Abstract
Advances in sequencing, bioinformatics and analytics now allow the structure, function and interrelations of whole microbial communities to be studied in greater detail. Collaborative efforts and multidisciplinary studies, crossing the boundary between environmental and medical microbiology, have allowed specific environmental, animal and human microbiomes to be characterized. One of the main challenges for microbial ecology is to link the phylogenetic diversity of host-associated microbes to their functional roles within the community. Much remains to be learned on the way microbes colonize the skin of different living organisms and the way the skin microbiome reacts to the surrounding environment (air, water, etc.). In this review, we discuss examples of recent studies that have used modern technology to provide insights into microbial communities in water and on skin, such as those in natural resources (thermal spring water), large mammals (humpback whales) and humans (the skin microbiome). The results of these studies demonstrate how a greater understanding of the structure and functioning of microbiota, together with their interactions with the environment, may facilitate the discovery of new probiotics or postbiotics, provide indicators for the quality of the environment, and show how changes in lifestyle and living environment, such as urbanization, can impact on the skin microbiome and skin health and disease in humans.
Key Points
| Multidisciplinary approaches and advances in sequencing, bioinformatics and analytics have allowed the diversity of whole microbial communities to be investigated. |
| Studies of the microbiomes of natural resources, such as spring water, have the potential to uncover targets for the development of new probiotics or postbiotics. |
| Analyses of the microbiomes of animals can provide vital information about the health status of the planet, whereas studies of the human microbiome can help determine the impact of living environments on human health and disease. |
Introduction on Microbial Communities and Their Study
Microbes (bacteria, viruses and fungi) are everywhere: among plants, animals and humans, and also in the environment (in the soil, water and air). They are often considered as pathogenic as they can be responsible for infections that may lead to severe health conditions or epidemics if they are not controlled. However, microbes also perform many valuable functions such as transforming food waste into compost, producing medicines, cleaning, and killing pests, and are involved in manufacturing processes (production of cheese, soy sauce, leather, etc.). They even play an important role in maintaining a good health status. Only a very small fraction of the global microbial population has been identified so far. Microbe identification has historically relied on isolation and culturing in laboratory conditions, which are not universal and are not suitable for the growth of the majority of microorganisms, thus limiting the identification of the whole diversity of a given microbial community.
New advanced molecular and bioinformatics tools have now allowed the identification and characterization of specific microbes on a larger scale, i.e. characterization of microbial communities (the microbiota) present in a particular area [1] and genomic identification of all microbes present in a specific environment (the microbiome). Metagenomic sequencing, allowing genome detection of all microbes present in a specific microbiome; metabolomic identification, through mass spectrometry of metabolites, lipids and peptides; bioinformatic softwares for analysis of phylogenetics, metaproteomics and metabolomics; and artificial intelligence-based data management and prediction of microbial interactions (i.e. artificially created microbial ecosystems [ACMEs]) are now being used in combination for the quantification and profiling of microorganisms, and their genetic material, environmental conditions, and interrelations [1–6].
The number of integrated studies on microorganisms using these techniques, referred to as microbiome studies, has been growing since the launch of the Human Microbiome Project (HMP) in 2007 [7]. The HMP is the largest study of microbial communities ever initiated in a healthy population. Another ongoing project is the Earth Microbiome Project (EMP, founded in 2010), which is a wide collaborative effort aiming to characterize microbial life on the planet and identify novel entities from a wide range of habitats, including air, water, sediments, plants, animals and humans [8] (http://www.earthmicrobiome.org). Such interdisciplinary and broad projects aim to provide a community-wide catalog of various microbiomes, using standardized protocols and analytical frameworks, to start answering many key questions about our microbial environment: What is the composition of a given microbiota (abundance, diversity of microbes)? What are their individual functions within the community? What are the interactions between them and their environment? How do they change and evolve? What is their role in health and disease?
In this review, we present a few examples of studies that are at the interface between environmental (water) and medical (health) microbiology, such as the analysis of a specific water microbiome, the skin microbiome of mammals living in the water, and the skin microbiome of humans. We address the influence of the urban environment on this specific ecosystem that protects our body from external threats.
A Specific Water Microbiome
Water is life and it is of crucial importance for all lifeforms on Earth, ranging from very small microorganisms to very large mammals, such as whales. Water helped to shape the first life forms on Earth, microorganisms. Water contains plethora of still unidentified species that are potential gold mines for chemicals and metabolites with medicinal, biotechnology, and energy applications.
Avène Thermal Spring Water (ATSW) from the Montagne Noire in France has been known for its therapeutic effects since the middle of the eighteenth century [9]. It has been extensively studied over several decades, using comprehensive clinical pharmacology approaches. ATSW from the Val d’Orb water catchment is a mineral groundwater. It has a residence time of over 200 years and arises through a dolomitic aquifer system. This thermal water, preserved from anthropogenic impact and pollution, has a constant temperature (21 °C) and a physicochemical composition characterized by a low mineral content [10], and contains around 1000 times less bacteria than surface water. The biogeochemical specificities and physicochemical characteristics of this deep aquifer system appear to have allowed a unique microbial community to develop in this water resource.
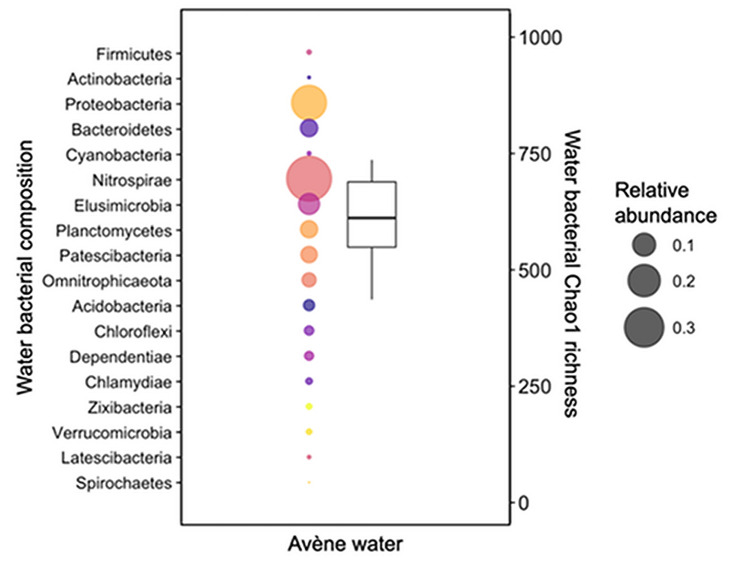
Metagenomic approaches have been used to analyze the microbiota of ATSW and potentially identify novel biosynthetic pathways and unknown metabolic features of this microbial consortium [11]. Over a 4-year sampling period, ATSW was shown to have a stable bacterial community, with a relatively constant richness of species and no major variations in diversity. Around 600 species were identified that were distributed over 39 phyla. The core bacteria of this ecosystem accounted for up to 76% of the total microbiota, in which Nitrospirae and Proteobacteria were the most prevalent phyla, accounting on average for 38% and 23% of the total core community, respectively (Fig. 1). The Nitrospirae phylum contains original phylotypes that develop only in the low mineral load (mainly composed of bicarbonate, magnesium, calcium and silicates) present within the water resource. Their metabolic pathways have not yet been characterized and need further investigation. Moreover, a significant number of variants present in this deep aquifer system (25–47% of all reads over the studied period) could not be assigned to an existing class, order, family or genus of bacteria, revealing the existence of a unique bacterial community in ATSW that merits further study. A novel microorganism, Aquaphilus dolomiae, has already been isolated and identified in ATSW [12]. This strain belongs to the family Neisseriaceae within the class Betaproteobacteria, and exhibits specific properties that are beneficial for the skin. A postbiotic, containing a biotechnological extract from this microorganism, was found to display immunomodulatory, anti-inflammatory, antipruritic and tolerogenic activities in pharmacological models of atopic dermatitis, and thus has been incorporated into a skin care emollient [13–16]. These activities included the modulation of innate immunity by agonizing toll-like receptor (TLR) 2, TLR4 and TLR5, the induction of antimicrobial peptides, the inhibition of cytokine production by T helper (Th) 1, Th2 and Th17 cells, and the inhibition of protease-activated receptor (PAR) 2 and thymic stromal lymphopoietin (TSLP). In addition, the extract was capable of inducing interleukin (IL)-10 secretion to activate regular T lymphocytes, and rendered human dendritic cells tolerogenic [14].
Fig. 1.
Bacterial composition and richness of Avène Thermal Spring Water. Sample composition and diversity are the average of samples taken between December 2014 and September 2018, with four samples taken per year. Bubble plots represent the bacterial composition (for simplicity, only major phyla shown). Each phylum is represented by a different color. The size of the bubble represents the relative abundance, as indicated in the legend. The y-axis for the bubble plots is on the left. The boxplot represents the bacterial Chao1 richness, with the y-axis on the right.
Data adapted from Bourrain et al. [11]
Further studies of the microbiome of natural resources, such as water, will likely uncover promising new targets for the development of microorganism-derived therapeutics, offering new approaches in medicine: in dermatology, for example, the design of new postbiotics could limit the need for topical and systemic use of antibiotics.
Microbiomes in Whales
Whales are the largest mammals on Earth and they live in and interact very closely with the aquatic, marine environment. Climate change can heavily impact their health and microbiome. The skin microbiome of whales can be an important biomarker for temperature changes, loss of diversity and changes in the marine environment. There are few studies on the cetacean microbiome and even fewer describing bacterial members on their skin [17].
Some recent microbiome studies explored the impact of the environment on the microbial communities present on these large marine mammals. For instance, aerial drone technology was used to allow noninvasive collection of the exhaled breath condensate from humpback whales for analysis of their respiratory microbiome [18]. This study identified a large core bacterial community from the whale blowhole, made up of 22 members. This core community was found to be shared by whales from two different geographic regions in distinct ocean basins (the North of Cape Cod and Vancouver Island coasts), and was distinct from that of the surface seawater. Two members were skin-associated bacteria, likely coming from the whale blowhole epithelium. The major part of this blow microbiome signature (20 bacterial members) was found to be closely related to microbial sequences isolated from other marine mammal microbiomes, in particular dolphins, and may serve as a feature for monitoring the respiratory health of whales and possibly other cetaceans worldwide. Furthermore, exploration of the gut microbiome of baleen whales—which prey on small fish and crustaceans with chitin-rich exoskeletons—revealed functional similarities with terrestrial herbivores, reflecting the shared role of these microbial communities in the fermentative metabolism of polysaccharides [19]. In contrast, the protein catabolism and essential amino acid synthesis pathways in the baleen whale microbiomes more closely resembled those of terrestrial carnivores. Thus, this study showed that diet, gastrointestinal physiology and evolutionary history shape the gut microbiota of whales. In particular, their multichambered foregut, serving as a preadapted fermentation chamber such as that found in ruminants, allows them to metabolize complex polysaccharides (such as chitin) from the exoskeletons of their prey. In contrast to the relative stability of large-whale blow microbiomes [18], the skin microbiome of humpback whales was found to differ temporally and regionally [20]. This was demonstrated in a study of the skin microbiome of 89 healthy humpback whales: a conserved core microbiome of six bacterial genera was identified but a shift in the relative abundances of these core bacteria was observed over time, along with the emergence of four additional core groups of bacteria during the late foraging season, likely reflecting seasonal changes in the Southern Ocean. Moreover, the skin microbiome composition differed between whales sampled at several regional locations along the Western Antarctic Peninsula. These findings suggest that temporal and environmental factors, in particular water temperature, and also the whale population itself, may influence their skin microbiome [20]. Skin diseases in cetaceans have been reported since at least the 1950s and have steadily increased since that time [21]. Species-specific assemblages on the whale’s skin can change seasonally, but these changes can also be linked to the presence of epiphytic diatoms and poor skin health [17]. These species have also been shown to be distinct from those found in surrounding planktonic samples [22]. These finding suggest that ecological and evolutionary forces, including the unique features of an animal’s epidermis, have sculpted cetacean skin microbiomes.
The microbiome of marine animals may therefore harbor important information on animal–environment interactions and on the ocean ecosystem. Whale-associated bacteria have been found to be enriched with genes associated with cell motility, enabling cells to move and colonize new skin surfaces, as well as with those associated with glycan biosynthesis and metabolism, potentially used to metabolize the whale-associated tissues or flocculating material that accumulates on the whale’s epidermis [21, 23]. It is likely that motility and chemotaxis are critical for microbial communities associated with skin surfaces, where strong chemical gradients are present. In addition, cell signaling pathways are important on skin surfaces, where bacteria may use signaling processes, such as quorum sensing, to organize cellular functions to colonize host organisms. Similar functions and processes may also apply to the human skin microbiome and its interaction with the air and water microbiomes.
The Human Skin Microbiome
As in whales, the skin microbiome of humans is of particular importance as it plays a major role in the homeostasis and protective function of the skin. Water likewise plays an important role in shaping the human skin microbiome, as it is in constant interaction with the skin, for instance through washing, hygiene habits, and clothes laundering. Identifying commensal and pathogenic microbes, and determining their relationship with the host, will help decipher their function in healthy skin, as well as in dermatologic disorders [1, 3, 24].
The topographical and temporal diversity of the human healthy skin microbiome has been thoroughly investigated [24–26], revealing that thousands of bacterial species are present on a single individual [25]. Bacteria are present on the skin surface and appendages, in deeper layers of the epidermis, and even in the dermis and dermal adipose tissue [27–29]. The total surface area of the skin in adult humans is usually estimated at around 2 m2; however, when all the skin appendages (hair follicles, and sweat and sebaceous glands) are considered, the actual surface area is more than 10 times larger, providing at least 30 m2 of epithelial surface for interaction with a multitude of microbes [30]. This larger estimated skin surface area suggests that microbial communities residing on the skin have the potential to greatly influence human health.
Most of the bacterial species on the skin are nonpathogenic and saprophytic, but some can become pathogenic depending on the host context [4]. Fungi, viruses and mites are also found on the skin, with fungi of the genus Malassezia predominating in seborrheic regions [24, 26]. Each of these microorganisms possesses distinct metabolic adaptations for survival in the different skin microenvironments [24].
The most abundant bacterial species found on human skin belong to only four phyla: Actinobacteria, Firmicutes, Proteobacteria and Bacteroidetes [25, 26]. The predominance of these phylotypes on specific skin areas depends on multiple factors, such as moisture levels, sebum content, temperature, pH, and UV exposure. Sebaceous sites are mainly colonized by Cutibacterium (formerly Propionibacterium [31]), and their microbiomes are generally less diverse, less evenly distributed, and less rich than those from moist areas harboring mainly Corynebacterium and Staphylococcus species. Dry sites additionally contain Micrococcus, Enhydrobacter and Streptococcus species [3, 25–27]. Adult skin bacterial composition remains relatively stable over a 2-year period [32]. However, temporal variations of the skin microbiome have been observed and appear to be site-dependent, with the popliteal fossa, volar forearm, and buttocks exhibiting the largest variations [25]. Sites that are at least partially occluded, such as the external auditory canal, the nares and the inguinal crease, are more stable over time in terms of community membership and structure [26]. Compared with other habitats (oral, gut or vagina), the skin microbiome displays the most between-subject variability [33] and shows the greatest variability over time [26]. It is generally agreed that more interpersonal and temporal variability is present among the less abundant and transient bacterial inhabitants than in the predominant and stable taxa [3]. Multiple host-specific factors, such as age, ethnicity and sex, also contribute to skin microbial variability. The effect of age is discussed further elsewhere in this supplement [34].
Altogether, environmental factors and intrinsic changes impact the structure and function of the skin microbiota, either preserving or impairing skin health.
Influence of the Urban Environment on the Microbiota
The environment where we live has a major impact on the skin microbiota, as the skin epithelial surface is in direct contact with the external environment. In particular, great variations in skin microbial communities might occur between people living in rural environments and those living in urban environments.
The study of Ying et al. on the skin bacterial community of healthy subjects aged 12–60 years, confirmed that body site is the major source of the variability in bacterial community structure (i.e. the relative abundance of different taxa), with the skin microenvironment (sebaceous, moist or dry) being the most influential factor [35]. The composition of the skin bacterial microbiota is also predominantly influenced by the place of residence, either urban or rural. Although the richness of the skin microbiome appeared similar between urban and rural populations, the intragroup variation in microbial community structure has been found to be significantly greater in rural subjects than in urban subjects [35]. Furthermore, urban populations exhibit an overall higher relative abundance of Trabulsiella, especially on the back of the hands, volar forearm and forehead (glabella). In addition, differences have been observed among women, with a higher content of Cutibacterium on glabella in urban residents than in rural residents, whereas the inverse was found for Corynebacterium. The impact of the living environment on the skin microbiota is also seen across age groups [35, 36], with the size of this effect varying with age, in particular among children [36]. The imprint of the land environment is indeed more prominent in toddlers (1–4 years of age) than in babies (< 1 year of age) and schoolchildren [36]. In contrast, living environment has no effect in teenagers [36]. Such variations in the skin microbial composition between rural and urban residents could be explained by different levels of exposure to soil, aquatic and host-associated microbial sources, stemming from urban inhabitants usually having more indoor occupations [35]. Different cultural habits and lifestyle-related factors might also account for the observed disparities in skin microbiomes [36].
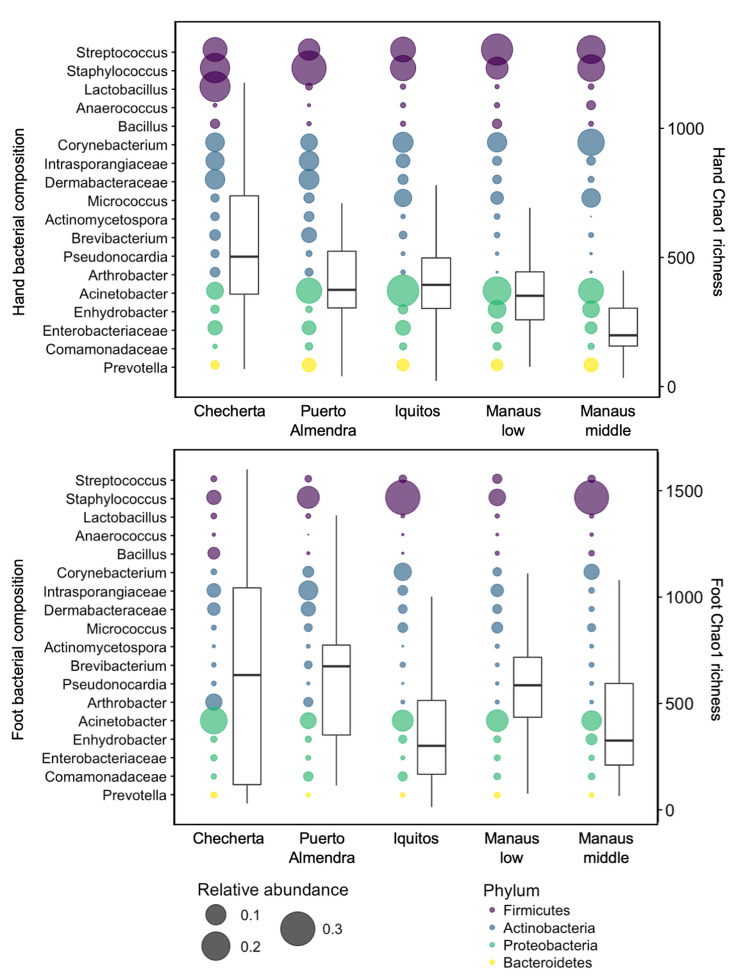
A recent study investigated the effects of urbanization on the house and human microbiomes [37]. A comprehensive chemical and microbial survey, spanning several areas of the same latitude in the Amazon rainforest, revealed a transition in the profile of these ecosystems along an urbanization gradient. Human, animal and household samples were collected from five different geographical locations: a remote village (Checherta), a rural village (Puerto Almendra), a large town (Iquitos), and low- and middle-class areas of a metropolis (Manaus). This study found that the number of chemicals in houses increased substantially with urbanization. Liquid chromatography with tandem mass spectrometry (LC–MS/MS) analysis of samples from each location showed that house surface chemicals, detergents, personal care and cleaning products, and medication-derived chemicals were more abundant in the more urbanized settings. Profiles of house surface chemicals in the metropolitan city of Manaus were similar regardless of the social level (lower or middle class), but were distinct from those observed in the rural populations. This trend was consistent with differences in access to cosmetic and cleaning products, which differed between Brazil and Peru. Furthermore, microbial profiles varied significantly between sampling locations. Increased urbanization was related to changes in the composition of house bacterial, fungal and microeukaryotic communities. Specifically, urbanization correlated with an increased relative abundance of human skin-associated fungi and bacteria in homes, and increased house and skin fungal diversity [37]. Among the fungi, potentially pathogenic strains, such as Aspergillus, Malassezia, Candida, and Eurotiales, were identified in urban skin and house samples. Meanwhile, the richness of the bacterial community on human skin (hands, arms and feet) decreased and its composition changed with urbanization (Fig. 2). Hand and arm bacterial richness in particular markedly decreased with urbanization. In addition, an increased relative abundance of Staphylococcus species and pathogenic fungi on the feet were associated with more frequent shoe use, whereas the microeukaryotic diversity on this skin surface decreased. The well-known skin bacteria, Corynebacterium, Micrococcus, Pseudomonas and Enhydrobacter, became more enriched on skin surfaces with urbanization, whereas the abundance of environmental bacteria (Dermabacteraceae and Intrasporangiaceae) decreased or nearly disappeared, along with a wide diversity of low-abundant bacteria and many other taxa that are generally not associated with humans [37]. A negative correlation between skin bacterial diversity and detergents was observed, suggesting that these products might account, at least in part, for the loss of diversity in the cutaneous bacterial communities in urban settings. A higher proportion of potentially pathogenic taxa, both bacterial and fungal, were found with increasing urbanization. This might explain the higher prevalence of typical Western diseases, such as acne, atopic dermatitis or Staphylococcus aureus infections. As an example, acne vulgaris is not found on the skin of indigenous tribes or on that of individuals from non-industrialized societies [38], whereas 79–95% of Western people are affected [39]. Only when indigenous people moved to Westernized cities did acne reportedly become a problem [40], proving the impact of environment and lifestyle on the skin disease. Overall, results from these studies indicate that urbanization has large-scale effects on chemical and microbial exposures, and hence on the human skin microbiota. Many factors are involved in these changes, such as lifestyle, house architecture, number of inhabitants per house, air pollution, detergents and cosmetic use, among many others [37]. Changes in skin microbiome composition between rural and urban populations might also stem from differences in microbial sources (soil, water, indoor versus outdoor occupations, etc.) [35]. These findings are useful to gain further insights into the health and disease states of the skin, especially in urban populations, where the prevalence and risk of atopic dermatitis have been reported to be higher than in rural populations [41–43]. As it is predicted that approximately two-thirds of the human population is going to live in cities by 2050 [44], we need to pursue maintenance of healthy human and home microbiomes to counteract the worldwide emergence of Western diseases.
Fig. 2.
Human bacterial composition across an urbanization gradient in the Amazon in South America. Sites ranged from a remote village isolated in the Amazon rainforest (Checherta), to a rural village (Puerto Almendra), a large town (Iquitos), and the lower and middle socioeconomic class of a metropolis city (Manaus). Samples presented per skin sample site: right hand (top panel), right foot (bottom panel). Bubble plots, with the y-axis on the left, represent the bacterial composition (for simplicity, only the major taxa are shown). The size of the bubble represents the relative abundance. Boxplots, with the y-axis on the right, represent the bacterial Chao1 richness. Firmicutes are depicted in purple, Actinobacteria in blue, Proteobacteria in green, and Bacteroides in yellow.
Data adapted from McCall et al. [37]
Conclusion
Microbiota present in the environment and on living organisms are very diverse with strong niche specialization. Sequencing technologies and bioinformatics enable the comprehensive study of different microbial ecosystems, allowing their complex structure and functioning to be unraveled. Microbiomes can serve as sensors for the health of the planet and its inhabitants. Understanding the interplay between our surrounding and intrinsic microbial communities is essential for accurately predicting the impact of perturbations or environmental changes on human health. In particular, increasing urbanization might disturb the skin ecosystem, and, as a consequence, human health, as the skin is the first line of physical protection from external aggressors. In addition, newly identified microbes could serve as novel probiotics or postbiotics, and provide medicinal solutions for combating human diseases such as dermatologic conditions. Our knowledge of the various microbiomes is constantly being updated, with substantial insights being gained over the last decade.
Acknowledgments
The authors thank Dr Muriel Bourrain, from Pierre Fabre Dermo-Cosmétique R&D center, for her large contribution to the study of ATSW microbiota, as well as Cécile Desjobert, PhD, Emma Pilling, PhD, and Marielle Romet, PhD (Sante Active Edition-Synergy Pharm), who provided medical writing assistance funded by Laboratoires dermatologiques Avène, Pierre Fabre Dermo-Cosmétique. The authors would also like to thank Maria Gloria Dominguez-Bello, who led the Amazon expeditions in 2012 and 2013, as well as the team of researchers, doctors, priests, and translators who joined and assisted in any possible way. The authors also thank the laboratories of Pieter C. Dorrestein and Rob Knight for the extensive analyses on the samples. The Amazon study was supported by the Sloan Foundation. Chris Callewaert was supported by the Belgian American Educational Foundation and the Research Foundation Flanders.
Author Contributions
All authors contributed to the conception of the article, performed literature searches and data analysis, and critically revised the manuscript.
Declarations
Funding
Medical writing assistance was funded by Laboratoires dermatologiques Avène, Pierre Fabre Dermo-Cosmétique.
Conflicts of interest
Chris Callewaert and Philippe Lebaron have no conflicts of interest to declare that might be relevant to the contents of this manuscript. Katia Ravard Helffer is employed by Pierre Fabre Dermo-Cosmétique.
Disclosure statement
This article has been published as part of a journal supplement wholly funded by Laboratoires dermatologiques Avène, Pierre Fabre Dermo-Cosmétique.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Code availability
Not applicable.
References
- 1.Grogan MD, Bartow-McKenney C, Flowers L, Knight SAB, Uberoi A, Grice EA. Research techniques made simple: profiling the skin microbiota. J Investig Dermatol. 2019;139(4):747–52 e1. [DOI] [PMC free article] [PubMed]
- 2.Geron A, Werner J, Wattiez R, Lebaron P, Matallana-Surget S. Deciphering the functioning of microbial communities: shedding light on the critical steps in metaproteomics. Front Microbiol. 2019;10:2395. doi: 10.3389/fmicb.2019.02395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice EA. The intersection of microbiome and host at the skin interface: genomic- and metagenomic-based insights. Genome Res. 2015;25(10):1514–1520. doi: 10.1101/gr.191320.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lebaron P, Bourrain M. The skin: a microbial ecosystem. Ann Dermatol Venereol. 2017;144:S35–S41. doi: 10.1016/S0151-9638(17)31041-4. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34(8):828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. Network relationships of bacteria in a stable mixed culture. Microb Ecol. 2008;56(3):403–411. doi: 10.1007/s00248-007-9357-4. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551(7681):457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selas B. History of thermalism at Avene-les-Bains and genesis of the Avene thermal spring water. Ann Dermatol Venereol. 2017;144(Suppl 1):S21–S26. doi: 10.1016/S0151-9638(17)31039-6. [DOI] [PubMed] [Google Scholar]
- 10.Lions J. Hydrochemistry of Avène thermal waters (Avène-les-Bains, France) J Eur Acad Dermatol Venereol. 2020;34(Suppl 5):4–7. doi: 10.1111/jdv.16336. [DOI] [PubMed] [Google Scholar]
- 11.Bourrain M, Suzuki MT, Calvez A, West NJ, Lions J, Lebaron P. In-depth prospection of avene thermal spring water reveals an uncommon and stable microbial community. J Eur Acad Dermatol Venereol. 2020;34(Suppl 5):8–14. doi: 10.1111/jdv.16599. [DOI] [PubMed] [Google Scholar]
- 12.Bourrain M, Villette C, Nguyen T, Lebaron P. Aquaphilus dolomiae gen. nov., sp. nov., isolated from a deep aquifer. Vie et Milieu. 2012;62(4):191–195. [Google Scholar]
- 13.Guerrero D, Garrigue E. Avène’s thermal water and atopic dermatitis. Ann Dermatol Venereol. 2017;144(Suppl 1):S27–S34. doi: 10.1016/S0151-9638(17)31040-2. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen T, Castex-Rizzi N, Redoulès D. Immunomodulatory, anti-inflammatory, anti-pruritus and tolerogenic activities induced by I-modulia®, an Aquaphilus dolomiae culture extract, in atopic dermatitis pharmacology models. Ann Dermatol Venereol. 2017;144:S42–S49. doi: 10.1016/S0151-9638(17)31042-6. [DOI] [PubMed] [Google Scholar]
- 15.Bianchi P, Theunis J, Casas C, Villeneuve C, Patrizi A, Phulpin C, et al. Effects of a new emollient-based treatment on skin microflora balance and barrier function in children with mild atopic dermatitis. Pediatr Dermatol. 2016;33(2):165–171. doi: 10.1111/pde.12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deleuran M, Georgescu V, Jean-Decoster C. An emollient containing Aquaphilus dolomiae extract is effective in the management of xerosis and pruritus: an international, real-world study. Dermatol Ther. 2020 doi: 10.1007/s13555-020-00415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooper R, Brealey JC, van der Valk T, Alberdi A, Durban JW, Fearnbach H, et al. Host-derived population genomics data provides insights into bacterial and diatom composition of the killer whale skin. Mol Ecol. 2019;28(2):484–502. doi: 10.1111/mec.14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apprill A, Miller CA, Moore MJ, Durban JW, Fearnbach H, Barrett-Lennard LG. Extensive core microbiome in drone-captured whale blow supports a framework for health monitoring. mSystems. 2017;2(5):e00119–17. doi: 10.1128/mSystems.00119-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders JG, Beichman AC, Roman J, Scott JJ, Emerson D, McCarthy JJ, et al. Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat Commun. 2015;6:8285. doi: 10.1038/ncomms9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bierlich KC, Miller C, DeForce E, Friedlaender AS, Johnston DW, Apprill A. Temporal and regional variability in the skin microbiome of humpback whales along the Western Antarctic Peninsula. Appl Environ Microbiol. 2018;84(5):e02574–17. doi: 10.1128/AEM.02574-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouton M, Botha A. Cutaneous lesions in cetaceans: an indicator of ecosystem status? In: Romero A, Keith EO, editors. New approaches to the study of marine mammals. 2012. pp. 123–50.
- 22.Chiarello M, Villeger S, Bouvier C, Auguet JC, Bouvier T. Captive bottlenose dolphins and killer whales harbor a species-specific skin microbiota that varies among individuals. Sci Rep. 2017;7(1):15269. doi: 10.1038/s41598-017-15220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apprill A, Robbins J, Eren AM, Pack AA, Reveillaud J, Mattila D, et al. Humpback whale populations share a core skin bacterial community: towards a health index for marine mammals? PLoS One. 2014;9(3):e90785. doi: 10.1371/journal.pone.0090785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16(3):143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 25.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreno B, Araviiskaia E, Berardesca E, Gontijo G, Sanchez Viera M, Xiang LF, et al. Microbiome in healthy skin, update for dermatologists. J Eur Acad Dermatol Venereol. 2016;30(12):2038–2047. doi: 10.1111/jdv.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bay L, Barnes CJ, Fritz BG, Thorsen J, Restrup MEM, Rasmussen L, et al. Universal dermal microbiome in human skin. mBio. 2020;11(1):e02945–19. doi: 10.1128/mBio.02945-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallo RL. Human skin is the largest epithelial surface for interaction with microbes. J Investig Dermatol. 2017;137(6):1213–1214. doi: 10.1016/j.jid.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholz CFP, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol. 2016;66(11):4422–4432. doi: 10.1099/ijsem.0.001367. [DOI] [PubMed] [Google Scholar]
- 32.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165(4):854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luna PC. Skin microbiome as years go by. Am J Clin Dermatol. 2020 doi: 10.1007/s40257-020-00549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying S, Zeng DN, Chi L, Tan Y, Galzote C, Cardona C, et al. The influence of age and gender on skin-associated microbial communities in urban and rural human populations. PLoS One. 2015;10(10):e0141842. doi: 10.1371/journal.pone.0141842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehtimaki J, Karkman A, Laatikainen T, Paalanen L, von Hertzen L, Haahtela T, et al. Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Sci Rep. 2017;7:45651. doi: 10.1038/srep45651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCall LI, Callewaert C, Zhu Q, Song SJ, Bouslimani A, Minich JJ, et al. Home chemical and microbial transitions across urbanization. Nat Microbiol. 2020;5(1):108–115. doi: 10.1038/s41564-019-0593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiner PE. Necropsies on Okinawans; anatomic and pathologic observations. Arch Pathol (Chic). 1946;42(4):359–380. [PubMed] [Google Scholar]
- 39.Cordain L, Lindeberg S, Hurtado M, Hill K, Eaton SB, Brand-Miller J. Acne vulgaris: a disease of Western civilization. Arch Dermatol. 2002;138(12):1584–1590. doi: 10.1001/archderm.138.12.1584. [DOI] [PubMed] [Google Scholar]
- 40.Cunliffe WJ, Cotterill JA. The acnes: clinical features, pathogenesis and treatment. In: Major problems in dermatology. vol. 6. Philadelphia: W.B. Saunders; 1975.
- 41.Schram ME, Tedja AM, Spijker R, Bos JD, Williams HC, Spuls PI. Is there a rural/urban gradient in the prevalence of eczema? A systematic review. Br J Dermatol. 2010;162(5):964–973. doi: 10.1111/j.1365-2133.2010.09689.x. [DOI] [PubMed] [Google Scholar]
- 42.Shaw TE, Currie GP, Koudelka CW, Simpson EL. Eczema prevalence in the United States: data from the 2003 National Survey of Children’s Health. J Investig Dermatol. 2011;131(1):67–73. doi: 10.1038/jid.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu F, Yan S, Li F, Cai M, Chai W, Wu M, et al. Prevalence of childhood atopic dermatitis: an urban and rural community-based study in Shanghai, China. PLoS One. 2012;7(5):e36174. doi: 10.1371/journal.pone.0036174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.United Nations. Department of Economic and Social Affairs, population division. World Urbanization Prospects: the 2018 revision. (ST/ESA/SER.A/420). New York: United Nations; 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.