the translation of mtDNA-encoded proteins. SHMT2 deletion significantly reduced the steady-state levels of the mature Complex I, which could be restored by exogenous supplementation with the one-carbon donor formate (74). These results suggest a novel role of the one-carbon metabolic intermediates derived from mitochondrial serine catabolism in the assembly of respiratory chain complex, possibly through supporting nuclear and/or cytosolic metabolic reactions instead of mitochondrion-specific processes, although the exact underlying mechanisms remain unclear (74).
Serine catabolism by the mitochondrial folate pathway was also identified as the major source of NAD(H) for cells with impaired electron transport chain activity (76). In the mitochondria, SHMT2 converts serine to glycine while producing 5,10-methylene-THF. MTHFD2 then produces 10-formyl-THF from 5,10-methylene-THF with concomitant generation of NAD(H). Using genetic manipulation, both of these enzymes were shown to be essential for folate-dependent NAD(H) production. When cellular respiration is impaired in HCT116 cells, NAD(H) production by the tricarboxylic acid (TCA) cycle is blocked. Therefore, the contribution of serine catabolism via SHMT2 and MTHFD2 to generate NAD(H) is increased to a level that it effectively becomes the major source of cellular NAD(H) (76). Several regulatory mechanisms actively promote this pathway in response to impaired respiration. For example, hypoxia induces SHMT2 via myelocytomatosis oncogene (MYC) and hypoxia-inducible factor (HIF), which also upregulates de novo serine synthesis and mitochondrial FOCM enzymes including MTHFD2 and MTHFD1L (80–83). Paradoxically in respiration-impaired HCT116 cells, loss of mitochondrial serine catabolism by SHMT2 inhibition or MTHFD2 deletion improved metabolic homeostasis and enhanced cell growth (76). The authors hypothesized that, in this case, NAD(H) generation via serine catabolism in response to impaired respiration decreased cell growth to prevent proliferation of respiration-impaired cells or to synthesize other products of FOCM at ischemic sites (76). Further studies are needed to solve this paradox and to understand the role of folate availability and/or metabolism in NAD(H) generation.
In addition to catalyzing mitochondrial folate metabolic reactions, the nonenzymatic functions of Mthfd2 have been reported in mouse embryonic stem cells (mESCs) (84). In the mitochondria, Mthfd2 maintains active OXPHOS system and improves the quality of mouse pluripotent stem cells (PSCs) through an indirect interaction between the MTHFD2 protein and mitochondrial electron transport chain complex III. In the nucleus, Mthfd2 regulates exonuclease 1 (EXO1) phosphorylation, thereby modulating homologous recombination (HR) repair and maintaining genome integrity in mESCs. Interestingly, the regulation of mESC pluripotency and HR repair by Mthfd2 was not mediated by the catalytic activity of MTHFD2, revealing a novel role of the mitochondrial FOCM enzyme in supporting cellular functions independent of its enzymatic functions.
In summary, mitochondrial FOCM may affect OXPHOS through several mechanisms, including the influence on 1) biosynthesis of nucleotides and therefore mtDNA replication (20, 85), 2) mRNA expression of the OXPHOS genes (27, 86), 3) biosynthesis of the mitochondria-encoded OXPHOS proteins (73, 77), and 4) regulation of cellular redox state (75, 76).
Mitochondrial FOCM and cancer
Aided by modern genomics and metabolomics tools, the roles of de novo serine synthesis and mitochondrial FOCM pathway in cancer have increasingly been appreciated (16, 87, 88). Given the increased requirement for nucleotides to support rapid proliferation of cancer cells, metabolism of one-carbon sources, serine and glycine, and mitochondrial folate metabolic enzymes are upregulated. Increased exogenous serine and glycine uptake was observed nearly universally across 60 cancer cell lines in the NCI-60 panel (89). However, another study using isotope tracers demonstrated that cancer cells selectively consumed exogenous serine and that cell proliferation was supported by consumption of serine instead of glycine (90). In addition to serine uptake, enzymes in the de novo serine synthesis pathway are highly expressed in some tumors. Phosphoglycerate dehydrogenase (PHGDH), the enzyme that catalyzes the first reaction in serine synthesis, exhibits gene copy-number gain in triple-negative breast cancer and melanoma (91, 92). Under stress conditions of cancer, all 3 enzymes [PHGDH, phosphoserine aminotransferase 1 (PSAT1), and phosphoserine phosphatase (PSPH)] involved in de novo serine biosynthesis from the glycolytic intermediate, 3-phosphoglycerate are upregulated (93). Concurrent with increased metabolism of one-carbon sources, enzymes of mitochondrial FOCM, in particular MTHFD2 and SHMT2 become the most consistently upregulated metabolic genes in cancer (69, 86, 94). In addition to transcriptional reprogramming, expression of SHMT2, MTHFD2, and ALDH1L2 is induced and activity of mitochondrial FOCM is increased over cytosolic FOCM in response to endoplasmic reticulum stress in cancer (93). SHMT2 expression is also induced upon hypoxic stress to maintain mitochondrial redox balance and prevent accumulation of reactive oxygen species (82, 95). The overexpression of MTHFD2 and SHMT2 is not only necessary for cancer cell survival and tumor progression (69, 86) but also predicts poor prognosis in multiple cancers, including breast cancer, gastrointestinal cancer, and intrahepatic cholangiocarcinoma (96–99). Metabolic reprogramming of mitochondrial FOCM induced by stress responses seems to adjust the microenvironment of cancer and render cancer cells resistant to folate-based antimetabolites (93). However, it is still unclear whether mitochondrial FOCM enzymes and serine metabolism are upregulated to support mitochondrial function or to provide formate for cytosolic and nuclear FOCM reactions in support of tumor growth. The understanding of the role of FOCM in cancer has led to advances in chemotherapy; however, our ability to selectively target FOCM for therapy is limited, due in large part to the lack of understanding of specific alterations of FOCM in cancer. Emerging studies that highlight the specific induction of mitochondrial FOCM enzymes in cancer may provide insights in targeting this pathway for cancer therapy.
Conclusions
There is a growing body of evidence demonstrating that mitochondrial FOCM plays a critical role in maintaining mtDNA integrity and mitochondrial function. Depending on the tissue/cell line, mitochondrial FOCM supports mitochondrial function by providing one-carbon units for mitochondrial tRNA taurinomethylation, methionyl-tRNA formylation, or de novo dTMP synthesis (Table 1). However, mitochondrial serine catabolism catalyzed by SHMT2 also generates formate, which readily traverses the cytosolic and nuclear compartments, and serves as the principal one-carbon source for all cytosolic and nuclear FOCM reactions (2). Therefore, whether it is the mitochondrial compartment–specific uses of one-carbon units or products from cytosolic or nuclear FOCM that affects mitochondrial function remains to be elucidated. Additionally, most of our knowledge regarding the contributions of mitochondrial FOCM to mtDNA maintenance and mitochondrial respiration is obtained from transformed cell lines. Given the remodeling of mitochondrial FOCM in cancer as well as the alterations in gene expression associated with cancer transformation that affect mitochondrial FOCM—for example, the significant downregulation of the normally abundant GNMT in human cancers (101)—further studies are required to investigate the role of mitochondrial FOCM in nontransformed tissues and in the context of diseases of aging. Understanding the regulation and complexity of mitochondrial folate metabolism could mechanistically promote effective prevention and treatment of folate-associated pathologies.
TABLE 1.
Tissue/cell line–specific effects of SHMT2 deficiency1
| Tissue/cell line | SHMT2 expression/activity | Biological effect | Reference |
|---|---|---|---|
| glyA CHO | Null | Increased uracil content in mtDNA | Anderson et al. (20) |
| 293A | Null | Impaired complex I assembly | Lucas et al. (74) |
| Jurkat | Null | Loss of fMet-tRNAMet | Minton et al. (78) |
| HCT116 | Null | Impaired tRNA taurinomethylation | Morscher et al. (73) |
| Mouse embryonic fibroblasts | Null | Mitochondrial respiration defects and growth retardation | Tani et al. (100) |
CHO, Chinese hamster ovary; fMet, N-formylmethionine; mtDNA, mitochondrial DNA; SHMT2, serine hydroxymethyltransferase 2.
ACKNOWLEDGEMENTS
The authors’ responsibilities were as follows—YX: drafted the initial manuscript; MSF: critically revised the manuscript; and both authors read and approved the final manuscript.
Notes
The authors reported no funding received for this study.
Author disclosures: The authors report no conflicts of interest.
Abbreviations used: ALDH1L2, aldehyde dehydrogenase 1 family member L2; CHO, Chinese hamster ovary; DHF, dihydrofolate; DHFR, dihydrofolate reductase; dNTP, deoxynucleotide triphosphate; dT, deoxythymidine; DSB, double-strand break; dTMP, deoxythymidine monophosphate; dTTP, deoxythymidine triphosphate; dUMP, deoxyuridine monophosphate; fMet, N-formylmethionine; FOCM, folate-mediated one-carbon metabolism; GCS, glycine cleavage system; GNMT, glycine N-methyltransferase; HR, homologous recombination; MDS, mtDNA-depletion syndrome; mESC, mouse embryonic stem cell; MPV17, mitochondrial inner membrane protein MPV17; mtDNA, mitochondrial DNA; MTFMT, mitochondrial methionyl-tRNA formyltransferase; MTHFD, methylenetetrahydrofolate dehydrogenase; MTHFD1L, methylenetetrahydrofolate dehydrogenase 1–like; NTD, neural tube defect; OXPHOS, oxidative phosphorylation; PHGDH, phosphoglycerate dehydrogenase; SAM, S-adenosylmethionine; SHMT, serine hydroxymethyltransferase; SUMO, small ubiquitin-like modifier; THF, tetrahydrofolate; TK, thymidine kinase; TYMS, thymidylate synthase; UNG, uracil-DNA N-glycosylase.
Contributor Information
Yuwen Xiu, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
Martha S Field, Email: mas246@cornell.edu, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
References
- 1. Tibbetts AS, Appling DR. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. [DOI] [PubMed] [Google Scholar]
- 2. Field MS, Kamynina E, Chon J, Stover PJ. Nuclear folate metabolism. Annu Rev Nutr. 2018;38:219–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hibbard BM, Hibbard ED, Jeffcoate TN. Folic acid and reproduction. Acta Obstet Gynecol Scand. 1965;44(3):375–400. [DOI] [PubMed] [Google Scholar]
- 4. Mills JL, Signore C. Neural tube defect rates before and after food fortification with folic acid. Birth Defect Res A. 2004;70(11):844–5. [DOI] [PubMed] [Google Scholar]
- 5. MacFarlane AJ, Stover PJ. Convergence of genetic, nutritional and inflammatory factors in gastrointestinal cancers. Nut Rev. 2007;65(12 Pt 2):157. [DOI] [PubMed] [Google Scholar]
- 6. Mason JB. Unraveling the complex relationship between folate and cancer risk. Biofactors. 2011;37(4):253–60. [DOI] [PubMed] [Google Scholar]
- 7. Kennedy DA, Stern SJ, Moretti M, Matok I, Sarkar M, Nickel C, Koren G. Folate intake and the risk of colorectal cancer: a systematic review and meta-analysis. Cancer Epidemiol. 2011;35(1):2–10. [DOI] [PubMed] [Google Scholar]
- 8. Lin HL, An QZ, Wang QZ, Liu CX. Folate intake and pancreatic cancer risk: an overall and dose-response meta-analysis. Public Health. 2013;127(7):607–13. [DOI] [PubMed] [Google Scholar]
- 9. Tio M, Andrici J, Eslick GD. Folate intake and the risk of breast cancer: a systematic review and meta-analysis. Breast Cancer Res Treat. 2014;145(2):513–24. [DOI] [PubMed] [Google Scholar]
- 10. Larsson SC, Giovannucci E, Wolk A. Folate and risk of breast cancer: a meta-analysis. J Natl Cancer Inst. 2007;99(1):64–76. [DOI] [PubMed] [Google Scholar]
- 11. Mason JB. Folate, cancer risk, and the Greek god, Proteus: a tale of two chameleons. Nutr Rev. 2009;67(4):206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Konno M, Asai A, Kawamoto K, Nishida N, Satoh T, Doki Y, Mori M, Ishii H. The one-carbon metabolism pathway highlights therapeutic targets for gastrointestinal cancer. Int J Oncol. 2017;50(4):1057–63. [DOI] [PubMed] [Google Scholar]
- 13. Goldin A, Venditti JM, Humphreys SR, Dennis D, Mantel N, Greenhouse SW. A quantitative comparison of the antileukemic effectiveness of two folic acid antagonists in mice. J Natl Cancer Inst. 1955;15(6):1657–64. [PubMed] [Google Scholar]
- 14. Gonen N, Assaraf YG. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resist Updat. 2012;15(4):183–210. [DOI] [PubMed] [Google Scholar]
- 15. Baggott JE, Tamura T. Folate-dependent purine nucleotide biosynthesis in humans. Adv Nutr. 2015;6(5):564–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yin J, Ren W, Huang X, Deng J, Li T, Yin Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front Immunol. 2018;9:1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25(1):27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gangjee A, Kurup S, Namjoshi O. Dihydrofolate reductase as a target for chemotherapy in parasites. Curr Pharm Des. 2007;13(6):609–39. [DOI] [PubMed] [Google Scholar]
- 19. Anderson DD, Stover PJ. SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS One. 2009;4(6):e5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anderson DD, Quintero CM, Stover PJ. Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc Natl Acad Sci. 2011;108(37):15163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci. 1997;94(7):3290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dianov GL, Timchenko TV, Sinitsina OI, Kuzminov AV, Medvedev OA, Salganik RI. Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Molec Gen Genet. 1991;225(3):448–52. [DOI] [PubMed] [Google Scholar]
- 23. Weeks LD, Zentner GE, Scacheri PC, Gerson SL. Uracil DNA glycosylase (UNG) loss enhances DNA double strand break formation in human cancer cells exposed to pemetrexed. Cell Death Dis. 2014;5(2):e1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stover PJ. One-carbon metabolism-genome interactions in folate-associated pathologies. J Nutr. 2009;139(12):2402–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Finkelstein JD. Methionine metabolism in mammals. J Nutr Biochem. 1990;1(5):228–37. [DOI] [PubMed] [Google Scholar]
- 26. Mudd SH, Brosnan JT, Brosnan ME, Jacobs RL, Stabler SP, Allen RH, Vance DE, Wagner C. Methyl balance and transmethylation fluxes in humans. Am J Clin Nutr. 2007;85(1):19–25. [DOI] [PubMed] [Google Scholar]
- 27. Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gómez Padilla P, Ables G, Bamman MM, Locasale JW et al. Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 2015;22(5):861–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anderson DD, Woeller CF, Chiang EP, Shane B, Stover PJ. Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J Biol Chem. 2012;287(10):7051–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacFarlane AJ, Liu X, Perry CA, Flodby P, Allen RH, Stabler SP, Stover PJ. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem. 2008;283(38):25846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sayed AR, Bourne D, Pattinson R, Nixon J, Henderson B. Decline in the prevalence of neural tube defects following folic acid fortification and its cost-benefit in South Africa. Birth Defect Res A. 2008;82(4):211–6. [DOI] [PubMed] [Google Scholar]
- 31. Oppenheim EW, Adelman C, Liu X, Stover PJ. Heavy chain ferritin enhances serine hydroxymethyltransferase expression and de novo thymidine biosynthesis. J Biol Chem. 2001;276(23):19855–61. [DOI] [PubMed] [Google Scholar]
- 32. Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem. 2002;277(41):38381–9. [DOI] [PubMed] [Google Scholar]
- 33. MacFarlane AJ, Anderson DD, Flodby P, Perry CA, Allen RH, Stabler SP, Stover PJ. Nuclear localization of de novo thymidylate biosynthesis pathway is required to prevent uracil accumulation in DNA. J Biol Chem. 2011;286(51):44015–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beaudin AE, Abarinov EV, Noden DM, Perry CA, Chu S, Stabler SP, Allen RH, Stover PJ. Shmt1 and de novo thymidylate biosynthesis underlie folate-responsive neural tube defects in mice. Am J Clin Nutr. 2011;93(4):789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Appling DR. Compartmentation of folate-mediated one-carbon metabolism in eukaryotes. FASEB J. 1991;5(12):2645–51. [DOI] [PubMed] [Google Scholar]
- 36. Barlowe CK, Appling DR. In vitro evidence for the involvement of mitochondrial folate metabolism in the supply of cytoplasmic one-carbon units. Biofactors. 1988;1(2):171–6. [PubMed] [Google Scholar]
- 37. Mackenzie CG, Frisell WR. The metabolism of dimethylglycine by liver mitochondria. J Biol Chem. 1958;232(1):417–27. [PubMed] [Google Scholar]
- 38. Davis SR, Stacpoole PW, Williamson J, Kick LS, Quinlivan EP, Coats BS, Shane B, Bailey LB, Gregory JF. Tracer-derived total and folate-dependent homocysteine remethylation and synthesis rates in humans indicate that serine is the main one-carbon donor. Am J Physiol Endocrinol Metab. 2004;286(2):E272–9. [DOI] [PubMed] [Google Scholar]
- 39. Lamers Y, Williamson J, Theriaque DW, Shuster JJ, Gilbert LR, Keeling C, Stacpoole PW, Gregory JF. Production of 1-carbon units from glycine is extensive in healthy men and women. J Nutr. 2009;139(4):666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad, Ser B. 2008;84(7):246–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salcedo E, Sims PF, Hyde JE. A glycine-cleavage complex as part of the folate one-carbon metabolism of Plasmodium falciparum. Trends Parasitol. 2005;21(9):406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad, Ser B. 2008;84(7):246–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lamers Y, Williamson J, Theriaque DW, Shuster JJ, Gilbert LR, Keeling C, Stacpoole PW, Gregory JF. Production of 1-carbon units from glycine is extensive in healthy men and women. J Nutr. 2009;139(4):666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7(4):1248–58. [DOI] [PubMed] [Google Scholar]
- 45. Heady JE, Kerr SJ. Purification and characterization of glycine N-methyltransferase. J Biol Chem. 1973;248(1):69–72. [PubMed] [Google Scholar]
- 46. Stover PJ, Chen LH, Suh JR, Stover DM, Keyomarsi K, Shane B. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J Biol Chem. 1997;272(3):1842–8. [DOI] [PubMed] [Google Scholar]
- 47. Sugiura T, Nagano Y, Inoue T, Hirotani K. A novel mitochondrial C1-tetrahydrofolate synthetase is upregulated in human colon adenocarcinoma. Biochem Biophys Res Commun. 2004;315(1):204–11. [DOI] [PubMed] [Google Scholar]
- 48. Walkup AS, Appling DR. Enzymatic characterization of human mitochondrial C1-tetrahydrofolate synthase. Arch Biochem Biophys. 2005;442(2):196–205. [DOI] [PubMed] [Google Scholar]
- 49. García-Martínez LF, Appling DR. Characterization of the folate-dependent mitochondrial oxidation of carbon 3 of serine. Biochemistry. 1993;32(17):4671–6. [DOI] [PubMed] [Google Scholar]
- 50. Li Y, Holmes WB, Appling DR, RajBhandary UL. Initiation of protein synthesis in Saccharomyces cerevisiae mitochondria without formylation of the initiator tRNA. J Bacteriol. 2000;182(10):2886–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Samsonoff WA, Reston J, McKee M, O'Connor B, Galivan J, Maley G, Maley F. Intracellular location of thymidylate synthase and its state of phosphorylation. J Biol Chem. 1997;272(20):13281–5. [DOI] [PubMed] [Google Scholar]
- 52. Zhou X, Solaroli N, Bjerke M, Stewart JB, Rozell B, Johansson M, Karlsson A. Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum Mol Genet. 2008;17(15):2329–35. [DOI] [PubMed] [Google Scholar]
- 53. Pontarin G, Ferraro P, Valentino ML, Hirano M, Reichard P, Bianchi V. Mitochondrial DNA depletion and thymidine phosphate pool dynamics in a cellular model of mitochondrial neurogastrointestinal encephalomyopathy. J Biol Chem. 2006;281(32):22720–8. [DOI] [PubMed] [Google Scholar]
- 54. Nishigaki Y, Marti R, Hirano M. ND5 is a hot-spot for multiple atypical mitochondrial DNA deletions in mitochondrial neurogastrointestinal encephalomyopathy. Hum Mol Genet. 2003;13(1):91–101. [DOI] [PubMed] [Google Scholar]
- 55. Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283(5402):689–92. [DOI] [PubMed] [Google Scholar]
- 56. Alonzo JR, Venkataraman C, Field MS, Stover PJ. The mitochondrial inner membrane protein MPV17 prevents uracil accumulation in mitochondrial DNA. J Biol Chem. 2018;293(52):20285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martiniova L, Field MS, Finkelstein JL, Perry CA, Stover PJ. Maternal dietary uridine causes, and deoxyuridine prevents, neural tube closure defects in a mouse model of folate-responsive neural tube defects. Am J Clin Nutr. 2015;101(4):860–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fenech M. Folate (vitamin B9) and vitamin B12 and their function in the maintenance of nuclear and mitochondrial genome integrity. Mutat Res. 2012;733(1–2):21–33. [DOI] [PubMed] [Google Scholar]
- 59. Branda RF, Brooks EM, Chen Z, Naud SJ, Nicklas JA. Dietary modulation of mitochondrial DNA deletions and copy number after chemotherapy in rats. Mutat Res. 2002;501(1–2):29–36. [DOI] [PubMed] [Google Scholar]
- 60. Chou YF, Yu CC, Huang RF. Changes in mitochondrial DNA deletion, content, and biogenesis in folate-deficient tissues of young rats depend on mitochondrial folate and oxidative DNA injuries. J Nutr. 2007;137(9):2036–42. [DOI] [PubMed] [Google Scholar]
- 61. Chou YF, Huang RF. Mitochondrial DNA deletions of blood lymphocytes as genetic markers of low folate-related mitochondrial genotoxicity in peripheral tissues. Eur J Nutr. 2009;48(7):429–36. [DOI] [PubMed] [Google Scholar]
- 62. Crott JW, Choi SW, Branda RF, Mason JB. Accumulation of mitochondrial DNA deletions is age, tissue and folate-dependent in rats. Mutat Res. 2005;570(1):63–70. [DOI] [PubMed] [Google Scholar]
- 63. Chen TF, Chiu MJ, Huang CT, Tang MC, Wang SJ, Wang CC, Huang RF. Changes in dietary folate intake differentially affect oxidised lipid and mitochondrial DNA damage in various brain regions of rats in the absence/presence of intracerebroventricularly injected amyloid β-peptide challenge. Br J Nutr. 2011;105(9):1294–302. [DOI] [PubMed] [Google Scholar]
- 64. Kronenberg G, Gertz K, Overall RW, Harms C, Klein J, Page MM, Stuart JA, Endres M. Folate deficiency increases mtDNA and D-1 mtDNA deletion in aged brain of mice lacking uracil-DNA glycosylase. Exp Neurol. 2011;228(2):253–8. [DOI] [PubMed] [Google Scholar]
- 65. Antonenkov VD, Isomursu A, Mennerich D, Vapola MH, Weiher H, Kietzmann T, Hiltunen JK. The human mitochondrial DNA depletion syndrome gene MPV17 encodes a non-selective channel that modulates membrane potential. J Biol Chem. 2015;290(22):13840–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. El-Hattab AW, Craigen WJ, Scaglia F. Mitochondrial DNA maintenance defects. Biochim Biophys Acta Mol Basis Dis. 2017;1863(6):1539–55. [DOI] [PubMed] [Google Scholar]
- 67. Kalko SG, Paco S, Jou C, Rodríguez MA, Meznaric M, Rogac M, Jekovec-Vrhovsek M, Sciacco M, Moggio M, Jimenez-Mallebrera C et al. Transcriptomic profiling of TK2 deficient human skeletal muscle suggests a role for the p53 signalling pathway and identifies growth and differentiation factor-15 as a potential novel biomarker for mitochondrial myopathies. BMC Genomics. 2014;15:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nikkanen J, Forsström S, Euro L, Paetau I, Kohnz RA, Wang L, Chilov D, Viinamäki J, Roivainen A, Suomalainen A et al. Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab. 2016;23(4):635–48. [DOI] [PubMed] [Google Scholar]
- 69. Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Haigis MC et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 2016;24(1):104–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Sabatini DM et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520(7547):363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tucker EJ, Hershman SG, Köhrer C, Belcher-Timme CA, Patel J, Goldberger OA, Christodoulou J, Silberstein JM, McKenzie M, Mootha VK et al. Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab. 2011;14(3):428–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morscher RJ, Ducker GS, Li SH, Mayer JA, Gitai Z, Sperl W, Rabinowitz JD. Mitochondrial translation requires folate-dependent tRNA methylation. Nature. 2018;554(7690):128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lucas S, Chen G, Aras S, Wang J. Serine catabolism is essential to maintain mitochondrial respiration in mammalian cells. Life Sci Alliance. 2018;1(2):e201800036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510(7504):298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang L, Garcia Canaveras JC, Chen Z, Wang L, Liang L, Jang C, Mayr JA, Zhang Z, Ghergurovich JM, Rabinowitz JD et al. Serine catabolism feeds NADH when respiration is impaired. Cell Metab. 2020;31(4):809–21.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Azize NA, Ngah WZ, Othman Z, Md Desa N, Chin CB, Md Yunus Z, Mohan A, Hean TS, Syed Zakaria SZ, Lock-Hock N. Mutation analysis of glycine decarboxylase, aminomethyltransferase and glycine cleavage system protein-H genes in 13 unrelated families with glycine encephalopathy. J Hum Genet. 2014;59(11):593–7. [DOI] [PubMed] [Google Scholar]
- 78. Minton DR, Nam M, McLaughlin DJ, Shin J, Bayraktar EC, Alvarez SW, Sviderskiy VO, Papagiannakopoulos T, Sabatini DM, Birsoy K et al. Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of formylmethionyl-tRNAs. Mol Cell. 2018;69(4):610–21.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yasukawa T, Suzuki T, Ishii N, Ueda T, Ohta S, Watanabe K. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNA(Lys) with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 2000;467(2–3):175–8. [DOI] [PubMed] [Google Scholar]
- 80. Yasukawa T, Suzuki T, Ueda T, Ohta S, Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem. 2000;275(6):4251–7. [DOI] [PubMed] [Google Scholar]
- 81. Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3(3):187–97. [DOI] [PubMed] [Google Scholar]
- 82. Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Thompson CB et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4(12):1406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Martínez-Reyes I, Chandel NS. Mitochondrial one-carbon metabolism maintains redox balance during hypoxia. Cancer Discov. 2014;4(12):1371–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yue L, Pei Y, Zhong L, Yang H, Wang Y, Zhang W, Chen N, Zhu Q, Gao J, Han J et al. Mthfd2 modulates mitochondrial function and DNA repair to maintain the pluripotency of mouse stem cells. Stem Cell Rep. 2020;15(2):529–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Mootha VK et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife. 2016;5:e10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Bardeesy N et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539(7629):390–5.. Erratum in: Nature 2019;575(7783):E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Reina-Campos M, Diaz-Meco MT, Moscat J. The complexity of the serine glycine one-carbon pathway in cancer. J Cell Biol. 2020;219(1):e201907022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rosenzweig A, Blenis J, Gomes AP. Beyond the Warburg effect: how do cancer cells regulate one-carbon metabolism?. Front Cell Dev Biol. 2018;6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7(4):1248–58. [DOI] [PubMed] [Google Scholar]
- 91. Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Vander Heiden MG et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Sabatini DM et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Reich S, Nguyen CDL, Has C, Steltgens S, Soni H, Coman C, Freyberg M, Bichler A, Seifert N, Medenbach J et al. A multi-omics analysis reveals the unfolded protein response regulon and stress-induced resistance to folate-based antimetabolites. Nat Commun. 2020;11(1):2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mehrmohamadi M, Liu X, Shestov AA, Locasale JW. Characterization of the usage of the serine metabolic network in human cancer. Cell Rep. 2014;9(4):1507–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tedeschi PM, Vazquez A, Kerrigan JE, Bertino JR. Mitochondrial methylenetetrahydrofolate dehydrogenase (MTHFD2) overexpression is associated with tumor cell proliferation and is a novel target for drug development. Mol Cancer Res. 2015;13(10):1361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu F, Liu Y, He C, Tao L, He X, Song H, Zhang G. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumor Biol. 2014;35(9):8685–90. [DOI] [PubMed] [Google Scholar]
- 98. Ning S, Ma S, Saleh AQ, Guo L, Zhao Z, Chen Y. SHMT2 overexpression predicts poor prognosis in intrahepatic cholangiocarcinoma. Gastroenterol Res Pract. 2018;2018:4369253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liu Y, Yin C, Deng MM, Wang Q, He XQ, Li MT, Li CP, Wu H. High expression of SHMT2 is correlated with tumor progression and predicts poor prognosis in gastrointestinal tumors. Eur Rev Med Pharmacol Sci. 2019;23(21):9379–92. [DOI] [PubMed] [Google Scholar]
- 100. Tani H, Ohnishi S, Shitara H, Mito T, Yamaguchi M, Yonekawa H, Hashizume O, Ishikawa K, Nakada K, Hayashi JI. Mice deficient in the Shmt2 gene have mitochondrial respiration defects and are embryonic lethal. Sci Rep. 2018;8(1):425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. DebRoy S, Kramarenko II, Ghose S, Oleinik NV, Krupenko SA, Krupenko NI. A novel tumor suppressor function of glycine N-methyltransferase is independent of its catalytic activity but requires nuclear localization. PLoS One. 2013;8(7):e70062. [DOI] [PMC free article] [PubMed] [Google Scholar]