Abstract
Recent advances in our understanding of the pathophysiology of atopic dermatitis (AD) have revealed that skin microbiome dysbiosis plays an important role in the disease. In this review, we describe how changes in the structure and function of the microbiome are involved in the pathogenesis of AD. We highlight recent data showing that differential changes in microbial diversity, both within and across communities from different body habitats (including the skin, gut, and oral mucosa), are associated with the development and severity of AD. We also describe recent evidence demonstrating that the metabolic activity of the skin microbiome can act as a regulator of inflammation, with alterations in the level of a skin microbiome-derived tryptophan metabolite, indole-3-aldehyde (IAId), being shown to play a role in AD. The various mechanisms by which interactions between the microbiome and components of the non-histaminergic pathway result in itch in AD are also discussed.
Key Points
| Patients with atopic dermatitis display lower levels of diversity in both the skin and oral microbiomes, with disease severity being negatively correlated with microbiome diversity in the skin but positively correlated with microbiome diversity in the oral cavity. |
| Alterations in skin microbiome function also play a role in the pathophysiology of atopic dermatitis, with alterations in the level of a key microbiome-derived tryptophan metabolite, indole-3-aldehyde, on the skin surface recently being implicated in the disease. |
| Recent advances in our understanding of the pathophysiology of itch indicate that multiple factors that perturb the normal functional interaction between the skin and the microbiome—including protease activity, neuropeptide levels, and skin pH—appear to contribute to the increased activation of the non-histaminergic itch pathway in patients with atopic dermatitis. |
Introduction
Atopic dermatitis (AD) is a multifactorial pruritic inflammatory skin disease involving genetic, environmental, and immune factors. Since the 1970s, the incidence of AD has increased two- to threefold in industrialized countries [1]. The short time span over which this increase has been observed makes it unlikely that genetic factors have played a major role, and environmental factors appear to have been more important in driving this rise in cases.
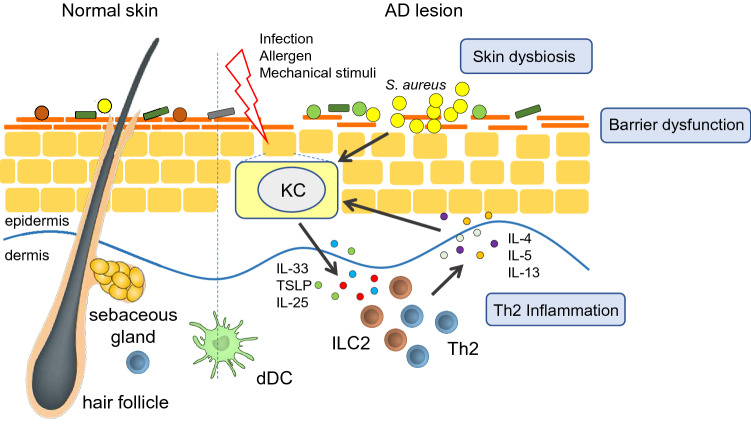
During recent years, great progress has been made toward increasing our understanding of the pathogenesis of AD. Skin barrier malfunction is a prominent feature of AD, but skin microbiome dysbiosis plays an equally important role in the disease mechanism [2]. Skin dysbiosis in AD is characterized by increased abundance of Staphylococcus aureus [3]. In the presence of this dysbiosis and barrier dysfunction, external stimuli such as infection, allergens, and mechanical injury lead to activation of skin keratinocytes to produce interleukin (IL)-33, IL-25, and thymic stromal lymphopoietin (TSLP), inducing differentiation of T helper type 2 (Th2) cells that secrete IL-4, IL-5, and IL-13 and resulting in Th2 inflammation [4, 5]. This inflammation can in turn activate keratinocytes, forming a vicious cycle that maintains skin inflammation in AD (Fig. 1). Thus, the skin microbiome, keratinocytes, Langerhans cells, and Th2 inflammation make up the skin microenvironment in AD. However, one of the features of AD is that it always precedes the onset of other atopic manifestations, including food allergies, rhinitis, and asthma, as part of a process that is referred to as the atopic march [6]. The local inflammation in AD therefore induces systemic immune responses, but the mechanism of interaction between the components of the skin microenvironment in the initiation and maintenance of these immune responses is unclear and requires further study.
Fig. 1.
The pathogenesis of AD. Skin barrier dysfunction and skin dysbiosis both play a key role in the pathogenesis of AD. External stimuli result in the release of cytokines from keratinocytes, inducing a T helper type 2 immune response that can in turn activate keratinocytes to form a cycle of skin inflammation. AD atopic dermatitis, dDC dermal dendritic cell, IL interleukin, ILC2 type 2 innate lymphoid cells, KC keratinocyte, S. aureus Staphylococcus aureus, Th2 T-helper type 2, TSLP thymic stromal lymphopoietin
This review focuses on the role of the microbiome in the pathophysiology of AD, describing first how recent studies have identified alterations in microbiome diversity and community structure and changes in the metabolic function of the microbiome that are associated with skin inflammation. We then summarize the mechanisms involved in the interactions between the microbiome and the peripheral sensory neurons that lead to itch in AD.
Role of Altered Microbiome Diversity and Community Structure in AD
The gut microbiome in infants with AD is significantly less diverse than that in healthy controls, and several signature microbes from the genera Lactobacillus, Bacteroides, and Clostridium have been identified [7, 8]. Similarly, studies have also revealed that skin microbial diversity is reduced in AD, with an increase in the abundance of S. aureus, particularly during AD flares, and a correlation between S. aureus abundance and disease severity [3]. Baurecht et al. [9] compared the configuration of the microbiome from skin at different sites in patients with AD and healthy individuals. Although bacterial composition was strongly dependent on the anatomic region, the relative abundance of staphylococci, particularly S. aureus, was enhanced across all skin sites in patients with AD, with the largest increases in abundance observed on chronic lesions compared with acute lesions or nonlesional skin [9]. This multisite study found that AD severity correlated with an increased abundance of the genus Corynebacterium and the phylum Proteobacteria [9].
Alterations in epidermal lipid metabolism leading to changes in the composition of all three of the major lipid classes in the stratum corneum—fatty acids (FAs), ceramides, and cholesterol—are well-recognized factors in the pathogenesis of AD [10–13]. However, few studies have investigated how these changes in lipid composition influence the structure of the microbiome in AD. Interestingly, the multisite study by Baurecht et al. [9] also identified specific correlations between microbiome structure and diversity and epidermal lipid composition, particularly at AD predilection sites: high levels of unsaturated long-chain FAs (LCFAs: FA20:1, FA20:2, FA22:1, and FA24:1) were associated with an increase in the abundance of Propionibacterium and Corynebacterium, whereas high levels of saturated short-chain FAs (SCFAs: FA16:0 and FA18:0) were associated with a decrease in the abundance of these genera. In addition, higher levels of the ceramide CER-[AS] were associated with an increase in the abundance of S. aureus [9]. Significant associations between lipid profiles and S. aureus colonization in patients with AD were also identified in a cross-sectional observational study conducted by Li et al. [14]. This study found that the levels of some ceramides, including CER[AH]C38–C52, CER[AP]C40, and some very-long-chain ceramides, CER[EOH]C66–C70 and CER[EOS]C70, were significantly lower in patients with AD and a positive S. aureus colonization status than in those without S. aureus colonization. Similarly, the levels of unsaturated FAs (16:1 and 18:1) and some triglycerides were significantly lower in patients with S. aureus colonization. These studies provide interesting insights into how changes in skin barrier composition can influence the structure and diversity of the microbiome in AD.
Besides the skin and the gut, other body sites—for example, the oral cavity and the respiratory tract—also harbor microbes [15, 16]. The composition, diversity, and interactions with the host of the microbial communities in these different body habitats are site specific, with alterations in the diversity or metabolic activity of these unique microbiomes resulting in inflammation, tissue damage, and atopic disease [15, 16].
The relationship between microbiomes across different body sites has recently been investigated in patients with AD. Li et al. [17] revealed differential alterations in community-level diversity between microbiomes from the same body habitats (alpha diversity) of patients with AD and healthy controls in a study comparing the structure and predicted functions of microbial communities from the skin, oral cavity, and gut. In patients with AD, the skin showed the most significant decrease in alpha diversity, followed by the oral cavity, whereas no differences in community structure were detected for the gut microbiome [17]. Differences in the extent of diversity between different habitats (beta diversity) were observed, with the microbial lineage between the skin and oral cavity being closer in patients with AD than in healthy controls. In addition, alterations in the level of enrichment of specific microbes at the different body habitats were observed in patients with AD compared with controls. At the genus level, Staphylococcus was enriched in the skin samples from patients with AD, whereas Halomonas was more abundant in the skin of healthy controls. In contrast, Halomonas was enriched in the oral cavity of patients with AD compared with in controls, showing that patterns of enrichment in AD vary according to body habitat. Levels of diversity in the different habitats were also differentially correlated with the clinical severity of AD: in the skin, microbiome alpha diversity was negatively correlated with skin disease severity, whereas a positive correlation was seen in the oral cavity. Finally, analysis of the predicted functional profiles of the microbiome from each habitat sampled, based on inferred Kyoto encyclopedia of genes and genomes pathways, revealed an inverse association between pathway enrichment in the skin and oral microbiomes: tryptophan metabolism was enriched in the oral cavity of patients with AD but attenuated in the skin. In summary, this study revealed differential changes in the structure of microbial communities across the different body habitats in patients with AD, with reduced levels of diversity both within and between the skin and oral microbiomes, together with inverse associations in predicted microbial function between these two microbial communities. Thus, in the future, control of skin inflammation in patients with AD might be achieved by manipulating the oral microbiome.
The Role of Altered Microbiome Function in AD
Microbiome Function in the Gut: The Role of Microbiome-Derived Metabolites
Nearly all our knowledge concerning the function of the microbiome comes from studies of the gut. Most of the functions of the microbiome are exerted through microbiome-derived metabolites. In the gut, SCFAs play the most important role in modulating host physiology. Gut commensal bacteria can digest complex carbohydrates, which the host itself cannot digest, to produce SCFAs that provide energy to host intestinal epithelial cells and play a role in preserving mucosal immunity [18]. Tryptophan is another important gut metabolite and is metabolized by three main pathways: the (1) kynurenine and (2) serotonin pathways that are predominantly, but not exclusively, used by the host and (3) the tryptamine pathway in the microbiome leading to the production of the majority of indole and indole derivatives [19]. These tryptophan metabolites act as ligands that can activate the aryl hydrocarbon receptor (AhR) on innate lymphoid cells to induce IL-22 secretion, driving the release of antimicrobial peptides (AMPs) and providing protection from infections by pathogens [18].
Altered Levels of Microbiome-Derived Tryptophan Metabolites May be a Key Feature in AD
Although the environment of the skin is very different from that in the gut (see Sect. 2), the discovery of the major role played by gut microbial metabolites in host physiology raised the question of whether there are microbial metabolites in the skin and, if so, whether these metabolites play a role in disease. The environment in the skin is dry, with high levels of salt, and is lacking in nutrition. However, although the surface of the skin is low in fiber, it is rich in protein derived from an abundance of dead keratinocytes and degenerating keratin filaments in the stratum corneum, which may act as a potential source of tryptophan metabolites for the microbiome [20].
In addition to its functions in the gut, the AhR also acts as an environmental sensor in the skin. It is activated by air pollutants and by 6-formylindolo[3,2-b] carbazole, a tryptophan metabolite released by keratinocytes in response to exposure to ultraviolet light [21]. AhR activation also occurs in response to tryptophan metabolites generated by the host and to indole-3-acetic acid and indole-3-aldehyde (IAId) generated by the microbial tryptamine pathway [21].
Several lines of evidence suggest that alterations in skin microbiome-derived tryptophan metabolism may play a role in AD. First, whole metagenome analysis of the skin microbiome led to the identification of several pathways that were perturbed in AD-prone skin compared with healthy skin, including lower-than-control levels of tryptophan metabolism [22]. Second, air pollutants have been shown to induce AD-like phenotypes in mice, and the AhR has been found to be activated in the lesional skin of patients with AD [23]. Third, coal tar has been used for hundreds of years to treat skin diseases and is an effective therapy for psoriasis and AD. In 2013, Van den Bogaard et al. [24] demonstrated that coal tar activates the AhR, resulting in induction of epidermal differentiation and restoration of major skin barrier proteins, including filaggrin.
To directly assess microbe-derived tryptophan metabolites on the skin surface and investigate their role in skin inflammation in patients with AD, Yu et al. [25] developed a gel patch method that was more efficient than conventional swabbing for collecting skin surface tryptophan and its metabolites. Using this new method coupled with liquid chromatography–tandem mass spectrometry analysis, Yu et al. [25] demonstrated that the levels of IAld in lesional and nonlesional skin of patients with AD were lower than those in healthy controls. The same authors used a mouse model of MC903-induced AD-like dermatitis to show that IAld played a key role in attenuating skin inflammation: topical administration of IAld to the ears of mice with AD-like dermatitis resulted in reduced skin redness, ear thickness, and scratching frequency and in lower levels of immunoglobulin E (IgE) and proinflammatory cytokines. This IAId-mediated attenuation of skin inflammation was abolished after topical application of an AhR antagonist and in AhR-null mice, demonstrating that IAId exerted its effects through activation of the AhR [25]. Upregulation of expression of factors further downstream in the AhR signaling pathway, such as cytochrome P450 (CYP)-1A1, in response to IAld application, provided confirmation of the involvement of the AhR in the mechanism of action of IAld. Application of IAld also led to a significant reduction in the levels of TSLP, with this inhibition being mediated by binding of AhR to two aryl hydrocarbon response elements (AhRE2 and AhRE4) in the promoter region of the TSLP gene. Thus, the findings from this study revealed for the first time that tryptophan metabolites on the skin surface can regulate skin inflammation by activation of the AhR and inhibition of TSLP production in keratinocytes. This study also highlighted the important role played by microbiome function in maintaining skin health by showing that alterations in the level of a key microbiome-derived metabolite were involved in the development of AD.
Role of Altered Microbiome Structure and Function in Itch in AD
Chronic itch is a very common feature in AD [26, 27]. It is defined as an itch that lasts longer than 6 weeks and is usually mediated by the non-histaminergic pathway. This non-histaminergic pathway plays a significant role in itch transmission and can be activated by proteases secreted by bacteria on the skin, such as S. aureus, which bind to the protease-activated receptors (PAR-2 and PAR-4) [28]. Multiple factors that perturb the normal functional interaction between the skin and the microbiome appear to contribute to the increased activation of the non-histaminergic itch pathway in patients with AD.
Protease Overactivity in the Skin of Patients with AD
Overactivity of proteases is a characteristic of AD. Comparative analysis of the complete transcriptome from skin biopsies from healthy individuals and patients with AD revealed that the genes encoding tryptase (TPSAB1) and PAR-2 and -4 (F2RL1 and F2RL3) were among those differentially expressed in itchy and healthy skin: levels of TPSAB1, F2RL1, and F2RL3 expression were higher in itchy skin from patients with AD than in skin from healthy controls [29]. Furthermore, immunohistochemical analysis showed that the number of tryptase-positive mast cells was not only higher than in healthy controls but also higher in itchy AD than in nonlesional skin [29]. Similarly, the largest increase in PAR-2 expression occurred in itchy AD skin, followed by a smaller increase in nonlesional skin. A similar pattern of higher levels of tryptase expression was observed in psoriatic skin; however, no differences in the levels of PAR-2 expression were observed between skin from patients with psoriasis and healthy skin [29].
The pH of the Skin Alters Microbiome Structure and Function, Promoting Itch
The pH of the skin has a significant impact on the structure and function of the microbiome and thus also on itch. The pH of healthy skin is acidic, ranging from pH 4 to 6. This acidic pH favors the growth of commensal bacteria, whereas pathogenic bacteria such as S. aureus thrive at neutral pHs [30]. In addition, the antibacterial activity of some of the AMPs secreted by the skin is higher at acidic pHs [30]. AD is associated with an increase in skin pH to ≥ 6.5, promoting the colonization of S. aureus at the skin surface, leading to dysbiosis and an increased abundance of microbiome-derived serine proteases [30]. In addition, this elevated pH also activates endogenous skin proteases, which have a pH optimum of 8, resulting in further activation of the keratinocyte PAR-2 receptors and the non-histaminergic pathway [30, 31].
Neuropeptides Secreted by the Skin Can Also Alter Microbiome Structure
Several neuropeptides secreted by the skin are known to be involved in the pathophysiology of itch in AD, including substance P (SP) [32]. Localized increases of SP in the skin have been shown to increase the virulence of some bacteria, including Bacillus and Staphylococci. Finally, the calcitonin gene-related peptide has been shown to modulate the virulence of another key component of the skin microbiome, S. epidermis. Thus, neuropeptides in the skin appear to act as host signals for the microbiome and play a key role in skin homeostasis and in the pathophysiology of disease [32].
Other Mechanisms of Microbial Activation of the Non-Histaminergic Itch Pathway
In addition to the PARs, a family of mas-related G protein-coupled receptors (MRGPRs) have been identified as itch receptors in peripheral sensory neurons [33]. Members of the human MRGPR family (MRGPRX) have been shown to be implicated in mediating non-histaminergic itch. MRGPRX1 has been shown to be activated by chloroquine, a well-known inducer of non-histaminergic itch [33]. MRGPRX2 is expressed in peripheral sensory neurons and on mast cells and has been shown to be activated by AMPs, produced in response to stimuli such as microbial pathogens, to promote the release of IL-31 [34]. IL-31 is a cytokine that plays an important role in itch, with overexpression of IL-31 in mice leading to the development of AD-like lesions and severe pruritus [34].
Toll-like receptors (TLRs) play an important role in initiating innate immune responses and in the interaction between the host and the microbiome. TLRs recognize a wide range of pathogens, via recognition of pathogen-associated molecular patterns, and are known to be expressed in primary sensory neurons. In particular, TLR7 has been found to be overexpressed in small dorsal root ganglion (DRG) neurons, especially in nociceptors expressing the transient receptor potential vanilloid type 1 (TRPV1) ion channel receptor [35]. TRPV1 is a key receptor in itch activation, and the coupling of TLRs with ion channels in primary sensory neurons may provide a mechanism by which TLR ligands can trigger immediate itch sensations [36]. In addition, studies of pain during S. aureus infection have indicated that pathogens are able to directly activate nociceptors through release of N-formyl peptides and the pore-forming toxin alpha hemolysin (Hlα) and modulation of ion channel activity [37].
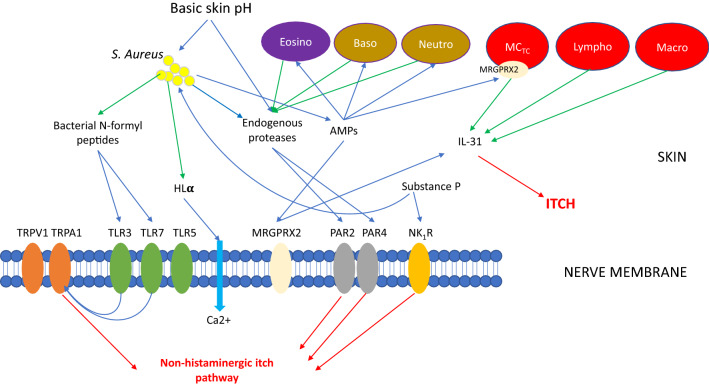
These recent developments in our understanding of itch have shown that nerve fibers are not just transmitters of itch but interact with the microbiome and the peptides involved in microbial function via an array of receptors (summarized in Fig. 2). Alterations in the interaction between microbes and components of the itch pathway are thus emerging as a further mechanism via which the microbiome is involved in the physiopathology of diseases such as AD.
Fig. 2.
Simplified schematic representation of the mechanism of itch induction via the non-histaminergic neural pathway in atopic dermatitis. Endogenous proteases may be secreted by basophils, neutrophils, eosinophils, mast cells, and the skin. AMPs antimicrobial peptides, Baso basophils, Eosino eosinophils, HLα alpha hemolysin, IL interleukin, Lympho lymphocyte, Macro macrophage, MCTc tryptase and chymase-expressing mast cell, MRGPRX human mas-related G protein-coupled receptors, Neutro neutrophils, NK1R neurokinin 1 receptor, PAR protease-activated receptor, S. aureus Staphylococcus aureus, TLR Toll-like receptors, TRPA1 transient receptor potential ankyrin 1, TRPV1 transient receptor potential vanilloid type 1; green arrows indicate secretion; blue arrows indicate activation
Treatment of Atopic Dermatitis by Modification of the Microbiome
There are many ways in which we can intervene in AD by modification of the microbiome, including using prebiotics (small molecules that regulate the bacteria), probiotics (live bacteria that regulate host responses), postbiotics (metabolites from bacteria), and antibiotics (AMPs from commensal bacteria). Although previous treatments have concentrated primarily on the use of probiotics, future therapeutic interventions involving postbiotics may prove to be safer and provide more easily commercialized new-generation treatments for AD and other atopic skin diseases. However, further understanding of the mechanism of action of skin metabolites in inflammation is needed to enable these molecules to be used effectively in future treatments. In addition, treatments for chronic itch in AD could involve treatments targeted toward reducing the activation of proteases or toward receptors and ligands involved in the interaction between microbes and the peripheral nerve system to minimize activation of the non-histaminergic pathway.
Conclusion
These recent advances in our understanding of the importance of the microbiome in maintaining skin health have revealed that alterations in the community structure of the microbiome, both within and across body habitats, and in the metabolic function of the skin microbiome are key factors in the pathophysiology of AD. Alterations in the function of the microbiome appear to play a role in skin inflammation and homeostasis, with reductions in the microbiome-derived tryptophan metabolite, IAId, being shown to be involved in AD. Further work is required to identify other potentially key microbiome-derived metabolites involved in maintaining skin health and to determine how these metabolites interact with the other cell types making up the skin microenvironment, such as keratinocytes, Langerhans cells, and peripheral nerves. Further insights into the interactions between the skin and microbiome will provide new targets for the next generation of treatments for barrier function, inflammation, and itch in AD.
Acknowledgements
The authors thank Emma Pilling, PhD, and Marielle Romet, PhD, (Synergy Pharm-Santé Active Edition) for medical writing assistance funded by Laboratoires Dermatologiques Avène–Pierre Fabre Dermo-Cosmétique.
Author Contributions
Both authors had the idea for the article, performed the literature search and data analysis, and critically revised the work.
Declarations
Funding
Medical writing assistance was funded by Laboratoires Dermatologiques Avène–Pierre Fabre Dermo-Cosmétique.
Conflict of interest
Gil Yosipovitch has acted as consultant and scientific advisory board member for Trevi, Pfizer, Galderma, Novartis, Sanofi Regeneron, Pierre Fabre, and LEO, and has had research sponsored by Pfizer, Sun Pharma, Novartis, LEO, and Sanofi Regeneron. Wei Li has received consultancy/speaker honoraria from Sanofi Regeneron, Pfizer, Lilly, Novartis, LEO, Pierre Fabre, MEDA Pharma (a Mylan Company), Astellas, and Galderma, and has participated as Principal Investigator in clinical trials sponsored by Sanofi Regeneron.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Disclosure statement
This article is published as part of a journal supplement wholly funded by Laboratoires Dermatologiques Avène–Pierre Fabre Dermo-Cosmétique.
Contributor Information
Wei Li, Email: liweiderma@163.com.
Gil Yosipovitch, Email: gyosipovitch@med.miami.edu.
References
- 1.Avena-Woods C. Overview of atopic dermatitis. Am J Manag Care. 2017;23(8 Suppl):S115–S123. [PubMed] [Google Scholar]
- 2.Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143(1):26–35. doi: 10.1016/j.jaci.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JE, Kim HS. Microbiome of the Skin and gut in atopic dermatitis (AD): understanding the pathophysiology and finding novel management strategies. J Clin Med. 2019;8(4):444. doi: 10.3390/jcm8040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefanovic N, Flohr C, Irvine AD. The exposome in atopic dermatitis. Allergy. 2020;75(1):63–74. doi: 10.1111/all.13946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czarnowicki T, Krueger JG, Guttman-Yassky E. Novel concepts of prevention and treatment of atopic dermatitis through barrier and immune manipulations with implications for the atopic march. J Allergy Clin Immunol. 2017;139(6):1723–1734. doi: 10.1016/j.jaci.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129(2):434–40, 40 e1-2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 8.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol. 2013;132(3):601–7 e8. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- 9.Baurecht H, Ruhlemann MC, Rodriguez E, Thielking F, Harder I, Erkens AS, et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J Allergy Clin Immunol. 2018;141(5):1668–76 e16. doi: 10.1016/j.jaci.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya N, Sato WJ, Kelly A, Ganguli-Indra G, Indra AK. Epidermal lipids: key mediators of atopic dermatitis pathogenesis. Trends Mol Med. 2019;25(6):551–562. doi: 10.1016/j.molmed.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park K, Lee S, Lee YM. Sphingolipids and antimicrobial peptides: function and roles in atopic dermatitis. Biomol Ther (Seoul) 2013;21(4):251–257. doi: 10.4062/biomolther.2013.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Fang H, Dang E, Wang G. The role of ceramides in skin homeostasis and inflammatory skin diseases. J Dermatol Sci. 2020;97(1):2–8. doi: 10.1016/j.jdermsci.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 13.van Smeden J, Bouwstra JA. Stratum corneum lipids: their role for the skin barrier function in healthy subjects and atopic dermatitis patients. Skin barrier function. Berlin: Karger Publishers; 2016. pp. 8–26. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Villarreal M, Stewart S, Choi J, Ganguli-Indra G, Babineau DC, et al. Altered composition of epidermal lipids correlates with Staphylococcus aureus colonization status in atopic dermatitis. Br J Dermatol. 2017;177(4):e125–e127. doi: 10.1111/bjd.15409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YJ, Marsland BJ, Bunyavanich S, O'Mahony L, Leung DY, Muraro A, et al. The microbiome in allergic disease: current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol. 2017;139(4):1099–1110. doi: 10.1016/j.jaci.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grice EA, Segre JA. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13(1):151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Xu X, Wen H, Wang Z, Ding C, Liu X, et al. Inverse association between the skin and oral microbiota in atopic dermatitis. J Invest Dermatol. 2019;139(8):1779–87 e12. doi: 10.1016/j.jid.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. 2017;35:8–15. doi: 10.1016/j.mib.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Gutierrez-Vazquez C, Quintana FJ. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity. 2018;48(1):19–33. doi: 10.1016/j.immuni.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YE, Fischbach MA, Belkaid Y. Skin microbiota-host interactions. Nature. 2018;553(7689):427–436. doi: 10.1038/nature25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colonna M. AHR: making the keratinocytes thick skinned. Immunity. 2014;40(6):863–864. doi: 10.1016/j.immuni.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Chng KR, Tay AS, Li C, Ng AH, Wang J, Suri BK, et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat Microbiol. 2016;1(9):16106. doi: 10.1038/nmicrobiol.2016.106. [DOI] [PubMed] [Google Scholar]
- 23.Hidaka T, Ogawa E, Kobayashi EH, Suzuki T, Funayama R, Nagashima T, et al. The aryl hydrocarbon receptor AhR links atopic dermatitis and air pollution via induction of the neurotrophic factor artemin. Nat Immunol. 2017;18(1):64–73. doi: 10.1038/ni.3614. [DOI] [PubMed] [Google Scholar]
- 24.van den Bogaard EH, Bergboer JG, Vonk-Bergers M, van Vlijmen-Willems IM, Hato SV, van der Valk PG, et al. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J Clin Invest. 2013;123(2):917–927. doi: 10.1172/JCI65642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Luo Y, Zhu Z, Zhou Y, Sun L, Gao J, et al. A tryptophan metabolite of the skin microbiota attenuates inflammation in patients with atopic dermatitis through the aryl hydrocarbon receptor. J Allergy Clin Immunol. 2019;143(6):2108–19 e12. doi: 10.1016/j.jaci.2018.11.036. [DOI] [PubMed] [Google Scholar]
- 26.Dawn A, Papoiu AD, Chan YH, Rapp SR, Rassette N, Yosipovitch G. Itch characteristics in atopic dermatitis: results of a web-based questionnaire. Br J Dermatol. 2009;160(3):642–644. doi: 10.1111/j.1365-2133.2008.08941.x. [DOI] [PubMed] [Google Scholar]
- 27.Yosipovitch G, Goon AT, Wee J, Chan YH, Zucker I, Goh CL. Itch characteristics in Chinese patients with atopic dermatitis using a new questionnaire for the assessment of pruritus. Int J Dermatol. 2002;41(4):212–216. doi: 10.1046/j.1365-4362.2002.01460.x. [DOI] [PubMed] [Google Scholar]
- 28.Yosipovitch G, Rosen JD, Hashimoto T. Itch: From mechanism to (novel) therapeutic approaches. J Allergy Clin Immunol. 2018;142(5):1375–1390. doi: 10.1016/j.jaci.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Nattkemper LA, Tey HL, Valdes-Rodriguez R, Lee H, Mollanazar NK, Albornoz C, et al. The genetics of chronic itch: gene expression in the skin of patients with atopic dermatitis and psoriasis with severe itch. J Invest Dermatol. 2018;138(6):1311–1317. doi: 10.1016/j.jid.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 30.Ali SM, Yosipovitch G. Skin pH: from basic science to basic skin care. Acta Derm Venereol. 2013;93(3):261–267. doi: 10.2340/00015555-1531. [DOI] [PubMed] [Google Scholar]
- 31.Jang H, Matsuda A, Jung K, Karasawa K, Matsuda K, Oida K, et al. Skin pH is the master switch of kallikrein 5-mediated skin barrier destruction in a murine atopic dermatitis model. J Invest Dermatol. 2016;136(1):127–135. doi: 10.1038/JID.2015.363. [DOI] [PubMed] [Google Scholar]
- 32.N'Diaye A, Gannesen A, Borrel V, Maillot O, Enault J, Racine PJ, et al. Substance P and calcitonin gene-related peptide: key regulators of cutaneous microbiota homeostasis. Front Endocrinol (Lausanne) 2017;8:15. doi: 10.3389/fendo.2017.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, et al. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139(7):1353–1365. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian H, Gupta K, Ali H. Roles of Mas-related G protein-coupled receptor X2 on mast cell-mediated host defense, pseudoallergic drug reactions, and chronic inflammatory diseases. J Allergy Clin Immunol. 2016;138(3):700–710. doi: 10.1016/j.jaci.2016.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu T, Gao YJ, Ji RR. Emerging role of Toll-like receptors in the control of pain and itch. Neurosci Bull. 2012;28(2):131–144. doi: 10.1007/s12264-012-1219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi J, Buzas K, Fan H, Cohen JI, Wang K, Mont E, et al. Painful pathways induced by TLR stimulation of dorsal root ganglion neurons. J Immunol. 2011;186(11):6417–6426. doi: 10.4049/jimmunol.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu IM, Heesters BA, Ghasemlou N, Von Hehn CA, Zhao F, Tran J, et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature. 2013;501(7465):52–57. doi: 10.1038/nature12479. [DOI] [PMC free article] [PubMed] [Google Scholar]