Abstract
The skin microbial communities, i.e., the microbiota, play a major role in skin barrier function so must remain dynamic to adapt to the changes in the niche environment that occur across the different body sites throughout the human lifespan. This review provides an overview of the major alterations occurring in the skin microbiome (microbial and genomic components) during the various stages of life, beginning with its establishment in the first weeks of life through to what is known about the microbiome in older populations. Studies that have helped identify the factors that most influence skin microbiome function, structure, and composition during the various life stages are highlighted, and how alterations affecting the delicate balance of the microbiota communities may contribute to variations in normal physiology and lead to skin disease is discussed. This review underlines the importance of improving our understanding of the skin microbiome in populations of all ages to gain insights into the pathophysiology of skin diseases and to allow better monitoring and targeted treatment of more vulnerable populations.
Key Points
| Studies of the cutaneous microbiome (microbial and genomic components) across different age groups have highlighted the dynamic nature of the skin microbial communities (the microbiota), beginning during the very early stages of life following initial exposure to the maternal microbiome and continuing with shifts in community structure and diversity through to old age. |
| Beyond the first few weeks of life, body site is a major determinant of skin microbiome structure and diversity, but many other factors that influence these site-specific communities during the different stages of life have been identified, with some alterations potentially being associated with skin disease. |
| Understanding the skin microbiome at all stages of life is important for determining the impact of early alterations in the microbiome on future health, gaining a better understanding of the pathophysiology of skin diseases, and aiding the development of age-adapted treatments. |
Introduction
The skin is the largest organ of the human body and is colonized by highly variable microbial communities, referred to as the microbiota, that are influenced by multiple factors. Different communities with distinct functions are present in the epidermal and dermal compartments, with the environment having a broader impact on the microbiota of the outermost layers of the epidermis [1, 2]. Skin-resident microbes contribute to the establishment of cutaneous homeostasis and can modulate inflammatory responses or trigger pathogenicity depending on the host context [3]. The mutualistic relationship between microbial communities and the host is essential for establishing the well-controlled and delicate balance needed for healthy skin. The skin microbiota plays a major role in the function of the skin as a barrier against environmental threats and infections. To fulfill this role, the microbiota remains dynamic throughout the human lifespan. Although the skin microbiome, comprising microbial and genomic components, has been well studied in adults [1, 2, 4–7], less information is available in children and the elderly. This article provides an overview of current knowledge on the main changes occurring in the skin microbiome throughout the different life stages, focusing on the epidermal microbiome, which is the most exposed to surrounding factors and is more diverse and varying than the dermal microbiome [1]. This knowledge is key for enabling the dermatologic specificities and needs of each age group to be better addressed.
Settlement at Birth
Birth marks a time of drastic change for the skin of the newborn as it undergoes a sudden transition from an almost bacteria-free aqueous environment to an atmospheric one with constant exposure to microbes. Immediately after birth, fetal skin is colonized by surrounding microorganisms. These microorganisms have been shown by Dominguez-Bello et al. [8] to differ according to the mode of delivery: the cutaneous bacterial signature (community diversity and structure) of vaginally delivered neonates resembles the mother’s vaginal bacteria, with Lactobacillus predominating, whereas that of neonates delivered by cesarean resembles the cutaneous bacteria, including Staphylococcus, Corynebacterium, and Cutibacterium (formerly Propionibacterium [9]). Furthermore, unlike their mothers, neonates have been shown to harbor bacterial communities that are undifferentiated across multiple body habitats (skin, oral, nasopharyngeal, gut, vagina, etc.), regardless of the delivery mode [8, 10]. The vertical transmission of the vaginal microbiota from the mother to her baby may serve an initial defensive role in neonatal skin. Conversely, reduced skin microbial diversity has been reported in neonates born by cesarean delivery [10]. This reduced diversity, combined with the lack of exposure to vaginal bacteria, may lead to delayed development of the nutritional and immune functions associated with the microbiota across the different body habitats [8], perhaps in part explaining the greater susceptibility of cesarean-born infants to certain pathogens and allergies. However, as cesarean delivery is usually medically indicated, the many potential confounders associated with the need for this procedure may also have an impact on the microbiome acquired by the offspring. Moreover, the skin microbial pattern has been found to differ depending on whether neonates were delivered by a labored or an unlabored cesarean [10].
Further analyses indicate that by 4–6 weeks after birth, infant skin microbiome structure and function have already significantly expanded and diversified, with prominent body site specificities similar to those of the maternal skin microbiome [10]. For instance, Chu et al. [10] showed that the microbiome of infant skin and nares was characterized by Staphylococcus (Firmicutes) and Corynebacterium (Actinobacteria), as in the mother. This reorganization appeared to be primarily driven by the body site and was no longer influenced by the mode of delivery. Hence, site-specific differences in skin microbial abundance and structure emerge very early in life, before the infant has had extensive contact with other environmental sources of microorganisms.
In preterm infants, physical and functional differences in the immature skin may alter the resident microbiome compared with that of full-term infants. Pammi et al. [11] found that overall skin bacterial richness (variety of species) and evenness (i.e., the relative abundance of the community) tended to be lower in preterm infants than in term infants, with no differences in richness or composition between surveyed skin sites observed in either group within the first 4 weeks of life. However, a trend was observed for increasing microbial diversification with increased gestational age at baseline and then during the first 4 weeks of life for both preterm and term neonates. Comparisons of bacterial composition revealed that Firmicutes (predominant genus Staphylococcus) were more abundant and Proteobacteria less abundant in preterm neonates than in term neonates. At 4 weeks after birth, the skin microbiome of the preterm infants did not differ according to the mode of delivery, similar to previous findings in term neonates [10], but was found to be altered by antibiotic exposure [11]. In preterm infants, immaturity of the skin structure and appendages, as well as the hospital environment, may explain the decreased skin microbial diversity (richness and evenness) as compared with term neonates, and partly explain the existence of gestational age-dependent differences. Comorbidities, including respiratory illness, sepsis, necrotizing enterocolitis, and invasive clinical procedures, may also affect the developing skin microbiome.
The first exposure of neonates to the vaginal microbiota appears to play an important role in the initial establishment of the cutaneous microbiota and may also have an impact on further bacterial colonization of the gut and other body habitats [8]. Differences in the initial skin microbiome may thus have broader impacts on infant health, and on health later in life, and therefore need to be more deeply investigated. Interventions aiming to restore the diversity of the cutaneous microbiome in preterm neonates could also be developed to prevent pathogen colonization and subsequent systemic infections in this more vulnerable population [11].
Infants
Gradual maturation of skin microbiome function, structure, and composition has been shown to continue throughout the first years of life [12]. Large interindividual variations in skin bacterial communities, along with tremendous diversity in both community composition and the timing of bacterial acquisition, have been observed in healthy infants by Capone et al. [12]. Although data indicate that the number and type of genera in a specific skin area do not significantly change over the first year, the relative abundance of the community has been reported to increase with age. A decline of Staphylococcus and Streptococcus species has been noted, which may contribute to the increased population richness and evenness observed by the end of the first year. The skin microbiome in infants stabilizes over time and, as in adults, becomes progressively site specific: this process begins within the first 3 months of life and reflects a balance between the influx of microbial strains and niche selection [12].
Bidirectional microbial transmission between the mother and infant contributes to the dynamics of skin microbial colonization over the first year postpartum. A longitudinal study of mother–infant dyads followed up to 12 months revealed the predominance of six bacterial genera (Staphylococcus, Pseudomonas, Enterobacter, Enterococcus, Proteus, and Klebsiella) belonging to two phyla (Firmicutes and Proteobacteria) in both mother and infant skin samples [13]. Peaks in the number of genera per individual were found at 6 and 9 months in both mothers and infants, corresponding to periods when physical activity started to increase and infant food sources began to expand. The abundance of four of the six most frequent genera varied significantly with time in both groups, without remarkable differences in the isolation rates between the mothers and their children [13], which is in agreement with the fact that by the fourth week of life the infant microbiota is already very similar to that of the adult [10]. Only Proteus and Staphylococcus aureus have been reported to remain largely constant over time, with S. aureus being more frequently found in mothers than in infants. This latter finding could predispose both mothers and newborns to skin infections and could also be linked to the development of atopic dermatitis in the children [14]. The role of enteric bacteria (Enterococcus, Enterobacter, Klebsiella), found in large numbers on the skin of both the infants and nursing mothers, remains to be addressed. Furthermore, Gaitanis et al. [13] revealed a decline over time in the concordance of isolated microbial species within the mother–infant dyad, suggesting that other factors beyond inheritance from the mother contribute to the progressive increase in infant skin microbial diversification with age.
Thus, the skin microbiome evolves over the first year, especially in the first weeks after birth, with this early colonization potentially affecting long-term microbiome stability and skin immune function. Infant skin is known to be sensitive and more prone to inflammatory conditions such as atopic dermatitis and diaper dermatitis and to infections such as candidiasis. Therefore, the timely and proper establishment of a healthy skin microbiome during early childhood has been suggested to have a pivotal role in preventing inflammatory skin diseases and access to potentially infectious microbes [12]. Bidirectional microbial transmission between the mother and infant appears to be of paramount importance in this process [13], but the role of microbiota inheritance from the father remains poorly documented.
Children Versus Adults
A shift in the microbiomes of the skin and nares has been shown to occur during the physiologic transition from childhood to adulthood. The site-specific composition of the skin bacterial community, observed from the first months of life [12], along with the overall increase in diversity with age, were confirmed in a study of 158 children aged between 1 and 10 years [15]. The major phyla retrieved from these children were similar to those found in adults, e.g., Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes, with essentially the same genera but at different ratios [4, 15] (Fig. 1). For instance, the skin of the children contained less lipophilic bacteria (Actinobacteria, predominantly Cutibacterium), as the sebum level tended to be lower than that in adults [15, 16]. It is noteworthy that Zhu et al. [15] found the mode of delivery to still have an impact on the skin microbiome in the 10-year-old children. In addition, the skin microbiome of each child was more similar to that of their own mother than to that of unrelated women (particularly in the case of the most abundant bacteria), likely because of the impact of shared living environments and lifestyles. Conversely, sex was not found to exert a notable effect on the skin microbial community during childhood [15].
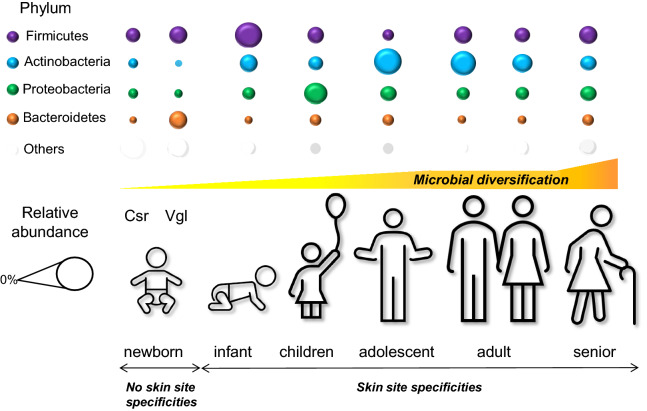
Fig. 1.
Skin microbiome evolution in healthy subjects as years go by. The graph provides a global overview of the relative abundance of major bacterial phyla of the human skin microbiota at different stages of life, measured by either 16S RNA or metagenomic approaches (DNA). Average data or main trends extracted from: newborns until 4 weeks (Dominguez-Bello et al. [8]), infants up to 12 months (Chu et al. [10] and Capone et al. [12]), children up to 10 years (Zhu et al. [15]), adolescents (Oh et al. [16]), adults (Ying et al. [17] and Grice et al. [4]), and older subjects above 60 years (Shibagaki et al. [23]). Csr cesarean delivery, Vgl vaginal delivery
In a comparison of bacterial communities at several body sites in children and young adults, significant differences were found between the groups, with communities at the nares exhibiting the most marked microbial shifts [16]. This site can harbor opportunistic pathogens, including S. aureus and Streptococcus pneumonia. Oh et al. [16] demonstrated that microbial diversity of the nares decreased with increasing sexual maturity, regardless of sex, indicating that stabilization and convergence of microbiomes in the nares occur in more mature subjects. In another study, the microbiome in the nares was found to cluster separately from that of other skin sites and to have a level of bacterial diversity that was lower than that of moist and dry sites [17]. Analysis of this bacterial community revealed that it contained Corynebacterium (35.5%) (Actinobacteria), Staphylococcus (17.7%) (Firmicutes), and Alloiococcus (5.2%) (Firmicutes), with these proportions being higher than those found at other skin sites [17]. In particular, S. aureus was overrepresented in the nares of younger subjects [16]. Moreover, a correlation between the incidence and abundance of this strain in the nares and at other distant skin sites has been found in both younger and older children, suggesting that the nares may serve as a reservoir for this pathogen.
This latter finding may have implications for the age-related incidence and severity of some cutaneous disorders such as atopic dermatitis. This cutaneous disorder involving skin colonization by S. aureus predominantly occurs in children and often resolves by adolescence and adulthood [14, 18]. Therefore, microbiome surveys of skin sites associated with atopic dermatitis (dry, moist, and sebaceous sites) are of particular interest for developing preventive or targeted therapeutic approaches for this skin disorder. Such investigations could benefit from new technologies, such as the 3D molecular mapping of the skin surface that allows correlation of the body distribution of skin molecules and bacteria [7].
Adolescence
During the transition through puberty, the skin microbiome has been shown to shift notably from a predominance of Firmicutes, Bacteroidetes, and Proteobacteria (beta- and gamma-proteobacteria) to the more lipophilic Actinobacteria (Corynebacterium and Cutibacterium) [16]. In particular, sebum overproduction has been linked to overcolonization by the commensal bacteria Cutibacterium acnes. This loss of strain diversity leads to an imbalance of the skin microbiota (dysbiosis) and potentially to acne, an inflammatory disorder of the pilosebaceous unit [6, 19]. Although these bacteria dominate the microbiota of pilosebaceous follicles, in subjects both with and without acne, recent studies have indicated that a higher abundance of some phylotypes with the potential to act as opportunistic pathogens is associated with acne lesions [19, 20].
Thus, changes in physiologic conditions at puberty may lead to an imbalance between the different skin community members and, in some cases, to the emergence of acne. The more pronounced physiological and anatomical differences between males and females that emerge at adolescence—such as sweat, sebum, and hormone production—have also been suggested to account for the cutaneous microbial differences seen between sexes in adults [17, 21].
Older Subjects
Although the skin microbial composition of healthy subjects has been found to remain largely stable over time during adulthood [22], age-related physiologic changes—particularly alterations in sebum secretion and immune function and a decrease in sweat—may affect the skin microbiome of older individuals. A marked modification in skin microbiome structure and composition has indeed been observed with aging [17], particularly in women [23]. Cutaneous microbial communities may also vary according to racial and ethnic backgrounds [24]. Yet, the most striking differences in bacterial content have been found to remain dependent on body site, influenced by their different physiologic characteristics, i.e., sebaceous, moist, or dry [17, 23, 24].
A comparative study of the skin microbiomes of healthy individuals in a Chinese cohort according to age revealed that older adults (50–60 years) had an overall lower bacterial richness than younger adults (25–35 years), regardless of sex, but exhibited greater intragroup variations [17] (Fig. 1). For instance, Enhydrobacter—a Proteobacteria deemed enriched in Chinese individuals [24]—was more abundant among older individuals than among adolescents and younger adults (5.8 vs. 3.3 and 2.4%, respectively), especially at sites such as the back and nares. In contrast, Cutibacterium was less abundant in older adults than in adolescents and younger adults (12.6 vs. 21.9 and 26.7%, respectively), especially on dry and moist sites [17]. Moreover, in another Chinese study [24], S. aureus was less abundant in older adults (51–90 years) than in children and adults (4.2 vs. 6.8 and 6.7%, respectively), with a higher relative abundance in males than in females. However, across skin sites and age groups, females were reported to present greater overall levels of bacterial diversity than males. Ying et al. [17] reported other sex differences among older individuals: species richness in older men has been found to be lower than that in older women, and women have also been shown to display greater intragroup variation than men. More specifically, Cutibacterium abundance was found to be greater in older men than in women, especially on glabellae and the back of the hands, likely because of the presence of relatively more terminal hair follicles including sebaceous glands in men. Moreover, the relative abundance of Staphylococcus and Alloiococcus in the nares of older individuals has been found to be greater in females than in males, whereas that of Anaerococcus (Firmicutes) is higher in males than in females (Fig. 1). These sex differences have been suggested to mainly stem from physiologic differences between male and female skin microenvironments, such as hormones, metabolism, perspiration rate, lipid content, and pH, but may also be associated with cosmetic use or sun exposure [17].
A comparison of the skin microbiomes across four skin sites between younger (< 40 years) and older (≥ 60 years) healthy Japanese women revealed that the richness of bacterial species was higher in the latter group for all tested skin sites [23]. Indeed, an overall lower abundance of Actinobacteria (predominant genus Cutibacterium) was observed in the older subjects, with a concurrent increase in other phyla (Firmicutes, Bacteroidetes, and Proteobacteria) in varying proportions depending on the skin site. The decrease in Cutibacterium might be related to the decrease in sebum secretion observed in aging skin, as this species is more lipophilic. In contrast, Corynebacterium (Actinobacteria), Acinetobacter (Proteobacteria), Streptococcus (Firmicutes), and Prevotella (Bacteroidetes) were more abundant in the older women than in their younger counterparts. The most striking alterations in bacterial structure between age groups were in the forearm (dry) and scalp (sebaceous) microbiomes. The diversification of the scalp microbiome in the older women was mainly attributed to a marked increase in the number of minor species. These changes in the abundance and structure of bacterial communities might contribute to the age-related diversification of the microbiomes in women specifically at these skin sites. However, it should be noted that changes in skin physiology, which might not exclusively depend upon aging, can also contribute to alterations of the skin microbiome. Detailed analysis by Shibagaki et al. [23] revealed that 31 bacterial species were overrepresented in older skin microbiomes, half of these species being normal oral bacteria. This finding suggested an age-dependent relationship between the skin and oral bacteria; however, the role played by this relationship in the bacterial diversification of older skin has yet to be clarified.
Comparative microbiome analyses according to the skin aging, mostly performed in healthy Asian women, displayed some differences in their conclusions and might not be generalizable. The main trend of a higher diversity and greater variation of bacterial species in the skin of older subjects should be confirmed in multiethnic studies taking into account sex specificities. Further investigations of these age-related changes also remain to be conducted at the functional level in healthy and pathogenic contexts.
Conclusion
This overview of the skin microbiome as years go by highlights the most striking known changes encountered by healthy skin during the main stages of life. As skin microbiota composition is crucial for developing adapted immune responses from a young age, differences in skin microbiome development may contribute to variations in normal physiology and to disease predisposition or inflammatory skin conditions. Therefore, gaining further insight into skin microbial settlement, and the variations and network of interactions that occur during different life stages, is essential for increasing our understanding of cutaneous disease pathogenesis and for the development of innovative treatments. Collecting information about the epidermal and dermal microbiomes could prove useful for monitoring the condition of the skin throughout life and for helping to predict the emergence of disease and response to treatments, ultimately guiding the choice of the most appropriate treatment approach or therapy. Modulating the microbiota will undoubtedly improve cutaneous health at all life stages.
Acknowledgements
The author thanks Cécile Desjobert, PhD, Emma Pilling, PhD, and Marielle Romet, PhD (Synergy Pharm-Santé Active Edition) who provided medical writing assistance funded by Laboratoires dermatologiques Avène, Pierre Fabre Dermo-Cosmétique.
Author Contributions
Paula Carolina Luna had the idea for the article, performed the literature search and data analysis, and critically revised the manuscript.
Disclosures
Funding
Medical writing assistance was funded by Laboratoires dermatologiques Avène, Pierre Fabre Dermo-Cosmétique.
Conflicts of interest
Paula Carolina Luna has no conflicts of interest that might be relevant to the content of this manuscript.
Disclosure statement
This article is published as part of a journal supplement wholly funded by Laboratoires dermatologiques Avène, Pierre Fabre Dermo-Cosmétique.
References
- 1.Bay L, Barnes CJ, Fritz BG, Thorsen J, Restrup MEM, Rasmussen L, et al. Universal dermal microbiome in human skin. mBio. 2020;11(1):e02945–19. doi: 10.1128/mBio.02945-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YE, Fischbach MA, Belkaid Y. Skin microbiota-host interactions. Nature. 2018;553(7689):427–436. doi: 10.1038/nature25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science (New York, NY). 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreno B, Araviiskaia E, Berardesca E, Gontijo G, Sanchez Viera M, Xiang LF, et al. Microbiome in healthy skin, update for dermatologists. J Eur Acad Dermatol Venereol. 2016;30(12):2038–2047. doi: 10.1111/jdv.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouslimani A, Porto C, Rath CM, Wang M, Guo Y, Gonzalez A, et al. Molecular cartography of the human skin surface in 3D. Proc Natl Acad Sci USA. 2015;112(17):E2120–E2129. doi: 10.1073/pnas.1424409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz CFP, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov. Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol. 2016;66(11):4422–4432. doi: 10.1099/ijsem.0.001367. [DOI] [PubMed] [Google Scholar]
- 10.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23(3):314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pammi M, O'Brien JL, Ajami NJ, Wong MC, Versalovic J, Petrosino JF. Development of the cutaneous microbiome in the preterm infant: a prospective longitudinal study. PLoS One. 2017;12(4):e0176669. doi: 10.1371/journal.pone.0176669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131(10):2026–2032. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaitanis G, Tsiouri G, Spyridonos P, Stefos T, Stamatas GN, Velegraki A, et al. Variation of cultured skin microbiota in mothers and their infants during the first year postpartum. Pediatr Dermatol. 2019;36(4):460–465. doi: 10.1111/pde.13829. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Yosipovitch G. The role of the microbiome and microbiome-derived metabolites in atopic dermatitis and non-histaminergic itch. Am J Clin Dermatol. 2020 doi: 10.1007/s40257-020-00538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu T, Liu X, Kong FQ, Duan YY, Yee AL, Kim M, et al. Age and mothers: potent influences of children's skin microbiota. J Invest Dermatol. 2019;139(12):2497 e6–2505 e6. doi: 10.1016/j.jid.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4(10):77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying S, Zeng DN, Chi L, Tan Y, Galzote C, Cardona C, et al. The influence of age and gender on skin-associated microbial communities in urban and rural human populations. PLoS One. 2015;10(10):e0141842. doi: 10.1371/journal.pone.0141842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dreno B, Pecastaings S, Corvec S, Veraldi S, Khammari A, Roques C. Cutibacterium acnes (Propionibacterium acnes) and acne vulgaris: a brief look at the latest updates. J Eur Acad Dermatol Venereol. 2018;32(Suppl 2):5–14. doi: 10.1111/jdv.15043. [DOI] [PubMed] [Google Scholar]
- 20.Dréno B, Dagnelie MA, Khammari A, Corvec S. The skin microbiome: a new actor in inflammatory acne. Am J Clin Dermatol. 2020 doi: 10.1007/s40257-020-00531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165(4):854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibagaki N, Suda W, Clavaud C, Bastien P, Takayasu L, Iioka E, et al. Aging-related changes in the diversity of women's skin microbiomes associated with oral bacteria. Sci Rep. 2017;7(1):10567. doi: 10.1038/s41598-017-10834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung MH, Wilkins D, Lee PK. Insights into the pan-microbiome: skin microbial communities of Chinese individuals differ from other racial groups. Sci Rep. 2015;16(5):11845. doi: 10.1038/srep11845. [DOI] [PMC free article] [PubMed] [Google Scholar]