Abstract
Many research teams all over the world focus their research on the SARS-CoV-2, the new coronavirus that causes the so-called COVID-19 disease. Most of the studies identify the main protease or 3C-like protease (Mpro/3CLpro) as a valid target for large-spectrum inhibitors. Also, the interaction of the human receptor angiotensin-converting enzyme 2 (ACE2) with the viral surface glycoprotein (S) is studied in depth. Structural studies tried to identify the residues responsible for enhancement/weaken virus-ACE2 interactions or the cross-reactivity of the neutralizing antibodies. Although the understanding of the immune system and the hyper-inflammatory process in COVID-19 are crucial for managing the immediate and the long-term consequences of the disease, not many X-ray/NMR/cryo-EM crystals are available. In addition to 3CLpro, the crystal structures of other nonstructural proteins offer valuable information for elucidating some aspects of the SARS-CoV-2 infection. Thus, the structural analysis of the SARS-CoV-2 is currently mainly focused on three directions—finding Mpro/3CLpro inhibitors, the virus-host cell invasion, and the virus-neutralizing antibody interaction.
Electronic supplementary material
The online version of this article (10.1007/s10930-020-09933-w) contains supplementary material, which is available to authorized users.
Keywords: COVID-19, Coronavirus, Spike, Inhibitors, Protein Data Bank, Molecular docking
Introduction
In late 2019, a new coronavirus (CoV) strain has passed from the animal host to human and caused the severe acute respiratory syndrome.[1]. The pathogen responsible for the epidemic of coronavirus disease 2019 (COVID-19) was classified as SARS-CoV-2 [2]. The current COVID-19 outbreak probably emerged from a local seafood marketin Wuhan, the capital of China's Hubei province [3]. At the time of writing the present paper (August 12, 2020), there are more than 20,000,000 confirmed cases and nearly 800,000 deaths reported on (https://www.who.int). According to Worldometers https://www.worldometers.info/coronavirus/, almost 14,000,000 persons have recovered. The new characteristics of the SARS-CoV-2 have brought it to the top of the list of the scientific community. The Lancet journal allowed free access for the articles focused on the SARS-CoV-2 and issued a special section Coronavirus Resource Centre (https://www.thelancet.com). Many other prestigious publishers devoted special free platforms to COVID-19—Centers for Disease Control and Prevention CDC https://www.cdc.gov and https://www.coronavirus.gov, National Institute of Health—LitCovid (go.nature.com/3almd5p), Latest Research and Reviews (https://www.nature.com), Coronavirus: Research, Commentary, and News (https://www.sciencemag.org), Corona-Webinar (https://www.springer.com), COVID-19/SARS-CoV-2 Resources (https://www.rcsb.org/), etc [4].
The SARS-CoV-2 is an enveloped RNA positive-sense single-stranded virus and belongs to the Coronaviridae family, the betacoronavirus group. The SARS-CoV-2 is the seventh human HCoV (HCoV) discovered until now. The other six HCoVs are HCoV-229E, HCoV-OC43, HCoV-NL63, HCoV-HKU1, severe acute respiratory syndrome coronavirus (SARS-CoV), and the Middle East respiratory syndrome coronavirus (MERS-CoV) [5]. There is the hypothesis that the origin of the SARS-CoV-2 strain is the bat [3]. The hypothesis that the intermediate host between the bat and the human is pangolin is not supported by the latest works [6, 7]. Much of the knowledge about the transmission of SARS-CoV-2 has gleaned from the SARS-CoV and MERS-CoV studies [5, 8]. There are some notable differences between tissue tropism of the HCoVs. Analysis of structural similarities and structural differences between HCoVs species could advance the understanding of SARS-CoV-2 pathogenesis [8–12].
The review aims to collect and synthesize the X-ray/NMR/cryo-EM structures of the SARS-Cov-2 deposited in the public database Protein Data Bank (PDB) (https://www.rcsb.org/pdb). Structures retrieved from PDB (August 12, 2020) were analyzed for relevant information on COVID-19 infection, synthesis of new inhibitors, SARS-CoV-2 interaction with host receptors, and the neutralizing antibodies interactions with spike glycoprotein. In this review, the SARS-CoV-2 related structures published in peer-reviewed papers are analyzed in depth. Very often the authors add minor/major revisions of the coordinate files after the PDB structure was submitted in the PDB. An association of scientists has created a public database in which CoVs structures are systematically validated (https://covid-19.bioreproducibility.org/) [13].
Objectives
The aim of this review is to provide an analysis of the SARS-CoV-2 structures deposited in the PDB. Every Wednesday, the COVID-19/SARS-CoV-2 Resources announce the new PDB structures (https://www.rcsb.org/). The rapid accumulation of the X-ray/NMR/cryo-EM structures of the SARS-Cov-2 in the Protein Data Base (PDB) needs an objective selection analysis of these crystal structures for further research. The COVID-19 is an ongoing pandemic and the virus undergoes mutations reflected in differences of the crystal structures. There are many crystal structures of the SARS-CoV-2 spike (S) co-crystallized with antibodies that further advance the understanding of the immunogenicity of the S [14–25]. Also, there is an intense work on designing effective inhibitors. There are many compounds mainly co-crystallized with the viral main protease (3CL-protease). The subgroup analysis of structural and non-structural protein (nsp) of the SARS-Cov-2 and other CoVs includes the multiple sequence alignment.
Methods
The X-ray/NMR/cryo-EM structures of the SARS-CoV-2 were retrieved by searching “SARS-CoV-2” in PDB (https://www.rcsb.org/pdb).
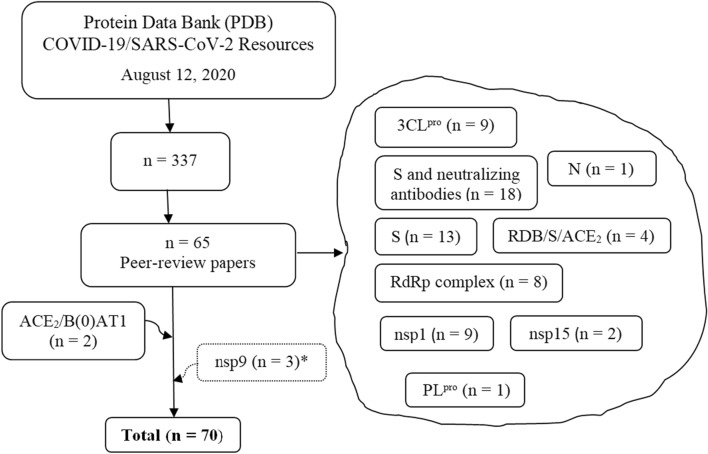
Inclusion criteria: The present review narrows the analysis of the SARS-CoV-2 crystal structures deposited in the PDB only for the crystal structures published in peer-reviewed journals (August 12, 2020). The further analysis of some important characteristics of the SARS-CoV-2 needs the inclusions of other five crystal structures (Fig. 1). The structures were checked in the Validated SARS-CoV-2 related structural models of potential drug targets site (https://covid-19.bioreproducibility.org/) [13].
Fig. 1.
The flow of the selection of the crystal structures retrieved from the PDB. *To be published
Exclusion criteria: The PDB SARS-CoV-2 related crystal structures non-published in peer-reviewed journals.
The structures were visualize and analyzed on Dassault Systèmes BIOVA program—Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes, 2016 (https://accelrys.com).
Sequence analysis: Multiple sequence alignment was made by the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo).
Molecular docking protocol: Protein–ligand docking experiments were performed with the AutoDock4.2 program, distributed as open source under a GPL license (https://autodock.scripps.edu) [26–28].
The SARS-CoV-2 Structures Analysed
The search of “SARS-CoV-2” in PDB retrieved 337 entries (August 12, 2020) (Table S1). Among them, only 65 of 2 X-ray/NMR/cryo-EM structures are published until now—PDB ID(s) 6LXT, 6VYB, 6VXX, 7BTF, 6M71, 6M0J, 6M3M, 6W01, 6VWW, 6VSB, 6LZG, 7BV1, 7BV2, 6YYT, 6M17, 6VW1,6XR8, 6XRA, 6LU7, 7BQY, 6LZE, 6M0K, 7BUY, 6Y2E, 6Y2G, 6Y2F, 6W41, 6YOR, 7BZ5, 7C01, 7BWJ, 7BYR, 6XCM, 6XCN, 6XCA, 7C2L, 7BZF, 7C2K, 6Z97, 6ZLW, 6ZM7, 6ZN5, 6ZON, 6ZP4, 6XDG, 6WQF, 7BW4, 7CAH, 6XEY, 6ZOX, 6ZOZ, 6ZOY, 6ZP0, 6ZP1, 6ZP2, 6YVA, 6ZCZ, 6ZDH, 6ZDG, 6ZER, 6ZFO, 6ZMI, 6ZMO, 6ZMT, and 6ZME [14–25, 29–50]. The first X-ray structure found (PDB ID 6LU7) belongs to the non-structural protein 5 (3C-like protease) of the SARS-CoV-2 in complex with the Michael acceptor-based inhibitor N3 (PRD_002214). The structure was deposited in PDB in 2020-01-26 and released in 2020-02-05 by the Liu X. et al. [41].
Two PDB entries (6M1D and 6M18) of the human ACE2 co-crystallized with the sodium-dependent neutral amino acid transporter B(0)AT1were added because these structures are part of the same work with the spike—receptor binding domain (RBD)/ACE2/ B(0)AT1 EM-structure (PDB ID 6M17) [39]. Three nsp9 structures non-published in the peer-reviewed papers were included in the study (PDB ID(s) 6W9Q, 6W4B, and 6WXD). Their sequences were analyzed by multiple sequence alignment and compared with previous works. The selection flow of the 70 PDB structures included in the present review is shown in Fig. 1.
SARS-CoV-2 Inhibitors
One of the most urgent aims of controlling the SARS-CoV-2 pandemic is to find out an efficient therapy. Healthcare professionals have established different clinical practice guidelines. The update of the clinical results helps the medical teams to monitor and refine their therapy, but a conclusion could be drawn only at the end of the SARS-CoV-2 pandemic. There is no specific cure for COVID-19, which explains the race to discover effective inhibitors and a universal vaccine. Despite numerous SARS-CoV-2 PDB structures co-crystallized with inhibitors, only a few of them have been publishing in peer-reviewed papers. Many research teams focus on designing inhibitors of viral proteases due to their role in viral replication [51]. There are two viral proteases: main protease/3C-like protease (Mpro/3CLpro) and papain-like protease (PLpro). These proteases cleave the polyprotein 1ab (UniProtKB P0C6X7) to yield the viral proteins [44]. There are seven PDB structures of SARS-CoV-2 3CLpro co-crystallized with antiviral drug candidates. Two structures are co-crystallized with N3 (PRD_002214) (PDB ID(s) 6LU7 and 7BQY) [41]. The 6LZE and 6M0K structures are co-crystallized with compounds 11a and 11b, respectively [42]. Two structures are co-crystallized with alpha-ketoamide 13b (PDB ID(s) 6Y2F and 6Y2G) [44]. The 7BUY structure is co-crystallized with the antineoplastic drug carmofur [43]. There is a cryo-EM crystal structure of the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) complex (nsp12/nsp8/nsp7) with the antiviral drug remdesivir (PDB ID 7BV2) [37]. However, these compounds are not optimal inhibitors of SARS-CoV-2. The comparison of the antiviral drug candidates against SARS-CoV-2 and other CoVs is shown in Table 1.
Table 1.
The comparison of the antiviral drug candidates against SARS-CoV-2 and other CoVs
| Compound | SARS-CoV-2 | Other CoVs | References | ||
|---|---|---|---|---|---|
| CC50 | EC50 | IC50 | IC50 | ||
| 3CLpro inhibitor candidates | |||||
| N3 | > 133 μM | 16.77 μM | – |
4.0 μM (HCoV-229E) 8.8 μM (FIPV) 2.7 μM (MHV-A59) |
[41, 51, 61, 63] |
| Ebselen | > 30 μM | 4.67 μM | 0.67 μM | - | [41, 51, 61, 63] |
| Cinaserin | > 200 μM | 20.61 μM | 124.93 μM |
31 μM (SARS-CoV) 19 μM (HCoV-229E) |
[41, 51, 61, 63] |
| 11a | > 100 μM | 0.53 μM | 0.053 μM | – | [42] |
| 11b | > 100 μM | 0.72 μM | 0.04 μM | – | |
| 13b | NA | 4–5 μM | 0.67 μM |
0.9 μM (SARS-CoV) 0.58 μM (MERS-CoV) |
[44] |
| Carmofur | 133.4 μM | 24.30 μM | 1.82 μM | NA | [43] |
|
Mean (SD) CI95 |
10.3 (9.23) 66.41 |
21.36 (46.31) 173.49 |
9.56 (10.57) 81.88 |
||
| PLpro inhibitor candidate | |||||
| GRL-0617 | – | – | 1.5 μM | 0.6 μM (SARS-CoV) | [49, 84] |
| RdRp inhibitor candidate | |||||
| Remdesivir | > 100 μM | 0.72 μM | – | – | [59] |
| Antiprotozoal drug | |||||
| Cloroquine | > 100 μM | 1.13 μM | – | – | [59] |
N3 (PRD_002214) (phenylmethyl) (4 ~ {S})-4-[[(2 ~ {S})-4-methyl-2-[[(2 ~ {S})-3-methyl-2-[[(2 ~ {S})-2-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]propanoyl]amino]butanoyl]amino]pentanoyl]amino]-5-[(3 ~ {S})-2-oxidanylidenepyrrolidin-3-yl]pent-2-enoate; 11a ~ {N}-[(2 ~ {S})-3-cyclohexyl-1-oxidanylidene-1-[[(2 ~ {S})-1-oxidanylidene-3-[(3 ~ {S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1 ~ {H}-indole-2-carboxamide; 11b ~ {N}-[(2 ~ {S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2 ~ {S})-1-oxidanylidene-3-[(3 ~ {S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1 ~ {H}-indole-2-carboxamide; 13b ~ {tert}-butyl ~ {N}-[1-[(2 ~ {S})-3-cyclopropyl-1-oxidanylidene-1-[[(2 ~ {S},3 ~ {R})-3-oxidanyl-4-oxidanylidene-1-[(3 ~ {S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate; GRL-0617 is naphthalene-based inhibitor, CC50 is the cytotoxic concentration of the extracts to cause death to 50% of viable cells in the host; EC50 is half maximal effective concentration; IC50 is half maximal inhibitory concentration, NA not available; SD is standard deviation, CI95 confidence interval 95%
SARS-CoV-2 3C-Like Protease Inhibitors
The 3CLpro (UniProtKB P0DTD1) is extensively studied for designing of new inhibitors due to its unique characteristics. It is also known as non-structural protein 5 (nsp5). It is a cysteine protease involved in the cleavage of the viral polyproteins 1a and 1ab. Moreover, there are no human counterparts for 3CLpro [52–57]. The 3CLpro is one of the targets for the control of zoonotic reservoir of CoVs. The peptidomimetics with 3-thiophene and 1-methylbenzotriazole backbones inhibit the bat CoVs HKU4-CoV and HKU5-CoV at sub-micromolar concentrations [58].
The need for specific treatment of COVID-19 had led to testing the inhibitory activity of other antiviral drugs. At the beginning of the SARS-CoV-2 pandemic, Wang et al. published the comparison of inhibitory activity of five antiviral drugs (ribavirin, penciclovir, favipiravir, nefamostat, and remdesivir) and two antiprotozoal drugs (nitazoxanide, and cloroquine). The EC50 of remdesivir and cloroquine against SARS-CoV-2 are 0.72 μM and 1.13 μM, respectively [59]. Dai et al. published the compounds 11b that showed EC50 of 0.72 μM, identical to the EC50 of remdesivir (Table 1) [42].
Compounds 11a and 11b (PDB ID(s) 6LZE and 6M0K) show good inhibitory activity at a concentration of 1 μM—100% and 96%, respectively [42]. Zhang et al. published three SARS-CoV-2 3CLpro—one free enzyme (PDB ID 6Y2E) and two crystal structures with a new inhibitor candidate—an alpha-ketoamide compound 13b (PDB ID(s) 6Y2G and 6Y2F) [44]. Compound 13b is a non-polymer compound derived from a peptidomimetic inhibitor against enterovirus proteases [60]. The X-ray crystal structures of the SARS-CoV-2 3CLpro co-crystallized with 13b demonstrated that the interactions from the dimer interface are essential for shaping the binding pocket. Also, the authors greatly improved the pharmacokinetic characteristics of the compound 13b. The compound 13b can be administered as an inhaler [44].
Two X-ray crystal structures of the SARS-CoV-2 3CLpro co-crystallized with Michael acceptor-based inhibitor (PRD_002214) were recently published: PDB ID(s) 6LU7 and 7BQY. The inhibitor PRD_002214, the inhibitor N3, is the result of the high-throughput screening of more 10,000 inhibitory compounds against SARS-CoV-2 3CLpro. The kinetic experiments showed that N3 is a time-dependent irreversible peptide-like inhibitor of the SARS-CoV-2 3CLpro. Molecular docking analysis demonstrated that N3 fit inside the substrate-binding pocket (Cys145-His41 catalytic dyad) highly conserved in CoVs [41, 61]. A recent structural study hypothesized that a cluster of residues outside the catalytic site are possibly conformationally relevant when bound to the N3 [62]. The inhibition of hydrolytic activity of the N3 was previously determined for other CoVs (HCoV-22E, FIPV, and MHV-A59) (Table 1). The overall analysis of the available data suggests that N3 is a wide-spectrum anti-CoV lead compound [63].
Another structural study demonstrated that the antineoplastic drug carmofur inhibits the 3CLpro SARS-CoV-2 (PDB ID 7BUY). Carmofur (PubChem CID 2577) (C11H16FN3O3) is an antimetabolite (pyrimidine analog) derivative of 5′-fluorouracil. Carmofur is used to treat colorectal cancer and breast cancer (DrugBank DB09010). The Jin et al. study shows that carmofur inhibits viral replication by forming a covalent bond with the cysteine residue from the catalytic site of the 3CLpro SARS-CoV-2 (Table 1) [64].
The 3CLpro is a particular cysteine protease that comprises three domains. The 3CLpro proteolytic activity requires the formation of the Cys(−)/His(+) zwitterionic state onto the catalytic dyad [65, 66]. Although the catalytic mechanism of CoV 3CLpro is not fully understood, extensive studies that connected the structural, computational, and biochemical approaches of different wild-type and mutated 3CLpro decipher important aspects of catalytic efficiency [67]. Thus, Wang et al. identify that, besides the cysteine from the catalytic dyad, the 3CLpro MERS-CoV has another cysteine nearby. The role of the second cysteine in catalysis is not fully understood. A conserved motif GSCGS forms consecutive three turns when starting the catalysis. Two structural characteristics are essential for catalysis. The first characteristic is a partial negative cluster formed by Arg-Tyr-Asp. Secondly, there is a conserved water molecule that mediates remote control between the partial negative charged cluster and the Cys-His dyad. Also, a conserved pair (Glu-His) very well conserved in 3CLpro CoVs forms a stable hydrogen-bond. The glutamine substrate recognizes the Glu-His pair by a steric effect. Three more residues have an essential role in the glutamine substrate interactions—the His166 and the nearby Tyr164 and Phe143. The tyrosine residue forms a strong hydrogen bond with His and the phenylalanine residue employs a steric effect to restrain the rotation of His [67].
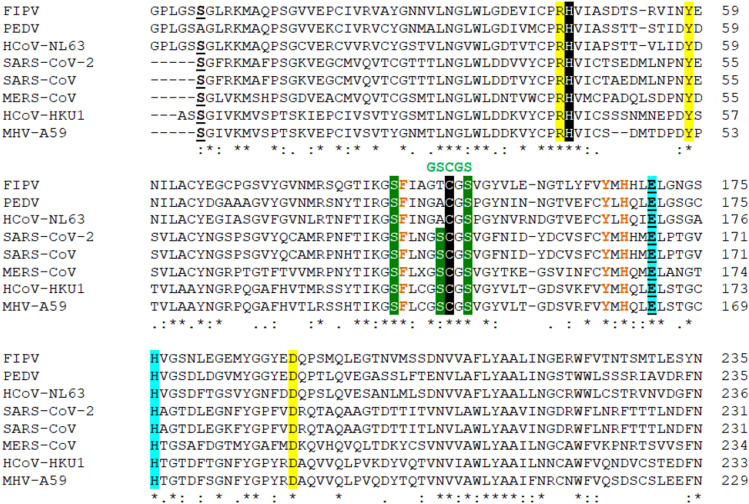
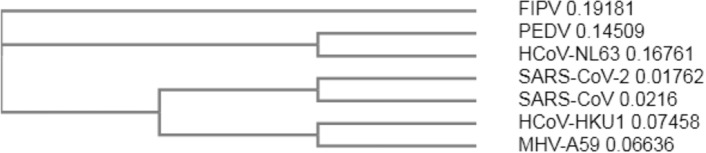
The multiple sequence alignment of the 3CLpro sequences from different CoVs allows an overall analysis. Thus seven 3CLpro from the following CoVs were aligned using Clustal Omega program: feline infectious peritonitis virus FIPV (PDB ID 5EU8), porcine epidemic diarrhea virus PEDV (PDB ID 5GWZ), human HCoV-NL63 (PDB ID 5GWY), SARS-CoV-2 (PDB ID 6LU7), SARS-CoV (PDB ID 3IWM), human HCoV-HKU1 (PDB ID 3D23), MERS-CoV (PDB ID 4RSP), and murine coronavirus (strain A59) MHV-A59 (PDB ID 6JIJ). The residues essential for catalysis—the Cys145-His41 dyad and the GSCGS motif highly conserved in CoVs—are shown in Fig. 2. The phylogenetic tree (cladogram) of the 3CLpro from different CoVs sequences analyzed confirms that SARS-CoV-2 is closer to the SARS-CoV (Fig. 3).
Fig. 2.
The multiple sequence alignment of 3CLpro of CoVs of different origin (using the Clustal Omega program). The Cys145 of SARS-CoV-2 3CLpro that interact with the compounds 11a and 11b is shown in black background along with the Cys145-His41 catalytic dyad highly conserved in 3CLpro from CoVs [42, 44, 59, 65, 66]; in green are marked the cluster of Ser with high affinity for small molecule inhibitors [44, 55]; in green it is shown the GSCGS motif essential for starting the catalysis; in light blue are marked the Glu-His residues critical for substrate binding by means of steric effect; in yellow background is marked the triad Arg-Tyr-Asp that forms a partial negative charge cluster that, by a conserved water molecule, mediates the interaction with Cys-His catalytic dyad; in brown are shown the residues involved in the glutamine substrate recognition—the conserved His and the conserved Tyr and Phe that interacts with by the phenolic hydroxyl group with His and employs a steric effect to restrain the rotation of His, respectively; in bold-underline are marked the Glu and Ser residues that are demonstrated to be essential in the dimer interactions in SARS-CoV-2 [60]; with dot “.” are marked the semi-conservative replacements; with colon “:” are marked the conservative replacements; with “*” are marked the identities of the residues
Fig. 3.
The phylogenetic tree (cladogram) of seven 3CLpro sequences of CoVs with different origin. feline infectious peritonitis virus (FIPV), porcine epidemic diarrhea virus (PEDV), HCoV-NL63, SARS-CoV-2, SARS-CoV, HCoV-HKU1, and coronavirus (strain A59) MHV-A59; performed by Clustal Omega program
SARS-CoV-2 RNA-Dependent RNA Polymerase Inhibitors
The RNA-dependent RNA polymerase (RdRp) (UniProtKB P0DTD1), also known as nsp12, is responsible for the replication and transcription of the viral genome. The RdRp complex is composed of the nsp12, nsp7, and nsp8. The nsp12 is the catalytic subunit and needs the nsp7 and nsp8 (eight subunits of each) to fulfill the replication process. The RdRp is considered an important target for new/already known drugs because of its evolutionary stability compared with the S glycoproteins that are more prone to mutations under selections of host immunity [48, 68]. A recent study investigated the feasibility of the RdRp to be targeted by novel nucleoside inhibitors or small molecules [69]. One previous work has shown that nsp7 and nsp8 of feline CoV form a 2:1 heterodimer (PDB ID 3UB0). The nsp7 and nsp8 from the SARS-CoV form an 8:8 hexadecamer (PDB ID 6NUR) and act as primase during viral replication [70–72].
Remdesivir (GS-5734) (PubChem CID 121304016) (DrugBank DB14761) is a prodrug of adenosine triphosphate (ATP) analog. Remdesivir is metabolized into the active form remdesivir-triphosphate (remdesivir-TP) (NCI Thesaurus GS-441524). The antiviral activity of the remdesivir-TP consists of the competition with ATP for RNA incorporation thus inhibiting viral RdRp. Remdesivir is a valuable therapeutic agent against Ebola virus infections [73]. Many studies investigated the potential therapeutic use of remdesivir against other viral infections including COVID-19 [74–77]. The therapeutic potency of the remdesivir in COVID-19 needs much more well-conducted studies and a thorough analysis of the clinical results [78]. The SARS-CoV-2 RdRp structure provides important structural details about RdRp -RNA interactions (PDB ID(s) 6YYT, 7BTF, 6M71, and 7BV1) [31, 37, 38]. The structures co-crystallized with remdesivir further advances the understanding of the mechanisms of viral RNA replication (PDB ID 7BV2) [37]. Yin et al. demonstrates that RdRp inhibition depends on the low remdesivir-triphosphate (RTP) concentration and low RTP/ATP ratio. Only RTP inhibits the RdRp polymerization activity. The structural studies have shown that the Asn691, Ser682, and Asp623 in the RdRp complex explain the binding of the RTP to the NTPs site [38]. However, the structural studies should be interpreted according to biochemical studies to further elucidate the incorporation of RTP into the growing RNA chain [38, 77, 79, 80].
The recent structural works provide insight into the nucleotide analog inhibitors’ ability to hamper SARS-CoV-2 RNA replication. The pre-translocated and post-translocated RdRp complex structures (PDB ID(s) 7C2K and 7BZF) show that an efficient inhibitory strategy could be the blocking of the interaction between nsp8 and nsp12 [45]. The biochemical and structural works of Peng et al. (PDB ID 7BW4) demonstrate that SARS-CoV-2 RdRp enzymatic activity is lower, about 35% for RNA synthesis than its SARS-CoV counterpart. The reason is the residue substitution in nsp12 and nsp8. Also, the thermal stability of the SARS-CoV-2 RdRp is lower compared to that of SARS-CoV RdRp The thermal stability of RdRp could advance the understanding of the adaptive evolution of SARS-CoV-2 in the human host that have a lower body temperature than bats [3, 48, 81].
SARS-CoV-2 Papain-Like Protease Inhibitors
The papain-like protease (PLpro) is a cysteine protease. PLpro, RNA binding domain, and membrane-anchoring domain form the non-structural protein 3 (nsp3). PLpro from different HCoVs has been screened for a large panel of chemicals because it is involved in CoV replication [82, 83]. The biochemical, structural, and functional studies advance the development of new inhibitors against SARS-CoV-2 PLpro. The naphthalene-based inhibitor GRL-0617 (PubChem CID 44828571) is an effective inhibitor of the SARS-CoV PLpro (Table 1) [84]. Shin et al. advances the hypothesis that the GRL-0617 has two effects against SARS-CoV-2 PLpro. First, the GRL-0617 binding to the amino acid Y268 was confirmed by the reduction of the inhibitory effect in mutated PLpro Y268T and Y268G. Second, the GRL-0617 promotes the antiviral interferon pathway and reduces viral replication (PDB ID 6YVA) [50].
SARS-CoV-2 Nucleocapsid Protein Inhibitors
The CoV nucleocapsid (N) is a highly immunogenic multifunctional protein. The CoV N binds the viral RNA strand into a long helical structure attached to the membrane (M) protein [85]. There are nine SARS-CoV-2 X-ray structures of N protein, but only one crystal structure is published until now (PDB ID 6M3M) [33]. Similar to the N proteins of other CoVs, the SARS-CoV-2N protein consists of the N-terminal RNA-binding domain (N-NTD), the C-terminal dimerization domain (N-CTD), and a Ser/Arg rich linker [86]. The ribonucleotide binding site of the CoV N-NTD domain was subject to structural studies to develop inhibitors that specifically reduce the RNA-binding affinity, thus altering the viral replication. The SARS-CoV-2N structures available are not co-crystallized with any inhibitor, but the Kang et al. structural study demonstrated that the SARS-CoV-2N employs a unique pattern for ribonucleotide binding, the residues Arg89 being one reason for the weak guanosine base recognition (KD for guanosine monophosphate GMP is 8 mmol/L) [33]. The Kang et al. claimed that these structural characteristics could be exploited for further investigation of inhibitor compounds, mainly the inhibitor PJ34 of the HCoV-OC43 N-NTD (PDB ID 4KXJ) which reduces the N protein’s RNA-binding affinity by 10% [86]. The PJ34 or N ~ 2 ~ ,N ~ 2 ~ -dimethyl-N ~ 1 ~ -(6-oxo-5,6-dihydrophenanthridin-2-yl)glycinamide (PubChem CID 4859, DrugBank DB 08,349) has the molecular weight 295.34 g/mol and molecular formula C17H17N3O2. The X-ray crystal structure deposited by Kang et al. (PDB ID 6M3M) was not co-crystallized with the inhibitor PJ34.
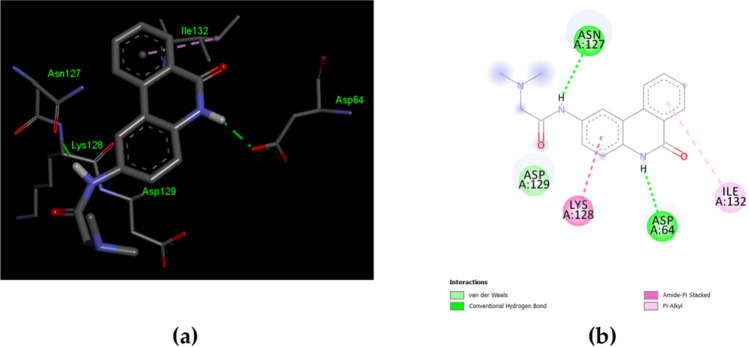
Herein, a further molecular docking of the inhibitor PJ34 with the SARS-CoV-2N-NTD (PDB ID 6M3M) was made. PJ34 interactions with SARS-CoV-2N proteins’ N-terminal domain (NTD) were compared with those of the HCoV-OC43 co-crystallized with the PJ34 inhibitor (PDB ID 4KXJ). The molecular docking of the compound PJ34 with the PDB ID 6M3M shows the following parameters: ∆G = − 5.66 and Ki = 71.34 μM. The interactions involved the residues that bind the PJ34 are different from the residues previously described at its counterpart from HCoV-OC43 (Fig. 4) [86]. The molecular docking offers only a quick view about ligand–protein interaction, deeper theoretical analysis, and experimental validation should be made. Sequence comparison of the N proteins' N-terminal domain (NTD) of the HCoV-OC43 and SARS-CoV-2 reveals some particularities that are worth further investigation. The sequence positions 48 to 51 (48-NNTA-51) allows easier access of the nucleotides. The threonine T55 and alanine A56 increase the steric clash with ribonucleotide phosphate moiety. The arginine R89 increases the polar features in the nitrogenous base binding site. (Fig. 5) [33].
Fig. 4.
The interactions of SARS-CoV-2 nucleocapsid (N) (PDB ID 6M3M) with the inhibitor PJ34. a 3D display of PJ34 interaction as ligand with the 6M3M residues; b 2D interactions diagram; the molecular docking results were visualized by Dassault Systèmes BIOVA program—Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes, 2016 (https://accelrys.com)
Fig. 5.
The multiple sequence alignment of SARS-CoV-2 nucleocapsid (N) (PDB ID 6M3M) and HCoV-OC43 (PDB ID 4KXJ) sequences (using the Clustal Omega program). The residues that interact with the compound PJ34 are shown in gray background, according to molecular docking results for 6M3M and according to Lin et al. findings 4KXJ; are considered all types of interactions—van der Waals, hydrogen bonds, carbon hydrogen bonds, and hydrophobic interactions; the striking differences between the two N sequences observed by the Kang et al. are marked in yellow background; the AMP-binding residues are underlined; in red is marked the Tyr residue whose mutation Y110A leads to a significant decrease of Kd for RNA binding [33]; with dot “.” are marked the semi-conservative replacements; with colon “:” are marked the conservative replacements; with “*” are marked the identities of the residues
SARS-CoV-2 Cell Entry
There are many works about the cell entry of CoVs because, on the one hand, this step is crucial for tissue and cell tropism, and on the other hand, it gains insight into the ability for interspecies transmission. The viral entry is crucial for viral replication. Therefore its deep understanding explains the SARS-CoV-2 pathogenesis and speeds up the finding of a specific treatment. The cell entry is a different multi-step process for each CoV. The first step of any viral infection is the presence of at least one suitable host receptor. The spike (S), a heavily glycosylated type I transmembrane glycoprotein, is the same to all HCoVs, but the host receptor is different [87]. Similar to the SARS-CoV and human coronavirus NL63 (HCoV-NL63), the SARS-CoV-2 attaches to host receptor ACE2 by S glycoprotein [3, 88, 89]. In contrast, the MERS-CoV uses cellular receptor dipeptidyl peptidase-4 (DPP4), known as CD2 receptor [90–92].
The S glycoprotein (UniProtKB P59594) is a homotrimer and each monomer contains two distinct functional subunits—S1 and S2—that are subject to proteolytic cleavage by cellular proteases. The host factors involved in priming the S glycoprotein are the type II transmembrane serine protease 2 (TMPRSS2) and furin [30, 93–95]. The TMPRSS2 is predominantly expressed in the prostate. Thus, the demonstration that its expression is androgen-dependent could explain the gender differences in the COVID-19 outcome. Also, the TMPRSS2 has a crucial role in other viral infections explaining the pneumotropism of H7N9 and some subtypes of influenza viruses [96]. The furin (E.C. 3.4.21.75) (PDB ID(s) 4OMC, 4OMD, etc.) (UniprotKB P09958) is a ubiquitous endoprotease that belongs to the family of proprotein convertases. The furin mediates the priming of various proteins—some bacterial toxins, H7N1and H5N1 influenza virus haemagglutinin [97, 98]. The proprotein convertases (PPC) cleave at single or paired basic residues within the motif R/K-(X)0,2,4,6-R/K (where X means any residue) [93, 99]. There is one cleavage at the S1/S2 boundary and the second one S2′ is within S2 upstream of the putative fusion protein [94, 96]. Some researchers claim that there are three cleavage sites (S1, S2, and S2′) [100]. The S1 subunit harbors the receptor binding domain (RBD) which contains the core and receptor-binding motif (RBM) [101]. The S1 subunit binds the peptidase domain of the human ACE2. Then, the internalization of the virus particle into the endosomes causes conformational changes of the S1 [35, 102]. The proteolysis by the cathepsin CTSL (E.C. 3.4.22.15; UniProtKB P07711) reveals the S2 subunit which mediates the virus-cell membrane fusion [2, 103–105].
Herein, a comparison of the S1/S2 and S2′ cleavage sites of the CoVs and SARS-CoV-2 was made [93]. The multiple sequence alignment of the different SARS-CoV-2 S glycoprotein with their counterparts of CoVs described by the Millet et al. allows the comparison of the furin cleavage sites. The following sequences were analyzed: nine SARS-CoV-2 sequences (PDB ID(s) 6XR8, 7C2L, 6VXX, 6WPS, 6VYB, 6XCN, 6XEY, 6X6P, and 6VSB), SARS-CoV [NP_828851.1], MERS-CoV [AFS88936.1], HCoV-HKU1 [AAT98580.1], HCoV-OC43 [AAA03055.1], HCoV-NL63 [AAS58177.1], HCoV-229E [BAL45637.1], canine CoV (CCoV-Elmo/02)[AAP72149.1], feline CoV (FCoV-RM)[ACT10854.1], Rousettus bat CoV (BatCoV-HKU10)[YP_006908642.1], Porcine epidemic diarrhea virus (PEDV-CV777)[AAK38656.1], TGEV Miller M60 [ABG89306.1], Murine hepatitis virus (MHV-A59)[AAA46455.1], Tylonycteris bat coronavirus HKU4 (BatCoV-HKU4)[YP_001039953.1], Pipistrellus bat coronavirus HKU5 (BatCoV-HKU5)[YP_001039962.1], and Infectious bronchitis virus (IBV-Beaudette)[NP_040831.1] (Figs. 6, 7).
Fig. 6.
The multiple sequence alignment of spike (S) S1/S2 cleavage site sequences using the Clustal Omega program. The S1/S2 sequences are bolded and the basic arginine and lysine residues are marked in red [93]
Fig. 7.
The multiple sequence alignment of spike (S) S2′ cleavage site sequences using the Clustal Omega program. The S1/S2 sequences are bolded and the basic arginine and lysine residues are marked in red [93]; in yellow background are marked the cysteine residues that form an internal disulfide bond (C840 and C851) and the residues that form a salt bridge that reinforces the previous disulfide bond (K835-D848); in light blue is shown the lysine (K854) that form a salt bridge with the aspartic acid (D614) (not shown) [40, 107–109]
The first striking difference between the analyzed S’ SARS-CoV-2 concerns the S1/S2 cleavage site. Thus, only some PDB entries (6XR8/6XRA, 7C2L, and 6XCN/6XCM) show the PPC or furin cleavage motif RRAR↓ [106]. Moreover, the furin cleavage motif from the S1/S2 sequence of the SARS-CoV-2 is more similar to its HCoV-OC43 counterpart than to furin cleavage motif of the SARS-CoV and MERS-CoV. In contrast, the S2′cleavage site sequence of all SARS-CoV-2 PDB entries analyzed has the furin cleavage motif similar to its SARS-CoV counterpart. The phylogenetic tree (cladogram) of the S sequences analyzed shows that the SARS-CoV-2 S is closer to SARS-CoV S, but the differences observed between the nine SARS-CoV-2 S sequences are reflected in the evolutionary relationship (Fig. 8).
Fig. 8.
The phylogenetic tree (cladogram) of the CoVs Spike (S) sequences of CoVs with different origin. feline infectious peritonitis virus (FIPV), porcine epidemic diarrhea virus (PEDV), HCoV-NL63, SARS-CoV-2, SARS-CoV, HCoV-HKU1, and coronavirus (strain A59) MHV-A59; performed by Clustal Omega program
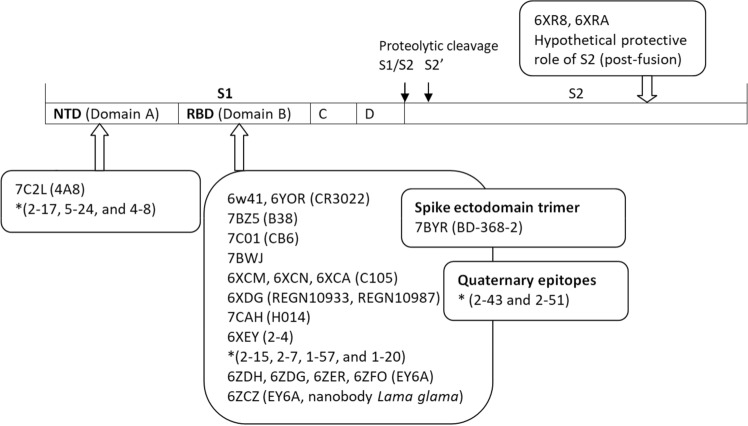
There are 22 published structures of SARS-CoV-2 S glycoprotein retrieved from the PDB. Among them, seven are S glycoprotein structures (PDD ID(s) 6VXX, 6VYB, 6VSB, 6LXT, 6Z97, 6XR8, and 6RA) and four structures are co-crystallized with the human ACE2 receptor (PDB ID(s) 6M0J, 6LZG, 6M17, and 6VW1). Out of 32 SARS-CoV-2 S glycoprotein structures co-crystallized with neutralizing antibodies, eleven has been published (PDB ID(s) 6W41, 7BZ5, 7C01, 7BWJ, 7BYR, 6XCM, 6XCN, 6XCA, 7C2L, 6XDG, and 6YOR) (Fig. 9).
Fig. 9.
The crystal structures co-crystallized with neutralizing antibodies. The epitopes of the SARS-CoV-2 spike are NTD (N terminal domain), RBD (receptor binding domain, quaternary epitopes, and ectodomain; there are indicated the PDB entries and in parenthesis the neutralizing antibody; * the most potent neutralizing antibodies from convalescent patients according to Liu et al. work [16]
Cai et al. (PDB ID(s) 6XR8 and 6XRA) assume a protective role of the S2 post-fusion structure that could elicit the non-neutralizing antibodies to evade the host immune system [40]. Also, the authors identify a fusion peptide proximal region (FPRP) with a critical role in rearrangements of S protein and membrane fusion, demonstrated by a mutation D614G that lead to more efficient cell entry. The aspartic acid (D614) forms a salt bridge with the lysine (K854) belonging to FPRP [40, 107–109]. Barnes et al. study demonstrates that the mutation D614G, a mutation that enhances the SARS-CoV-2 transmissibility, is unlikely to affect antibodies from recovering COVID-19 patients included in the study.[24].
The ACE2 (E.C.3.4.17.23; UniProtKB Q9BYF1) is a homodimer that catalyzes the reaction: angiotensin II + H2O = angiotensin-(1–7) + L-phenylalanine [39, 110]. The human ACE2 gene is expressed in a large panel of organs—lungs (type II pneumocytes), heart, kidney, intestine, cerebral neurons—or immune cells—alveolar monocytes/macrophages. The ACE2 expression is up-regulated by interferon and influenza A virus in human nasal epithelia and lung cells [111, 112]. Shang et al. recently published the structure of the complex SARS-CoV-2 RBD/human ACE2 (PDB ID 6VW1). A comparison with the SARS-CoV highlighted that SARS-CoV-2 RBM forms larger binding interface and more contacts with human ACE2. The authors focused on the receptor-binding motif (RBM) of the viral RBD (S1 subunit) and the two virus-binding hotspots—31 and 353—of the human ACE2 that are stabilized by the viral residues Q493 and L455 and N501, respectively. The study successfully determined that the tight SARS-CoV-2 binding onto the human ACE2 due to a four residue motif 482 GVEG 485 and to the mutation L486F in the RBM sequence [101].
Yan et al. have recently published the cryo-electron microscopy structures of the full-length human ACE2-B(0)AT1 complex and RBD-ACE2-B(0)AT1 (PDB ID 6M17, 6M1D, 6M18) [39]. The authors have shown that two S glycoprotein trimers simultaneously bind to an ACE2 homodimer. Moreover, the authors highlighted some residues that could explain the difference between the cell entry of SARS-CoV-2 and SARS-CoV. Two mutations (Val404 to Lys317 and Leu472 to Phe486) are responsible for an enhancement interaction of SARS-CoV-2-RBD and ACE2. In contrast, the mutation Arg436 to Asn439 weakens the SARS-CoV-2-RBD and hACE2 interaction [39]. The sodium-dependent neutral amino acid transporter B(0)AT1 (SLC6A19) (UniProtKB Q695T7) mediates absorption of neutral amino acids across the membrane of the renal and intestinal epithelial cells and mutations in B(0)AT1 (such as A69T and R240Q) may cause Harnup disorder [113–116]. The B(0)AT1 expression onto the small intestinal cells depends on the co-expression of the ACE2 and is enhanced by aminopeptidase N (CD13) [117]. The aminopeptidase N (E.C. 3.4.11.2; UniProt UK P15114) mediates other viral infections—transmissible gastroenteritis virus responsible for diarrheal disease in piglets, cytomegalovirus or human CoV-229E [118–120]. Lan J. et al. compared the SARS-CoV and SARS-CoV-2 RBD bound to hACE2 and suggested a convergent evolution between the two coronaviruses. They further suggest that a unique “RRAR” cleavage site at the S1/S2 boundary of the SARS-Cov-2 has a crucial role in rapid inter-human transmission (PDB ID 6M0J). The authors emphasize the importance of neutralizing antibodies in the evolution of SARS-CoV-2 infection. Their structural study suggests that the lack of cross-reactivity by m396 and 80R antibodies that neutralizes SARS-CoV resides in several residue changes in the SARS-CoV-2 RBD [121].
Wang et al. firstly identify that S1 C-terminal domain (SARS-CoV-2 S-CTD) is the key region that interacts with the human ACE2. A comprehensive comparison between SARS-CoV-2 (PDB ID 6LZG) with similar SARS-CoV, reveals that the former has a higher affinity for the receptor hACE2. Moreover, the authors advance the hypothesis of the “hotspot” region in S for receptor binding. Further experiments with the polyclonal antibodies, confirm the differences between SARS-CoV-2 and SARS-CoV’ S glycoprotein [36].
SARS-CoV-2 and Host Immune System
The COVID-19 is the disease due to the interaction of the pathogen SARS-CoV-2 with the human host. Consequently, all aspects of this interaction draw the picture of the COVID-19. The interaction of SARS-CoV-2 with the human host is dynamic and has new features compared to other pandemics in human history. First, immunity developed after the SARS-CoV-2 infection is not yet well understood [6, 122, 123]. Then, the mechanisms of tissue damage in COVID-19 are not fully understood. The most striking characteristic of COVID-19 is the storm of pro-inflammatory cytokines that ultimately is responsible for the vast majority of the death [124]. The biggest challenge is the best protocol to treat critically ill patients. Methylprednisolone has already been reported to reduce the worst outcomes (https://advaitabio.com/news/covid19-analysis/) [125]. The managing of the hyper-inflammatory process in COVID-19 proved to be crucial for the evolution of the disease. In SARS-CoV infections, some pro-inflammatory cytokines (IP-10, IL-8, and MCP-1) are elevated. Meanwhile, there is no antiviral (IFN) response [126]. In many diseases associated with systemic inflammatory response syndrome, the control of damage-associated molecular patterns (DAMPs) by their counteracting molecules suppressing/inhibiting DAMPs (SAMPs) is crucial [127]. Previous studies on the crystal structures of SARS-CoV S glycoprotein mutants neutralized by 80R-specific antibodies have been considered a hope for the immunotherapeutic strategy for the future outbreak of SARS (PDB ID 2GHV) [128].
There are 32 structures of SARS-CoV-2 about the immune system published in peer-reviewed papers (PDB ID9s) 6W41, 7BZ5, 7C01, 7BWJ, 7BYR, 6XCM, 6XCN, 7C2L, 6XDG, 6YOR, 7CAH, 6XEY, 6ZCZ, 6ZDH, 6ZDG, 6ZER, and 6ZFO). More than eight months after the onset of the pandemic, health care professionals and researchers are able to analyze the clinical evolution of many patients. The host immune response to SARS-CoV-2 infection is perhaps the most controversial issue. Most of the last PDB entries are about the SARS-CoV-2 neutralizing antibodies. The patients recovered with COVID-19 rapidly advanced studies with plasma samples from convalescents. Numerous research teams already identified numerous potent neutralizing antibodies against multiple epitopes on the SARS-CoV-2 spike (Fig. 9). Yuan et al. published the cryo-EM crystal structure of the SARS-CoV-2 S1 with the CR3022 antibody as an effort to understand the antigenicity of the SARS-CoV-2 (PDB ID 6W41). Their results show that CR3022 Fab binds to SARS-CoV RBD with higher affinity (Table 2). In the selection process of the selection of the neutralizing antibodies against SARS-CoV-2, Ju et al. provide evidence that binding affinity does not predict ACE2 competing capacity [20]. The CR3022 antibodies fail to neutralize SARS-CoV-2 in vitro. The CR3022 epitope does not overlap with the ACE2 binding site in SARS -CoV-2 S-RBD. However, SARS-CoV and SARS-CoV-2 have a conserved, but cryptic epitope that could be worth considering for a vaccine [14]. Moreover, the Huo et al. suggest that CR3022 binding facilitates conversion to the fusion-incompetent post-fusion state (PDB ID 6YOR) [15]. These findings offer promising perspectives on COVID-19 therapy with neutralizing antibodies.
Table 2.
Comparison of the neutralizing antibodies against SARS-CoV-2 and SARS-CoV
| Antibody/epitope | SARS-CoV-2 | SARS-CoV | References | ||
|---|---|---|---|---|---|
| IC50 | KD | KD | |||
| S-RBD epitope | |||||
| CR3022 Fab | NA | 115 ± 3 nM | 1.0 ± 0.1 nM | [14] | |
| CR3022 Ig | NA | < 0.1 nM | < 0.1 nM | [14] | |
| CB6 | 0.036 ± 0.007 μg /ml | 2.49 ± 1.65 nM | NA | [19] | |
| C105 | 0.0261 μg /ml | NA | NA | [24] | |
| B38 | 0.177 μg/ml | NA | NA | [18] | |
| H4 | 0.896 μg/ml | NA | NA | [18] | |
| B5 | 1.375 μg/ml | NA | NA | [18] | |
| H2 | 1.000 μg/ml | NA | NA | [18] | |
| H014 | 3 nM* | NA | NA | [17] | |
| 4A8 | 0.39 μg/ml | 92.7 nM | NA | [25] | |
|
REGN10933 REGN10987 |
0.0374 nM 0.0421 nM |
0.56 nM | NA | [23] | |
| EY6A | 20.7 μg/ml | 2 nM | NA | [22] | |
|
2–15, 1–57, 2–7, 4–20, 2–36, 1–20, 2–38, 2–4, 2–30 |
0.0007 to 0.209 μg/ml | NA | NA | [16] | |
| BD-217, BD-218, BD-236, BD-361, BD-368, BD-368–2, BD-395 | 0.015 to 1.6 μg/ml | 0.039 to 2.8 nM | NA | [21] | |
| S-NTD epitope | |||||
| 5–24, 4–18, 4–8, 5–7, 4–19, 1–87, 2–17, 1–68 | 0.007 to 0.109 μg/ml | NA | NA | [16] | |
| Other epitopes (quaternary epitopes) | |||||
|
2–43 2–51 |
0.003 μg/ml 0.007 μg/ml |
NA | NA | [16] | |
*pseudovirus neutralization assay, NA- not available
The challenge of the therapy with neutralizing antibodies is to overcome the virus mutants. The therapeutic cocktail of neutralizing antibodies against SARS-CoV-2 is an efficient approach. The Hansen et al. team discovered a pair of non-competitive neutralizing antibodies that simultaneously bind to S-RBD SARS-CoV-2 (namely REGN10933 and REGN10987) (PDB ID 6XDG) [23]. The mapping of the SARS-CoV-2 epitopes targeted by potent neutralizing mAbs demonstrated the diversity of the neutralizing antibodies directed against the S-RBD, S-NTD, and the epitopes that do not map to the S-RBD or S-NTD. Even though the authors Liu et al. do not establish the role of S-NTD in blocking SARS-CoV-2 infection, these findings are crucial for vaccine development [16].
The studies about the host immune system are, on the one hand, about the production of neutralizing antibodies, and on the other hand, about the virus’s ability to hamper defense immunity. The CoVs have an impressive ability to suppress the IFN response. An antagonist of the IFN response is the nsp15 [129]. The viral nsp3 has an active role in suppressing innate immunity by blocking the interferon regulatory factor (IRF3) (Gene ID 3661) and altering the NF-kappaB signaling, which controls the expression of some inflammatory cytokine genes [130, 131]. The nsp3 has a crucial role in damaging the host’s first line of defense against SARS-CoV-2. Nsp3 probably has a role in producing the unusual inflammation described in patients with severe COVID-19 [132]. The SARS-CoV-2 nsp3 has no crystal structures published until now. The nsp3 (~ 200 kDa) is a large multi-domain protein (E.C. 3.4.19.12) (UniProtUK P0DTD1) responsible for the cleavage of the replicase polyprotein 1ab at the N-terminus [133]. The papain-like protease (PLpro) domain of the viral nsp3 exhibits deubiquitinating activity [134]. A recent study has shown that deubiquitinating/deISGylating (DUB/deISG) nsp3 deficient mutants (H1652R, V1691K, and V1691R) result in attenuating the MERS virus [135]. A recent study identified specific mutations in nsp3 corroborated with nsp2 that suggest potential mechanisms that explain higher contagiousness of SARS-CoV-2 compared to SARS-CoV [136].
Thoms et al. published nine cryo-EM crystal structures of the SARS-CoV-2 nsp1 (PDB ID(s) 6ZLW, 6ZM7, 6ZN5, 6ZON, 6ZP4, 6ZMI, 6ZMO, 6ZMT, and 6ZME). The nsp1 (E.C. 3.4.19.12., UniProtKB P0DTD1) is a host translation inhibitor that interacts with the 40S ribosomal subunit. There are two consequences of the suppressing of the host gene expression: the viral gene expression and evasion of the virus from the host immune response. The SARS-CoV-2 nsp1 prevents translation of the interferon, some pro-inflammatory cytokines, and interferon-stimulated antiviral ISGs [46].
Other Non-structural Proteins of SARS-CoV-2
The organization of the non-structural (nsp) proteins in CoV is not fully understood. The CoV has two overlapping open reading frames (ORFs) ORF 1a and ORF 1b, and by 1 ribosomal frameshifting—a translational regulation mechanism described in retroviruses—there is a regulation of relative ratio of structural to enzymatic proteins [137, 138]. The CoV synthesizes two polyproteins that are further cleaved by the viral proteases. The replicase polyprotein 1ab (UniProtKB P0DTD1) also contains the proteases responsible for the cleavage [139]. The viral protease 3CLpro is extensively studied for the designing of new drugs to control the spread of human and zoonotic CoVs [140].
The nsp9 is an RNA-replicase which binds the viral RNA and has a regulatory effect in viral replication [141]. Three unpublished crystal structures of nsp9 from SARS-CoV-2 were retrieved from the PDB (PDB ID(s) 6W9Q, 6W4B, and 6WXD) (Litter et al., and Tan et al.). Previous studies focused on nsp9 from other CoVs, demonstrated that the residues from the dimer interface greatly influence their nucleic acid binding affinity [142]. According to Zeng et al. findings, the dimerization greatly depends on the N-finger and the two glycine residues from a conserved region GXXXG [142]. Eight nsp9 sequences were analyzed—porcine coronavirus HKU15 [PDB D5YM8] (PDCoV); human coronavirus HKU1[YP_459943](HCoV-HKU1); murine hepatitis virus strain A59 [GeneBank ACO72881.1](MHV); porcine epidemic diarrhea virus CV777 [PDB ID 5HIZ](PEDV); bat coronavirus CDPHE15/USA/2006 [GeneBank YP_008439220.1] (BatCoV-CDPHE15); Middle East respiratory syndrome-related coronavirus [GeneBank AHI48749.1](MERS-CoV); Severe acute respiratory syndrome coronavirus 2 [PDB ID 6W4B](SARS-CoV-2); SARS coronavirus Shanhgai LY [GeneBank AAP82976.1](SARS-CoV). The multiple sequence alignment of the eight nsp9 proteins is slightly different to Zeng et al. findings in respect of the PDCoV nsp9. The N-finger sequence with the consensus sequence with the polar residues NNE (Asn-Asn-Glu) is noticed in all nsp9 sequences except the PDCoV nsp9 (PDB D5YM8) (Fig. 10).
Fig. 10.
The multiple sequence alignment of nsp9 sequences using the Clustal Omega program. The N-finger (in red) and GXXXG motif (grey background) are important for dimerization. The conserved residues of N-finger are underlined
The nsp15 is uridylate-specific endoribonulcease (NendoU) (E.C. 3.1.-.-) highly conserved among vertebrates coronaviruses. The nsp15 plays an important role in the life cycle of CoVs [143, 144]. The recent SARS-CoV-2 nsp15 structures show that it is a hexamer and the catalytic site belong to the C-terminal domain (PDB ID(s) 6W01 and 6VWW). The study concludes that the SARS-CoV-2 nsp15 differs by MERS-CoV nsp15 by residues that coordinate the manganese ion [145].
Discussion
The SARS-CoV-2 emerged at the late of the year 2019. The epidemiology of COVID-19 pandemic is beyond the subject of this paper. However, we can learn how other epidemics have been handled throughout history. There are many historical records, but an episode about the plaque epidemic, brilliantly described by Irving Stone in The Agony and the Ecstasy, could be a school-case. In the Florence transformed in morgue and deserted by people, Michelangelo—Michelangelo di Lodovico Buonarroti Simoni (1475-1564)—chose to stay in the city and take care of his ill younger brother until the last moment and doing the best for a decent funeral. Strikingly, in the XVI century, Michelangelo applied the recommendation for COVID-19 – hygiene and social distance. He thoroughly brushed himself into a bathtub with hot water and sent away his sister-in-law and his nephews. Even he was asymptomatic, in our days terms, he refused Granacci’s wine. Unlike the plague described by Irving Stone, COVID-19 pandemic is considerably longer and for the time being there is no hope for its end soon.
Until the emergence of SARS-CoV in 2003 and MERS in 2012, the CoVs infections do not pay much attention. Now, facing the SARS-CoV-2, the spectrum of re-emergence of the SARS-CoV-2 or the animal-human transmission of a new CoV strain is more real than ever. So, the understanding of all aspects of the SARS-Cov-2 is of great interest. The rapid involvement of the scientific communities greatly advances the elucidation of some aspects of COVID-19 pandemic to be prepared in the event of a new contagious and fatal CoV disease.
The structural studies focused on designing specific inhibitors to gain knowledge about the COVID-19 treatment, and further studies involving biophysical methods will speed up the SARS-CoV-2 cure [146, 147]. Likewise, many research teams advance their studies on understanding the most subtle details of the virus-host cell interaction, the emergence of the neutralizing antibodies, or the designing of new inhibitors. The virus and host factors are both relevant for the initiation and evolution of a viral infection. The SARS-CoV-2 shares epidemiological patterns with SARS-CoV and MERS, but there are some structural differences. Thus, even though SARS-CoV and SARS-CoV-2 use the same host receptor, SARS-CoV-2 binds to the ACE2 with higher affinity [101].
Two main features are associated with the worst outcome in SARS-CoV-2 infection—co-morbidities and elderly. The traits of immune system at different ages could be a strong explanation about the accompanying symptoms of various viruses when they present with an underlying disease. For example, episodes of wheezing in young children have been observed in respiratory syncytial virus (RSV) infections. In contrast, in adults and older children, the episodes of wheezing have been observed in human rhinoviruses (HRV) and HCoVs (HCoV-NL63 and HCoV-HKU1) infections [148]. Recent studies identify ACE-2 maturation stages, and the hypertension treatment with ACE inhibitors could be an explanation for mild disease in children [149, 150]. Thus, structural studies about spike -ACE-2 and ACE2-neutralizing antibodies are of great interest. The crystal structures of SARS-CoV-2 reveal important details about the viral infection. First, the full-length human ACE2 structure reveals that viral cell entry involves the simultaneous binding of two S glycoproteins to ACE2. Also, the use of B(0)AT1 in stabilization of the full-length human ACE2 structure advances the hypothesis that B(0)AT1 may play a role in enteric infections of some CoVs [39].
Certain virus components trigger the immune system and limit the interspecies transmission of different CoVs. The structural studies greatly advance the selection of the best immunogens for SARS-CoV-2 prophylaxis. A comparison of the HCoV suggests that the spike’s SARS-CoV-2-CTD domain could be a valid immunogen for a future vaccine [36]. The viral immune evasion depends on more than one factor and structural work of many research teams greatly advances the understanding of immune response. The SARS-CoV-2 nsp1 is one of the major immune factors that impedes the host immune clearance. A recent structural work demonstrated that SARS-CoV-2 nsp1 blocks RIG-I-dependent innate immune response [46].
The hypothesis of the bats being at the origin of the SARS-CoV-2 is, until now, not either demonstrated or rejected. However, it is about 96% identity between the genome of SARS-CoV-2 and the genome of BatCov RaTG13 [3]. Even the genome identity between SARS-CoV-2 and SARS-CoV is about 80%—recent studies claim a close evolutionary relationship between the two viruses [3, 101]. The results of the structural studies could bring valuable perspective of the host specificity. Thus, Ziegler et al. work about the human and mouse ACE2 demonstrated that different to mouse ACE2, the ACE2 is an interferon-stimulated gene (ISG) in human primary upper airway epithelial basal cells [112]. These results are useful not only for using an appropriate animal/cellular model in experimental research but for screening for the potential animal host of the SARS-CoV-2, which could elucidate the chain of the virus passing from the animal host to human. Also, the differences of an essential step in CoVs replication, the priming of S glycoprotein by host cell proteases, could be exploited to elucidate the zoonotic potential of the SARS-CoV-2. The findings of Hoffmann et al. highlighted that the S1/S2 cleavage site sequence of SARS-CoV-2 S glycoprotein harbors several arginine residues (multibasic) on the contrary to its closely related the bat CoV RaTG13 [94].
Current theory holds that most of the deaths are due to an excessive level of pro-inflammatory cytokines in circulation. The humoral immune response is most studied and structural studies add important results that further explain biochemical findings [121]. The most optimistic outcome is to obtain a vaccine that elicits strong and long-term immunity. Until then, understanding the evolutions of phenotypic changes of the SARS-CoV-2 and elucidation of epidemiological aspects of the COVID-19 pandemic greatly depends on the analysis of all available research results.
Conclusions
There is a huge effort in deciphering the SARS-CoV-2 pandemic, leading to many studies conducted around the world. The scientific work relies on observation, experiments, and structural studies to provide a deep view of SARS-CoV-2. The current structural analysis of the SARS-CoV-2 is mainly focused on three major lines—finding new hydrolase inhibitors, the virus-host cell invasion, and the virus-neutralizing antibody interaction. There is an intense work on structural studies of SARS-CoV-2 proteins, mainly for S glycoprotein that is crucial for the pathogenesis of all CoVs, but Totum est majus sua parte (The whole is bigger than the part). The convalescent patients’ B cells demonstrated that not a single spike epitope elicits the neutralizing antibodies. The host-virus interaction is a dynamic process, and the host ‘s defense mechanisms are crucial not only to cure the COVID-19 infection but to prevent further re-infection. In this sense, structural studies involving neutralizing antibodies gain a perspective in deciphering the immunity of cured persons. Also, the works about the viral nsp(s) aid the understanding of the viral immune evasion. Despite the limits of X-ray/cryo-EM crystal structure studies – mainly the lack of information about the cell-mediated immunity and time-consuming experiments for clinical validation of the new inhibitors – the thorough analysis of the PDB entries is a powerful tool for further understanding of the human CoVs infections.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Many thanks to the Editorial of The Protein Journal for the flexible approach during the COVID-19 pandemic. I would like to thank Professor Lawrence Berliner and the referees for their comments that much improved the quality of the manuscript. Many thanks to Tudor Constantin Badea for the fruitful discussions during the mobility project.
Funding
This work was supported by a Grant of Ministry of Research and Innovation, CNCS - UEFISCDI, Project Number PN-IIIP4-ID-PCCF-2016-0016, within PNCDI III and by a mobility project of the Romanian Ministry of Research and Innovation, CNCS - UEFISCDI, the Project Number PN-III-P1-1.1-MCD-2018-0108, within PNCDI III.
Compliance with Ethical standards
Conflict of interest
The author declares no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Biswas A, Bhattacharjee U, Chakrabarti AK, et al. Emergence of novel coronavirus and COVID-19: whether to stay or die out? Crit Rev Microbiol. 2020 doi: 10.1080/1040841X.2020.1739001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak: an update on the status. Mil Med Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Lou YX, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, Allot A, Lu Z. Keep up with the latest coronavirus research. Nature. 2020;579:193. doi: 10.1038/d41586-020-00694-1. [DOI] [PubMed] [Google Scholar]
- 5.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z, Xiao X, Wei X, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020 doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Zai J, Zhao Q, et al. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J Med Virol. 2020 doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JFW, Lau SKP, To KKW, et al. Middle East Respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menachery VD, Yount BL, Debbink K, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackay IM, Arden KE. MERS coronavirus: diagnostics, epidemiology and transmission. Virol J. 2015;12:1–21. doi: 10.1186/s12985-015-0439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Qi J, Yuan Y, et al. Bat origins of MERS-CoV supported by bat Coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond RG, Tan X, Johnson MA. SARS-unique fold in the Rousettus bat coronavirus HKU9. Protein Sci. 2017;26:1726–1737. doi: 10.1002/pro.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wlodawer A, Dauter Z, Shabalin I, et al. Ligand-centered assessment of SARS-CoV-2 drug target models in the Protein Data Bank. FEBS J. 2020 doi: 10.1111/febs.15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;80-(368):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huo J, Zhao Y, Ren J, et al. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies directed to multiple epitopes on SARS-CoV-2 spike. Nature. 2020 doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 17.Lv Z, Deng YQ, Ye Q, et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;80-(5881):eabc5881. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Wang F, Shen C, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;80-(368):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 20.Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020 doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 21.Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020 doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou D, Duyvesteyn HME, Chen CP, et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat Struct Mol Biol. 2020 doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 23.Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;80-:eabd0827. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes CO, West AP, Huey-Tubman KE, et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;80-(369):eabc6952. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodsell DS, Olson AJ. Automated docking of substrates to proteins by simulated annealing. Proteins Struct Funct Genet. 1990;8:195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- 27.Ravindranath PA, Forli S, Goodsell DS, et al. AutoDockFR: advances in protein-ligand docking with explicitly specified binding site flexibility. PLoS Comput Biol. 2015;11:e1004586. doi: 10.1371/journal.pcbi.1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizvi SMD, Shakil S, Haneef M. A simple click by click protocol to perform docking: autoDock 4.2 made easy for non-bioinformaticians. Excli J. 2013;12:831–857. [PMC free article] [PubMed] [Google Scholar]
- 29.Xia S, Liu M, Wang C, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Yan L, Huang Y, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;80-(782):eabb7498. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 33.Kang S, Yang M, Hong Z, et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020 doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin W, Mao C, Luan X, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;80-(1560):eabc1560. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hillen HS, Kokic G, Farnung L, et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020 doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 39.Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;80-(367):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Y, Zhang J, Xiao T, et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;80-:eabd4251. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 42.Dai W, Zhang B, Su H, et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;80-(4489):eabb4489. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Z, Zhao Y, Sun Y, et al. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Lin D, Sun X, et al (2020) Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science 80-: 368:409–412. doi: 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed]
- 45.Wang Q, Wu J, Wang H, et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020 doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thoms M, Buschauer R, Ameismeier M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;8665:1–11. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kneller DW, Phillips G, O’Neill HM, et al. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat Commun. 2020 doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Q, Peng R, Yuan B, et al. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong X, Qu K, Ciazynska KA, et al. A thermostable, closed SARS-CoV-2 spike protein trimer. Nat Struct Mol Biol. 2020 doi: 10.1038/s41594-020-0478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin D, Mukherjee R, Grewe D, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L, Gui C, Luo X, et al. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J Virol. 2005;79:7095–7103. doi: 10.1128/jvi.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Q, Weber E, Yang H. Recent developments on coronavirus main protease/3C like protease inhibitors. Recent Pat Antiinfect Drug Discov. 2013;8:150–156. doi: 10.2174/1574891x113089990017. [DOI] [PubMed] [Google Scholar]
- 53.Xue X, Yang H, Shen W, et al. Production of authentic SARS-CoV Mpro with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J Mol Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pillaiyar T, Manickam M, Namasivayam V, et al. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, Yang M, Ding Y, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Báez-Santos YM, Barraza SJ, Wilson MW, et al. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J Med Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Wang XJ. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J Genet Genomics. 2020;47:119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.St. John SE, Tomar S, Stauffer SR, Mesecar AD. Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4 - The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS) Bioorganic Med Chem. 2015;23:6036–6048. doi: 10.1016/j.bmc.2015.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Lin D, Kusov Y, et al. Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem. 2020 doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 61.Thenin-Houssier S, De Vera IMS, Pedro-Rosa L, et al. Ebselen, a small-molecule capsid inhibitor of HIV-1 replication. Antimicrob Agents Chemother. 2016;60:2195–2208. doi: 10.1128/AAC.02574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffin JWD. SARS-CoV and SARS-CoV-2 main protease residue interaction networks change when bound to inhibitor N3. J Struct Biol. 2020;211:107575. doi: 10.1016/j.jsb.2020.107575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H, Xie W, Xue X, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005 doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin Z, Zhao Y, Sun Y, et al. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020 doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 65.Anand K, Palm GJ, Mesters JR, et al. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paasche A, Zipper A, Schafer S, et al. Evidence for substrate binding-induced zwitterion formation in the catalytic Cys-His dyad of the SARS-CoV main protease. Biochemistry. 2014;53:5930–5946. doi: 10.1021/bi400604t. [DOI] [PubMed] [Google Scholar]
- 67.Wang H, He S, Deng W, et al. Comprehensive insights into the catalytic mechanism of Middle East Respiratory Syndrome 3C-like protease and severe acute respiratory syndrome 3C-like protease. ACS Catal. 2020;10:5871–5890. doi: 10.1021/acscatal.0c00110. [DOI] [PubMed] [Google Scholar]
- 68.Shi F, Xie Y, Shi L, Xu W. Viral RNA polymerase: a promising antiviral target for influenza A virus. Curr Med Chem. 2013;20:3923–3934. doi: 10.2174/09298673113209990208. [DOI] [PubMed] [Google Scholar]
- 69.Neogi U, Hill KJ, Ambikan AT, et al. Feasibility of known RNA polymerase inhibitors as anti-SARS-CoV-2 Drugs. Pathogens. 2020 doi: 10.3390/pathogens9050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao Y, Ma Q, Restle T, et al. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J Virol. 2012;86:4444–4454. doi: 10.1128/jvi.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.te Velthuis AJW, van den Worm SHE, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Warren TK, Jordan R, Lo MK, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gordon CJ, Tchesnokov EP, Woolner E, et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295:6785–6797. doi: 10.1074/jbc.ra120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agostini ML, Andres EL, Sims AC, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gordon CJ, Tchesnokov EP, Feng JY, et al. The antiviral compound remdesivir potently inhibits RNAdependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Norrie JD. Remdesivir for COVID-19: challenges of underpowered studies. Lancet. 2020;395:1525–1527. doi: 10.1016/S0140-6736(20)31023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bouvet M, Imbert I, Subissi L, et al. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci USA. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tchesnokov E, Feng J, Porter D, Götte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Shea TJ, Cryan PM, Cunningham AA, et al. Bat flight and zoonotic viruses. Emerg Infect Dis. 2014;20:741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deng X, Mettelman RC, O’Brien A, et al. Analysis of coronavirus temperature-sensitive mutants reveals an interplay between the macrodomain and papain-like protease impacting replication and pathogenesis. J Virol. 2019 doi: 10.1128/JVI.02140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghosh AK, Takayama J, Rao KV, et al. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation. J Med Chem. 2010;53:4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ratia K, Pegan S, Takayama J, et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci USA. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang TK, Wu MPJ, Chen ST, et al. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin SY, Liu CL, Chang YM, et al. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xia S, Yan L, Xu W, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019 doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Milewska A, Nowak P, Owczarek K, et al. Entry of human coronavirus NL63 into the cell. J Virol. 2018 doi: 10.1128/JVI.01933-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu G, Hu Y, Wang Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scobey T, Yount BL, Sims AC, et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020 doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen LW, Mao HJ, Wu YL, et al. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1–10. doi: 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dahms SO, Hardes K, Becker GL, et al. X-ray structures of human furin in complex with competitive inhibitors. ACS Chem Biol. 2014;9:1113–1118. doi: 10.1021/cb500087x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hardes K, Becker GL, Lu Y, et al. Novel furin inhibitors with potent anti-infectious activity. ChemMedChem. 2015;10:1218–1231. doi: 10.1002/cmdc.201500103. [DOI] [PubMed] [Google Scholar]
- 99.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 100.Barile E, Baggio C, Gambini L, et al. Potential therapeutic targeting of coronavirus spike glycoprotein priming. Molecules. 2020;25:2424. doi: 10.3390/molecules25102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 103.Wong SK, Li W, Moore MJ, et al. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jeffers SA, Tusell SM, Gillim-Ross L, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tian S, Huajun W, Wu J. Computational prediction of furin cleavage sites by a hybrid method and understanding mechanism underlying diseases. Sci Rep. 2012 doi: 10.1038/srep00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang L, Jackson CB, Mou H, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Benvenuto D, Demir AB, Giovanetti M, et al. Evidence for mutations in SARS-CoV-2 Italian isolates potentially affecting virus transmission. J Med Virol. 2020 doi: 10.1002/jmv.26104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Daniloski Z, Jordan T, Ilmain J, et al. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. bioRxiv. 2020 doi: 10.1101/2020.06.14.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin Q, Keller RS, Weaver B, Zisman LS. Interaction of ACE2 and integrin beta1 in failing human heart. Biochim Biophys Acta. 2004;1689:175–178. doi: 10.1016/j.bbadis.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 111.Magrone T, Magrone M, Jirillo E. Focus on receptors for coronaviruses with special reference to angiotensin-converting enzyme 2 as a potential drug target - a perspective. Endocr Metab Immune Disord Drug Targets. 2020 doi: 10.2174/1871530320666200427112902. [DOI] [PubMed] [Google Scholar]
- 112.Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020 doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kowalczuk S, Broer A, Tietze N, et al. A protein complex in the brush-border membrane explains a Hartnup disorder allele. FASEB J. 2008;22:2880–2887. doi: 10.1096/fj.08-107300. [DOI] [PubMed] [Google Scholar]
- 114.Azmanov DN, Kowalczuk S, Rodgers H, et al. Further evidence for allelic heterogeneity in Hartnup disorder. Hum Mutat. 2008;29:1217–1221. doi: 10.1002/humu.20777. [DOI] [PubMed] [Google Scholar]
- 115.Camargo SMR, Singer D, Makrides V, et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009;136:872–882. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fairweather SJ, Broer A, Subramanian N, et al. Molecular basis for the interaction of the mammalian amino acid transporters B0AT1 and B0AT3 with their ancillary protein collectrin. J Biol Chem. 2015;290:24308–24325. doi: 10.1074/jbc.M115.648519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jando J, Camargo SMR, Herzog B, Verrey F. Expression and regulation of the neutral amino acid transporter B0AT1 in rat small intestine. PLoS ONE. 2017;12:e0184845. doi: 10.1371/journal.pone.0184845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu W, Zhang S, Shen Y, Yang Q. Epidermal growth factor receptor is a co-factor for transmissible gastroenteritis virus entry. Virology. 2018;521:33–43. doi: 10.1016/j.virol.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li Z, Tomlinson AC, Wong AH, et al. The human coronavirus HCoV-229E S-protein structure and receptor binding. Elife. 2019 doi: 10.7554/eLife.51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Soderberg C, Giugni TD, Zaia JA, et al. CD13 (human aminopeptidase N) mediates human cytomegalovirus infection. J Virol. 1993;67:6576–6585. doi: 10.1128/JVI.67.11.6576-6585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 122.Dhama K, Sharun K, Tiwari R, et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccines Immunother. 2020 doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Han X, Qi J, Song H, et al. Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein. Virology. 2017;507:101–109. doi: 10.1016/j.virol.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saghazadeh A, Rezaei N. Immune-epidemiological parameters of the novel coronavirus–a perspective. Expert Rev Clin Immunol. 2020 doi: 10.1080/1744666X.2020.1750954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang Y, Jiang W, He Q, et al. A retrospective cohort study of methylprednisolone therapy in severe patients with COVID-19 pneumonia. Signal Transduct Target Ther. 2020;5:57. doi: 10.1038/s41392-020-0158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen J, Subbarao K. The immunobiology of SARS*. Annu Rev Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- 127.Land WG. Use of DAMPs and SAMPs as therapeutic targets or therapeutics: a note of caution. Mol Diagn Ther. 2020 doi: 10.1007/s40291-020-00460-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hwang WC, Lin Y, Santelli E, et al. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J Biol Chem. 2006;281:34610–34616. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Deng X, Baker SC. An “Old” protein with a new story: coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018;517:157–163. doi: 10.1016/j.virol.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kawai T, Akira S. Signaling to NF-kappaB by toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 131.Razani B, Whang MI, Kim FS, et al. Non-catalytic ubiquitin binding by A20 prevents psoriatic arthritis-like disease and inflammation. Nat Immunol. 2020;21:422–433. doi: 10.1038/s41590-020-0634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang W, Zhao Y, Zhang F, et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 ( COVID-19): the perspectives of clinical immunologists from China. Clin Immunol. 2020;214:108393. doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bailey-elkin BA, Knaap RCM, Johnson GG, et al. Crystal structure of the Middle East Respiratory Syndrome Coronavirus ( MERS-CoV ) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J Biol Chem. 2014;289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clasman JR, Everett RK, Srinivasan K, Mesecar AD. Decoupling deISGylating and deubiquitinating activities of the MERS virus papain-like protease. Antiviral Res. 2020;174:104661. doi: 10.1016/j.antiviral.2019.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Angeletti S, Benvenuto D, Bianchi M, et al. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J Med Virol. 2020 doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Plant EP, Dinman JD. The role of programmed-1 ribosomal frameshifting in coronavirus propagation. Front Biosci. 2008;13:4873–4881. doi: 10.2741/3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yoshimoto FK. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ponnusamy R, Moll R, Weimar T, et al. Variable oligomerization modes in coronavirus non-structural protein 9. J Mol Biol. 2008;383:1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zeng Z, Deng F, Shi K, et al. Dimerization of coronavirus nsp9 with diverse modes enhances its nucleic acid binding affinity. J Virol. 2018 doi: 10.1128/JVI.00692-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu X, Fang P, Fang L, et al. Porcine deltacoronavirus nsp15 antagonizes interferon-beta production independently of its endoribonuclease activity. Mol Immunol. 2019;114:100–107. doi: 10.1016/j.molimm.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang L, Li L, Yan L, et al. Structural and biochemical characterization of endoribonuclease Nsp15 encoded by middle east respiratory syndrome coronavirus. J Virol. 2018 doi: 10.1128/JVI.00893-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV -2. Protein Sci. 2020 doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Holdgate G, Embrey K, Milbradt A, Davies G. Biophysical methods in early drug discovery. ADMET DMPK. 2019;7:222–241. doi: 10.5599/admet.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lundqvist T. The devil is still in the details–driving early drug discovery forward with biophysical experimental methods. Curr Opin Drug Discov Dev. 2005;8:513–519. [PubMed] [Google Scholar]
- 148.Kistler A, Avila PC, Rouskin S, et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Brodin P. Why is COVID-19 so mild in children? Acta Paediatr Int J Paediatr. 2020;109:1082–1083. doi: 10.1111/apa.15271. [DOI] [PubMed] [Google Scholar]
- 150.Brough HA, Kalayci O, Sediva A, et al. Managing childhood allergies and immunodeficiencies during respiratory virus epidemics – the 2020 COVID-19 pandemic. Pediatr Allergy Immunol. 2020 doi: 10.1111/pai.13262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.