Abstract
Acquisition of a lipid-laden phenotype by immune cells has been defined in infectious diseases and atherosclerosis, but remains largely uncharacterized in cancer. Here, in breast cancer models we found that neutrophils are induced to accumulate neutral lipids upon interaction with resident mesenchymal cells (MCs) in the pre-metastatic lung. Lung MCs elicit this process through repressing the adipose triglyceride lipase (ATGL) activity in neutrophils in prostaglandin E2-dependent and -independent manners. In vivo, neutrophil-specific deletion of genes encoding ATGL or ATGL inhibitory factors altered neutrophil lipid profiles and breast tumor lung metastasis in mice. Mechanistically, lipids stored in lung neutrophils are transported to metastatic tumor cells through a macropinocytosis-lysosome pathway, endowing the tumor cells with augmented survival and proliferative capacities. Pharmacological inhibition of macropinocytosis significantly reduced metastatic colonization by breast tumor cells in vivo. Collectively, our work reveals that neutrophils serve as an energy reservoir to fuel breast cancer lung metastasis.
Introduction
Metastatic disease remains the major cause of cancer related death. Among the vital organs to which solid tumors metastasize, the lung is one of the most common sites. In the past two decades, significant advances in our understanding of lung metastasis have revealed intricate interactions between disseminated tumor cells (DTCs) and the lung resident immune microenvironment that are essential for the development of metastatic lung lesions1, 2. Within the lung immune microenvironment, bone marrow (BM)-derived neutrophils have been reported as an indispensable component which facilitates solid tumor metastasis1, 2. Accumulating evidence suggests that neutrophils regulate lung metastasis as tumor cells colonize the lung, where neutrophils function to suppress anti-tumor immunity, accelerate DTC extravasation and proliferation, and awaken dormant DTCs via neutrophil extracellular traps (NETs)3, 4, 5, 6. Through these effects, neutrophils act in concert with other organ-resident stromal cells contributing to formation of the pre-metastatic and metastatic niches1, 2. However, the DTC supportive effects of neutrophils and other lung resident stromal cells were mostly studied at transcriptional and protein levels, and the metabolic crosstalk between the lung microenvironment and DTCs is largely undefined.
In fact, solid tumor metastasis is a highly inefficient process7. DTCs need to undergo metabolic alterations to adapt to the new environment and successfully survive and colonize the metastatic sites8, 9. Recent studies showed that DTCs can fully benefit from local resources including the metabolic energetics of the organ microenvironment10, 11. In ovarian cancer metastasis to omentum, ovarian cancer cells are capable of acquiring lipids from omental adipocytes to support their proliferation through accelerated β-oxidation11. In colon cancer liver metastasis, colon cancer cells take up extracellular phosphocreatine in the liver microenvironment to generate adenosine triphosphate for their metastatic survival10. Therefore, the organ microenvironment serves to metabolically support DTCs during metastasis, however, a metabolic-based regulatory role for immune cells in this context has not been systematically examined.
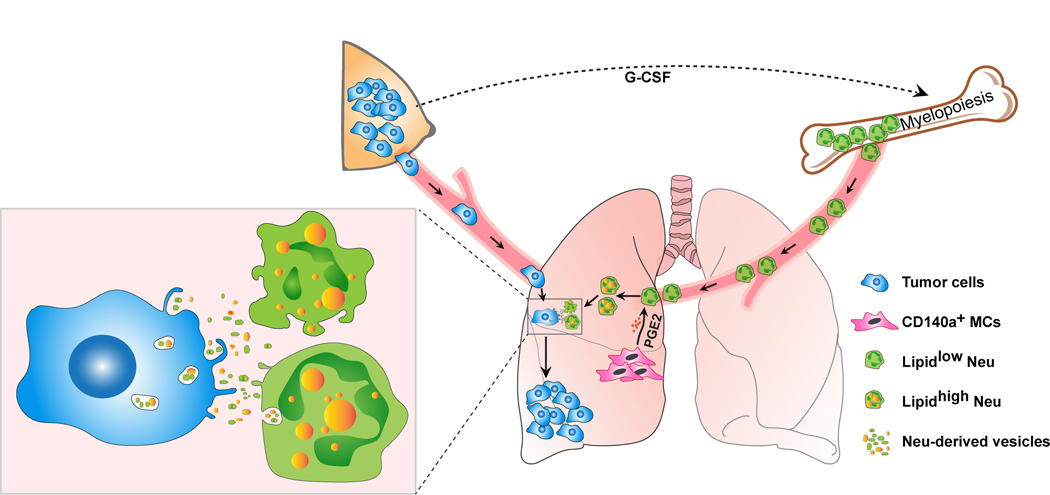
In this study, we show that lung-infiltrating neutrophils function as a nutrient source to fuel DTCs during lung metastasis of breast cancer in mouse models. Neutrophils potently accumulate neutral lipids starting from the pre-metastatic stage, which is stimulated by the lung resident mesenchymal cells (MCs). Upon metastatic breast tumor cells colonize the lung, they will take up the lipids from the lung neutrophils and acquire elevated levels of survival and proliferation. Our results therefore revealed an unrecognized role of neutrophils to metabolically regulate breast cancer lung metastasis.
Results
Neutrophils have a lipid-laden phenotype in the lung
Preceding tumor metastasis, in response to tumor- and host-derived factors, organ-infiltrating neutrophils are generated in hematopoietic tissues and organs such as bone marrow (BM) and spleen12. While neutrophils have been extensively studied for their metastasis-modulating effects, it remains largely unknown whether these effects are intrinsic or if they are acquired as neutrophils transit specific tissue environments. Using the mouse 4T1 orthotopic breast tumor model, we compared the transcriptional profiles of neutrophils isolated from BM, peripheral blood (PB) and lung by RNA sequencing (RNA-seq). At the pre-metastatic stage, a fundamental difference in gene expression was detected between lung neutrophils and those isolated from BM or PB (Fig. 1a and Extended Data Fig. 1a, b).
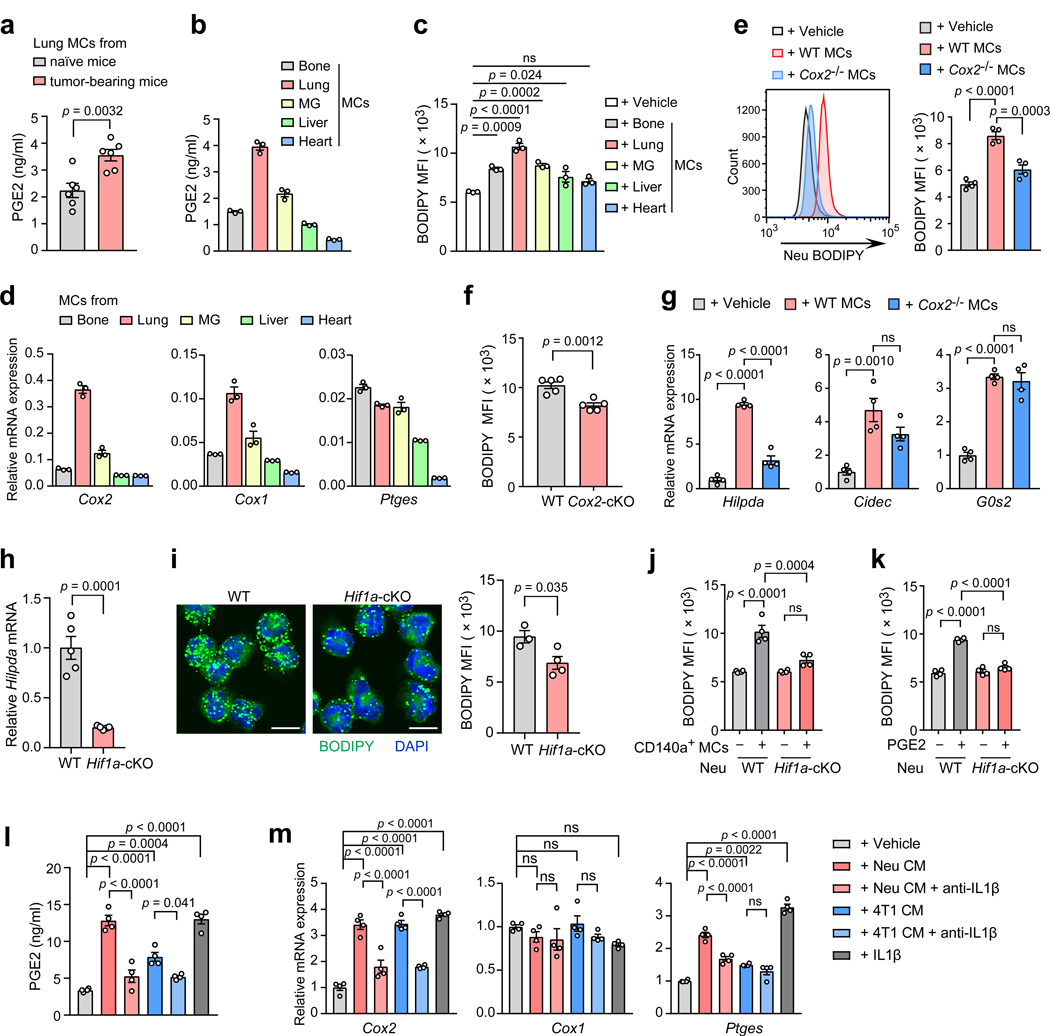
Fig. 1. Neutrophils acquire a lipid-laden phenotype in the pre-metastatic lung.
a, Volcano plots showing fold-change and p-value for the comparison of lung neutrophils versus PB neutrophils based on the RNA-seq data from the 4T1 orthotopic model (n = 3). b, Intracellular lipids in neutrophils detected under confocal microscopy (upper; scale bars, 5 μm) or transmission electron microscopy (lower; scale bars, 0.5 μm). Arrowheads indicate lipid droplets (LDs) (representative of n = 3 mice). c, Immunostaining of lung section of MMTV-PyMT mice showing in situ LDs (BODIPY) in neutrophils (Ly6G). Scale bars, 5 μm. (representative of n = 3 mice). d, Lipid quantification in neutrophils by flow cytometry (n = 6). e, Cellular triglyceride (TG) contents in neutrophils (n=5). f, Relative levels of the indicated types of lipids in PB and lung neutrophils measured by mass spectrometry (n = 5). See Extended Data Fig. 2a–h. g, A diagram showing ATGL and its regulatory factors in TG hydrolysis. h, Relative mRNA expression (as compared to the control gene Rps18, encoding ribosomal protein S18) of the indicated genes in neutrophils determined by qRT-PCR (n = 5). i, The indicated protein expression in neutrophils detected by Western blotting with GAPDH as a loading control (n = 3). j, Correlation between expression of human LD-associated genes and that of neutrophil signature genes in human breast cancer lung metastasis samples (GSE14018, n = 16), analyzed by linear regression with Pearson’s correlation (See Methods). k, Relative TG hydrolase activity in neutrophils (n = 6 and 5 for PB and lung, respectively). l, Cellular lipase activity in neutrophils (n = 5). In b, d, e, h, i, k and l, neutrophils (Neu) were isolated from the indicated tissues from tumor-bearing MMTV-PyMT mice at the pre-metastatic stage. n represents biologically independent animals except j for independent human patients. Data are mean ± s.e.m. and P values in animal experiments were determined by one-way ANOVA with Tukey’s multiple comparisons test except a and k by unpaired two-tailed t-test. ns, not significant.
Two lipid droplet (LD)-associated genes, Hilpda and G0s213, 14, were listed among the top differential genes in lung neutrophils versus BM neutrophils (Extended Data Fig. 1a) or PB neutrophils (Fig. 1a), whereas their expression levels were comparable between BM and PB neutrophils (Extended Data Fig. 1b). Further inspection of the genes involving in the main lipid metabolic pathways15 (Extended Data Fig. 1c) revealed that a portion of genes associated with lipid absorption and LDs formation, but not LDs degradation and β-oxidation, are expressed at higher levels in lung neutrophils in comparison to their BM and PB counterparts (Extended Data Fig. 1d). Therefore, neutrophils tend to upregulate lipid storage genes in the pre-metastatic lung.
Consistent with the gene expression profiles, neutrophils isolated from, or residing in the pre-metastatic lungs, were indeed found to contain a greater abundance of LD-like structures than those from BM and PB in both orthotopic 4T1 model (Extended Data Fig. 1e, f) and the genetically engineered MMTV-PyMT metastatic mammary cancer model (Fig. 1b, c). Quantitatively, the total lipids in lung neutrophils were also much higher than in neutrophils isolated from other tissues including BM, PB, spleen and primary tumors in both the orthotopic 4T1 model (Extended Data Fig. 1g, h) and the MMTV-PyMT model (Fig. 1d). The lung-associated lipid-laden phenotype of neutrophils was not only detected under tumor-bearing conditions, but it also existed in tumor-free naïve mice though the contrast observed between lung neutrophils and their counterparts in other tissues was less marked (Extended Data Fig. 1h). Hence lung resident neutrophils intrinsically tend to accumulate lipids, which is further reinforced during breast cancer progression.
As the major intracellular organelles storing neutral lipids, especially triglyceride (TG), LDs are mostly studied in adipocytes and have recently been recognized as a critical metabolic player in other types of cells such as macrophages in infectious diseases and atherosclerosis16, 17. In accord with their high LD content, lung neutrophils contain higher levels of TG, than their BM and PB counterparts in both the MMTV-PyMT and the orthotopic 4T1 models (Fig. 1e and Extended Data Fig. 1i). Additional mass spectrometric analysis revealed that all TG species were detected at much higher levels in lung neutrophils than in PB neutrophils, while lipid types other than TG did not show such a notable difference (Fig. 1f and Extended Data Fig. 2a–h). Thus, neutrophils acquire a TG-enriched lipid-laden phenotype in the pre-metastatic lung.
By probing into the TG metabolic genes from the RNA-seq data, the genes governing TG hydrolysis were found to differ greatly between lung neutrophils and BM or PB neutrophils (Extended Data Fig. 1d). Hydrolysis of TG is primarily catalyzed by adipose triglyceride lipase (ATGL), which is regulated by its activator protein α-β hydrolase domain-containing protein 5 (ABHD5) and its inhibitory factors HILPDA, CIDEC and G0S218, 19 (Fig. 1g). At both transcriptional and protein levels, all three ATGL inhibitory factors HILPDA, CIDEC and G0S2 were each more highly expressed in lung neutrophils compared to those isolated from BM or PB in both the MMTV-PyMT (Fig. 1h, i) and the 4T1 models (Extended Data Fig. 2i, j). In contrast, the genes encoding ATGL and its activator ABHD5 did not show such a tissue-specific difference in expression (Fig. 1h and Extended Data Fig. 2i). By analyzing a transcriptome dataset (GSE14018) derived from human breast cancer lung metastasis samples20, the expression of HILPDA, CIDEC and G0S2 was also found to be positively correlated with that of neutrophil signature genes21 (Fig. 1j) suggesting that human neutrophils may undergo similar metabolic changes during lung metastatic progression of human breast cancer.
As a result of the high expression of ATGL inhibitory factors, the TG hydrolase activity of lung neutrophils was expectedly lower than that in PB neutrophils (Fig. 1k for the MMTV-PyMT model; Extended Data Fig. 2k for the 4T1 model). A similar pattern was also shown for the total lipase activity (Fig. 1l and Extended Data Fig. 2l). Therefore, the lipid-laden phenotype in lung neutrophils is likely caused by their elevated expression of ATGL inhibitory factors, which in turn repress their TG hydrolysis leading to intracellular TG accumulation.
Lung resident mesenchymal cells (MCs) drive neutrophils to accumulate lipids
To directly determine if the lung is a unique site for neutrophils to accumulate lipids, we intravenously implanted fluorescent dye-labeled, BM-isolated neutrophils. In both tumor-free and tumor-bearing (pre-metastatic stage) recipient mice, the implanted neutrophils that infiltrated the lung, but not other tissues, substantially increased their lipid levels (Fig. 2a). We then ex vivo co-cultured various types of lung stromal cells with BM-isolated neutrophils to identify whether certain lung stromal cells drive neutrophils to accumulate lipids. Among the lung stromal cells examined, the CD140a+ MCs were found to significantly upregulate the neutrophil lipids (Fig. 2b). By immunostaining of the pre-metastatic lung sections from MC-reporter CD140a-GFP mice, we observed a close proximity of in situ neutrophils to lung MCs within the lung interstitium (Fig. 2c). Hence, when neutrophils infiltrate the lung interstitium, they could be elicited by their proximal resident MCs to start the process of lipid storage.
Fig. 2. Lung CD140a+ mesenchymal cells (MCs) drive lipid accumulation in neutrophils.
a, Comparison of the lipid levels in the implanted neutrophils in the indicated organs or tissues of recipient naïve or 4T1 tumor-bearing mice (see Methods, n = 6). b, BM neutrophils (Neu) were mono-cultured or co-cultured with the indicated stromal cells sorted from naïve BALB/cJ mouse lungs. Lipid levels of neutrophils were quantitated by flow cytometry 16 hours later. c, Immunostaining of lung section showing the localization of MCs and neutrophils in AT3-g-csf orthotopic tumor-bearing CD140a-GFP mice (pre-metastatic stage) (representative of n = 4 mice). Receptor for advanced glycation end products (RAGE) was also stained to indicate the lung epithelium. Scale bar, 20 μm. d, Relative mRNA expression of the indicated genes in BM neutrophils after their mono-culture or co-culture with CD140a+ lung MCs or lung MC-conditioned medium. e, An upstream regulator (soluble proteins or chemicals) prediction was performed using the ingenuity pathway analysis based on the differentially expressed genes between lung and BM neutrophils, using the RNA-seq data as in Extended Data Fig. 1a. f, The lipid levels of BM neutrophils stimulated with the indicated candidate regulators, as measured by flow cytometry (see Methods). g, Relative mRNA expression of the indicated genes in BM neutrophils stimulated with PGE2 (10 ng ml−1). h, The lipid levels of BM neutrophils co-cultured with CD140a+ MCs in the presence and absence of the PGE2 receptor (EP) antagonists were quantitated by flow cytometry (see Methods). i, Measurement of the lung neutrophil lipid levels in 4T1 tumor-bearing mice without and with administration with the EP2 antagonist (PF-04418948, 10 mg kg−1) (n = 6). The BM neutrophils used in b, d and f-h were isolated from the 4T1 tumor-bearing BALB/cJ mice (n = 4 except g with n = 3). n represents biologically independent animals. Data are mean ± s.e.m. and P values were determined by one-way ANOVA with Tukey’s multiple comparisons test except g and i by unpaired two-tailed t-test. ns, not significant.
As HILPDA, CIDEC and G0S2 were among the most up-regulated TG-regulating factors in lung neutrophils, we speculated whether lung MCs trigger neutrophils to accumulate lipids through regulating these ATGL inhibitory factors. Indeed, lung MCs were able to stimulate the expression of all three genes encoding ATGL inhibitory factors, while the lung MC-conditioned medium could only up-regulate Hilpda expression in neutrophils (Fig. 2d). Therefore, lung MCs modulate the lipid metabolic gene expression in neutrophils through both soluble factor(s) and cell-cell contact dependent mechanisms.
Through a screening for the possible lung MC-derived candidate soluble factors (Fig. 2e), as predicted by ingenuity pathway analysis of the RNA-seq data from lung versus BM neutrophils in the 4T1 model, prostaglandin E2 (PGE2) was found to be superior to other candidate factors in elevating the neutrophil lipid level (Fig. 2f). Consistently, PGE2 was able to upregulate Hilpda but not Cidec or G0s2 confirming that PGE2 is at least one soluble factor mechanism underlying lung MCs-triggered neutrophil lipid-laden phenotype (Fig. 2g). PGE2, a bioactive lipid belonging to the family of eicosanoids, has been extensively reported to play multifaceted roles in inflammation and cancer22. PGE2 exerts its effects through binding to PGE2 receptors (EP1-4) expressed on the target cell surface23. By further ex vivo and in vivo work, EP2 was determined as a primary PGE2 receptor to respond to lung MCs (Fig. 2h, i). Taken together, PGE2 was revealed as a possible lung MC-derived soluble factor which modulates the lipid profiles in neutrophils.
A PGE2-HIF1α-HILPDA axis is revealed for lung MC-driven neutrophil lipid accumulation
To better understand the role of PGE2 in endowing the lung-associated neutrophil lipid-laden phenotype, we next characterized the PGE2 production by different tissue-derived MCs. Lung MCs isolated from naive mice, were shown to intrinsically produce a considerable amount of PGE2 (1~4 ng per 105 cells in 24 hours), whereas this capacity was further increased under tumor-bearing conditions (Fig. 3a). Furthermore, lung MCs produced apparently higher levels of PGE2 than those isolated from the other tissues examined, including bone, mammary gland, liver and heart (Fig. 3b). This differential pattern in PGE2 production among tissue-specific MCs, as expected, corresponded well with their abilities to stimulate neutrophils for lipid accumulation (Fig. 3c).
Fig. 3. A critical role of the PGE2-HIF1α-HILPDA axis in lung MC-triggered lipid storage in neutrophils.
a, The PGE2 levels in lung MCs (105 cells per ml in 24 hours) from naïve and AT3-g-csf tumor bearing CD140a-GFP mice (n = 6). b-d, The CD140a+ MCs were freshly isolated from the indicated tissues of naïve CD140a-GFP mice (n = 3). Their PGE2 secretion (b), abilities to stimulate lipid accumulation in BM neutrophils (c), and relative mRNA expression (to Rps18) of the indicated genes (d) were assessed. MG, mammary gland. e-g, Lung MC-specific COX2 in eliciting the lipid-laden neutrophil phenotype. In vitro, BM neutrophils were mono-cultured or co-cultured with lung MCs isolated from WT or Cox2-cKO (CD140a-Cre; Cox2flox/flox) mice. Then the lipid levels (e) and relative mRNA expression of the indicated genes (g) in neutrophils were determined. The BM neutrophils were isolated from the AT3-g-csf tumor-bearing mice (n = 4). In vivo, the lipid levels in lung neutrophils isolated from the AT3-g-csf tumor-bearing WT and Cox2-cKO mice (n = 5) were compared (f). h, Relative mRNA levels of Hilpda in lung neutrophils isolated from AT3-g-csf tumor-bearing WT and Hif1a-cKO (S100a8-Cre; Hif1aflox/flox) mice (n = 5). i, Lipid levels in lung neutrophils from AT3-g-csf-bearing WT (n = 3) and Hif1a-cKO (n = 4) mice, were detected by confocal microscopy (left; scale bars, 5 μm) or flow cytometry (right). j, k, BM neutrophils isolated from AT3-g-csf tumor-bearing WT or Hif1α-cKO mice (n = 4) were mono-cultured or co-cultured with lung MCs (j), or stimulated with exogenous PGE2 (10 ng ml−1) (k). Then the lipid levels in neutrophils were determined. l, m, Lung MCs were cultured with the indicated conditioned medium (CM) or IL-1β (20 ng ml−1), with and without anti-IL1β (1 μg ml−1). The PGE2 levels (l) and the relative mRNA expression of the indicated genes (m) were measured. The lung MCs were isolated from naïve CD140a-GFP mice (n = 4). n represents biologically independent animals. Data are mean ± s.e.m. and P values were determined by one-way ANOVA with Tukey’s multiple comparisons test except a, f, h, and i by unpaired two-tailed t-test. ns, not significant.
PGE2 is synthesized from arachidonic acid via the actions of several key enzymes including cyclooxygenase 2 (COX2), cyclooxygenase 1 (COX1) and prostaglandin E synthases (PTGES)24. Among the three enzyme-encoding genes, Cox2 and Cox1, but not Ptges, in lung MCs were expressed at higher levels than in those isolated from other tissues examined (Fig. 3d). In light of the essential functions of COX2 in inflammation and cancer22, 25, we next characterized the role of mesenchymal cell-specific COX2 in the lung pre-metastatic niche. In vitro, Cox2 ablation in lung MCs indeed reduced their capacity to stimulate lipid accumulation in neutrophils (Fig. 3e). In vivo, conditional knockout (cKO) of Cox2 in CD140a+ MCs significantly alleviated the lipid-laden phenotype of lung neutrophils (Fig. 3f). Thus, lung MC Cox2-PGE2 signaling is critical for inducing the neutrophil lipid-laden phenotype.
Among the three ATGL inhibitory factors, only Hilpda was found to be dependent on the lung MC Cox2-PGE2 signaling (Fig. 3g). HILPDA, also known as hypoxia-inducible gene 2 (HIG2), was recently reported as a hypoxia-induced endogenous inhibitor of ATGL in cancer cells and macrophages26, 27. Conditional knockout of hypoxia-inducible factor 1-alpha (Hif1a), the master transcriptional regulator of cellular response to hypoxia28, in neutrophils (S100a8-Cre; Hif1a flox/flox)29 led to diminished expression of Hilpda (Fig. 3h) and mitigated lipid-laden phenotype in lung neutrophils (Fig. 3i). Ex vivo, loss of Hif1a in neutrophils abated their lipid accumulating capacity in response to lung MCs or exogenous PGE2 (Fig. 3j, k). Collectively, a COX2-PGE2 → HIF1α→ HILPDA axis was revealed as a mechanism linking lung MCs to lipid-laden lung neutrophils.
Lastly, we sought to determine how tumor-bearing conditions upregulate PGE2 production in lung MCs. It is well known that tumor cells themselves and the host immune cells can remotely regulate organ stromal cells via soluble factors2. Indeed, tumor cell and lung neutrophil-derived conditioned media were able to upregulate Cox2 gene expression and PGE2 production in lung MCs (Fig. 3l, m). To identify such tumor cell- and/or neutrophil- derived soluble factor(s), we focused on inflammatory cytokines that are known to be pivotal in formation of the pre-metastatic niche1, 2. Reexamination of the neutrophil RNA-seq data (Extended Data Fig. 1a) revealed that Il1b was one of the most highly expressed inflammatory cytokine genes in lung neutrophils versus BM neutrophils. Neutralization of IL1β indeed reduced neutrophil or tumor cell CM-induced Cox2 expression and PGE2 production in lung MCs ex vivo (Fig. 3l, m). Addition of exogenous IL1β was similarly effective to the CMs in induction of Cox2 and PGE2 in lung MCs (Fig. 3l, m). Therefore a neutrophil-IL1β → lung MC-PGE2 → neutrophil-lipids amplifying loop serves to reinforce the neutrophil lipid-laden phenotype in the lung pre-metastatic niche.
Lipids stored in neutrophils function to promote lung metastasis of breast cancer
LDs formation has been documented in macrophages in infectious diseases and atherosclerosis in which LDs serve as an energy source for pathogens, elicit inflammatory responses by producing eicosanoids and mediate atherosclerotic plaque formation17, 30. To understand the functional consequence of LDs formation in pre-metastatic lung neutrophils, we specifically deleted the LD-associated genes, including Atgl and ATGL inhibitory genes Hilpda and Cidec, in neutrophils. Upon crossing S100a8-Cre driver mice targeting neutrophils29, 31 with genetically modified mice that have loxP-flanked Atgl, Hilpda or Cidec, the resultant cKO strains (S100a8-Cre; Atglflox/flox; S100a8-Cre; Hilpdaflox/flox; and S100a8-Cre; Cidecflox/flox) were exploited in the orthotopic AT3-g-csf tumor model. The AT3-g-csf line is based on the murine breast cancer cell line AT3 originated from a MMTV-PyMT tumor in the C57BL/6J background32, and further constructed to overexpress granulocyte-colony stimulating factor (G-CSF) for induction of the host inflammatory condition6 at a similar level to the 4T1 model.
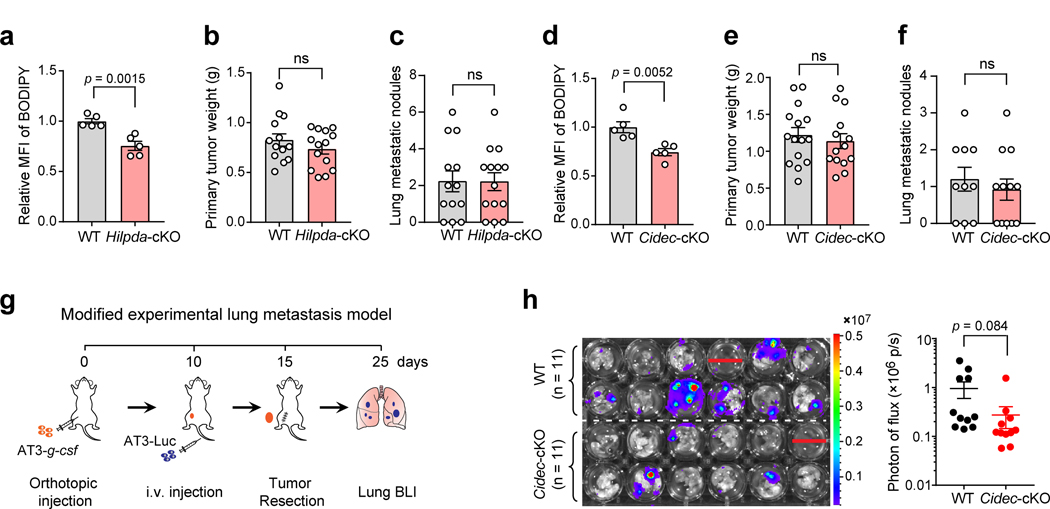
Corresponding to the elevated lipid level in lung neutrophils (Fig. 4a), more robust spontaneous lung metastases were detected in Atgl-cKO mice, compared to their wild type (WT) controls, while the primary tumor growth remained unaltered (Fig. 4b, c). In contrast, genetic ablation of either Hilpda or Cidec in neutrophils was not sufficient to influence the spontaneous lung metastases although each deficiency did cause a significant reduction of lipid levels in lung neutrophils (Extended Data Fig. 3a–f). We reasoned that the three ATGL inhibitory factors in neutrophils may compensate each other for their tumor-modulating effects when a single factor was defective, while loss of Atgl in neutrophils caused fundamental alterations of their lipid-laden phenotype and metastasis-regulating ability.
Fig. 4. Genetic ablation of Atgl in host neutrophils leads to more aggressive lung metastasis of breast cancer.
a, Left: Representative images showing LDs in lung neutrophils from AT3-g-csf-bearing WT and Atgl-cKO mice. Scale bars, 5 μm. Right: Lipid levels of neutrophils from above mice (n = 6) were quantitated by flow cytometry. b, c, WT (n = 15) and Atgl-cKO mice (n = 16) were orthotopically implanted with AT3-g-csf cells. The primary tumors were resected and weighed on day 15 (n = 10) (b). At the end point (day 30), spontaneous lung metastases were assessed. Representative histological images of the lung sections stained with H&E are shown, and arrowheads indicate metastatic lesions (c, left). Scale bars, 1 mm. The number of metastatic nodules on the lungs were counted (c, right). d-f, Following the modified experimental lung metastasis model (see Extended Data Fig. 3g and Methods), the lung metastatic progression of AT3-Luc cells in Atgl-cKO recipient mice (d) and Hilpda-cKO recipient mice (e), as well as E0771-Luc cells in Atgl-cKO recipient mice (f) was determined by ex vivo bioluminescence imaging (BLI), with their WT littermates as controls. Representative BLI images of the harvested lungs are shown (d-f, left). n = 11 (WT) and 10 (Atgl-cKO) for d, n = 13 (WT) and 14 (Hilpda-cKO) for e, and n =10 per group for f. g, Comparison of the spontaneous lung metastases developed in MMTV-PyMT; Atgl-cKO mice (n = 8) and their WT littermates (MMTV-PyMT mice) (n = 9). Representative histological lung sections stained with H&E are shown, and arrowheads indicate metastatic nodules. Scale bars, 1 mm. n represents biologically independent animals. Data are mean ± s.e.m. and P values were determined by unpaired two-tailed t-test; ns, not significant.
Neutrophils play a critical role in the metastatic colonization step and are a key component of the lung pre-metastatic niche in breast cancer models1, 2, 3. To specifically assess the functional contribution of lipid-laden neutrophils in lung metastatic colonization and avoid the potential confound of similar effects that could occur in the primary tumor microenvironment, we next adopted a modified experimental lung metastasis model (Extended Data Fig. 3g). In this model, AT3-g-csf tumor cells were first orthotopically implanted in WT and the lipid metabolic gene cKO mice to develop host conditions with varying neutrophil lipid storage statuses. At the pre-metastatic stage for orthotopic tumors, the luciferase-labeled AT3 cells (AT3-Luc) were intravenously injected in these WT and cKO mice. By monitoring the injected tumor cells with bioluminescence imaging (BLI), the impact of host neutrophil lipid status on metastatic colonization can be quantitatively evaluated. As expected, the neutrophil lipid-high host condition (Atgl-cKO) apparently favored the AT3 tumor colonization and outgrowth in the lung (Fig. 4d). Conversely, metastasis colonization was significantly inhibited by the neutrophil lipid-low host environment (Hilpda-cKO) (Fig. 4e). Comparing this result to the insignificant effect of Hilpda-cKO in the spontaneous metastasis model (Extended Data Fig. 3c), it is plausible to speculate that HILPDA-associated lipid metabolism in neutrophils exerts a more prominent effect in certain metastatic step(s) like colonization, but is insufficient to alter the entire process of metastasis in vivo. Furthermore, genetic ablation of Cidec in host neutrophils, also showed a tendency to suppress the metastatic colonization of AT3 cells (Extended Data Fig. 3h).
Using another syngeneic breast tumor cell line E0771, which is genetically distinct from the AT3 line, a similar metastasis-promoting effect was found when the neutrophil lipid-laden phenotype was reinforced by Atgl-cKO in the experimental metastasis model (Fig. 4f). Moreover, targeted deletion of neutrophil Atgl in the MMTV-PyMT transgenic strain (MMTV-PyMT; S100a8-Cre; Atglflox/flox) significantly augmented spontaneous lung metastasis (Fig. 4g). Thus, neutrophil-specific ablation of LD-associated genes modulated lipid storage in lung neutrophils and consequently led to altered lung metastasis of breast tumors.
Neutrophils transfer their stored lipids to metastatic tumor cells via release of vesicles
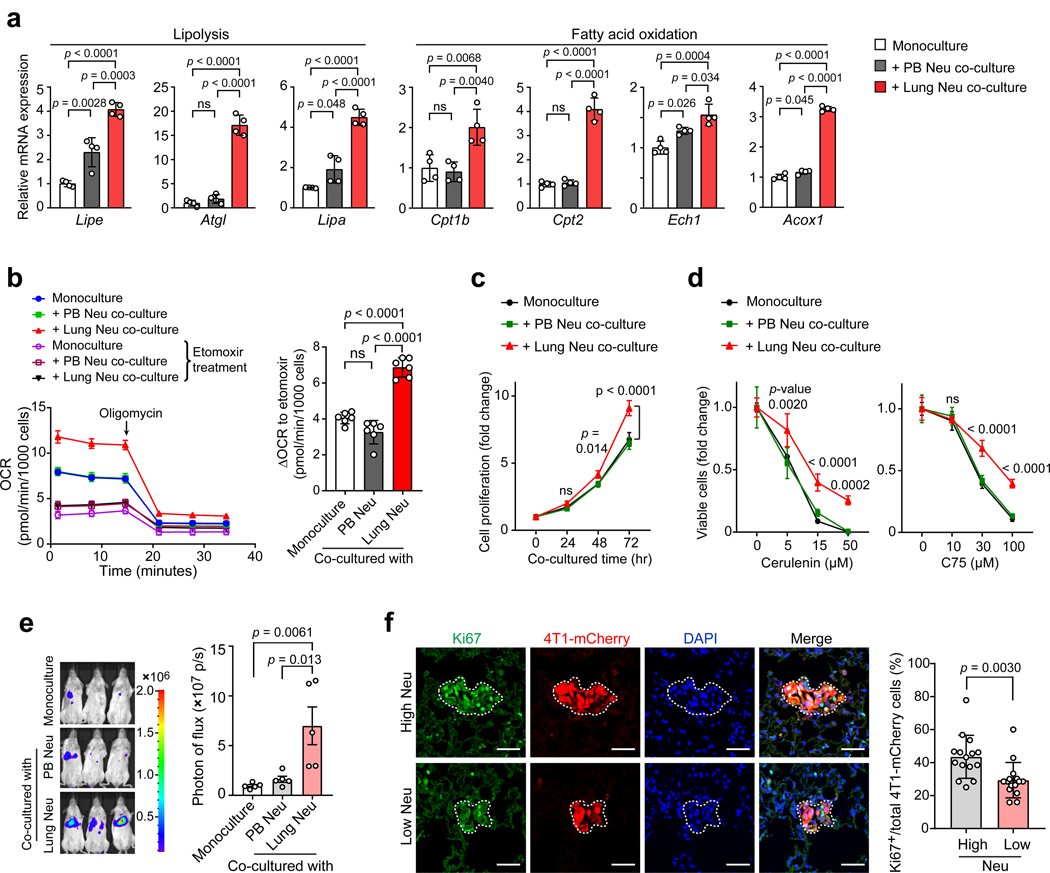
Lipid metabolism has been extensively studied in tumor cells themselves33, whereas the metabolic crosstalk between the tumor microenvironment and tumor cells remains less characterized. Limited evidence about such a crosstalk has been documented between tumor cells and adipose tissues at the primary and metastatic sites11, 34, 35, 36. We suspected that the lipid-laden neutrophils might play an “adipose” role in fueling DTCs based on our observation that the early lung-colonizing tumor cells were exclusively embraced by the Ly6G+ neutrophils (Fig. 5a). We first tested this possibility by ex vivo co-culturing tumor cells with neutrophils pre-loaded with fluorescence-labeled palmitic fatty acid. Following the co-culture, the labeled lipids originally only present in neutrophils were detected in tumor cells, suggesting possible lipid transport from neutrophils to tumor cells (Extended Data Fig. 4a). We further compared the PB and lung neutrophils, derived from tumor-bearing mice, for their capacities to transfer lipids to tumor cells. As lung neutrophils contained intrinsically higher levels of total lipids and TGs than PB neutrophils, consistently, tumor cells acquired more abundant lipids and TGs after co-culture with lung neutrophils than those with PB neutrophils (Fig. 5b and Extended Data Fig. 4b, c).
Fig. 5. Lung-infiltrating neutrophils transfer their stored lipid to metastatic tumor cells.

a, Adjacent localization of early seeded 4T1-mCherry cells and neutrophils (Ly6G) in the lung (see Methods). Scale bars, 20 μm (representative of n = 3 mice). b, The cellular TG contents of 4T1-mCherry cells after mono-culture or co-culture with PB or lung neutrophils (see Methods). c, Measurement of the lipids in early lung-colonizing 4T1-mCherry cells in G-CSF-pretreated mice without and with anti-Ly6G-based neutrophil depletion (see Methods) (n = 5). d, The lipid transfer from BODIPY-loaded lung neutrophils to tumor cells in vivo as determined by flow cytometry (see Methods) (n = 4). e, 4T1 cells were fed with the un-separated conditioned media (CM), small-sized (<100 kDa) or large-sized (>100 kDa) CM portion prepared from BODIPY-loaded lung neutrophils. Then lipids incorporated into the tumor cells were detected 4 hours later. Scale bars, 20 μm. f, Protein and TG levels in the large-sized lung or PB-derived neutrophil CM portions (n = 6). g, Localization of the absorbed lipids from BODIPY-loaded lung neutrophils with lysosomes (Lysotracker) in 4T1 tumor cells. Scale bars, 5 μm. h, The effects of EIPA, genistein and chlorpromazine in blockage of lipid transport from lipid-laden lung neutrophils to tumor cells (see Methods). Scale bars, 20 μm. i, The impact of EIPA on the capacity of 4T1 cells to acquire lipids from lung neutrophils. j, 4T1 cells were co-cultured with lung neutrophil-derived vesicles and imaged by transmission electron microscopy (see Methods). Red arrowheads indicate the engulfment of the lipid-like vesicles, and yellow arrowheads indicate macropinosome-like structures. Scale bars, 0.5 μm. The lung and PB neutrophils throughout this figure were isolated from the pre-metastatic stage of 4T1 tumor-bearing mice. n represents biologically independent animals. Data are mean ± s.e.m. for c and f, and mean ± s.d. from 3 (b) or 4 (i) biologically independent cell cultures. For e, g, h and j, data are representative of 3 biologically independent experiments. P values were determined by one-way ANOVA with Tukey’s multiple comparisons test (b, c and i) or unpaired two-tailed t-test (f). ns, not significant.
To gain insight into the in vivo lipid transport from neutrophils to tumor cells, we pre-treated naïve mice with recombinant mouse G-CSF to induce a host condition with high neutrophil infiltration before infusing mCherry-labeled tumor cells. In both 4T1 and AT3 models, a higher lipid level was detected in tumor cells colonized in G-CSF-pretreated mouse lungs than those in control mice (Fig. 5c and Extended Data Fig. 4d). Such a lipid elevation in injected tumor cells was nearly abolished by neutrophil depletion, confirming a specific role of neutrophils as the lipid source fueling tumor cells in the lung microenvironment (Fig. 5c and Extended Data Fig. 4d). To obtain more direct in vivo evidence of neutrophil-tumor cell lipid transfer, neutrophils pre-labeled with fluorescent lipids and tumor cells were successively infused into the recipient mice. By analyzing the freshly isolated lung tissues, the fluorescent lipids originally present in neutrophils were indeed detected in tumor cells (Fig. 5d and Extended Data Fig. 4e). Therefore, lipid-laden neutrophils were indeed capable of transferring their stored lipids to metastatic tumor cells. Moreover, this lipid transfer was further reinforced when neutrophils underwent apoptosis upon activation (Extended Data Fig. 4f). Considering that neutrophils are characterized by a short lifespan37, the rapid neutrophil turnover may expedite their lipid transfer to early arrived DTCs to satisfy their high energy demands7, 38.
Free fatty acids (FFAs) have been regarded as the main form of lipid transfer from adipose tissues to tumor cells11, 34, 35, 36. More recently, an alternative lipid transfer form, exosome-sized lipid-filled vesicles, was shown to mediate lipid transport from adipocytes to macrophages39. To determine the lipid transfer form from lung neutrophils to tumor cells in our study, we divided the BODIPY-labeled lung neutrophil-derived conditioned medium (CM) into a portion that would contain FFAs and most soluble proteins (<100 kDa) and a portion containing non-FFA vesicles and larger proteins (>100 kDa). Upon incubating these two CM portions with tumor cells, the large-sized CM portion was determined to be the primary lipid contributor for tumor cells (Fig. 5e). In the large-sized CM portion prepared from the pre-metastatic lung neutrophils, there was a much higher level of TG, but not proteins, compared to that prepared from the PB neutrophils (Fig. 5f). Therefore, the lipid transport from neutrophils to tumor cells is mainly via large-sized vesicles, and not FFAs.
Engulfment of large-sized extracellular vesicles by mammalian cells is usually achieved via a process called macropinocytosis40. Under nutrient deprivation stress, tumor cells are able to exploit macropinocytosis to take up extracellular nutrients to support their growth using the lysosomal pathway41, 42. In our study, upon lipid transport from neutrophils to tumor cells, the lipids were indeed detected to be localized within tumor cell lysosomes (Fig. 5g). Furthermore, selective inhibition of macropinocytosis by 5-(N-ethyl-N-isopropyl) amiloride (EIPA)42, but not inhibition of clathrin- or caveolae-dependent endocytosis43, completely abolished the lipid acquisition in tumor cells from the lipid-laden lung neutrophils (Fig. 5h, i). Under transmission electron microscopy, the membrane protrusions and retractions, and subsequent formation of large macropinosome-like vacuoles in tumor cells could be readily detected when neutrophil-derived vesicles were fed to tumor cells (Fig. 5j). Taken together, these data suggested that a macropinocytosis-lysosome pathway governs the lipid transport from lung neutrophils to metastatic tumor cells.
Tumor cells engulfing neutrophil-derived lipids acquire an augmented proliferative capacity
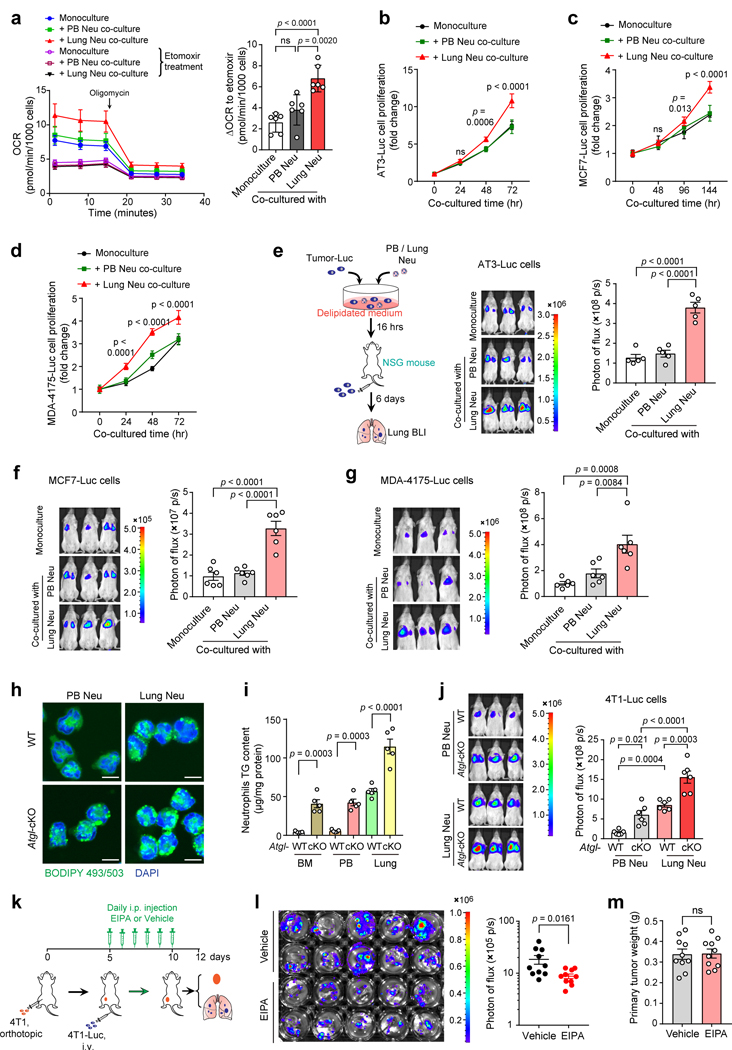
We next determined how tumor cell behaviors are altered in tumor cells after they ingest lipids from neutrophils. In vitro, tumor cells co-cultured with pre-metastatic lung neutrophils significantly upregulated their expression of key lipolysis genes—lipase E (Lipe), Atgl and lipase A (Lipa), as well as fatty acid oxidation-associated genes including carnitine palmitoyltransferase 1B (Cpt1b), carnitine palmitoyltransferase II (Cpt2), enoyl-CoA hydratase 1 (Ech1) and acyl-CoA oxidase 1 (Acox1) (Fig. 6a). Corresponding to the upregulation of these lipid utilization genes, lung neutrophil-educated tumor cells acquired an elevated oxygen consumption rate (OCR) indicating increased oxidative phosphorylation, which was significantly suppressed by etomoxir, a CPT1 inhibitor (Fig. 6b and Extended Data Fig. 5a). Therefore, acquisition of neutrophil-derived lipids drives tumor cells to undergo a metabolic shift towards lipid utilization.
Fig. 6. Neutrophil-derived lipids enhance the proliferative capacity of metastatic tumor cells.
a, Relative mRNA expression of the indicated genes in 4T1 cells upon a mono-culture or co-culture with PB or lung neutrophils. b, Oxygen consumption rates (OCR) of 4T1 cells, upon a mono-culture or co-culture with PB or lung neutrophils, were measured by Seahorse XFe96 Analyzer (left). The amount of OCR derived from fatty acid oxidation was quantified as the magnitude of the response to etomoxir treatment (right). c, Measurement of the proliferative capacities of 4T1-Luc cells without and with co-culture with PB or lung neutrophils (see Methods). d, Comparison of the pro-survival abilities of 4T1-Luc cells without and with co-culture with PB or lung neutrophils, when de novo lipogenesis was inhibited by cerulenin or C75 (see Methods). e, Comparison of the metastatic colonization potentials of 4T1-Luc cells without and without co-culture with PB or lung neutrophils (see Methods). Representative BLI images of the recipient mice and quantification of BLI signals within the lung areas are shown (n = 5). f, Detection of Ki67+ lung-colonizing 4T1-mCherry cells in control (high Neu) and neutrophil-depleted (Low Neu) 4T1-bearing mice (see Methods). Left: Representative microscopy images; Scale bars, 50 μm. Right: the percentages of Ki67+ cells in total mCherry+ 4T1 cells. Five pictures were chosen from each mouse lung section. n = 3 mice per group. Data are mean ± s.d. Neutrophils throughout this figure were isolated from the pre-metastatic stage of 4T1 tumor-bearing mice. Data are mean ± s.e.m. for e, and mean ± s.d. from 4 (a, c and d) or 6 (b) biologically independent cell cultures. P values were determined by one-way ANOVA with Tukey’s multiple comparisons test (a, b and e), two-way ANOVA with Sidak’s multiple comparisons test (c and d, mono-culture versus lung Neu co-culture) or unpaired two-tailed t-test (f). ns, not significant.
In accord with above metabolic changes, tumor cells, upon co-culture with lung neutrophils, had a higher proliferative capacity under nutrient deprivation (Fig. 6c and Extended Data Fig. 5b–d) and a more potent pro-survival ability when de novo lipogenesis was inhibited in vitro (Fig. 6d), compared to tumor cells after co-culture with PB neutrophils. In vivo, tumor cells educated by lung neutrophils gained a higher potential for metastatic colonization than those educated by PB neutrophils (Fig. 6e and Extended Data Fig. 5e–g). In situ, the proportion of Ki67+ proliferating lung-colonizing tumor cells was significantly reduced when neutrophils were depleted (Fig. 6f). Therefore, uptake of lung neutrophil-derived lipids endowed tumor cells with more robust proliferative and survival capacities.
Lipid-laden neutrophils fuel breast cancer lung metastasis in vivo
Neutrophils are well-known to nourish tumor cells via their secretome including a variety of cytokines, chemokines, growth factors and extracellular matrix44. To exclude the possibility that non-lipid neutrophil-derived factors drive tumor cell modulation, we further compared the tumor-regulatory effects of neutrophils isolated from Atgl-cKO mice and their WT littermates. In line with Atgl loss-caused lipid-laden reinforcement (Fig. 7a and Extended Data Fig. 5h, i), both lung and PB neutrophils from the Atgl-cKO mice were superior to their respective WT counterparts in amplifying the tumor cell colonization in the lung (Fig. 7b and Extended Data Fig. 5j). These data indicated a specific role of neutrophil-derived lipids to endow tumor cells with an increased capacity for metastatic colonization.
Fig. 7. Blockage of macropinocytosis inhibits metastatic colonization in vivo.
a, Transmission electron microscopy images of neutrophils from AT3-g-csf tumor-bearing WT and Atgl-cKO mice (representative of n = 4 mice). Arrowheads indicate lipid droplets. Scale bars, 0.5 μm. b, AT3-Luc cells, upon co-culture with PB or lung neutrophils isolated from AT3-g-csf tumor-bearing WT or Atgl-cKO mice, were intravenously injected into NSG mice (see Methods). Representative BLI images of the recipient mice and quantification of BLI signals within the lung areas are shown (n = 6). c, The impacts of host neutrophil changes in quantity and in their lipid-laden phenotype on the proliferation of lung-colonizing tumor cells, as measured by BrdU incorporation (see Methods) (n = 5). d-e, The effects of EIPA treatment on lung colonization by AT3 tumor cells (d) and on primary tumor growth (e) were determined following the experimental design as in Extended Data Fig. 5k (also see Methods). n = 9 (vehicle group) or 10 (EIPA treatment group). n represents biologically independent animals. Data are mean ± s.e.m. and P values were determined by one-way ANOVA with Tukey’s multiple comparisons test (b and c) or by unpaired two-tailed t-test (d and e). ns, not significant.
To further illustrate how host neutrophils quantitatively and qualitatively modulate the colonized tumor cells in the lung microenvironment in vivo, AT3-g-csf and AT3 parental cell lines were employed to induce neutrophil-high versus neutrophil-low host conditions, while Atgl-cKO and WT mice were utilized for generating neutrophil lipid-high versus lipid-low host conditions. Upon successful colonization in the lung, the tumor cell proliferation under above different host conditions was compared and it was clearly shown that an increase in either host neutrophil number, or lipid content, was sufficient to accelerate tumor cell proliferation in the lung (Fig. 7c). Therefore, neutrophils quantitatively and qualitatively (metabolically) dominate the tumor cell fates within the lung microenvironment.
As macropinocytosis was revealed as a main pathway for tumor cells to ingest neutrophil-derived lipids, we next explored whether pharmacological inhibition of macropinocytosis represents an effective approach to restrain metastatic colonization. In both 4T1 and AT3-g-csf tumor models, administration of macropinocytosis inhibitor EIPA in tumor-bearing mice significantly reduced tumor cell colonization in the lung (Fig. 7d and Extended Data Fig. 5k, l). Such EIPA-mediated blockage of lipid transport, however, did not alter the primary tumor growth suggesting that the lipid-laden neutrophils may exert effects specifically at the metastatic sites (Fig. 7e and Extended Data Fig. 5m). Hence, targeting the lipid metabolism of the lung microenvironment is indeed effective in controlling breast cancer lung metastasis.
Data from above mouse models revealed that lung MCs trigger formation of a lipid-rich pre-metastatic niche in the lung, and we therefore speculated that primary tumor cells with a relatively higher capacity to absorb and utilize lipids might adapt more favorably to the lipid-rich lung environment that leads to lung metastasis. To address this, we analyzed a transcriptome dataset (GSE2603) derived from human primary breast tumor samples in which patients’ lung relapse information was available45. Indeed, the primary tumors expressing higher levels of genes associated with lipid utilization pathways, including lipid catabolism, lipid transport and lipid oxidation, showed a high risk for developing lung metastases in breast cancer patients (Extended Data Fig. 6). In addition, the gene expression levels of neutrophil chemotaxis, migration and extravasation pathways were also shown to be related to lung metastasis of those patients (Extended Data Fig. 6). This result suggested that the capacity of primary tumors to elicit neutrophil trafficking to the organs, such as the lung, is critical for triggering the subsequent metastatic cascade that occurs in the lung. A further analysis on these lipid- and neutrophil-associated pathways will help develop signature markers with prognostic value for breast cancer patients.
Overall, our work revealed a lung MC→ neutrophil→ tumor cell metabolic axis (Extended Data Fig. 7) which dominates extent of metastatic colonization, the most rate-limiting step of the metastatic process in solid cancers7.
Discussion
In this study, we demonstrated that neutrophils serve as an energy source to fuel metastatic tumor cells. At the pre-metastatic stage of breast cancer, after their expansion in hematopoietic tissues, neutrophils infiltrate the peripheral organs and tissues. At the lung, resident MCs trigger the lipid storage in infiltrating neutrophils. When DTCs arrive at the lung, these lipid-laden neutrophils then transfer their stored lipids to DTCs for their survival and proliferation leading to metastatic colonization and outgrowth.
Our results suggested that lung resident stromal cells, specifically CD140a+ MCs, act as an initiator driving the metabolic dynamics in infiltrating neutrophils and DTCs. Although we have characterized PGE2 as a primary lung MC-derived soluble factor and PGE2→ HIF1α→ HILPDA as the subsequent molecular cascade in neutrophils, ongoing efforts are underway to further define the cell-cell contact mechanisms underlying CIDEC and G0S2 induction in neutrophils. In vivo, a single depletion of ATGL inhibitory genes in neutrophils only caused a mild or insignificant reduction in their metastasis-regulating capacity, while genetic ablation of neutrophil Atgl led to substantially elevated lung metastases of breast tumor cells. A better understanding of the molecular mechanisms behind induction of ATGL inhibitory factors will potentially allow targeting of multiple LD-associated factors that could provide more effective approaches for abolishing the neutrophil lipid-laden phenotype and its detrimental effects.
In mammals, TGs are superior to carbohydrates and proteins for energy storage due to their reduced and anhydrous characteristics46. In our study, TGs are stored as lipid droplets in neutrophils, a cell type with a fast turnover rate, in the lung microenvironment. This may facilitate a timely release of TGs from neutrophils to the early DTCs which have high energy demands47. As macropinocytosis is known as a rapid nutrient-scavenging pathway in cancer cells independent of protein synthesis48, this endocytic pathway would further accelerate energy acquisition by DTCs from the lung microenvironment. Additional work is needed to further characterize the nature of lipid vesicles, the molecular bases of vesicle release by neutrophils and macropinocytosis by tumor cells.
In addition to energy storage, lipids are also serve as precursors of various bioactive lipid mediators such as prostaglandins24, 25. In a recent report, neutrophils were shown to take up lipids, particularly arachidonic acids, for synthesis of PGE2 within the primary tumor microenvironment and resulted in enhanced immunosuppressive effects31. We did compare the immunosuppression by WT and Atgl-cKO mice-derived lung neutrophils, however, they were found to be comparable in suppression of natural killer cells (data not shown), the reported major metastatic colonization-restraining immune cells6, 49. It will be intriguing to further elucidate the roles of lung neutrophil-derived bioactive lipid mediators in the lung metastatic niche, as well as a possible lipid transfer from neutrophils to tumor cells within the primary tumor and other tissue microenvironments. Such work will help to develop a systemic understanding, from a metabolic perspective, of neutrophil-tumor cell crosstalk in solid tumor metastasis.
Neutrophils are known to have multifaceted functions in metastasis including both pro-tumoral and anti-tumoral effects2, 44. Through expression of immunoregulatory factors, growth factors, inflammatory mediators and proteinases, as well as by forming NETs, neutrophils have been mostly reported to support metastatic progression1, 44. On the other hand, through production of reactive oxygen species, neutrophils were also indicated to be anti-metastatic through a direct cytotoxic effect on tumor cells44. Our results add a new layer of complexity to the functions of neutrophils in metastasis by revealing a modulatory role for neutrophil metabolism and energy availability. Metabolic adaptation of DTCs in their metastasized organs is emerging as an increasingly important topic for understanding organ-tropic metastases38, 50. Our findings will advance a deeper understanding of the metabolic regulation of DTCs by organ resident stromal cells, and point to the metabolism of organ-specific stroma as a novel and promising therapeutic target for prevention and treatment of solid tumor metastases.
Methods
Animal Studies
For all animal studies, mice of similar age were block randomized in an unblinded fashion. Female mice aged 8–10 weeks were used unless indicated otherwise. Sample sizes were estimated based on pilot experiments and were selected to provide sufficient numbers of mice in each group for statistical analysis. Animal handing and experimental protocols were reviewed and approved by the Institutional Animal Care and Use Committee of The Jackson Laboratory. The mice were fed on a chow diet ad libitum and housed in a specific pathogen-free facility in plastic cages at 22 °C and 40–50% humidity, with a daylight cycle from 6 a.m. to 6 p.m. The mouse strains used in this study are as follows: BALB/cJ, C57BL/6J, NOD-scid (NOD.Cg-Prkdcscid/J), NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ), MMTV-PyMT (B6.FVB-Tg(MMTV-PyVT)634Mul/LellJ), CD140a-GFP (B6.129S4-Pdgfratm11(EGFP)Sor/J) mice were obtained from The Jackson Laboratory. Cox2 flox/flox (B6;129S4-Ptgs2tm1Hahe/J) and CD140a-cre+/− (C57BL/6-Tg(Pdgfra-cre)1Clc/J) mice were obtained from The Jackson Laboratory and were crossed to generate Cox2 conditional knockout (CD140a-cre+/−Cox2 flox/flox) mice. Atglflox/flox (B6N.129S-Pnpla2tm1Eek/J), Cidecflox/flox (B6.Cg-Cidectm1.1Gonz/J), Hilpdaflox/flox (Hilpdatm1.1Nat/J), Hif1aflox/flox (B6.129-Hif1αtm3Rsjo/J), S100a8-cre+/− (B6.Cg-Tg(S100A8-cre,-EGFP)1Ilw/J) mice were obtained from The Jackson Laboratory. Each of these 4 floxed strains were crossed with S100a8-cre+/− transgenic mice to generate conditional knockout (cKO) mice. For example, Atglflox/flox mice were bred to S100a8-cre+/− mice to generate S100a8-cre+/−Atglflox/WT mice, which were backcrossed onto Atglflox/flox mice to generate S100a8-cre+/−Atglflox/flox mice (Atgl-cKO). Age-matched littermate Atglflox/flox mice were used as WT controls.
Male MMTV-PyMT mice were crossed with female S100a8-cre+/−Atglflox/flox mice to generate MMTV-PyMT; S100a8-cre+/−Atglflox/WT mice, which were backcrossed onto Atglflox/flox mice to generate MMTV-PyMT; S100a8-cre+/−Atglflox/flox mice (MMTV-PyMT; Atgl-cKO). Age-matched littermate MMTV-PyMT; Atglflox/flox mice were used as controls.
Spontaneous breast cancer metastasis models
1) Orthotopic models: cultured 4T1, AT3, E0771, AT3-g-csf or E0771-g-csf tumor cells (5×105 cells per mouse) were resuspended in 50 μl growth factor-reduced Matrigel (Corning) and injected into the fourth mammary fat pads of female syngeneic BALB/cJ or C57BL/6J mice. The lung metastases in these mice were then monitored by histological analysis and macroscopic examination. Day 10 and earlier was determined as the pre-metastatic stage for the 4T1 model, whereas day 12 and earlier as the pre-metastatic stage for the AT3-g-csf and E0771-g-csf model. Unless otherwise stated, neutrophils used for ex vivo cultures and assays were isolated from the tissues (bone marrow, peripheral blood, lungs, spleen and primary tumors) of tumor-bearing mice at the pre-metastatic stage in each model.
2) MMTV-PyMT transgenic model: this model was determined to be pre-metastatic prior to 5 months of age. At 5 months of age, MMTV-PyMT (C57BL/6 background) mice had spontaneously developed primary tumors (weight 0.5–1.0 g) but had no detectable lung metastases by histological analysis and macroscopic examination.
3) Spontaneous metastasis model in WT and the lipid metabolic gene cKO mice (Atgl-cKO, Cidec-cKO and Hilpda-cKO): mice were orthotopically implanted with AT3-g-csf cells. On day 15, primary tumors were surgically resected. On day 30, mice were euthanized and their lungs were excised, inflated with Bouin’s solution, and the numbers of metastatic nodules were counted under a dissecting microscope.
4) Spontaneous metastasis model in MMTV-PyMT and MMTV-PyMT; Atgl-cKO mice: 6 month-old mice that had spontaneously developed primary tumors (2.5–3.0 g) were euthanized. Lungs were excised and inflated with Bouin’s solution and the numbers of metastatic nodules were counted under a dissecting microscope.
Experimental breast cancer metastasis model
C57BL/6J mice or lipid metabolic gene cKO mice were first orthotopically injected with AT3-g-csf (or E0771-g-csf) (5×105 cells) to generate the neutrophil-high host conditions. Luciferase-labeled AT3-Luc cells (or E0771-Luc) (5×105 cells per mouse) were then intravenously injected on day 10, a time point within the pre-metastatic stage. On day 15, primary tumors were surgically resected. At the end point (day 25), mice were euthanized and the lungs were rapidly excised for ex vivo bioluminescence imaging (BLI). Isolated lungs were placed in PBS buffer supplemented with D-luciferin (150 μg ml−1; Gold Biotechnology) in a 24-well plate and the bioluminescence signals were detected with the Xenogen IVIS system. Image exposure times were between 10 s and 30 s, depending on the signal strength. Photon emission was quantified using Living Images software as the sum of all detected photon counts per second within the specified area after subtracting background luminescence.
To explore the effect of EIPA using above experimental metastasis model, C57BL/6J (or BALB/CJ) mice were first orthotopically implanted with AT3-g-csf (or 4T1) to induce a neutrophil-high host condition. Luciferase-labeled AT3-Luc (or 4T1-Luc) cells were intravenously injected in the above mice at their pre-metastatic stage. Then EIPA (10 mg kg−1) or vehicle was given daily by intraperitoneal injection for 8 (C57BL/6J mice) or 6 (BALB/CJ mice) consecutive days. At the end point, the metastatic progression of AT3-Luc (or 4T1-Luc) cells in the lung was detected by ex vivo BLI.
Cell lines
4T1 cells and MCF7 cells were purchased from the ATCC, and E0771 cells were purchased from CH3 Biosystems. MDA-4175 cell line, originally generated from Dr. Joan Massagué’s lab (Memorial Sloan-Kettering Cancer Center)45, was kindly provided by Dr. Yibin Kang (Princeton University). AT3 cell line was kindly provided by Dr. Scott I. Abrams (Roswell Park Comprehensive Cancer Center), which was originally generated from Dr. Abrams’s lab32. E0771 cells were maintained in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) and all other cell lines were maintained in DMEM supplemented with 10% FBS. All cells were cultured in a 5% CO2 humidified incubator at 37 °C. No further authentication of these cell lines was performed. Cell morphology and growth characteristics were monitored during the study and compared with published reports to ensure their authenticity. All cell lines used in this study were determined to be negative for Mycoplasma prior to experiments, and were used within 6 passages after thawing.
Lentivirus-mediated tumor cell labeling or gene overexpression
For luciferase labeling, parental 4T1, AT3, E0771, MCF7 and MDA-4175 cells were infected with luciferase-expressing lentivirus (Addgene #17477) in order to generate 4T1-Luc, AT3-Luc, E0771-Luc, MCF7-Luc and MDA-4175-Luc cell lines, respectively. Construct-positive cells were selected with G418 Sulfate. For mCherry labeling, parental 4T1, AT3 and MDA-4175 cells were infected with mCherry-expressing lentivirus (Addgene #36084) to generate 4T1-mCherry, AT3-mCherry and MDA-4175-mCherry cells, respectively. After 48 hours, the mCherry+ cells were sorted out from the infected cells by fluorescence-activated Cell Sorting (FACS). In order to overexpress G-CSF, AT3 and E0771 cells were infected with g-csf (Csf3)-expressing lentivirus (the vector was a gift from Dr. Robert A. Weinberg, Massachusetts Institute of Technology). Construct-positive cells (AT3-g-csf and E0771-g-csf) were selected with G418 Sulfate.
Tissue dissociation
Euthanized mice were perfused with 5 mL PBS through the right ventricle. Lungs were inflated with 1–2 ml protease solution [1.5 mg ml−1 collagenase type I (Thermo Fisher), 0.1 mg ml−1 elastase (STEMCELL Technologies), 5 U ml−1 dispase II (Thermo Fisher) and 0.1 mg ml−1 DNase I (Sigma) in Hank’s Balanced Salt Solution (HBSS)], cut into small pieces (< 2 mm2), and incubated in 2 ml protease solution for 30 min at 37 °C with frequent agitation. RPMI-1640 + 5% fetal calf serum (FCS) was added, and tissues were disrupted by pipetting and washed with RPMI-1640. After filtering through a 70 μm cell strainer, cells were washed with 5 mL RPMI-1640 + 5% FCS, incubated at room temperature for 1.5 min in 2 mL ACK red blood cell lysis solution, filtered through a 40 μm strainer, centrifuged, and resuspended in RPMI-1640 + 5% FCS.
Primary tumors were dissected, minced, and digested with enzyme mix (1.5 mg ml−1 collagenase I, 0.75 mg ml−1 hyaluronidase and 0.1 mg ml−1 DNase I) in HBSS for 1 hour at 37 °C. Enzymatic reactions were stopped by addition of 5% FCS in DMEM, and suspensions were dispersed through a 70 μm cell strainer; Spleen was removed into a petri-dish to be smashed by full frosted microscope slides gently, then the suspension was filtered through a 70 μm cell strainer; Bone marrow was collected from the tibias and femurs of both hind legs of the mice and flushed using RPMI-1640 + 5% FCS through a 70 μm cell strainer; Peripheral blood was collected by cardiac puncture with 5 mM EDTA in PBS as an anticoagulant. For flow cytometric analysis or further purification, single-cell suspensions of tumor, spleen, bone marrow and peripheral blood were subjected to hypotonic lysis (ACK red blood cell lysis solution) to remove erythrocytes and washed with 1× PBS containing 0.5% BSA and 2 mM EDTA.
Purification and in vitro culture of neutrophils
Neutrophils were purified from the digested tissues by using anti-Ly6G MicroBead Kit (Miltenyi Biotec), following manufacturer’s instructions. The isolated neutrophils were analyzed by flow cytometry and the cells with a > 95% purity were used for the next procedure.
The purified neutrophils were cultured in RPMI-1640 supplemented with 10% FBS. To screen the lung MC-derived candidate soluble factors, BM neutrophils were stimulated by the candidate regulators for 16 hours and then the lipid levels of neutrophils were detected by BODIPY 493/503 staining and flow cytometry. CD40 ligand (CD40LG), insulin like growth factor 2 (IGF2): 100 ng ml−1; colony-stimulating factor 2 (CSF2), hepatocyte growth factor (HGF), TNF superfamily member 11 (TNFSF11), interleukin 6 (IL6), interleukin 4 (IL4), interleukin 33 (IL33), prostaglandin E2 (PGE2): 10 ng ml−1; transforming growth factor beta 1 (TGFβ1): 1 ng ml−1; hydrogen peroxide (H2O2): 5 μM.
To determine the EP receptor(s) used by neutrophils to respond to lung MCs, selective EP receptor antagonists were added to BM neutrophils and lung MC co-culture system for 16 hours, then the lipid levels of neutrophils were detected by BODIPY 493/503 staining and flow cytometry. Antagonists for EP inhibitors: EP1, SC-51089; EP2, PF-04418948; EP3, L-798106; EP4, MF-498; each at a final concentration of 1 μg ml−1.
Isolation and primary culture of CD140a+ MCs
The lung, liver, mammary gland tissue and heart were cut into small pieces (<1 mm3), and the tissue pieces were dissociated with protease solution [1.5 mg ml−1 collagenase type I, 0.1 mg ml−1 elastase, 5 U ml−1 dispase II and 0.1 mg ml−1 DNase I in HBSS] for 1 hour. Cells were then resuspended, passed through a 70 μm cell strainer, and finally plated into culture dishes with MC culture medium (MesenCult™ Expansion Kit, Catalog #05513, STEMCELL Technologies). For isolating mouse bone MCs, the bone marrow cavities were flushed at least three times in order to thoroughly deplete hematopoietic cells. Then the compact bones were dissected into fragments of 1–3 mm3 and digested with collagenase type II (1 mg ml−1). The released cells were discarded and the digested bone fragments were cultivated in the MC culture medium. After 5 days of expansion, CD45−CD140a+ cells (from BALB/cJ mice) or GFP+ cells (from CD140a-GFP mice) were sorted by FACS. The first to third passages of the primary MCs were used in the next co-culture experiments.
Flow cytometry and cell sorting
Prepared single-cell suspensions from mouse tissues or ex vivo cultured cells were incubated with fluorescently labeled antibodies (1 : 200 in 2% FCS in PBS) directed against cell surface markers. Cell surface labeling was performed on ice for 30 min. All antibodies were purchased from Biolegend (molecule, fluorophore): CD45, Alexa Fluor 700; CD11b, BV650; Ly6c, BV570; Ly6G, Pacific Blue; CD326, APC/Fire™ 750; CD31, FITC; CD140a, Brilliant Violet 421. Cells were run on FACSymphony A5 cytometer (BD Biosciences) and data were analyzed by FlowJo (Tristar). Further information about the above antibodies, as well as other research design, is available in the Nature Research Reporting Summary linked to this article.
For cell sorting, a low-pressure sort was performed on the BD FACSAria II cell sorter with a 100 μm nozzle. Dead cells were stained with 4, 6-diamidino-2-phenylindole (DAPI) or propidium iodide (PI).
Measurement of lipid levels in neutrophils by flow cytometry
For the measurement of cellular lipid contents, after cell surface labeling, cells were washed and resuspended in 0.5 μg ml−1 BODIPY 493/503 for 15 min at room temperature in the dark before the analysis.
For directly determine if the lung is a unique site for neutrophils to accumulate lipids, neutrophils isolated from the BM of naïve mice were labeled with a cell dye-CMTMR, then intravenously infused into naïve mice or pre-metastatic orthotopic 4T1 tumor-bearing BALB/cJ mice. After 4 hours, lipid levels of the CMTMR-labeled neutrophils from different tissues were determined by BODIPY 493/503 staining and flow cytometry.
In vivo BrdU incorporation
WT and Atgl-cKO mice were first orthotopically implanted with AT3 or AT3-g-csf tumor cells to generate the neutrophil-low (AT3 implantation) or neutrophil-high host condition (AT3-g-csf implantation). At the pre-metastatic stage for orthotopic tumors, AT3-mCherry cells were intravenously injected. After 6 days, the in vivo tumor cell proliferation was determined using an FITC-BrdU Flow Kit (BD Biosciences # 559619). Briefly, the mice received a single dose of BrdU (1 mg per mouse) by intraperitoneal injection. Four hours later, the mice were euthanized and the single cell suspensions of lungs were prepared for flow cytometry analysis following the manufacturer’s instructions.
Tissue staining, immunofluorescence and light microscopy
For preparation of paraffin-embedded sections, dissected tissues were fixed in 10% formalin for 24 hours at room temperature, washed twice with PBS, and then stored in 70% ethanol solution at 4 °C until they were embedded in paraffin and sectioned at 4 μm. H&E staining was performed according to standard histopathological procedures.
For preparation of cryosections, formalin-fixed dissected tissues were embedded in Tissue-Tek O.C.T. (Electron Microscopy Sciences) and frozen on dry ice. The frozen tissues were stored at –80 °C until they were sectioned at 8 μm. The cryosections were incubated with mouse FcR Blocking Reagent (Miltenyi) followed by incubation with specific primary antibodies and fluorescently labelled secondary antibodies (Thermo Fisher). The cell nuclei were stained with DAPI.
To detect the lipid droplets under light microscopy, the purified neutrophils were first stained with BODIPY 493/503 at 1 μg ml−1 in PBS for 20 min. Then the cells were cytospun onto glass slides and mounted for imaging.
To determine the localization of early seeded metastatic tumor cells and neutrophils in the lung, NOD-scid IL2rgnull (NSG) mice were orthotopically implanted with unlabeled 4T1 tumor cells and at their pre-metastatic stage these mice received intravenous injection of mCherry-labeled 4T1 cells (4T1-mCherry). On day 2 and day 6 following 4T1-mCherry cell injection, the lungs were dissected and sectioned for immunostaining of neutrophils (Ly6G), the early seeded 4T1-mCherry cells and the epithelium (RAGE).
Unless otherwise stated, more than 6 images per sample were acquired on a Leica SP8 inverted confocal microscope with a 40× or 63× objective and Leica Application Suite X software platform. All images were taken with identical laser setup and voltage settings.
Ki67 staining
NOD-scid mice were first orthotopically implanted with 4T1 cells (5×105 per mouse). After 5 days, the mice received daily intraperitoneal injections of anti-Ly6G (clone 1A8; Bio X Cell, 12.5 μg per mouse) or rat IgG2a isotype control (Clone 2A3; Bio X Cell, 12.5 μg per mouse) to develop a neutrophil-low or neutrophil-high host condition, respectively. After an additional 5 days, 4T1-mCherry cells (5×105 per mouse) were intravenously injected in the same mice, and 6 days after these injections the mice were euthanized and 8 μm cryosections of lung tissues were prepared. The cryosections were then incubated with mouse FcR Blocking Reagent (Miltenyi Biotec) followed by incubation with Ki67 monoclonal antibody (clone SolA15, 0.5 μg ml−1) and Alexa Fluor 488-labeled secondary antibody (Invitrogen, 0.1 μg ml−1). The cell nuclei were stained with DAPI. Images were acquired on a Leica SP8 inverted confocal microscope. The percentages of Ki67+ cells in total mCherry+ 4T1 cells were counted.
Transmission Electron Microscopy
Neutrophils were freshly isolated from BM, PB and lungs of tumor-bearing BALB/cJ, Atgl-cKO or MMTV-PyMT mice at the pre-metastatic stage. 4T1 tumor cells were co-cultured with neutrophil-derived vesicles for 4 hours in delipidated medium (DMEM containing 10% delipidized fetal bovine serum (Gemini Bio)). The neutrophils or tumor cells were first fixed with 4% paraformaldehyde. The fixed cells were then embedded, sectioned and stained with osmium tetroxide and uranyl acetate. Images were captured by JEOL 1230 Transmission Electron Microscope with an AMT 2K Digital Camera.
RNA sequencing analysis
To compare the gene expression of neutrophils residing in BM, PB and lung, 4T1 cells (5×105 cells) were re-suspended in 50 μl growth factor-reduced Matrigel (Corning) and injected into the fourth mammary fat pads of female syngeneic BALB/cJ mice (8-week-old, n = 3). On day 10 (pre-metastatic stage), the mice were euthanized and then CD45+CD11b+Ly6GhighLy6Cmed neutrophils from BM, PB and lungs were sorted by FACS.
Total RNA was isolated from purified neutrophils using the miRNeasy Mini kit (Qiagen), according to manufacturers’ protocols, including the optional DNase digest step. Sample concentration and quality were assessed using the Nanodrop 2000 spectrophotometer (Thermo Scientific) and the Total RNA Nano assay (Agilent Technologies). Libraries were prepared using the KAPA mRNA HyperPrep Kit (KAPA Biosystems), according to the manufacturer’s instructions. Briefly, the protocol entails isolation of polyA containing mRNA using oligo-dT magnetic beads, RNA fragmentation, first and second strand cDNA synthesis, ligation of Illumina-specific adapters containing a unique barcode sequence for each library, and PCR amplification. Libraries were checked for quality and concentration using the D5000 assay on the TapeStation (Agilent Technologies) and quantitative PCR (KAPA Biosystems), according to the manufacturers’ instructions. Libraries were pooled and sequenced, 100 bp paired-end on the HiSeq 4000 (Illumina) using HiSeq 3000/4000 SBS Kit reagents (Illumina).
Paired end, Illumina-sequenced stranded RNA-Seq reads were filtered and trimmed for quality scores > 30 using a custom python script. The filtered reads were aligned to Mus musculus GRCm38 using RSEM (v1.2.12) to launch Bowtie2 (v2.2.0) (command: rsem-calculate-expression -p 12 --phred33-quals --seed-length 25 --forward-prob 0 --time --output-genome-bam -- bowtie2 --paired-end). RSEM calculates expected counts and transcript per million (TPM). The expected counts from RSEM were used in the edgeR package (v3.20.9) to determine differentially expressed (DE) genes. Core analysis was performed using Ingenuity Pathway Analysis (QIAGEN Inc.). The analysis was restricted to DE genes with absolute value of log(fold change) > 5.
RNA isolation and qRT-PCR
Total RNA was isolated directly from cells using the Direct-zol RNA Miniprep Plus Kit (Zymo). Reverse transcription was performed with High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher). Gene expression levels were quantified by qRT-PCR on the ViiA 7 Real-Time PCR System (ThermoFisher). Threshold cycle values (Ct) were normalized to Rps18 (ΔCt), and these values across samples were compared (ΔΔCt) to quantify relative gene expression. Primer sequences are listed in Supplementary Table 1.
Western blotting
Total protein was extracted by lysing neutrophils in RIPA buffer containing protease inhibitor. Protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and were then transferred onto polyvinylidene fluoride (PVDF) membranes. After blocking with 5% non-fat milk in TBS-T, membranes were incubated with the primary antibody. These primary antibodies were used: G0S2 polyclonal antibody (Thermo Fisher, Catalog # PA5-97764); CIDEC polyclonal antibody (Novus Biologicals, Catalog # NB100-430); HILPDA polyclonal antibody (Antibodies-online, Catalog # ABIN1030425); GAPDH monoclonal antibody (D16H11) (Cell Signaling Technology, Catalog # 5174). Goat-anti-rabbit IgG conjugated to horseradish peroxidase (Cell Signaling Technology) was used as the secondary antibody. Protein signal was detected with Image Acquisition using Azure c600 (Azure Biosystems).
Determination of TG hydrolase activity in neutrophils
Purified neutrophils were washed twice in ice-cold PBS and then disrupted on ice by passing 20 times through a 22 G needle in 0.2 ml of cell extraction buffer (0.25 M sucrose, 1 mM EDTA, 1 mM DTT, and protease inhibitors, pH 7.0). The cells were clarified by centrifugation at 1, 000 g for 15 min. The TG hydrolase activity against 3H-labeled triolein was measured as described previously14. Briefly, 25 μl of cell extracts were mixed with 25 μl of substrate solution (418 μM triolein, and 48 μM of phosphatidylcholine/phosphatidylinositol in a ratio of 3:1 are sonicated in PBS). Before using, 0.5 μCi [9, 10-3H]-triolein (Perkin-Elmer) are added to the TG substrate and vortex to completely mix and incubated in a water bath at 37°C for 60 min. The reaction was terminated by directly adding 650 μl of methanol/chloroform/heptane (10: 9: 7) and 200 μl of 0.1 M potassium carbonate, 0.1 M boric acid, pH 10.5. After centrifugation (800 g, 20 min) the radioactivity in 200 μl of the upper phase was determined by liquid scintillation counting.
Determination of lipid contents and total lipase activity
The triglyceride contents of freshly isolated neutrophils were determined by Triglyceride Quantification Colorimetric/Fluorometric Kit (Biovision), and the total lipase activity of freshly isolated neutrophils were measured by Lipase Activity Assay Kit (Cayman chemical), following their respective manufacturer’s instructions.
Determination of PGE2 levels in MC culture supernatants
MCs were seeded at 1×105 per ml medium in the absence or presence of exogenous stimulations and incubated for 24 hours. PGE2 concentrations in supernatants were measured by using the PGE2 ELISA Kit (Cayman chemical), according to the manufacturer’s instructions. The net concentration of PGE2 was calculated upon subtracting the quantity of PGE2 from medium control.
Liquid chromatography–mass spectrometry (LC-MS)
The freshly isolated neutrophils were immediately frozen in liquid nitrogen and stored at −80°C until use. Untargeted lipidomics was performed by Cayman Chemical. In brief, lipids were extracted from cell pellets using the Bligh and Dyer method and lipidomics analysis was conducted following the protocol described by Breitkopf et al.51 with some modifications, using an ultra-performance liquid chromatography (UPLC) system (UltiMate 3000, Thermo Fisher Scientific) coupled with a hybrid Quadrupole-Orbitrap mass spectrometer (Q‐Exactive Plus, Thermo Fisher Scientific). Analysis of LC-MS data was performed using Lipostar software (version 1.0.5; Molecular Discovery). Lipid identification was provided by the software using the Lipid Maps database as reference. The peak area of each feature was normalized to peak area per 1 million cells. The lipid types measured include: triglyceride (TG), cholesteryl ester (CE), phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylinositol (PI), Cer, ceramides (Cer), phosphatidylglycerol (PG).
Tumor cell lipid uptake assay
4T1-mCherry or AT3-mCherry tumor cells were seeded in a 6-well plate in 10% FBS/DMEM medium. Cells were allowed to adhere and grow to 40% to 60% confluence for 24 hours. Then the cells were starved for 2 hours in delipidated media. The freshly isolated neutrophils were added to the seeded tumor cells (the ratio of neutrophils: tumor cells = 20: 1) in delipidated media in a direct cell–cell contact manner for 16 hours. For determination of the TG content in tumor cells, the mCherry+ tumor cells were sorted and TG content was measured by Triglyceride Quantification Colorimetric/Fluorometric Kit (Biovision). For lipid content analysis by flow cytometry, all the cells were stained with BODIPY 493/503 and anti-CD45 antibody and analyzed by flow cytometry. The mCherry+CD45− cells were gated as tumor cells. The relative mean fluorescence intensity (MFI) of BODIPY 493/503 was quantified and calculated as fold change in co-cultured cells compared with monoculture controls.
To directly observe lipid transfer between neutrophils and tumor cells, the neutrophils were pre-incubated with fluorescently labeled palmitic fatty acid (BODIPY FL C16 from Thermo Fisher, 5 μM) at 37 °C in RPMI-1640 media for 1 hour. Then the extracellular BODIPY was removed from the cells by washing 4 times with 1× HBSS with 0.2% fatty acid–free BSA. After washing, the labeled neutrophils were added to the dishes where 4T1-mCherry cells had been seeded, and these cells were co-cultured in a direct cell–cell contact manner for 4 hours. The cells were then fixed and imaged on a microscopy platform. When the endocytic inhibitors (EIPA: 50 μM; Genistein: 50 μM; Chlorpromazine: 15 μM) were used, they were separately added 1 hour prior to neutrophil addition.
In vivo lipid transport
To detect lipid transport from lung neutrophils to tumor cells in vivo, NOD-scid mice were used so that a sufficient number of tumor cells could be seeded in the lung to meet the requirements of the lipid content analysis by flow cytometry, in the experimental metastasis model.
In one experiment designed to compare the lipid levels in implanted tumor cells in the absence and presence of host neutrophils, NOD-scid mice received daily intraperitoneal injection of recombinant mouse G-CSF (PeproTech, 2.5 μg per mouse) without and with intraperitoneal injection of anti-Ly6G (clone 1A8; Bio X Cell, 12.5 μg per mouse) to generate the neutrophil-high and neutrophil-low host conditions, respectively. One week later, 4T1-mCherry or AT3-mCherry cells (5×105 cells per mouse) were intravenously injected into these mice. After 24 hours, the lungs were dissected, stained with anti-CD45, DAPI and BODIPY 493/503 and analyzed by flow cytometry. The lipid levels of tumor cells (CD45−mCherry+) seeded in the lungs were quantitated as MFI of BODIPY 493/503.
In another experiment designed to provide a direct in vivo evidence for the lipid transfer from neutrophils to tumor cells, lung neutrophils were first prepared from the pre-metastatic stage of 4T1 tumor-bearing mice, and then incubated with fluorescently labeled palmitic fatty acid (BODIPY FL C16, 5 μM) in RPMI-1640 medium at 37 °C for 1 hour. Extracellular BODIPY was then removed by washing the cells 5 times with 1× HBSS containing 0.2% fatty acid–free BSA. NOD-scid mice were then intravenously injected with these BODIPY FL C16-loaded lung neutrophils or unlabeled neutrophils (5×106 cells per mouse). Two hours later, these mice received a second intravenous injection of mCherry-labelled tumor cells (4T1-mCherry, AT3-mCherry or MDA-4175-mCherry, 5×105 cells per mouse). After another 4 hours, the mouse lungs were dissected for measurement of the BODIPY FL C16 levels in tumor cells (CD45−mCherry+) by flow cytometry.
Cell proliferation assay
Luciferase labeled cells (4T1-Luc, AT3-Luc, MCF7-Luc or MDA-4175-Luc) were seeded in a 24-well plate in 10% FBS/DMEM medium. Cells were allowed to adhere overnight, and were mono-cultured or co-cultured with freshly isolated neutrophils (neutrophils: tumor cells = 20 : 1) in DMEM supplemented with 10% FBS in a direct cell–cell contact manner for 24 hours. The cell culture media was then changed to delipidated media, and the luciferase activity was measured at different time points by using the SpectraMax i3 plate reader (Molecular Devices). For drug treatment experiments, 4T1-Luc cells were seeded and co-cultured with neutrophils as mentioned above. The cell culture medium was then changed to fresh delipidated media containing either DMSO, cerulenin (Cayman chemical) or C75 (Cayman Chemical) at the indicated doses. The final concentration of all wells contained equivalent amounts of DMSO solvent (0.2%). After a 48 hour treatment, the cells were harvested and the luciferase activity was measured by using the SpectraMax i3 plate reader. All values were normalized to the DMSO control sample and performed in triplicate for all cell lines.
Extracellular flux analysis of tumor cells
Fatty acid oxidation of tumor cells pre-cultured with neutrophils were determined through real-time measurement of the oxygen consumption rate (OCR) using the Seahorse XFe96 Analyzer (Agilent Technologies), according to the manufacturer’s instructions for the XF Fatty Acid Oxidation Assay. Briefly, FACS-isolated tumor cells were seeded into XFp 96-well microplate wells at a density of 30,000 cells per well and spun for 5 min at 1400 rpm to settle cells. The cells were maintained in XF assay media (XF DMEM medium (Agilent Technologies) supplemented with 1 mM pyruvate, 2 mM glutamine and 10 mM glucose) in a CO2-free 37 °C incubator for 30 minutes to 1 hour. Fifteen minutes prior to plate loading, etomoxir (or vehicle) was added to cells to a final concentration of 20 μM.
Neutrophil vesicle isolation and labeling
Neutrophils were incubated with fluorescently labeled palmitic fatty acid (BODIPY FL C16, 5 μM) at 37 °C in RPMI-1640 media for 1 hour. Then the extracellular BODIPY was removed from the cells by washing 4 times with 1× HBSS with 0.2% fatty acid–free BSA. The labeled neutrophils were incubated in RPMI 1640 media without serum at 37 °C at a density of 1 million cells per ml. After 16 hours, the conditioned medium was collected and centrifuged at 1,200 g for 15 minutes to remove cell debris and apoptotic bodies. The resulting supernatant was then passed through a 0.8 μm syringe filter (Sigma) and then concentrated via 100 kDa centrifuge filters (Millipore Amicon Ultra-15 mL Centrifuge Filters). The filtrated medium (<100 kDa) and concentrated vesicle-enriched medium (>100 kDa, washed 2 times with RPMI 1640) were used to feed tumor cells at a concentration of 1 million cell equivalents per ml. The tumor cells were fed for 4 hours prior to imaging. Lysotracker (LysoTracker™ Red DND-99, Thermo Fisher) was added for the last 30 minutes of incubation to label lysosome.
Measurement of the metastasis-initiating potentials of neutrophil-educated tumor cells
The luciferase-labeled tumor cells (AT3-Luc, MCF7-Luc, MDA-4175-Luc or 4T1-Luc) were mono-cultured or co-cultured with different neutrophils for 24 hours in delipidated medium, then the tumor cells were purified by depleting neutrophils using anti-Ly6G microbeads. The purified tumor cells (5×104 cells per injection) were then intravenously injected into NSG mice. Six days later, the mice were anesthetized under 2.5% isoflurane and administered with D-luciferin (150 mg kg−1) by intraperitoneal injection. The bioluminescence signals were detected with the Xenogen IVIS system. Four hours later, the mice were euthanized and the lungs were rapidly excised for ex vivo BLI analysis as described above. Photon emission from each lung tissue in ex vivo BLI analysis was quantified and shown.
Correlation analysis of LD-associated genes and human neutrophil signature genes using published microarray data
To evaluate whether the lipid gene expression is correlated with neutrophil signature genes in human lung metastasis samples, we analyzed a published microarray dataset (GSE1401820). The LD signature genes (HILPDA, CIDEC and G0S2) as revealed in this study, and the reported human neutrophil signature genes (CREB5, CDA, CHST15, S100A12, APOBEC3A,CASP5, MMP25, HAL, C1orf183, FFAR2, MAK, CXCR1, STEAP4, MGAM, BTNL8, CXCR2, TNFRSF10C and VNN3)21 were used for correlation analysis. For each signature, we averaged the expression level of the genes in each of the 16 individual samples. Then the Pearson correlation coefficient between LD signature and neutrophil signature gene expressions was calculated.
Survival analysis of genes related with lipid catabolism or neutrophil trafficking
In order to determine whether neutrophil gene signature and lipid catabolism gene signature are associated with lung metastasis-free survival, we utilized a microarray dataset profiled by Affymetrix Human Genome U133A Array from 99 primary breast cancer patients (GEO accession number: GSE2603)45. After excluding those with incomplete clinical annotations, it results in 82 analyzable samples. First, a univariate Cox regression analysis was performed to evaluate the association between expression levels of genes and samples’ lung metastasis-free survival (LMS). Then, we calculated a risk score for each sample which was defined as a linear combination of expression values of genes in one signature set weighted by their estimated Cox model regression coefficients. If the risk score for one sample was in the top 20th percentile of the risk scores, then it was classified into high-risk group, otherwise into low-risk group. The top 20th percentile was used as a threshold because about 20 percent of the cases were expected to eventually develop lung metastasis45. Finally, Kaplan-Meier survival curves and log-rank test were applied to evaluate the differences in sample’ LMS between high-risk group and low-risk group.
Quantification and statistical analysis
Data are presented as mean ± standard error of mean (s.e.m.) or mean ± standard deviation (s.d.) as indicated. P values were calculated using Prism 7.04 statistical software (GraphPad Software Inc.). The details of the statistical tests carried out are indicated in the respective figure legends and statistical tests were two-tailed unless otherwise indicated. Values were compared using an unpaired two-tailed t-test to compare two groups or ordinary one-way ANOVA with Tukey’s multiple comparisons test to compare the variance in three or more groups with one independent factor (e.g. treatment group). When there were effects of two factors (e.g. treatment and time) on a dependent variable, two-way ANOVA with Sidak’s multiple comparisons test was employed. For Kaplan–Meier analyses of breast cancer patients (GSE2603), statistical differences in survival curves were calculated by log-rank (Mantel–Cox) test. Correlation analysis of gene expression values of each signature gene group within each sample was performed by linear regression with Pearson’s correlation coefficient (r). Unless indicated in the figure legends, three independent experiments were performed and similar results were obtained. P value smaller than 0.05 was considered significant, whereas greater than 0.05 was assigned as not significant (ns). Exact P values are provided in each figure.
Data availability
The RNA-sequencing data are deposited to the ArrayExpress and are available under accession E-MTAB-9128. For lung metastasis-free survival analysis of breast cancer patients, the published microarray dataset was used (GSE2603, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2603). For correlation analysis of signature genes in breast cancer lung metastasis samples, the published microarray dataset (GSE14018, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14018) was used. All other data supporting the findings of this study are available within the article and its supplementary information files, and on reasonable request from the corresponding author. A Nature Research Reporting Summary for this article is available as a Supplementary Information file. Source data for Figs. 1–7 and Extended Data Figs. 1–5 are provided with the article.
Extended Data
Extended Data Fig. 1. Lung neutrophils acquire a lipid-laden phenotype in the orthotopic 4T1 tumor model.
a-d, Transcriptional profiles of BM, PB and lung neutrophils sorted from the orthotopic 4T1 tumor-bearing BALB/cJ mice (n = 3). Volcano plots showing fold-change and p-value for the comparison of lung versus BM neutrophils (a), and PB versus BM neutrophils (b) based on the RNA-seq data. P values were determined by unpaired two-tailed t-test and smaller than 0.05 were considered significant. A schematic diagram showing the major pathway and key factors in lipid metabolism (c). A heat map depicts expression levels of the major lipid metabolic genes (d). e, Upper: a diagram showing the pre-metastatic and metastatic stages in the orthotopic 4T1 tumor model; lower: Representative immunostaining of neutrophils (Ly6G) in lung sections of naïve and 4T1 tumor-bearing BALB/cJ mice (n = 4 from 2 independent experiments). Scale bars, 20 μm. f, Intracellular lipids in neutrophils detected by BODIPY 493/503 staining under microscope (n = 4 mice from 2 independent experiments). Scale bars, 5 μm. g, The flow cytometry gating strategy is shown: neutrophils were identified as the Ly6G+Ly6Clow cell population which was gated on CD45+CD11b+ cells. h, Measurement of total lipid contents in neutrophils isolated from naïve and 4T1 tumor-bearing mice by BODIPY 493/503 staining and flow cytometry (n = 6). i, Cellular TG contents in neutrophils (n = 5). In f and i, neutrophils were isolated from the indicated tissues and organs of orthotopic 4T1 tumor-bearing mice at the pre-metastatic stage. n represents biologically independent animals. Data are mean ± s.e.m. and P values were determined by one-way ANOVA with Tukey’s multiple comparisons test (h and i). ns, not significant.
Extended Data Fig. 2. Up-regulation of genes encoding ATGL inhibitory factors led to reduced TG hydrolysis in pre-metastatic lung-infiltrating neutrophils.
a-h, Levels of total triglyceride (TG) (a), cholesteryl ester (CE) (b), phosphatidylethanolamine (PE) (c), phosphatidylcholine (PC) (d), phosphatidylserine (PS) (e), phosphatidylinositol (PI) (f), ceramides (Cer) (g) and phosphatidylglycerol (PG) (h) in PB and lung neutrophils isolated from the pre-metastatic stage of orthotopic 4T1 tumor-bearing mice (n = 5), were determined by liquid chromatography-mass spectrometry. Relative levels of these lipid were also shown in Fig. 1f. The exact values of specific chemical species for each lipid type were provided in Source Data file. i-l, Neutrophils were isolated from the BM, PB and lung tissues of orthotopic 4T1 tumor-bearing mice (n = 5) at the pre-metastatic stage and compared for relative mRNA expression (to Rps18) of Hilpda, Cidec, G0s2, Atgl and Abhd5 by qRT-PCR (i); HILPDA, CIDEC and G0S2 protein expression by Western blotting with GAPDH as a loading control (j); relative TG hydrolase activity (k); and cellular lipase activity (l). n represents biologically independent animals. Data are mean ± s.e.m. and P values were determined by one-way ANOVA with Tukey’s multiple comparisons test (i and l) or unpaired two-tailed t-test (k). ns, not significant.
Extended Data Fig. 3. Neutrophil-specific genetic ablation of Hilpda or Cidec did not lead to significant changes on lung metastases of breast cancer in vivo.
a-f, The neutrophil-targeting cKO mice (a-c, Hilpda-cKO; d-f, Cidec-cKO), and their wild type littermates were orthotopically implanted with AT3-g-csf cells. On day 15, the relative lipid levels in lung neutrophils were determined by BODIPY 493/503 staining and flow cytometry (a and d) (n = 5 per group), and the resected primary tumors were weighed (b and e). At the end point (day 30), spontaneous lung metastases of WT and cKO mice were quantified (c and f). n = 13 (WT group) and 14 (Hilpda-cKO group) for b and c; n = 15 (WT group) and 14 (Cidec-cKO group) for e, n = 10 (WT group) and 12 (Cidec-cKO group) for f. g, A schematic diagram showing the modified experimental lung metastasis model employed in this study. Mice were first orthotopically implanted with non-labeled AT3-g-csf cells to induce a neutrophil-high host condition. Luciferase-labeled AT3-Luc cells were then implanted intravenously on day 10, a time point within the pre-metastatic stage. On day 15, the primary tumors were resected. At the end point, the lung metastatic progression of AT3-Luc cells was detected by ex vivo BLI. h, Following the above modified experimental lung metastasis model, the lung metastatic progression of AT3-Luc cells in WT and Cidec-cKO recipient mice was determined by ex vivo BLI. Representative BLI images of the harvested lungs are shown and red lines indicate blank wells without lung tissues (left) (n = 11). n represents biologically independent animals. Data are mean ± s.e.m. and P values were determined by unpaired two-tailed t-test. ns, not significant.
Extended Data Fig. 4. Tumor cells absorb lipids from lipid-laden lung neutrophils.
a, Representative images from 2 biologically independent experiments showing lipid transfer from BODIPY FL C16-loaded neutrophils to 4T1-mCherry cells. Arrowheads indicate neutrophils. Scale bars, 20 μm. b, 4T1-mCherry or AT3-mCherry cells were mono-cultured or co-cultured with their respective tumor-bearing mice-derived neutrophils that were pre-loaded with BODIPY FL C16. Then the intensity of BODIPY in tumor cells was examined by flow cytometry. Representative of 3 biologically independent experiments. c, The cellular TG contents of AT3-mCherry cells after mono-culture or co-culture with neutrophils. Data are mean ± s.d. from 3 biologically independent cell cultures. d, Measurement of the lipids in early lung-colonizing AT3-mCherry cells in G-CSF-pretreated mice without and with anti-Ly6G-based neutrophil depletion (see Methods) (n = 5). Data are mean ± s.e.m. e, The lipid transfer from BODIPY FL C16-loaded lung neutrophils to the indicated tumor cells in vivo as determined by flow cytometry (see Methods) (n = 4). Representative flow cytometry histograms are shown. f, As depicted in the left panel, AT3-mCherry or 4T1-mCherry cells were mono-cultured or co-cultured with PB or lung neutrophils. In one experimental group, lung neutrophils were pre-activated by phorbol 12-myristate 13-acetate (PMA) and hydrogen peroxide (H2O2). The total lipid levels in tumor cells were determined by BODIPY 493/503 staining and flow cytometry. Data are mean ± s.d. from 5 (4T1-mCherry) or 3 (AT3-mCherry) biologically independent cell cultures. Neutrophils used throughout this figure were isolated from the pre-metastatic stage of 4T1 tumor-bearing mice or AT3-g-csf tumor-bearing mice. n represents biologically independent animals. P values were determined by one-way ANOVA with Tukey’s multiple comparisons test. ns, not significant.
Extended Data Fig. 5. Neutrophil-derived lipids enhance the proliferative capacity of metastatic tumor cells.
a, Oxygen consumption rates (OCR) of AT3 cells, upon mono-culture or co-culture with PB or lung neutrophils (left). The amount of OCR derived from fatty acid oxidation was quantified as the magnitude of the response to etomoxir (right). b-d, Measurement of the proliferative capacities of the indicated tumor cells without and with co-culture with PB or lung neutrophils (see Methods). e-g, Comparison of the metastatic colonization potentials of the indicated tumor cells (e, AT3-Luc; f, MCF7-Luc; and g, MDA-4175-Luc) without and with co-culture with PB or lung neutrophils (see Methods). Representative BLI images of the recipient mice and quantification of BLI signals within the lung areas are shown (n = 5 for e and n = 6 for f, g). h, i, PB and lung neutrophils were isolated from AT3-g-csf tumor-bearing WT or Atgl-cKO mice (pre-metastatic stage). The intracellular lipids in neutrophils were detected by BODIPY 493/503 staining under microscope (representative of n = 5 mice). Scale bars, 5 μm (h). The cellular TG contents of neutrophils were measured (n = 5) (i). j, 4T1-Luc cells, upon co-culture with PB or lung neutrophils isolated from AT3-g-csf tumor-bearing WT or Atgl-cKO mice (pre-metastatic stage), were intravenously injected into NSG mice (see Methods). Representative BLI images of the recipient mice and quantification of BLI signals within the lung areas are shown (n = 6). k-m, As depicted in k, the effects of EIPA treatment on lung colonization by 4T1 tumor cells (l) and on primary tumor growth (m) were determined (see Methods) (n = 10). Neutrophils throughout this figure were isolated from the pre-metastatic stage of 4T1 tumor-bearing mice (c, d, f and g) or AT3-g-csf tumor-bearing mice (a, b and e), except otherwise stated. n represents biologically independent animals. Data are mean ± s.d. from 6 (a) or 4 (b-d) biologically independent cell cultures, and mean ± s.e.m. for e-g, i, j, l and m. P values were determined by two-way ANOVA with Sidak’s multiple comparisons test (b-d, mono-culture versus lung Neu co-culture), one-way ANOVA with Tukey’s multiple comparisons test (a, e-g, i and j) or unpaired two-tailed t-test (l and m). ns, not significant.
Extended Data Fig. 6. Both lipid- and neutrophil-associated gene expression signatures are related with lung metastasis in human breast cancer patients.
Kaplan–Meier plots of lung metastasis-free survival of breast cancer patients, stratified by expression of indicated gene signature sets in their primary tumors (GEO accession number: GSE2603, n = 82 patients). A risk score was calculated for each sample which was defined as a linear combination of expression values of genes in one signature set weighted by their estimated Cox model regression coefficients. If the risk score for one sample was in the top 20th percentile of the risk scores, then it was classified into high-risk group, otherwise into low-risk group. P values were calculated by a 2-sided log-rank test. The used gene signature sets are derived from Gene Ontology (GO): Biological Process of MsigDB v.7.1, including GO_POSITIVE_REGULATION_OF_LIPID_CATABOLIC_PROCESS (M14107, 25 genes); GO_POSITIVE_REGULATION_OF_LIPID_TRANSPORT (M11731, 61 genes); GO_LIPID_OXIDATION (M15880, 101 genes); GO_NEUTROPHIL_MIGRATION (M25402, 119 genes); GO_REGULATION_OF_NEUTROPHIL_CHEMOTAXIS (M19283, 30 genes); and GO_NEUTROPHIL_EXTRAVASATION (M24616, 13 genes).
Extended Data Fig. 7. A lung MC → neutrophil → tumor cell metabolic axis in the lung metastatic niche.
A schematic diagram depicts how lung neutrophils are stimulated by lung resident MCs to accumulate lipids, and in turn transport their stored lipids to metastatic tumor cells for survival and proliferation leading to accelerated lung metastasis in a breast cancer model.
Supplementary Material
Acknowledgments
We would like to thank Dr. Scott I. Abrams (Roswell Park Comprehensive Cancer Center) for providing the AT3 breast cancer cell line, Dr. Yibin Kang (Princeton University) for providing the MDA-4175 breast cancer cell line, and thank Dr. Robert A. Weinberg (Whitehead Institute for Biomedical Research) for providing the lentiviral vector expressing mouse Csf3. This work was supported by grants from the National Institutes of Health (R00-CA188093 to G.R., R37-CA237307 to G.R., P30-CA034196 to G.R., L.D.S. and S.L., R35-GM133562 to S.L., and R24-OD026440 and R01-AI132963 to L.D.S.), the U.S. Department of Defense (W81XWH-18-1-0013 to G.R.), Leukemia Research Foundation New Investigator Grant (to S.L.) and The Jackson Laboratory Director’s Innovation Fund (19000-17-31 to S.L.). Q.L. is supported by the Pyewacket Fund at The Jackson Laboratory. We acknowledge critical comments from Drs. Edison T. Liu, Nadia A. Rosenthal, Kevin Seburn and Carmen Robinett. We also thank Dr. Grace Stafford for RNA-seq analysis, and Will Schott for cell sorting, as well as assistance from The Jackson Laboratory Scientific Service.
Footnotes
Competing interests
The authors declare no competing of interest.
References
- 1.Liu Y, Cao X. Characteristics and Significance of the Pre-metastatic Niche. Cancer cell 2016, 30(5): 668–681. [DOI] [PubMed] [Google Scholar]
- 2.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nature reviews Cancer 2017, 17(5): 302–317. [DOI] [PubMed] [Google Scholar]
- 3.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 2015, 528(7582): 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau CS, et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 2015, 522(7556): 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 2018, 361(6409). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiegel A, Brooks MW, Houshyar S, Reinhardt F, Ardolino M, Fessler E, et al. Neutrophils Suppress Intraluminal NK Cell-Mediated Tumor Cell Clearance and Enhance Extravasation of Disseminated Carcinoma Cells. Cancer discovery 2016, 6(6): 630–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massague J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature 2016, 529(7586): 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupuy F, Tabaries S, Andrzejewski S, Dong Z, Blagih J, Annis MG, et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell metabolism 2015, 22(4): 577–589. [DOI] [PubMed] [Google Scholar]
- 9.Christen S, Lorendeau D, Schmieder R, Broekaert D, Metzger K, Veys K, et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell reports 2016, 17(3): 837–848. [DOI] [PubMed] [Google Scholar]
- 10.Loo JM, Scherl A, Nguyen A, Man FY, Weinberg E, Zeng Z, et al. Extracellular metabolic energetics can promote cancer progression. Cell 2015, 160(3): 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature medicine 2011, 17(11): 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messmer MN, Netherby CS, Banik D, Abrams SI. Tumor-induced myeloid dysfunction and its implications for cancer immunotherapy. Cancer immunology, immunotherapy : CII 2015, 64(1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heckmann BL, Zhang X, Xie X, Liu J. The G0/G1 switch gene 2 (G0S2): regulating metabolism and beyond. Biochimica et biophysica acta 2013, 1831(2): 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmanabha Das KM, Wechselberger L, Liziczai M, De la Rosa Rodriguez M, Grabner GF, Heier C, et al. Hypoxia-inducible lipid droplet-associated protein inhibits adipose triglyceride lipase. Journal of lipid research 2018, 59(3): 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munir R, Lisec J, Swinnen JV, Zaidi N. Lipid metabolism in cancer cells under metabolic stress. British journal of cancer 2019, 120(12): 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallochi AL, Teixeira L, Oliveira KDS, Maya-Monteiro CM, Bozza PT. Lipid Droplet, a Key Player in Host-Parasite Interactions. Frontiers in immunology 2018, 9: 1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chistiakov DA, Melnichenko AA, Myasoedova VA, Grechko AV, Orekhov AN. Mechanisms of foam cell formation in atherosclerosis. Journal of molecular medicine 2017, 95(11): 1153–1165. [DOI] [PubMed] [Google Scholar]
- 18.Missaglia S, Coleman RA, Mordente A, Tavian D. Neutral Lipid Storage Diseases as Cellular Model to Study Lipid Droplet Function. Cells 2019, 8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen TS, Jessen N, Jorgensen JO, Moller N, Lund S. Dissecting adipose tissue lipolysis: molecular regulation and implications for metabolic disease. Journal of molecular endocrinology 2014, 52(3): R199–222. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer cell 2009, 16(1): 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell reports 2017, 18(1): 248–262. [DOI] [PubMed] [Google Scholar]
- 22.Nakanishi M, Rosenberg DW. Multifaceted roles of PGE2 in inflammation and cancer. Seminars in immunopathology 2013, 35(2): 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugimoto Y, Narumiya S. Prostaglandin E receptors. The Journal of biological chemistry 2007, 282(16): 11613–11617. [DOI] [PubMed] [Google Scholar]
- 24.Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nature reviews Immunology 2015, 15(8): 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Dubois RN. Eicosanoids and cancer. Nature reviews Cancer 2010, 10(3): 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Saarinen AM, Hitosugi T, Wang Z, Wang L, Ho TH, et al. Inhibition of intracellular lipolysis promotes human cancer cell adaptation to hypoxia. eLife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight M, Braverman J, Asfaha K, Gronert K, Stanley S. Lipid droplet formation in Mycobacterium tuberculosis infected macrophages requires IFN-gamma/HIF-1alpha signaling and supports host defense. PLoS pathogens 2018, 14(1): e1006874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palazon A, Goldrath AW, Nizet V, Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity 2014, 41(4): 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 2004, 119(3): 431–443. [DOI] [PubMed] [Google Scholar]
- 30.den Brok MH, Raaijmakers TK, Collado-Camps E, Adema GJ. Lipid Droplets as Immune Modulators in Myeloid Cells. Trends in immunology 2018, 39(5): 380–392. [DOI] [PubMed] [Google Scholar]
- 31.Veglia F, Tyurin VA, Blasi M, De Leo A, Kossenkov AV, Donthireddy L, et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature 2019, 569(7754): 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart TJ, Abrams SI. Altered immune function during long-term host-tumor interactions can be modulated to retard autochthonous neoplastic growth. Journal of immunology 2007, 179(5): 2851–2859. [DOI] [PubMed] [Google Scholar]
- 33.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism 2008, 7(1): 11–20. [DOI] [PubMed] [Google Scholar]
- 34.Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017, 541(7635): 41–45. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Di Martino JS, Bowman RL, Campbell NR, Baksh SC, Simon-Vermot T, et al. Adipocyte-Derived Lipids Mediate Melanoma Progression via FATP Proteins. Cancer discovery 2018, 8(8): 1006–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang YY, Attane C, Milhas D, Dirat B, Dauvillier S, Guerard A, et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI insight 2017, 2(4): e87489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adrover JM, Nicolas-Avila JA, Hidalgo A. Aging: A Temporal Dimension for Neutrophils. Trends in immunology 2016, 37(5): 334–345. [DOI] [PubMed] [Google Scholar]
- 38.Schild T, Low V, Blenis J, Gomes AP. Unique Metabolic Adaptations Dictate Distal Organ-Specific Metastatic Colonization. Cancer cell 2018, 33(3): 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flaherty SE 3rd, Grijalva A, Xu X, Ables E, Nomani A, Ferrante AW Jr. A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science 2019, 363(6430): 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim JP, Gleeson PA. Macropinocytosis: an endocytic pathway for internalising large gulps. Immunology and cell biology 2011, 89(8): 836–843. [DOI] [PubMed] [Google Scholar]
- 41.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013, 497(7451): 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim SM, Nguyen TT, Ravi A, Kubiniok P, Finicle BT, Jayashankar V, et al. PTEN Deficiency and AMPK Activation Promote Nutrient Scavenging and Anabolism in Prostate Cancer Cells. Cancer discovery 2018, 8(7): 866–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Molecular therapy : the journal of the American Society of Gene Therapy 2005, 12(3): 468–474. [DOI] [PubMed] [Google Scholar]
- 44.Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nature reviews Clinical oncology 2019, 16(10): 601–620. [DOI] [PubMed] [Google Scholar]
- 45.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature 2005, 436(7050): 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell 2001, 104(4): 531–543. [DOI] [PubMed] [Google Scholar]
- 47.Lehuede C, Dupuy F, Rabinovitch R, Jones RG, Siegel PM. Metabolic Plasticity as a Determinant of Tumor Growth and Metastasis. Cancer research 2016, 76(18): 5201–5208. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Commisso C. Macropinocytosis in Cancer: A Complex Signaling Network. Trends in cancer 2019, 5(6): 332–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohs I, Ducimetiere L, Marinho J, Kulig P, Becher B, Tugues S. Restoration of Natural Killer Cell Antimetastatic Activity by IL12 and Checkpoint Blockade. Cancer research 2017, 77(24): 7059–7071. [DOI] [PubMed] [Google Scholar]
- 50.Basnet H, Tian L, Ganesh K, Huang YH, Macalinao DG, Brogi E, et al. Flura-seq identifies organ-specific metabolic adaptations during early metastatic colonization. eLife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 51.Breitkopf SB, Ricoult SJH, Yuan M, Xu Y, Peake DA, Manning BD, et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics 2017, 13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing data are deposited to the ArrayExpress and are available under accession E-MTAB-9128. For lung metastasis-free survival analysis of breast cancer patients, the published microarray dataset was used (GSE2603, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2603). For correlation analysis of signature genes in breast cancer lung metastasis samples, the published microarray dataset (GSE14018, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14018) was used. All other data supporting the findings of this study are available within the article and its supplementary information files, and on reasonable request from the corresponding author. A Nature Research Reporting Summary for this article is available as a Supplementary Information file. Source data for Figs. 1–7 and Extended Data Figs. 1–5 are provided with the article.