Significance
Tospoviruses are major field crop pathogens that cause significant economic damage, raising global food security concerns. However, their assembly, attachment to their host cells, and genome delivery mechanisms remain unclear. This paper provides detailed structural insights on tospovirus glycoprotein N (GN). The significant knowledge gap regarding the structure and function of envelope glycoproteins from plant viruses in general and tospoviruses in particular impedes our fundamental understanding of these viruses’ host cell recognition and entry mechanisms. Our structural analysis of the tomato spotted wilt virus glycoprotein N (TSWV GN) reveals an evolutionary link between plant- and animal-infecting viruses that extends beyond the Bunyavirales order, describes a dimerization interface, and provides possible strategies for countermeasures against these important agricultural threats.
Keywords: virion assembly, envelope glycoproteins, X-ray crystallography, plant viruses, virus structure
Abstract
Tospoviridae is a family of enveloped RNA plant viruses that infect many field crops, inflicting a heavy global economic burden. These tripartite, single-stranded, negative-sense RNA viruses are transmitted from plant to plant by thrips as the insect vector. The medium (M) segment of the viral genome encodes two envelope glycoproteins, GN and GC, which together form the envelope spikes. GC is considered the virus fusogen, while the accompanying GN protein serves as an attachment protein that binds to a yet unknown receptor, mediating the virus acquisition by the thrips carrier. Here we present the crystal structure of glycoprotein N (GN) from the tomato spotted wilt virus (TSWV), a representative member of the Tospoviridae family. The structure suggests that GN is organized as dimers on TSWV’s outer shell. Our structural data also suggest that this dimerization is required for maintaining GN structural integrity. Although the structure of the TSWV GN is different from other bunyavirus GN proteins, they all share similar domain connectivity that resembles glycoproteins from unrelated animal-infecting viruses, suggesting a common ancestor for these accompanying proteins.
Enveloped plant viruses belong to three major families: Tospoviridae, Fimoviridae, and Rhabdoviridae. The first two families are members of the order Bunyavirales, which are single-stranded, negative-sense RNA viruses, capable of infecting a wide range of hosts from plants to animals, including humans.
Tomato spotted wilt virus (TSWV), first identified in Australia in 1930 (1), is a representative member of the family Tospoviridae, infecting hundreds of plant species, and is ultimately responsible for the loss of many different field crops, including food, fiber, and ornamental crops. Therefore, TSWV infection is a major global agricultural concern that results in heavy economic burden, with serious implications for food security (2, 3). Currently, prevention measures are focused on eliminating TSWV’s insect vector, the western flower thrips (Frankliniella occidentalis), in the field. However, these pesticides are largely inefficient and raise environmental concerns. These challenges, together with the lack of a rapid detection method, have a devastating impact on agricultural crops and the global economy, stressing the need for developing new TSWV countermeasures.
There is a delicate interplay between TSWV and its thrips vector, with a narrow window of opportunity for virus acquisition during larval development (4). Specifically, TSWV acquisition is accomplished when the first and second instar larvae of the thrips feed on an infected plant (5, 6), followed by entry of the virus into the thrips’ anterior midgut epithelial cells. The virus spreads to surrounding gut cells and crosses the basal lamina to infect the circular and longitudinal muscle cells (5). The virus’s route of dissemination and accumulation is tightly linked to the insect’s development. The virus spreads in the insect through pupal stages and accumulates in the salivary glands of adults, where major viral replication occurs (7–9). When an adult thrips feeds on an uninfected plant, the virus is released to the wounded plant tissue directly into the cell’s cytoplasm, and from there, the virus spreads to the entire plant via its movement protein, using the plasmodesmata for short distances and the phloem–xylem system to spread to organs and periphery tissues (10). It was shown that both TSWV envelope glycoproteins (GN and GC) are critical for thrips infection, but they are not required once the virus is mechanically introduced within a wounded tissue (11, 12). Therefore, once a TSWV is delivered into a susceptible plant host, the cell wall and plasma membrane are no longer barriers to the virus’s entry.
In thrips, TSWV is thought to enter the gut cells via receptor-mediated endocytosis, yet the identity of such a thrips receptor has remained elusive. An effort to identify thrips proteins that copurify with TSWV particles found distinct bands in gel overlay experiments, but the lack of genome sequence data and the limited proteomics technologies at the time prevented the characterization of these proteins (13, 14). Recently, the first thrips proteins that bind to TSWV particles were identified, using electrospray ionization mass spectrometry (15).
The TSWV genome is composed of three RNA segments (large, medium, and small), enclosed in a bilayered lipid membrane. The medium (M) segment encodes for two envelope glycoproteins, GN and GC. These are translated as a one-glycoprotein precursor that is cotranslationally processed by host proteases in the absence of other viral proteins to yield the two mature glycoproteins (16–19). Similar to what was previously demonstrated for other bunyaviruses (20–23), the TSWV mature glycoproteins travel from the endoplasmic reticulum (ER) to the Golgi apparatus, the site of virion morphogenesis (24). GC localization to the Golgi was shown to depend on its interaction with GN, its accompanying protein, which includes a Golgi-targeting signal that was found to be functional in both plant and animal cells (25–27). To date, the organization and stoichiometry of TSWV glycoproteins on the membrane of the mature virion are yet unknown, but it was previously demonstrated that both GN homodimers and GN/GC heterodimers exist on membranes of purified TSWV virions (28).
GN and GC are embedded in the viral lipid membrane, with the bulk of the glycoprotein facing outward, and are ∼58 and 95 kDa, respectively (29, 30). It was first predicted and later demonstrated for other members of the Bunyavirales order that GC, the viral fusogen, is a class II membrane fusion protein (31–35), resembling other fusogens from other unrelated RNA viruses (36, 37), and it was inferred by sequence homology to also be the case with tospoviruses (32). In contrast, when it comes to the fusogen-accompanying protein (i.e., GN), there is structural diversity in domain folds and organization on the mature virions among different members of the Bunyavirales order. For example, the GN from phleboviruses assembles in either hexameric or pentameric rings as part of the pseudoequivalent T = 12 icosahedral assembly of the virion (38), whereas the hantaviruses’ GN is organized in a pseudofourfold local symmetry with no apparent higher organization (39). An electron cryotomography study of the orthobunyavirus envelope spike complex implies that GN is positioned in proximity to the viral membrane, whereas GC trimerizes in a tripod-like ultrastructure (40) that was later shown to be mediated through GC’s N-terminal α-helical head domain (41). Moreover, recently published atomic resolution structures of GN proteins from phleboviruses (family: Phenuiviridae) and hantaviruses (family: Hantaviridae) (38, 39, 42) show structural diversity in their fold and epitope distribution. This structural diversity of GN proteins is probably a key to their ability to adopt different modes of assembly on the viral envelope.
Here we present an atomic resolution structure of glycoprotein N (GN) from TSWV, a structure of an envelope glycoprotein from a plant virus. The structure reveals a unique protein domain fold with an evolutionary link to animal-infecting viruses, a dimer complex formation, and additional molecular determinants that are vital for rationalized design of entry inhibitors against TSWV.
Results and Discussion
The Soluble Core of TSWV GN Has a Unique Fold.
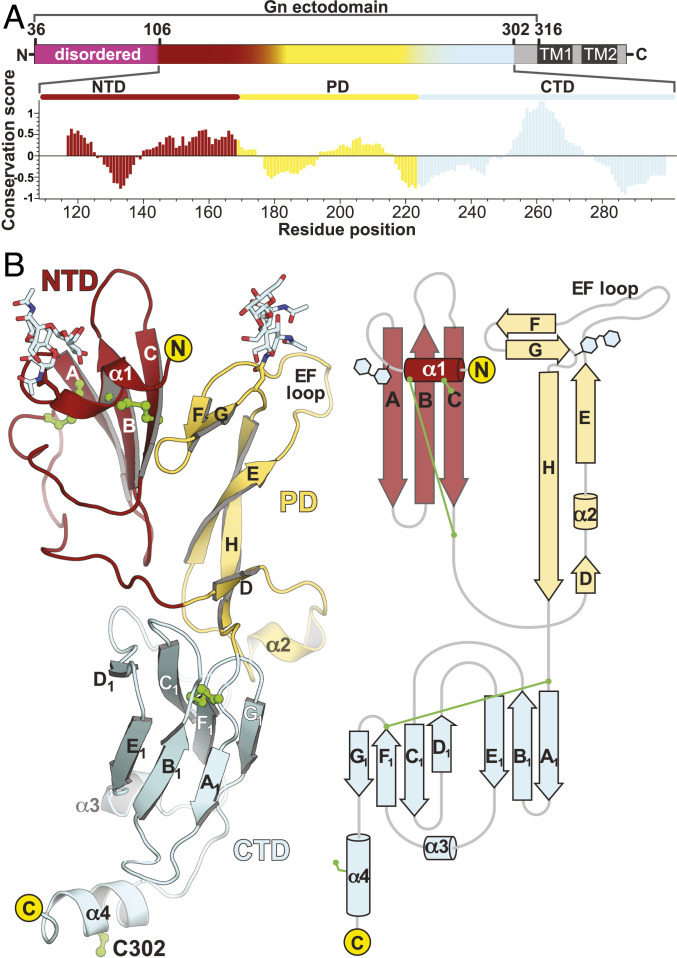
TSWV GN is a type I transmembrane protein with an N-terminal ectodomain followed by a transmembrane segment anchored on the viral membrane envelope. A previous study showed that the ectopic expression of TSWV GN residues 37 to 313 (also termed soluble GN, or sGN, Fig. 1A) in Sf9 insect cells produces a soluble, secreted protein (29). sGN followed by a C-terminal 8×His-tag was expressed using a baculovirus insect cells system and purified to homogeneity. To reveal the mode of assembly and the functional landscape of TSWV GN, we determined the crystal structure of sGN to 4.1 Å resolution (SI Appendix, Table S1). The visible part of the structure spans residues 107 to 305 (Fig. 1B), with missing electron density at both N and C termini (residues 36 to 107 and 306 to 310, respectively).
Fig. 1.
Overview of sGN crystal structure. (A, Top) A linear representation of the TSWV GN gene product. Magenta represents the disordered N-terminal extension, red shows NTD, yellow shows PD, and pale cyan shows CTD. In gray are regions that were not included in the expressed construct. Dark gray are the putative transmembrane domains. (Bottom) A graphical representation of CONSURF (43) conservation analysis. Scores account for evolutionary distance and are normalized to units of SD. Zero represents the average evolutionary rate, and <0 indicates increasing conservation. The scores were then averaged in a window of ±7 residues (i.e., 15 total residues) for every residue and plotted as seen in the histogram. The color scheme represents the three domains of sGN’s structure. (B) A cartoon representation (Left) and topology depiction (Right) of sGN crystal structure. Cysteine residue side chains are in green.
The overall structure of TSWV sGN has a three-domain fold with a short C-terminal α-helical extension (Fig. 1). The N-terminal domain (NTD, residues 107 to 169) encompasses a short helix (α1) parallel to the plane of the subsequent three-stranded β-sheet. The polypeptide chain then continues as a long coiled loop toward the pincer domain (PD; residues 170 to 222), a novel structure that consists of the EH β-sheet, a short helix located between two β-strands (D and E), the EF loop, and the FG β-sheet. The C-terminal domain (CTD) has a β-sandwich fold made of A1B1E1 and C1D1F1G1 β-sheets followed by a C-terminal α-helical extension (helix α4). Two N-glycosylation sites are observed in the electron density map at N116 and N210 in the NTD and PD, respectively. These glycosylations sites are likely projecting outward from the viral membrane, while the CTD is juxtamembrane oriented.
TSWV GN Has the Same Domain Connectivity as Other Accompanying Proteins from Animal-Infecting RNA Viruses.
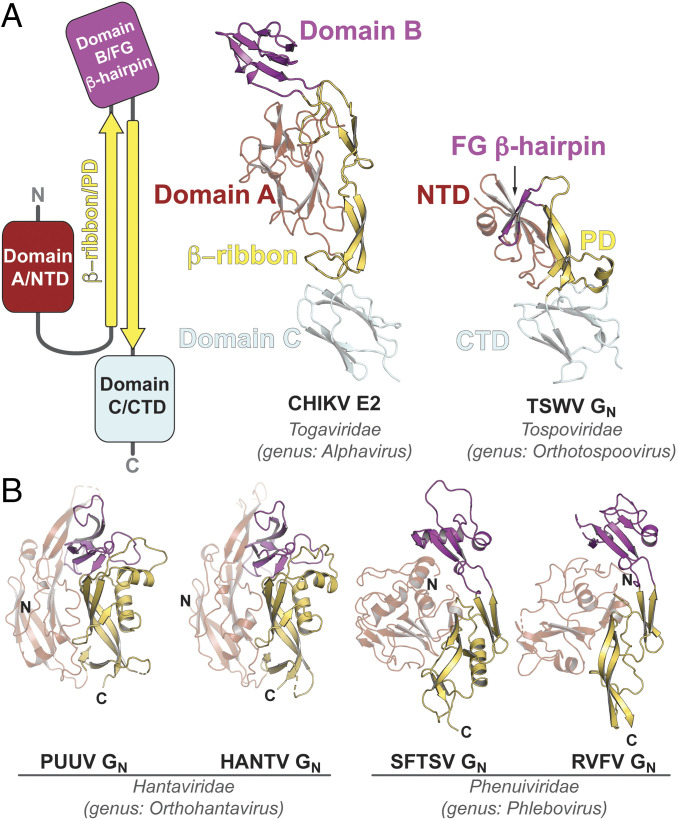
Recently, the crystal structures of GN from members of the Phenuiviridae and Hantaviridae families (all members of the Bunyavirales order) (38, 39, 42, 44) were determined. It was implied that GN from hantaviruses resembles the domain organization and connectivity of the E2 envelope glycoprotein from alphaviruses, an unrelated positive-sense, single-stranded RNA (ssRNA) virus (45). Unlike TSWV sGN, GN proteins from the Phenuiviridae family members display a unique α/β-fold that includes a β-arch motif, possibly a remnant of the β-ribbon motif found in alphavirus E2. Although the CTD β-sandwich of TSWV GN is homologous to domain C found in the alphavirus E2 (SI Appendix, Fig. S1), searches for structural homologs and classification in automated structure comparison programs such as the DALI server (46–48), SCOP (Structural Classification of Proteins) (49) and ECOD (Evolutionary Classification of Domains) (50, 51) did not reveal any similar structures or structural motifs for the NTD and PD. Nevertheless, we see a strong similarity between TSWV’s GN and the domain topology of the alphavirus E2 protein (Fig. 2A): The first domain (NTD) is connected via an arch, which in the tospovirus extends to contain β-strand D (Fig. 1) and the helix α2. At the other end of the “β-arch,” the FG β-hairpin represents a minimal version of the alphavirus E2 domain B (Fig. 2A). Intriguingly, unlike the animal-infecting bunyaviruses, tospoviruses keeps the juxtamembrane β-sandwich domain (e.g., domain C in E2), while in the other GN proteins, this domain is replaced by what appears to be a linker that is followed by the transmembrane domain (Fig. 2B). It is plausible that in the case of tospoviruses, evolution has replaced domains A and B by a simpler fold (NTD and FG β-hairpin, respectively) and kept domain C (CTD) downstream to a β-arch (the PD), a remnant of the β-ribbon motif of alphaviruses E2. This striking similarity in the topological organization of all these accompanying proteins suggests a common ancestor that strongly diverged, compared to the fusion proteins they escort.
Fig. 2.
The domain topology of TSWV GN resembles that of animal-infecting enveloped RNA viruses. (A) On the Left, a topology of domain organization of the alphavirus E2. Domain labels are of alphavirus E2 and the equivalent in TSWV GN. In the Middle, the crystal structure of the chikungunya virus (CHIKV) E2 (PDB ID code 3N43) compared to TSWV GN (Right). Both structures are to scale. (B) Crystal structures of GN proteins from other members of the Bunyavirales order. PDB ID codes from the Left: 5FXU (Puumala virus, PUUV), 5OPG (Hantaan virus, HANTV), 5Y10 (severe fever with thrombocytopenia syndrom virus, SFTSV), and 5Y0W (Rift Valley fever virus, RVFV). The color scheme is as in Fig. 1 and A. Note the lack of domain C/CTD in all these structures.
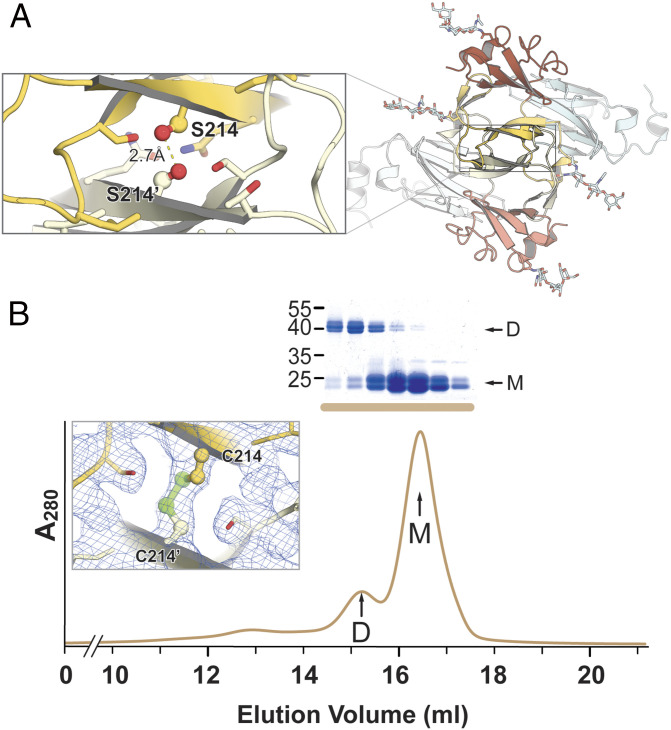
sGN Forms a Covalently Linked Dimer in Solution.
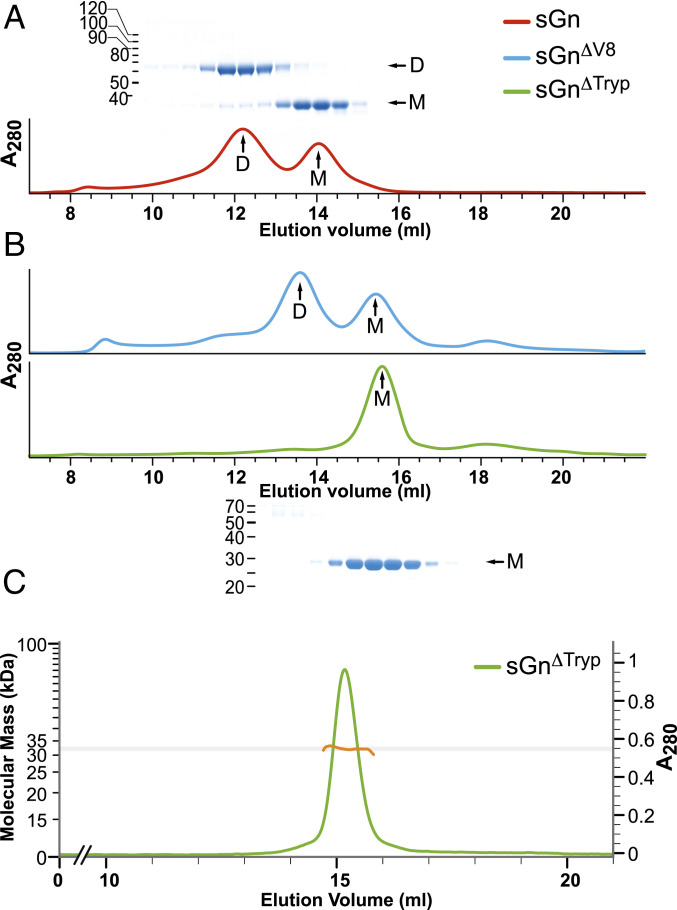
GN proteins from bunyavirales members usually exhibit a high cysteine content (e.g., 27 to 28 cysteines in phenuiviruses, 22 in orthohantavirus). In contrast, TSWV GN has only eight cysteine residues, seven of which were included in our sGN construct. Four cysteines are engaged in disulfide bonds that stabilize the NTD (C119–C150 and C127–C161), two form a disulfide bond located at the CTD (C224–C285), and the seventh cysteine (C302) is at the C-terminal α4 helix extension (Fig. 1B). A size exclusion chromatography (SEC) profile of the purified sGN shows two distinct elution peaks that correspond to two molecular weight species (Fig. 3A). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis under reducing and nonreducing conditions revealed that the higher molecular weight population represents a dimer covalently linked by an intermolecular disulfide bridge (Fig. 3A). This observation is consistent with a previous report which detected, in addition to disulfide-linked GN dimers on the virus envelope, a monomeric GN (28), implying a transient disulfide bond formation.
Fig. 3.
GN partially dimerizes in solution. (A) SEC profile of sGN with the corresponding nonreducing SDS-PAGE. D: dimer; M: monomer. Colors are as per the legend. (B) SEC profile of sGN∆V8 and sGN∆tryp. On the bottom is the nonreducing SDS-PAGE of the sGN∆tryp peak fractions, indicating the removal of the intermolecular disulfide bond. (C) SEC-MALS profile of sGN∆tryp confirming the molecular weight of a monomeric sGN∆tryp.
To confirm that cysteine 302 is engaged in intermolecular disulfide dimerization, we subjected sGN to limited proteolysis with V8 (Glu-C) and trypsin proteases. Both yielded resistant fragments with a similar molecular weight, as observed by SDS-PAGE (SI Appendix, Fig. S2A). However, while the V8-treated sGN (sGN∆V8) maintained disulfide-dependent dimerization and exhibited a SEC elution profile similar to that of sGN, the trypsin-treated sGN (sGN∆tryp) eluted exclusively as a monomer (Fig. 3 B–D). A Lys-Arg-rich region at sGN’s C terminus, upstream to C302 (Fig. 1A), was predicted as trypsin’s cleavage site. This was validated by Western blot analysis with an anti-His antibody, showing the removal of the C-terminal 8×His-tag (SI Appendix, Fig. S2B) and demonstrating that C302 is, indeed, involved in the intermolecular disulfide formation in solution.
Oligomerization Interfaces of TSWV GN.
As demonstrated by the SEC analysis, both sGN and sGN∆V8 partially dimerize in solution through a disulfide bridge. This was previously also shown for the recombinant ectodomain of GN from Rift Valley fever virus (RVFV) and severe fever with thrombocytopenia syndrome virus (SFTSV), bunyaviruses that are distantly related to the Tospoviridae family (42). In these cases, the C-terminal cysteine-rich region (stem region) was removed to improve solubility and crystallization. These crystal structures, however, show only one molecule of GN in the asymmetric unit and are lacking structural information regarding their mode of assembly and oligomerization. This is not the case for the TSWV sGN crystal structure. To further investigate the GN intermolecular mode of assembly, we also crystallized sGN∆V8 and sGN∆tryp and determined their structure to 3.4 and 2.8 Å resolutions, respectively (SI Appendix, Table S1).
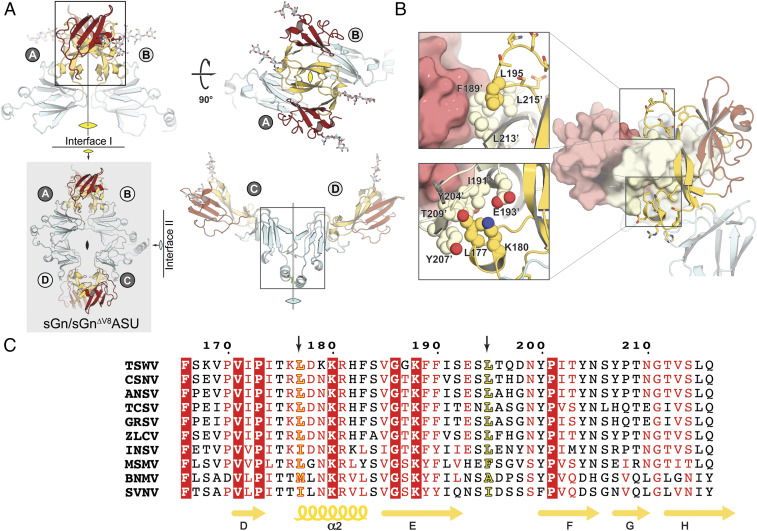
The crystal structures of sGN and sGN∆V8 belong to the same space group and have an identical crystal packing (SI Appendix, Fig. S3 and Table S1). In these structures, there are four copies of GN in the asymmetric unit, organized in a noncrystallographic pseudodihedral (D2) symmetry (Fig. 4, gray box). The four molecules can be arranged in two types of dimers, each using a distinct dimerization interface: a covalently linked dimer through C302–C′302 disulfide (copies A–C and B–D) and a noncovalent interface using a PD–PD interaction (copies A–B and C–D) (Fig. 4A). We observed electron density for the C302–C′302 disulfide only between A and C copies but not for B and D, suggesting that a sGN–sGN covalent interaction is not mandatory for crystal packing and appears to be transient. Furthermore, the observation that GN on the mature virion’s envelope only partially participates in the disulfide dependent dimerization supports this notion (28). Calculations of the buried surface area (BSA) at the noncovalent interface results in an area of ∼100 Å2 (Fig. 4, Right), an indication of a weak protein–protein interaction. In contrast, the BSA at the noncovalent PD–PD interaction is ∼1,800 Å2 (Fig. 4 A, Top), implying a biologically relevant interface (53).
Fig. 4.
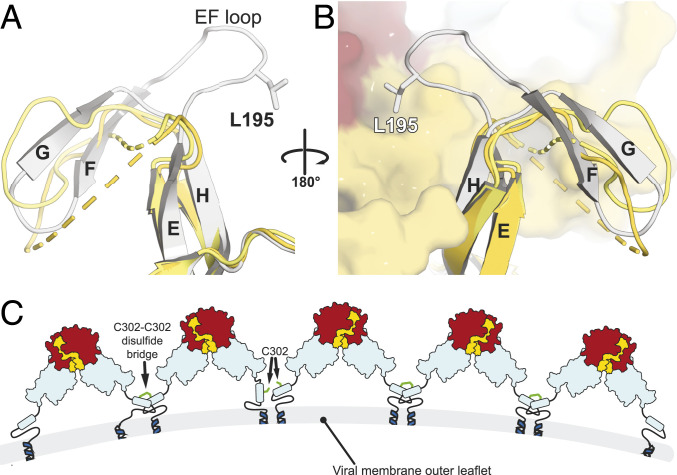
The sGN/sGN∆V8 asymmetric unit contains four molecules organized in two types of dimers. (A) The sGN/sGN∆V8 asymmetric unit (gray box) shows dihedral symmetry (D2) composed of two type of dimers, a noncovalent dimer (A–B and C–D interfaces) and disulfide linked dimers (A–D and B–C interfaces). (B) The unique interactions of the leucine residues at the tips of the pincer. Color scheme is as before. Light colors (light red, pale yellow, and pale cyan) represent the reciprocal protomer in the dimer. (C) Multiple sequence alignments at the PD region show that L177 and L195 are conserved. MUSCLE (52) was used for the multiple alignment of tospovirus GN, including TSWV (APG79624.1), CSNV (BAF62146.1, chrysanthemum stem necrosis virus), ANSV (AYJ76754.1, alstroemeria necrotic streak virus), TCSV (AAU10599.2, tomato chlorotic spot virus), GRSV (AAU10600.2, groundnut ringspot virus), ZLCV (BAF62147.1, zucchini lethal chlorosis virus), INSV (AFP20544.1, impatiens necrotic spot virus), MSMV (YP_009346016.1, melon severe mosaic tospovirus), BNMV (YP_006468902.1, bean necrotic mosaic virus), and SVNV (AMB72701.1, soybean vein necrosis virus).
The sGN–sGN Noncovalent Dimer Interface Is Partially Conserved.
The noncovalent interface of sGN–sGN interaction is mediated through a unique protein fold that we designated “the pincer domain.” PDs from each protomer grip each other in a pincerlike manner. Two leucine residues, the conserved L177 and the less conserved L195, are positioned at the tips of the pincer and are involved in van der Waals interactions with the reciprocal PD (Fig. 4 B and C). Conservation analysis of the PD–PD interface shows a conserved surface generated by the PD domain (SI Appendix, Fig. S4A). In addition, a complementary electrostatic charge is generated by the conserved K180 on one protomer and the partially conserved E193 on the reciprocal one (Fig. 3B and SI Appendix, Fig. S4B). This charge complementarity can induce an electrostatic steering effect during the association of the two protomers, as was seen in other cases (54–56).
Dimerization through the PD Is Independent of the C-Terminal C302 Disulfide Bond.
To confirm the existence of the PD–PD interaction in solution, we dissolved the sGN∆V8 crystals in water (ddH2O) and subjected the sample to a matrix assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectroscopy measurement (SI Appendix, Fig. S5). We assigned average masses for the sGN∆V8 monomer, dimer, and tetramer, indicating that not all sGN∆V8 copies in the crystal are involved in a covalent disulfide bond and that also noncovalent interactions occur in solution since sGN∆V8 tetramers could not be assembled otherwise. Taken together (the larger BSA, the limited involvement of sGN in the intermolecular disulfide bridge, and our MALDI-TOF results), these findings suggest that the noncovalent interface through the PD domain is the main dimerization interface in GN–GN interactions.
As mentioned before, we also determined the crystal structure of sGN∆tryp. Unlike the sGN and sGN∆V8 crystals, sGN∆tryp crystals belong to a different space group and present a different molecular packing (SI Appendix, Fig. S3 and Table S1). Surprisingly, in this crystal form, we found only three sGN∆tryp copies in the asymmetric unit, of which two associate through the previously described noncovalent PD–PD interface (SI Appendix, Fig. S3). The third copy, however, loosely interacts with the dimer through the CTD of one protomer in what appears to be a crystal contact interaction (SI Appendix, Fig. S3). The recurring presence of the same noncovalent interface in all crystal forms strengthens our previous premise regarding the physiological relevance of the PD–PD interface.
Although our SEC experiments determine that sGN∆tryp is a monomer in solution (Fig. 2 B and C), we sought to explore whether PD–PD interactions occur, to some extent, in solution too. To address this question, we generated an sGN∆tryp−S214C mutant. S214 is located at the core of the PD–PD dimer interface with a distance of 2.7 Å from the same serine of the reciprocal protomer (S214’, Fig. 5A). While the sGN∆tryp SEC profile showed exclusively a monomer peak, in the SEC elution profile of sGN∆tryp−S214C we observed an additional population, corresponding to a dimer which was then confirmed by a nonreducing SDS-PAGE analysis (Fig. 5B). Deconvolution of the SEC elution profile, followed by a calculation of the area under the curve, shows that only 19% of the sample is “locked” in a disulfide-dependent dimer, implying a weak interaction in solution. Moreover, we determined the crystal structure of the sGN∆tryp−S214C, confirmed its proper fold, and demonstrated the existence of the artificial C214–C214′ disulfide bond (Fig. 5B and SI Appendix, Fig. S3 and Table S1). In this crystal form, there are also three molecules in the asymmetric unit, two of which are dimerized through PD–PD interaction (SI Appendix, Fig. S3). Therefore, in the context of the sGN∆tryp and sGN∆tryp−S214C crystals, 66% of sGN∆tryp−S214C molecules are in a dimeric conformation, implying that this dimerization is concentration dependent.
Fig. 5.
sGN∆tryp dimerizes weakly in solution. (A) A top view of the noncovalent dimerization interface. In the magnified box in the Top Left is a close-up view of the S214 that was mutated to cysteine. (B) SEC elution profile of purified sGN∆tryp−S241C and the corresponding nonreducing SDS-PAGE (Coomassie brilliant blue stained) of the peak fractions. The Inset shows a 2Fobserved-Fcalculated (2Fo-Fc) electron density map obtained from the sGN∆tryp−S241C crystals, confirming the existence of the C214–C214′ disulfide bond.
Since the virus envelope is often considered a dense lattice composed of the envelope glycoproteins, we suggest that GN’s behavior on the viral envelope might be similar to the one in our sGN/sGN∆V8/sGN∆tryp crystals. Moreover, it was previously shown that molecular crowding enhances virus assembly in vitro (57) and that crystallization conditions, such as high molecular weight polyethylene glycol (PEG) at high concentration, mimic the crowded viral envelope and the cell interior (58), which can therefore drive the equilibrium toward the sGN–sGN complex.
The Pincer Domain Fold Is Stabilized through Dimerization.
Various protein folds are stabilized through intermolecular interactions. Intriguingly, the monomeric conformation in the sGN∆tryp and sGN∆tryp−S214C crystal structures displays a major change in the protein’s fold (Fig. 6). Superposition of this “orphan” copy onto a copy that is involved in a dimeric interaction shows a major divergence between the Cα backbone traces. As a result, in the monomeric sGN∆tryp, residues 195 to 204 are completely untraceable in the electron density map as the small FG β-sheet and the EF loop are completely misfolded (Fig. 6A). This strongly indicates that in the monomeric form of sGN, the PD cannot adopt its characteristic fold. Thus, it is likely that upon homodimerization, this region assumes its secondary structure and final fold through an induced fit mechanism and gains stability through the PD–PD interactions (Fig. 6B).
Fig. 6.
The fold of TSWV GN is stabilized through the PD–PD dimerization interface. (A) A superposition of the monomeric copies found in sGN∆tryp and sGN∆tryp−S214C asymmetric units (yellow and pale yellow, respectively) onto a sGN∆tryp copy that is involved in PD–PD interaction (white). The NTD was hidden to simplify the view. (B) The same superposition in 180° view, this time in the context of the dimer. (C) A hypothetical model for the organization of GN on the membrane of TSWV. GN is organized in dimers through the PD, and in addition, some of the dimers are covalently linked through the nonobligatory C302–C302′ disulfide bond. Further structural investigations are required to locate GC in this model.
A Model for GN Assembly on the Viral Membrane.
Bunyaviruses diverge in their glycoprotein organization on the viral envelope. In phleboviruses, the GN proteins form either hexones (six-copy rings) or pentones (five-copy rings) (38, 59), while in hantaviruses, like the Puumala virus (PUUV) and the Hantaan virus (HANV), GN is organized in a pseudofourfold symmetry (39, 60). These two GN protein types have a similar molecular weight, and both are positioned distally to the viral membrane, while GC is juxtaposed to it (38, 39). In contrast, GN of orthobunyaviruses is a significantly smaller protein and was suggested to be localized in close proximity to the membrane (40). However, none of the available GN structures provide atomic resolution details for the interacting interfaces that are relevant to the assembly of the virus (39, 42). Our structural and biochemical studies, together with previous studies on the intact virions (28), suggest that the minimal building block of the TSWV GN on the viral membrane might include GN homodimers that associate through the PD domain (Fig. 6C). This minimal building block could further assemble into higher oligomers through a homotypic, nonobligatory disulfide bond through the C-terminal C302. The crowded environment of the virus membrane and the restraints that the transmembrane domain force upon the GN ectodomain promote a favorable orientation that steers GN into dimerizing.
Also present on the viral membrane is GC, the proposed fusogen of TSWV. In the absence of structural information on the holo-spike complex of GN and GC, it is difficult to position GC in our model with high confidence, especially when the N terminus of GN (residues 37 to 107) is disordered. Nevertheless, GN–GC heterodimers were reported before and were demonstrated to be also covalently linked through a disulfide bond on the viral membrane (28). Therefore, we propose that disulfide-linked homo- and heterodimers (GN–GN and GN–GC, respectively) play a role in the assembly of the TSWV virion and are essential for orienting GN’s noncovalent dimerization through the PD domain. However, further investigations using integrated structural methodologies are needed to determine the complete assembly mode of the glycoproteins on the envelope of this important plant pathogen.
Materials and Methods
Protein Expression and Purification.
For protein expression, the open reading frame (ORF) encoding sGN (M segment residues 37 to 317) was PCR amplified from the complementary DNA (cDNA) of the M segment of the Brazilian Br-01 strain (GenBank accession no. AGM53737.1, kindly provided by Aviv Dombrovsky, Volcani Center, Agricultural Research Organization (ARO), Rishon LeZion, Israel. The fragment was then subcloned into the pAcGP67 vector (BD biosciences) in frame with the baculovirus gp67 signal sequence and a C-terminal eight-histidine purification tag.
pAcGP67-sGN construct and ProGreen, linearized baculovirus genomic DNA (AB vector), were cotransfected into Sf9 insect cells to produce recombinant baculovirus-expressing sGN. Virus stocks were amplified with three sequential infections of Sf9 cells. For sGN expression, Tni insect cells (expression systems) were expanded to a 4 L suspension at 27 °C until infected at a density of 2 × 106 cells/mL with 1% (vol/vol) of third-passage baculovirus stock. Immediately after infection, the temperature was lowered to 20 °C, and the cell media was collected 96 h postinfection. Following media concentration and buffer exchange to nickel-nitrilotriacetic acid (Ni-NTA) binding buffer (50 mM Na-phosphate, pH 8.0, 300 mM NaCl, 10 mM imidazole), secreted sGN was purified by nickel affinity chromatography (Ni-NTA agarose, QIAgen) following the manufacturer’s instructions. For the His-tag removal by carboxypeptidase A (CPA), a desalting chromatography (HiTrap Desalting, GE Healthcare) was performed followed by a CPA treatment at 4 °C for 16 h (1 mU CPA per 1 μg protein). To generate sGN∆V8 or sGN∆tryp, at that stage we also added V8 (Glu-C) or trypsin proteases (1 mU/μg), respectively. All protein preparations went through a final SEC step, using 10/300 GL Superdex200 (GE Healthcare) preequilibrated with 20 mM Tris (pH 8) and 200 mM NaCl. Relevant fractions were pooled (for sGN and sGN∆V8 that eluted in two distinct peaks, each population was pooled separately), concentrated to 3.4 to 6 g/L, aliquoted, and then flash frozen in liquid nitrogen (LN2). Samples were stored at −80 °C until use.
Crystallization of the Different sGN Constructs.
All crystals were grown by the hanging drop vapor diffusion method at 16 °C. TSWV sGN (3.4 g/L) was mixed with a reservoir solution (0.1 M Tris, pH 8.5, and 1.8 M ammonium sulfate) in a 1:1 protein to reservoir ratio. Rod-shaped crystals (400 × 75 × 75 µm) with hexagonal cross-section appeared ∼2 to 3 wk after the drop setup and were found to belong to the P6222 space group. Fortunately, in the same conditions, a diamond-shaped crystal was also observed 4 to 8 wk after the drop setup and belonged to the C2221 space group. For sGN∆V8 crystals, protein at 6 g/L was mixed with a reservoir solution (0.1 M Tris, pH 8.5, and 25% PEG 4000 [wt/vol]) in a 1:1 protein to reservoir ratio. Diamond-shaped crystals with dimensions of 400 × 200 × 250 µm were observed in the following 2 to 4 wk and also belonged to the C2221 space group (SI Appendix, Table S1). To obtain sGN∆tryp crystals, one volume of sGN∆tryp (6 g/L) was mixed with one volume of reservoir solution containing 28% PEG 3350 (wt/vol), 0.1 M Tris (pH 7.5), and 2% ethanol. Crystals with a plate morphology that belonged to the P21 space group grew in clusters and reached their final size after 3 to 5 d. sGN∆trypS214C crystals were obtained using sGN∆trypS214C at 6.4 g/L mixed with a reservoir solution (19% PEG 8000 [wt/vol], 0.1 M Tris, pH 7.5, and 0.2 M magnesium chloride hexahydrate) in a 1:1 protein to reservoir ratio. Crystals with a rod morphology grew in clusters and reached their final size after 1 to 3 d. For the purpose of cryoprotection, all crystals were transferred to a solution containing the original reservoir conditions supplemented with 30% glycerol as a cryoprotectant agent.
For derivatization, selected diamond-shaped crystals of sGN were soaked in 0.1 M Tris (pH 8.5) and 1.8 M ammonium sulfate, 1 mM potassium osmate, and 30% glycerol as a cryoprotectant for 3 wk and flash frozen in liquid nitrogen. Data from all crystals were collected at 100 K on a PILATUS detector at the European Synchrotron Radiation Facility (ESRF). We used XDS (X-ray Detector Software) (61, 62) to index and integrate the data and obtained phase information using single-wavelength anomalous dispersion with the PHENIX (Python-based Hierarchical ENvironment for Integrated Xtallography) suite (63). The structure of sGN was manually built using COOT (Crystallographic Object-Oriented Toolkit) (64). To improve the map quality, we used rigid-body refinement with PHENIX. Iterative cycles of manual building and refinements resulted in a model of sGN to 4.24 Å resolution. This model was used for molecular replacement in PHENIX as a search model to determine the crystal structure of sGN∆V8 to 3.4 Å resolution. Subsequently, a single chain of sGN∆V8 structure was used as a search model of sGN∆tryp that was refined to a resolution of 2.8 Å with excellent statistics (SI Appendix, Table S1). All molecular graphics were produced using PyMOL (PyMOL Molecular Graphics System, Version 1.8, Schrödinger).
Hydrodynamic and Multiangle Scattering Analysis of sGN∆tryp.
Analytical SEC and multiangle light scattering (MALS) experiments were performed in 20 mM Tris (pH 8.0) or Tris⋅HCl (pH 8.0) and 0.2 M NaCl. A total of 0.1 mL sGN∆tryp at 1 g/L was loaded onto a Superdex 200 (10/300) column coupled to a mini DAWN TREOS spectrometer and Optilab T-rEX (Wyatt Ttechnology) refractometer at a flow rate of 0.7 mL/min. TSWV sGN∆tryp was detected as it eluted from the column, with a ultraviolet detector at 280 nm, a light scattering detector at 690 nm, and a refractive index detector. The molar mass of TSWV sGN∆tryp was determined from the Debye plot of light scattering intensity versus scattering angle. Data processing was performed using ASTRA software (Wyatt Technology).
Matrix-Assisted Laser Desorption/Ionization: Time of Flight.
Samples were prepared from 10 to 15 sGN∆V8 crystals which were dissolved and dialyzed overnight in double distilled water to remove salts. Serial dilutions of pure sGN∆V8 using a sinapinic acid matrix were made using 1% sinapinic acid, 50% acetonitrile, and 0.3% trifluoroacetic acid in water. Samples were analyzed using a Bruker MALDI-TOF Autoflex Speed mass spectrometer.
Conservation Analysis.
For multiple sequence alignment (MSA) we used Basic Local Alignment Search Tool (BLAST) against a no-redundant sequences database. To obtain unique sequences of orthotospoviruses, we excluded TSWV in our search. For the MSA in MUSCLE (Multiple Sequence Comparison by Log-Expectation) (52), the following sequences were used: BAF62146.1 (chrysanthemum stem necrosis virus, CSNV), AYJ76754.1 (alstroemeria necrotic streak virus, ANSV), BAF62147.1 (zucchini lethal chlorosis virus, ZLCV), AAU10600.2 (groundnut ringspot virus, GRSV), AAU10599.2 (tomato chlorotic spot virus, TCSV), AFP20544.1 (impatiens necrotic spot virus, INSV), YP_009346016.1 (melon severe mosaic tospovirus, MSMV), AMB72701.1 (soybean vein necrosis virus, SVNV), AMY62789.1 (tomato necrotic spots associated virus, TNSaV), YP_009259254.1 (kiwi fruit tospovirus, KFTV), ACO52398.1 (calla lily chlorotic spot virus, CLCSV), AOO95321.1 (polygonum ringspot virus, PRSV), ADD14053.1 (watermelon bud necrosis virus, WBNV), AFX74696.1 (spider lily necrotic spot virus, SLNV), YP_009345144.1 (pepper chlorotic spot virus, PCSV), YP_006468902.1 (bean necrotic mosaic virus, BNMV), AWM95330.1 (hippeastrum chlorotic ringspot virus, HCRV), AAZ81611.1 (watermelon silver mottle virus, WSMoV), AEX09313.1 (tomato yellow ring virus, TYRV), QIC52037.1 (mulberry vein banding virus, MVBV), QHU78624.1 (capsicum chlorosis virus, CCV), and YP_009505290.1 (iris yellow spot virus, IYSV). The MSA was then submitted to the CONSURF server (43, 65) for conservation analysis.
Supplementary Material
Acknowledgments
We thank Dr. Aviv Dombrovski from the Volcani Center (ARO, Israel) for providing us with TSWV M segment cDNA. We also thank Dr. Mario Lebendiker from the Wolfson Center for Applied Structural Biology at the Hebrew University in Jerusalem for the SEC-MALS analysis. Special thanks go to the staff of the ESRF Grenoble ID23 and ID29 and the staff of the Berlin Electron Storage Ring Society for Synchrotron Radiation (BESSY II) at beamlines 14.1 and 14.2 for their valuable assistance in data collection and beamline operation. We also thank Prof. Yarden Opatowsky for the useful discussion and proofreading of the manuscript. This project was partially funded of by Israel Science Foundation Grant 401/18 to M.D.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004657117/-/DCSupplemental.
Data Availability.
Structures reported in this paper have been deposited in the Protein Data Bank (PDB), under accession codes 6Y9L, 6Y9M, 6YA0, and 6YA2 (sGN, sGN∆V8, sGN∆tryp, sGN∆tryp−S214C, respectively). All study data are included in the article and SI Appendix.
References
- 1.Samuel G., Bald J. G., Pittman H. A., Investigations on Spotted Wilt of Tomatoes, (Council for Scientific and Industrial Research, Melbourne, 1930). [Google Scholar]
- 2.Pappu H. R., Jones R. A., Jain R. K., Global status of tospovirus epidemics in diverse cropping systems: Successes achieved and challenges ahead. Virus Res. 141, 219–236 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Oliver J. E., Whitfield A. E., The genus tospovirus: Emerging bunyaviruses that threaten food security. Annu. Rev. Virol. 3, 101–124 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Whitfield A. E., Falk B. W., Rotenberg D., Insect vector-mediated transmission of plant viruses. Virology 479–480, 278–289 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Ullman D. E., John C. J., Mau R. L. F., Westcot D. M., Custer D. M., A midgut barrier to tomato spotted wilt virus acquisition by adult western flower thrips. Phytopathology 82, 1333–1342 (1992). [Google Scholar]
- 6.van de Wetering F., Goldbach R., Peters D., Tomato spotted wilt tospovirus ingestion by first instar larvae of Frankliniella occidentalis is a prerequisite for transmission. Phytopathology 86, 900–905 (1996). [Google Scholar]
- 7.Nagata T., Inoue-Nagata A. K., van Lent J., Goldbach R., Peters D., Factors determining vector competence and specificity for transmission of tomato spotted wilt virus. J. Gen. Virol. 83, 663–671 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Ullman D. E., German T. L., Sherwood J. L., Westcot D. M., Cantone F. A., Tospovirus replication in insect vector cells: Immunocytochemical evidence that the nonstructural protein encoded by the S RNA of tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology 83, 456–463 (1993). [Google Scholar]
- 9.Montero-Astúa M., Ullman D. E., Whitfield A. E., Salivary gland morphology, tissue tropism and the progression of tospovirus infection in Frankliniella occidentalis. Virology 493, 39–51 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Heinlein M., Plant virus replication and movement. Virology 479–480, 657–671 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Nagata T., Inoue-Nagata A. K., Prins M., Goldbach R., Peters D., Impeded thrips transmission of defective tomato spotted wilt virus isolates. Phytopathology 90, 454–459 (2000). [DOI] [PubMed] [Google Scholar]
- 12.de Oliveira Resende R. et al., Generation of envelope and defective interfering RNA mutants of tomato spotted wilt virus by mechanical passage. J. Gen. Virol. 72, 2375–2383 (1991). [DOI] [PubMed] [Google Scholar]
- 13.Kikkert M. et al., Binding of tomato spotted wilt virus to a 94-kDa thrips protein. Phytopathology 88, 63–69 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Bandla M. D., Campbell L. R., Ullman D. E., Sherwood J. L., Interaction of tomato spotted wilt tospovirus (TSWV) glycoproteins with a thrips midgut protein, a potential cellular receptor for TSWV. Phytopathology 88, 98–104 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Badillo-Vargas I. E., Chen Y., Martin K. M., Rotenberg D., Whitfield A. E., Discovery of novel thrips vector proteins that bind to the viral attachment protein of the plant bunyavirus tomato spotted wilt virus. J. Virol. 93, e00699-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adkins S. et al., Baculovirus expression and processing of tomato spotted wilt tospovirus glycoproteins. Phytopathology 86, 849–855 (1996). [Google Scholar]
- 17.Kormelink R., de Haan P., Meurs C., Peters D., Goldbach R., The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J. Gen. Virol. 74, 790 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Kormelink R., de Haan P., Meurs C., Peters D., Goldbach R., The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J. Gen. Virol. 73, 2795–2804 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Adkins S. et al., Baculovirus expression and processing of tomato spotted wilted tospovirus glycoproteins. 86, 849–855 (1996). [Google Scholar]
- 20.Antic D., Wright K. E., Kang C. Y., Maturation of Hantaan virus glycoproteins G1 and G2. Virology 189, 324–328 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Matsuoka Y., Ihara T., Bishop D. H., Compans R. W., Intracellular accumulation of Punta Toro virus glycoproteins expressed from cloned cDNA. Virology 167, 251–260 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuismanen E., Posttranslational processing of Uukuniemi virus glycoproteins G1 and G2. J. Virol. 51, 806–812 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi X., Lappin D. F., Elliott R. M., Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins. J. Virol. 78, 10793–10802 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kikkert M. et al., Tomato spotted wilt virus particle morphogenesis in plant cells. J. Virol. 73, 2288–2297 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kikkert M., Verschoor A., Kormelink R., Rottier P., Goldbach R., Tomato spotted wilt virus glycoproteins exhibit trafficking and localization signals that are functional in mammalian cells. J. Virol. 75, 1004–1012 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerrard S. R., Nichol S. T., Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J. Virol. 76, 12200–12210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeiro D. et al., Tomato spotted wilt virus glycoproteins induce the formation of endoplasmic reticulum- and Golgi-derived pleomorphic membrane structures in plant cells. J. Gen. Virol. 89, 1811–1818 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Whitfield A. E., Ullman D. E., German T. L., Expression and characterization of a soluble form of tomato spotted wilt virus glycoprotein GN. J. Virol. 78, 13197–13206 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohamed N. A., Isolation and characterization of subviral structures from tomato spotted wilt virus. J. Gen. Virol. 53, 197–206 (1981). [Google Scholar]
- 30.Tas P. W. L., Boerjan M. L., Peters D., Structural proteins of tomato spotted wilt virus. J. Gen. Virol. 36, 267–279 (1977). [Google Scholar]
- 31.Willensky S. et al., Crystal structure of glycoprotein C from a hantavirus in the post-fusion conformation. PLoS Pathog. 12, e1005948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guardado-Calvo P. et al., Mechanistic insight into bunyavirus-induced membrane fusion from structure-function analyses of the hantavirus envelope glycoprotein Gc. PLoS Pathog. 12, e1005813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dessau M., Modis Y., Crystal structure of glycoprotein C from Rift Valley fever virus. Proc. Natl. Acad. Sci. U.S.A. 110, 1696–1701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tischler N. D., Gonzalez A., Perez-Acle T., Rosemblatt M., Valenzuela P. D. T., Hantavirus Gc glycoprotein: Evidence for a class II fusion protein. J. Gen. Virol. 86, 2937–2947 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Cortez I. et al., Genetic organisation of iris yellow spot virus M RNA: Indications for functional homology between the G(C) glycoproteins of tospoviruses and animal-infecting bunyaviruses. Arch. Virol. 147, 2313–2325 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Rey F. A., Heinz F. X., Mandl C., Kunz C., Harrison S. C., The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375, 291–298 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Roussel A. et al., Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14, 75–86 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Halldorsson S. et al., Shielding and activation of a viral membrane fusion protein. Nat. Commun. 9, 349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S. et al., A molecular-level account of the antigenic hantaviral surface. Cell Rep. 15, 959–967 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowden T. A. et al., Orthobunyavirus ultrastructure and the curious tripodal glycoprotein spike. PLoS Pathog. 9, e1003374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hellert J. et al., Orthobunyavirus spike architecture and recognition by neutralizing antibodies. Nat. Commun. 10, 879 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y. et al., Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope. Proc. Natl. Acad. Sci. U.S.A. 114, E7564–E7573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N., ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rissanen I. et al., Structural transitions of the conserved and metastable hantaviral glycoprotein envelope. J. Virol. 91, e00378-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guardado-Calvo P., Rey F. A., The envelope proteins of the bunyavirales. Adv. Virus Res. 98, 83–118 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Holm L., Kaariainen S., Wilton C., Plewczynski D., Using Dali for structural comparison of proteins. Curr. Protoc. Bioinformatics 14, 5.5.1-5.5.24 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Holm L., Rosenström P., Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holm L., Sander C., Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Murzin A. G., Brenner S. E., Hubbard T., Chothia C., SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Cheng H. et al., ECOD: An evolutionary classification of protein domains. PLoS Comput. Biol. 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng H., Liao Y., Schaeffer R. D., Grishin N. V., Manual classification strategies in the ECOD database. Proteins 83, 1238–1251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ran X., Gestwicki J. E., Inhibitors of protein-protein interactions (PPIs): An analysis of scaffold choices and buried surface area. Curr. Opin. Chem. Biol. 44, 75–86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuzawa S. et al., Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell 130, 323–334 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Mueller T. D., Zhang J. L., Sebald W., Duschl A., Structure, binding, and antagonists in the IL-4/IL-13 receptor system. Biochim. Biophys. Acta 1592, 237–250 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Sporny M. et al., Structural evidence for an octameric ring arrangement of SARM1. J. Mol. Biol. 431, 3591–3605 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Smith G. R., Xie L., Lee B., Schwartz R., Applying molecular crowding models to simulations of virus capsid assembly in vitro. Biophys. J. 106, 310–320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyrrell J., Weeks K. M., Pielak G. J., Challenge of mimicking the influences of the cellular environment on RNA structure by PEG-induced macromolecular crowding. Biochemistry 54, 6447–6453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huiskonen J. T., Overby A. K., Weber F., Grünewald K., Electron cryo-microscopy and single-particle averaging of rift valley fever virus: Evidence for GN-GC glycoprotein heterodimers. J. Virol. 83, 3762–3769 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Battisti A. J. et al., Structural studies of Hantaan virus. J. Virol. 85, 835–841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liebschner D. et al., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Landau M. et al., ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structures reported in this paper have been deposited in the Protein Data Bank (PDB), under accession codes 6Y9L, 6Y9M, 6YA0, and 6YA2 (sGN, sGN∆V8, sGN∆tryp, sGN∆tryp−S214C, respectively). All study data are included in the article and SI Appendix.