Abstract
Intramammary infections (IMI) with Staphylococcus aureus are a common cause of bovine mastitis and can result in both clinical (CM) or subclinical mastitis (SCM). Although bacterial isolates of S. aureus differ in their virulence potential it is largely unclear which bacterial virulence factors are responsible for increased clinical severity. We performed a genome wide association study and used a generalized linear mixed model to investigate the correlation between gene carriage, lineage and clinical outcome of IMI in a collection of S. aureus isolates from cattle with CM (n = 125) and SCM (n = 151) from 11 European countries. An additional aim was to describe the genetic variation of bovine S. aureus in Europa. The dominant lineages in our collection were clonal complex (CC) 151 (81/276, 29.3%), CC97 (54/276, 19.6%), CC479 (32/276, 11.6%) and CC398 (19/276, 6.9%). Virulence and antimicrobial resistance (AMR) gene carriage was highly associated with CC. Among a selection of nine virulence and AMR genes, CC151, CC479 and CC133 carried more virulence genes than other CCs, and CC398 was associated with AMR gene carriage. Whereas CC151, CC97 were widespread in Europe, CC479, CC398 and CC8 were only found in specific countries. Compared to CC151, CC479 was associated with CM rather than SCM (OR 3.62; 95% CI 1.38–9.50) and the other CCs were not. Multiple genes were associated with CM, but due to the clustering within CC of carriage of these genes, it was not possible to differentiate between the effect of gene carriage and CC on clinical outcome of IMI. Nevertheless, this study demonstrates that characterization of S. aureus CC and virulence genes helps to predict the likelihood of the occurrence of CM following S. aureus IMI and highlights the potential benefit of diagnostics tools to identify S. aureus CC during bovine mastitis.
Subject terms: Genetics, Microbiology, Diseases
Introduction
Mastitis is responsible for significant financial losses on dairy farms due to reduced milk yield, milk unsuitable for consumption, treatment costs and culling of animals1,2. The main causes of bovine mastitis are bacterial intramammary infections (IMI), with Staphylococcus aureus being one of the most relevant pathogens3. Infections with S. aureus mostly result in subclinical mastitis (SCM), but can also lead to clinical mastitis (CM)4.
The S. aureus clones responsible for bovine mastitis predominantly belong to bovine-associated clonal complexes (CC), such as CC151, CC97, CC133, CC479 and CC7715,6. The genomic content of S. aureus clones can differ greatly due to their accessory genome, which makes up approximately 25% of the total genome7. The accessory genome of S. aureus predominantly consists of genes introduced by horizontal gene transfer (HGT), with these transferred genes often being phage genes, virulence and antimicrobial resistance (AMR) genes, expected to affect the pathogenesis of S. aureus IMI8. A large number of virulence genes have been identified in S. aureus isolates obtained from bovine mastitis cases, several of which are involved in evasion of the host immune response during infection8,9. Leukocidins, such as LukMF’, can directly attack immune cells in the lumen of the mammary gland and superantigens (SAs), such as enterotoxins seA–seQ, or toxic shock syndrome toxin 1 (tsst-1), massively activate T lymphocytes and antigen-presenting cells, interfering with the buildup of a proper adaptive immune response10. Furthermore, Staphylococcal superantigen-like proteins (SSLs) disrupt different pathways of the immune response, such as pathogen recognition by TLR-2 (SSL-3, SSL-4), neutrophil function and recruitment (SSL-1, SSL-5, SLL-7, SSL-13) and the opsonization of bacteria (SSL-7)10,11. Some of these immune evasion factors are considered ruminant adapted10, such as LukMF’ and the S. aureus pathogenicity island encoded variant of the Von Willebrand factor‐binding protein (SaPI vWFbp)12,13. Furthermore, S. aureus can carry AMR encoding genes, including blaZ, tetM and mecA, which reduce effectiveness of antimicrobial treatment of S. aureus mastitis14. The majority of virulence and AMR genes are located on mobile genetic elements and S. aureus belonging to the same CC have specific virulence gene signatures due to clonal expansion6,15,16 and restriction-modification systems that reduce HGT between different S. aureus lineages17,18.
Several studies investigated CCs and virulence/AMR gene carriage among bovine S. aureus isolates responsible for IMI6,9,19, but only few directly compare isolates from CM and SCM cases. Recently, we identified a specific bovine S. aureus lineage (CC479) that was associated with CM in The Netherlands20. In addition, carriage of certain genes (lukM-lukF′, seO) was overrepresented among CM isolates compared to SCM isolates21,22. This suggests that some S. aureus lineages are more likely to cause CM than others. Although both CM and SCM lead to financial losses for the farm and are associated with animal disease and welfare, the costs of treatment of CM are substantially higher than of SCM1,23. Also, the effect of CM on animal welfare obviously is considerable larger than for SCM24. Therefore, identification of S. aureus isolates with increased risk of developing into CM is likely valuable to better support farm health management decisions. However, studies comparing isolates from CM and SCM have been mainly on a national scale20–22, whereas the genetic makeup of bovine S. aureus likely differs between countries. Therefore, in this study we performed whole-genome sequencing (WGS) of 276 S. aureus isolates from cattle with CM or SCM in 11 European countries.
The primary aim of this study was to investigate differences in lineage and genomic content between S. aureus isolates responsible for CM and SCM. A second aim was to describe the variation in lineages, virulence gene and AMR gene carriage of bovine S. aureus isolates in Europe.
Results
Bovine S. aureus CCs differ in their carriage of immune evasion and AMR genes
After selection of isolates and quality control of WGS results, 276 genomes were available of S. aureus isolates obtained from bovine mastitis cases originating from 254 unique herds in eleven different European countries. There was an approximately even distribution of CM (125/276, 45%) and SCM (151/276, 55%) isolates, and S. aureus in the collection belonged to eighteen different CCs. The most prevalent CCs were CC151 (81/276, 29.3%), CC97 (54/276, 19.6%), CC479 (32/276, 11.6%), CC133 (25/276, 9.1%), CC398 (19/276, 6.9%), CC1 (14/276, 5.1%), CC20 (11/276, 4.0%) and CC8 (11/276, 4.0%), and carriage of a selection of nine key virulence and AMR genes among isolates was related to CC (Table 1). The CC151, CC479 and CC133 strains had high carriage of virulence genes but lacked AMR genes. All these three CCs carried the lukM-lukF′ genes and CC479, CC151 also possessed SA genes. In addition, CC479, CC133 S. aureus carried the SaPI encoded vWFbp gene (Table 1). In contrast, CC398 S. aureus lacked all these virulence factors but did have a high carriage rate of the AMR genes blaZ (8/19, 42%), tetM (19/19, 100%) and mecA (11/19, 58%) (Table 1). The CC97 displayed a moderate carriage of both virulence gene lukM-lukF’ (16/54, 30%) and the AMR gene blaZ (16/54, 30%) (Table 1).
Table 1.
Number and percentage of isolates positive for a selection of virulence and antimicrobial resistance genes per clonal complex and number and proportion of clinical mastitis of 276 S. aureus isolates obtained from bovine clinical and subclinical mastitis in 11 European countries.
| CCa | n | % | Virulence genes n (%) | Antimicrobial resistance genes n (%) | Manifestation of mastitis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| lukM-lukF′b | scn | SaPI vWFbp | seI | seL | tsst-1 | blaZ | tetM | mecA | CMc | Odds ratiod (95% CI) | Pg | |||
| 151 | 81 | 29.3 | 81 (100) | 0 (0) | 0 (0) | 80 (99) | 19 (23) | 19 (23) | 1 (1) | 0 (0) | 0 (0) | 41 (51) | Refe | Ref |
| 97 | 54 | 19.6 | 16 (30) | 7 (13) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 16 (30) | 0 (0) | 0 (0) | 23 (42) | 0.75 (0.37–1.54) | 0.44 |
| 479 | 32 | 11.6 | 30 (94) | 0 (0) | 32 (100) | 32 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (3) | 0 (0) | 25 (78) | 3.62 (1.38–9.50) | < 0.01 |
| 133 | 25 | 9.1 | 23 (92) | 21 (84) | 25 (100) | 0 (0) | 2 (8) | 2 (8) | 0 (0) | 0 (0) | 0 (0) | 13 (52) | 1.05 (0.42–2.61) | 0.92 |
| 398 | 19 | 6.9 | 0 (0) | 10 (52) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 8 (42) | 19 (100) | 11 (58) | 5 (27) | 0.41 (0.12–1.48) | 0.17 |
| 1 | 14 | 5.1 | 9 (64) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (21) | 0.30 (0.07–1.23 | 0.10 |
| 20 | 11 | 4.0 | 0 (0) | 0 (0) | 0 (0) | 7 (64) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0.34 (0.08–1.45) | 0.15 |
| 8 | 11 | 4.0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (54) | 0 (0) | 0 (0) | 2 (18) | 0.21 (0.43–1.10) | 0.06 |
| 9 | 5 | 1.8 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (66) | 0 (0) | 0 (0) | 2 (33) | NAf | NA |
| 50 | 5 | 1.8 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (20) | 0 (0) | 0 (0) | 1 (20) | NA | NA |
| 49 | 4 | 1.4 | 1 (25) | 0 (0) | 0 (0) | 0 (0) | 1 (25) | 0 (0) | 1 (25) | 0 (0) | 0 (0) | 1 (25) | NA | NA |
| 7 | 4 | 1.4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (50) | NA | NA |
| 5 | 4 | 1.4 | 0 (0) | 0 (0) | 0 (0) | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (75) | NA | NA |
| 45 | 3 | 1.1 | 0 (0) | 0 (0) | 0 (0) | 1 (33) | 1 (25) | 0 (0) | 1 (33) | 0 (0) | 0 (0) | 0 (0) | NA | NA |
| 101 | 1 | 0.4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | NA | NA |
| 22 | 1 | 0.4 | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | NA | NA |
| 30 | 1 | 0.4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | NA | NA |
| 425 | 1 | 0.4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | NA | NA |
| Total | 276 | 160 (58) | 38 (14) | 57 (21) | 125 (45) | 24 (9) | 21 (8) | 40 (15) | 20 (7) | 11 (4) | 125 (45) | |||
aClonal complex.
bCarriage of genes determined using pangenome data from roary.
cNumber and percentage of clinical mastitis cases per CC.
dCluster-specific odds ratio of the isolate being cultured from CM versus SCM from a generalized linear mixed model with country as a random effect.
eReference class for variable CC within the generalized linear mixed model.
fOnly CCs with n > 10 were included in model.
gSignificance of class within the variable CC.
Heatmaps of the BLAST score ratio (BSR)25 of all S. aureus genes annotated as (putative) SAs or SSLs by prokka26 demonstrated that bovine S. aureus CCs differ in their carriage of these immune evasion factors (Supplementary Figs. S1, S2). Notable differences in SA carriage were the high number of SAs (up to 12 different SAs) genes carried by CC151, whereas CC398 isolates lacked SAs. Although most SSLs were detected in all S. aureus, the BSR of these SSLs differed between S. aureus CCs. However, the SSL-7, SSL-8, SSL-9 genes were only absent in CC479 S. aureus. Furthermore, two genes that were annotated as unnamed SSLs (GenBank references: WP_143564871.1 and WP_124375191) were exclusively found among CC97 isolates (Supplementary Fig. S2).
To screen for target genes that could differentiate between major ruminant CCs in a PCR-based assay, the presence of potential CC exclusive genes was also investigated (Supplementary Fig. S3, Supplementary Table S1). The highest number of CC-exclusive genes were found for CC479 (n = 17), followed by CC20 (n = 4), CC151 (n = 3), CC8 (n = 2) and CC133 (n = 2). For CC97, CC398, only a single unique gene was identified and no CC exclusive genes were found for CC1 isolates.
Heterogeneous spatial distribution of CCs across Europe
There was a significant difference in the distribution of S. aureus CCs between different countries (Fisher's Exact Test, p < 0.001). Whereas CC151 (10 out 11 countries) and CC97 (9 out 11) S. aureus were detected in almost all countries, CC398 (6 out 11) and CC479 (5 out 11) were considerably less widespread in our collection (Table 2). The CC398 lineage was predominantly found in isolates from Poland and Spain. Isolates belonging to CC8 originated exclusively from either Italy or Bavaria region in Germany. Also, all CC50 isolates originated from Denmark (Table 2).
Table 2.
Number and percentage of bovine S. aureus of 276 S. aureus isolates per clonal complex ranked per country or contributing region obtained from bovine clinical and subclinical mastitis in 11 European countries.
| CCa | N (%) | Be n (%) |
Dk n (%) |
Fr n (%) |
Ge (Lsb) n (%) |
Ge (Byc) n (%) |
Hu n (%) |
It n (%) |
NL n (%) |
Po n (%) |
Pt n (%) |
Es n (%) |
Uk n (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 151 | 81 (29) | 13 (43) | 5 (31) | 4 (17) | 10 (34) | 8 (27) | 3 (16) | 6 (20) | 14 (64) | 0 (0) | 2 (29) | 2 (17) | 14 (52) |
| 97 | 54 (20) | 4 (13) | 1 (6) | 7 (30) | 4 (14) | 6 (20) | 7 (37) | 9 (30) | 0 (0) | 5 (16) | 0 (0) | 2 (17) | 9 (33) |
| 479 | 32 (12) | 8 (27) | 0 (0) | 0 (0) | 2 (7) | 8 (27) | 0 (0) | 3 (10) | 8 (36) | 0 (0) | 3 (43) | 0 (0) | 0 (0) |
| 133 | 25 (9) | 4 (13) | 4 (25) | 2 (9) | 7 (24) | 2 (7) | 0 (0) | 2 (7) | 0 (0) | 1 (3) | 0 (0) | 3 (25) | 0 (0) |
| 398 | 19 (7) | 0 (0) | 0 (0) | 1 (4) | 2 (7) | 0 (0) | 1 (5) | 1 (3) | 0 (0) | 10 (32) | 0 (0) | 4 (33) | 0 (0) |
| 1 | 14 (5) | 1 (3) | 0 (0) | 1 (4) | 0 (0) | 0 (0) | 2 (11) | 1 (3) | 0 (0) | 7 (23) | 0 (0) | 0 (0) | 2 (7) |
| 20 | 11 (4) | 0 (0) | 0 (0) | 4 (17) | 1 (3) | 0 (0) | 5 (26) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (8) | 0 (0) |
| 8 | 11 (4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (17) | 0 (0) | 6 (20) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 9 | 5 (2) | 0 (0) | 1 (6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3) | 0 (0) | 1 (3) | 2 (29) | 0 (0) | 0 (0) |
| 50 | 5 (2) | 0 (0) | 5 (31) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 5 | 4 (1) | 0 (0) | 0 (0) | 2 (9) | 0 (0) | 0 (0) | 0 (0) | 1 (3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4) |
| 49 | 4 (1) | 0 (0) | 0 (0) | 1 (4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (10) | 0 (0) | 0 (0) | 0 (0) |
| 7 | 4 (1) | 0 (0) | 0 (0) | 0 (0) | 3 (10) | 0 (0) | 1 (5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 45 | 3 (1) | 0 (0) | 0 (0) | 1 (4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (6) | 0 (0) | 0 (0) | 0 (0) |
| 101 | 1 (0.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| 22 | 1 (0.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3) | 0 (0) | 0 (0) | 0 (0) |
| 30 | 1 (0.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (3) | 0 (0) | 0 (0) | 0 (0) |
| 425 | 1 (0.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (4) |
| Total | 276 | 30 | 16 | 24 | 29 | 30 | 19 | 30 | 22 | 31 | 7 | 12 | 27 |
aClonal complex.
bLower Saxony region in Germany.
cBavaria region in Germany.
Pangenome of bovine S. aureus and phylogenetic analysis
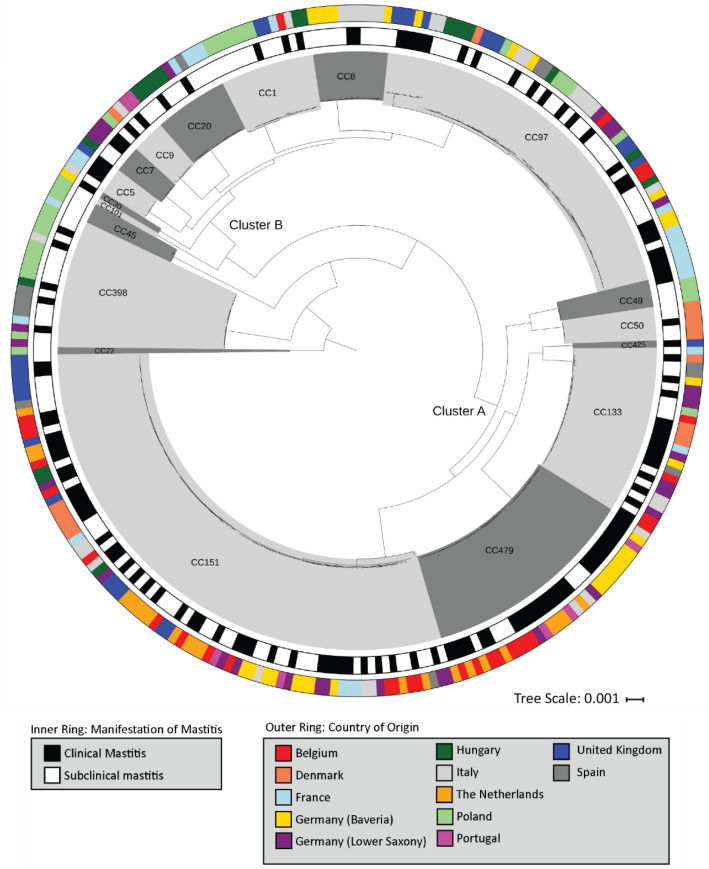
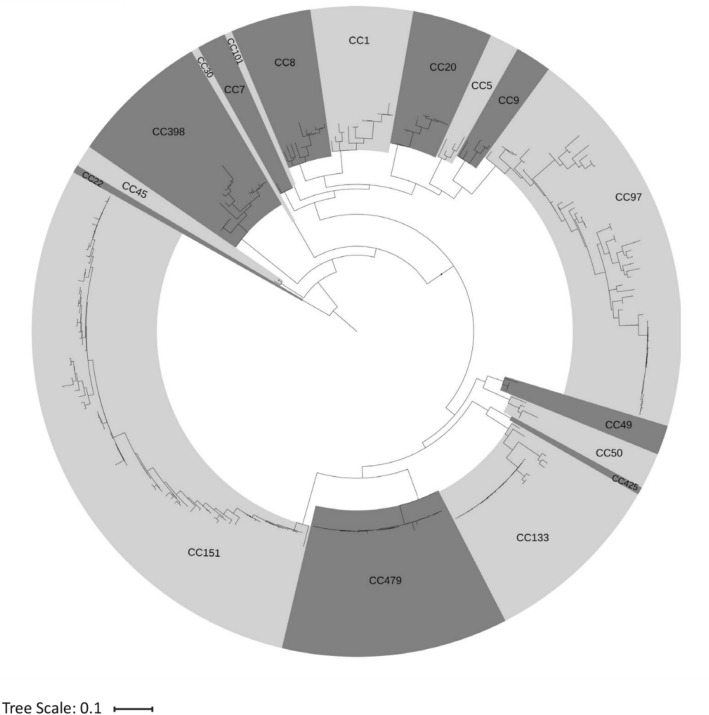
To investigate the phylogeny of our bovine S. aureus isolates, the pangenome of the entire collection was determined (n = 5720 genes) and a phylogenetic tree was constructed based on a super-alignment of the 1953 genes in the core genome (genes present in > 99% of isolates) (Fig. 1). A second phylogenetic tree was built based on the binary presence or absence of genes in the accessory genome (Fig. 2). Within both phylogenetic trees, isolates clustered within CC and two major clusters of CCs could be identified. The largest cluster, labeled as cluster A consisted of CC151, CC479, CC133, CC425, CC49 and CC50 isolates and second largest cluster B included CC97, CC8, CC1, CC20, CC9, CC7, CC5, CC30 and CC101. (Figs. 1, 2). There was uneven distribution of CM and SCM isolates between clusters, with more CM isolates being present in Cluster A (81/148, 55%) compared to cluster B (38/103, 36%) (Pearson's Chi-squared test p < 0.01).
Figure 1.
Maximum-likelihood phylogenetic tree based on the alignment of core genome (genes n = 1953) performed using roary v3.1239 and MAFFT of 276 S. aureus isolates obtained from bovine clinical and subclinical mastitis cases from 11 European countries. Dark and light grey shading displays Clonal Complex (CC), the outer ring represents the country of origin and the inner ring the clinical versus subclinical mastitis of each isolate. The phylogenetic tree was rooted with the CC22 clade and visualized using iTOL v3.641.
Figure 2.
Maximum-likelihood phylogenetic tree based on presence or absence of genes of the accessory genome (n = 2023 genes) of 276 S. aureus isolates obtained from bovine clinical or subclinical mastitis from 11 European countries. Dark and light grey shading displays Clonal Complex (CC). The phylogenetic tree was rooted with the CC22 clade and visualized using iTOL v3.641.
In general, the phylogenetic tree based on the core genome was similar to the tree based on the accessory genome. In addition, the phylogenetic tree shows considerable variation in accessory gene content, most notably within CC151 and CC97, but almost no variation in accessory gene carriage was observed among CC479 and CC133 S. aureus isolates (Fig. 2).
Staphylococcus aureus CC479 is associated with CM
In order to further study the association between S. aureus CC and clinical outcome of IMI, a generalised linear mixed model (GLMM) was built with CM versus SCM as the outcome variable, CC as predictor variable and the country of orgin of isolates as a random effect. For this model, isolates belonging to CCs with n < 10 (CC9, CC50, CC49, CC7, CC45, CC5, CC101, CC22, CC30, and CC425) were clustered together in a single category labeled as ‘other’. Including CC improved the fit over the model compared to the model with only random effects (ANOVA, p = 0.003) and isolates belonging to CC479 were more likely to originate from CM than from SCM (OR 3.62; 95% CI 1.38–9.50, p < 0.01) compared to the reference category (CC151). In addition, there was a trend for CC8 (OR 0.22; 95% CI 0.04–1.1; p = 0.06) and CC1 (OR 0.30; 95% CI 0.07–1.23; p = 0.10) to originate from SCM (Table 1).
Clinical manifestation of mastitis is associated with a large number of different genes
To study genetic differences that could underlay variation in pathogenity of bovine S. aureus isolates, we performed a genome wide association study (GWAS) and 153 genes were associated with CM isolates. In agreement with the results from the GLMM, genes exclusively present in the CC479 isolates (n = 59) had the highest OR, ranging from 4.6 to 5.6 [Benjamini–Hochberg Procedure (BHP); p-values < 0.05]. Among the CM-associated genes, a total of 20 genes matched our inclusion criteria for significant genes of interest (best pairwise comparision p < 0.1; BHP p < 0.1) and the OR of these genes ranged from 1.95 to 3.42 (Table 3). Among these genes, the hypothetical potentially bacteriophage derived DUF3310 domain-containing protein (OR 2.35) was the only gene associated with CM that was detected among all main lineages. Carriage of SA genes seM, seN, seI, seG, seO and seU, as well as the accessory gene regulator (agr) D type II gene was associated with CM and these genes were present in all CC479 isolates and the majority of CC151 isolates (Table 3). In addition, three genes from within the S. aureus pathogenicity island bovine 3 (SaPIbov3) were also associated with CM and were carried by all CC151, CC479 and 77% of CC97 isolates.
Table 3.
Odds Ratio, carriage rate per clonal complex and GenBank references of genes associated with clinical mastitis based on a genome wide association study performed on 276 S. aureus isolates obtained from bovine clinical and subclinical mastitis in 11 European countries.
| Genea | Odds Ratiob | Best pairwise comparision pc | CC151 (%)d | CC97 (%) | CC479 (%) | CC133 (%) | CC398 (%) | CC1 (%) | CC20 (%) | CC8 (%) | Other CCs (%)5 | Weighted average (%) | GenBank reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DUF3310 domain-containing protein | 2.35 | < 0.001 | 82 | 60 | 97 | 74 | 95 | 79 | 80 | 30 | 50 | 75 | WP_001624706.1 |
| DUF2483 domain-containing phage protein | 2.36 | 0.01 | 43 | 35 | 97 | 12 | 0 | 7 | 0 | 11 | 10 | 33 | WP_001077279.1 |
| NAD-specific glutamate dehydrogenase | 2.83 | 0.01 | 100 | 0 | 97 | 60 | 0 | 0 | 0 | 0 | 30 | 50 | SAO39934.1 |
| phiPVL ORF050-like protein | 2.23 | 0.02 | 98 | 77 | 100 | 0 | 0 | 14 | 30 | 0 | 17 | 59 | ABD21348.1 |
| Staphylococcal enterotoxin type C1/U | 2.21 | 0.03 | 98 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 66 | 44 | WP_109162118.1 |
| Staphylococcal enterotoxin type G | 1.95 | 0.04 | 98 | 0 | 100 | 0 | 0 | 0 | 100 | 0 | 43 | 49 | WP_141060424.1 |
| Staphylococcal enterotoxin type N | 1.95 | 0.04 | 98 | 0 | 100 | 0 | 0 | 0 | 100 | 0 | 43 | 49 | WP_109162119.1 |
| Staphylococcal enterotoxin type I | 1.95 | 0.04 | 98 | 0 | 100 | 0 | 0 | 0 | 100 | 0 | 43 | 49 | QCW39073.1 |
| Multidrug transporter protein (SaPIbov3) | 2.31 | 0.04 | 99 | 77 | 100 | 0 | 0 | 14 | 30 | 0 | 17 | 59 | WP_065315972.1 |
| Intramembrane metalloprotease/(SaPIbov3) | 2.31 | 0.04 | 99 | 77 | 100 | 0 | 0 | 14 | 30 | 0 | 17 | 59 | WP_070008570.1 |
| Hypothetical protein (SaPIbov3) | 2.31 | 0.04 | 99 | 77 | 100 | 0 | 0 | 14 | 30 | 0 | 17 | 59 | WP_000921697.1 |
| Staphylococcal enterotoxin type M | 2.07 | 0.06 | 98 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 20 | 43 | WP_109162116.1 |
| SAS066 AgrD ( type II) | 2.52 | 0.06 | 98 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 47 | 46 | SCU54394.1 |
| Accessory gene regulator protein B4 (type II) | 2.52 | 0.06 | 98 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 47 | 46 | CZQ66246.1 |
| Hypothetical protein | 2.83 | 0.06 | 100 | 13 | 100 | 0 | 0 | 0 | 0 | 0 | 3 | 44 | WP_000389772.1 |
| TfoX/Sxy family protein | 2.83 | 0.06 | 100 | 13 | 100 | 0 | 0 | 0 | 0 | 0 | 3 | 44 | WP_000179903.1 |
| Arsenate reductase | 1.97 | 0.06 | 99 | 0 | 100 | 100 | 0 | 0 | 0 | 100 | 66 | 61 | WP_000163240.1 |
| Trypsin-like serine protease | 2.61 | 0.06 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 50 | 17 | WP_043054986.1 |
| SACOL0901 pathogenicity island protein | 2.82 | 0.07 | 98 | 88 | 100 | 84 | 0 | 14 | 30 | 28 | 27 | 71 | WP_109183239.1 |
| DUF1433 domain-containing protein | 3.42 | 0.07 | 98 | 98 | 100 | 100 | 0 | 0 | 100 | 0 | 63 | 79 | WP_031900638.1 |
aGene annotation by prokka and confirmed using BLAST search of gene sequence 4. Carriage rate of gene per clonal complex.
bOdds Ratio of causing clinical rather than subclinical mastitis from the Genome wide association study 5. Includes isolates belonging to CC9, CC50, CC5, CC49, CC7, CC45, CC101, CC20, CC30 and CC425.
cBest pairwise comparison P value from GWAS performed using scoary.
Furthermore, 61 genes were associated with SCM (i.e. an OR < 1), from which 10 genes matched our selection criteria and the OR of these genes ranged between 0.09 and 0.44 (Table 4). Most genes (8/10) coded for hypothetical proteins and the genes with predicted function were identified as an antitoxin YezG family protein (OR 0.32) and a putative DNA binding protein (OR 0.29). The SCM-associated genes were mostly carried by CC1, CC20, and CC8 S. aureus, but were always absent from CC479 and most CC151 isolates (Table 4).
Table 4.
Odds Ratio, carriage rate per clonal complex and GenBank references of genes associated with subclinical mastitis based on a genome wide association study performed on 276 S. aureus isolates obtained from bovine clinical and subclinical mastitis in 11 European countries.
| Genea | Odds ratiob | Best pairwise comparision pc | CC151 (%)d | CC97 (%) | CC479 (%) | CC133 (%) | CC398 (%) | CC1 (%) | CC20 (%) | CC8 (%) | Other CCs (%)e | Weighted average (%) | GenBank reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Putative DNA-binding protein | 0.29 | < 0.01 | 2 | 22 | 0 | 16 | 42 | 64 | 82 | 18 | 7 | 18 | AUM57693.1 |
| Hypothetical protein | 0.40 | < 0.01 | 0 | 94 | 0 | 0 | 100 | 100 | 100 | 100 | 34 | 42 | WP_000375476.1 |
| Hypothetical protein | 0.27 | 0.02 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 34 | 24 | WP_000070812.1 |
| Probable antitoxin YezG | 0.32 | 0.03 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 3 | 15 | WP_000142094.1 |
| Hypothetical protein | 0.38 | 0.03 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 62 | 27 | WP_078068548.1 |
| Hypothetical protein | 0.44 | 0.03 | 0 | 98 | 0 | 0 | 0 | 100 | 100 | 100 | 62 | 39 | WP_072426418.1 |
| Hypothetical protein | 0.32 | 0.04 | 0 | 50 | 0 | 0 | 100 | 7 | 0 | 91 | 52 | 26 | WP_000431307.1 |
| Hypothetical protein | 0.09 | 0.06 | 0 | 0 | 0 | 8 | 32 | 0 | 0 | 9 | 14 | 5 | WP_000993183.1 |
| Hypothetical protein | 0.26 | 0.06 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 100 | 34 | 13 | ETO57257.1 |
| Hypothetical protein | 0.43 | 0.06 | 1 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 86 | 39 | WP_078370397.1 |
aGene annotation by prokka and confirmed using BLAST search of gene sequence.
bOdds Ratio of causing clinical rather than subclinical mastitis from the Genome wide association study.
cBest pairwise comparison P value from GWAS performed using scoary.
dCarriage rate of gene per clonal complex.
eIncludes isolates belonging to CC9, CC50, CC5, CC49, CC7, CC45, CC101, CC20, CC30 and CC425.
Discussion
There is a large diversity in the carriage of virulence and AMR genes between bovine S. aureus lineages6,9, but it is unclear which genetic differences underly the observed variation in pathogenicity following bovine IMI. Therefore, this study aimed to identify genetic differences between S. aureus isolated from CM and SCM in dairy cattle. A secondary goal of the study was to describe the diversity of bovine S. aureus lineages in Europe and their carriage of immune evasion factors. We found CC479 to be strongly associated with CM rather than with SCM cases.
Although eighteen different CCs were present in our isolate collection, most S. aureus belonged to a limited number of CCs, with the five CCs (CC151, CC97, CC479, CC133 and CC398) making up more than 75% of all isolates. All these CCs have previously been associated with bovine mastitis6,19,27,28. The distribution of S. aureus CCs differed between geographical locations. Although the isolates in our collection were not a random sample, they did originate from 254 unique herds with a maximum of one CM and one SCM isolate from the same herd. The prevalence of S. aureus CCs per country from our study is in line with studies performed in Denmark29, Germany6 and The Netherlands30. Nevertheless, these isolates cannot be expected to fully represent the genetic diversity of bovine S. aureus on a national level. Because in addition to a relative low number of isolates, there likely was some clustering within certain regions in several of the countries, which may have resulted in an underestimation of the true genetic diversity in the population. We must also note that the aim of our sampling design was to collect isolates from an equal number of clinical and subclinical cases. This does not reflect the true population of S. aureus isolates in dairy herds, as the prevalence of subclinical infections is substantially higher than of clinical infections. Since CM and SCM are both expressions of S. aureus IMI and as SCM can develop into CM and vice versa31, isolates may have been misclassified, biasing any associations between CM versus SCM and S. aureus genotype towards the null-effect. Still, our study, as well as other studies using a similar classification of mastitis20,21, did identify genetic differences between CM and SCM S. aureus isolates, but possibly, some other associations may have been missed in our approach.
A key finding of the current study was the association between S. aureus belonging to CC479 and CM. This corresponds with a previous study which reported that experimental infection with CC479 S. aureus results in more severe clinical signs and a higher bacterial load than infection with a CC151 S. aureus strain32. In addition, the association between CC479 and CM was also observed in our previous analysis of Dutch mastitis isolates20. The underlaying mechanisms for this apparent increased virulence of CC479 remains unknown, but we have suggested a SNP in the repressor of toxins (rot) gene, resulting in an increased production of LukMF’ by CC479 isolates as a possible cause20. Our GWAS identified several genes associated with clinical outcome of IMI, but as S. aureus belonging to the same CC share a similar (virulence) gene profile, there was a strong link between carriage of these genes and lineage. Therefore, it was not possible to differentiate between the effect of individual genes/sets of genes and S. aureus lineage on the clinical outcome of IMI. Interestingly, genes associated with CM by GWAS which were all present in CC479 isolates, were also almost all present in CC151 S. aureus (Table 3), whereas the latter CC had an approximately even distribution of isolates originating from CM (51%) and SCM (49%). Since CC151 and CC479 share most CM-associated genes, these genes are likely spuriously associated to CM, due to confounding by other factors within CC479. Differences in gene expression are more likely causes of the increased virulence of CC479, in line with our hypothesis regarding the non-functional rot gene18. We confirmed that this mutation was present in all CC479 isolates within our collection (results not shown). It is likely that the absence of functional rot affects the expression levels of multiple virulence genes within CC479 S. aureus20, likely leading to substantially increased probability of causing CM. Future research, e.g. infection studies with genetically modified CC479 isolates lacking potential virulence factors or with restored rot function, are required to identify these potential causal mechanisms.
The results from our GLMM suggest that CC8 and CC1 are less likely to cause CM in cows and most of the genes that were associated with CM were carried by CC8 and CC1 isolates. Previous work identified that CC8 predominantly make up the S. aureus genotype B, a highly contagious subtype of S. aureus28,33. We only detected CC8 S. aureus among isolates from Italy and Germany, suggesting that this lineage is not widespread throughout Europe. Indeed, reports on CC8/GTB S. aureus primarily originate from Switzerland and Austria28,34,35. In contrast, the CC1 lineage was detected in six different countries in this study, although 50% of the CC1 isolates were collected in Poland.
Phylogenetic trees constructed based on core genome alignment and binary presence and absence of accessory genes clustered the isolates perfectly within the assigned CC. This demonstrates that MLST is an adequate genotyping technique, but Fig. 2 shows that considerable variation exists in the accessory genome among isolates within a CC. Interestingly, there was very limited variation in accessory gene carriage among CC479 and CC133 S. aureus isolates and this suggests a rapid clonal expansion of these specific S. aureus clones. The CC133 lineage is considered a ruminant-adapted lineage but is mostly associated with small ruminants16. Therefore, it is possible that the highly similar bovine CC133 S. aureus isolates represent a subgroup of CC133 S. aureus that is adapted to cattle and this subgroup, as well as the highly similar CC479 isolates, is considerably more successful at infecting the bovine mammary gland than other CC133, CC479 S. aureus.
Although our analyses showed associations between specific genes and CC with CM, it is important to note that none of these genes were essential for CM in our collection. Clones that were associated with CM cases based on CC or gene carriage were also cultured from SCM and vice versa. Besides pathogen factors, the clinical outcome of an IMI depends on host factors, such as breed, stage of lactation, environmental factors and their interaction31,36. Nevertheless, from a diagnostic perspective, the identification of S. aureus isolates with an increased risk of developing into CM (e.g. CC479) or highly contagious isolates (e.g. CC8) can support farmers and veterinarians in deciding on the most appropriate intervention strategy to control S. aureus in a herd. For Streptococcus uberis, it has been demonstrated that MALDI-TOF MS performed on milk samples can identify S. uberis isolates with increased CM risk37. However, it is not certain if this technique is able to distinguish between different CCs of S. aureus. There are several PCR based test that detect pathogen-specific genes in milk and an assay for the detection of CC8 (genotype B) S. aureus has already been developed44. We identified multiple CC-specific genes that could be employed to differentiate between common ruminant-associated S. aureus lineages (Supplementary Table S1).
In summary, this study identified that a limited number of S. aureus CCs are responsible for bovine mastitis in Europe and that CC479 is strongly associated with CM. Although our analysis shows specific genes are associated with CM, the presence of these genes in CC associated with SCM indicates it is likely that differential gene expression rather than gene carriage affects clinical presentation of IMI. This study shows that the type of infecting S. aureus influences the clinical outcome of IMI in dairy cattle. Therefore, identification of S. aureus CC can help predict the likelihood of the occurrence of CM following S. aureus IMI and highlights the potential benefit of diagnostics tools to identify S. aureus CC during bovine mastitis.
Methods
Bovine mastitis isolates
Twelve mastitis research groups or diagnostic labs from eleven countries (Belgium, Denmark, France, Germany, Hungary, Italy, Poland, Portugal, Spain, The Netherlands and United Kingdom) were asked to submit a convenience sample of approximately 30 S. aureus isolates obtained from cases of bovine mastitis. Participants were asked to submit a maximum of one CM and one SCM isolate per herd with as much as possible an equal number of CM and SCM isolates, coming from different regions in their country. The sample size was based on what was considered feasible to be collected within a reasonable period of time, while giving a good impression on the situation in a country. No formal sample size calculations were performed. For each isolate the sampling date, farm ID, geographical location of farm (city and/or region), cow ID, clinical manifestation (CM/SCM) was reported in a pre-structured Excel workbook (Microsoft Corp., Redmond, WA, USA).
Clinical mastitis was defined as visible signs of inflammation of the udder, indicated by a hard swollen quarter and/or abnormal milk; SCM was defined as a high somatic cell count (> 200,000 cells/mL) while no clinical signs of mastitis could be observed.. The definitions were communicated to all participating researchers and were stated in a workbook to ensure that a uniform definition of CM and SCM was applied by all participants. No constraints were put on duration or chronicity of infections. The clinical status (CM or SCM) was observed and recorded at the moment of sampling.
Isolates were recultured from their transport media onto to sheep blood agar plates, and incubated overnight at 37 °C. Next, single colonies were picked and grown in 2 mL Todd Hewitt Broth (THB) (Sigma, St. Louis, MO, USA) for 16 h at 37 °C with agitation. Bacterial glycerol stocks (25% glycerol) were made by adding 0.5 mL of bacterial broth to 0.5 mL 50% glycerol solution in distilled water. Furthermore, DNA was extracted using a simple boiling protocol to confirm bacterial species by PCR targeting the S. aureus specific femA gene, as described by Hoekstra et al.20. Isolates that were negative for the femA PCR or lacked mandatory metadata were excluded from the final collection. For each herd, a maximum of one isolate from a CM case and one from a SCM case was allowed and if multiple CM or SCM isolates were donated from the same herd, one was selected using the random number function of Excel 2015 (Microsoft, Redmond, WA, USA).
DNA extraction, genome sequencing and multilocus sequence typing
DNA for whole genome sequencing was extracted using the DNeasy UltraClean Microbial Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s instructions. Purity and DNA yield were measured by spectrophotometry and whole-genome sequencing was performed using Illumina HiSeq sequencing (Illumina Inc., San Diego, CA, United States). Multilocus sequence type was determined based on genome data. Each sequence type was assignment to a CC based on eBURST analysis using PHYLOViZ Online38.
Annotation of genomes, pan-genome analysis and phylogenetic analyses
After quality control of whole genome sequence results, genomes were annotated using prokka v1.1126 and the pan/core genome (genes present in 99% < of genomes) determined using roary v3.1239. Alignment of the core genome was performed using MAFFT v7.407 and the phylogenetic trees was built with Fasttree v2.140. Subsequently, trees were visualized using iTOL v3.641 and trees were rooted using the CC22 clade. The large-scale BLAST score ratio (LS-BSR) pipeline was used to obtain matrixes with BLAST score ratio of each annotated gene25. For each isolate, the presence of the genes encoding leukocidin LukMF’ (lukM, GenBank accession: 1262967; lukF′, GenBank accession: 1262954), ruminant-specific Staphylococcal complement inhibitor variant (scn, Genbank accession: ADN53656.1), SaPI encoded ruminant specific vWFbp variant (SaPI vWFbp, GenBank accession: HM234507.1), enterotoxin type I (seI, GenBank accession: EFC00985.1), enterotoxin L (seL, GenBank accession: BAO65763.1), toxic shock syndrome toxin 1 (tsst-1, GenBank accession: WP_001035596.1), penicillin-hydrolyzing class A beta-lactamase (blaZ, GenBank accession: WP_000733621.1), tetracycline resistance protein type M (tetM, GenBank accession: AKI94996.1) and penicillin binding protein 2A (mecA, GenBank accession: WP_104447100.1) was determined using LS-BSR output. Genes were identified using a threshold value of BSR of > 0.9 compared to the reference gene. In addition, a heatmap of LS-BSR score of isolates was created for genes annotated as SLL or SA by prokka26 using the pheatmap package42 of the R statistical software version v3.5.443.
Statistical analysis
The GLMM analysis was performed using the lme4 package44 of the R statistical software version v3.5.443. The model used clinical manifestation of mastitis (SCM, CM) as outcome variable and CC of S. aureus was a fixed effect. The country of origin of isolates was used as a random effect in the model. To reduce the number of levels within the variable CC, CCs represented by < 10 isolates were grouped together into a category ‘Other’. Furthermore, the association between CC and country of origin of mastitis isolates was investigated by Fisher’s exact test and association between genetic cluster and clinical manifestation was investigated using the Pearson's Chi-squared test. Both tests were performed using the R statistical software version v3.5.443.
GWAS
Based on the pangenome analysis in roary, GWAS was performed using scoary v1.6.1645. Default settings of scoary were used during our analysis with an initial threshold of naive p < 0.01. Genes that matched our inclusion criteria (BHP p < 0.1; best pairwise comparison p value of p < 0.1) were considered significant results of interest.
Supplementary information
Acknowledgements
This study was supported by the Medical Research Council (UK) grant MR/N002660/1.
Torben W. Bennedsgaard, Andrew Biggs, Sarne De Vliegher, Demetrio Herrera Mateo, Reglindis Huber-Schlenstedt, Jørgen Katholm, Péter Kovács, Volker Krömker, Guillaume Lequeux, Paolo Moroni, Luís Pinho, Sebastian Smulski, Karlien Supré, Jantijn M. Swinkels, Theo J. G. M. Lam and Gerrit Koop are members of the European Mastitis Panel.
Author contributions
G.K. and T.L. conceived and supervised the study project. T.B., A.B, S.V., D.M., R.H., J.K., P.K., V.K., G.L., P.M., L.P., S.S., K.S., J.H. and T.L. collected bovine S. aureus isolates from their respective countries. M.P. and J.H. carried out the experimental work and M.H. assisted in the whole genome sequencing of bacterial isolates. J.H. performed analysis on the genomic data and visualized results. A.Z. assisted with the interpretation of results and provided additional computational expertise. J.H. wrote the original manuscript and G.K., T.L., L.B., V.R., A.S., P.M., A.Z. and J.S. provided a critical review of the manuscript. T.L. and G.K. contributed equally to the study project.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-75179-2.
References
- 1.Aghamohammadi M, et al. Herd-level mastitis-associated costs on Canadian dairy farms. Front. Vet. Sci. 2018;5:100. doi: 10.3389/fvets.2018.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halasa T, Huijps K, Østerås O, Hogeveen H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007;29:18–31. doi: 10.1080/01652176.2007.9695224. [DOI] [PubMed] [Google Scholar]
- 3.Peton V, Le Loir Y. Staphylococcus aureus in veterinary medicine. Infect. Genet. Evol. 2014;21:602–615. doi: 10.1016/j.meegid.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Wellnitz O, Bruckmaier RM. The innate immune response of the bovine mammary gland to bacterial infection. Vet. J. 2012;192:148–152. doi: 10.1016/j.tvjl.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Zadoks RN, Middleton JR, McDougall S, Katholm J, Schukken YH. Molecular epidemiology of mastitis pathogens of dairy cattle and comparative relevance to humans. J. Mammary Gland Biol. Neoplasia. 2011;16:357–372. doi: 10.1007/s10911-011-9236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlotter K, et al. Leukocidin genes lukF-P83 and lukM are associated with Taphylococcus aureus clonal complexes 151, 479 and 133 isolated from bovine udder infections in Thuringia, Germany. Vet. Res. 2012;43:42. doi: 10.1186/1297-9716-43-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsay JA, Holden MTG. Understanding the rise of the superbug: Investigation of the evolution and genomic variation of Staphylococcus aureus. Funct. Integr. Genom. 2006;6:186–201. doi: 10.1007/s10142-005-0019-7. [DOI] [PubMed] [Google Scholar]
- 8.Magro G, et al. Virulence genes of S. aureus from dairy cow mastitis and contagiousness risk. Toxins (Basel). 2017;9:195. doi: 10.3390/toxins9060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monistero V, et al. Staphylococcus aureus isolates from bovine mastitis in eight countries: Genotypes, detection of genes encoding different toxins and other virulence genes. Toxins (Basel). 2018;10:247. doi: 10.3390/toxins10060247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koymans KJ, Vrieling M, Gorham RD, Jr, van Strijp JA. Staphylococcal immune evasion proteins: Structure, function, and host adaptation. Curr. Top. Microbiol. Immunol. 2016 doi: 10.1007/82_2015_5017. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y, et al. Staphylococcal superantigen-like protein 13 activates neutrophils via formyl peptide receptor 2. Cell. Microbiol. 2018;20:e12941. doi: 10.1111/cmi.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viana D, et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 2010;77:1583–1594. doi: 10.1111/j.1365-2958.2010.07312.x. [DOI] [PubMed] [Google Scholar]
- 13.Vrieling M, et al. Bovine Staphylococcus aureus secretes the leukocidin LukMF′ to kill migrating neutrophils through CCR1. MBio. 2015;6:e00335. doi: 10.1128/mBio.00335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliver SP, Murinda SE. Antimicrobial resistance of mastitis pathogens. Vet. Clin. N. Am. Food Anim. Pract. 2012;28:165–185. doi: 10.1016/j.cvfa.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Grumann D, Nübel U, Bröker BM. Staphylococcus aureus toxins—Their functions and genetics. Infect. Genet. Evol. 2014;21:583–592. doi: 10.1016/j.meegid.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Bar-Gal GK, et al. Host-specificity of Staphylococcus aureus causing intramammary infections in dairy animals assessed by genotyping and virulence genes. Vet. Microbiol. 2015;176:143–154. doi: 10.1016/j.vetmic.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Moller AG, Lindsay JA, Read TD. Determinants of phage host range in Staphylococcus species. Appl. Environ. Microbiol. 2019;85:e00209-19. doi: 10.1128/AEM.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper LP, et al. DNA target recognition domains in the Type I restriction and modification systems of Staphylococcus aureus. Nucleic Acids Res. 2017;45:3395–3406. doi: 10.1093/nar/gkx067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt T, Kock MM, Ehlers MM. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: Genetic diversity and inter-species host transmission. Front. Microbiol. 2017;8:511. doi: 10.3389/fmicb.2017.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoekstra J, et al. High production of LukMF′ in Staphylococcus aureus field strains is associated with clinical bovine mastitis. Toxins (Basel). 2018;10:200. doi: 10.3390/toxins10050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haveri M, Roslöf A, Rantala L, Pyörälä S. Virulence genes of bovine Staphylococcus aureus from persistent and nonpersistent intramammary infections with different clinical characteristics. J. Appl. Microbiol. 2007;103:993–1000. doi: 10.1111/j.1365-2672.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- 22.Pichette-Jolette S, et al. Partial prediction of the duration and the clinical status of Staphylococcus aureus bovine intramammary infections based on the phenotypic and genotypic analysis of isolates. Vet. Microbiol. 2019;228:188–195. doi: 10.1016/j.vetmic.2018.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Huijps K, Lam TJGM, Hogeveen H. Costs of mastitis: Facts and perception. J. Dairy Res. 2008;75:113–120. doi: 10.1017/S0022029907002932. [DOI] [PubMed] [Google Scholar]
- 24.Peters MDP, Silveira IDB, Fischer V. Impact of subclinical and clinical mastitis on sensitivity to pain of dairy cows. Animal. 2015;9:2024–2028. doi: 10.1017/S1751731115001391. [DOI] [PubMed] [Google Scholar]
- 25.Sahl JW, Caporaso JG, Rasko DA, Keim P. The large-scale blast score ratio (LS-BSR) pipeline: A method to rapidly compare genetic content between bacterial genomes. PeerJ. 2014;2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 27.Artursson K, Söderlund R, Liu L, Monecke S, Schelin J. Genotyping of Staphylococcus aureus in bovine mastitis and correlation to phenotypic characteristics. Vet. Microbiol. 2016;193:156–161. doi: 10.1016/j.vetmic.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 28.Boss R, et al. Bovine Staphylococcus aureus: Subtyping, evolution, and zoonotic transfer. J. Dairy Sci. 2016;99:515–528. doi: 10.3168/jds.2015-9589. [DOI] [PubMed] [Google Scholar]
- 29.Ronco T, et al. Genomic investigation of Staphylococcus aureus isolates from bulk tank milk and dairy cows with clinical mastitis. Vet. Microbiol. 2018;215:35–42. doi: 10.1016/j.vetmic.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Ikawaty R, et al. Characterization of Dutch Staphylococcus aureus from bovine mastitis using a multiple locus variable number tandem repeat analysis. Vet. Microbiol. 2009;136:277–284. doi: 10.1016/j.vetmic.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 31.Rainard P, et al. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transbound. Emerg. Dis. 2017;65:149–165. doi: 10.1111/tbed.12698. [DOI] [PubMed] [Google Scholar]
- 32.Vrieling M, et al. LukMF′ is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci. Rep. 2016;6:37759. doi: 10.1038/srep37759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leuenberger A, et al. Genotypes of Staphylococcus aureus: On-farm epidemiology and the consequences for prevention of intramammary infections. J. Dairy Sci. 2019;102:3295–3309. doi: 10.3168/jds.2018-15181. [DOI] [PubMed] [Google Scholar]
- 34.van den Borne BHP, et al. A longitudinal study on transmission of Staphylococcus aureus genotype B in Swiss communal dairy herds. Prev. Vet. Med. 2017;136:65–68. doi: 10.1016/j.prevetmed.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Cosandey A, et al. Staphylococcus aureus genotype B and other genotypes isolated from cow milk in European countries. J. Dairy Sci. 2016;99:529–540. doi: 10.3168/jds.2015-9587. [DOI] [PubMed] [Google Scholar]
- 36.De Vliegher S, Fox LK, Piepers S, McDougall S, Barkema HW. Invited review: Mastitis in dairy heifers: Nature of the disease, potential impact, prevention, and control. J. Dairy Sci. 2012;95:1025–1040. doi: 10.3168/jds.2010-4074. [DOI] [PubMed] [Google Scholar]
- 37.Archer SC, Bradley AJ, Cooper S, Davies PL, Green MJ. Prediction of Streptococcus uberis clinical mastitis risk using Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) in dairy herds. Prev. Vet. Med. 2017;144:1–6. doi: 10.1016/j.prevetmed.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ribeiro-Gonçalves B, Francisco AP, Vaz C, Ramirez M, Carriço JA. PHYLOViZ Online: Web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 2016;44:W246–W251. doi: 10.1093/nar/gkw359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Page AJ, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I, Bork P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolde, R. CRAN—Package pheatmap. https://cran.r-project.org/web/packages/pheatmap/index.html (2019).
- 43.R Development Core Team 3.0.1. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing vol. 2. https://www.r-project.org (2013).
- 44.Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 45.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17:238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.