Abstract
Background:
There is a growing recognition of the inherent limitations of the use of the left ventricular ejection fraction (LVEF) to accurately phenotype patients with heart failure (HF).
Objectives:
To identify unique proteomic signatures for patients with HFrEF, HFmrEF and HFpEF, as well as to identify molecular differences between ischemic and non-ischemic HF patients.
Methods:
We used high content aptamer-based proteomics technology (SOMAscan) to interrogate the blood proteome of age and gender matched HF patients within different LVEF groups.
Results:
Within the Washington University Heart Failure registry, we identified age/sex matched patients within three LVEF categories: HFrEF (LVEF < 40%), HF with a mid-range LVEF (HFmrEF [LVEF 40–50%]) and HF with a preserved LVEF (HFpEF [LVEF > 50%]). We found that patients with HFrEF, HFmrEF, and HFpEF had unique variations in circulating proteins which reflected distinct biological pathophysiologies. Bioinformatics analysis revealed that there were biological themes that were unique to HFrEF, HFpEF and HFmrEF patients. Comparative analyses of patients with HFmrEF with improved LVEF and patients with HFmrEF with unchanged LVEF revealed marked differences between these two patient populations and indicated that patients with recovered LVEF are more similar to patients with HFpEF than to patients with HFrEF. Moreover, there were marked difference in the proteomic signatures of patients with ischemic and non-ischemic HF.
Conclusions:
Viewed together these findings suggest that it may be possible to use high-content multiplexed proteomics assays in combination with the clinical assessment of LVEF to more accurately identify clinical phenotypes of HF patients.
Keywords: heart failure, proteomics, left ventricular ejection fraction
Condensed Abstract:
High content aptamer-based proteomics technology (SOMAscan) was used to interrogate the blood proteome of age and gender matched HF patients who were classified based on their LVEF into HFrEF (LVEF < 40%), HF with a mid-range LVEF (HFmrEF [LVEF 40–50%]) and HF with a preserved LVEF (HFpEF [LVEF > 50%]). Patients with HFrEF, HFmrEF, and HFpEF had unique variations in circulating proteins which reflected distinct biological pathophysiologies. Bioinformatics analysis revealed biological themes that were unique to HFrEF, HFpEF and HFmrEF patients, suggesting that it may be possible to use proteomics assays to more accurately predict clinical phenotypes of HF patients.
INTRODUCTION
The use of left ventricular ejection fraction (LVEF) to stratify patients with heart failure (HF) is currently the most accurate clinical method to identify those HF patients who are more likely to respond favorably to neurohormonal antagonists and medical devices (1). Nonetheless, there is growing awareness in the field of the need to phenotype HF patients beyond the conventional assessment of LVEF (2). As one example, patients with HF with a history of a reduced LVEF ≤ 40% (HFrEF) who recover LV function (LVEF ≥ 50%) on evidence based medical and device therapies would no longer qualify for the use of diuretics and neurohormonal antagonists based on the current guideline recommendations for the treating patients with HFrEF (3). However, the result of the TRED-HF trial demonstrated that discontinuation of evidence based medical therapies in HFrEF patients with a recovered LVEF ≥ 50% culminates in worsening HF (4). Thus, in the modern era of HF therapeutics the assessment of LVEF in isolation does not necessarily provide accurate information with respect to a HF patient’s phenotype.
The development of high-throughput molecular biology techniques (omics) provides the opportunity to deep phenotype HF patients and to increase our understanding of the pathophysiology of HF (reviewed in (5)). In the current study we have used an aptamer-based proteomics technology, SOMAscan, to interrogate the blood proteome of age and gender matched HF patients who were classified based on their LVEF into HFrEF (LVEF < 40%), HF with a mid-range LVEF (HFmrEF [LVEF 40–50%]) and HF with preserved LVEF (HFpEF [LVEF > 50%]). Here we demonstrate that is feasible to use a high-content multiplexed assay to identify distinct proteomic signatures for patients with HFrEF, HFmrEF and HFpEF, as well as to identify molecular differences between ischemic and non-ischemic HF patients.
MATERIALS AND METHODS
Subjects and samples:
The patients for these studies represent a subset of patients with HFrEF (LVEF <40% [n=47]), HFpEF (LVEF >50% [n=43]), and HFmrEF (LVEF 40–50% [n=83]), who were selected randomly from a larger cohort of previously reported (6) age (± 5 years) and gender matched patients with HFrEF (LVEF <40% [n= 102]), HFpEF (LVEF >50% [82]), and HFmrEF (LVEF 40–50% [n=168]) enrolled in the Washington University Heart Failure Registry, using a computer generated random selection algorithm. The Washington University Heart Failure Registry is a prospective registry of inpatients and outpatients with clinical evidence of HF, irrespective of LVEF (7). Detailed patient information along with a blood sample was prospectively collected at the time of enrollment (March 2010 – August 2013) into the registry, and patient vital status was followed for 2 years after enrollment (7). The registry and the analysis included in this study were approved by the Washington University Internal Review Board under the IRB entitled “Washington University Heart Failure Registry.” As described previously, the HFmrEF patients were further categorized into HFmrEF improved (n = 59), HFmrEF unchanged (n = 8) or HFmrEF deteriorated (n = 16), based on whether the LVEF at the time of enrollment into the registry was improved, worsened, or the same as a prior assessment of LVEF that was obtained at the time that the patient was first diagnosed with HF, which was determined from a retrospective chart review. Because there were too few patients in the HFmrEF deteriorated and HFmrEF unchanged groups, for the purpose of this for subgroup analysis, we combined these patients into HFmrEF unimproved group (n = 24). The cohorts of HFrEF and HFpEF patients in this study were propensity matched for sex and age with the HFmrEF cohort, as described (6).
Plasma Proteomic Profiling, Data Preprocessing and Bioinformatics Analyses
Statistical Analysis
RESULTS
Internal and Biological Validation of the SOMAscan Protein Expression Data
The results of the SOMAscan protein expression data were internally validated for precision and accuracy. Precision was assessed based on replicate analysis and accuracy was assessed based on expected changes in plasma proteins across specific patient subgroups. We ran 5 replicates of a vendor-provided calibrator sample and 3 replicates of pooled plasma on each of the 8 assay plates we analyzed. The coefficient of variation (CV) for the vendor provided calibrators was less than 3.5% among 50% of SOMAmer reagents, and less than 12.4% among 95% of SOMAmer reagents. The median CV from pooled plasma samples was 5.2% across the 8 plates of the SOMAscan assays. This analysis confirmed the reproducibility (precision) of the measurements reported herein.
To assess the accuracy and biological relevance of protein measurements, we analyzed the expression of coagulation factors in patients treated with Coumadin, a compound that inhibits the synthesis of vitamin K dependent clotting factors, as well as the relationship between the putative kidney function marker CST3 and plasma creatinine. Supplemental Figure 1A–B illustrates that patients treated with Coumadin, as expected, had significantly lower levels of the vitamin K dependent coagulation factors F2, F7, F9, F10, proteins C and protein S. In addition, Supplemental Figure 1C–D demonstrates that the plasma levels of the putative kidney function marker CST3 correlated well with plasma creatinine levels.
The verified positive response to Coumadin elucidated the need for a source of variation analysis to identify potential confounders for the heart failure cohort analysis. As anticipated, age and gender, which were matched across the cohorts, contributed relatively little to the overall variation (Supplemental Figure 2). Similarly the use Coumadin, calcium channel blockers, and other related medications (ACE inhibitors, beta-blockers, dyslipidemia drugs) contributed little to the signal between cohorts. Further, LVEF as a continuous variable contributed ~ 1% to the overall variability of the HF proteome. In contrast, heart failure etiology (ischemic vs. non-ischemic) contributed between 5.8 – 14.1 % of the observed variance. The largest component of the variance in the analysis was unassigned, indicating an as-of-yet undetermined source.
To further validate the accuracy of the SOMAscan analysis, we performed ELISAs on three proteins whose expression levels varied among the HFrEF, HFmrEF and HFpEF patients, namely cystatin C, soluble ST2 (sST2), and C-reactive protein. As shown in Supplemental Figure 1E the expression levels of the analytes determined by the SOMAscan and the protein values determined by ELISA were closely correlated across the range of proteins that were tested: cystatin C, r = 0.94 (p < 0.001); ST2, r = 0.95 (p < 0.001) and C-reactive protein, r = 0.95 (p < 0.001).
Patient Characteristics
Table 1 displays the baseline characteristics of the 173 subjects included in the study. As shown, the age for the entire cohort of HF patients was 55.3 ± 12.6 years, with 58% male patients, 73 % Caucasian and 24% African Americans. The major co-morbidities included hyperlipidemia (43%), hypertension (55%), diabetes (25%), coronary artery disease (24%), obstructive sleep apnea (27%) and chronic obstructive pulmonary disease (14%). The distribution of NYHA functional class for the entire cohort was: 14 % class I, 55% class II, 23% class III, and 8% class IV. 86% of patients were on a beta-blocker and 85% of the patients were on an ACE inhibitor or an angiotensin receptor blocker. The baseline demographics for the HFrEF, HFmrEF, and HFpEF were relatively well matched, with the exception that there were more patients with ischemic heart disease in the HFrEF subgroup.
Table 1:
Demographics of Patients with HFrEF, HFmrEF and HFpEF
| Overall (n = 173) | HFrEF (n = 47) | HFmrEF (n = 83) | HFpEF (n = 43) | P‐value | |
|---|---|---|---|---|---|
| Age, years | 55 ± 12.55 | 56 ± 12.59 | 54 ± 11.97 | 56 ± 13.70 | 0.61 |
| Caucasian | 127 (73%) | 35 (74%) | 62 (75%) | 30 (70%) | 0.83 |
| African American | 46 (27%) | 12 (26%) | 21 (25%) | 13 (30%) | 0.83 |
| Male sex | 100 (58%) | 27 (57%) | 49 (59%) | 24 (56%) | 0.94 |
| Diabetes | 44 (25%) | 12 (26%) | 23 (28%) | 9 (21%) | 0.71 |
| High blood pressure | 96 (55%) | 26 (55%) | 42 (51%) | 28 (65%) | 0.30 |
| Coronary Artery Disease (CAD) | 41 (24%) | 21 (45%) | 14 (17%) | 6 (14%) | <0.01 |
| Emphysema (COPD) | 25 (14%) | 5 (11%) | 10 (12%) | 10 (23%) | 0.16 |
| PVD | 1 (1%) | 1 (2%) | 0 (0%) | 0 (0%) | 0.26 |
| CVA/TIA | 16 (9%) | 5 (11%) | 4 (5%) | 7 (16%) | 0.10 |
| High cholesterol (HLD) | 75 (43%) | 28 (60%) | 28 (34%) | 19 (44%) | 0.02 |
| Obstructive sleep apnea (OSA) | 47 (27%) | 15 (32%) | 18 (22%) | 14 (33%) | 0.30 |
| AF/atrial flutter | 9 (5%) | 3 (6%) | 1 (1%) | 5 (12%) | 0.04 |
| ICD/CRT | 86 (50%) | 35 (74%) | 41 (49%) | 10 (23%) | <0.01 |
| Heart Rate, b.p.m | 76 ± 13.39 | 82 ± 13.98 | 74 ± 12.30 | 74 ± 13.22 | <0.01 |
| SBP, mmHg | 118 ± 19.79 | 111 ± 19.77 | 120 ± 16.29 | 123 ± 23.77 | 0.01 |
| DBP, mmHg | 72 ± 11.12 | 69 ± 11.28 | 75 ± 8.87 | 72 ± 13.90 | 0.03 |
| Mean arterial pressure, mmHg | 88 ± 13.08 | 83 ± 13.62 | 90 ± 10.54 | 89 ± 15.74 | 0.02 |
| Creatinine, mg/dL, median (Q1, Q3) | 1.15 (0.86, 1.42) | 1.22 (0.94, 1.53) | 1.09 (0.89, 1.37) | 0.98 (0.81, 1.49) | 0.18 |
| Sodium, mmol/L | 139 ± 3.36 | 138 ± 3.27 | 140 ± 3.19 | 140 ± 3.66 | 0.12 |
| eGFR, mL/min/1.73 m2 | 72 ± 28.96 | 65 ± 30.64 | 74 ± 25.67 | 74 ± 32.56 | 0.23 |
| ACC/AHA Stage | <0.01 | ||||
| B | 5 (3%) | 0 (0%) | 2 (2%) | 3 (7%) | 0.14 |
| C | 159 (92%) | 39 (83%) | 80 (96%) | 40 (93%) | 0.02 |
| D | 9 (5%) | 8 (17%) | 1 (1%) | 0 (0%) | <0.01 |
| NYHA Class | <0.01 | ||||
| I | 24 (14%) | 1 (2%) | 16 (19%) | 7 (16%) | |
| II | 96 (55%) | 25 (53%) | 50 (60%) | 21 (49%) | |
| III | 40 (23%) | 13 (28%) | 14 (17%) | 13 (30%) | |
| IV | 13 (8%) | 8 (17%) | 3 (4%) | 2 (5%) | |
| Beta Blockers | 149 (86%) | 38 (81%) | 75 (90%) | 36 (84%) | 0.28 |
| ACEi | 115 (66%) | 26 (55%) | 63 (76%) | 26 (60%) | 0.04 |
| ARB | 33 (19%) | 13 (28%) | 11 (13%) | 9 (21%) | 0.13 |
| Diuretics | 137 (79%) | 37 (79%) | 70 (84%) | 30 (70%) | 0.16 |
| Digoxin | 54 (31%) | 18 (38%) | 28 (34%) | 8 (19%) | 0.10 |
| CCB | 25 (14%) | 3 (6%) | 11 (13%) | 11 (26%) | 0.03 |
| Nitrates | 48 (28%) | 17 (36%) | 18 (22%) | 13 (30%) | 0.19 |
| Anti-HLD | 95 (55%) | 33 (70%) | 40 (48%) | 22 (51%) | 0.04 |
| Hydralazine | 31 (18%) | 13 (28%) | 11 (13%) | 7 (16%) | 0.12 |
| Anti-platelet | 112 (65%) | 33 (70%) | 54 (65%) | 25 (58%) | 0.49 |
| Coumadin | 51 (29%) | 21 (45%) | 21 (25%) | 9 (21%) | 0.02 |
| MRAs | 63 (36%) | 20 (43%) | 31 (37%) | 12 (28%) | 0.35 |
Values are listed as number of observations or average ± standard deviation. Percentages for each ejection fraction group are reported in parenthesis. Abbreviations: ACEi: angiotensin converting enzyme inhibitor; ARB: angiotensin receptor blocker; AF: atrial fibrillation; CCB: calcium channel blocker; CAD: coronary artery disease; COPD: chronic obstructive pulmonary disease; CVA: cerebrovascular accident; GFR: glomerular filtration rate; HFrEF: heart failure with reduced ejection fraction; HFmrEF: heart failure with mid-range ejection fraction; HFpEF: heart failure with preserved ejection fraction; HLD: hyperlipidemia; ICD/CRT: implantable cardioverter defibrillator/cardiac resynchronization therapy; MRAs: mineralocorticoid receptor antagonists; PVD: peripheral vascular disease; TIA: transient ischemic attack.
Proteomic Profiling of Heart Failure Patients Stratified by LV Ejection Fraction
We performed a 3-dimensional principal component analysis (PCA) of the proteomic profiles for the HFrEF, HFmrEF, and HFpEF patients. As shown in Figure 1, the HFpEF, HFrEF and HFmrEF patients partitioned into 3 partially overlapping clusters (8). The centroid location was then computed for each of the three cohort groups within the first three component space, and the inter-cohort Euclidean distance of separation between the centroids of the HFrEF, HFmrEF and HFpEF was determined. As shown in Figure 1A, the distance between the cluster centroids was 4.2 units for HFmrEF vs HFpEF, 5.2 units for HFrEF vs HFpEF, and 6.2 units for HFmrEF vs HFrEF. The unique distances between the centroids for the different clusters suggests that the plasma proteomic profiles were most similar for patients with HFmrEF and HFpEF, and were most dissimilar for HFmrEF and HFrEF patients. These latter findings were intriguing, insofar as ~ 70% of the patients in the HFmrEF group were HFrEF patients whose LVEF increased on medical and/or device therapies. To further explore these findings, we stratified the HFmrEF patients into HFmrEF-improved and HFmrEF-unimproved subgroups (see Supplemental Table S-1 for demographics of these subgroups). Figure 1B shows that the centroid for the cluster of the HFmrEF-unimproved patients was approximately equidistant between the centroids for the HFrEF cluster at 3.3 units and the HFpEF cluster at 3.2 units, whereas the centroid for the HFmrEF-improved cluster was furthest in distance from the HFrEF at 7.7 units and closer to the centroid for the are HFrEF cluster at 5.1 units, suggesting that there was greater overlap of the proteomic profiles for HFmrEF-improved patients and HFpEF patients than for HFmrEF-improved and HFrEF patients.
Figure 1. Three-dimensional PCA analysis of the spatial relationship of the serum proteome profiles of different EF groups.

The serum proteome of patients with different LVEF at the time of enrollment is depicted on a three-dimensional PCA plot. (A) The 3 major EF groups are identified, and (B) 4 different groups are identified because the HFmrEF group is divided into HFmrEF-improved and HFmrEF-unimproved. A centroid (small round dot within an ellipse) was calculated based on the PCA vectors for each group and the spatial distance between centroids was calculated.
Comparative heat maps were also utilized to visualize the differences and similarities in the proteomic signatures for subjects with HFrEF, HFmrEF, and HFpEF. As illustrated in Figure 2, 120 proteins were differentially expressed (62 increased/58 decreased) between HFrEF vs. HFpEF patients, 183 were differentially expressed (96 increased/58 decreased) between HFrEF vs. HFmrEF patients, and 54 proteins were differentially expressed (31 increased, 23 decreased) between HFmrEF vs. HFpEF patients. These heat map data are internally consistent with the analyses of centroid clusters (Figure 1A). Viewed together, these data suggest that the molecular similarity, as represented by plasma protein levels, is greatest for HFmrEF and HFpEF patients and least for HFmrEF and HFrEF patients.
Figure 2. Heat maps of differentially expressed serum proteins for patients with HFrEF, HFmrEF and HFpEF.
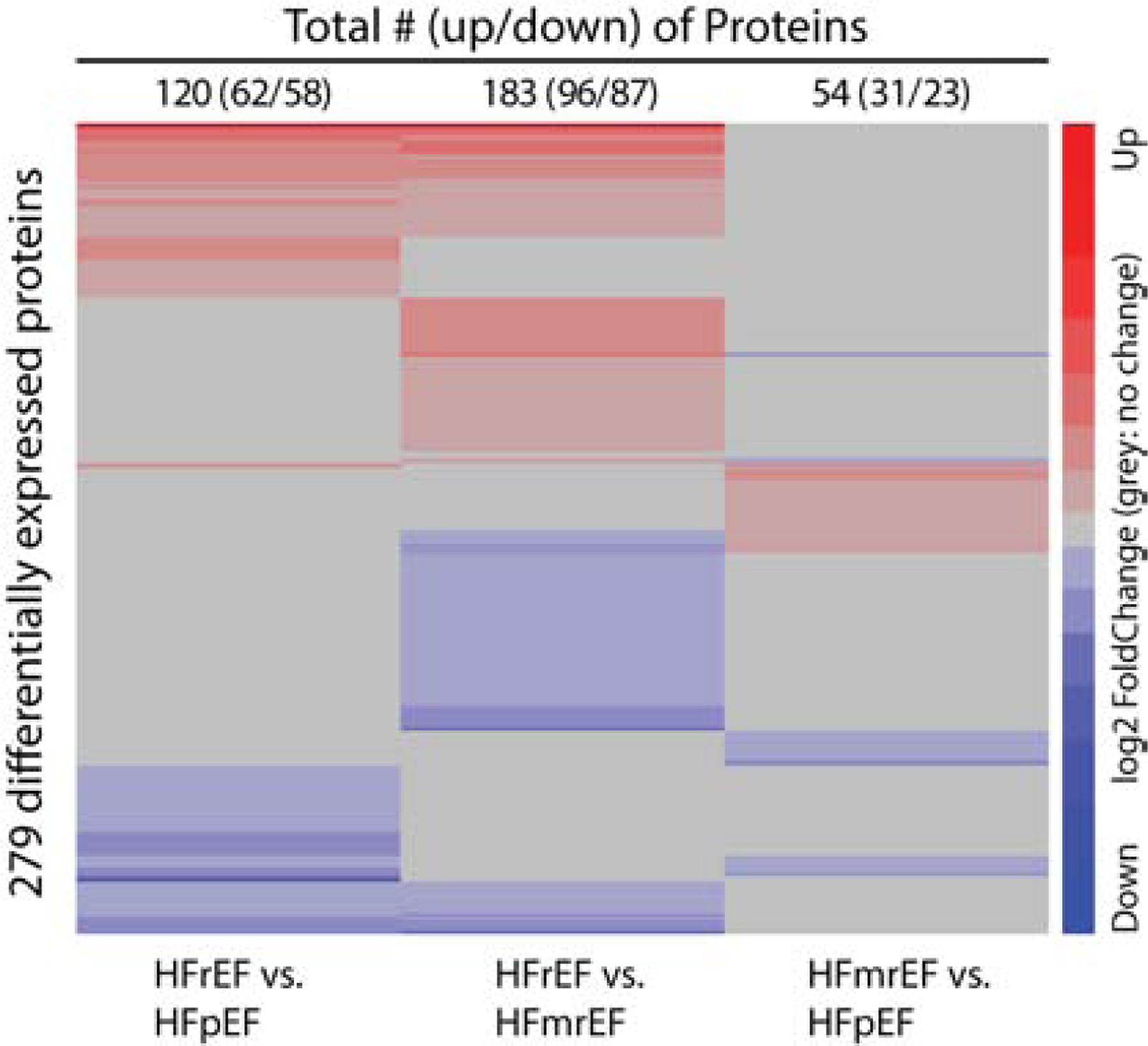
We clustered proteins with the Euclidean distance and the Ward clustering algorithm. The proteins non-differentially expressed between the two groups of interest are shown in gray to highlight the differences and similarities between groups. Key: red = increased expression; blue = decreased relative expression.
Given that the PCA analysis of HFmrEF-improved and HFmrEF-unimproved cohorts revealed minimal overlap of their protein profiles, additional heat maps were generated for the protein levels in both groups, which were then compared to HFrEF and HFpEF patients. Figures 3A and 3B demonstrate that the greatest differences in relative protein levels (ranked from greatest to least), were observed for HFmrEF-improved vs HFrEF patients (226 proteins [114increased/ 112 decreased] > HFmrEF-improved vs. HFpEF patients (81 proteins [52 increased/ 21 decreased]) > HFmrEF-unimproved vs. HFrEF (35 proteins [18 increased/ 17 decreased) ≈ HFmrEF-unimproved vs. HFpEF patients (26 proteins [16increased/ 10 decreased]). These data are fully consistent with the analyses of the distances between the centroid clusters shown in Figure 1B, and suggest that the similarity of the plasma protein levels are greatest for HFmrEF-unimproved and HFrEF patients and least for HFmrEF-improved and HFrEF patients.
Figure 3. Heat maps of differential representation of serum proteins comparing HFrEF and HFpEF vs HFmrEF-improved and HFmrEF-unimproved.
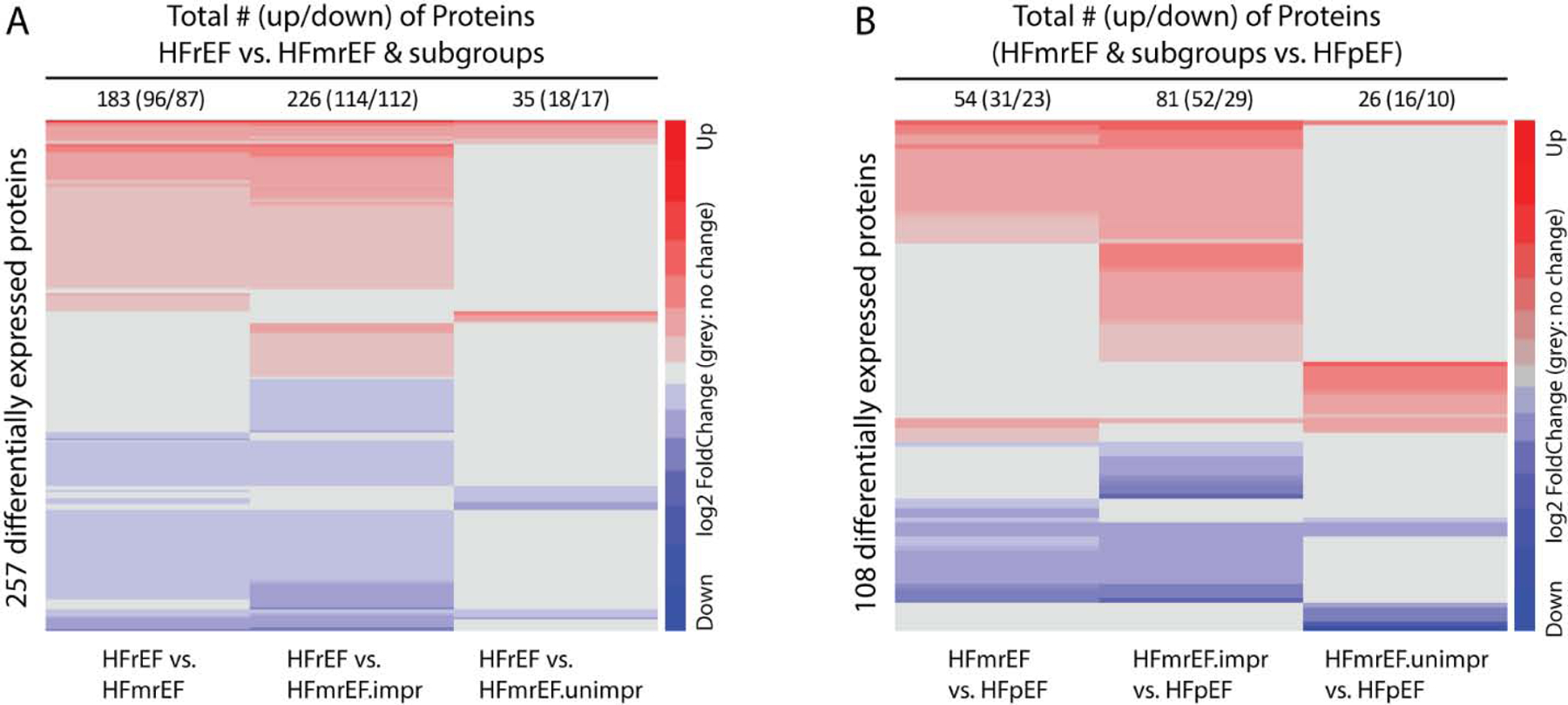
We clustered proteins with the Euclidean distance and the Ward clustering algorithm. The proteins non-differentially expressed between the two groups of interest are shown in gray to highlight the differences and similarities between groups. A: comparisons between HFmrEF and its subgroups versus HFrEF. B: comparisons between HFmrEF and its subgroups versus HFpEF. (Key: red = increased expression; blue = decreased relative expression)
To determine whether the blood proteome reflected proteins that were expressed in the heart, we compared the 280 proteins that were differentially expressed in the HFrEF, HFmrEF and the HFpEF patients with those known in the heart-proteome (https://www.proteinatlas.org/humanproteome/tissue/heart). Of the 280 proteins that were differentially expressed among the different LVEF classifications in the present study, 6 proteins were specific to the heart, 139 were expressed in the heart as well as other tissues, and 135 were either not expressed in the heart, or were expressed with low tissue specificity. Thus, ~ 51% of the circulating proteins that were detected in the present study had either specific to the heart, or the heart as well as other tissues. (Supplemental Table S-2).
Effect of HF Etiology on Protein Levels in LVEF Subgroups
The covariate analysis shown in Supplemental Figure 2 revealed that the etiology of heart failure contributed ~ 6 – 14% of the variability of the signal values for the plasma proteomic profiles. Given this, an analysis was completed to determine if the differential protein expression in patients with ischemic and non-ischemic HF stratified by LVEF. Figure 4 shows that the greatest differences in protein expression levels for the pairwise comparison of ischemic vs. non-ischemic HF etiology were (in rank order) for HFrEF vs HFpEF (184 proteins) ≈ HFrEF vs HFmrEF (172 proteins) > HFmrEF vs HFpEF (94 proteins). Intriguingly, the pairwise comparisons of differentially expressed proteins revealed that there was surprisingly little overlap of the proteins between patients with an ischemic or non-ischemic etiology of HF, regardless of the LVEF stratification. The Venn diagrams in Figure 4 show that the overlap was greatest for the comparison of HFrEF vs HFmrEF (8.1%), followed by HFrEF vs HFpEF (3.3%), and was least for HFmrEF vs HFpEF (1.1%). The sample size was considered too small to investigate the effects of the etiology of heart failure within the HFmrEF subgroups, so this analysis was not performed.
Figure 4. Heat maps and Venn diagrams of serum proteins differentially expressed across major LVEF subgroups categorized by heart failure etiology.

Heat maps of proteins differentially expressed in the pairwise comparisons for each of the 3 major EF groups stratified by ischemic vs non-ischemic etiology. (A) HFrEF vs. HFpEF, (B) HFmrEF vs. HFpEF, and (C) HFmrEF vs. HFpEF. Proteins were hierarchically clustered with the Euclidean distance and the Ward cluster method. The proteins non-differentially expressed between the two groups of interest are shown in gray to highlight the differences and similarities between groups (Key: red = increased expression; blue = decreased relative expression)
Given the differences in plasma proteome observed between patients with ischemic and non-ischemic cardiomyopathy, we asked whether the difference in serum proteome profile observed between patients with HFrEF, HFmrEF-improved, HFmrEF-unimproved and HFpEF were the result of differences between ischemic and non-ischemic heart failure. Accordingly, we repeated the analysis shown in Figure 1 by determining the centroids for the ischemic and non-ischemic patients in the HFrEF, HFmrEF (HFmrEF-improved and HFmrEF-unimproved) and HFpEF subgroups. As shown in Supplemental Figure 3, the centroids for ischemic and non-ischemic patients were in close proximity, suggesting that differences in proteomic profiles for ischemic vs non-ischemic patients did not explain the differences in the proteome between the different LVEF classifications.
Comprehensive Biological Theme Analysis
A biological theme analysis (pathways, processes, cellular signaling) was performed using the CompBio platform to evaluate circulating proteins that were differentially expressed in HFrEF, HFmrEF, and HFpEF patients. An example of an annotated output from the specific comparison of HFrEF vs HFmrEF is illustrated in Supplemental Figure 4, with a summarization of themes from all comparisons shown in Supplemental Figure 5A–C. Substantial heterogeneity was observed with respect to the categories of enriched proteins for each of the LVEF comparisons. The top five thematic biological clusters that were differentially expressed between HFrEF and HFpEF patients were related to increased WNT/IGF signaling, increased TDGF/morphogenic regulation, increased BMP/morphogenic regulation, increased VEGF-A/Angiogenesis and decreased JAK/STAT signal transduction. The 5 most prominent themes for the differences between HFrEF and HFmrEF patients were related to increased IGF/IGBP signaling, increased MMP activity, increased chemokine induction, increased VEGF-A/Angiogenesis and increased follistatin myogenic regulation. In contrast, the top 5 thematic biological differences between HFmrEF and HFpEF patients were related to increased lymphotoxin signaling, increased LPS-TLR signaling, increased complement/chemoattractant signaling, and decreased cathepsin lysosomal activity. Data for key proteins associated with biological themes of interest illustrating the differential expression patterns across the 3 cohorts are shown in Supplemental Figure 5D.
To provide a simple format for visualizing the differences in biological themes between the LVEF groups, the theme lists provided in Supplemental Figure 5A–C were used to generate a pseudo heat map of the most salient biological themes that were differently expressed between the HFrEF, HFmrEF, and HFpEF cohorts (Figure 5). These themes were further grouped into broader biological categories to facilitate comparisons between groups. Within the HFrEF cohort, 5 categories contained themes that were differentially expressed as compared to both the HFmrEF and HFpEF cohorts. The HFrEF increased-expression categories included growth factor signaling, inflammation, neurotrophic signaling, remodeling/hypertrophy and angiogenesis (VEGF-A related), whereas decreased-expression themes included those related to coagulation and myeloproliferation (“other”). Biological themes differentially expressed in HFmrEF patients as compared to HFrEF and HFpEF patients, included decreased inflammation and increased signal transduction. Finally, themes expressed differently in HFpEF patients as compared to HFrEF and HFmrEF patients, included growth factor signaling, inflammation, angiogenesis (VEGF-C related). Viewed together, this bioinformatics analysis suggests that HFrEF, HFmrEF, and HFpEF have proteomic signatures that reflect distinct pathophysiologies.
Figure 5. Pseudo heat map of differentially expressed proteins for patients with HFrEF, HFmrEF, and HFpEF.

A pseudo heat map was constructed from the comparisons of differentially expressed proteins illustrated in Supplemental Figure S-5A–C for patients with HFrEF, HFmrEF, and HFpEF. Related biological themes from the list of differentially expressed proteins in Figure S-5A–C were grouped into higher-level biological groupings, in order to permit comparisons between the various HF subgroups. (Key: red = increased expression; blue = decreased relative expression)
We also created a pseudo heat map to delineate the salient biological themes that were differently expressed between ischemic and nonischemic heart failure patients. As shown in Supplemental Figure 6, biological themes related to increased growth factor signaling were numerically more predominant in ischemic vs non-ischemic HF, whereas the number of themes related to decreased inflammation were numerically less prominent in ischemic vs non-ischemic HF. However, to our surprise, there were no striking differences in the biological themes between ischemic vs non-ischemic HF.
DISCUSSION
The results of this study, in which we used a high-content proteomics assay to analyze the blood proteome of HF patients stratified by their LV ejection fraction, shows that while changes in LVEF capture a small portion of the proteome variability across heart failure patients, patients with HFrEF, HFmrEF, and HFpEF have specific proteomic signatures that reflect distinct pathobiologies (Central Illustration). Accordingly, the proteomic signatures for patients with HFrEF and HFpEF were different (Figures 1, 2 and 5), consistent with the prevailing view that these two different classifications of HF have a different underlying pathophysiologies and respond differently to medical therapies. Bioinformatics analysis revealed that there were biological themes that were unique to HFrEF patients when compared to HFpEF and HFmrEF patients, including increased growth factor signaling, increased ECM remodeling, increased angiogenesis (Figure 5), reflecting the greater degree of LV remodeling (dilation) and LV systolic dysfunction in HFrEF patients. Intriguingly, there was an increase in circulating proteins related to innate immunity and a decrease in the expression of proteins related to humoral immunity in HFrEF patients compared with HFpEF and HFmrEF patients. The biological themes that were unique to HFpEF patients when compared to HFrEF and HFmrEF patients were enriched for proteins reflecting growth factor signaling, increased humoral immunity, and increased angiogenesis (Figure 5).
Central Illustration. Heart Failure Patients Stratified By Their Left Ventricular Ejection Fraction Have Distinct Proteomic Signatures.
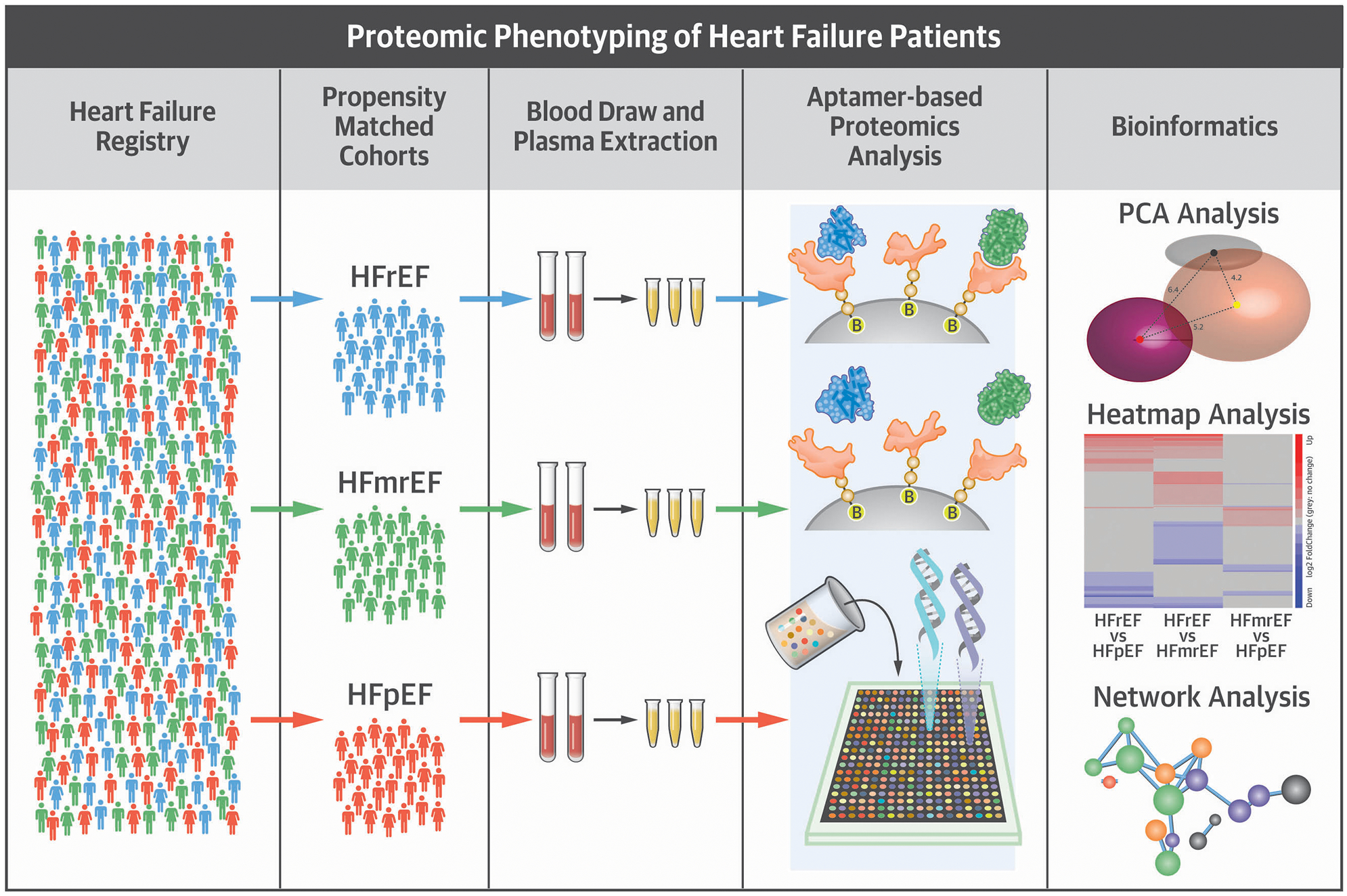
A cohort of age and gender matched patients were randomly selected from the Washington University Heart Failure registry. Patients were stratified into 3 HF cohorts were identified based on their left ventricular ejection fraction: heart failure with reduced ejection fraction (HFrEF; LVEF <40%), heart failure with a preserved ejection fraction (HFpEF; LVEF >50%), and heart failure with a mid-range ejection fraction (HFmrEF; LVEF 40–50%). A high-content aptamer-based proteomics assay was used to analyze the blood proteome of the patients in each cohort. Bioinformatics analysis showed that patients with HFrEF, HFmrEF, and HFpEF have specific proteomic signatures that reflect distinct pathobiologies.
Patients with midrange LVEF represent a heterogeneous patient population comprised of a large proportion of HFrEF patients whose LVEF partially recovered on evidence based medical therapies, and a smaller proportion of HFpEF patients whose LVEF declined. Remarkably, the plasma proteome of patients with HFmrEF was distinct from the plasma proteome of HFrEF patients (Figure 1A, Figure 2), and partially overlapped that of HFpEF patients, in agreement with prior clinical observations that HFmrEF patients have a functional capacity and clinical outcomes that are similar to that of HFpEF patient (6,9). Bioinformatics analysis suggested that the circulating proteins in HFmrEF patients were enriched for themes reflecting changes in adaptive immune responses and increased signal transduction (Figure 5). When we further sub-classified the HFmrEF patients in the HFmrEF-improved and HFmrEF-unimproved, we observed that the protein signature of HFmrEF-unimproved patients partially overlapped that of HFrEF and HFpEF patients, whereas the proteomic signature for HFmrEF-improved patients was completely distinct from HFrEF patients, and partially overlapped that of HFpEF patients (Figures 1 and 2). To the extent that levels of proteins in the circulatory system reflect the physiology of a given individual, the observation that the plasma proteomic profile of HFmrEF-improved patients was distinct from that of HFrEF patients suggests that patients who partially recover their LVEF on evidence based medical therapies are biologically unique and are different than HFrEF patients whose LVEF did not recover on medical therapy. The finding that the plasma proteomic profile of HFmrEF-improved patients overlaps that of HFpEF patients is consistent with the concept that recovery of LV function, although associated with improved event-free survival, is still associated with a persistent HF risk and recurrent HF hospitalizations (i.e. “myocardial remission”) (6,10).
An unanticipated finding of these studies was the marked difference in the proteomic signatures of patients with ischemic and non-ischemic HF. Indeed, there was < 10% overlap of the plasma proteomic profiles of ischemic and non-ischemic patients, regardless of the LVEF stratification (Figure 3). While it has long been recognized that the clinical outcomes for patients with ischemic and non-ischemic HF are different (11). Our data suggest that biological differences in ischemic and non-ischemic HF patients are greater than previously supposed.
Profiling the Blood Proteome in Heart Failure
The use of protein biomarkers (e.g. BNP or NT-proBNP) has become routine in the clinical diagnosis and management of patients with HF (reviewed in (12,13)). The appreciation that single biomarker strategies do not reflect all aspects of the complex pathophysiology of HF has given rise to the use of multi-biomarker testing panels in order to improve medical care (12,14). However, current multi-biomarker approaches employ multiplexed immunoassays that have inherent technical limitations, such as lack of specificity for different protein isoforms, insensitivity to detect lower abundance proteins, and antibody cross-reactivity. Moreover, while multi-maker strategies are able to measure more aspects of the pathophysiology of HF than single biomarkers, multi-biomarker strategies have inherent limitations in terms of revealing the complex physiology of HF, insofar as they measure predefined proteins in known pathways, and thus are not able to detect clinically-relevant proteins in unrecognized biological pathways.
Although high-throughput, quantitative analysis using mass-spectrometry (MS)-based proteomics of blood, plasma, and urine is an extremely attractive unbiased approach to assess the circulating proteome, the application of untargeted MS has been challenging thus far because of the high dynamic range of protein abundances (5), as well as the lack of reproducible, robust, high-throughput workflows to reliably identify and verify potential biomarkers. As a result, untargeted MS-based proteomic approaches have not been utilized in HF. Emerging technologies that address these issues include nucleotide-labeled immunoassays and aptamer reagents, which can be automated for efficient multiplexing of thousands of proteins with high throughput. Aptamer proteomics have been applied to large cohorts of patients to identify hundreds of novel protein biomarkers for coronary artery disease (15) and myocardial injury (16).Furthermore, aptamer proteomics have been used alongside ELISA to validate novel HF biomarkers (17). Germane to this discussion, a recent study using aptamer based proteomics in 43 patients with HF (median LVEF 52 [IQR 32–55]) and 43 control subjects identified 9 candidate HF biomarkers. Two of these proteins, angiopoietin-2 and thrombospondin-2, were subsequently confirmed in separate validation cohorts as robust biomarkers for detecting acute and pre-clinical HF (18). Although the scope of this study was not intended to identify specific HF biomarkers within each LVEF classification, we did observe significantly (p < 0.05) increased levels of both angiopoietin-2 and thrombospondin-2 in HFrEF patients when compared to HFmrEF and HFpEF subgroups (Supplemental Figure 5D).
Our findings are in overall agreement with a prior study that measured 92 biomarkers in the plasma of HFrEF, HFmrEF, and HFpEF patients enrolled in the Scottish Cohort of the BIOSTAT-CHF study, which used proximity extension assay technology (Olink Proteomics, Uppsala Sweden) (18), and concluded that their study provided “biological context for the presence of clearly distinct syndromes” (i.e. HFrEF, HFmrEF, and HFpEF). The results of the present study both confirm and expand upon this study, which reported upregulation of biological pathways related to cellular growth, including GDF-15 (Supplemental Figure 5) in HFrEF patients, and increased inflammation in HFpEF (18). Because of the 14-fold broader coverage with the SOMAscan (1,3000 proteins vs 92 proteins) we were able to identify 279 differentially expressed proteins among the HF groups, as opposed the 14 unique proteins in the BIOSTAT-CHF registry. The broader coverage allowed us to identify novel biological themes in HFmrEF (decreased inflammation and increased signal transduction) and HFpEF (growth factor signaling, angiogenesis) that were not detected in the study by Tromp et al., as well as expand upon themes in HFrEF (remodeling/hypertrophy and angiogenesis). We were also able to identify unique differences between HFmrEF-improved and HFmrEF-unimproved, which was not explored in the baseline samples BIOSTAT-CHF registry, whose samples were obtained (by design) from suboptimally treated HF patients.
Limitations
Our study has several limitations that warrant discussion. First, although the samples that were used for this study were obtained from a well-phenotyped cohort of HF patients, the number of patients that were studied is relatively small, drawn from a single center, and because of the use of propensity matching the age of the HFpEF patients is younger than traditional HFpEF cohorts. For these reasons, the results of this study should be regarded as provisional, pending validation in separate cohorts of HF patients from different centers. Second, only 29% of the patients with HFmrEF where in the HFmrEF-unimproved subgroup. The imbalance in sample size between the HFmrEF-unimproved and HFmrEF-improved groups has the potential to introduce a bias in the differential protein expression analysis reported. Given that the subgroups analyzed were not perfectly matched in terms of medical therapy, we cannot exclude that the differences in serum proteome profile were, at least in part, the result of differences in medical treatment among the different groups.
Conclusions
High-throughput proteomic approaches allow for the detection of thousands of circulating proteins in real time, and thus have the potential to provide a broader and more inclusive mechanistic understanding of the dysregulation of the molecular pathways in HF than is currently possible using single or multi-biomarker panels. The results of this study demonstrate that is feasible to use multiplex aptamer proteomics to identify distinct proteomic signatures for patients with HFrEF, HFmrEF and HFpEF, as well as to identify important differences in the proteome of ischemic and non-ischemic HF patients. The current work is hypothesis generating and it should be viewed as a first step towards potential deeper phenotyping of HF patients that goes beyond the conventional assessment of LVEF. Our findings will require verification in separate and larger cohorts of HF patients. Nonetheless, the results of this study suggest that high-content proteomics can be used to identify important biological differences between HF patients that are sub-classified according to conventional LVEF classifications (Figure 5). Although the exact clinical applicability of these findings remains to be determined, there are several potential applications that can be envisioned. First, given that the LVEF cut-offs that have been used were assigned arbitrarily based on the need to identify enriched cohorts of patients for clinical trials, it may be possible to use proteomic approaches to better define conventional LVEF cut-offs. For example, the proper LVEF cutoff to define HFpEF is unknown with values ranging from > 45%, >50%, > 60 % proposed by different groups. Indeed, the recent results of the PARAGON trial (19), wherein HFpEF patients with an LVEF of 45–50% appeared to respond differently to sacubitril/valsartan than HFpEF with an LVEF ≥ 50%, suggests that there are biologically meaningful differences among patients with HFpEF. It may be possible to use high content proteomic approaches to better identify these patients. Second, our findings with respect to the biological differences of HFmrEF-improved patients, suggest that it may be possible to utilize proteomic strategies to distinguish HFrEF patients with a recovered LVEF from HFpEF patients, and perhaps identify those patient who can be weaned from medical therapy and those that should remain on evidence based medical therapies. Moreover, the results of this study raise the exciting possibility that it may be possible in the not too distant future to combine the longitudinal assessment of high-content proteomics assays with machine learning algorithms, in order to predict the response of HF patients to medical therapies, as well as predict changes in the health care status of patients with HF. Finally, it remains to be determined whether the variations in proteomic profile that we observed across groups were a cause or effect of the different heart failure phenotypes. Whether proteomics-based approaches will be superior to single or multi-biomarker panel approaches for personalizing the care of HF patients is an important question that will require a deeper understanding of the limitations, costs and the clinical applicability of these different approaches.
Supplementary Material
Table 2.
Etiologies of Heart Failure for the Patient Cohorts
| Etiology | Total (n=173) | HFrEF (n=47) | HFmrEF-improved (n=59) | HFmrEF-unimproved (n=24) | HFpEF (n=43) | P-value |
|---|---|---|---|---|---|---|
| Ischemic (CAD) | 41 (24%) | 21 (45%) | 9 (15%) | 5 (21%) | 6 (14%) | 0.57 |
| Non-ischemic | 132 (76%) | 26 (55%) | 50 (85%) | 19 (79%) | 37 (86%) | |
| Idiopathic | 96 (56%) | 22 (47%) | 46 (78%) | 11 (46%) | 17 (40%) | <0.01 |
| Amyloid | 1 (1%) | 1 (5%) | ||||
| Congenital | 6 (4%) | 1 (5%) | 5 (12%) | |||
| Hypertrophic | 5 (3%) | 1 (3%) | 2 (9%) | 2 (5%) | ||
| Peripartum | 7 (5%) | 2 (5%) | 1 (2%) | 2 (9%) | 2 (5%) | |
| Restrictive | 1 (1%) | 1 (3%) | ||||
| Valvular | 2 (2%) | 1 (3%) | 1 (5%) | |||
| Other | 13 (8%) | 2 (4%) | 1 (5%) | 10 (24%) | ||
| Mixed | 1 (1%) | 1 (2%) |
CAD: coronary artery disease; EF: ejection fraction; HF: heart failure. Percentages for each EF group are reported in parenthesis. P-values shown are for the difference in frequency between HFmrEF-improved and HFmrEF-unimproved subgroups.
Clinical Perspectives.
Competency in Medical Knowledge:
There are variations in circulating proteins in patients with heart failure (HF) across a range of left ventricular ejection fraction (EF) related to differing pathophysiology that is not entirely captured by categorizations as HF with preserved, reduced or minimally reduced EF (HFpEF, HFrEF or HFmrEF). Serum proteomic profiles differ in patients with ischemic vs. non-ischemic cardiomyopathies and in those with static vs. improved EF.
Translational Outlook:
Integration of LVEF with multiplex proteomics assays holds the potential to improve identification of clinical phenotypes of patients with HF.
Central Illustration Highlights:
A high content proteomics assay was used to analyze the blood proteome of patients with HFrEF (LVEF <40%), HFmrEF (LVEF 40–50%), and HFpEF (LVEF > 50%)
Patients with HFrEF, HFmrEF, and HFpEF had unique variations in circulating proteins which reflected distinct biological pathophysiologies
Bioinformatics analysis revealed biological themes that were unique to HFrEF, HFpEF and HFmrEF patients, suggesting that it may be possible to use proteomics assays to more accurately predict clinical phenotypes of HF patients
Acknowledgments
This study was supported by NIH U10 HL110309 and NIH RC2-HL102222, as well as support from the Department of Medicine at Washington University School of Medicine and The Barnes-Jewish Foundation. LA is supported by K08 HL1945108 and AJ is supported by NIH K08 HL138262. The Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1TR000448 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
DISCLOSURES: R.D. Head may receive royalty income based on the CompBio technology developed by R.D. Head and licensed by Washington University to PercayAI. All other authors have declared that no conflict of interest exist.
Abbreviations
- ACE
angiotensin converting enzyme
- HF
heart failure
- HFmrEF
heart failure with a mid-range ejection fraction
- HFpEF
heart failure with a preserved ejection fraction
- HFrEF
heart failure with a reduced ejection fraction
- LVEF
left ventricular ejection fraction
- PCA
Principal Components Analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bristow MR, Kao DP, Breathett KK et al. Structural and Functional Phenotyping of the Failing Heart: Is the Left Ventricular Ejection Fraction Obsolete? JACC Heart Fail 2017;5:772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mann DL. Is It Time for a New Taxonomy for Heart Failure? J Card Fail 2016;22:710–2. [DOI] [PubMed] [Google Scholar]
- 3.Yancy CW, Jessup M, Bozkurt B et al. 2016 ACC/AHA/HFSA Focused Update on New Pharmacological Therapy for Heart Failure: An Update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. J Am Coll Cardiol 2016. [Google Scholar]
- 4.Halliday BP, Wassall R, Lota AS et al. Withdrawal of pharmacological treatment for heart failure in patients with recovered dilated cardiomyopathy (TRED-HF): an open-label, pilot, randomised trial. Lancet 2019;393:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith JG. Molecular Epidemiology of Heart Failure: Translational Challenges and Opportunities. JACC Basic Transl Sci 2017;2:757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rastogi A, Novak E, Platts AE, Mann DL. Epidemiology, pathophysiology and clinical outcomes for heart failure patients with a mid-range ejection fraction. Eur J Heart Fail 2017;19:1597–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joseph SM, Novak E, Arnold SV et al. Comparable Performance of the Kansas City Cardiomyopathy Questionnaire in Heart Failure Patients with Preserved and Reduced Ejection Fraction. Circ Heart Fail 2013;6:1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodpaster AM, Kennedy MA. Quantification and statistical significance analysis of group separation in NMR-based metabonomics studies. Chemometr Intell Lab Syst 2011;109:162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadruz W Jr. West E, Santos M et al. Heart Failure and Midrange Ejection Fraction: Implications of Recovered Ejection Fraction for Exercise Tolerance and Outcomes. Circ Heart Fail 2016;9:e002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esler M, Jennings G, Lambert G, Meredith I, Horne M, Eisenhofer G. Overflow of catecholamine neurotransmitters to the circulation: source, fate, and functions. Physiol Rev 1990;70:963–985. [DOI] [PubMed] [Google Scholar]
- 11.Ehlert FA, Cannom DS, Renfroe EG et al. Comparison of dilated cardiomyopathy and coronary artery disease in patients with life-threatening ventricular arrhythmias: Differences in presentation and outcome in the AVID registry. Am Heart J 2001;142:816–22. [DOI] [PubMed] [Google Scholar]
- 12.Ibrahim NE, Januzzi JL Jr. Established and Emerging Roles of Biomarkers in Heart Failure. Circ Res 2018;123:614–629. [DOI] [PubMed] [Google Scholar]
- 13.Sarhene M, Wang Y, Wei J et al. Biomarkers in heart failure: the past, current and future. Heart Fail Rev 2019. [DOI] [PubMed] [Google Scholar]
- 14.Tromp J, Khan MAF, Mentz RJ et al. Biomarker Profiles of Acute Heart Failure Patients With a Mid-Range Ejection Fraction. JACC Heart Fail 2017;5:507–517. [DOI] [PubMed] [Google Scholar]
- 15.Ganz P, Heidecker B, Hveem K et al. Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients With Stable Coronary Heart Disease. JAMA 2016;315:2532–41. [DOI] [PubMed] [Google Scholar]
- 16.Smith JG, Gerszten RE. Emerging Affinity-Based Proteomic Technologies for Large-Scale Plasma Profiling in Cardiovascular Disease. Circulation 2017;135:1651–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chirinos JA, Zhao L, Jia Y et al. Reduced Apolipoprotein M and Adverse Outcomes Across the Spectrum of Human Heart Failure. Circulation 2020;141:1463–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells QS, Gupta DK, Smith JG et al. Accelerating Biomarker Discovery Through Electronic Health Records, Automated Biobanking, and Proteomics. J Am Coll Cardiol 2019;73:2195–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solomon SD, McMurray JJV, Anand IS et al. Angiotensin-Neprilysin Inhibition in Heart Failure with Preserved Ejection Fraction. N Engl J Med 2019;381:1609–1620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


