Abstract
The sarcomere is the functional unit of cardiac muscle, essential for normal heart function. To date, it has not been possible to study, in real time, thin filament-based activation dynamics in live cardiac muscle. We report here results from a cardiac troponin C (TnC) FRET-based biosensor integrated into the cardiac sarcomere via stoichiometric replacement of endogenous TnC. The TnC biosensor provides, for the first time, evidence of multiple thin filament activating ligands, including troponin I interfacing with TnC and cycling myosin, during a cardiac twitch. Results show that the TnC FRET biosensor transient significantly precedes that of peak twitch force. Using small molecules and genetic modifiers known to alter sarcomere activation, independently of the intracellular Ca2+ transient, the data show that the TnC biosensor detects significant effects of the troponin I switch domain as a sarcomere-activating ligand. Interestingly, the TnC biosensor also detected the effects of load-dependent altered myosin cycling, as shown by a significant delay in TnC biosensor transient inactivation during the isometric twitch. In addition, the TnC biosensor detected the effects of myosin as an activating ligand during the twitch by using a small molecule that directly alters cross-bridge cycling, independently of the intracellular Ca2+ transient. Collectively, these results aid in illuminating the basis of cardiac muscle contractile activation with implications for gene, protein, and small molecule-based strategies designed to target the sarcomere in regulating beat-to-beat heart performance in health and disease.
Keywords: myofilament, troponin, myosin, contraction, calcium
Introduction
The cardiac sarcomere is a multimeric contractile apparatus consisting of a thin myofilament-based allosteric regulatory complex together with the myosin-based thick myofilament that generates force [1]. Interlacing myofilaments operate in synchrony to regulate and generate the forces necessary for heart performance. Beat-to-beat control of cardiac sarcomere activation refers to the status of the thin filament regulatory system in controlling the degree to which contraction is turned on and off during a twitch. Sarcomere activation control mechanisms involve a dynamic interplay between thin filament-localized Ca2+ in concert with regulatory elements in the thick and thin myofilaments, including troponin, tropomyosin, and myosin [1, 2]. Sarcomere activation can be dramatically altered by factors intrinsic to myofilaments, including posttranslational modifications, the environmental milieu (ischemia/hypoxia), and by inherited mutations in sarcomeric proteins [3-5]. Despite great efforts, it has not been possible to directly investigate and monitor sarcomere activation in the physiological setting of live cardiac muscle during a twitch.
The thin filament regulatory system, which is at the center of sarcomere activation dynamics and its beat-to-beat regulation during heart contraction, is composed of actin, tropomyosin (Tm), and the troponin complex (Tn)[1, 6]. Regulation of sarcomere activation refers to the processes that control Tm's position on the thin filament[1, 2]. Cardiac troponin is a heterotrimeric complex comprised of the Tm binding subunit, troponin T (cTnT), the calcium binding subunit, troponin C (cTnC), and the inhibitory subunit, troponin I (cTnI). Cardiac TnI (cTnI) is the molecular switch of the thin filament regulatory system [6, 7]. Prior to contraction, the C-terminus of cTnI is strongly bound to actin, inhibiting Tm from rotating to a position favorable for strong myosin binding to actin. Upon Ca2+ binding to cTnC, cTnI rapidly switches from strong binding to actin to strong binding to cTnC. This cTnI switch mechanism is essential for opening the TnC hydrophobic patch, and then permitting Tm to translocate on the thin filament for active contraction to commence [2, 8, 9].
The prevailing model of sarcomere activation posits that Ca2+, cTnI switching,[9] and strong myosin binding serve as activating ligands to regulate Tm’s position on actin from an inhibiting to a disinhibiting position[1]. A wealth of biochemical studies indicate that Ca2+, cTnI, and myosin are the key effectors in orchestrating the sequential blocked, closed, and open state transitions in the myofilament regulatory system, as governed by their effects on Tm displacement [10, 11]. A central feature of this model is that strong myosin binding is required to displace Tm to form the open state of thin filament regulatory apparatus in initiating contraction [1, 11, 12]. While highly informative, it is presently unknown whether this model holds in the dynamic physiological setting of live, intact muscle.
Well established approaches are in place to quantitatively detect and monitor the electrical and intracellular Ca2+ handling properties of live cardiac muscle [13]. However, to date, it has not been possible to investigate thin filament activation dynamics in real time under physiological conditions of intact excitation-contraction coupling and under load, all of which dramatically affect overall contractile performance [1]. To address this, we implement here a live cardiac muscle thin filament activation biosensor. The FRET-based biosensor was genetically engineered into full length human cTnC and incorporated into the myofilaments. The rationale is based on extensive in vitro works documenting that cTnC's global conformation changes upon Ca2+ activation in vitro. Data show that cTnC becomes more compact, with the N and C terminal lobes of TnC coming closer together, going from the apo to Ca2+ activated state [14-17]. Our premise of engineering a TnC conformation-dependent FRET-based biosensor integrated into the sarcomere of intact cardiac muscle derives from these studies. We use this TnC probe with the aim to detect global TnC conformational changes during a single cardiac twitch contraction.
In addition, the premise that TnC’s conformation provides a nexus point in responding to and integrating near and long-range thin filament activating ligands, including Ca2+ binding to TnC, cTnI binding to TnC, Tm and myosin, is predicated on extensive in vitro works, as has been shown during steady-state conditions [1, 2, 6, 18-20]. In permeabilized cardiac muscle reconstituted with cTnC conformational state-sensitive fluorescent probes, it has been shown that myosin cross-bridge binding to actin can induce TnC conformational changes under steady-state conditions, even in the absence of Ca2+ [19]. This is evidence that myosin binding causes Tm displacement and that this can be detected by TnC conformational changes, at least under steady-state conditions [1, 19]. More recently, in vitro demonstration of FRET-based probes on TnC to detect Ca2+-dependent TnC conformational changes have been established using the isolated troponin complex and in reconstituted permeabilized preparations under steady-state activating conditions, with recordings on the tens of seconds to minutes time-scale [17]. It is presently unknown whether these foundational steady-state results will translate to transient conditions in intact cardiac muscle in real time, which operates on the milliseconds time-scale. We tested here the hypothesis that TnC’s conformation can serve as a real-time biosensor and report the effects of multiple thin filament activating ligands during a single cardiac twitch contraction in live muscle.
Material and Methods
Animal Work
The procedures used in this study were approved under guidelines of the University of Minnesota Committee on the Use and Care of Animals. Adult ventricular cardiac myocytes were isolated from adult female Sprague Dawley rats or adult C57/Bl6 mice of both sexes.
FRET pair, gene construct, recombinant vectors, and transgenesis
An intramolecular Clover/mRuby2 FRET pair was engineered into cardiac troponin C (details below). The Clover and mRuby2 FRET pair was used because, at the time of study design, this FRET pair was superior to any previous FRET pair in terms of fluorescence intensity brightness and photostability, with the highest Förster radius of any ratiometric FRET pair yet described [17, 21, 22]. Clover (λexcitation max/ λemission max = 505/515 nm) and mRuby2 (λexcitation max/λemission max = 559/600 nm) were used as mRuby2 is an outstanding FRET partner for Clover because of its high extinction coefficient, extensive excitation overlap with Clover emission, high quantum yield, large Stokes shift, fast maturation, and high photostability[21, 22]. Excellent overlap between donor emission and acceptor absorbance yields a Clover-mRuby2 R0 of 6.3 nm—the highest R0 of any previous FRET pair (at that time of study initiation). Thus, the Clover-mRuby2 pair allows FRET imaging with less phototoxicity and a higher signal/background fluorescence ratio than any previous CFP and YFP at the time of study [21, 22].
The 480bp full-length (less the starting methionine) human cardiac Troponin C (hcTnC) cDNA was synthesized de novo and engineered to be flanked on the 5’ end by a 19 amino acid flexible linker peptide enriched in leucine, glycine, serine, and alanine; and flanked on the 3’ end by an 8 amino acid flexible linker enriched in leucine and alanine. Linker design was based in part on previous works showing a range of linker lengths attaching CFP/YFP fluors to cTnC not affecting the Ca2+/Mg2+ binding characteristics of isolated TnC [17]. The 19 amino acid linker is inserted between a PasI restriction site at the 5’ end and an EcoRV restriction site at the 3’ end. The 8 amino acid linker is inserted between a PfoI restriction site at the 5’ end and a BclI restriction site at the 3’ end. On the 5’ end of the 19 amino acid flexible linker is the 684bp cDNA of the green fluorescent protein “Clover”. The Clover cDNA is inserted between a SalI restriction site at the 5’ end and the aforementioned PasI restriction site at the 3’ end. On the 3’ end of the 8 amino acid flexible linker is the 714bp cDNA of the red fluorescent protein “mRuby2”. The mRuby2 cDNA is inserted between the aforementioned BclI restriction site at the 5’ end and an FseI restriction site at the 3’ end.
The human cardiac TnC biosensor gene construct was inserted in a DNA vector downstream of an alpha myosin heavy chain (αMHC) promoter element for robust and cardiac specific expression in a mammalian system. Human cardiac TnC is highly conserved between mammalian species and used to aid in comparing to other studies that used human cTnC[17]. This αMHC TnC biosensor vector was inserted by pronuclear injection into donor egg cells and implanted in C57/Bl6 murine surrogate mothers. Live pups were genotyped using the PCR sense primer located in mRuby2: 5’-CAACATCAAGATGCCCGGCA-3’, and the PCR antisense primer located in the SV40 PolyA region: 5’-CACTGCATTCTAGTTGTGGTTTGTCC-3’ with an annealing temperature of 62°C. Of the live pups, 5 founders carrying the transgene were identified and the line with greatest breeding vigor was selected for further validation.
For recombinant adenovirus, a similar DNA constructed was synthesized de novo but containing HindIII and EcoRI at the 5’ and 3’ ends respectively for insertion into a pDC316 recombinant adenovirus production vector. pDC316-TnC biosensor and an adenovirus genome cosmid were co-transfected by calcium phosphate into confluent HEK293 cells. Noble agar was overlaid after transfection and clonal viral plaques were amplified. High-titer plaque-purified stocks were produced and viral aliquots were stored at −80°C [23].
Heart morphology and echocardiography
After initial weigh-in, eight to twenty week old NTG and TG mice of both sexes were euthanized by sodium pentobarbital injection. Hearts were excised and rinsed of excess blood and weighed. Hindlimbs were degloved and the tibia was exposed by blunt dissection prior to measurement.
For Sirius red/fast green staining, fresh frozen slides were fixed with chilled acetone then washed in 70% ethanol followed by water. Slides were then stained with Sirius red/fast green for 15 minutes and rinsed in water. After rinsing Sirius red/fast green stain, slides were dehydrated with 70% and 100% ethanol and then Formula 83 (CBG Biotech cat# CH0104).
Eight to twenty week old Ntg and TnC biosensor Tg mice of both sexes were lightly anesthetized by 2.5% Isoflurane and prepared for echocardiography. M-mode imaging was performed with a Vevo 2100 (Visual Sonics) at 1000fps with the MS550D 22–55 MHz transducer. The parasternal short axis view was used to assess systolic and diastolic ventricle size. M-mode imaging was also used to calculate fractional shortening.
Myocyte Isolation and Culture
Eight to twelve week old mice of both sexes were injected with 0.25cc 10,000U/ml heparin and anesthetized 10 minutes later with 0.15cc 50mg/ml sodium pentobarbital. The heart was rapidly excised and placed in ice cold Krebs-Henseleit buffer modified with 25mM HEPES, 10mM 2,3-Butanedione monoxime, 30mM Taurine and the aorta was cannulated. The mounted heart was placed on a perfusion system at 37°C. The heart was perfused with calcium free modified Krebs-Henseleit buffer with 2mg/ml crudely purified collagenase for 10 minutes. The heart was then removed from the cannula, the atria were removed and the ventricles were dissected into pieces approximately 2mm in diameter. The dissected ventricular tissue was then gently mechanically triturated in collagenase to liberate isolated myocytes. After brief low-speed centrifugation, isolated myocytes were resuspended in nominal calcium and .025mg/ml bovine serum albumin in a modified Kreb-Henseleit buffer. After the final trituration step, calcium was added back to the buffer step wise until reaching a final concentration of 2mM CaCl2. Myocytes were plated on laminin-coated (ThermoFisher cat# 23017015) glass coverslips in M199 media (Gibco cat# 31100-035) containing 5% fetal bovine serum and incubated for one hour at 37°C with 5% CO2. After the one-hour incubation, the media was changed to serum free M199 media and the myocytes were used for experiments within 5 hours. In a subset of studies, cardiac myocytes were also isolated from adult rats and transduced with recombinant adenovirus containing TnC FRET biosensor expression cassette (driven by CMV), using methods detailed previously [3, 24].
Small molecules.
For the small molecule studies in isolated myocytes, sarcomere length and fluorescence were first recorded under baseline conditions as outlined above. Following baseline recordings, the myocytes were then treated with one of the following small molecules for 10 minutes for pairwise analysis: Isoproterenol, a β adrenoreceptor agonist [13] (50nM, Sigma), W7, a TnC-TnI uncoupling drug [25, 26] (10μM, Sigma), Pimobendan, a troponin Ca2+ sensitizer and positive inotrope [27, 28] (10μM, Sigma), Omecamtiv mercarbil, a selective cardiac myosin modulator [29, 30] (250nM, Selleckchem), or para-Nitroblebbistatin, a myosin II inhibitor [31] (10μM; kind gift from A. Málnási-Csizmadia). After treatment, sarcomere length and fluorescence were recorded again.
Cardiac papillary isolation
Eight to twelve week old mice of both sexes were injected with 0.15cc 10,000U/ml heparin and then euthanized by cervical dislocation. The hearts were rapidly excised and rinsed in Krebs–Henseleit Buffer (KHB) supplemented with 40 mM BDM. Intact anterior and posterior papillary muscles were carefully dissected from the left ventricle in oxygenated (95% O2/5% CO2) BDM–KHB pH 7.4 at 4°C with minimal cutting and stretching injury. Papillary muscles were attached to platinum foil (0.001x0.02”, 4N) clips in the shape of an Ω using 7-0 silk suture. Papillary muscles were then mounted by the platinum clips in a flow chamber (IonOptix) filled with BDM-KHB between a hook connected to length-controlled micromanipulator and a force transducer (MyoTronic) via platinum foil (0.001x0.02”, 4N) clips. Preoxygenated (95% O2, 5% CO2) KHB pH 7.4 with 2mM CaCl2 was perfused into the chamber at a flow rate of about 2 mL/min. The temperature in the muscle chamber was set by ambient room temperature at 22°C. After a 10-20min BDM washout protocol, the muscle was placed under load and stretched to an optimum physiological length where the developed isometric twitch force was maximal. Stimulus was applied through platinum at 4V with either 0.2Hz or 1Hz pacing frequency with an 8ms pulse width. Force and fluorescence recording were then made concurrently using the IonOptix Calcium and Contractility System as described below.
Real-time calcium, contractility, and sarcomere biosensor measurements
For real-time simultaneous sarcomere length and TnC biosensor measurements, a modified Ionoptix Calcium and Contractility system was used. Sarcomere length changes were detected at 240Hz sampling rate and fluorescence measurements were detected at 1000Hz sampling rate with two photomultipler tubes (Fig. S2). Measurements were made at 37°C and a stimulation frequency of 0.2Hz, unless specified otherwise. This pacing is used by many groups and allowed SL transient comparison to past works on isolated myocytes [3]. For TnC biosensor measurements, a Xenon arc lamp was used and filtered with a 500/10nm hard-coated bandpass filter before entering the microscope turret. The microscope turret featured a 505 nm dichroic long-pass filter. Fluorescent light from the sample was shunted through microscope side port where it was split by a dichroic mirror with light above 650nm going toward the Myocam for sarcomere length detection and light below 650nm being further shunted to an additional filter cube. In this final filter cube, a 565nm dichroic long-pass mirror split the light again. Light with wavelengths below 565nm corresponding to the Clover protein was further refined by a 534/20nm hard-coated bandpass filter before reaching the photomultiplier tuber and light with wavelengths above 565nm was further cleaned up by a 592/43nm hard-coated bandpass filter before reaching the photomultiplier tube. For calcium fluorescence measurements, the mouse myocytes were pre-loaded for 20 minutes in M199 solution containing 1 μM Fura-2AM (ThermoFisher Cat #F1221), and a ten-minute period was allowed for deesterification. For calcium fluorescence measurements the 360/380nm ratio was determined. For each myocyte, several contractions were measured, averaged, and analyzed using the IonWizard software.
For adenovirus transduced biosensor experiments, adult cardiac myocytes were isolated as previously described24. Briefly, the myocytes were isolated by collagenase digestion and mechanical trituration prior to plating on laminin coated coverslips. Myocytes were treated with AdTnC biosensor at a multiplicity of infection of 100 for 48hrs prior to measurement with the aforementioned IonOptix calcium and contractility system.
For the Omecamtiv mecarbil (OM) studies in the intact papillary muscle, force and fluorescence were first recorded under baseline conditions as outlined above. Following the collection of baseline recordings, buffer, as described above, with a final concentration of 250nM OM was perfused into the chamber for 10 minutes. After 10 minutes force and fluorescence were recorded again. For both single myocyte and papillary muscle experiments, a paired experimental design was used (each prep compared before and after) to evaluate small molecules and drugs which is a powerful experimental design to quantify statistically significant differences.
Steady-state force and TnC biosensor signal in permeabilized cardiac muscle
Skinned Papillary Strips. Papillary muscles were isolated as described above. Once the muscles were dissected from the left ventricle, they were cut in half longitudinally and platinum foil clips in the shape of an omega (Ω) were attached to each end of each individual strip using 7-0 silk suture. Papillary muscles were then incubated in skinning solution (100mM KCl, 10mM Imidazole, 2mM EGTA, 1mM MgCl2 (Fluka Analytical), 4mM ATP, 50% glycerol, protease inhibitor (Pierce Protease Inhibitor Tablets A32963), 30mM BDM, 1% w/v Triton X-100) at room temperature for 2 hours. After incubation, papillary muscles were individually placed in 1.5ml microcentrifuge tubes filled with approximately 1ml of storage solution (100mM KCl, 10mM Imidazole, 2mM EGTA, 1mM MgCl2 (Fluka Analytical), 4mM ATP, 50% glycerol, and protease inhibitor (Pierce Protease Inhibitor Tablets A32963)) and stored at −20°C for no more than 7 days. At the time of the experiment, a papillary strip was mounted by the platinum clips in a flow chamber (Ionoptix) filled with relaxing solution (100mM KCl, 10mM Imidazole, 2mM EGTA, 1mM MgCl2 (Fluka Analytical), 4mM ATP) between hooks connected to a length-controlled micromanipulator and a force transducer (MyoTronic). The temperature of the chamber was set by ambient room temperature at 22°C. The strip was stretched to an optimum physiological sarcomere length of approximately 2.0μm as detected by a MyoCam (Ionoptix). Strips were calcium activated using solutions of pCa 9, pCa 7, pCa 6.5, pCa 6, pCa 5.7, pCa 5, and pCa4 and changes in developed tension and fluorescence intensity were simultaneously recorded using the Ionoptix Calcium and Contractility system. Recorded forces and fluorescence intensities were normalized to pCa4 values for each individual preparation and then fit to the Hill equation:
with pCa50 being the calcium concentration at half activation and nHill representing the Hill coefficient, using non-linear least squares regression in R statistical programming language.
Permeabilized adult cardiac myocytes. Cardiac myocytes were mechanically isolated from the ventricles of whole hearts from adult Tg and Ntg littermate mice, as described [32, 33]. The experimental apparatus and the permeabilized single cardiac myocyte attachment and tension-pCa determination procedures are as detailed previously[32, 33]. Relaxing and activating solutions contained (mM): 7 EGTA, 1 free Mg2+, 4 MgATP, 14-5 creatine phosphate, 20 imidazole, and sufficient KCl to yield a total ionic strength of 180 mm. Solution pH was adjusted to pH 7.00 using KOH/HCl at 15 °C.
Western Blotting and Immunofluorescence
The primary antibodies used were to α-actinin (immunofluorescence) (1:500, Abcam, EA-53), and cardiac troponin C (western blot) (1:2500, Novus Biologicals, 1A2). The secondary antibodies were conjugated to Alexa Fluor 647 (ThermoFisher) and IRDye Alexa Fluor 680 (ThermoFisher) respectively. For Western blots signals were visualized and quantified using the Odyssey system (Licor). Laminin coated coverslips containing isolated myocytes were treated with 4% paraformaldehyde in phosphate buffered saline for five minutes followed by two washes in phosphate buffered saline. Samples were blocked with 5% bovine serum albumin with 0.5% Triton X-100 in phosphate buffered saline for at least one hour. Primary antibody was applied at room temperature for one hour in phosphate buffered saline with 0.5% Triton X-100 and 0.5% BSA. Several washes were performed in PBS with 0.5% Triton X-100 and 5% BSA, followed by one-hour incubation of the secondary antibody at room temperature in PBS with 0.5 Triton X-100 and 0.5% BSA. Coverslips were mounted on microscope slides with ProLong Diamond mounting medium. Myocytes were imaged using a Nikon A1R confocal microscope with 60x water-immersion objective; myocytes were illuminated sequentially in line scanning mode by 488nm, 561nm, and 640nm lasers.
Analysis and Statistics
For contractility and calcium analysis, curve fitting was performed on unfiltered, averaged single myocyte or papillary transients (IonWizard 6.5; IonOptix) using a truncated Taylor series expansion of any analytical function to represent the signal within a phase of the transient, additional details on the fitting methodology is available from IonOptix.
To account for changes in the fluorophore concentration during papillary isometric twitches, ratiometric analyses were performed, taking advantage of the intramolecular nature of the FRET biosensor and its 1:1 stoichiometry.
Where I refers to the fluorophore intensity counted using the PMT. Here, the intensity modulation of the mRuby2 fluorophore signal is ratioed against the intensity of the Clover fluorophore signal. Increases in mRuby2 intensity greater than the proportional increase in Clover intensity are assumed to be due to an increase the degree of FRET between the two fluorophores. [34]
Mean ± S.E.M. are presented in histogram summary figures. For isoproterenol, W7, pimobendan, omecamtiv mecarbil, and para-nitroblebbistatin studies, two-tailed paired t-tests were performed. For all other summary histograms, two-tailed unpaired t-tests were performed (GraphPad PRISM version 6.0; GraphPad Software Inc.). In figures 2, 3, and 4 the Clover intensity transient is overlaid with a LOWESS curve the aid visual representation of the single cell fluorescent traces[35]. No analyses were performed on these curves as they are only for ease of visualization[35]. The parameters of the LOWESS curve were Q=20 and λ=1 with a tri-cube weighting function of: where d is the distance of a given data point from the point on the curve being fitted. Sample sizes were established on the basis of Power analysis, as informed by previous experience in the laboratory with respect to biological variability. P values <0.05 were considered statistically significant and indicated by asterisks in figures.
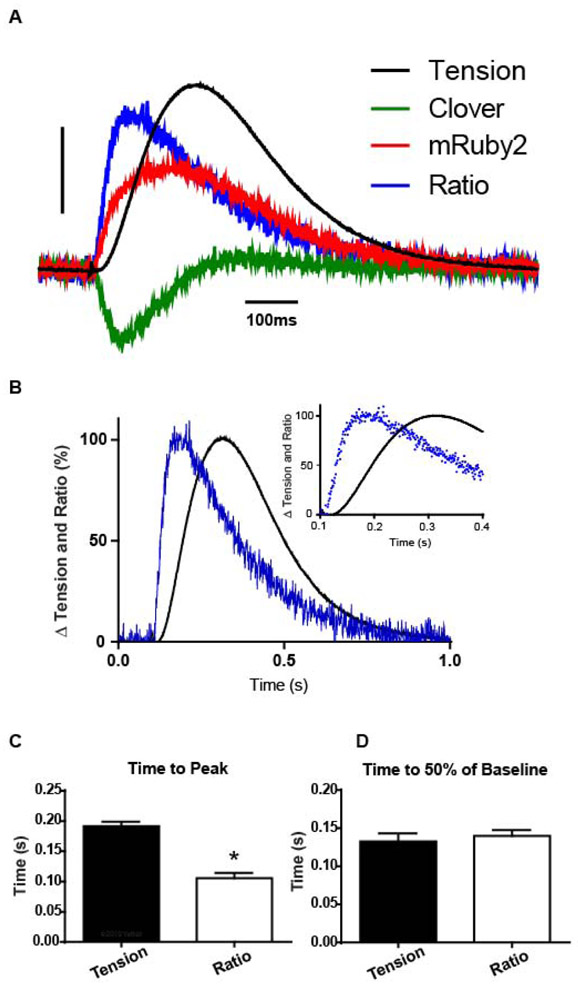
Figure 2. TnC biosensor transient in unloaded intact single adult cardiac myocytes.
A, Representative single cardiac myocyte traces of simultaneous sarcomere length, and the Clover, and mRuby2 flurorescence transients, with Clover fluorescence decreasing and simultaneously mRuby2 fluorescence increasing, indicating FRET. Scale bar represents a 5% change from baseline. B, Amplitude normalized Clover (green) and sarcomere length (black) dynamics from ensemble averaged myocytes (transients normalized from 0 to 100% to compare kinetics) show TnC biosensor Clover transient preceding sarcomere length shortening, inset shows the same data, highlighting the peak timing differences. C, Summary statistics from sarcomere length dynamics and Clover intensity show that peak activation of the TnC biosensor precedes sarcomere length change, and biosensor inactivation follows relaxation of the myocyte. Myocytes are unloaded and measured at 37°C with 0.2Hz stimulation, n=14 myocytes in each group. Mean ± S.E.M. are presented, unpaired two-tailed t-test: *P < 0.05.
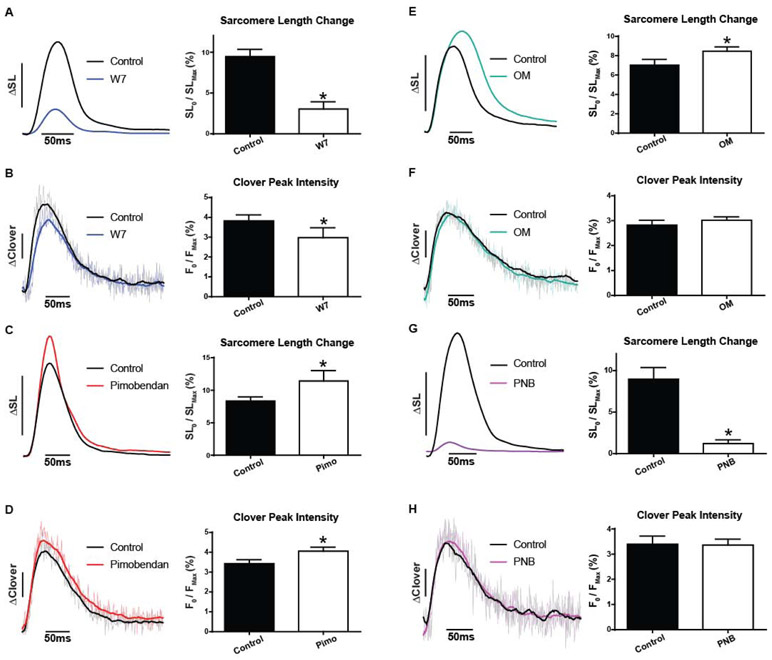
Figure 3. Myofilament-targeted small molecules probe function in live cardiac myocytes. TnC-TnI modulation.
A, Ensemble average traces of SL shortening (left) in isolated myocytes before and after treatment with 10 μM W7 and summary statistics (right) from sarcomere dynamics reveal decreased contractile amplitude (n=8 pairs). B, Ensemble average traces of Clover transient (left) in isolated myocytes before and after treatment with 10 μM W7 and summary statistics (right) of Clover signal reveal decreased peak fluorescence intensity (n=8 pairs). C, Ensemble average traces of SL shortening (left) in isolated myocytes before and after treatment with 10 μM Pimobendan with summary statistics (right) from sarcomere dynamics showing increased contractile amplitude (n=11 pairs). D, Ensemble average traces of Clover signal (left) in isolated myocytes before and after treatment with 10 μM Pimobendan and summary statistics (right) of Clover intensity changes reveal increased peak fluorescence intensity (n=11 pairs). Myocytes measured at 37°C with 0.2Hz stimulation; ΔSL bar represents a 5% change from baseline, ΔClover bar represents a 1% change from baseline. Mean ± S.E.M are presented, Paired two-tailed t-test: *P < 0.05. Myosin targeted small molecules probe sarcomere activation in live cardiac myocytes. E, Ensemble average traces of SL shortening (left) in isolated myocytes before and after treatment with 250 nM Omecamtiv mecarbil and summary statistics (right) from sarcomere dynamics reveal increased contractile amplitude (n=14 pairs). F, Ensemble average traces of Clover signal (left) in isolated myocytes before and after treatment with 250nM Omecamtiv mecarbil and summary statistics (right) of Clover intensity changes reveal no difference in peak fluorescence intensity (P = 0.2847) (n=17 pairs). G, Ensemble average traces of SL shortening (left) in isolated myocytes before and after treatment with 10μM para-Nitroblebbistatin and summary statistics (right) from sarcomere dynamics reveal decreased contractile amplitude (n=8 pairs). H, Ensemble average traces of Clover signal (left) in isolated myocytes before and after treatment with 10μM para-Nitroblebbistatin and summary statistics (right) of Clover intensity changes reveal no difference in peak fluorescence intensity (P = 0.8930) (n=8 pairs). Myocytes were measured at 37°C with 0.2Hz stimulation; ΔSL bar represents a 5% change from baseline, ΔClover bar represents a 1% change from baseline. Mean ± S.E.M are presented, Paired two-tailed t-test: *P < 0.05.
Results
Real-time detection of TnC-based thin filament activation in intact adult cardiac myocytes
As detailed below, the troponin C FRET-based biosensor probe was integrated into the sarcomere and shown to function normally in cardiac muscle, in terms of myofilament activation and contractile function from cell to whole animal. Data support that the well-known activation-dependent conformational changes in TnC (compacted global TnC structure during activation [15, 17, 36]) bring the Clover and mRuby2 FRET pair closer together (in the vicinity of the R0 of 6.3 nm) to increase FRET and produce a transient that is detected with millisecond time resolution during a single twitch (e.g., Fig. 1).
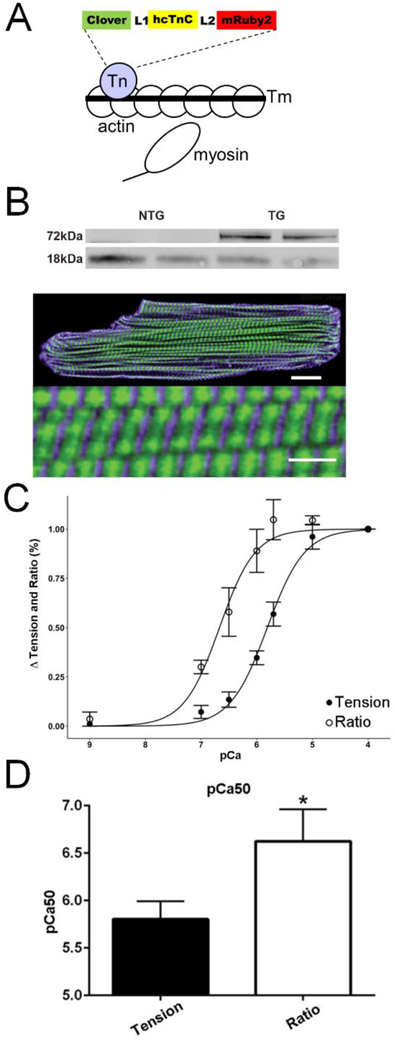
Figure 1. Expression and function of the cardiac sarcomere incorporated TnC biosensor in intact cardiac myocytes.
A, Model of the troponin C sarcomere biosensor, where FRET occurs by conformational changes in TnC during sarcomere activation. B, Western blot of cTnC expression probed with an anti-cTnC antibody in transgenic mouse hearts reveals stoichiometric replacement of endogenous cTnC by the higher molecular weight biosensor cTnC, blot is spliced horizontally to remove intervening non-specific bands. Full blot is in the Supplement. Below. Immunofluorescence with confocal microscopy images showing localization of Clover (green) is restricted between α-actinin (purple) delineated Z lines, scale bar is 25μm, bottom of panel shows magnified detail of the same cell, Scale bar is 2.5μm. C. Measurements of the Ca2+-sensitivity of steady-state isometric tension development and simultaneous FRET ratiometric fluorescence intensity changes in skinned papillary strips exposed to solutions of increasing Ca2+ concentration (FRET ratio defined in Methods). Tension was normalized to the pCa 4.0 value obtained in each preparation (n=6). D. Summary statistics of Ca2+ concentration at half maximal activation (pCa50) for tension and FRET fluorescence ratio demonstrate a significantly higher pCa50 for the fluorescence ratio than tension (n=6). Summary statistics of the Hill coefficient (nHill) for tension (1.33 ± 0.11) and fluorescence ratio (1.77 ± 0.27) show no significant difference (n=6). Mean ± S.E.M are presented, Unpaired two-tailed t-test: *P < 0.05.
The TnC biosensor was composed of fluorescent proteins Clover (donor) and mRuby2 (acceptor) engineered at the N- and C-terminal ends of cardiac TnC respectively (see Methods for details), and cardiac-directed TnC biosensor transgenic mouse lines were then established (Methods; Figs. 1a; S1,2). In independent Tg lines, the TnC biosensor stoichiometrically replaced 47% of the endogenous cTnC (Fig. 1b). Immunofluorescence detection of the TnC biosensor in intact cardiac myocytes demonstrated correct localization of the TnC biosensor in the thin filaments between the α-actinin labeled Z-lines (Fig. 1b, bottom). Similar findings were obtained in rat adult cardiac myocytes using acute gene transfer (Fig. S10). Thus, consistent with numerous past sarcomere-engineering studies, we find the TnC biosensor is incorporated correctly into the sarcomere without evidence of accumulation in the cytosol [3, 23, 37-41].
The troponin C biosensor derives from extensive in vitro studies showing steady-state Ca2+ responsive FRET upon TnC probe reconstitution into myofilaments [17]. To characterize the TnC biosensor we developed, biophysical analysis of Ca2+ titrations in permeabilized cardiac muscle preparations was conducted by simultaneously measuring force and TnC conformation-dependent FRET signals under steady-state Ca2+ activating conditions (Fig. 1C,D). Data show, qualitatively similar to that of Badr et al.[17], sigmoidal tension-pCa and TnC FRET ratio-pCa relationships, with the TnC FRET ratio-pCa curve shifted to the left of the tension-pCa curve, under steady state Ca2+ activating conditions. These biophysical data show that the sarcomere integrated TnC FRET biosensor detects Ca2+ -induced conformational changes in TnC in a Ca2+ concentration-dependent manner such that steady-state FRET increased as a function of increased activating levels of Ca2+ (Fig. 1C). In addition, data show that TnC biosensor-derived FRET was significantly more sensitive to Ca2+ than isometric tension (Fig. 1C,D).
In addition, as inherited mutations in the sarcomere can directly cause cardiomyopathy [42], we validated that the TnC biosensor itself had no significant effects on heart structure or function. Tension-pCa relationship studies in permeabilized single adult cardiac myocytes with (Tg) and without (Ntg littermate) the TnC biosensor incorporated into the sarcomeres were conducted (Fig. S3). Data show no change in pCa50, indicating that the design and sarcomere integration of the TnC biosensor had no effect on the steady state Ca2+ activating function of the muscle. This differs and significantly improves upon earlier studies in which the TnC FRET probe caused a significant alteration in skinned muscle pCa50 values [17]. Also, heart histology, heart morphology, or in vivo heart function were not different than controls, nor did the TnC biosensor have any effect on intact cardiac myocyte function (also confirmed by Ad5 acute gene transfer; Fig.S10), as compared to myocytes from non-transgenic littermates (Figs. S4-7). Because the cardiac muscle is highly sensitive to even small perturbations in sarcomere function, these are direct physiological evidence that the TnC biosensor confers normal TnC function, from the isolated myocyte to the intact heart level in vivo. These in vivo results are in agreement with in vitro work showing various CFP/YFP FRET pairs engineered cTnC having normal Ca2+/Mg2+ binding properties [17]. Collectively, these data show the TnC biosensor integrated normally into the sarcomere and confers normal TnC sarcomeric function that is indistinguishable from control. The sarcomere-integrated TnC biosensor also differs significantly from other probes that were designed specifically as cytoplasmic or myofilament localized Ca2+ indicators [43, 44].
We next sought to advance steady-state studies by us (Fig. 1), and others [16, 17, 36, 45], to dynamic twitch contractions in live cardiac muscle. To enable this, we leveraged this TnC FRET pair, optimized for enhanced photostability and brightness [21, 22]. In intact single adult cardiac myocytes, TnC biosensor Clover and mRuby2 signals were resolved in the millisecond time domain and were anti-correlated, indicating FRET (Fig. 1; Movie 1). This finding, together with work in reconstituted systems (Fig. 1), and with other FRET-based TnC pairs activated by direct Ca2+ titrations in vitro [17], is evidence that the TnC biosensor is capable of reporting in real-time the global conformational changes in TnC that occur during the cardiac twitch. Cardiac myocytes are small enough (lengths ~110 um) for the optical system to collect all the emitted fluorescence across the entire contracting myocyte thus, with this technique, possible motion artifacts of fluorophores moving in and out of the plane of detection, are highly mitigated, making it feasible to track only Clover. To validate Clover signal versus FRET pair ratio data, in a subset of experiments, FRET ratios (mRuby/Clover; defined in Methods) were determined for the isolated cells and shown to be no different than the Clover only transient.
Thin filament activating roles of cTnI and myosin in live cardiac myocytes
We investigated thin filament activation dynamics in intact and unloaded adult cardiac myocytes previously estimated from modeling studies [46]. Results showed that the peak amplitude of the TnC biosensor transient significantly preceded peak contraction (sarcomere length/SL)(Fig. 2, Fig. S8). The TnC biosensor transient was temperature-sensitive, paralleling the temperature dependence of contraction (Fig. S9), and, while temperature has myriad effects on muscle, this is consistent with the temperature dependence of the troponin-Tm regulatory system [1].
Next we tested the effects of enhanced cardiac myocyte excitation-contraction coupling on sarcomere activation via β-adrenergic stimulation (isoproterenol/Iso)[13]. Here, the TnC biosensor peak amplitude was increased and the TnC biosensor decay rate was significantly faster with Iso, paralleling the effects of Iso to increase contraction (positive inotropy) and to speed relaxation (positive lusitropy)(Fig. S8). The increased amplitude and faster decay rate of the TnC biosensor transient by Iso can be explained by the effects of Iso to increase the Ca2+ amplitude and by the combined effects of cTnI phosphorylation and faster Ca2+ transient decay to cause faster myofilament inactivation [47]. MyBP-C, titin, and multiple other molecules could also have roles, and it will be interesting to dissect these in future studies.
We examined the role of cTnI as an activating ligand [6, 9]. In vitro studies have well demonstrated that the cTnI switch peptide binding to cTnC is required for the full activation of cTnC, unlike in skeletal muscle where Ca2+ binding alone appears sufficient for full TnC activation [9, 48]. We first used N-aminohexyl-5-chloro-1-naphthalenesulfonamide (W7), a small molecule, originally discovered in binding to calmodulin, that does bind directly to the cTnC hydrophobic patch and markedly decreases cTnI switch peptide binding to the N-terminus of cTnC [25]. Thus, W7 was used to test the effects of inhibiting cTnC-cTnI interaction and consequently this disrupts sarcomere activation in live myocytes [26]. Here, the TnC biosensor amplitude was significantly depressed by W7 (Fig. 3) accompanying the effect of W7 to reduce myocyte SL amplitude (Fig. 3a). Because the Ca2+ transient is not altered by W7 (Fig. S11), this is evidence that the TnC biosensor is not a myoplasmic Ca2+ sensor. In conjunction with the switch-peptide occlusion mechanism [9, 48] and the effects of W7 on contraction [26], this is evidence that the TnC biosensor directly detects the key role of cTnC-cTnI interaction in live cells [1, 2, 9]. Next, we used Pimobendan, a small molecule positive inotrope [27] shown to directly enhance the affinity of cTnI for cTnC [28]. Here, Pimobendan caused a significant increase in the TnC biosensor FRET amplitude that paralleled the increase in SL amplitude (Fig. 3). Thus, the TnC biosensor reports the enhanced sarcomere activation elicited by increasing the affinity of cTnI for cTnC’s hydrophobic cleft. Collectively, these findings, by titrating cTnC-cTnI interactions, demonstrate that cTnI engagement with cTnC is a key effector of cardiac sarcomere activation in the live myocyte.
Present models suggest that strong myosin binding is the essential ligand causing the open state of the myofilament regulatory system [1, 10, 11]. To test this theory in intact cardiac myocytes, we first used Omecamtiv mecarbil (OM), a myosin-modulating drug shown to increase the rate of cross-bridge cycling by accelerating phosphate release, with no effects on intracellular Ca2+ handling [29, 30]. OM caused a significant increase in SL amplitude in adult cardiac myocytes but, surprisingly, had no effect on TnC biosensor amplitude or kinetics (Fig. 3). This result was unexpected, given the extensive literature on strong myosin binding as the required effector of sarcomere activation [1, 11]. To test more thoroughly this hypothesis, we investigated whether fully blocking strong myosin binding would depress the TnC biosensor signal, as predicted by current theory [1, 11]. TnC biosensor transgenic cardiac myocytes were exposed to para-nitroblebbistatin (PNB), a non-fluorescent small molecule that selectively disables myosin function by blocking myosin in an actin-detached state and rendering myosin incapable of producing force [31]. Surprisingly, whereas PNB effectively blocked unloaded myocyte contraction (Fig. 3), there was no effect of PNB on TnC biosensor transient amplitude or kinetics (Fig. 3).
Mechanical load and sarcomere activation
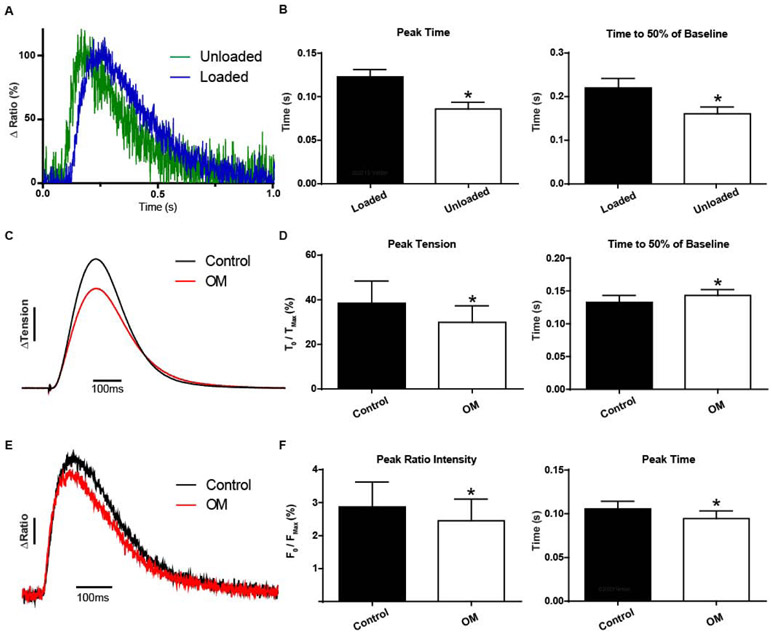
Mechanical load is well known to markedly alter myosin function, and load is predicted to have an impact on sarcomere activation in live muscle [1]. To compare to the results above using unloaded conditions, we next studied intact cardiac muscles under mechanical load. Cardiac papillary muscles were attached to a recording apparatus, and load was imposed via stepwise muscle length extension to set the optimal peak isometric twitch force amplitude for each preparation [1] (experimental apparatus shown in Fig. S2). Thus, for all experiments below, mindful of potential inhomogenities, muscles were studied at the physiologically relevant optimal length that produced peak isometric twitch contractions. Results showed that the TnC biosensor Clover and mRuby2 transients were anti-correlated, indicating FRET (Fig. 4). Cardiac muscle under mechanical load showed the TnC biosensor transient [herein, the mRuby2/Clover ratio to account for motion effects [34]] reached peak significantly before the development of peak isometric twitch force (Fig. 4). For these papillary experiments, mRuby2/Clover ratios are the necessary and well-validated approach to effectively address motion artifacts of the larger multicellular cardiac muscles [34]. In addition, motion artifacts are also mitigated because of the intramolecular FRET probe strategy we used, in that motion would change equally the numbers of Clover and mRuby2 fluorophores detected by the PMT. Interestingly, alignment of the loaded and unloaded TnC biosensor FRET ratio traces showed a prolongation of the loaded transient (by ~28%) that was due to a significant delay in the inactivation of the TnC biosensor transient (Fig. 5a,b).
Figure 4. TnC biosensor transient in live intact cardiac muscle under physiological load.
A, Representative single cardiac papillary muscle traces of simultaneous twitch force, Clover, mRuby2, and ratiometric calculation of fluorescent intensity. Scale bar represents a 10% force change or a 2% change in fluorescence intensity ratio. B, Representative single cardiac papillary of amplitude normalized FRET ratio (blue) and tension (black) dynamics at 22°C. Inset shows the same data highlighting the peak timing difference. C, D, Summary statistics of tension and FRET ratio reveals that the TnC biosensor conformational change precedes peak tension development (n=9 pairs). Experiments were at room temperature and 1.0Hz stimulation; ΔTension bar represents a 10% change from baseline, ΔClover bar represents a 2% change from baseline. Mean ± S.E.M are presented, Unpaired two-tailed t-test: *P < 0.05.
Figure 5. Load-dependent effects of myosin on sarcomere activation in live cardiac muscle.
A, Ensemble average of single myocytes (green) and papillaries (blue) reveals a load dependence to sarcomere activation (22°C and 0.2Hz). Note, FRET ratio-derived transients obtained in single myocytes were not different from Clover transient traces in the same single myocytes.
B, Summary statistics of sarcomere biosensor kinetics reveals increasing load and physiological complexity significantly delays peak activation and time to inactivation (n=8 per group). C, Ensemble average traces of tension recordings (non normalized) in papillaries before and after treatment with 250 nM Omecamtiv mecarbil shows alterations in tension development and kinetics, ΔTension bar represents a 10% change from baseline. D, Summary statistics of papillary twitch tension with 250nM Omecamtiv mecarbil reduces the change in peak tension and slows the relaxation kinetics (n=14 pairs). E, Ensemble average traces of FRET ratio recordings (non-normalized) in papillaries before and after treatment with 250 nM Omecamtiv mecarbil shows alterations in peak intensity and kinetics. ΔRatio bar represents a 5% change in FRET from baseline. F, Summary statistics of ensemble traces from e show Omecamtiv mecarbil reduces the peak FRET ratio intensity change while shortening time to peak activation (n=14 pairs). C-F, Data obtained at 22°C with 1.0Hz stimulation. Mean ± S.E.M are presented. Unpaired and paired two-tailed t-tests were used, as appropriate: *P < 0.05.
To investigate mechanism, we probed sarcomere activation in the presence of the direct myosin modulatory small molecule OM [30]. Under load, OM caused a more rapid upstroke of the TnC biosensor transient, as quantified by the time to peak signal intensity being significantly faster with OM (Fig. 5e,f). In addition, OM decreased twitch force as reported by a significantly depressed peak amplitude of the TnC biosensor transient (Fig. 5c,d). These results, together with the loaded vs. unloaded findings above (Fig. 5a,b), provide direct evidence in live cardiac muscle that the troponin-located TnC biosensor is sensitive to detecting the effects of cycling myosin as a sarcomere activating ligand.
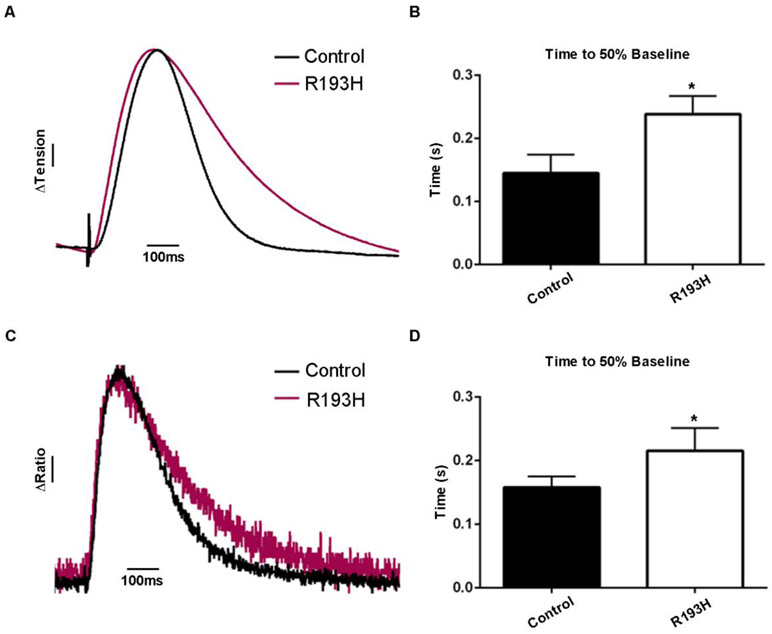
Next, we examined the effects on sarcomere activation due to an inherited sarcomeric mutation known to cause significant cardiomyopathy in patients [3]. The cTnI mutation R193H was used here as it has been shown to cause heightened myofilament Ca2+ sensitivity and organ-level diastolic dysfunction in animal models and in humans [24, 40, 49-51]. Here, cTnI R193H mice [40] were crossed with TnC biosensor mice to generate double transgenic lines. In loaded cardiac muscle from cTnI R193H Tg mice, results showed a significant delay in twitch force decay, and this was paralleled by a significant delay in TnC biosensor transient inactivation (Fig. 6). This finding, together with results showing no effect of cTnI R193H on the amplitude or kinetics of the intracellular Ca2+ transient [51], is evidence that cTnI R193H prolongs sarcomere activation by increasing the lifetime of cTnC-cTnI interaction. This prolonged cTnC-cTnI interaction is predicted to contribute to inappropriately increased myofilament-based Ca2+ buffering in the late phase of twitch contraction [42]. This sarcomere-based dysfunction, in turn, has been modeled as a key substrate for producing the fatal arrhythmias characteristic of the inherited cardiomyopathies [52].
Figure 6. Altered sarcomere activation due to cardiomyopathic mutant sarcomeric protein expression in live cardiac muscle.
A, Ensemble average traces of tension recordings in papillaries show restrictive cardiomyopathy cTnI mutant R193H have alterations in relaxation kinetics, ΔTension bar represents a 10% change from baseline. B, Summary statistics of papillary twitch tension shows R193H slows relaxation kinetics (n=6 for control and n=5 for R193H mutants). C, Ensemble average traces of FRET ratio recordings in papillaries show R193H mutants have slowed relaxation kinetics, ΔRatio bar represents a 10% change in FRET from baseline. D, Summary statistics of papillary FRET ratio shows R193H mutation slows relaxation kinetics (n=6 for control and n=4 for R193H mutants). A-D, Data obtained at 22°C with 1.0Hz stimulation. Mean ± S.E.M are presented. Unpaired t-tests were used, as appropriate: *P < 0.05.
Discussion
Elucidating the physiological basis of thin filament activation in live cardiac muscle is essential to understanding beat-to-beat regulation of heart performance in vivo. We sought here to design and test a sarcomere-incorporated biosensor capable of detecting thin myofilament activation status in live cardiac muscle. Towards this goal, we implemented a cardiac TnC FRET-based biosensor to test the hypothesis that TnC’s conformation can serve as a real-time biosensor and report the effects of multiple thin filament activating ligands during a single cardiac twitch contraction in live muscle.
Data show that the TnC biosensor is incorporated normally into the myofilaments by stoichiometric replacement of endogenous TnC, and that the biosensor itself has no untoward effects on cardiac myocyte or heart functional performance in vitro or in vivo. Because even slight changes in troponin Ca2+ binding kinetics cause marked alterations in cardiac muscle function, this is physiologically relevant functional evidence that the Ca2+ activating properties of the TnC biosensor functions as endogenous TnC. This is in agreement with in vitro work showing various CYP/YFP FRET pairs engineered in cTnC did not alter Ca2+/Mg2+ binding properties of isolated TnC [17]. We therefore posit that this TnC biosensor detects, for the first time to our knowledge, activation-dependent changes in cardiac TnC’s conformation, as determined by intramolecular FRET in real time during a twitch. Results show a well-delineated TnC conformation-dependent FRET transient, wherein the biosensor signal rises and reaches a peak prior to peak force during a twitch contraction. In addition, data derived from small molecule and genetic approaches, shown previously to alter thin filament activation [3, 26], provide evidence that the TnC biosensor can detect the effects of multiple thin filament activating ligands, including effects due to Ca2+, the TnI switch domain interfacing with TnC, and cycling cross-bridges altering TnC’s conformation. This is a significant physiological advance, as it was not a foregone conclusion that past biophysical studies probing TnC and sarcomere function under steady-state activating conditions[15, 16, 36], which can take minutes to collect data, would translate to the physiological setting of intact excitation-contraction coupling in live cardiac muscle, wherein the dynamics of the contraction transient are on the millisecond time scale.
We interpret the biosensor FRET transient as reporting TnC’s global conformational change, from extended to compact, during the twitch, as based on extensive in vitro work on TnC[17, 36, 53, 54]. During a twitch, we posit that the TnC-tethered FRET probes Clover (donor) and mRuby2 (acceptor) move closer together, consistent with the increased FRET signal observed. Based on the Clover/mRuby2 R0 of 6.3 nm, together with understanding of the useful FRET pair spacing range of 0.5 – 1.5 Ro[21, 22, 55], we estimate that the Clover to mRuby2 fluorophore distance varies within a range of ~3 nm (high FRET) to >~9 nm (no detectable FRET) during the twitch. Thus, with this biosensor probe, we report the presence and absence of FRET for this interprobe spacing range. In addition, given that each TnC biosensor is integrated into the sarcomere at a spacing no closer than every seven actin monomers, and together with the Clover/mRuby2 R0, we conclude that each TnC biosensor operates independently from neighboring biosensors, substantiating intramolecular FRET and not inter-molecular FRET.
In addition, given the experimental limitations of the contracting myocyte, in which the number of fluorophores in the detected volume varies in time and from sample to sample, it is virtually impossible to measure FRET efficiency values, or the precise corresponding inter-probe distances, accurately. This would require having simultaneous measurements of the fluorescence of donor-acceptor and donor-only samples[56], which is not feasible. However, there is enough information to show that: (a) the changes in FRET are within the expected range for this system (lobes of TnC)[36, 53], (b) the fluorescence signals are detected with sufficient precision to detect subnanometer changes in distance, and therefore (c) the time dependence of the FRET signal change reflects accurately the time dependence of TnC structural changes during the myocyte twitch. Given uncertainties regarding the conformations and orientations of the tethered fluorescent proteins relative to the attached sites on TnC[55], the FRET values observed are consistent with distances within the sensitivity range of this donor-acceptor pair, which has an R0 value of 6.3 nm [21, 22]. Thus, they are within the range of Ca2+-dependent FRET values and FRET changes that have been previously reported by others for cardiac TnC [17].
There is strong rationale for proposing that TnC’s conformation serves as a nexus in detecting the effects of multiple ligands that regulate thin filament activation. Extensive in vitro data demonstrate that TnC’s conformation is sensitive to and can integrate near and far myofilament activating ligands across the sarcomere, including Ca2+ binding to TnC, cTnI binding to cTnC, and myosin binding to actin [1, 12, 18, 19, 36]. Studies in reconstituted myofilament systems show that fluorescent probes placed on skeletal TnC can detect the effects of cycling myosin to displace Tm and induce thin filament activation, when studied under steady-state activating conditions [18]. This effect was also seen in cardiac TnC conformational changes in reconstituted cardiac muscles[19]. In addition, recent in vitro studies have engineered FRET pairs into cardiac TnC to successfully monitor TnC conformational changes under steady-state Ca2+-activating conditions [17]. While foundational, an important caveat with these previous studies is that data are collected under steady state conditions, operating on the seconds-to-minutes time scale. Because live cardiac muscle operates under dynamic conditions, with excitation-contraction coupling occurring on the 10s of millisecond timescale, it is unclear whether previous steady-state findings would translate to the physiological setting of live muscle. Accordingly, the findings reported here are significant in showing evidence of TnC-localized biosensing of multiple activating ligands, including cycling myosin, under conditions of physiological excitation-contraction coupling and mechanical load in live cardiac muscle.
Data shows that the TnC biosensor is capable of detecting the effects of the TnI switch domain as an activating ligand. Small-molecule and genetic approaches were used that specifically disrupt TnI’s interaction with TnC, independent of effects on the Ca2+ transient. In addition, under physiological loading conditions, the TnC-localized biosensor detects the effects of small molecules that specifically alter myosin cross-bridge cycling rate. To our knowledge, these are the first data providing evidence in live cardiac muscle that TnI and cycling myosin cross-bridges can transduce thin filament activation, as detected by TnC conformational changes during a physiological twitch contraction. Based on extensive in vitro data that TnI-TnC and myosin transduce sarcomere-activating effects via Tm displacement [1, 6, 57, 58], the present data suggest that the TnC biosensor transient reports effects of Tm displacement in regulating thin filament activation. The potential implications of these findings, in further understanding the mechanism of sarcomere function in intact cardiac muscle, are discussed further below.
It is well established that tropomyosin’s (Tm) position on the thin filament governs the overall activation status of the cardiac sarcomere [1, 2]. In diastole, Tm blocks strong crossbridge formation to actin, and this inhibition is relieved by a Ca2+-dependent switch in troponin. Here, the C-terminus of TnI switches its strong binding to actin to strong binding to TnC [9], thus permitting Tm displacement and force production [8]. In addition, structural studies provide compelling evidence that strong myosin binding is required to fully displace Tm and accomplish complete activation of the thin filament regulatory system [57, 58]. Thus, in cardiac muscle both TnI switching, from actin to TnC, and myosin binding to actin serve as activating ligands by orchestrating Tm displacement on the thin filament. The present findings provide new evidence of the dynamics of TnI and myosin activating ligands during a twitch, with evidence that cycling myosin effects are highly dependent upon mechanical loading in live cardiac muscle.
Our findings can be discussed in the context of current models of sarcomere activation. Sarcomere activation is defined as the functional status of the thin filament regulatory system, whereby the open state position of Tm is permissive for strong cross-bridge formation with actin [1, 2]. It follows that regulation of sarcomere activation entails the processes that control Tm's position on the thin filament to control contraction. Sarcomere activation has been modeled by Tm orchestrating blocked, closed, and open states on the thin filament, and this formalism has guided the field for decades [1, 11]. However, assigning the relative roles of cTnI and myosin as activating ligands, during the non-steady-state conditions of the cardiac twitch, has remained elusive. This is an important void to address, as sarcomere dysfunction underlies multiple forms of acquired and inherited heart disease [3, 42]. Biochemical studies posit strong myosin binding to actin as the essential activating ligand required for the open state of the thin filament regulatory system [11]. However, recent cryo-EM studies suggest that the open state, as discerned by the disposition of Tm on the actin filament, is highly dependent on striated muscle-lineage [57]. Specifically, in cardiac muscle, structural-based results suggest that the Ca2+-dependent cTnC-cTnI complex is the dominant activating ligand translocating Tm into the force-permissive open state, with little to no significant role of myosin to acquire the open state [57]. Results from the present study find support from these structural findings. Under unloaded and loaded conditions, cTnI appears sufficient as an activating ligand. Results further suggest that under loaded conditions, myosin has a primary role to maintain Tm's position in the open state.
This study provides multiple lines of evidence that the cTnC-cTnI interface is a key sarcomere activating ligand during the cardiac twitch. Results with small molecules, which directly target modification of the cTnC-cTnI interface, [9] indicate that the initiation of the sarcomere open state during the cardiac twitch is due to the effects of the cTnC-cTnI as an activating ligand regulatory complex. Strengthening (pimobendan) or weakening (W7) the cTnC-cTnI interface [9, 25, 48], independently of Ca2+ handling, causes a corresponding increase or decrease in the TnC biosensor transient amplitude, respectively. In addition, the biosensor transient is significantly prolonged and decays more slowly due to a disease-causing cTnI R193H mutant, known to prolong cTnC-cTnI association and delay cardiac relaxation, all without altering the Ca2+ transient [3]. Because sarcomere-modifying small molecules (e.g., W7, OM) and mutant cTnI R193H do not alter the intracellular Ca2+ transient, these are evidence that the TnC biosensor transient is reflective of sarcomere-intrinsic regulatory processes and is not a myoplasmic Ca2+ sensor. This distinguishes the TnC biosensor from other genetically encoded sensors that specifically report Ca2+ content in cardiac myocytes, and not sarcomere regulatory dynamics per se [59]. This distinction is important because sarcomere performance can be altered dramatically in cardiac diseases independent of changes in the intracellular Ca2+ transient [3, 51].
Because cTnI's switching mechanism, from actin to TnC, is well known from in vitro studies to control Tm's translocation on the actin filament [1, 2], it follows that the troponin-localized biosensor is sensitive to, and provides an indirect measure of, Tm's disposition (e.g., Blocked/Closed to Open state) during the twitch. Upon muscle stimulation, data here support a model whereby the formation of the cTnCCa2+ state causes the regulatory switch peptide domain of cTnI to rapidly reverse its strong binding to actin and to then become strongly bound to cTnC. Here, formation of the cTnC - cTnI complex is stabilized, under either loaded or unloaded conditions, and this appears to dominate the initiation of the sarcomere-activating open state required for cycling myosin interactions with actin (Fig. S12). In the setting of the cardiac twitch, intracellular Ca2+ rises and significantly falls before peak force development [1], underscoring the key role of the proximity of the cTnI switch peptide to the N-terminal domain of cTnC to sustain Tm in a dis-inhibiting position, permitting the sarcomere activation open state [9]. This is supported by noting the TnC biosensor transient reaches its peak prior to development of peak force. Results using W7 or pimobendan further underscore how inhibiting or enhancing cTnC-cTnI interactions, respectively, ultimately affects the population of cTnI available to re-bind to actin and shift Tm's position from open to closed/blocked states.
The TnC biosensor detected the thin filament activating effects of cycling myosin crossbridge under conditions of mechanical load. In 1972, Bremel and Weber first suggested the role of myosin as a key regulator of sarcomere activation in muscle [10]. In the following decades, numerous studies have supported the critical role of myosin as an actuator of the troponin-tropomyosin allosteric regulatory system to initiate sarcomere activation [1, 2, 6, 11, 20]. Compared to unloaded conditions, mechanical loading demonstrated prolonged sarcomere activation, as manifest by a significant delay in the inactivation of the TnC biosensor transient. Because mechanical loading causes a significant slowing in the cross-bridge cycling rate and increases the myosin duty cycle [1], this is evidence that the troponin-localized biosensor can detect, and is sensitive to, the load-dependent effect of altered myosin cycling on the TnC biosensor during a twitch, as predicted from in vitro studies [1, 10, 18, 19]. The data further suggest that during the decay phase of the cardiac twitch, cycling myosin attachment to actin sustains Tm in the open state, which, in a reciprocal manner, facilitates the maintaining of the cTnI switch peptide domain at or near the cTnC hydrophobic patch [9]. Therefore, we posit that cycling myosin effectively increases the lifetime of the cTnC-cTnI activating ligand complex during the relaxation phase of the twitch (Fig. S12). The TnC biosensor detected this effect of cycling myosin, demonstrating for the first time in live muscle, the dynamic interconnection between the thin filament and myosin in orchestrating sarcomere activation.
The TnC biosensor also detected the effects of modifying cycling myosin using a myosin-specific small molecule approach. The myosin-modulating drug OM depressed the TnC biosensor peak amplitude while the TnC biosensor transient had a faster time to peak. These findings are consistent with studies showing OM accelerating the phosphate release step from myosin while suppressing myosin's working stroke [29]. Myosin-targeted small-molecule effects on the TnC biosensor transient are of further interest as disease-causing myosin mutations can produce similar alterations in myosin function [3, 5, 42]. We posit that other sarcomere mutants, including those in myosin heavy and light chains, MyBP-C, titin, and others that modify myosin cycling [3], would affect sarcomere activation as well. In unloaded conditions, completely blocking myosin function did not affect the TnC biosensor transient. The force-velocity relationship dictates that under unloaded conditions the cross-bridge cycling rate increases and the number of strong binding cross-bridges significantly decreases, as compared to loaded contractions [1]. This can account for the lack of a detected effect of myosin on the TnC biosensor during unloaded contractions. It then follows that myosin's effect to activate the sarcomere is highly load dependent, as now shown here in live cardiac muscle via the TnC biosensor. Taken together, these data support a primary role for cycling myosin under baseline physiological conditions to sustain sarcomere activation, rather than to initiate sarcomere activation, most evident in the late phase of contraction under load, as supported by recent cryo-EM studies [57]. Collectively, cTnI and strong myosin binding each have roles in sarcomere activation/inactivation that are time and load dependent in the context of the cardiac twitch (Fig. S12).
Taken together, these findings have implications for understanding cardiac sarcomere activation mechanisms that govern beat-to-beat control of heart performance. For example, in several inherited and acquired cardiac diseases, sarcomeric dysfunction, and not Ca2+ mishandling, underlies disease initiation, and this is termed EC-uncoupling [42, 51, 60]. Here, disease severity couples closely with the degree of sarcomere-based EC-uncoupling. In acute ischemia, hypoxia, and acidosis, the intracellular Ca2+ transient is normal when sarcomere-based heart pump performance is severely depressed [60]. Furthermore, in inherited cardiomyopathies premature sudden cardiac death can result directly from sarcomeric mutations, uncoupled from alterations in the Ca2+ transient [42]. These underscore the critical role of sarcomere performance in cardiac health and disease. Advances here may provide further insights into the function of the sarcomere in health and disease, including elucidating the roles of new sarcomere-modifying drugs [27], myofilament post-translational modifications [47], and the effects of an expanding catalog of sarcomere mutations that cause human disease [3, 42].
Future investigations of sarcomere regulation under varying physiological conditions including stress and across species will be of interest. In addition, this new biosensor should be of value in discerning mechanisms of inter-myofilament signaling, including new small molecules, disease models, and the roles of MyBP-C, titin, and others. Collectively, these advances further highlight the sarcomere as an excellent therapeutic target. Finally, it would be of interest to develop a TnC biosensor for live skeletal muscle study. We speculate, owing to the significant differing mechanism of troponin-based sarcomere regulation in heart versus skeletal muscle [48], that ligands of sarcomere activation will differ significantly between the striated muscle lineages when studied in the physiological context of intact muscle.
Supplementary Material
Movie S1. Online TnC biosensor transient recordings showing single cardiac myocyte visualization of Clover and mRuby2 transients during the cardiac twitch in adult cardiac myocytes.
Highlights.
Cardiac troponin C (TnC) FRET-based biosensor was integrated into the sarcomere
TnC biosensor detected thin filament activation in real time in live cardiac muscle during twitch contractions
The TnC biosensor detected multiple ligands of activation, including Ca2+, TnI, myosin and the effects of physiological loading
Acknowledgements
We thank Drs. J.M. Muretta and M.A. Sanders for helpful discussions and K.W. Prins and H. Cohen for assistance. We thank A. Málnási-Csizmadia for the gift of para-nitroblebbistatin. This work was supported by grants from NIH, AHA and UMN LHI and IBP.
Footnotes
Disclosures
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gordon AM, Homsher E, Regnier M, Regulation of contraction in striated muscle, Physiol Rev 80(2) (2000) 853–924. [DOI] [PubMed] [Google Scholar]
- [2].Tobacman LS, Thin filament-mediated regulation of cardiac contraction, Annu. Rev. Physiol 58 (1996) 447–481. [DOI] [PubMed] [Google Scholar]
- [3].Davis J, Westfall MV, Townsend D, Blankinship M, Herron TJ, Guerrero-Serna G, Wang W, Devaney E, Metzger JM, Designing heart performance by gene transfer, Physiol Rev 88(4) (2008) 1567–1651. [DOI] [PubMed] [Google Scholar]
- [4].Metzger JM, Westfall MV, Covalent and noncovalent modification of thin filament action: the essential role of troponin in cardiac muscle regulation, Circ. Res 94(2) (2004) 146–158. [DOI] [PubMed] [Google Scholar]
- [5].Seidman C, Genetic causes of inherited cardiac hypertrophy: Robert L. Frye Lecture, Mayo Clin. Proc 77(12) (2002) 1315–1319. [DOI] [PubMed] [Google Scholar]
- [6].Farah CS, Reinach FC, The troponin complex and regulation of muscle contraction, FASEB J 9(9) (1995) 755–767. [DOI] [PubMed] [Google Scholar]
- [7].Takeda S, Yamashita A, Maeda K, Maeda Y, Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form, Nature 424(6944) (2003) 35–41. [DOI] [PubMed] [Google Scholar]
- [8].Farah CS, Miyamoto CA, Ramos CH, da Silva AC, Quaggio RB, Fujimori K, Smillie LB, Reinach FC, Structural and regulatory functions of the NH2- and COOH-terminal regions of skeletal muscle troponin I, J. Biol. Chem 269(7) (1994) 5230–5240. [PubMed] [Google Scholar]
- [9].Sykes BD, Pulling the calcium trigger, Nat. Struct. Biol 10(8) (2003) 588–589. [DOI] [PubMed] [Google Scholar]
- [10].Bremel RD, Weber A, Cooperation within actin filament in vertebrate skeletal muscle, Nat. New Biol 238(82) (1972) 97–101. [DOI] [PubMed] [Google Scholar]
- [11].McKillop DF, Geeves MA, Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament, Biophys. J 65(2) (1993) 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Geeves MA, Holmes KC, The molecular mechanism of muscle contraction, Adv. Protein Chem 71 (2005) 161–193. [DOI] [PubMed] [Google Scholar]
- [13].Bers DM, Cardiac excitation-contraction coupling, Nature 415(6868) (2002) 198–205. [DOI] [PubMed] [Google Scholar]
- [14].Sevrieva I, Knowles AC, Kampourakis T, Sun YB, Regulatory domain of troponin moves dynamically during activation of cardiac muscle, J Mol Cell Cardiol 75 (2014) 181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li KL, Ghashghaee NB, Solaro RJ, Dong W, Sarcomere length dependent effects on the interaction between cTnC and cTnI in skinned papillary muscle strips, Arch Biochem Biophys 601 (2016) 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li KL, Rieck D, Solaro RJ, Dong W, In situ time-resolved FRET reveals effects of sarcomere length on cardiac thin-filament activation, Biophys J 107(3) (2014) 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Badr MA, Pinto JR, Davidson MW, Chase PB, Fluorescent Protein-Based Ca2+ Sensor Reveals Global, Divalent Cation-Dependent Conformational Changes in Cardiac Troponin C, PLoS. One 11(10) (2016) e0164222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zot AS, Potter JD, Reciprocal coupling between troponin C and myosin crossbridge attachment, Biochemistry 28(16) (1989) 6751–6756. [DOI] [PubMed] [Google Scholar]
- [19].Hannon JD, Martyn DA, Gordon AM, Effects of cycling and rigor crossbridges on the conformation of cardiac troponin C, Circ. Res 71(4) (1992) 984–991. [DOI] [PubMed] [Google Scholar]
- [20].Moss RL, Razumova M, Fitzsimons DP, Myosin crossbridge activation of cardiac thin filaments: implications for myocardial function in health and disease, Circ Res 94(10) (2004) 1290–300. [DOI] [PubMed] [Google Scholar]
- [21].Bajar BT, Wang ES, Zhang S, Lin MZ, Chu J, A Guide to Fluorescent Protein FRET Pairs, Sensors (Basel) 16(9) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, Tsien RY, Lin MZ , Improving FRET dynamic range with bright green and red fluorescent proteins, Nat. Methods 9(10) (2012) 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Westfall MV, Rust EM, Albayya F, Metzger JM, Adenovirus-mediated myofilament gene transfer into adult cardiac myocytes, Methods Cell Biol 52 (1997) 307–322. [PubMed] [Google Scholar]
- [24].Davis J, Wen H, Edwards T, Metzger JM, Allele and species dependent contractile defects by restrictive and hypertrophic cardiomyopathy-linked troponin I mutants, J. Mol. Cell Cardiol 44(5) (2008) 891–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Oleszczuk M, Robertson IM, Li MX, Sykes BD, Solution structure of the regulatory domain of human cardiac troponin C in complex with the switch region of cardiac troponin I and W7: the basis of W7 as an inhibitor of cardiac muscle contraction, J. Mol. Cell Cardiol 48(5) (2010) 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Thompson BR, Martindale J, Metzger JM, Sarcomere neutralization in inherited cardiomyopathy: Small molecule proof-of-concept to correct hyper-Ca2+ sensitive myofilaments, Am. J. Physiol Heart Circ. Physiol (2016) ajpheart. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hwang PM, Sykes BD, Targeting the sarcomere to correct muscle function, Nat. Rev. Drug Discov 14(5) (2015) 313–328. [DOI] [PubMed] [Google Scholar]
- [28].Schlecht W, Li KL, Hu D, Dong W, Fluorescence Based Characterization of Calcium Sensitizer Action on the Troponin Complex, Chem. Biol. Drug Des 87(2) (2016) 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Woody MS, Greenberg MJ, Barua B, Winkelmann DA, Goldman YE, Ostap EM, Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke, Nat. Commun 9(1) (2018) 3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Malik FI, Hartman JJ, Elias KA, Morgan BP, Rodriguez H, Brejc K, Anderson RL, Sueoka SH, Lee KH, Finer JT, Sakowicz R, Baliga R, Cox DR, Garard M, Godinez G, Kawas R, Kraynack E, Lenzi D, Lu PP, Muci A, Niu C, Qian X, Pierce DW, Pokrovskii M, Suehiro I, Sylvester S, Tochimoto T, Valdez C, Wang W, Katori T, Kass DA, Shen YT, Vatner SF, Morgans DJ, Cardiac myosin activation: a potential therapeutic approach for systolic heart failure, Science 331(6023) (2011) 1439–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kepiro M, Varkuti BH, Vegner L, Voros G, Hegyi G, Varga M, Malnasi-Csizmadia A, para-Nitroblebbistatin, the non-cytotoxic and photostable myosin II inhibitor, Angew. Chem. Int. Ed Engl 53(31) (2014) 8211–8215. [DOI] [PubMed] [Google Scholar]
- [32].Metzger JM, Myosin binding-induced cooperative activation of the thin filament in cardiac myocytes and skeletal muscle fibers, Biophys. J 68(4) (1995) 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Metzger JM, Parmacek MS, Barr E, Pasyk K, Lin WI, Cochrane KL, Field LJ, Leiden JM, Skeletal troponin C reduces contractile sensitivity to acidosis in cardiac myocytes from transgenic mice, Proc. Natl. Acad. Sci. U. S. A 90(19) (1993) 9036–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ding Y, Li J, Enterina JR, Shen Y, Zhang I, Tewson PH, Mo GC, Zhang J, Quinn AM, Hughes TE, Maysinger D, Alford SC, Zhang Y, Campbell RE, Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange, Nat. Methods 12(3) (2015) 195–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cleveland W, Robust Locally Weighted Regression and Smoothing Scatterplots., Journal of the American Statistical Association 74(368) (1979) 829–836. [Google Scholar]
- [36].Zhang X, Kampourakis T, Yan Z, Sevrieva I, Irving M, Sun YB, Distinct contributions of the thin and thick filaments to length-dependent activation in heart muscle, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Westfall MV, Rust EM, Metzger JM, Slow skeletal troponin I gene transfer, expression, and myofilament incorporation enhances adult cardiac myocyte contractile function, Proc. Natl. Acad. Sci. U. S. A 94(10) (1997) 5444–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Michele DE, Albayya FP, Metzger JM, Thin filament protein dynamics in fully differentiated adult cardiac myocytes: toward a model of sarcomere maintenance, J. Cell Biol 145(7) (1999) 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Day SM, Westfall MV, Fomicheva EV, Hoyer K, Yasuda S, Cross NC, D’Alecy LG, Ingwall JS, Metzger JM, Histidine button engineered into cardiac troponin I protects the ischemic and failing heart, Nat. Med 12(2) (2006) 181–189. [DOI] [PubMed] [Google Scholar]
- [40].Davis J, Wen H, Edwards T, Metzger JM, Thin filament disinhibition by restrictive cardiomyopathy mutant R193H troponin I induces Ca2+-independent mechanical tone and acute myocyte remodeling, Circ. Res 100(10) (2007) 1494–1502. [DOI] [PubMed] [Google Scholar]
- [41].Palermo J, Gulick J, Ng W, Grupp IL, Grupp G, Robbins J, Remodeling the mammalian heart using transgenesis, Cell Mol. Biol. Res 41(6) (1995) 501–509. [PubMed] [Google Scholar]
- [42].Seidman JG, Seidman C, The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms, Cell 104(4) (2001) 557–567. [DOI] [PubMed] [Google Scholar]
- [43].Sparrow AJ, Sievert K, Patel S, Chang YF, Broyles CN, Brook FA, Watkins H, Geeves MA, Redwood CS, Robinson P, Daniels MJ, Measurement of Myofilament-Localized Calcium Dynamics in Adult Cardiomyocytes and the Effect of Hypertrophic Cardiomyopathy Mutations, Circ. Res 124(8) (2019) 1228–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Heim N, Garaschuk O, Friedrich MW, Mank M, Milos RI, Kovalchuk Y, Konnerth A, Griesbeck O, Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor, Nat Methods 4(2) (2007) 127–9. [DOI] [PubMed] [Google Scholar]
- [45].Rieck DC, Li KL, Ouyang Y, Solaro RJ, Dong WJ, Structural basis for the in situ Ca(2+) sensitization of cardiac troponin C by positive feedback from force-generating myosin cross-bridges, Arch Biochem Biophys 537(2) (2013) 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Robertson SP, Johnson JD, Potter JD, The time-course of Ca2+ exchange with calmodulin, troponin, parvalbumin, and myosin in response to transient increases in Ca2+, Biophys. J 34(3) (1981) 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yasuda S, Coutu P, Sadayappan S, Robbins J, Metzger JM, Cardiac transgenic and gene transfer strategies converge to support an important role for troponin I in regulating relaxation in cardiac myocytes, Circ. Res 101(4) (2007) 377–386. [DOI] [PubMed] [Google Scholar]
- [48].Sia SK, Li MX, Spyracopoulos L, Gagne SM, Liu W, Putkey JA, Sykes BD, Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain, J. Biol. Chem 272(29) (1997) 18216–18221. [DOI] [PubMed] [Google Scholar]
- [49].Mogensen J, Kubo T, Duque M, Uribe W, Shaw A, Murphy R, Gimeno JR, Elliott P, McKenna WJ, Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations, J. Clin. Invest 111(2) (2003) 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Davis J, Metzger JM, Combinatorial effects of double cardiomyopathy mutant alleles in rodent myocytes: a predictive cellular model of myofilament dysregulation in disease, PLoS. One 5(2) (2010) e9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Davis J, Yasuda S, Palpant NJ, Martindale J, Stevenson T, Converso K, Metzger JM, Diastolic dysfunction and thin filament dysregulation resulting from excitation-contraction uncoupling in a mouse model of restrictive cardiomyopathy, J. Mol. Cell Cardiol 53(3) (2012) 446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Baudenbacher F, Schober T, Pinto JR, Sidorov VY, Hilliard F, Solaro RJ, Potter JD, Knollmann BC, Myofilament Ca2+ sensitization causes susceptibility to cardiac arrhythmia in mice, J. Clin. Invest 118(12) (2008) 3893–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Li KL, Methawasin M, Tanner BCW, Granzier HL, Solaro RJ, Dong WJ, Sarcomere length-dependent effects on Ca(2+)-troponin regulation in myocardium expressing compliant titin, J Gen Physiol 151(1) (2019) 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sevrieva IR, Brandmeier B, Ponnam S, Gautel M, Irving M, Campbell KS, Sun YB, Kampourakis T, Cardiac myosin regulatory light chain kinase modulates cardiac contractility by phosphorylating both myosin regulatory light chain and troponin I, J Biol Chem (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Day RN, Fluorescent proteins for FRET: monitoring protein interactions in living cells, in: Day RN, Davidson MW (Eds.), The fluorescent protein revolution, CRC Press, Boca Raton, Florida, 2014, pp. 245–278. [Google Scholar]
- [56].Thomas DD, Muretta JM, Colson BA, Mello RN, Kast D, Spectroscopic Probes of Muscle Proteins, Comprehensive Biophysics, Vol 4: Molecular Motors and Motility (2011) 226–250. [Google Scholar]
- [57].Risi C, Eisner J, Belknap B, Heeley DH, White HD, Schroder GF, Galkin VE, Ca(2+)- induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy, Proc. Natl. Acad. Sci. U. S. A 114(26) (2017) 6782–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lehman W, Craig R, Tropomyosin and the steric mechanism of muscle regulation, Adv. Exp. Med. Biol 644 (2008) 95–109. [DOI] [PubMed] [Google Scholar]
- [59].Kaestner L, Scholz A, Tian Q, Ruppenthal S, Tabellion W, Wiesen K, Katus HA, Muller OJ, Kotlikoff MI, Lipp P, Genetically encoded Ca2+ indicators in cardiac myocytes, Circ. Res 114(10) (2014) 1623–1639. [DOI] [PubMed] [Google Scholar]
- [60].Lee JA, Allen DG, Mechanisms of acute ischemic contractile failure of the heart. Role of intracellular calcium, J. Clin. Invest 88(2) (1991) 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Online TnC biosensor transient recordings showing single cardiac myocyte visualization of Clover and mRuby2 transients during the cardiac twitch in adult cardiac myocytes.