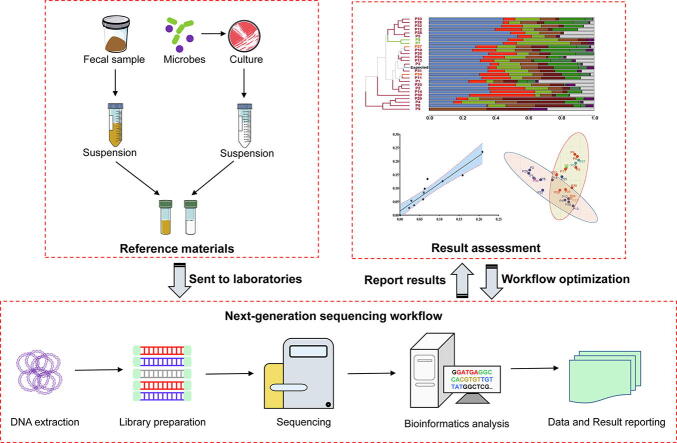
Graphical abstract
Keywords: Shotgun metagenomic sequencing, 16S rRNA gene sequencing, Microbial community profiling, Microbiome, Microbiota
Abstract
Introduction
Microbiome research based on high-throughput sequencing has grown exponentially in recent years, but methodological variations can easily undermine the reproducibility across studies.
Objectives
To systematically evaluate the comparability of sequencing results of 16S rRNA gene sequencing (16Ss)- and shotgun metagenomic sequencing (SMs)-based microbial community profiling in laboratories under routine conditions.
Methods
We designed a multicenter study across 35 participating laboratories in China using designed mock communities and homogenized fecal samples.
Results
A wide range of practices and approaches was reported by the participating laboratories. The observed microbial compositions of the mock communities in 46.2% (12/26) of the 16Ss and 82.6% (19/23) of the SMs laboratories had significant correlations with the expected result (Spearman r>0.59, P <0.05). The results from laboratories with near-identical protocols showed slight interlaboratory deviations. However, a high degree of interlaboratory deviation was found in the observed abundances of specific taxa, such as Bacteroides spp. (range: 0.3%-53.5%), Enterococci spp. (range: 0.8%-43.9%) and Fusobacterium spp. (range: 0.1%-39.8%). SMs performed better than 16Ss in detecting low-abundance bacteria (B. bifidum). The differences in DNA extraction methods, amplified regions and bioinformatics analysis tools (taxonomic classifiers and database) were important factors causing interlaboratory deviations. Addressing laboratory contamination is an urgent task because various sources of unexpected microbes were found in negative control samples.
Conclusions
Well-defined control samples, such as the mock communities in this study, should be routinely used in microbiome research for monitoring potential biases. The findings in this study will provide guidance in the choice of more reasonable operating procedures to minimize potential methodological biases in revealing human microbiota composition.
Introduction
Due to the ever-evolving development of analytical methodologies, the microbiome, especially the gut microbiome, has become a hot topic in biomedical research and now represents one of the most studied and interesting fields in medicine. Mounting evidence has linked changes in the composition and activity of the microbiota (especially the gut microbiota) to a wide range of diseases and ecological phenotypes, such as diabetes [1], obesity [2], colorectal cancer [3], liver cirrhosis [4], rheumatoid arthritis [5] and severe depression [6]. Although the knowledge to date of a causative role for any of the microbial members detected in these approaches is still very limited, it can be expected that human microbiome data will be transformed into biomarkers related to the diagnosis or prognosis of human diseases in the near future [7].
Currently, most bacterial and archaeal taxa across diverse biomes remain uncultured [8], [9], restricting the possibility of characterizing the full picture of environmental microbial communities through culture techniques. Fortunately, increasingly powerful next-generation sequencing (NGS) technologies are allowing us to pry deeper and more clearly into the structure, function and diversity of the human microbiome without prior culturing [10]. 16S rRNA gene sequencing (16Ss) and shotgun metagenomic sequencing (SMs) are the two main NGS tools implemented for microbial community profiling. 16Ss is used to identify and classify microbes by selectively amplifying and sequencing the hypervariable regions of the 16S rRNA gene. As 16Ss is high throughput (ten to hundreds of microbiotas in a single sequencing run) [11], is cost effective [12] and has increasingly accessible bioinformatics tools [13], it has become a widely deployed method for profiling complex microbial communities [14], [15]. SMs sequences the genomes of all the microbes isolated from the entire microbial community. Its advantage lies in the capacity for strain-level reconstruction in the taxonomic analysis and for the functional annotation with pathway predictions of the studied microbiome [16], [17], [18].
Unfortunately, microbiome studies based on these sequencing strategies published at an exponential rate over the past several years have been documented to be difficult to reproduce across independent studies [16], [17], [18]. Significant variation has been reported even across studies of the same disease [20], [21]. For example, multiple studies demonstrated an elevated Firmicutes/Bacteroidetes ratio in obese subjects [22], [23], [24], but some other studies reported weak associations between this ratio and obesity [25], [26], [27]. Regarding the previous microbiome and colorectal cancer (CRC) studies, Feng et al. found that the gene and genus richness of the gut microbiota in CRC patients was significantly higher than that in the control group [28], while, in another study, CRC patient microbiomes exhibited reduced gene richness and gene alpha diversity [3]. A recent systematic review that was based on 13 case-control studies investigating gut microbiota differences between Parkinson's disease (PD) patients and controls showed that the abundance of butyrate-producing species was decreased in the PD group compared to that in controls in only 9 studies [29]. Moreover, several subversive views on the human microbiome, such as the presence of microbes in the blood and placenta, have been strongly questioned by different researchers [29]. These conflicting reports have triggered a “reproducibility crisis” in the microbiome field, undermining the credibility of microbiome science and delaying its translation [33], [34]. In addition to differences in study populations, the irreproducibility of metagenome analysis stems mainly from a wide range of experimental variables at all steps of the experiment workflow, including sample handling and nucleic acid extraction [35], [36], primer choice (for 16Ss) [36], sequencing strategies [37] and bioinformatics analysis [38].
With the rapid development of sequencing technology, an increasing number of laboratories have established private procedures for microbial community profiling, providing us with additional choices of sequencing platforms for the study of the relationship between the human microbiome and diseases. However, with a lack of standards in metagenomic data generation and processing, do their results still have high interlaboratory variability when analyzing identical specimens as that reported in previous studies [37], [38]? To systematically evaluate the comparability and accuracy of microbial community profiling detected with 16Ss and SMs techniques in different laboratories and discover sources of variation that affect the accuracy of test results, we conducted a multicenter evaluation study in 2019 among 35 individual laboratories in China using designed mock samples and fecal samples. The results objectively reflect the existing issues in current microbial community diversity analysis.
Materials and methods
Study design
The National Center for Clinical Laboratories (NCCL) organized this multicenter quality evaluation study. A total of 35 laboratories from China that had developed workflows and routinely performed 16Ss- and/or shotgun metagenomic sequencing-based microbial community profiling volunteered to participate in this activity. Among these laboratories, 14 participated in the quality assessment of both methods, 9 laboratories participated in only the quality assessment of the 16S rRNA gene sequencing method, and the other 12 laboratories participated in only the quality assessment of the metagenomic sequencing method. Each of the participating laboratories was sent one or two sample sets on dry ice. One sample set contained five types of physical samples prepared by the NCCL: (1) Sample 201901 (total volume, 1 ml; microbial count, 6 × 108 cell). A microbial cell mock community consisted of 6 g-positive bacterial species (Bifidobacterium bifidum, Clostridium beijerinckii, Clostridium butyricum, Cutibacterium acnes, Enterococcus faecalis and Lactobacillus gasseri) and 5 g-negative bacterial species (Bacteroides thetaiotaomicron, Bacteroides fragilis, Enterobacter hormaechei, Escherichia coli and Fusobacterium nucleatum). These 11 bacteria belong to five phyla and eight genera. Except for one microbe, Cutibacterium (formerly Propionibacterium) acnes, which is a member of the normal human skin microbiota, the other 10 bacterial strains are generally present in the healthy gut microflora; (2) Sample 201902 (total volume, 100 ul; DNA amount, 2.8 μg). A DNA mixture of the 11 bacteria in sample 201901. The DNA amount (ng) of each bacterium was theoretically consistent with that in 201901 (approximately 2.8 μg); (3) Sample 201903 (total volume, 100 ul; microbial count, approximately 1.17 × 109 cell). A homogenized human stool sample; (4) Sample 201904 (total volume, 100 ul). A sample with quantitative amounts (3 × 107 cells) of F. nucleatum species added to Sample 201903; and (5) Sample 2019NC (total volume, 1 ml). A negative sample consisted of 1 ml sterile PBS buffer. All the details of sample preparation (e.g., bacterial culture and spiking, stool sample collection and handling, Sample aliquoting and storage) and microbial quantification strategies (e.g., droplet digital PCR (ddPCR), high-sensitivity dsDNA assay, agarose gel electrophoresis) are available in the supplementary material (Supplementary methods).
A questionnaire focused on the methodological aspects of their operating procedures for microbiome profiling was also delivered to the laboratories alongside the sample sets. All the samples were stored at −80 °C prior to handling in the participating laboratories and were tested and analyzed within one month. The results of taxonomic classification of each sample were reported to the NCCL. The NCCL compared the results reported by each laboratory with the expected results to evaluate their accuracy and comparability and further analyzed the methodological differences in different laboratories to find the factors that caused the interlaboratory deviations.
Statistical analysis
We calculated the Spearman rank correlation coefficient to identify the correlation between the results reported in participating laboratories and the theoretical results in mock communities (samples 2001901 and 201902). A P value < 0.05 indicates a correlation between them. The higher the r value is, the stronger the correlation. To evaluate the factors that may cause interlaboratory deviations (e.g., DNA extraction methods, PCR amplified regions, sequencers and bioinformatics analysis tools), principal component analysis (PCA) and permutational multivariate analysis of variance (PERMANOVA) over the Bray-Curtis distances were performed based on the relative microbial abundance data of the feces sample 201903 generated by participants using 16S rRNA gene sequencing at the genus level and shotgun metagenomic sequencing at the species level.
Results
Methodological variance in participating laboratories
To assess the level of methodological variance in the routine testing process of participating laboratories, each laboratory was required to complete predesigned electronic questionnaires for both sequencing methods, recording the details of reagents, instruments, and software and their corresponding parameters in performing tests. We summarized the recorded data from all the participants (Table 1). The detailed findings are summarized in the Supplementary material (Supplementary Results). Briefly, the operation procedures established for microbiome analysis among the participants are very different, including a wide variety of different methodological approaches for DNA extraction (with or without a bead-beating step), different PCR primers for 16Ss, multiple types of sequencing platforms (Illumina, MGI, Ion Torrent, PacBio and others), and various bioinformatics pipelines and reference databases. Based on their specific standard operating procedures (SOPs), the final sequencing data generated by each laboratory were significantly different. For 16Ss, the median was 0.07 G (interquartile range (IQR) 0.0034G-7G), that for SMs was approximately 8G (IQR 0.23G-19.4G).
Table 1.
Methodological variance reported by the participating laboratories.
| 16Ss (26 laboratories) |
SMs (23 laboratories) |
||
|---|---|---|---|
| N | N | ||
| 1. DNA extraction kit manufacture | 1. DNA extraction kit manufacture | ||
| Qiagen | 9 | Qiagen | 7 |
| Tiangen | 3 | Tiangen | 3 |
| Zymo Research | 3 | Zymo Research | 2 |
| Omega | 3 | Omega | 3 |
| Other/Custom | 8 | Other/Custom | 8 |
| 2. bead-beating included in cell wall disruption? | 2. Bead-beating included in cell wall disruption? | ||
| Yes | 19 | Yes | 14 |
| No | 7 | No | 9 |
| 3. Amplified region of 16S rRNA gene | 3. DNA fragmentation method | ||
| V3-4 | 13 | Enzymatic | 13 |
| V4 | 6 | Physical (ultrasound) | 10 |
| V1-9 | 3 | 4. Sequencing Platform | |
| V4-5 | 1 | Illumina NovaSeq | 9 |
| V1-2 | 1 | Illumina Hiseq | 4 |
| V2, V3, V4, V6-7, V8, V9 | 2 | MGISEQ-2000 | 3 |
| 4. Sequencing Platform | Illumina MiSeq | 2 | |
| Illumina NovaSeq | 3 | DA8600 | 2 |
| Illumina MiSeq | 14 | NextSeq CN500 | 2 |
| Illumina Hiseq | 5 | MGISEQ-200RS | 1 |
| Ion Torrent PGM | 3 | 5. Sequencing Mode | |
| PacBio Sequel | 1 | Paired-end | 18 |
| 5. Sequencing Mode | Single-end | 5 | |
| Paired-end | 22 | 6. Sequencing read length | |
| Single-end | 4 | 100 | 4 |
| 6. Sequencing read length | 150 | 15 | |
| 150 | 5 | 200 | 2 |
| 250 | 10 | 300 | 2 |
| 300 | 7 | 7. Taxonomic classifier | |
| 400 | 1 | MetaPhlAn2 | 12 |
| 600 | 2 | Kraken 2 | 5 |
| 68,866a | 1 | SOAPaligner/soap2 | 3 |
| 7. Data Analysis Pipeline | Diamond 0.9.27 | 2 | |
| Qiime | 11 | Explify V2.1.0 (IDbyDNA Inc.) | 1 |
| USEARCH | 7 | 8. Reference Databases | |
| Parallel-META Pipeline | 1 | Default database in taxonomic classifiers or NCBI nr database | 20 |
| Mother | 3 | MetaHIT database | 3 |
| EzBioCloud | 1 | ||
| Ion Reporter™ | 3 | ||
| 8. Reference Databases | |||
| Greengenes | 11 | ||
| SILVA | 10 | ||
| NCBI 16S rRNA database | 1 | ||
| PrecisionGene Database (PRS-DB) | 2 | ||
| EzBioCloud 16S database | 1 | ||
| Custom | 1 | ||
Generated using a third generation of sequencer (PacBio Sequel) by P24.
Variations in mock communities at the genus level by 16Ss
Overview of the reported data
In sample 201901, only 7 laboratories (26.9%, 7/26) detected all 9 genera of bacteria. In addition to the low-abundance Bifidobacterium spp., which was undetected by 50% (13/26) of laboratories, Enterobacter spp. had the lowest detection rate (61.5%, 16/26), followed by Escherichia spp. (unreported in 2 laboratories) and Clostridium spp. (unreported in 1 laboratory) (Supplementary Dataset 1).
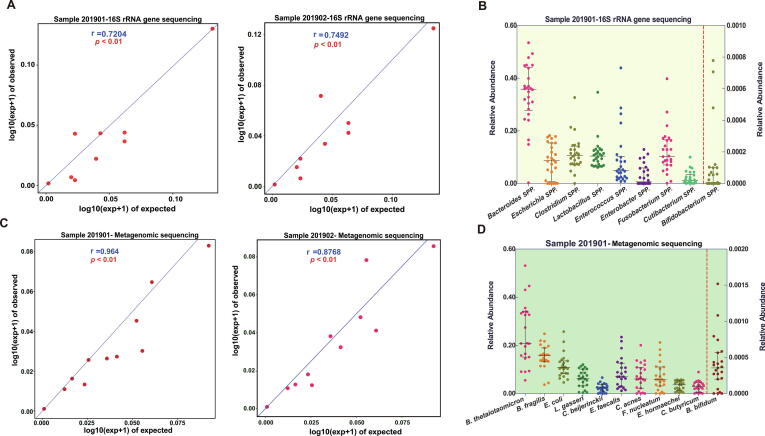
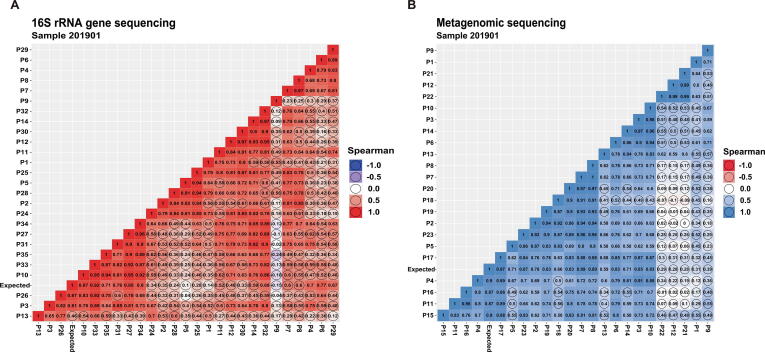
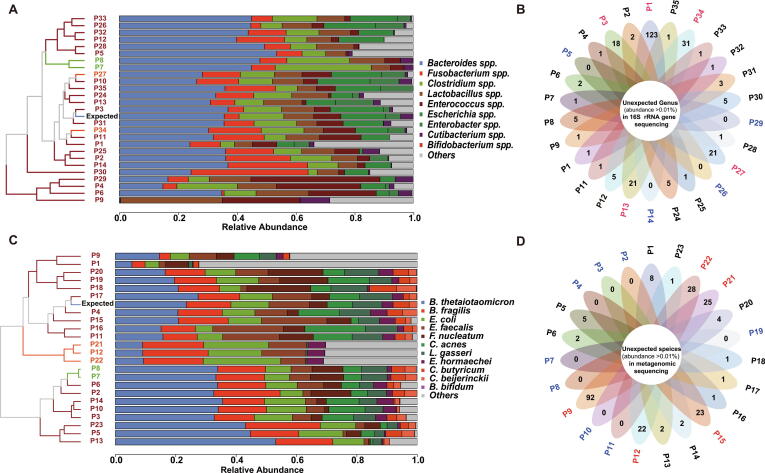
Taking the reported results of all laboratories as a whole, a moderate correlation (Spearman rank correlation coefficient r > 0.72, P < 0.01) was found between the median observed microbial abundances and the expected microbial abundances in samples 201901 or 201902 (Fig. 1A). However, the observed relative abundance of each bacterium varied greatly from laboratory to laboratory, especially that of Bacteroides spp. (range: 0.3% (P9)–53.5% (P5)), Enterococcus spp. (range: 0.8% (P5)–43.9% (P29)) and Fusobacterium spp. (range: 0.1% (P9)–39.8% (P30)) (Fig. 1B, Supplementary Dataset 1). Spearman's rank correlation showed moderate or strong significant correlations between the reported results of 46.2% (12/26) of laboratories and the expected result (range of r value: 0.67–0.92, Fig. 2A). The interlaboratory differences in reported microbiota composition could be intuitively reflected in a clustered histogram (Fig. 3A). P7 and P8 performed microbiome analysis using the same protocol, and the final results were very related (r = 0.97) (Figs. 2A, 3A). P27 and P34 utilized similar procedures as well. Their results were highly correlated (r = 0.96) and very close to the expected composition (r > 0.8). In contrast, several laboratories reported microbial compositions that were very different from the expected results due to insufficient detection of certain bacteria. For example, the results from P9 did not show any significant correlation with those of the other laboratories (all P values >0.05, Fig. 2A). This finding was likely due to the almost complete lack of Bacteroides spp. (0.1%) and the complete lack of Clostridium spp. (Fig. 3A, Supplementary Dataset 1).
Fig. 1.
The analysis of the observed results of mock communities in 35 laboratories. Spearman rank correlation coefficients were calculated for both the 16Ss (A) and SMs laboratories (C) to identify the correlation between the median observed microbial abundances and the expected microbial abundances in the samples 201901 and 201902. The scatter plot shows that the observed relative abundance of each bacterium at the genus level for 16Ss (B) and at the species level for SMs (D) varied greatly among laboratories. The observed relative abundance of the designed low-abundance Bifidobacterium spp. in 16Ss or B. bifidum in SMs is displayed on the right axis. The line displays the interquartile range (lower quartile to upper quartile).
Fig. 2.
The correlation between the results for the sample 201901 reported by any two 16Ss laboratories (A) or SMs laboratories (B) was evaluated by Spearman's rank correlation coefficient. The number in every square represents the Spearman r value. The higher the r value is, the stronger the correlation. A square marked with a black circle means that there is no correlation between the results of the two corresponding laboratories (P value >0.05). “Expected” indicates the designed value by the NCCL.
Fig. 3.
The clustered histogram intuitively reflects the interlaboratory differences in the reported microbiota composition in the sample 201901 (A for 16Ss at the genus level and C for SMs at the species level). The number of unexpected bacteria with a relative abundance >0.01% was counted in every laboratory (B for 16Ss at the genus level and D for SMs at the species level). The number on each petal refers to the identified unexpected bacteria for the corresponding laboratory.
The presence of unexpected bacteria is a matter of concern. In this study, 25 laboratories reported unexpected genera (Supplementary Dataset 2). The number of bacteria with an abundance >0.01% in every laboratory varied from 1 to 123 (Fig. 3B). The most frequent was Kocuria spp. (Table 2), which were reported by 17 (65.4%, 17/26) laboratories and were further aligned to the species Kocuria kristinae in most of the laboratories. This bacterium is a normal skin-resident gram-positive (G+) facultative anaerobic bacterium [37], [38], indicating the possibility of the presence of exogenous contamination during sample processing. The other unexpected bacteria were mostly intestinal bacteria. They might have originated from mismatches in sequence alignment, as the sequenced 16S rRNA gene is very similar in multiple bacteria. Notably, the occurrence of laboratory cross-contamination among samples processed in the same batch might be another cause.
Table 2.
Unexpected bacteria found in sample 201901.
| 16 s rRNA gene sequencing |
Shotgun metagenomic sequencing |
||
|---|---|---|---|
| Bacterium (Genus) | No. of Laboratory | Bacterium (Species) | No. of Laboratory |
| Kocuria sp. | 17 | Klebsiella pneumoniae | 10 |
| Citrobacter sp. | 9 | Bacteroides ovatus | 7 |
| Blautia sp. | 8 | Bacteroides vulgatus | 7 |
| Klebsiella sp. | 8 | Bacteroides caccae | 6 |
| Collinsella sp. | 7 | Bacteroides cellulosilyticus | 6 |
| Salmonella sp. | 7 | Lactobacillus johnsonii | 6 |
| Roseburia sp. | 6 | Parabacteroides distasonis | 6 |
| Ruminococcus sp. | 6 | Others | ≤5 |
| Othersa | ≤5 | ||
Full list is showed in supplementary Table S2.
Potential sources of variation
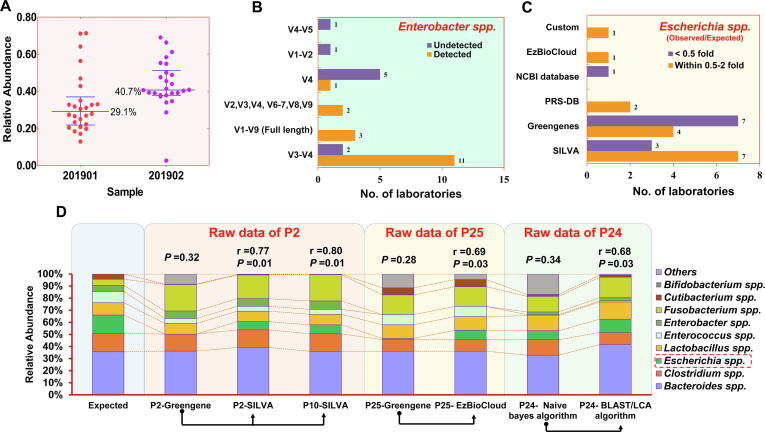
Previous studies have shown that different DNA extraction approaches (kits) vary in efficiency for different types of microbes, especially for hard-to-lyse microbes (such as G+ bacteria), leading to significant deviations in DNA yield and bacterial composition [40], [41], [42]. This issue is ubiquitous in the laboratories participating in this evaluation study. The cumulative proportion of G+ bacteria detected in the genome mixed sample (201902) in 84.6% (22/26) of the laboratories was higher than that in the corresponding whole-cell mixed sample (201901), and the median abundance among all laboratories was also higher (40.7% vs 29.1%) (Supplementary Dataset 1, Fig. 4A), indicating that the yield of G+ bacteria lost more than that of gram-negative bacteria during the extraction process.
Fig. 4.
Potential sources of variation in 16Ss. (A) The cumulative relative abundance of G+ bacteria detected in the samples 201901 and 201902. Each point represents a participating laboratory. The line displays the interquartile range (lower quartile to upper quartile). The performance of the 16Ss laboratories in detecting Enterobacter spp. using different amplification regions (B) and in identifying Enterobacter spp. using different reference databases (C). (D) More accurate results were obtained by reanalyzing the raw data of the sample 201901 reported by several laboratories (P2, P25 and P24). The raw data of P2 were reanalyzed by P2 and P10 with pipelines including the SILVA database. The raw data of P25 were reanalyzed using the EzBioCloud database. The raw data of P24 were reanalyzed by changing the annotation algorithm. The correlations between the observed microbial abundances and the expected microbial abundances were calculated, and the corresponding Spearman's R and P values are shown above the histograms.
The chosen PCR primers are also critical determinants of the final bacterial sequence profiles [43]. In this study, primers targeting multiple different regions of the 16S rRNA gene were used. From the reported results, we observed that Enterobacter spp. might be significantly affected by amplification region bias. This genus was identified in 11 (84.6%) of the 13 laboratories that used V3-V4 region amplification primers and in all laboratories using the full length (V1-V9 regions; P6, P24, and P26) or nearly full length (7 hypervariable regions; P7 and P8) of the 16S rRNA gene. However, no Enterobacter spp. was detected in 4 (80%) of the 5 laboratories using the V4 primer alone in all samples. The primers for the V1-V2 or V4-V5 regions failed to detect this genus as well (Fig. 4B).
Escherichia coli is the most common microbe in gut microbiota. However, Escherichia spp. were underrepresented <2-fold in as many as 42.3% (11/26) of laboratories when using 16Ss in this study. In contrast, with SMs, only 2 laboratories had abundances slightly less than 1/2 of the expected 12.6% (0.37-fold in P1, 0.46-fold in P16) (Supplementary Dataset 3). We reviewed the results reported by all laboratories and found that different databases performed differently in the accurate identification of Escherichia spp. (Fig. 4C). Through replacing the initially used reference database, we reanalyzed the sequencing data of two laboratories where Escherichia spp. was completely omitted in all of the sample sets (P2) or strongly underestimated (1%; <10-fold) (P25). The final results showed that the relative abundance of Escherichia spp. increased, and importantly, the reanalysis did not introduce bias to other species (Fig. 4D, Supplementary Dataset 4–5), indicating that the choice of reference database does affect the effective identification of certain taxa. Thus, we highlight that when analyzing the taxa of interest, researchers should take into account the potential analysis errors and bias caused by the untimely updating of the database (such as the Greengenes database, which has not been updated science 2013).
In addition, P24 effectively reduced the proportion of unclassified sequences by changing the annotation algorithm (Fig. 4D). In summary, we found the above factors affecting the reproducibility of 16Ss by analyzing the reported data. Although these findings need further verification, they can still play a guiding role in the laboratory optimization of process variables.
Variations in mock communities at the species level by SMs
Overview of the reported data
Compared with 16Ss, SMs was more sensitive in species identification. Eighteen (78.3%) of the 23 SMs laboratories were able to detect all 11 target bacteria mixed in the sample 201901 (Supplementary Dataset 1). A total of 91.3% (21/23) SMs laboratories successfully detected low-abundance B. bifidum (0.02%), while as described above, up to half of the 16Ss laboratories failed to detect this bacterium even at the genus level. The median observed microbial composition in the mock sample 201901 or 201902 by SMs showed a higher correlation (Spearman r > 0.87; P < 0.01) with the expected value than that analyzed by 16Ss (Fig. 1C). The observed relative abundance variation of B. thetaiotaomicron was the largest, ranging from 5.5% (P1) to 53% (P13) (Fig. 1D). The results of 82.6% (19/23) of laboratories showed significant correlations with the expected result (range of r values: 0.59–0.97; P < 0.05) (Fig. 2B). Similar to those of 16Ss, the results analyzed by the near-identical SMs workflow were very similar (P7 and P8; P12, P21 and P22) (Fig. 3C). Due to the existence of a high proportion of unclassified sequences or unexpected species (“others” in Fig. 4B), the final results of 5 laboratories (P1, P9, P12, P21 and P22) were very different from the expected result.
The issue of false positive results was also observed in SMs (Supplementary Dataset 2). We counted the reported species with a relative abundance >0.01% and found that 65.2% (15/23) of laboratories had unexpected bacteria, of which 5 laboratories reported more than 20 species (range: 22–92 species) (Fig. 3D). Overall, Streptococcus pneumoniae was the most frequent unexpected species reported by 15 (43%, 10/23) laboratories (Table 2). The following were 4 Bacteroides species, B. ovatus, B. vulgatus, B. caccae and B. cellulosilyticus, which are part of the indigenous microflora of the gastrointestinal tract and closely genetically related to the Bacteroides species (B. thetaiotaomicron and B. fragilis) mixed in the sample 201901 [44]. Thus, it cannot be ruled out that the unexpected annotation of the four bacteria was caused by mismatches of similar sequences from B. thetaiotaomicron or B. fragilis.
Potential sources of variation
As observed with the 16Ss, SMs results were also affected by biases caused by nucleic acid extraction (Fig. S1). Laboratories need to design experiments to optimize the sample handling and nucleic acid extraction processes and avoid significant differences in results [45].
As described above, the sequences that failed to be assigned to the 11 target bacteria in the sample 201901 accounted for a high proportion (from 30.3% to 72.3%) in five laboratories (P1, P9, P12, P21 and P22). We compared their protocols and found that the deviation might be caused by the taxonomic classifier and database used. In detail, P1 and P9 carried out the taxonomic classification using DIAMOND software, which is a DNA-to-protein classifier [46]. Broadly, protein classifiers have many more unclassified reads than DNA-to-DNA classifiers (such as Kraken 2) because the former target only the coding sequence of the genome and, therefore, will not be able to classify noncoding sequencing reads [47]. P12, P21 and P22 used exactly the same protocol for microbiome analysis. The reference database was the MetaHIT gene catalog, which was established by the MetaHIT research team and included only human gut nonredundant microbial genes [48]. Thus, compared with the more comprehensive NCBI taxonomy database, this database will also miss noncoding sequences and cannot identify microbes other than intestinal bacteria (e.g., Cutibacterium acnes was not detected in this study).
Detection of the spiked F. nucleatum in fecal samples
Seven (26.9%) of the 16Ss laboratories reported Fusobacterium spp. in the fecal sample 201903, with the relative abundance in each laboratory lower than 0.15%. By contrast, all laboratories could detect Fusobacterium spp. in the sample 201904, with F. nucleatum species added in advance, and the abundance was higher than that in the paired sample 201903 (Supplementary Dataset 6). However, the relative abundance varied greatly among laboratories, ranging from 0.015% (P9) to 17.1% (P24) (Supplementary Fig. S2A). Further analysis revealed that only 13 (50%) laboratories were able to identify F. nucleatum to the species level in the sample 201904 (Supplementary Dataset 7). The workflows of other laboratories did not show species-level identification capabilities for the colorectal cancer-related bacterium.
Through SMs, low-abundance F. nucleatum in the sample 201903 could be identified in 11 laboratories. In the sample 201904, each laboratory reported F. nucleatum. Although the range of calculated abundances was wide, as it was with 16Ss, the median value (2.35%) was closer than that for 16Ss (3.9%) to the estimated value of 2.5%, as shown in Supplementary Fig. S2B.
Variations in fecal samples
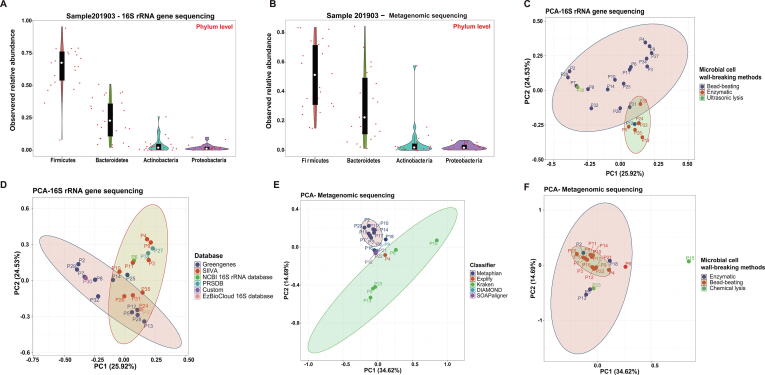
Overall, the detection results by the two methods in fecal samples (201903) were relatively consistent at the phylum level. However, the observed relative abundances of the dominant two bacterial phyla (Firmicutes and Bacteroides) in each laboratory responded differently to the protocols they used (Fig. 5A and B). To investigate potential causes that led to interlaboratory deviations in the final microbial compositions, principal component analysis (PCA) was conducted on the relative microbial abundance data of the sample 201903 generated by participants using 16Ss at the genus level and SMs at the species level. For 16Ss, PCA was able to visibly discriminate the profiles originating from laboratories adding a bead-beating step and those using only enzymatic lysis methods for microbial cell wall disruption (Fig. 5C). The results generated by the SILVA database and those by the Greengenes database also tended to cluster in two distinct groups (Fig. 5D). However, other factors, such as the DNA extraction kit, amplification primers, sequencing technology, and taxonomic classifier, did not seem to exhibit a clear direct impact on the microbial dissimilarities (Supplementary Fig. S3) since the grouped results were irregularly distributed on the ordination plot. These findings were further confirmed by PERMANOVA. Reference databases (R2 = 0.40, P = 0.001) and microbial cell wall disruption methods (R2 = 0.18, P = 0.008) were both factors affecting interlaboratory reproducibility (Table 3). With the same statistical analysis, we found that the interlaboratory variations that occurred among the microbial taxa identified by SMs could be explained by multiple factors, such as DNA extraction methods (wall disruption methods and extraction kits), taxonomic classifiers (MetaPhlAn and Kraken) and databases (Fig. 5E, F; Table 3). However, we did not observe significant variation introduced by different types of sequencers (P = 0.17) (Table 3).
Fig. 5.
Variations detected in the fecal sample 201903. Violin plot showing the results reported by 16Ss (A) and SMs (B) laboratories for the dominant 4 phyla. PCA analysis was applied for the evaluation of variations introduced by microbial cell wall disruption methods (C) and reference databases (D) in 16Ss laboratories and taxonomic classifiers (E) and microbial cell wall disruption methods (F) in SMs laboratories. For 16Ss, the results of bead-beating strategies could be clearly distinguished from those of enzymatic lysis methods (C) and the results generated by the SILVA database and those by the Greengenes database also tended to cluster into distinct groups (D). For SMs, the results generated by two taxonomic classifiers (MetaPhlAn and Kraken) were visibly discriminated (E) and the results produced using bead-beating methods are clustered together (F).
Table 3.
Effect sizes and explained variances of the main factors assessed by PERMANOVA analysis.
| Factors | Sums Of Sqs | Mean Sqs | F. Model | R2 | P value | |
|---|---|---|---|---|---|---|
| 16S rRNA gene sequencing | Databases | 1.91 | 0.38 | 2.67 | 0.40 | 0.00 |
| Microbial cell wall-breaking methods | 0.84 | 0.42 | 2.45 | 0.18 | 0.01 | |
| Classifiers | 1.40 | 0.23 | 1.31 | 0.29 | 0.11 | |
| DNA extraction kits | 1.25 | 0.21 | 1.13 | 0.26 | 0.27 | |
| Sequencers | 0.78 | 0.19 | 1.02 | 0.16 | 0.44 | |
| Primers (amplified regions) | 0.81 | 0.16 | 0.81 | 0.17 | 0.79 | |
| Shotgun metagenomic sequencing | Classifiers | 2.96 | 0.74 | 4.69 | 0.51 | 0.00 |
| Databases | 0.78 | 0.78 | 3.28 | 0.13 | 0.00 | |
| DNA ectraction kits | 2.49 | 0.36 | 1.62 | 0.43 | 0.01 | |
| Microbial cell wall-breaking methods | 0.84 | 0.42 | 1.69 | 0.14 | 0.04 | |
| Sequencers | 1.81 | 0.30 | 1.21 | 0.31 | 0.17 | |
Negative control
We included a PBS buffer aliquot as a negative control sample, which was required to be tested using the same procedure as that used for the other samples. At least one bacterium was reported in 10 (38.5%) of the 16Ss laboratories and 7 (30.4%, 7/23) of the SMs laboratories, with observed numbers ranging from 1 to 22 and 2 to 46, respectively (Supplementary Dataset 8). Overall, Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria were the dominant bacterial phyla, accounting for 42.7% (41/96), 25% (24/96), 14.6% (14/96) and 13.5% (13/96), respectively, in the 7 SMs laboratories. The sources of these detected contaminating microbes varied. For example, the species C. acnes, Moraxella osloensis, Staphylococcus hominis and Staphylococcus epidermidis are common bacteria living on the skin, on mucous membranes or in the surrounding environment. Dozens of bacteria belonging to the gut microbiota, such as Bacteroides dorei, B. thetaiotaomicron and Prevotella copri, might be derived from cross-contamination of gut-derived samples during sample preparation. The genera Stenotrophomonas, Pseudomonas and Xanthomonas were possibly derived from reagent contamination [49].
Discussion
Our study reflects the fact that there is great methodological variation in the current microbiome research in different laboratories. Thirty-five participating laboratories have established what they believe is the most reasonable way to carry out microbial analysis, according to local conditions. Only a few laboratories used near-identical protocols (P7 and P8; P12, P21 and P22; P27 and P34). High interlaboratory variability was found in the reported relative abundance of each bacterium in the mock communities. Even though the results from laboratories with near-identical protocols showed good precision (slight intracenter deviations), they were not necessarily accurate (such as the results of P12, P21 and P22). Further statistical analysis demonstrated that multiple factors, such as the DNA extraction method, amplification region and bioinformatic protocol choice, could introduce interlaboratory bias that affects the perception of community diversity in the analyzed samples.
Of note, because of the complexity of microbial communities as well as uncontrollable technical biases, no normalization approaches or guidelines have been established to improve the reproducibility of microbiome research in different laboratories. Even attempting to find consensus “best practices” for microbiome studies is challenging [50], [51], and a great deal of research has been conducted to find solutions to these problems [42], [52], [53]. DNA extraction contributes a majority of the experimental variability. We observed in most laboratories that the relative abundance of G+ bacteria in the whole-cell sample was decreased compared with that in the corresponding genome sample, which does not require DNA extraction. This issue occurred in many previous studies as well [40], [41], [54]. Introducing a bead-beating homogenization step was considered a practical method to increase the yield of hard-to-lyse microbes (such as G+ bacteria) [45]. However, approaches/kits based on bead-beating vary in efficiency [40]. The parameters used (bead size, grinding time, etc.) need to be validated to avoid producing shortened DNA fragments, which can contribute to DNA loss during subsequent library preparation [51]. To ensure consistency and reproducibility of DNA extraction in different laboratories, Greathouse et al. recommended three minimal standards that should be followed: (1) provide the details of the DNA extraction process in a study so that other studies can reproduce the entire process to the greatest extent; (2) introduce appropriate quality control materials to detect DNA extraction bias and possible contamination; and (3) utilize the same DNA extraction protocol across studies for multicenter studies [51]. Regarding the third recommendation, we emphasize that the method adopted should be a pre-evaluated DNA extraction protocol with minimal deviation, similar to what the International Human Microbiome Standards (IHMS) group recommended [45].
Likewise, the choice of 16S rRNA gene variable region primers is one essential aspect demanding careful consideration when performing 16Ss because primer bias toward particular taxonomic groups has been widely reported [55], [56], [57]. For example, a gut microbiota analysis showed that the genera Sphingomonas, Roseburia, and Bilophila were detectable only with V3-V4 sequencing, whereas the genera Clostridium and Lactococcus could be detected with only V4-V5 sequencing [58]. Fouhy et al. demonstrated that the V4-V5 primers gave the most comparable results across platforms (MiSeq and Ion PGM) [59]. In V4 sequencing, the genus Cutibacterium (formerly Propionibacterium) was almost completely undetected [60]. The widely used “universal” (27f and 1492r) primers for targeting the full-length 16S rRNA gene (V1-V9) failed to amplify more than 40% of purified Actinobacteria isolates [61]. In this study, we found that the V3-V4 region primers are superior to the V4, V1-V2 and V4-V5 region primers in identifying Enterobacter spp. These observed differences highlight the importance of careful selection of the 16S rRNA gene-targeting primers and emphasize that extreme caution should be taken when comparing studies conducted with different 16S rRNA gene-sequencing methods. Using two or more primer sets to completely cover the bacterial diversity in complex samples may be a viable solution for 16Ss [62].
At present, there are many bioinformatics tools can be selected for taxonomic identification and classification. However, the lack of consistency in bioinformatic processing steps has a significant effect on the comparability of results between individual studies and sometimes leads to erroneous conclusions [38]. A previous study showed that the number of species identified by different metagenomic classifiers can differ by over 3 orders of magnitude on the same simulated datasets [63]. DNA-to-DNA classifiers (Kraken-like) tend to perform better than DNA-to-protein tools (DIAMOND-like) for taxonomic classification in SMs [47]. In this study, it was found that nearly 70% (18) of the 26 16Ss laboratories use Qiime or USEARCH for taxonomic classification. Further PCA analysis also found no visible difference between the results using Qiime and USEARCH analysis (Supplementary Fig. S3). PERMANOVA analysis confirmed that the bioinformatics tool was not the main driver of inter-laboratory differences in 16Ss laboratories (P = 0.11) but was a factor causing differences among SMs laboratories (P < 0.01). MetaPhlAn2 was the most frequently used tool in the SMs laboratories (52%, 12/23). In PCA plot, the results of laboratories using MetaPhlAn2 were clustered together (Fig. 5E), indicating that the results using this tool tend to be more consistent. However, based on this analysis, we couldn’t determine that which metagenomic classifier is better, because in a real-world scenario, there is many other experimental factors that affect the accuracy of the final result. In order to accurately assess which bioinformatics tool is more accurate and reliable, further experiments are needed to evaluate them with real sequencing dataset of known characteristics or simulated sequencing data, to avoid the bias introduced in the wet lab experiments (sampling, nucleic acid extraction, library construction and sequencing). Thus, we suggest that before choosing any bioinformatics pipeline, a laboratory should evaluate its performance in taxonomic classification using simulated and experimental datasets to ensure that no analytical bias is produced for certain taxa.
Regardless of 16Ss or SMs, the contamination of exogenous microorganisms is always an inevitable problem [49]. The results for the mock communities (201901 and 201902) and the negative control (2019NC) indicated that the presence of contaminating microorganisms varied between laboratories. Similar to previous findings, the sources of these occult contaminants may include molecular biology regents (e.g., DNA extraction kits, PCR reagents, etc.) [64], researchers, plastic consumables [65], cross-contamination between samples and laboratory environments [50], [66]. The effects of unexpected contaminants are particularly problematic in low-biomass samples that contain very little endogenous DNA [53]. There are numerous options to minimize the effects of contamination in microbiome analysis. For example, during sampling and processing, experimenters should wear protective clothing and equipment (i.e., laboratory coats, face masks, hairnets, sleeves, and clean disposable gloves) to cover all exposed skin if possible to reduce the introduction of contaminants into the samples. As many procedures as possible, such as DNA extraction, library preparation, and sequencing, should be completed in a cleaned, isolated working environment with appropriately treated equipment and consumables [66]. To monitor and minimize regent contamination, it is wise to choose one kit type for all of the samples in a microbiome study. If multiple kits are used, a record should be made of which samples were processed with which kit so that contamination of a particular kit lot number can be traced through to the final dataset [49], [53]. Importantly, concurrent sequencing of negative control samples (sampling blank control, DNA extraction blank control, and no-template amplification control) is strongly advised in every analysis for detecting contamination and assessing the levels of cross-contamination between samples [49], [66]. Furthermore, positive controls (microbial whole-cell or DNA mock communities) should be processed alongside samples to assess biomass and contamination levels and ensure that contaminants do not drive the results of the study [64], [66]. Additionally, contamination assessment is suggested to be a minimal publication requirement of a high-throughput metagenomic study. The approach taken to identify and minimize the effects of contaminants during analysis should be clearly reported to enhance reproducibility and allow such approaches to be critically evaluated by others [67].
Mock communities (reference samples with known microbial cell or DNA compositions) are necessary for standardizing analyses [68]. Using such well-defined samples, we can discover possible biases during the experiment (such as the DNA extraction, amplification and data analysis biases found in this study) to benchmark different technologies and to verify that the analysis in each run is within acceptable bias limits [55], [69], [70]. A valuable example is that Yeh and colleagues discovered highly aberrant and strong biases from marine microbiome analysis by the routine use of mock communities as internal standards [71]. Thus, when planning a 16Ss- or SMs-based microbiome study, the inclusion of a mock community is strongly encouraged. Ideally, the mock communities should include more than just a few members that are representative of the samples being analyzed to detect problems that occur in some interesting taxa and to help validate clustering [71]. At present, commercial mock communities are available from the American Type Culture Collection (ATCC) (www.atcc.org), BEI resource (www.beiresources.org) and Zymo Research (www.zymoresearch.com). Notably, as in this and previous studies, the use of in-house developed mock communities is also encouraged because they can more accurately reflect the variability of interesting or important bacteria than the commercially available communities [13], [38], [72].
The current study is not without limitations. First, for 16Ss, considering that the purpose of microbial community profiling is to find the relationship between the final taxonomic annotation results (microbes and their relative abundances) and human health, we evaluated the reproducibility of different laboratories based on an annotated species checklist rather than operational taxonomic unit (OTU) data. We could not find a uniform standard to compare the OTU data reported by each laboratory because their amplification regions, clustering standards and naming rules were different. In addition, we did not distribute duplicate samples to evaluate intralaboratory consistency, as previous studies did [37], [38]. However, in fact, we found that the results reported by laboratories with near-identical protocols showed slight interlaboratory deviations. Finally, another important value of SMs is to reveal the functional characteristics of the microbiome. However, the samples synthesized in this study were not useable for the assessment of the reproducibility of functional analysis. If further research finds a suitable way to carry out this work, it will be of enhanced significance for the standardization of microbiome research.
Conclusion
Microbiome research holds great promise for multiple fields, but methodological variations can easily undermine the progress and reputation of this developing research area. Therefore, it is necessary for researchers to carry out reasonable investigations and research to recognize these variations and provide data support for addressing them scientifically. High interlaboratory deviations were found in this multicenter quality assessment of 16Ss- and SMs-based microbiota profiling. To improve the comparability of interlaboratory results and to carry out scientific and reasonable metaanalyses of microbiome studies, methodologies in routine sequencing laboratories urgently need to be optimized and standardized, quality control materials (such as mock communities) should be routinely used, and external quality assessment (EQA) programs should be established and gradually improved. In addition, studies should now be encouraged to accurately report methodological details (their SOPs) while publishing research results to achieve traceability of methodological deviations and further improve the interlaboratory comparability of microbiome analysis data.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank the 35 laboratories for participating actively in our research and for reporting the test results to NCCL on time.
This work was supported by the “AIDS and Hepatitis, and Other Major Infectious Disease Control and Prevention” Program of China under Grant [No. 2018ZX10102001] and the National Natural Science Foundation of China under Grant [No. 81703276]. The funder had no role in study design, data collection, analysis, interpretation, or writing of the paper.
Author contributions were as follows: study design, J.L., and R.Z.; Data collection, P.G., R.L., and D.H.; Data analysis, D.H., R.L., R.Z., and P.T.; Wrote the paper: D.H., J.L., and R.Z. All authors have read and approved the final version of the manuscript.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.07.010.
Contributor Information
Rui Zhang, Email: ruizhang@nccl.org.cn.
Jinming Li, Email: jmli@nccl.org.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 3.Yu J., Feng Q., Wong S.H., Zhang D., Liang Q.Y., Qin Y. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2016;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 4.Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 5.McInnes I.B., Schett G. The Pathogenesis of Rheumatoid Arthritis. New Engl. J. Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 6.Naseribafrouei A., Hestad K., Avershina E., Sekelja M., Linløkken A., Wilson R. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motility. 2014;26:1155–1162. doi: 10.1111/nmo.12378. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach M.A. Microbiome: focus on causation and mechanism. Cell. 2018;174:785–790. doi: 10.1016/j.cell.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steen A.D., Crits-Christoph A., Carini P., DeAngelis K.M., Fierer N., Lloyd K.G. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019;13:3126–3130. doi: 10.1038/s41396-019-0484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen-Vercoe E. Bringing the gut microbiota into focus through microbial culture: recent progress and future perspective. Curr Opin Microbiol. 2013;16:625–629. doi: 10.1016/j.mib.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Structure, function and diversity of the healthy human microbiome. Nature 2012; 486: 207–14. [DOI] [PMC free article] [PubMed]
- 11.Tourlousse D.M., Yoshiike S., Ohashi A., Matsukura S., Noda N., Sekiguchi Y. Synthetic spike-in standards for high-throughput 16s rrna gene amplicon sequencing. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw984. w984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 13.Kioroglou D., Mas A., Portillo M.D.C. Evaluating the effect of qiime balanced default parameters on metataxonomic analysis workflows with a mock community. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 15.Hansen M.E.B., Rubel M.A., Bailey A.G., Ranciaro A., Thompson S.R., Campbell M.C. Population structure of human gut bacteria in a diverse cohort from rural tanzania and botswana. Genome Biol. 2019;20 doi: 10.1186/s13059-018-1616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forster S.C., Kumar N., Anonye B.O., Almeida A., Viciani E., Stares M.D. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol. 2019;37:186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proctor L.M., Creasy H.H., Fettweis J.M., Lloyd-Price J., Mahurkar A., Zhou W. The integrative human microbiome project. Nature. 2019;569:641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wirbel J., Pyl P.T., Kartal E., Zych K., Kashani A., Milanese A. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riva A., Borgo F., Lassandro C., Verduci E., Morace G., Borghi E. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in firmicutes populations. Environ Microbiol. 2017;19:95–105. doi: 10.1111/1462-2920.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patrone V., Vajana E., Minuti A., Callegari M.L., Federico A., Loguercio C. Postoperative changes in fecal bacterial communities and fermentation products in obese patients undergoing bilio-intestinal bypass. Front Microbiol. 2016;7:200. doi: 10.3389/fmicb.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu D., Xia Z., Deng J., Jiao X., Liu L., Li J. Glucorticoid-induced obesity individuals have distinct signatures of the gut microbiome. Biofactors. 2019;45:892–901. doi: 10.1002/biof.1565. [DOI] [PubMed] [Google Scholar]
- 25.Chen X., Sun H., Jiang F., Shen Y., Li X., Hu X. Alteration of the gut microbiota associated with childhood obesity by 16s rrna gene sequencing. Peerj. 2020;8:e8317. doi: 10.7717/peerj.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walters W.A., Xu Z., Knight R. Meta-analyses of human gut microbes associated with obesity and ibd. Febs Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallianou N., Stratigou T., Christodoulatos G.S., Dalamaga M. Understanding the role of the gut microbiome and microbial metabolites in obesity and obesity-associated metabolic disorders: current evidence and perspectives. Curr Obesity Rep. 2019;8:317–332. doi: 10.1007/s13679-019-00352-2. [DOI] [PubMed] [Google Scholar]
- 28.Feng Q., Liang S., Jia H., Stadlmayr A., Tang L., Lan Z. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 29.Nuzum N.D., Loughman A., Szymlek-Gay E.A., Hendy A., Teo W.P., Macpherson H. Gut microbiota differences between healthy older adults and individuals with parkinson's disease: a systematic review. Neurosci Biobehav Rev. 2020 doi: 10.1016/j.neubiorev.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Hornung B.V.H., Zwittink R.D., Kuijper E.J. Issues and current standards of controls in microbiome research. FEMS Microbiol Ecol. 2019;95 doi: 10.1093/femsec/fiz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter J., Armet A.M., Finlay B.B., Shanahan F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell. 2020;180:221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 35.Greathouse K.L., Sinha R., Vogtmann E. Dna extraction for human microbiome studies: the issue of standardization. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z., Hui P.C., Hui M., Yeoh Y.K., Wong P.Y., Chan M. Impact of preservation method and 16s rrna hypervariable region on gut microbiota profiling. Msystems. 2019;4 doi: 10.1128/mSystems.00271-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiergeist A., Reischl U., Gessner A. Multicenter quality assessment of 16s ribosomal dna-sequencing for microbiome analyses reveals high inter-center variability. Int J Med Microbiol. 2016;306:334–342. doi: 10.1016/j.ijmm.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Sinha R., Abu-Ali G., Vogtmann E., Fodor A.A., Ren B., Amir A. Assessment of variation in microbial community amplicon sequencing by the microbiome quality control (mbqc) project consortium. Nat Biotechnol. 2017;35:1077–1086. doi: 10.1038/nbt.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy N.A., Walker A.W., Berry S.H., Duncan S.H., Farquarson F.M., Louis P. The impact of different dna extraction kits and laboratories upon the assessment of human gut microbiota composition by 16s rrna gene sequencing. Plos One. 2014;9:e88982. doi: 10.1371/journal.pone.0088982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Z., Sun J., Lv A., Wang A. Biases from different dna extraction methods in intestine microbiome research based on 16s rdna sequencing: a case in the koi carp, cyprinus carpio var. Koi Microbiologyopen. 2019;8:e626. doi: 10.1002/mbo3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ducarmon Q.R., Hornung B.V.H., Geelen A.R., Kuijper E.J., Zwittink R.D. Toward standards in clinical microbiota studies: comparison of three dna extraction methods and two bioinformatic pipelines. Msystems. 2020;5 doi: 10.1128/mSystems.00547-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M. Evaluation of general 16s ribosomal rna gene pcr primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng L.J., Hsueh P.R., Huang Y.H., Tsai J.C. Identification of bacteroides thetaiotaomicron on the basis of an unexpected specific amplicon of universal 16s ribosomal dna pcr. J Clin Microbiol. 2004;42:1727–1730. doi: 10.1128/JCM.42.4.1727-1730.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costea P.I., Zeller G., Sunagawa S., Pelletier E., Alberti A., Levenez F. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol. 2017 doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 46.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using diamond. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 47.Ye S.H., Siddle K.J., Park D.J., Sabeti P.C. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178:779–794. doi: 10.1016/j.cell.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin J., Li R., Raes J., Arumugam M., Tims S., Vos D.W.M. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollock J., Glendinning L., Wisedchanwet T., Watson M. The madness of microbiome: attempting to find consensus “best practice” for 16s microbiome studies. Appl Environ Microbiol. 2018;84 doi: 10.1128/AEM.02627-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 52.Zinter M.S., Mayday M.Y., Ryckman K.K., Jelliffe-Pawlowski L.L., DeRisi J.L. Towards precision quantification of contamination in metagenomic sequencing experiments. Microbiome. 2019;7:62. doi: 10.1186/s40168-019-0678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D., Hofstaedter C.E., Zhao C., Mattei L., Tanes C., Clarke E. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:14–52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan S., Cohen D.B., Ravel J., Abdo Z., Forney L.J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. Plos One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parada A.E., Needham D.M., Fuhrman J.A. Every base matters: assessing small subunit rrna primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 56.D'Amore R., Ijaz U.Z., Schirmer M., Kenny J.G., Gregory R., Darby A.C. A comprehensive benchmarking study of protocols and sequencing platforms for 16s rrna community profiling. BMC Genomics. 2016;17:55. doi: 10.1186/s12864-015-2194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghyselinck J., Pfeiffer S., Heylen K., Sessitsch A., De Vos P. The effect of primer choice and short read sequences on the outcome of 16s rrna gene based diversity studies. Plos One. 2013;8:e71360. doi: 10.1371/journal.pone.0071360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rintala A., Pietilä S., Munukka E., Eerola E., Pursiheimo J., Laiho A. Gut microbiota analysis results are highly dependent on the 16s rrna gene target region, whereas the impact of dna extraction is minor. J Biomol Tech: JBT. 2017;28:19–30. doi: 10.7171/jbt.17-2801-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fouhy F., Clooney A.G., Stanton C., Claesson M.J., Cotter P.D. 16s rrna gene sequencing of mock microbial populations- impact of dna extraction method, primer choice and sequencing platform. BMC Microbiol. 2016;16:123. doi: 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson M.C., Morrison H.G., Benjamino J., Grim S.L., Graf J. Analysis, optimization and verification of illumina-generated 16s rrna gene amplicon surveys. Plos One. 2014;9:e94249. doi: 10.1371/journal.pone.0094249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farris M.H., Olson J.B. Detection of actinobacteria cultivated from environmental samples reveals bias in universal primers. Lett Appl Microbiol. 2007;45:376–381. doi: 10.1111/j.1472-765X.2007.02198.x. [DOI] [PubMed] [Google Scholar]
- 62.Starke I.C., Vahjen W., Pieper R., Zentek J. The influence of dna extraction procedure and primer set on the bacterial community analysis by pyrosequencing of barcoded 16s rrna gene amplicons. Mol Biol Int. 2014;2014:548610–548683. doi: 10.1155/2014/548683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McIntyre A.B.R., Ounit R., Afshinnekoo E., Prill R.J., Hénaff E., Alexander N. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol. 2017;18 doi: 10.1186/s13059-017-1299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Goffau M.C., Lager S., Salter S.J., Wagner J., Kronbichler A., Charnock-Jones D.S. Recognizing the reagent microbiome. Nat Microbiol. 2018;3:851–853. doi: 10.1038/s41564-018-0202-y. [DOI] [PubMed] [Google Scholar]
- 65.Motley S.T., Picuri J.M., Crowder C.D., Minich J.J., Hofstadler S.A., Eshoo M.W. Improved multiple displacement amplification (imda) and ultraclean reagents. BMC Genomics. 2014;15:443. doi: 10.1186/1471-2164-15-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eisenhofer R., Minich J.J., Marotz C., Cooper A., Knight R., Weyrich L.S. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 2019;27:105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 67.Weyrich L.S., Farrer A.G., Eisenhofer R., Arriola L.A., Young J., Selway C.A. Laboratory contamination over time during low-biomass sample analysis. Mol Ecol Resour. 2019;19:982–996. doi: 10.1111/1755-0998.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knight R., Vrbanac A., Taylor B.C., Aksenov A., Callewaert C., Debelius J. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 69.Culbreath K., Melanson S., Gale J., Baker J., Li F., Saebo O. Validation and retrospective clinical evaluation of a quantitative 16s rrna gene metagenomic sequencing assay for bacterial pathogen detection in body fluids. J Mol Diag. 2019;21:913–923. doi: 10.1016/j.jmoldx.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 70.Bowers R.M., Clum A., Tice H., Lim J., Singh K., Ciobanu D. Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genomics. 2015;16:856. doi: 10.1186/s12864-015-2063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeh Y., Needham D.M., Sieradzki E.T., Fuhrman J.A. Taxon disappearance from microbiome analysis reinforces the value of mock communities as a standard in every sequencing run. Msystems. 2018;3:e18–e23. doi: 10.1128/mSystems.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brooks J.P., Edwards D.J., Harwich M.D., Rivera M.C., Fettweis J.M., Serrano M.G. The truth about metagenomics: quantifying and counteracting bias in 16s rrna studies. BMC Microbiol. 2015;15 doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
Further reading
- 19.Schloss P.D. Identifying and overcoming threats to reproducibility, replicability, robustness, and generalizability in microbiome research. Mbio. 2018;9 doi: 10.1128/mBio.00525-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theis K.R., Romero R., Winters A.D., Greenberg J.M., Gomez-Lopez N., Alhousseini A. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time pcr, 16s rrna gene sequencing, and metagenomics. Am J Obstet Gynecol. 2019;220:261–267. doi: 10.1016/j.ajog.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornung B.V.H., Zwittink R.D., Ducarmon Q.R., Kuijper E.J. Response to: 'circulating microbiome in blood of different circulatory compartments' by Schierwagen et al. Gut. 2019 doi: 10.1136/gutjnl-2019-318601. 2019-318601. [DOI] [PubMed] [Google Scholar]
- 32.Segata N. No bacteria found in healthy placentas. Nature. 2019;572:317–318. doi: 10.1038/d41586-019-02262-8. [DOI] [PubMed] [Google Scholar]
- 39.Chen H., Chi H., Chiu N., Huang F. Kocuria kristinae: a true pathogen in pediatric patients. J Microbiol Immunol Infect. 2015;48:80–84. doi: 10.1016/j.jmii.2013.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.