Abstract
Fibroblast growth factor receptor 4 (FGFR4) is a cell surface receptor tyrosine kinases (RTKs) for FGFs.
Several studies have focused on the association between FGFR4 polymorphisms and cancer development. This meta-analysis aimed to estimate the association between FGFR4 rs351855 (Gly388Arg), rs1966265 (Val10Ile), rs7708357, rs2011077, and rs376618 polymorphisms and cancer risk. Eligible studies were identified from electronic databases. All statistical analyses were achieved with the STATA 14.0 software. Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were used to quantitatively estimate the association. Overall, no significant association was found among rs351855, rs2011077, and rs376618 polymorphisms with the risk of overall cancer. The rs1966265 polymorphism significantly decreased the risk of cancer in recessive (OR = 0.87, 95% CI = 0.78–0.97, P=0.009, TT vs CT+CC) genetic model. Whereas the rs7708357 polymorphism was positively associated with cancer risk in dominant (OR = 1.17, 95% CI = 1.02–1.36, P=0.028) genetic model. Stratified analysis revealed that rs351855 variant significantly increased the risk of prostate cancer in heterozygous (OR = 1.16, 95% CI = 1.02–1.32, P=0.025 AG vs GG), dominant (OR = 1.20, 95% CI = 1.06–1.35, P=0.004, AG+AA vs GG), and allele (OR = 1.22, 95% CI = 1.06–1.41, P=0.005, A vs G) genetic models.
In summary, the findings of this meta-analysis indicate that rs1966265, rs7708357, and rs351855 polymorphisms are correlated to cancer development. Further well-designed studies are necessary to draw more precise conclusions.
Keywords: Cancer, FGFR4, Meta-analysis, Polymorphism, Susceptibility
Introduction
Cancer poses a major health problem in both developing and developed countries [1–3]. There were approximately 18.1 million new cases and 9.6 million cancer deaths in 2018 [4]. The exact mechanism of cancer development is not clear yet. Mounting evidence have indicated that cancer development and progression is influenced by environmental and genetic factors [3,5–7].
The human fibroblast growth factor receptors (FGFRs), a subfamily of cell surface receptor tyrosine kinases (RTKs), consist of four closely related family members (FGFR1–4) [8]. FGFR activation by a various fibroblast growth factors (FGFs) triggers a cascade that leads to the activation of multiple signal transduction pathways, including the Ras/Raf/MapK, PI3K/Akt, STAT, and PLCγ, which can promote cell survival, cell proliferation, tissue development, differentiation, angiogenesis, epithelial-to-mesenchymal transition (EMT), angiogenesis, and can thereby involve in carcinogenesis [9–11].
The human FGFR4 gene, also termed as cluster of differentiation 334 (CD334), is mapped to chromosome 5 (5q 35.1) [12] and is highly polymorphic. A common nonsynonymous single nucleotide polymorphism (SNP) at codon 388 (rs351855 G>A) in exon 9, which results in a change of glycine to arginine (Gly388Arg), was recognized in the transmembrane domain of the EGFR4 receptor [13]. Several studies inspected the relationship between FGFR4 gene rs351855 G>A polymorphism and numerous types of cancer including breast cancer [13–18], cervical cancer [19–21], colon cancer [13,18,22], gastric cancer [23], prostate cancer [24–27], head and neck squamous cell carcinoma (HNSCC) [28,29], oral squamous cell carcinoma (OSCC) [30,31], lung cancer [32–34], hepatocellular carcinoma [35–37], sarcoma [38], skin cancer [39], neuroblastoma [40], non-Hodgkin’s lymphoma [41], and glioma [42]. There are few direct reports about the effect of FGFR4 polymorphism on the gene expression. FGFR4 rs351855 polymorphism induced higher expression of FGFR4 protein and worse prognosis in breast cancer [43]. It has been reported that the rate of degradation of the Arg388 receptor was slower than the Gly388 receptor in neuroblastoma cells and also initiated internalization of the receptor into multivesicular structures (Rev1-1) [40]. In another investigation, the researchers showed that expression of the FGFR4 Arg388 protein activated the extracellular signal-related kinase pathway with subsequent expression of several genes which were associated with the aggressive form of prostate cancer [44], (Rev1-1). Researchers have reported that there was not any significant difference between different genotypes of FGFR4 in gastric cancer [45]. Interestingly in the lung normal tissue, genotype-dependent transcriptional profile is present [46]. In the past few years, there were few epidemiological analysis and meta-analysis focusing on FGFR4 in uterine leiomyomata [47], hip bone geometry [48–50], and all types of cancer [31,51]. Our current meta-analysis covers Gly388Arg rs351855 G>A and Val10Ile rs1966265 polymorphism in FGFR4 polymorphisms to cancer susceptibility and provide wider information in this important regulator of cancers (Rev 1-2).
Methods
Literature search and inclusion criteria
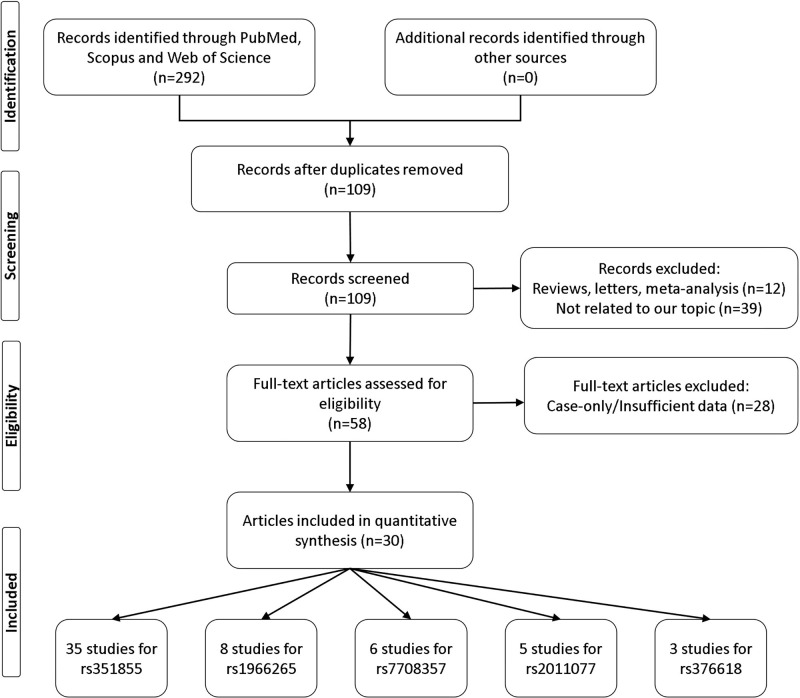
We performed a literature research for all eligible articles regarding the association between FGFR4 polymorphisms on multiple electronic databases including Web of Science, PubMed, Scopus, and Google Scholar databases through using the following terms: ‘FGFR4 OR CD334’ AND ‘polymorphism OR, SNP, OR variation OR mutation’ AND ‘cancer OR carcinoma OR neoplasm OR tumor’ up to 10 May 2020. Besides, we also screened references of the included studies. Figure 1 shows the process of studies selection. Relevant studies included the meta-analysis if they met the following inclusion criteria: (1) original case–control studies addressing the correlation between FGFR4 polymorphisms; (2) studies containing sufficient genotype data in both cases and controls; (3) the largest sample sizes were selected when repeatedly published articles by the same team. The exclusion criteria were: (1) conference abstract, case reports, reviews, duplication data; (2) insufficient genotype data provided.
Figure 1. Flow chart illustrates the detailed study selection process of this meta-analysis.
Data extraction
Two investigators independently screened the literature and extracted data from eligible studies according to exclusion and inclusion criteria. The following data were collected from each study including the first author’s name, publication year, country, ethnicity of participants, cancer type, genotyping methods, the sample size, and the genotype and allele frequencies of cases and controls (Table 1).
Table 1. Characteristics of the studies eligible for meta-analysis.
| First author | Year | Country | Ethnicity | Type of cancer | Source of control | Genotyping method | Case/control | Cases | Controls | HWE | Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gly388Arg rs351855 G>A | GG | AG | AA | G | A | GG | AG | AA | G | A | |||||||||
| Ansell | 2009 | Sweden | Caucasian | HNSCC | PB | PCR-RFLP | 110/192 | 61 | 49 | - | - | 81 | 111 | - | - | - | 9 | ||
| Bange | 2002 | Russia | Caucasian | Breast | PB | PCR-RFLP | 61/123 | 26 | 28 | 7 | 80 | 42 | 55 | 60 | 8 | 170 | 76 | 0.114 | 7 |
| Bange | 2002 | Germany | Caucasian | Breast | PB | PCR-RFLP | 84/123 | 41 | 34 | 9 | 116 | 52 | 55 | 60 | 8 | 170 | 76 | 0.114 | 8 |
| Bange | 2002 | Italy | Caucasian | Colon cancer | PB | PCR-RFLP | 82/123 | 37 | 38 | 7 | 112 | 52 | 55 | 60 | 8 | 170 | 76 | 0.114 | 8 |
| Batschauer | 2011 | Brazil | Caucasian | Breast | PB | PCR-RFLP | 68/85 | 39 | 26 | 3 | 104 | 32 | 47 | 35 | 3 | 129 | 41 | 0.249 | 9 |
| Chen | 2018 | Taiwan | Asian | Cervical cancer | HB | TaqMan | 226/335 | 69 | 101 | 56 | 239 | 213 | 96 | 165 | 74 | 357 | 313 | 0.845 | 9 |
| Chou | 2017 | Taiwan | Asian | OSCC | PB | TaqMan | 955/1191 | 225 | 524 | 206 | 974 | 936 | 334 | 596 | 261 | 1264 | 1118 | 0.873 | 11 |
| Fang | 2013 | China | Asian | NSCLC | HB | Sequencing | 629/729 | 193 | 331 | 105 | 717 | 541 | 163 | 391 | 175 | 717 | 741 | 0.049 | 9 |
| FitzGerald | 2009 | U.S.A. | Caucasian | Prostate | PB | SNPlex | 1254/1251 | 587 | 544 | 123 | 1718 | 790 | 631 | 496 | 124 | 1758 | 744 | 0.070 | 15 |
| FitzGerald | 2009 | U.S.A. | African | Prostate | PB | SNPlex | 146/80 | 104 | 39 | 3 | 247 | 45 | 60 | 18 | 2 | 138 | 22 | 0.646 | 13 |
| Gao | 2014 | China | Asian | NHL | NA | PCR-RFLP | 421/486 | 117 | 189 | 115 | 423 | 419 | 171 | 240 | 75 | 582 | 390 | 0.541 | 11 |
| Heinzle | 2012 | Austria | Caucasian | Colon cancer | PB | TaqMan | 85/1660 | 42 | 33 | 10 | 117 | 53 | 802 | 723 | 135 | 2327 | 993 | 0.114 | 10 |
| Ho | 2009 | Singapore | Asian | HCC | PB | Sequencing | 58/88 | 27 | 17 | 14 | 71 | 45 | 30 | 38 | 20 | 98 | 78 | 0.241 | 6 |
| Ho | 2010 | U.K. | Caucasian | Prostate | PB | TaqMan | 397/291 | 183 | 182 | 32 | 548 | 246 | 150 | 117 | 24 | 417 | 165 | 0.860 | 11 |
| Hosseini | 2017 | Iran | Asian | Breast Cancer | PB | PCR-RFLP | 126/160 | 87 | 33 | 6 | 207 | 45 | 54 | 57 | 49 | 165 | 155 | <0.001 | 6 |
| Jiang | 2015 | China | Asian | Breast cancer | NA | Snapshot | 747/716 | 205 | 404 | 138 | 814 | 680 | 270 | 348 | 98 | 888 | 544 | 0.398 | 12 |
| Li | 2017 | China | Asian | Cervical Cancer | HB | PCR-RFLP | 162/162 | 35 | 79 | 48 | 149 | 175 | 50 | 72 | 40 | 172 | 152 | 0.170 | 8 |
| Ma | 2008 | Japan | Asian | Prostate | HB | PCR-RFLP | 492/179 | 163 | 196 | 133 | 522 | 462 | 67 | 87 | 25 | 221 | 137 | 0.701 | 10 |
| Mawrin | 2006 | Germany | Caucasian | Glioma | HB | PCR-RFLP | 94/25 | 39 | 51 | 4 | 129 | 59 | 10 | 13 | 2 | 33 | 17 | 0.428 | 6 |
| Morimoto | 2003 | Japan | Asian | Sarcomas | NA | PCR-RFLP | 143/102 | 54 | 72 | 17 | 180 | 106 | 39 | 50 | 13 | 128 | 76 | 0.624 | 10 |
| Naidu | 2009 | Malaysia | Asian | Breast | HB | PCR-RFLP | 387/252 | 179 | 172 | 36 | 530 | 244 | 132 | 105 | 15 | 369 | 135 | 0.322 | 9 |
| Nan | 2009 | U.S.A. | Caucasian | Skin cancer | PB | TaqMan | 768/833 | 365 | 325 | 78 | 1055 | 481 | 406 | 343 | 84 | 1155 | 511 | 0.359 | 11 |
| Shen | 2013 | China | Asian | Gastric cancer | PB | Sequencing | 304/392 | 118 | 124 | 62 | 360 | 248 | 132 | 188 | 72 | 452 | 332 | 0.724 | 11 |
| Sheu | 2015 | China | Asian | HCC | HB | TaqMan | 289/595 | 82 | 150 | 57 | 314 | 264 | 159 | 314 | 122 | 632 | 558 | 0.146 | 8 |
| Spinola | 2005 | Italy | Caucasian | Lung | HB | Pyrosequencing | 274/401 | 148 | 104 | 22 | 400 | 148 | 193 | 168 | 40 | 554 | 248 | 0.699 | 9 |
| Spinola | 2005 | Italy | Caucasian | Breast | HB | Pyrosequencing | 142/220 | 67 | 55 | 20 | 189 | 95 | 112 | 83 | 25 | 307 | 133 | 0.117 | 8 |
| Spinola | 2005 | Italy | Caucasian | CRC | HB | Pyrosequencing | 179/220 | 98 | 63 | 18 | 259 | 99 | 112 | 83 | 25 | 307 | 133 | 0.117 | 8 |
| Tanuma | 2010 | Japan | Asian | OSCC | HB | PCR-SSCP | 150/100 | 69 | 53 | 28 | 191 | 109 | 42 | 48 | 10 | 132 | 68 | 0.487 | 8 |
| Tsay | 2020 | Taiwan | Asian | Cervical cancer | HB | TaqMan | 428/856 | 114 | 222 | 92 | 450 | 406 | 242 | 426 | 188 | 910 | 802 | 0.984 | 10 |
| Ture | 2015 | Turkey | Asian | Lung cancer | HB | PCR-RFLP | 124/100 | 66 | 47 | 11 | 179 | 69 | 48 | 46 | 6 | 142 | 58 | 0.242 | 7 |
| Wang | 2004 | U.S.A. | Caucasian | Prostate | PB | PCR-RFLP | 284/97 | 125 | 117 | 42 | 367 | 201 | 53 | 40 | 4 | 146 | 48 | 0.291 | 8 |
| Wang | 2004 | U.S.A. | African | Prostate | PB | PCR-RFLP | 45/94 | 37 | 6 | 2 | 80 | 10 | 76 | 18 | 0 | 170 | 18 | 0.305 | 7 |
| Whittle | 2016 | U.S.A. | Caucasian | Neuroblastoma | NA | PCR-RFLP | 126/114 | 45 | 69 | 12 | 159 | 93 | 50 | 60 | 4 | 160 | 68 | 0.006 | 9 |
| Wimmer | 2019 | Germany | Caucasian | HNSCC | PB | PCR-RFLP | 284/123 | 188 | 84 | 12 | 460 | 108 | 55 | 60 | 8 | 170 | 76 | 0.114 | 9 |
| Yang | 2012 | China | Asian | HCC | HB | TaqMan | 711/740 | 216 | 351 | 144 | 783 | 639 | 247 | 361 | 132 | 855 | 625 | 0.996 | 10 |
| First author | Year | Country | Ethnicity | Type of Cancer | Source of control | Genotyping method | Case/control | Cases | Controls | HWE | Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Val10Ile rs1966265 | CC | CT | TT | C | T | CC | CT | TT | C | T | |||||||||
| Chen | 2018 | Taiwan | Asian | Uterine Cervical | HB | TaqMan | 227/335 | 61 | 105 | 61 | 227 | 227 | 91 | 168 | 76 | 350 | 320 | 0.927 | 9 |
| Chou | 2017 | Taiwan | Asian | OSCC | PB | TaqMan | 955/1191 | 213 | 514 | 228 | 940 | 970 | 285 | 580 | 326 | 1150 | 1232 | 0.391 | 11 |
| FitzGerald | 2009 | U.S.A. | Caucasian | Prostate | PB | SNPlex | 1259/1254 | 782 | 405 | 72 | 1969 | 549 | 742 | 447 | 65 | 1931 | 577 | 0.827 | 15 |
| FitzGerald | 2009 | U.S.A. | African | Prostate | PB | SNPlex | 147/80 | 132 | 15 | 0 | 279 | 15 | 70 | 10 | 0 | 150 | 10 | 0.551 | 13 |
| Jiang | 2015 | China | Asian | Breast | NA | Snapshot | 747/716 | 171 | 408 | 168 | 750 | 744 | 126 | 364 | 226 | 616 | 816 | 0.322 | 12 |
| Nan | 2009 | U.S.A. | Caucasian | Skin cancer | PB | TaqMan | 753/821 | 461 | 251 | 41 | 1173 | 333 | 507 | 271 | 43 | 1285 | 357 | 0.390 | 11 |
| Sheu | 2015 | China | Asian | HCC | HB | TaqMan | 289/595 | 65 | 160 | 64 | 290 | 288 | 151 | 300 | 144 | 602 | 588 | 0.835 | 8 |
| Tsay | 2020 | Taiwan | Asian | Cervical cancer | HB | TaqMan | 428/856 | 95 | 226 | 107 | 416 | 440 | 215 | 420 | 221 | 850 | 862 | 0.585 | 10 |
| First author | Year | Country | Ethnicity | Type of cancer | Source of control | Genotyping method | Case/control | Cases | Controls | HWE | Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7708357 | GG | GA | AA | G | A | GG | GA | AA | G | A | |||||||||
| Chen | 2018 | Taiwan | Asian | Uterine cervical | HB | TaqMan | 227/335 | 222 | 4 | 1 | 448 | 6 | 321 | 13 | 1 | 655 | 15 | 0.038 | 9 |
| Chou | 2017 | Taiwan | Asian | OSCC | PB | TaqMan | 955/1191 | 932 | 22 | 1 | 1886 | 24 | 1167 | 23 | 1 | 2357 | 25 | 0.015 | 11 |
| FitzGerald | 2009 | U.S.A. | Caucasian | Prostate | PB | SNPlex | 1258/1254 | 459 | 632 | 167 | 1550 | 966 | 507 | 569 | 178 | 1583 | 925 | 0.368 | 15 |
| FitzGerald | 2009 | U.S.A. | African | Prostate | PB | SNPlex | 146/78 | 49 | 74 | 23 | 172 | 120 | 34 | 35 | 9 | 103 | 53 | 0.999 | 13 |
| Sheu | 2015 | China | Asian | HCC | HB | TaqMan | 289/595 | 283 | 5 | 1 | 571 | 7 | 577 | 18 | 0 | 1172 | 18 | 0.708 | 8 |
| Tsay | 2020 | Taiwan | Asian | Cervical cancer | HB | TaqMan | 428/856 | 416 | 10 | 2 | 842 | 14 | 838 | 17 | 1 | 1693 | 19 | 0.005 | 9 |
| First author | Year | Country | Ethnicity | Type of cancer | Source of control | Genotyping method | Case/control | Cases | Controls | HWE | Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2011077 | CC | CT | TT | C | T | CC | CT | TT | C | T | |||||||||
| Chen | 2018 | Taiwan | Asian | UT-cervical | HB | TaqMan | 227/335 | 63 | 102 | 62 | 228 | 226 | 94 | 163 | 78 | 351 | 319 | 0.652 | 9 |
| Chou | 2017 | Taiwan | Asian | OSCC | PB | TaqMan | 955/1191 | 210 | 509 | 236 | 929 | 981 | 288 | 577 | 326 | 1153 | 1229 | 0.299 | 11 |
| Ma | 2008 | Japan | Asian | Prostate | HB | PCR-RFLP | 492/179 | 94 | 285 | 113 | 473 | 511 | 11 | 85 | 83 | 107 | 251 | 0.075 | 10 |
| Sheu | 2015 | China | Asian | HCC | HB | TaqMan | 289/595 | 66 | 159 | 64 | 291 | 287 | 147 | 297 | 151 | 591 | 599 | 0.968 | 8 |
| Tsay | 2020 | Taiwan | Asian | Cervical cancer | HB | TaqMan | 428/856 | 94 | 224 | 110 | 412 | 444 | 217 | 418 | 221 | 852 | 860 | 0.495 | 10 |
| First author | Year | Country | Ethnicity | Type of cancer | Source of control | Genotyping method | Case/control | Cases | Controls | HWE | Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs376618 | AA | AG | GG | A | G | AA | AG | GG | A | G | |||||||||
| FitzGerald | 2009 | U.S.A. | Caucasian | Prostate | PB | SNPlex | 1238/1245 | 703 | 448 | 87 | 1854 | 622 | 712 | 437 | 96 | 1861 | 629 | 0.013 | 15 |
| FitzGerald | 2009 | U.S.A. | African | Prostate | PB | SNPlex | 145/80 | 65 | 59 | 21 | 189 | 101 | 38 | 38 | 4 | 114 | 46 | 0.154 | 13 |
| Nan | 2009 | U.S.A. | Caucasian | Skin cancer | PB | TaqMan | 762/830 | 451 | 273 | 38 | 1175 | 349 | 468 | 326 | 36 | 1262 | 398 | 0.026 | 11 |
Quality assessment
Two investigators evaluated the quality of each study using the quality assessment criteria [52]. Quality scores of studies ranged from 0 (lowest) to 15 (highest). Studies with scores ≤9 were considered as low quality, while those with scores > 9 were considered as high quality.
Statistical analysis
Meta-analysis was carried out using STATA 14.0 software (Stata Corporation, College Station, TX, U.S.A.). The Hardy–Weinberg equilibrium (HWE) of control genotypes was determined by the chi-square test.
The strength of the association between FGFR4 polymorphisms and cancer susceptibility was evaluated by pooled odds ratios (ORs) and their 95% confidence intervals (CIs) in five (heterozygous, homozygous, dominant, recessive, and allele) genetic models. The significance of the pooled OR was assessed by the Z-test, and P<0.05 was considered to be statistically significant. The between-study heterogeneity was evaluated by the Q statistic. When the PQ < 0.1, indicating the presence of heterogeneity, the random-effects model was selected, otherwise, the fixed-effects model was chosen.
Publication bias was inspected by using Begg’s funnel plots and the asymmetric plots implied potential publication bias. Egger’s test was used to measure the degree of asymmetry. A P<0.05 indicated significant publication bias.
Sensitivity analyses was done to evaluate whether a single study influenced the overall pooled results by omitting each study in turn.
Results
Study characteristics
A total of 57 case–control studies from 30 published articles [13–42] that met the inclusion criteria were included in our meta‐analyses. Of these 57 studies, the FGFR4 rs351855 in 35 studies, rs1966265 in 8 studies, rs7708357 in 6 studies, rs2011077 in 5 studies, and rs376618 in 3 studies were analyzed, respectively. The characteristics and relevant data of the included studies are presented in Table 1.
Meta-analysis results
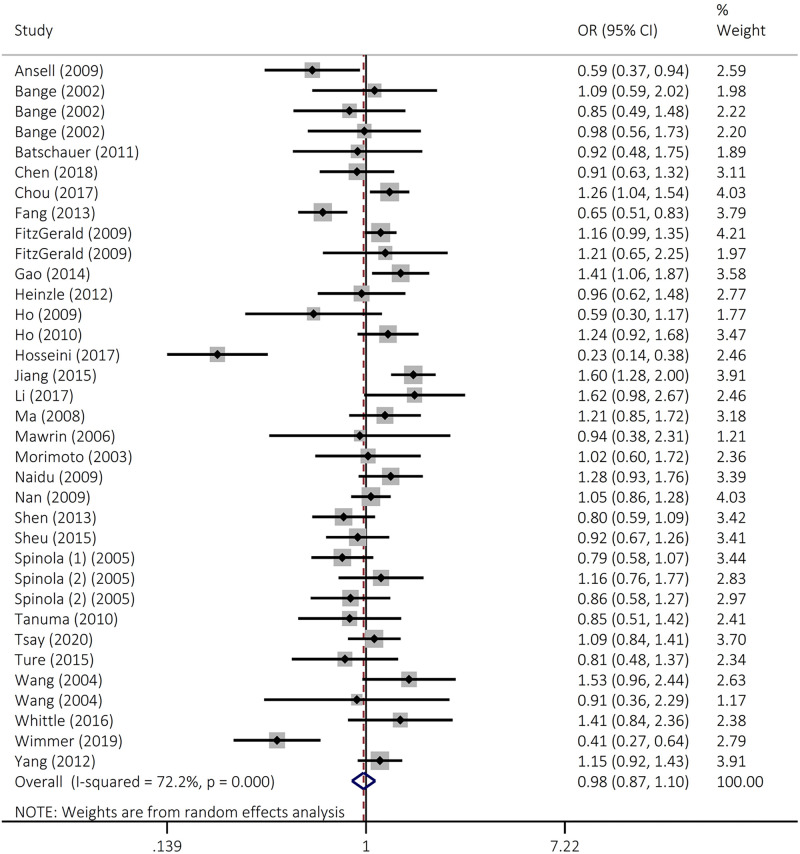
The findings did not support an association between FGFR4 rs351855 polymorphism and overall cancer susceptibility in heterozygous (OR = 0.97, 95% CI = 0.87–1.07, P=0.514, AG vs GG), homozygous (OR = 1.14, 95% CI = 0.95–1.37, P=0.166, AA vs GG), dominant (OR = 0.98, 95% CI = 0.87–1.10, P=0.686, AG+AA vs GG), recessive (OR = 1.15, 95% CI = 0.98–1.33, P=0.79, AA vs AG+GG), and allele (OR = 1.02, 95% CI = 0.93–1.12, P=0.663, A vs G) genetic models (Figure 2 and Table 2). Stratified analysis was achieved by ethnicity and cancer type (Table 3 and Figure 3). The results indicated that rs351855 variant significantly increased the risk of prostate cancer in heterozygous (OR = 1.16, 95% CI = 1.02–1.32, P=0.025 AG vs GG), dominant (OR = 1.20, 95% CI = 1.06–1.35, P=0.004, AG+AA vs GG), and allele (OR = 1.22, 95% CI = 1.06–1.41, P=0.005, A vs G) genetic models.
Figure 2. Forest plot for the association of the FGFR4 rs351855 polymorphism with overall cancer susceptibility in codominant (AG+AA vs GG).
Table 2. The pooled ORs and 95% CIs for the association between FGFR4 polymorphisms and cancer susceptibility.
| n | Genetic model | Association test | Heterogeneity test | Egger’s test P | Begg’s test P | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Z | P | χ2 | I2 (%) | P | |||||
| Overall | ||||||||||
| rs351855 G>A | 35 | AG vs GG | 0.97 (0.87–1.07) | 0.65 | 0.514 | 82.87 | 60.2 | <0.0001 | 0.012 | 0.064 |
| AA vs GG | 1.14 (0.95–1.37) | 1.39 | 0.166 | 116.25 | 71.6 | <0.0001 | 0.966 | 0.975 | ||
| AG+AA vs GG | 0.98 (0.87–1.10) | 0.40 | 0.686 | 122.33 | 72.2 | <0.0001 | 0.061 | 0.129 | ||
| AA vs AG+GG | 1.15 (0.98–1.33) | 1.76 | 0.79 | 94.88 | 65.2 | <0.0001 | 0.476 | 0.306 | ||
| A vs G | 1.02 (0.93–1.12) | 0.47 | 0.639 | 150.59 | 78.1 | <0.0001 | 0.416 | 0.293 | ||
| rs1966265 C>T | 8 | CT vs CC | 1.01 (0.89–1.14) | 0.14 | 0.891 | 11.12 | 37.1 | 0.133 | 0.739 | 1.000 |
| TT vs CC | 0.94 (0.77–1.16) | 0.56 | 0.574 | 14.60 | 58.9 | 0.024 | 0.373 | 0.176 | ||
| CT+TT vs CC | 0.98 (0.87–1.11) | 0.31 | 0.759 | 11.52 | 39.2 | 0.118 | 0.810 | 0.805 | ||
| TT vs CT+CC | 0.87 (0.78–0.97) | 2.61 | 0.009 | 14.07 | 57.3 | 0.029 | 0.094 | 0.051 | ||
| T vs C | 0.95 (0.87–1.04) | 1.03 | 0.303 | 14.24 | 50.8 | 0.047 | 0.722 | 0.805 | ||
| rs7708357 G>A | 6 | AG vs GG | 1.17 (0.95–1.44) | 1.45 | 0.146 | 5.61 | 10.9 | 0.346 | 0.221 | 0.039 |
| AA vs GG | 1.10 (0.87–1.40) | 0.83 | 0.406 | 3.61 | 0.0 | 0.607 | 0.143 | 1.000 | ||
| AG+AA vs GG | 1.17 (1.02–1.36) | 2.19 | 0.028 | 4.72 | 0.0 | 0.451 | 0.467 | 0.091 | ||
| AA vs AG+GG | 0.98 (0.79–1.21) | 0.20 | 0.840 | 3.77 | 0.0 | 0.583 | 0.097 | 0.624 | ||
| A vs G | 1.08 (0.98–1.20) | 1.51 | 0.132 | 4.22 | 0.0 | 0.518 | 0.964 | 0.348 | ||
| rs2011077 C>T | 5 | CT vs CC | 1.03 (0.79–1.33) | 0.21 | 0.831 | 11.30 | 64.6 | 0.023 | 0.054 | 0.014 |
| TT vs CC | 0.79 (0.49–1.25) | 1.02 | 0.309 | 28.54 | 86.0 | <0.0001 | 0.228 | 0.327 | ||
| CT+TT vs CC | 0.94 (0.69–1.28) | 0.39 | 0.695 | 17.85 | 77.6 | 0.001 | 0.091 | 0.050 | ||
| TT vs CT+CC | 0.79 (0.56–1.13) | 1.27 | 0.203 | 28.84 | 86.1 | <0.0001 | 0.681 | 1.000 | ||
| T vs C | 0.89 (0.70–1.13) | 0.97 | 0.332 | 33.89 | 88.2 | <0.0001 | 0.380 | 0.327 | ||
| rs376618 A>G | 3 | AG vs AA | 0.95 (0.85–1.09) | 0.56 | 0.753 | 1.76 | 0.0 | 0.414 | 0.761 | 0.602 |
| GG vs AA | 1.04 (0.81–1.33) | 0.29 | 0.771 | 4.12 | 51.5 | 0127 | 0.067 | 0.117 | ||
| AG+GG vs AA | 0.97 (0.76–1.10) | 0.45 | 0.654 | 1.27 | 0.0 | 0.531 | 0.858 | 0.602 | ||
| GG vs AG+AA | 1.19 (0.74–1.93) | 0.71 | 0.476 | 5.04 | 60.3 | 0.080 | 0.014 | 0.117 | ||
| G vs A | 0.99 (0.90–1.09) | 0.20 | 0.841 | 2.21 | 9.5 | 0.331 | 0.383 | 0.602 | ||
Table 3. Stratified analysis of rs351855 polymorphisms by ethnicity and cancer type.
| n | Genetic model | Association test | Heterogeneity test | Egger’s test P | Begg’s test P | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Z | P | χ2 | I2 (%) | P | |||||
| Caucasian | 15 | AG vs GG | 0.97 (0.84–1.12) | 0.42 | 0.672 | 26.56 | 47.3 | 0.002 | 0.162 | 0.586 |
| AA vs GG | 1.11 (0.95–1.29) | 1.30 | 0.193 | 19.36 | 27.7 | 0.152 | 0.331 | 0.216 | ||
| AG+AA vs GG | 0.96 (0.83–1.12) | 0.47 | 0.636 | 35.15 | 57.3 | 0.002 | 0.257 | 0.471 | ||
| AA vs AG+GG | 1.09 (0.94–1.26) | 1.09 | 0.278 | 16.49 | 15.1 | 0.284 | 0.118 | 0.125 | ||
| A vs G | 1.03 (0.92–1.15) | 0.43 | 0.666 | 30.34 | 53.9 | 0.007 | 0.789 | 0.458 | ||
| Asian | 17 | AG vs GG | 0.96 (0.82–1.12) | 0.56 | 0.572 | 55.29 | 71.1 | 0.000 | 0.023 | 0.039 |
| AA vs GG | 1.1 (0.84–1.44) | 0.72 | 0.470 | 94.65 | 83.1 | 0.000 | 0.636 | 0.510 | ||
| AG+AA vs GG | 0.98 (0.81–1.17) | 0.27 | 0.786 | 86.32 | 81.5 | 0.000 | 0.092 | 0.070 | ||
| AA vs AG+GG | 1.13 (0.91–1.40) | 1.12 | 0.262 | 76.08 | 79.0 | 0.000 | 0.832 | 0.458 | ||
| A vs G | 1.01 (0.87–1.16) | 0.11 | 0.913 | 119.83 | 86.6 | 0.000 | 0.352 | 0.217 | ||
| Breast cancer | 7 | AG vs GG | 0.94 (0.66–1.33) | 0.35 | 0.729 | 26.00 | 76.9 | 0.000 | 0.050 | 0.099 |
| AA vs GG | 1.03 (0.48–2.22) | 0.08 | 0.939 | 44.25 | 86.4 | 0.000 | 0.358 | 0.186 | ||
| AG+AA vs GG | 0.91 (0.58–1.44) | 0.40 | 0.691 | 50.88 | 88.2 | 0.000 | 0.135 | 0.099 | ||
| AA vs AG+GG | 1.05 (0.56–1.96) | 0.16 | 0.877 | 31.50 | 81.0 | 0.000 | 0.540 | 0.176 | ||
| A vs G | 0.92 (0.61–1.38) | 0.40 | 0.690 | 71.30 | 91.6 | 0.000 | 0.233 | 0.072 | ||
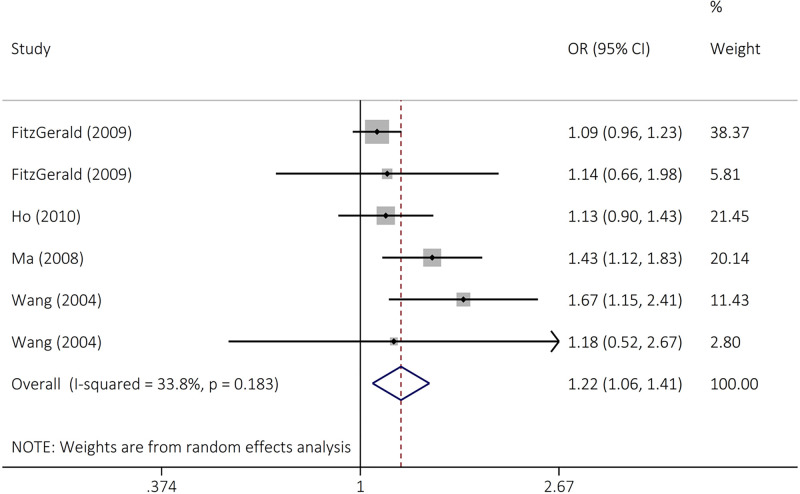
| Prostate cancer | 6 | AG vs GG | 1.16 (1.02–1.32) | 2.25 | 0.025 | 2.91 | 0.0 | 0.714 | 0.422 | 0.188 |
| AA vs GG | 1.60 (0.98–2.61) | 1.90 | 0.058 | 13.39 | 62.7 | 0.020 | 0.378 | 0.462 | ||
| AG+AA vs GG | 1.20 (1.06–1.35) | 2.89 | 0.004 | 1.67 | 0.0 | 0.892 | 0.639 | 0.851 | ||
| AA vs AG+GG | 1.56 (0.92–2.65) | 1.63 | 0.103 | 17.29 | 71.1 | 0.004 | 0.452 | 0.624 | ||
| A vs G | 1.22 (1.06–1.41) | 2.81 | 0.005 | 7.55 | 33.8 | 0.183 | 0.279 | 0.260 | ||
| Gastrointestinal cancer | 7 | AG vs GG | 0.92 (0.80–1.06) | 1.17 | 0.243 | 6.73 | 10.9 | 0.346 | 0.071 | 0.090 |
| AA vs GG | 1.06 (0.88–1.28) | 0.63 | 0.528 | 3.72 | 0.0 | 0.715 | 0.581 | 0.881 | ||
| AG+AA vs GG | 0.95 (0.84–1.09) | 0.70 | 0.487 | 6.14 | 2.2 | 0.408 | 0.093 | 0.652 | ||
| AA vs AG+GG | 1.10 (0.94–1.30) | 1.17 | 0.241 | 2.34 | 0.0 | 0.886 | 0.824 | 0.762 | ||
| A vs G | 1.01 (0.92–1.10) | 0.16 | 0.873 | 4.37 | 0.0 | 0.627 | 0.172 | 0.230 | ||
Bold values denote statistical significance at the P <0.05 level.
Figure 3. Forest plot for the association of the FGFR4 rs351855 polymorphism with prostate cancer susceptibility (A vs G).
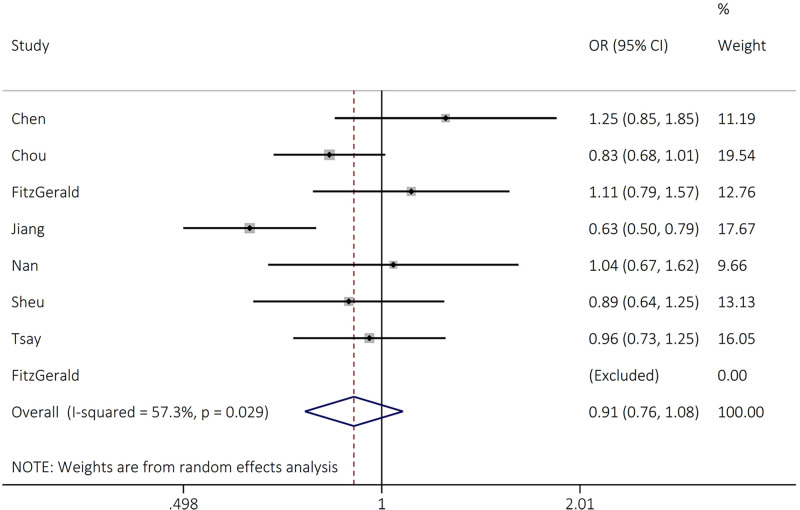
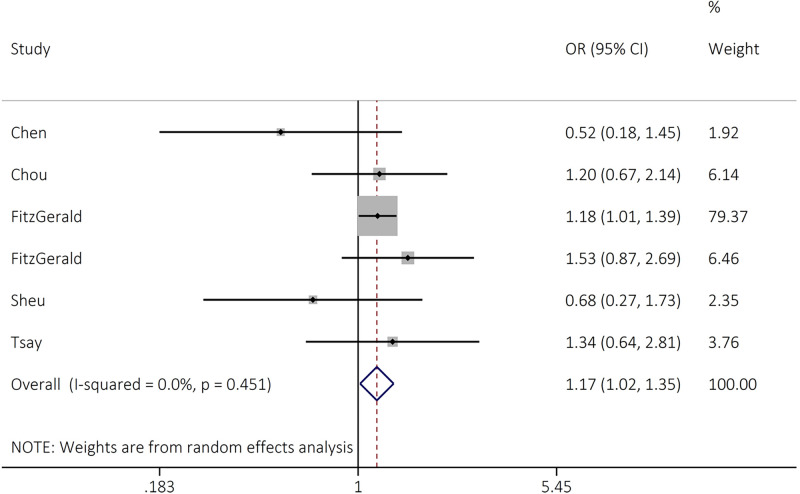
For FGFR4 rs1966265 polymorphism, the findings revealed that this variant significantly reduced the risk of cancer susceptibility in recessive (OR = 0.87, 95% CI = 0.78–0.97, P=0.009, TT vs CT+CC) model (Table 2 and Figure 4).
Figure 4. Forest plot for the association between FGFR4 rs1966265 and overall cancer risk in recessive (TT vs CT+CC) models.
The rs7708357 variant of FGFR4 significantly increased the risk of cancer development in dominant (OR = 1.17, 95% CI = 1.02–1.36, P=0.028, AG+AA GG) genetic model (Table 2 and Figure 5).
Figure 5. Forest plot for the association between FGFR4 rs7708357 and overall cancer risk in dominant model (AG+AA vs GG).
The rs2011077 and rs376618 variants were not associated with overall cancer risk in any genetic models tested (Table 2).
Heterogeneity and publication bias
As shown in Table 2, heterogeneity among the studies was observed in all genetic comparisons for rs351855 and rs2011077. For rs1966265, heterogeneity was not found in heterozygous and dominant genetic models. While, heterogeneity was not detected in all genetic models for rs7708357 and rs376618.
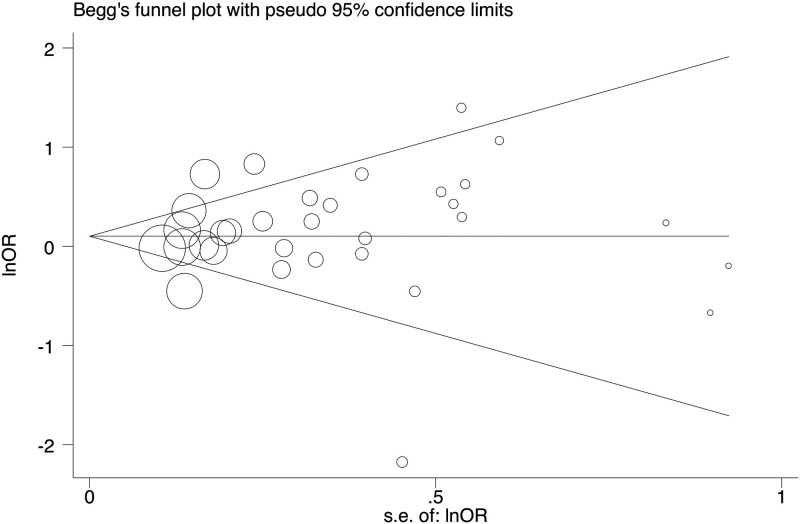
The potential publication bias was evaluated using Begg’s funnel plot and Egger’s test. The shape of funnel plots was symmetrical and the Egger’s test supported no existence of publication bias in all comparison except rs351855 polymorphism in heterozygous and rs376618 polymorphism in recessive genetic model (Table 2 and Figure 6).
Figure 6. Begg’s funnel plot for the test of publication bias for FGFR4 rs351855 in recessive model (AA vs AG+GG).
Sensitivity analysis
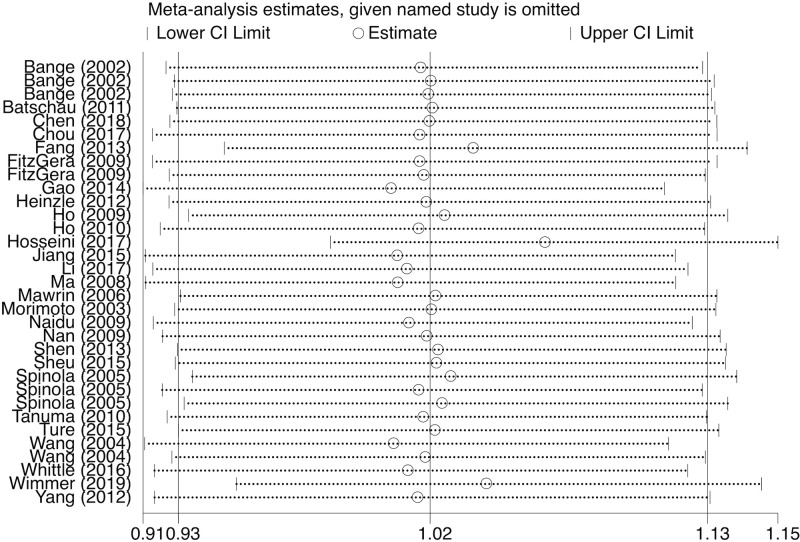
We performed sensitivity analysis to assess the effect of a specific publication on the overall estimate. For rs351855, the pooled ORs showed no significant change appeared when each study was neglected, one at a time, in heterozygous, dominant, and allele genetic models (Figure 7). For rs1966265, sensitivity analysis indicated no changes of results in heterozygous, homozygous, dominant, recessive, and allele genetic models. For rs7708357, no alterations of results were detected in homozygous, recessive, and allele genetic models. Thus, the final pooled results are both stable and reliable.
Figure 7. Sensitivity analysis on the association between the rs351855 polymorphism and susceptibility of overall cancer in allele genetic model (A vs G).
Discussion
FGFs and their receptors (FGFRs) regulate numerous cellular processes including the regulation of cell proliferation, differentiation, migration, and metabolism [12]. Deregulation of FGFRs signaling have been found to play an important role in cancer development and progression as well as resistance to anticancer [53–55]. Overexpression of FGFR4 predict metastasis and poor survival outcome in various cancers [56–58]. Blocking FGFR4 significantly suppresses the cancer and indicates that FGFR4 is a potential target for the cancer treatment [59]. Polymorphisms in the FGFR4 rs351855 (Gly388Arg) polymorphism, is positioned in the transmembrane domain of the EGFR4. It has been found that Arg388 allele causes increased receptor stability and prolonged receptor activation [60].
Several reports have examined the relationship between FGFR4 gene polymorphisms and diverse cancer types [13–20,22–42]. However, the findings were inconsistent. Therefore, this updated meta-analysis including more eligible studies was performed to evaluate the impact of FGFR4 polymorphisms on cancer susceptibility. For FGFR4 rs351855 polymorphism, the findings from 34 studies including 10407 cases and 12382 controls did not support an association between this polymorphism and overall cancer susceptibility. Stratified analyses showed that this SNP significantly increased the risk of prostate cancer (n=6) in heterozygous, homozygous, dominant, and allele genetic models. The variant was not related to breast cancer as well as gastrointestinal cancer. Furthermore, the variant was not correlated with ethnicity. A meta-analysis performed by Xiong et al. [51] from 27 studies indicated a significant association between FGFR4 rs351855 polymorphism and overall cancer risk in recessive genetic model. Stratified analysis showed that rs351855 SNP significantly increased the risk of prostate cancer. A meta-analysis performed by Shu et al. [61] on 14 studies investigated the association between FGFR4 rs351855 polymorphism and various cancer risks indicated a significant association between this SNP and risk of overall cancer in all heterozygous, homozygous, dominant, recessive, and allele tested genetic models.
FGFR4 rs1966265 changes chemotherapy response in breast cancer [62], higher risk of oral squamous cell carcinoma susceptibility [31], initiation of cervical cancer (Taiwanese women) [19], and higher risk of breast cancer in Chinese women of Heilongjiang province [16]. FGFR4 rs2011077 TC+CC polymorphism is associated with higher tumor stage, tumor size, and grading in urothelial cell carcinoma [21]. FGFR4 rs2011077 with the GG genotype also increased the risk of prostate cancer in Japanese population [26].
To the best of our knowledge, for the first time, we performed pooled analysis to inspect the impact of rs1966265, rs7708357, rs2011077, and rs376618 polymorphisms and overall cancer risk.
For FGFR4 rs1966265 polymorphism, the findings revealed that this variant significantly reduced the risk of cancer susceptibility in recessive (OR = 0.87, 95% CI = 0.78–0.97, P=0.009, TT vs CT+CC) model (Table 2 and Figure 3). Regarding rs7708357 polymorphism, the finding indicated that the rs1966265 variant significantly increased the risk of overall cancer in dominant (OR = 1.17, 95% CI = 1.02–1.36, P=0.028, AG+AA GG) genetic model (Table 2 and Figure 4). While, the rs2011077 and rs376618 polymorphisms were not associated with cancer risk in any genetic models tested (Table 2).
Some limitations of this meta-analysis should be taken into account. First, the sample sizes of this meta-analysis were not large especially for rs1966265 (n=7 studies), rs7708357 (n=5 studies), rs2011077 (n=4 studies), and rs376618 (n=3 studies) polymorphisms as well as in stratified analyses, which may lead to reduced statistical power. Second, the strength of the association were measured by unadjusted ORs for confounding factors due to the lack of demographic and environmental factors, which might have affected the results. Third, publication bias may be unavoidable since we were only able to acquire data from published articles. Finally, the meta-analysis was associated with a significant heterogeneity in some polymorphisms.
The current investigation provided a source for basic medical scientist and clinician to understand the importance of FGFR4 in different types of cancers and use the results as potential biomarkers for susceptibility to cancers. It also provided a collection of previous investigation on this gene to help epidemiologist scientists for their future investigations (Rev 1-4).
In summary, this meta-analysis revealed that FGFR4 rs351855 (Gly388Arg) polymorphism might be a marker for susceptibility to prostate cancer. The rs1966265 polymorphism significantly decreased and rs1966265 polymorphism significantly increased the risk of overall cancer. No significant associations were found for the FGFR4 rs2011077 and rs376618 polymorphisms. However, these findings need to be further confirmed through large samples and different ethnic populations.
Acknowledgements
We would like to dedicate this article to Professor Mohammad Hashemi who passed away recently after the submission of this work. He was a pioneer in genetic studies.
Abbreviations
- CD334
cluster of differentiation 334
- CI
confidence interval
- EGFR
epidermal growth factor receptor
- EMT
epithelial-to-mesenchymal transition
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- HNSCC
head and neck squamous cell carcinoma
- MAPK
mitogen-activated protein kinase
- OR
odds ratio
- PI3K
Phosphoinositide 3-kinases
- PLCγ
phospholipase C gamma
- RTK
receptor tyrosine kinases
- SNP
single nucleotide polymorphism
- STAT
signal transducer and activator of transcription
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
Abdolkarim Moazeni-Roodi did the analysis, participated in revision and updating the manuscript. Sahel Sarabandi and Shima Karami did the literature review and helped in data analysis. Mohammad Hashemi prepared the final draft of manuscript and supervised the analysis. Saeid Ghavami prepared the final edit of manuscript, supervised the whole project and led the revision after passing away of Professor Mohammad Hashemi.
References
- 1.Moazeni-Roodi A., Aftabi S., Sarabandi S., Karami S., Hashemi M. and Ghavami S. (2020) Genetic association between HOTAIR gene and the risk of cancer: an updated meta-analysis. J. Genet. 99, 48 10.1007/s12041-020-01214-w [DOI] [PubMed] [Google Scholar]
- 2.Hashemi M., Aftabi S., Moazeni-Roodi A., Sarani H., Wiechec E. and Ghavami S. (2020) Association of CASP8 polymorphisms and cancer susceptibility: a meta-analysis. Eur. J. Pharmacol. 881, 173201 10.1016/j.ejphar.2020.173201 [DOI] [PubMed] [Google Scholar]
- 3.Hashemi M., Karami S., Sarabandi S., Moazeni-Roodi A., Malecki A., Ghavami S. et al. (2019) Association between PD-1 and PD-L1 polymorphisms and the risk of cancer: a meta-analysis of case-control studies. Cancers (Basel) 11, 10.3390/cancers11081150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A. and Jemal A. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 5.Moazeni-Roodi A., Ghavami S. and Hashemi M. (2019) Survivin rs9904341 polymorphism significantly increased the risk of cancer: evidence from an updated meta-analysis of case-control studies. Int. J. Clin. Oncol. 24, 335–349 10.1007/s10147-019-01408-y [DOI] [PubMed] [Google Scholar]
- 6.Hashemi M., Sarabandi S., Karami S., Smieja J., Moazeni-Roodi A., Ghavami S. et al. (2020) LMO1 polymorphisms and the risk of neuroblastoma: Assessment of meta-analysis of case-control studies. J. Cell. Mol. Med. 24, 1160–1168 10.1111/jcmm.14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shojaei S., Koleini N., Samiei E., Aghaei M., Cole L.K., Alizadeh J. et al. (2020) Simvastatin increases temozolomide-induced cell death by targeting the fusion of autophagosomes and lysosomes. FEBS J. 287, 1005–1034 10.1111/febs.15069 [DOI] [PubMed] [Google Scholar]
- 8.Burke D., Wilkes D., Blundell T.L. and Malcolm S. (1998) Fibroblast growth factor receptors: lessons from the genes. Trends Biochem. Sci. 23, 59–62 10.1016/S0968-0004(97)01170-5 [DOI] [PubMed] [Google Scholar]
- 9.Semrad T.J. and Mack P.C. (2012) Fibroblast growth factor signaling in non-small-cell lung cancer. Clin. Lung Cancer 13, 90–95 10.1016/j.cllc.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 10.Powers C.J., McLeskey S.W. and Wellstein A. (2000) Fibroblast growth factors, their receptors and signaling. Endocr. Relat. Cancer 7, 165–197 10.1677/erc.0.0070165 [DOI] [PubMed] [Google Scholar]
- 11.Turner N. and Grose R. (2010) Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10, 116–129 10.1038/nrc2780 [DOI] [PubMed] [Google Scholar]
- 12.Lang L. and Teng Y. (2019) Fibroblast growth factor receptor 4 targeting in cancer: new insights into mechanisms and therapeutic strategies. Cells. 8, 31 10.3390/cells8010031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bange J., Prechtl D., Cheburkin Y., Specht K., Harbeck N., Schmitt M. et al. (2002) Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 62, 840–847 [PubMed] [Google Scholar]
- 14.Batschauer A.P., Cruz N.G., Oliveira V.C., Coelho F.F., Santos I.R., Alves M.T. et al. (2011) HFE, MTHFR, and FGFR4 genes polymorphisms and breast cancer in Brazilian women. Mol. Cell. Biochem. 357, 247–253 10.1007/s11010-011-0895-1 [DOI] [PubMed] [Google Scholar]
- 15.Hosseini M. (2017) The relationship between polymorphic fibroblast growth factor receptor (FGFR) gene and breast cancer risk. Arch. Breast Cancer 4, 24–27 [Google Scholar]
- 16.Jiang Y., Sun S., Wei W., Ren Y., Liu J. and Pang D. (2015) Association of FGFR3 and FGFR4 gene polymorphisms with breast cancer in Chinese women of Heilongjiang province. Oncotarget 6, 34023–34029 10.18632/oncotarget.5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naidu R., Har Y.C. and Taib N.A. (2009) Polymorphism of FGFR4 Gly388Arg does not confer an increased risk to breast cancer development. Oncol. Res. 18, 65–71 10.3727/096504009789954609 [DOI] [PubMed] [Google Scholar]
- 18.Spinola M., Leoni V.P., Tanuma J., Pettinicchio A., Frattini M., Signoroni S. et al. (2005) FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol. Rep. 14, 415–419 [PubMed] [Google Scholar]
- 19.Chen T.H., Yang S.F., Liu Y.F., Lin W.L., Han C.P. and Wang P.H. (2018) Association of fibroblast growth factor receptor 4 genetic polymorphisms with the development of uterine cervical cancer and patient prognosis. Reprod. Sci. 25, 86–93 10.1177/1933719117702250 [DOI] [PubMed] [Google Scholar]
- 20.Li Y.P., Zhang L., Zou Y.L. and Yu Y. (2017) Association between FGFR4 gene polymorphism and high-risk HPV infection cervical cancer. Asian Pac. J. Trop. Med. 10, 680–684 10.1016/j.apjtm.2017.07.008 [DOI] [PubMed] [Google Scholar]
- 21.Tsay M.-D., Hsieh M.-J., Lee C.-Y., Wang S.-S., Chen C.-S., Hung S.-C. et al. (2020) Involvement of FGFR4 gene variants on the clinicopathological severity in urothelial cell carcinoma. Int. J. Environ. Res. Public Health 17, 129 10.3390/ijerph17010129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinzle C., Gsur A., Hunjadi M., Erdem Z., Gauglhofer C., Stattner S. et al. (2012) Differential effects of polymorphic alleles of FGF receptor 4 on colon cancer growth and metastasis. Cancer Res. 72, 5767–5777 10.1158/0008-5472.CAN-11-3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y.Y., Lu Y.C., Shen D.P., Liu Y.J., Su X.Y., Zhu G.S. et al. (2013) Fibroblast growth factor receptor 4 Gly388Arg polymorphism in Chinese gastric cancer patients. World J. Gastroenterol. 19, 4568–4575 10.3748/wjg.v19.i28.4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.FitzGerald L.M., Karlins E., Karyadi D.M., Kwon E.M., Koopmeiners J.S., Stanford J.L. et al. (2009) Association of FGFR4 genetic polymorphisms with prostate cancer risk and prognosis. Prostate Cancer Prostatic Dis. 12, 192–197 10.1038/pcan.2008.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho C.K., Anwar S., Nanda J. and Habib F.K. (2010) FGFR4 Gly388Arg polymorphism and prostate cancer risk in Scottish men. Prostate Cancer Prostatic Dis. 13, 94–96 10.1038/pcan.2009.49 [DOI] [PubMed] [Google Scholar]
- 26.Ma Z., Tsuchiya N., Yuasa T., Inoue T., Kumazawa T., Narita S. et al. (2008) Polymorphisms of fibroblast growth factor receptor 4 have association with the development of prostate cancer and benign prostatic hyperplasia and the progression of prostate cancer in a Japanese population. Int. J. Cancer 123, 2574–2579 10.1002/ijc.23578 [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Stockton D.W. and Ittmann M. (2004) The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clin. Cancer Res. 10, 6169–6178 10.1158/1078-0432.CCR-04-0408 [DOI] [PubMed] [Google Scholar]
- 28.Ansell A., Farnebo L., Grenman R., Roberg K. and Thunell L.K. (2009) Polymorphism of FGFR4 in cancer development and sensitivity to cisplatin and radiation in head and neck cancer. Oral Oncol. 45, 23–29 10.1016/j.oraloncology.2008.03.007 [DOI] [PubMed] [Google Scholar]
- 29.Wimmer E., Ihrler S., Gires O., Streit S., Issing W. and Bergmann C. (2019) Fibroblast growth factor receptor 4 single nucleotide polymorphism Gly388Arg in head and neck carcinomas. World J. Clin. Oncol. 10, 136–148 10.5306/wjco.v10.i3.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanuma J., Izumo T., Hirano M., Oyazato Y., Hori F., Umemura E. et al. (2010) FGFR4 polymorphism, TP53 mutation, and their combinations are prognostic factors for oral squamous cell carcinoma. Oncol. Rep. 23, 739–744 [PubMed] [Google Scholar]
- 31.Chou C.H., Hsieh M.J., Chuang C.Y., Lin J.T., Yeh C.M., Tseng P.Y. et al. (2017) Functional FGFR4 Gly388Arg polymorphism contributes to oral squamous cell carcinoma susceptibility. Oncotarget 8, 96225–96238 10.18632/oncotarget.21958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spinola M., Leoni V., Pignatiello C., Conti B., Ravagnani F., Pastorino U. et al. (2005) Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J. Clin. Oncol. 23, 7307–7311 10.1200/JCO.2005.17.350 [DOI] [PubMed] [Google Scholar]
- 33.Ture M., Yakut T., Deligonul A., Karkucak M., Sag S.O., Hartavi M. et al. (2015) Investigation of FGFR4 (Gly388Arg) gene polymorphism in primary lung cancer patients. Int. J. Hum. Genet. 15, 7–12 10.1080/09723757.2015.11886245 [DOI] [Google Scholar]
- 34.Fang H.M., Tian G., Zhou L.J., Zhou H.Y. and Fang Y.Z. (2013) FGFR4 genetic polymorphisms determine the chemotherapy response of Chinese patients with non-small cell lung cancer. Acta Pharmacol. Sin. 34, 549–554 10.1038/aps.2012.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho H.K., Pok S., Streit S., Ruhe J.E., Hart S., Lim K.S. et al. (2009) Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J. Hepatol. 50, 118–127 10.1016/j.jhep.2008.08.015 [DOI] [PubMed] [Google Scholar]
- 36.Sheu M.J., Hsieh M.J., Chiang W.L., Yang S.F., Lee H.L., Lee L.M. et al. (2015) Fibroblast growth factor receptor 4 polymorphism is associated with liver cirrhosis in hepatocarcinoma. PLoS ONE 10, e0122961 10.1371/journal.pone.0122961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Zhou Y., Lu M., An Y., Li R., Chen Y. et al. (2012) Association between fibroblast growth factor receptor 4 polymorphisms and risk of hepatocellular carcinoma. Mol. Carcinog. 51, 515–521 10.1002/mc.20805 [DOI] [PubMed] [Google Scholar]
- 38.Morimoto Y., Ozaki T., Ouchida M., Umehara N., Ohata N., Yoshida A. et al. (2003) Single nucleotide polymorphism in fibroblast growth factor receptor 4 at codon 388 is associated with prognosis in high-grade soft tissue sarcoma. Cancer 98, 2245–2250 10.1002/cncr.11778 [DOI] [PubMed] [Google Scholar]
- 39.Nan H., Qureshi A.A., Hunter D.J. and Han J. (2009) Genetic variants in FGFR2 and FGFR4 genes and skin cancer risk in the Nurses’ Health Study. BMC Cancer 9, 172 10.1186/1471-2407-9-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittle S.B., Reyes S., Du M., Gireud M., Zhang L., Woodfield S.E. et al. (2016) A polymorphism in the FGFR4 gene is associated with risk of neuroblastoma and altered receptor degradation. J. Pediatr. Hematol. Oncol. 38, 131–138 10.1097/MPH.0000000000000506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao L., Feng Z., Li Q., Li L., Chen L. and Xiao T. (2014) Fibroblast growth factor receptor 4 polymorphism is associated with increased risk and poor prognosis of non-Hodgkin’s lymphoma. Tumour Biol. 35, 2997–3002 10.1007/s13277-013-1386-7 [DOI] [PubMed] [Google Scholar]
- 42.Mawrin C., Kirches E., Diete S., Wiedemann F.R., Schneider T., Firsching R. et al. (2006) Analysis of a single nucleotide polymorphism in codon 388 of the FGFR4 gene in malignant gliomas. Cancer Lett. 239, 239–245 10.1016/j.canlet.2005.08.013 [DOI] [PubMed] [Google Scholar]
- 43.Wei W., You Z., Sun S., Wang Y., Zhang X., Pang D. et al. (2018) Prognostic implications of fibroblast growth factor receptor 4 polymorphisms in primary breast cancer. Mol. Carcinog. 57, 988–996 10.1002/mc.22819 [DOI] [PubMed] [Google Scholar]
- 44.Yu W., Feng S., Dakhova O., Creighton C.J., Cai Y., Wang J. et al. (2011) FGFR-4 Arg(3)(8)(8) enhances prostate cancer progression via extracellular signal-related kinase and serum response factor signaling. Clin. Cancer Res. 17, 4355–4366 10.1158/1078-0432.CCR-10-2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye Y., Shi Y., Zhou Y., Du C., Wang C., Zhan H. et al. (2010) The fibroblast growth factor receptor-4 Arg388 allele is associated with gastric cancer progression. Ann. Surg. Oncol. 17, 3354–3361 10.1245/s10434-010-1323-6 [DOI] [PubMed] [Google Scholar]
- 46.Falvella F.S., Frullanti E., Galvan A., Spinola M., Noci S., De Cecco L. et al. (2009) FGFR4 Gly388Arg polymorphism may affect the clinical stage of patients with lung cancer by modulating the transcriptional profile of normal lung. Int. J. Cancer 124, 2880–2885 10.1002/ijc.24302 [DOI] [PubMed] [Google Scholar]
- 47.Gallagher C.S., Makinen N., Harris H.R., Rahmioglu N., Uimari O., Cook J.P. et al. (2019) Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nat. Commun. 10, 4857 10.1038/s41467-019-12536-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu Y.H., Estrada K., Evangelou E., Ackert-Bicknell C., Akesson K., Beck T. et al. (2019) Meta-analysis of genomewide association studies reveals genetic variants for hip bone geometry. J. Bone Miner. Res. 34, 1284–1296 10.1002/jbmr.3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baird D.A., Evans D.S., Kamanu F.K., Gregory J.S., Saunders F.R., Giuraniuc C.V. et al. (2019) Identification of novel loci associated with hip shape: a meta-analysis of genomewide association studies. J. Bone Miner. Res. 34, 241–251 10.1002/jbmr.3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J., Liu Q., Yuan S., Xie W., Liu Y., Xiang Y. et al. (2017) Genetic predisposition to lung cancer: comprehensive literature integration, meta-analysis, and multiple evidence assessment of candidate-gene association studies. Sci. Rep. 7, 8371 10.1038/s41598-017-07737-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong S.W., Ma J., Feng F., Fu W., Shu S.R., Ma T. et al. (2017) Functional FGFR4 Gly388Arg polymorphism contributes to cancer susceptibility: Evidence from meta-analysis. Oncotarget 8, 25300–25309 10.18632/oncotarget.15811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He J., Liao X.Y., Zhu J.H., Xue W.Q., Shen G.P., Huang S.Y. et al. (2014) Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci. Rep. 4, 6159 10.1038/srep06159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Babina I.S. and Turner N.C. (2017) Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 17, 318–332 10.1038/nrc.2017.8 [DOI] [PubMed] [Google Scholar]
- 54.Porta R., Borea R., Coelho A., Khan S., Araujo A., Reclusa P. et al. (2017) FGFR a promising druggable target in cancer: Molecular biology and new drugs. Crit. Rev. Oncol. Hematol. 113, 256–267 10.1016/j.critrevonc.2017.02.018 [DOI] [PubMed] [Google Scholar]
- 55.Dienstmann R., Rodon J., Prat A., Perez-Garcia J., Adamo B., Felip E. et al. (2014) Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann. Oncol. 25, 552–563 10.1093/annonc/mdt419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye Y.W., Zhang X., Zhou Y., Wu J., Zhao C., Yuan L. et al. (2012) The correlations between the expression of FGFR4 protein and clinicopathological parameters as well as prognosis of gastric cancer patients. J. Surg. Oncol. 106, 872–879 10.1002/jso.23153 [DOI] [PubMed] [Google Scholar]
- 57.Li J., Ye Y., Wang M., Lu L., Han C., Zhou Y. et al. (2016) The over-expression of FGFR4 could influence the features of gastric cancer cells and inhibit the efficacy of PD173074 and 5-fluorouracil towards gastric cancer. Tumour Biol. 37, 6881–6891 10.1007/s13277-015-4411-1 [DOI] [PubMed] [Google Scholar]
- 58.Sahadevan K., Darby S., Leung H.Y., Mathers M.E., Robson C.N. and Gnanapragasam V.J. (2007) Selective over-expression of fibroblast growth factor receptors 1 and 4 in clinical prostate cancer. J. Pathol. 213, 82–90 10.1002/path.2205 [DOI] [PubMed] [Google Scholar]
- 59.Xin Z., Song X., Jiang B., Gongsun X., Song L., Qin Q. et al. (2018) Blocking FGFR4 exerts distinct anti-tumorigenic effects in esophageal squamous cell carcinoma. Thorac. Cancer 9, 1687–1698 10.1111/1759-7714.12883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J., Yu W., Cai Y., Ren C. and Ittmann M.M. (2008) Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia 10, 847–856 10.1593/neo.08450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shu C. and Wang J. (2017) Association between FGFR4 Gly388Arg polymorphism (rs351855) and cancer risk: A meta analysis including 10,584 subjects. Meta Gene 13, 32–37 10.1016/j.mgene.2017.04.003 [DOI] [Google Scholar]
- 62.Chen L., Qi H., Zhang L., Li H., Shao J., Chen H. et al. (2018) Effects of FGFR gene polymorphisms on response and toxicity of cyclophosphamide-epirubicin-docetaxel-based chemotherapy in breast cancer patients. BMC Cancer 18, 1038 10.1186/s12885-018-4951-z [DOI] [PMC free article] [PubMed] [Google Scholar]