Abstract
Rosacea is a complex facial skin condition associated with abnormal inflammation and vascular dysfunction. Next to the known trigger factors, the role of microbiota in the development and aggravation of rosacea continues to raise interest. Demodex folliculorum mites, Helicobacter pylori, Staphylococcus epidermidis, Chlamydia pneumoniae, and the Demodex-associated bacterium, Bacillus oleronius are microbes that have been linked with rosacea. However, the results of studies which assessed their involvement in the disease have been inconsistent and inconclusive. Microbiological research in many different disciplines exploded in recent years as methods to analyze complex microbial communities at the taxonomic and phylogenetic levels became available. Here, we provide an update on the microorganisms implicated in rosacea and review the potential pathogenic role of microbes in the development of rosacea.
Key Points
| Microbes have long been suspected to play a role in rosacea. |
| Better understanding of the microbiota and its role in rosacea pathophysiology may help determine the impact of microbial dysbiosis and host reactivity in rosacea populations. |
| Deeper knowledge of microbe–host interactions in rosacea will allow targeted patient care and aid the development of innovative therapies. |
Introduction
Rosacea is a chronic inflammatory skin disease affecting the central face. It was first described medically in the 14th century by Dr. Guy de Chauliac, a French surgeon [1]. The reported prevalence of rosacea is quite high and varies considerably between study populations [2] and geographical areas [3]. Rosacea is reported less frequently in people with highly pigmented skin than in those with a fair complexion, which is likely due to under-detection of the disease in individuals with a dark skin tone [3]. In a recent meta-analysis of studies of the general population (N = 26,519,836 individuals) and dermatology outpatients (N = 18,483), the pooled proportion of subjects with rosacea was 5.46% (estimated range 0.09–22.41%) in the general adult population and 2.39% (estimated range 0.00–23.14%) among dermatology outpatients [4]. Based on these findings, the National Rosacea Society (NRS) estimates that 415 million people suffer from rosacea worldwide [5]. Adults of 45–60 years of age are primarily affected, with no significant difference between women and men [4].
Although the pathophysiology of rosacea is poorly understood, the efficacy of antibiotics in rosacea treatment suggests that microbes are a pathogenic factor. All microorganisms (bacteria, viruses, fungi, and mites) present in/on the skin constitute the skin microbiota, including resident microorganisms (the core microbiota), which are symbionts, and transient microorganisms (the ‘tourists’), which arise from the environment and persist for hours to days before disappearing [6]. The microbiota acts as a barrier and has a protective role in the skin. Its composition depends on several factors, including skin pH, temperature, lipid composition, and humidity [7], as well as sex, age ([6, 7] and Luna [43] in this journal issue), stress, ethnicity [6], and environment (air pollution, detergents, cosmetics, etc. [7]; Callewaert et al. [44] in this journal issue). Ethnicity is thought to be less important than the local human body environment (sebaceous, dry, or moist) in determining cutaneous microbiota composition [8]. Under normal conditions, both the resident and transient microorganisms are nonpathogenic, and a balance is reached [6]. If this balance is altered, dysbiosis occurs and may aggravate skin diseases [6, 9].
Thanks to genomic advances, microbial research in many different disciplines has exploded in recent years [10]. Historically, bacteria from the human microbiota were studied using a culture-based approach. However, this method only allowed the identification of bacteria that could grow under standard laboratory conditions, therefore underestimating the microbial diversity of human-associated microbiota [10]. Analyses of complex microbial communities at the taxonomic and phylogenetic levels are now possible through the sequencing of bacteria-specific small subunit ribosomal RNAs (16S rRNAs) [10]. Compared with traditional culture-based methods, these molecular analyses have allowed the greater bacterial diversity on the human skin to be uncovered [11]. Corynebacteria, Propionibacteria (both Actinobacteria), and Staphylococci (Firmicutes) are the three most common genera identified in healthy subjects [11]. Microorganisms other than bacteria, such as Malassezia (a polymorphic yeast) and Demodex (a parasitic arthropod), are also found on healthy skin [6]. Both symbiotic microbiota (i.e., Demodex folliculorum and Staphylococcus epidermidis) and potentially pathogenic agents (i.e., Helicobacter pylori, Bacillus oleronius, and Chlamydia pneumoniae) are implicated in rosacea, although their precise roles remain unclear [9, 12].
After a short summary of the current knowledge on rosacea, this review discusses the role of the skin and gut microbiota in the pathophysiology of this disease. With this aim, a PubMed search of the English language literature was performed on January 31, 2020 and updated on May 30, 2020, without any limits on date of publication. Search terms included “rosacea,” “microbiome,” “microbiota,” “skin,” “gut,” and “antibiotics.”
Rosacea: Description, Classification, and Pathophysiology
Description and Classification
Rosacea often begins with a tendency to blush (as in response to emotionally stressful situations) or to flush (as after eating spicy food). Other symptoms include persistent erythema, telangiectasia, papules, pustules, phyma, and edema [2]. Pain, stinging, and burning have also been reported [2]. In most cases, the central face (nose, chin, central cheeks, and glabella) is affected in a symmetrical manner; eyes can also be involved [2]. The symptoms of rosacea are reminiscent of those of alcohol abuse; thus, they can be wrongly interpreted and lead to patients being stigmatized. In Asia, rosacea is called “Ju Sa” (酒皻). The Chinese character “Ju” 酒 means alcohol, which happens to have the same pronunciation as redness, and “Sa” 皻 means enlargement of the nose.
Rosacea was classified into four clinical subtypes in 2002 by an NRS expert committee [13]: (1) erythematotelangiectatic rosacea (ETR) dominated by facial redness, (2) papulopustular rosacea (PPR) featuring bumps and pimples, (3) phymatous rosacea characterized by skin thickening—usually most prominent on the nose—and (4) ocular rosacea involving eye irritation. However, this first standardized classification of rosacea, solely based on the morphologic characteristics, was not optimal as many cases showed an admixture of multiple subtypes. An updated classification, based on phenotypes and an increased understanding of the disease pathophysiology, was published in 2018 [14]. The two diagnostic features are (1) fixed centrofacial erythema in a characteristic pattern that may periodically intensify and (2) phymatous changes. Signs and symptoms defined as major features include flushing, papules and pustules, telangiectasia, and the following ocular manifestations: lid margin telangiectasia, interpalpebral conjunctival injection, spade-shaped infiltrates in the cornea, and scleritis and sclerokeratitis. Secondary features, which may accompany diagnostic or major features, include burning or stinging sensations, edema, dryness, and the following ocular manifestations: “honey crust” and collarette accumulation at the base of the lashes, irregularity of the lid margin, and evaporative tear dysfunction (rapid tear breakup time) (Fig. 1). A diagnosis of rosacea may be made in the presence of at least one diagnostic feature or at least two major features in the absence of any diagnostic feature [14]. The fixed centrofacial erythema of rosacea can be induced or aggravated by triggering factors, which include sun exposure, emotional stress, hot/cold weather, alcohol consumption, hot beverages, spicy food, and microorganisms [2]. To optimize the diagnosis of ocular rosacea and improve the description of its features, the global ROSacea COnsensus (ROSCO) 2019 Panel [15] recently updated the description of ocular rosacea to include lid margin telangiectasia, blepharitis, keratitis, conjunctivitis, and anterior uveitis (Fig. 2).
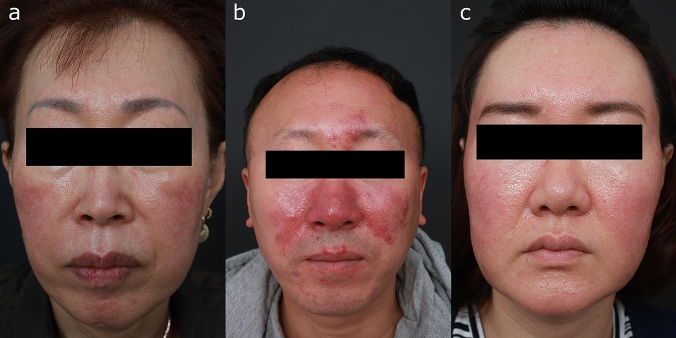
Fig. 1.
Clinical phenotypes of rosacea. a Fixed centrofacial erythema and few papules. b Centrofacial erythema, phymatous changes of the nose, and papules. c Centrofacial erythema, mild phymatous changes, papules, and edema (published with the kind permission of the patients)
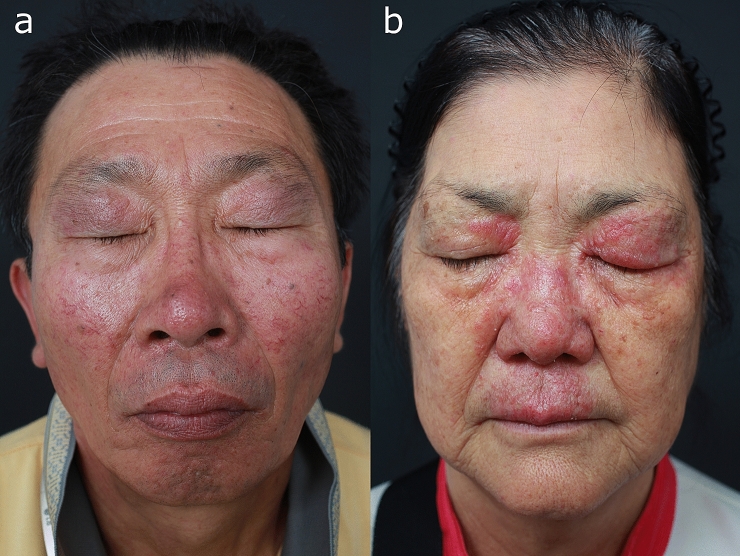
Fig. 2.
Patients with concurrent ocular manifestations. a Fixed centrofacial erythema, mild phymatous changes of the nose, telangiectasia, and mild blepharitis. b Centrofacial erythema, phymatous changes, papules, blepharitis, and edema (published with the kind permission of the patients)
Pathophysiology
The pathophysiology of rosacea is not fully understood, but involves dysregulation of the neurovascular and immune systems [2]. Data also suggest that the disease has a genetic component; no causative gene has been identified, but the expression of a number of genes involved in both the innate and adaptive immune systems has been found to be higher in patients with rosacea [12]. The immune dimension is of particular interest because the microbiota is thought to contribute to the pathophysiology of rosacea at this level.
Similar to acne, dysregulation of the innate immune system has been reported in rosacea: increased baseline expression of (1) cathelicidin, an anti-microbial peptide; (2) kallikrein 5 (KLK5), a serine protease that cleaves cathelicidin into its active peptide form, LL-37; (3) toll-like receptor 2 (TLR-2); and (4) matrix metalloproteinases (MMPs), including MMP-2 and MMP-9. MMP-9 activates KLK5 by cleaving its preproenzyme form, and TLR-2 activation on keratinocytes increases KLK5 expression and activity, thus leading to higher expression of LL-37 [12]. Besides LL-37, other cathelicidin peptides have been found in rosacea affected skin, but not in that of healthy subjects [16]. These abnormal cathelicidin peptides have been shown to play a role in inflammation and angiogenesis, and could thus contribute to rosacea pathophysiology [12]. Mast cells may also be involved because they are upregulated in patients with rosacea and release both MMP-9 and LL-37 [12, 17].
The Skin Microbiota is Potentially Associated with Rosacea
Several studies [18–20] recently used the microbial 16S rRNA polymerase chain reaction (PCR) amplification and sequencing technique to assess potential differences in the skin microbiota between patients with rosacea and healthy controls (Table 1). In a comparative study performed in 60 twins discordant for rosacea [18], the four most abundant phyla retrieved from the facial skin (assessed as the relative mean percentage abundance) were Firmicutes (42.98%), Proteobacteria (39.29%), Actinobacteria (15.88%) and Bacteroidetes (1.04%). No significant difference in the mean abundance of phyla was observed between groups of individuals with or without rosacea, or between different areas of the face [18]. In contrast, significant differences in skin microbiota were identified in a case–control observational study comparing 19 patients with mild-to-moderate rosacea (ETR, PPR, or both) with 19 matched healthy subjects [19]. Compared with healthy subjects, six species were depleted in ETR patients, and five species were enriched and six depleted in PPR patients. Despite these differences, Cutibacterium acnes (formerly called Propionibacterium acnes) was the most abundant species in all subjects [19]. In another study, in which the microbiota of Demodex mites were examined, C. acnes was also the predominant species in both ETR and PPR patients, and in sex- and age-matched healthy subjects [20]. Five other species (S. epidermidis, Corynebacterium kroppenstedtii, Streptococcus mitis, Cutibacterium granulosum and Snodgrassella alvi) were common to all subjects, but 72 species were only retrieved from the skin biopsies of patients with rosacea (with 37 and 31 species being specific to ETR and PPR, respectively) [20]. This study also found that the proportions of Proteobacteria and Firmicutes were higher in the PPR group compared with the ETR and control groups, whereas the proportion of Actinobacteria was lower in the PPR group than in the ETR and control groups [20]. Interestingly, B. oleronius was not identified in this study, even though this bacterium was suggested to be involved in rosacea after it was cultured from a D. folliculorum mite of a patient with PPR [21].
Table 1.
Recent comparative studies using 16S rRNA gene sequencing to analyze microbiota in rosacea
| Title (author, year of publication) | Number of subjects/study comparison | Race | Age (years) mean ± SD (range) | 16S rRNA gene sequencing and analysis | Outcome |
|---|---|---|---|---|---|
| Skin microbiota | |||||
| Characterization of the facial microbiome in twins discordant for rosacea (Zaidi et al. 2018) [18] |
60 subjects (42 healthy subjects, 18 patients with rosacea) Rosacea vs control |
White (82.5%) Black (6.3%) Other (11.2%) |
Rosacea: 31.3 ± 10.2 Healthy: 36.4 ± 17.3 |
V3–V4 region of the 16S rRNA gene Greengenes database |
Top four most abundant phyla: Firmicutes (43%), Proteobacteria (39%), Actinobacteria (16%), and Bacteroidetes (1%) No significant difference in mean abundance for all phyla between left vs right cheek nor between healthy subjects and rosacea patients Comparable alpha diversity between monozygotic twin pairs with and without rosacea Two genera (Gordonia and Geobacillus) and an age of 30–60 years (vs 0–30 years) were significantly predictive of NRS score |
| Characterization and analysis of the skin microbiota in rosacea: a case–control study (Rainer et al. 2019) [19] |
19 subjects with rosacea (ETR, PPR, or both); 19 rosacea-free controls Rosacea vs control |
All Caucasian |
Rosacea: age-matched to control Control: (23–65) |
V3–V4 hypervariable region of the 16S rRNA gene Greengenes database |
Weak negative correlation between the relative abundance of Cutibacterium acnes and NRS scores (R2=0.0378, P = 0.124). Significantly enriched and depleted species in ETR: (enriched) none; (depleted) Actinomyces europaeus, Prevotella tannerae, Prevotella intermedia, Campylobacter ureolyticus, Corynebacterium kroppenstedtii Significantly enriched and depleted species in PPR: (enriched) Porphyromonas endodontalis, Ruminococcus gnavus, Azorhizobium doebereinerae, Shewanella algae, Providencia stuartii, Roseomonas mucosa; (depleted) Dysgonomonas gadei, Azorhizobium doebereinerae, Shewanella algae, Providencia stuartii, Cutibacterium granulosum, Anoxybacillus kestanbolensis |
| Characterization and analysis of the skin microbiota in rosacea: impact of systemic antibiotics (Woo et al. 2020) [22] |
12 subjects with rosacea Rosacea before vs after 6 weeks of oral doxycycline |
Asian (Korean) |
49.2 ± 18.6 (20–79) Median: 51 |
V3–V4 hypervariable region of the 16S rRNA gene NCBI taxonomy |
3.43-fold increase in Weissella confusa relative abundance after oral doxycycline treatment (P = 0.008) Higher relative abundance of Cutibacterium acnes in patients aged 60 and under vs those aged over 60 (P = 0.018) Lower mean relative abundance of Cutibacterium acnes (P = 0.081) and higher relative abundance of Snodgrassella alvi (P = 0.087) in IGA4 vs IGA3 skin Comparable alpha diversity between the before-treatment and the after-treatment groups |
| Gut microbiota | |||||
| Rosacea and its association with enteral microbiota in Korean females (Nam et al. 2018) [37] |
12 rosacea patients; 251 control subjects Rosacea vs control |
Asian (Korean) |
Rosacea: 42.6 Control: 43.0 |
V3–V4 region of the 16S rRNA gene Greengenes database |
Reduced abundance of Peptococcaceae family and Methanobrevibacter genus, but increased abundance of Acidaminococcus and Megasphaera genera in patients with rosacea vs control subjects, with significance retained after adjustment |
| An altered fecal microbial profiling in rosacea patients compared to matched controls (Chen et al. 2020) [38] |
11 rosacea patients; 110 control subjects Rosacea vs control |
Asian (Taiwanese) |
Rosacea: 49.9 ± 11.3 Control: 50.6 ± 10.2 |
V3–V4 hypervariable region of the 16S rRNA gene Greengenes database |
Significant reduction of fecal microbial richness in rosacea patients Enriched Rhabdochlamydia, CF231, Bifidobacterium, Sarcina, and Ruminococcus genera in rosacea patients. Reduced abundance of Lactobacillus, Megasphaerae, Acidaminococcus, Hemophilus, Roseburia, and Clostridium genera in rosacea patients |
16S rRNA bacteria-specific small subunit ribosomal RNA, ETR erythematotelangiectatic rosacea, IGA Investigator’s Global Assessment, NCBI National Center for Biotechnology Information, NRS National Rosacea Society, PPR papulopustular rosacea, SD standard deviation
Thus, the comparative studies conducted so far have produced discordant results concerning the differences in the overall microbiota composition between rosacea patients and healthy subjects and the type of bacteria potentially involved in rosacea. These differences may be due to disparities in patient age, ethnicity, environment, and geographical location, as such factors are known to affect skin microbiota composition.
Factors Influencing Skin Microbiota in Patients with Rosacea
Patient Age
The relative abundance of C. acnes in Caucasians with rosacea has been found to be lower in older patients (females aged 50 years and over, and males aged over 55) than in their younger counterparts [19]. My team also confirmed that the relative abundance of C. acnes in Asian rosacea patients was lower in those aged over 60 compared to those aged 60 and under (18-fold difference, P = 0.018) [22]. When the cut-off age was set at 50 years, S. epidermidis was the predominant species in Asians aged under 50 (42%), followed by C. acnes (16%), and Acinetobacter haemolyticus (13%). Although less abundant, S. epidermidis was also the predominant species in patients aged over 50 (21%), followed by Pseudomonas koreensis (12%), C. acnes (11%), and S. alvi (9%). Additional analyses of the same cohort showed a significantly higher relative abundance of S. alvi (P = 0.022) in rosacea patients aged over 50 compared to those under 50.
It should be noted that the relative abundance of C. acnes has also been reported to vary with age in the general population [23], with it being the least abundant during childhood (4–6 years; 2.1%), having a dramatic surge at puberty (11–13 years; 13.5%) and a peak in young adults (25–34 years; 40.3%), followed by a decline in middle-age (37–53 years; 27.2%), and a subsequent larger fall in abundance in the elderly (62–74 years; 8.7%).
Rosacea Severity
The microbial composition of the skin has been examined according to rosacea severity. My group [22] found S. epidermidis to be predominant in patients with both moderate (38%) and severe (17%) rosacea, with respective grades of 3 and 4 on the 5-point Investigator’s Global Assessment (IGA) grading scale. This was followed by C. acnes (22%) in patients with severe rosacea, and by P. koreensis (16%), S. alvi (12%), and Corynebacterium tuberculostearicum (11%) in patients with moderate rosacea. S. alvi was significantly more abundant (P = 0.0081) [22] in patients with severe rosacea than in those with moderate rosacea. Conversely, a lower relative abundance of C. acnes (P = 0.0087) [22] was seen in patients with severe rosacea. Although not statistically significant, a linear regression analyses performed by Rainer et al. [19] showed a weak negative correlation between C. acnes abundance (P = 0.124) and rosacea severity (NRS), together with a positive correlation between C. kroppenstedtii abundance (P = 0.065) and severity. According to a study by Zaidi et al. [18], the relative abundance of nine genera significantly correlated with the NRS severity scores (Pearson correlation coefficient). While there was a negative correlation between Geobacillus and rosacea severity (P = 0.041), a positive correlation was noted with Gordonia (P = 0.003), Janibacter (P = 0.002), Jeotgalicoccus (P = 0.001), Tepidimonas (P = 0.010), Dietzia (P = 0.012), Wautersiella (P = 0.017), Mycobacterium (P = 0.032), and Weissella (P = 0.042). Only Geobacillus and Gordonia remained significantly associated with rosacea severity after a multivariate random effect Poisson regression analysis.
Unlike acne, rosacea usually progresses/worsens over time [18] and it is likely that the negative correlation of C. acnes and the positive association of S. alvi with rosacea severity [22] is in part due to age. Interestingly, S. alvi has been identified in Demodex mites from both rosacea patients (ETR and PPR) and healthy controls [20]. S. alvi is a member of the bee gut flora [24] and has been associated with inflammatory acne; its relative abundance was found to decrease significantly after a 6-week oral doxycycline treatment (H.S. Kim; unpublished data).
Antibiotics
Although antibiotics/antiprotozoal drugs are widely used to control the inflammatory papules and pustules of rosacea [15], few studies have investigated their influence on the composition and diversity of the skin microbiota. Using a culture-based approach in the late 1980s, Eriksson and Nord demonstrated that topical metronidazole (1% cream applied twice daily for 1 month) does not alter the skin microbial composition [25]. Using the 16S rRNA PCR amplification and sequencing technique, my team recently examined the effect of oral doxycycline (100 mg, twice daily for 6 weeks) on the skin microbiota in 12 rosacea patients [22]. We found that the treatment did not significantly affect bacterial alpha diversity, but an increase in the relative abundance of Weissella confusa (P = 0.008) was noted [22]. In our study, the 6-week oral doxycycline treatment decreased rosacea severity from IGA grades 3–4 (i.e., moderate to severe, median 3) to IGA grades 2–3 (i.e., mild to moderate, median 2). The increase in the relative abundance of W. confusa upon oral doxycycline treatment is in contrast with the results of Zaidi et al. [18], where Weissella positively correlated with rosacea severity. Thus, further studies are needed to properly interpret the exact role of W. confusa in rosacea.
Cell-free culture supernatants of W. confusa have displayed antibacterial potential and anti-inflammatory properties, and some strains have been classed as probiotics [26, 27]. However, the clinical relevance of W. confusa in the context of polymicrobial infections remains unclear.
Skin Temperature
Beyond microbial composition, the activity of certain bacteria appears to be affected by facial skin temperature, which is higher in patients with rosacea than in healthy subjects. In a comparative pilot study [28], S. epidermidis strains isolated from the skin of four patients with untreated rosacea were found to be consistently β-hemolytic, whereas those isolated from the skin of four control subjects were nonhemolytic. Moreover, although bacteria from both groups grew at the same rate at 30 °C and at 37 °C, those isolated from rosacea patients produced a significantly wider range of proteins, in larger amounts, at 37 °C than they did at 30 °C.
Involvement of Skin Microbiota in Rosacea Pathophysiology
Pattern recognition receptors (PRRs) expressed on the skin participate in a continuous immune surveillance that allows symbiont microorganisms to thrive while eliminating potential pathogens [6, 9]. Two of these PRRs—TLR-2 and nucleotide-binding oligomerization domain (NOD)-like receptor family, pyrin domain-containing 3 (NLRP3, also called NALP3)—are upregulated in rosacea patients, and their activation by Demodex mites are thought to trigger inflammation in rosacea (summarized in Fig. 3) [2, 9, 12]. Indeed, chitin (from the mite exoskeleton) can stimulate the pro-inflammatory response of keratinocytes through TLR-2 [12], and mite allergens have been shown to activate NOD-like receptors in vitro [9]. Microbiota residing on Demodex mites may also be involved in this process: antigens from B. oleronius reportedly induced the proliferation of peripheral blood mononuclear cells from rosacea patients [21], and stimulated the production of cathelicidin, MMP-9, tumor necrosis factor (TNF) and interleukin (IL)-8 by neutrophils from healthy subjects [9, 12]. In patients with rosacea, this inflammatory state causes an increase in the facial skin temperature [28]. This in turn can affect microbiota growth and balance [9], and modify the behavior of S. epidermidis so that it secretes more proteins [28] (see Sect. 4.4). Moreover, as S. epidermidis antigens are recognized by TLR-2, the bacterium also participates in skin inflammation [12].
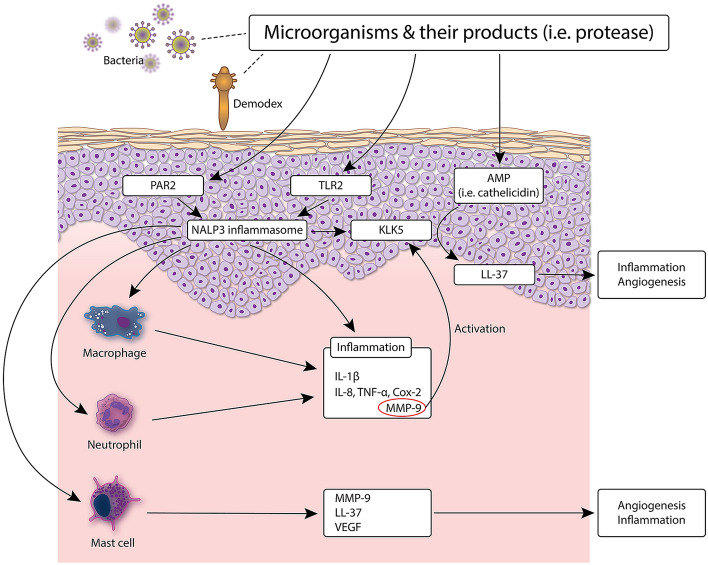
Fig. 3.
Involvement of the skin microbiota in rosacea pathophysiology. Pattern recognition receptors (PRRs) expressed on the skin participate in a continuous immune surveillance that allows symbiont microorganisms to thrive while eliminating potential pathogens. Two of these PRRs—TLR-2 and NLRP3 (also called NALP3)—are upregulated in rosacea patients, and their activation by Demodex mites is thought to trigger inflammation in rosacea. Microbiota residing on Demodex mites may also be involved in this process: antigens from Bacillus oleronius reportedly induced the proliferation of peripheral blood mononuclear cells from rosacea patients and stimulated the production of cathelicidin, MMP-9, TNF, and IL-8 by neutrophils from healthy subjects. AMP anti-microbial peptide, Cox cyclooxygenase, IL interleukin, KLK kallikrein, LL-37 active peptide form of cathelicidin, MMP matrix metalloproteinase, NALP Nacht leucine-rich repeat protein, NLRP nucleotide-binding oligomerization domain (NOD)-like receptor family, pyrin domain-containing, PAR protease-activated receptor, TLR toll-like receptor, TNF tumor necrosis factor, VEGF vascular endothelial growth factor
Unlike in acne, C. acnes does not seem to play a major role in the pathogenesis of rosacea [29]. In fact, here C. acnes is suspected to have a protective effect in healthy skin by breaking down sebum into free fatty acids (FFAs), which can prevent the growth of pathogens [30].
Does the Gut Microbiota Play a Role in Rosacea?
Patients with rosacea may have systemic comorbidities, including diseases of the gastrointestinal tract (e.g., gastroesophageal reflux disease, gastritis, Crohn’s disease, ulcerative colitis, celiac disease, and small intestinal bacterial overgrowth [SIBO] syndrome), suggesting a link between rosacea and the gut [31–33]. Furthermore, the gut microbiota has been shown to influence skin homeostasis: either directly in case of intestinal barrier disruption, or indirectly through the modulation of systemic immunity or by generating short-chain fatty acids [7]. The gut is inhabited by a huge number of microorganisms (> 1014) constituting the gut microbiota, and several factors including the mode of birth, food, age, stress, and antibiotics may affect its composition and may therefore trigger rosacea [34]. Another argument is the positive effect of oral probiotics observed in patients with rosacea. In a randomized, controlled, non-blinded trial, patients with papulopustular exanthema (including 36% with rosacea) who received the bacteria Escherichia coli Nissle 1917 as an oral probiotic as well as a standard topical therapy had a better outcome than patients who only received the standard treatment (P < 0.01) [35]. Fortuna et al. [36] also reported a case of rosacea with scalp involvement that was successfully treated with low-dose doxycycline (40 mg/day) combined with oral probiotics (Bifidobacterium breve BR03 and Lactobacillus salivarius LS01) for 8 weeks, followed by probiotics alone. No relapse or flare-up of disease was observed during 6 months of follow-up [36]. However, the mechanisms underlying the involvement of gut bacteria in rosacea pathophysiology have not yet been elucidated. Although studies have failed to confirm an association between H. pylori and rosacea, antibodies against cytotoxin-associated gene A (CagA), an H. pylori virulent factor, have been identified in most patients with rosacea [9, 12]. The gut microbiota in rosacea patients was recently investigated in two Asian metagenomic studies [37, 38]. In the study by Nam et al. [37] performed in Korean women (12 rosacea patients and 251 controls), bacteria from the Peptococcaceae family and the Methanobrevibacter genus were detected in control subjects but not in rosacea patients, whereas bacteria from the Acidaminococcus and Megasphaera genera were significantly more abundant in rosacea patients than in controls [37]. In contrast, Chen et al. [38] observed a lower abundance of Acidaminococcus and Megasphaera genera in Taiwanese subjects (11 rosacea patients and 110 controls, 90.9% women). Lactobacillus, Hemophilus, Roseburia, and Clostridium genera were also less abundant in rosacea patients than in controls, whereas Rhabdochlamydia, CF231, Bifidobacterium, Sarcina, and Ruminococcus genera were significantly more abundant [38]. Therefore, differences between rosacea patients and healthy subjects were observed in both studies, although the results were discordant. Further studies with higher numbers of rosacea patients are required. As rosacea is induced or aggravated by emotional stress [2], it is likely that the brain is involved in cross-talk between the skin and gut microbiota in rosacea, as in acne, through the gut-brain-skin axis (Fig. 4) [7, 9, 39–42].
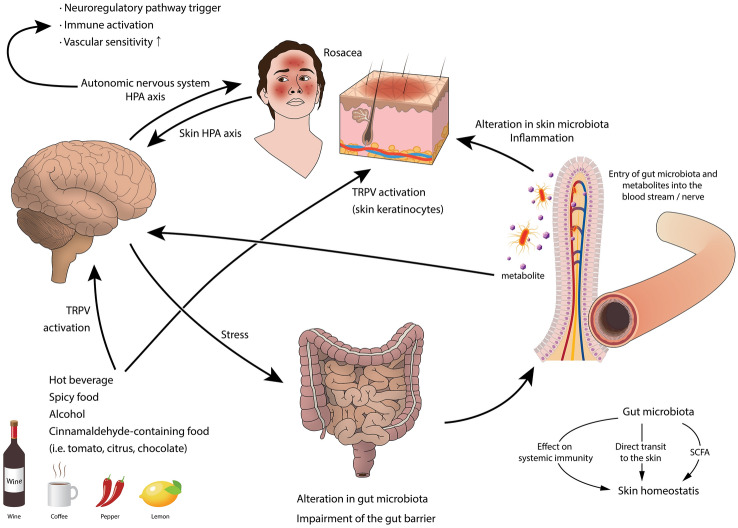
Fig. 4.
A proposed model of the gut-brain-skin axis in rosacea. HPA hypothalamic pituitary adrenal, SCFA short-chain fatty acid, TRPV transient receptor potential vanilloid
Conclusion and Perspectives
The 16s rRNA studies highlighted in this review suggest a link between the skin and gut microbiota and rosacea; however, the role of these microorganisms in the disease pathophysiology is yet to be determined. With limited data, it is not known whether dysbiosis and altered microbiota metabolism are potentiators of inflammation or secondary outcomes in response to changes in the skin microenvironment. Microbial 16S rRNA PCR amplification and sequencing have enabled us to analyze complex microbial communities at the taxonomic and phylogenetic levels. However, to our disappointment, results from microbial studies in rosacea patients have so far been discordant at the level of both the skin [18–20] and the gut [37, 38]. It is especially difficult to interpret the findings from these studies because skin and gut microbial composition depends on numerous factors, including food, age, stress, and environment. There are also great interpersonal differences in the human microbiota, a problem that can only be overcome by conducting studies using paired samples from the same patient [22]. Although data are scarce, the reports of successful rosacea treatment with oral probiotics combined with conventional therapy are promising [35, 36] and provide evidence for the role of the gut microbiota in rosacea pathogenesis. In acne, several clinical trials reported positive results with oral or topical probiotics used alone or in combination. Further investigations are necessary to assess the efficacy of oral/topical probiotics and/or prebiotics in rosacea and identify their exact mechanisms of action. We should be aware that such treatments may require customization due to great interpersonal differences in the skin and gut microbiota. Moreover, as food affects gut microbiota composition [34], further research in this domain could lead to adapted dietary advice being provided to patients with rosacea.
Another exciting field of research is investigation of the gut-brain-skin axis in rosacea; a greater understanding of how the brain (i.e., stress) influences the gut and skin microbiota could also provide better therapeutic strategies (i.e., stress-relieving practices).
Acknowledgements
The author thanks Laurence Rous, PhD, Emma Pilling, PhD, and Marielle Romet, PhD (Synergy Pharm-Santé Active Edition) for medical writing assistance funded by Laboratoires Dermatologiques Avène–Pierre Fabre Dermo-Cosmétique.
Author contributions
The author had the idea for the article, performed literature searches and data analysis, contributed to the drafting of the manuscript, and critically reviewed and approved the final version of the paper.
Declarations
Funding
Hei Sung Kim’s work was supported by a National Research Foundation of Korea (NRF) grant funded by the South Korean Government (2017R1C1B5016144 and 2020R1F1A1048238).
Conflict of interest
The Author has no conflict of interest to declare.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Disclosure statement
This article is part of a Supplement wholly funded by Laboratoires Dermatologiques Avène–Pierre Fabre Dermo-Cosmétique
References
- 1.de Bersaques J. Historical notes on (acne) rosacea. EJD Eur J Dermatol. 1995;5(1):16–22. [Google Scholar]
- 2.Steinhoff MBJ. Chapter 79: Rosacea. In: Kang SAM, Bruckner AL, Enk AH. Margolis DJ, McMichael AJ, Orringer JS editor. Fitzpatrick’s dermatology, 9e: Mc Graw Hill Education; 2019.
- 3.Alexis AF, Callender VD, Baldwin HE, Desai SR, Rendon MI, Taylor SC. Global epidemiology and clinical spectrum of rosacea, highlighting skin of color: review and clinical practice experience. J Am Acad Dermatol. 2019;80(6):1722-9.e7. doi: 10.1016/j.jaad.2018.08.049. [DOI] [PubMed] [Google Scholar]
- 4.Gether L, Overgaard LK, Egeberg A, Thyssen JP. Incidence and prevalence of rosacea: a systematic review and meta-analysis. Br J Dermatol. 2018;179(2):282–289. doi: 10.1111/bjd.16481. [DOI] [PubMed] [Google Scholar]
- 5.National Rosacea Society. New study finds 415 million people may suffer from rosacea worldwide. 2018. https://www.rosacea.org/press/2018/july/new-study-finds-415-million-people-may-suffer-from-rosacea-worldwide. Accessed 11 Mar 2020.
- 6.Dreno B, Araviiskaia E, Berardesca E, Gontijo G, Sanchez Viera M, Xiang LF, et al. Microbiome in healthy skin, update for dermatologists. J Eur Acad Dermatol Venereol. 2016;30(12):2038–2047. doi: 10.1111/jdv.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YB, Byun EJ, Kim HS. Potential role of the microbiome in acne: a comprehensive review. J Clin Med. 2019 8(7). [DOI] [PMC free article] [PubMed]
- 8.Perez Perez GI, Gao Z, Jourdain R, Ramirez J, Gany F, Clavaud C, et al. Body site is a more determinant factor than human population diversity in the healthy skin microbiome. PLoS One. 2016;11(4):e0151990. doi: 10.1371/journal.pone.0151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes AD. Potential role of microorganisms in the pathogenesis of rosacea. J Am Acad Dermatol. 2013;69(6):1025–1032. doi: 10.1016/j.jaad.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Tang L. Sequence-based identification of human-associated microbiota. In: Milestones in Human Microbiota Research. Nature. 2019:S10.
- 11.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Two AM, Wu W, Gallo RL, Hata TR. Rosacea: part I. Introduction, categorization, histology, pathogenesis, and risk factors. J Am Acad Dermatol. 2015;72(5):749–758. doi: 10.1016/j.jaad.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 13.Wilkin J, Dahl M, Detmar M, Drake L, Feinstein A, Odom R, et al. Standard classification of rosacea: report of the National Rosacea Society Expert Committee on the Classification and Staging of Rosacea. J Am Acad Dermatol. 2002;46(4):584–587. doi: 10.1067/mjd.2002.120625. [DOI] [PubMed] [Google Scholar]
- 14.Gallo RL, Granstein RD, Kang S, Mannis M, Steinhoff M, Tan J, et al. Standard classification and pathophysiology of rosacea: the 2017 update by the National Rosacea Society Expert Committee. J Am Acad Dermatol. 2018;78(1):148–155. doi: 10.1016/j.jaad.2017.08.037. [DOI] [PubMed] [Google Scholar]
- 15.Schaller M, Almeida LM, Bewley A, Cribier B, Dlova NC, Kautz G, et al. Rosacea treatment update: recommendations from the global ROSacea COnsensus (ROSCO) panel. Br J Dermatol. 2017;176(2):465–471. doi: 10.1111/bjd.15173. [DOI] [PubMed] [Google Scholar]
- 16.Yamasaki K, Di Nardo A, Bardan A, Murakami M, Ohtake T, Coda A, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat Med. 2007;13(8):975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- 17.Muto Y, Wang Z, Vanderberghe M, Two A, Gallo RL, Di Nardo A. Mast cells are key mediators of cathelicidin-initiated skin inflammation in rosacea. J Invest Dermatol. 2014;134(11):2728–2736. doi: 10.1038/jid.2014.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaidi AK, Spaunhurst K, Sprockett D, Thomason Y, Mann MW, Fu P, et al. Characterization of the facial microbiome in twins discordant for rosacea. Exp Dermatol. 2018;27(3):295–298. doi: 10.1111/exd.13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainer BM, Thompson KG, Antonescu C, Florea L, Mongodin EF, Bui J, et al. Characterization and analysis of the skin microbiota in rosacea: a case-control study. Am J Clin Dermatol. 2020;21(1):139–147. doi: 10.1007/s40257-019-00471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murillo N, Aubert J, Raoult D. Microbiota of Demodex mites from rosacea patients and controls. Microb Pathog. 2014;71–72:37–40. doi: 10.1016/j.micpath.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Lacey N, Delaney S, Kavanagh K, Powell FC. Mite-related bacterial antigens stimulate inflammatory cells in rosacea. Br J Dermatol. 2007;157(3):474–481. doi: 10.1111/j.1365-2133.2007.08028.x. [DOI] [PubMed] [Google Scholar]
- 22.Woo YR, Lee SH, Cho SH, Lee JD, Kim HS. Characterization and analysis of the skin microbiota in rosacea: impact of systemic antibiotics. J Clin Med. 2020 9(1). [DOI] [PMC free article] [PubMed]
- 23.Zhai W, Huang Y, Zhang X, Fei W, Chang Y, Cheng S, et al. Profile of the skin microbiota in a healthy Chinese population. J Dermatol. 2018;45(11):1289–1300. doi: 10.1111/1346-8138.14594. [DOI] [PubMed] [Google Scholar]
- 24.Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2013 63(6):2008–18. [DOI] [PubMed]
- 25.Eriksson G, Nord CE. Impact of topical metronidazole on the skin and colon microflora in patients with rosacea. Infection. 1987;15(1):8–10. doi: 10.1007/BF01646108. [DOI] [PubMed] [Google Scholar]
- 26.Lee KW, Park JY, Jeong HR, Heo HJ, Han NS, Kim JH. Probiotic properties of Weissella strains isolated from human faeces. Anaerobe. 2012;18(1):96–102. doi: 10.1016/j.anaerobe.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 27.Dey DK, Khan I, Kang SC. Anti-bacterial susceptibility profiling of Weissella confusa DD_A7 against the multidrug-resistant ESBL-positive E. coli. Microb Pathog. 2019;128:119–130. doi: 10.1016/j.micpath.2018.12.048. [DOI] [PubMed] [Google Scholar]
- 28.Dahl MV, Ross AJ, Schlievert PM. Temperature regulates bacterial protein production: possible role in rosacea. J Am Acad Dermatol. 2004;50(2):266–272. doi: 10.1016/j.jaad.2003.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Jahns AC, Lundskog B, Dahlberg I, Tamayo NC, McDowell A, Patrick S, et al. No link between rosacea and Propionibacterium acnes. Apmis. 2012;120(11):922–925. doi: 10.1111/j.1600-0463.2012.02920.x. [DOI] [PubMed] [Google Scholar]
- 30.Marples RR, Downing DT, Kligman AM. Control of free fatty acids in human surface lipids by Corynebacterium acnes. J Invest Dermatol. 1971;56(2):127–131. doi: 10.1111/1523-1747.ep12260695. [DOI] [PubMed] [Google Scholar]
- 31.Rainer BM, Fischer AH, Luz Felipe da Silva D, Kang S, Chien AL. Rosacea is associated with chronic systemic diseases in a skin severity-dependent manner: results of a case-control study. J Am Acad Dermatol. 2015;73(4):604–608. doi: 10.1016/j.jaad.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Egeberg A, Weinstock LB, Thyssen EP, Gislason GH, Thyssen JP. Rosacea and gastrointestinal disorders: a population-based cohort study. Br J Dermatol. 2017;176(1):100–106. doi: 10.1111/bjd.14930. [DOI] [PubMed] [Google Scholar]
- 33.Thompson KG, Rainer BM, Kang S, Chien AL. The skin microbiota as a link between rosacea and its systemic comorbidities. Int J Dermatol. 2020;59(4):513–514. doi: 10.1111/ijd.14802. [DOI] [PubMed] [Google Scholar]
- 34.Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manzhalii E, Hornuss D, Stremmel W. Intestinal-borne dermatoses significantly improved by oral application of Escherichia coli Nissle 1917. World J Gastroenterol. 2016;22(23):5415–5421. doi: 10.3748/wjg.v22.i23.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fortuna MC, Garelli V, Pranteda G, Romaniello F, Cardone M, Carlesimo M, et al. A case of scalp rosacea treated with low dose doxycycline and probiotic therapy and literature review on therapeutic options. Dermatol Ther. 2016;29(4):249–251. doi: 10.1111/dth.12355. [DOI] [PubMed] [Google Scholar]
- 37.Nam JH, Yun Y, Kim HS, Kim HN, Jung HJ, Chang Y, et al. Rosacea and its association with enteral microbiota in Korean females. Exp Dermatol. 2018;27(1):37–42. doi: 10.1111/exd.13398. [DOI] [PubMed] [Google Scholar]
- 38.Chen YJ, Lee WH, Ho HJ, Tseng CH, Wu CY. An altered fecal microbial profiling in rosacea patients compared to matched controls. J Formos Med Assoc. 2020. [DOI] [PubMed]
- 39.Stokes JH, Pillsbury DM. The effect on the skin of emotional and nervous states: iII. Theoretical and practical consideration of a gastro-intestinal mechanism. Arch Dermatol Syphilol. 1930;22(6):962–993. [Google Scholar]
- 40.Arck P, Handjiski B, Hagen E, Pincus M, Bruenahl C, Bienenstock J, et al. Is there a ‘gut-brain-skin axis’? Exp Dermatol. 2010;19(5):401–405. doi: 10.1111/j.1600-0625.2009.01060.x. [DOI] [PubMed] [Google Scholar]
- 41.Bowe WP, Logan AC. Acne vulgaris, probiotics and the gut-brain-skin axis—back to the future? Gut Pathog. 2011;3(1):1. doi: 10.1186/1757-4749-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowe W, Patel NB, Logan AC. Acne vulgaris, probiotics and the gut-brain-skin axis: from anecdote to translational medicine. Benef Microbes. 2014;5(2):185–199. doi: 10.3920/BM2012.0060. [DOI] [PubMed] [Google Scholar]
- 43.Luna PC. Skin microbiome as years go by. Am J Clin Dermatol. 2020 doi: 10.1007/s40257-020-00549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Callewaert C, Helffer KR, Lebaron P. Skin microbiome and its interplay with the environment. Am J Clin Dermatol. 2020 doi: 10.1007/s40257-020-00551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]