Significance
[PSI+] is a prion (infectious protein) form of the yeast Sup35 protein, propagating as an amyloid filamentous polymer. We find that each of the ribosome-associated chaperones—Ssb1/2p, Zuo1p, and Ssz1p—at their normal expression levels, blocks [PSI+] generation and the propagation of most new [PSI+] prion variants generated in their absence. The curing mechanism involves the functional triad of ribosome-associated chaperones. Our results suggest that cells do not want to have a prion: Cells reduce the chance of a prion emerging at the polypeptide level, and usually immediately cure those prions that do arise perhaps by limiting fiber growth.
Keywords: prion, antiprion system, [PSI+], Ssb, ribosome-associated complex
Abstract
The yeast prion [PSI+] is a self-propagating amyloid of the translation termination factor, Sup35p. For known pathogenic prions, such as [PSI+], a single protein can form an array of different amyloid structures (prion variants) each stably inherited and with differing biological properties. The ribosome-associated chaperones, Ssb1/2p (Hsp70s), and RAC (Zuo1p (Hsp40) and Ssz1p (Hsp70)), enhance de novo protein folding by protecting nascent polypeptide chains from misfolding and maintain translational fidelity by involvement in translation termination. Ssb1/2p and RAC chaperones were previously found to inhibit [PSI+] prion generation. We find that most [PSI+] variants arising in the absence of each chaperone were cured by restoring normal levels of that protein. [PSI+] variants hypersensitive to Ssb1/2p have distinguishable biological properties from those hypersensitive to Zuo1p or Ssz1p. The elevated [PSI+] generation frequency in each deletion strain is not due to an altered [PIN+], another prion that primes [PSI+] generation. [PSI+] prion generation/propagation may be inhibited by Ssb1/2/RAC chaperones by ensuring proper folding of nascent Sup35p, thus preventing its joining amyloid fibers. Alternatively, the effect of RAC/Ssb mutations on translation termination and the absence of an effect on the [URE3] prion suggest an effect on the mature Sup35p such that it does not readily join amyloid filaments. Ssz1p is degraded in zuo1Δ [psi-] cells, but not if the cells carry any of several [PSI+] variants. Our results imply that prions arise more frequently than had been thought but the cell has evolved exquisite antiprion systems that rapidly eliminate most variants.
Like the [PSI+] prion, an amyloid of the translation termination factor Sup35p, [URE3] is a prion of Ure2p, a repressor of transcription of genes for catabolism of poor nitrogen sources (1–16). The majority of prion variants/strains are lethal or highly pathogenic (17), and the absence of these prions in 70 wild yeast strains indicates that even the mildest variants are net deleterious to the host (18, 19). Evidently, the loss of precise translation termination (in [PSI+] strains) or of intrinsic nitrogen regulation (as in [URE3] cells) by conversion of soluble Sup35p or Ure2p to prion amyloid aggregates, is detrimental to the host cell. In addition, most variants of these two prions have detrimental effects beyond mere deficiency of the normal form (17; reviewed in ref. 20). To prevent these harmful diseases, yeast has evolved systems to repress prion generation, to cure prions after they arise (“antiprion systems”), or to limit prion toxicity, like the antipathogen systems in mammals targeting fungi, bacteria, and viruses. But, unlike invaders from outside, most cases of prion disease are sporadic, arising spontaneously for no known reason. So, it is inferred that, in a normal cell, cellular antiprion systems are continuously blocking prion generation, propagation, and toxicity (reviewed in refs. 20–22). In the absence of an antiprion factor prions will appear with elevated frequency and many of those prion variants that arose in its absence are cured when the antiprion factor is restored to normal levels.
The disaggregating chaperone Hsp104 was first found to function in the propagation of [PSI+] (23) and other amyloid-based prions (24), and in the curing of [PSI+] by its overproduction (23). These two different activities of Hsp104 were then separated by showing that disruption of the Hsp104 N-terminal region eliminates the Hsp104 overproduction curing ability without any effect on [PSI+] propagation (25). In fact, spontaneous [PSI+] generation was elevated by over 10-fold in an N-terminal mutant, hsp104T160M, and the normal level of WT Hsp104 was able to cure many of the [PSI+] variants arising in the mutant strain (26).
Overproduced endosomal sorting factor Btn2p and its paralogue Cur1p were shown to cure the [URE3] prion, in the case of Btn2p by sequestering [URE3] prion aggregates, preventing distribution of aggregates to daughter cells (27, 28). It was then found that most of the [URE3] prion variants arising in a btn2Δ cur1Δ strain were cured by reintroduction of normal levels of either Btn2p or Cur1p or both (29). Although these three prion-curing systems were initially identified by using overproduction curing, it was then shown that these systems are working constantly in a normal cell whose cellular environment is not altered by intentional overproduction or deficiency of any components.
Most recently, a simple genetic screen for finding antiprion factors identified two novel anti-[PSI+] components, Siw14p, a pyrophosphatase specific for 5PP-IP5 (5-diphosphoinositol pentakisphosphate) in the inositol polyphosphate synthesis pathway, and Upf proteins (Upf1p, Upf2p and Upf3p), core components of the nonsense-mediated mRNA decay (NMD) apparatus, respectively (30, 31). By lowering the levels of some inositol poly-/pyro-phosphates (PP-IPs), restored normal levels of Siw14p cure many [PSI+] prion variants that were generated in its absence and are dependent on the resulting higher PP-IP levels (30). Normal levels of Upf proteins constantly repress [PSI+] occurrence and block the propagation of most [PSI+] prion variants formed in their absence. The curing mechanism involves Sup35p-binding by each Upf protein and by the trimeric Upf complex (31). These studies of Btn2p, Cur1p, Hsp104, Siw14p, and the Upf proteins have uncovered new classes of prion variants, unable to arise and further propagate in normal strains with the respective antiprion systems, but able to arise and further propagate in cells without those systems (reviewed in ref. 20).
The Hsp70 chaperones Ssb1p and Ssb2p (Ssb1/2p) bind directly to translating ribosomes and newly synthesized nascent polypeptide chains (32, 33). Zuo1p (Hsp40/DnaJ homolog) and Ssz1p (Hsp70/DnaK homolog) form a stable heterodimeric ribosome-associated complex (RAC) that is necessary for the ribosome-association of Ssb1/2p (34–38). Moreover, deletion of both SSB1 and SSB2 or of ZUO1 or SSZ1 showed similar phenotypes, such as growth defects, cold sensitivity, and severe sensitivity to translation inhibitors (32, 34, 36). Ssb1/2p and RAC act in concert on nascent polypeptides and protect them from misfolding and aggregation (39, 40; reviewed in refs. 41 and 42).
Chernoff aptly described ssb1/2 mutations as “protein mutators,” when he showed that they increased the frequency of the cytoplasmic gene [PSI+] arising (43). In accordance with the cellular functions of the Ssb1/2p-RAC system, deletion of SSB1/2 or ZUO1 or SSZ1 lead to increased frequency of [PSI+] generation, either spontaneously or induced by Sup35p overproduction (43–46). Restored Ssb1p on a CEN plasmid was unable to cure [PSI+] variants arising in an ssb1/2Δ strain (43). Curing of [PSI+] by overproduction of Hsp104 was impaired by ssb1/2Δ, but made more efficient in ssz1Δ or zuo1Δ (43, 44). The observed release of Ssb1/2p from ribosomes in ssz1Δ or zuo1Δ mutants led to the inference that soluble Ssb1/2p impairs [PSI+] propagation, while ribosome-bound Ssb1/2p impairs [PSI+] generation (44).
Here, we find that most [PSI+] prion variants arising in the absence of Ssb1p or RAC are stably propagated in the mutant strain, but can be cured by restoring normal levels. Furthermore, we show that different [PSI+] variants arise in mutants of different ribosome-associated chaperones.
Results
Spontaneous and Induced [PSI+] Generation Is Elevated in Ribosome-Associated Chaperone-Deficient Mutants.
Both spontaneous and induced [PSI+] generation (by Sup35 N or NM overproduction) were previously reported to be elevated in ssb1/2∆, zuo1∆, or ssz1∆ strains (43–47). To confirm these results and obtain [PSI+] isolates in each mutant strain for further use, [PSI+] generation was investigated. [PIN+] ([PSI]-inducibility) is a prion form of Rnq1p that strongly increases the frequency of spontaneous or induced [PSI+] appearance (48–51). Because [PIN+] variants differ dramatically in their ability to stimulate [PSI+] appearance (52), comparison of [PSI+] generation rates must be done in cells that have the same [PIN+] variant. For these experiments, each strain to be tested for [PSI+] generation was cured of prions by growth on 5 mM guanidine HCl (GuHCl), an inhibitor of Hsp104 that cures all amyloid-based prions (53–56). Into each resulting [psi−][pin−] recipient strain (WT and mutants), [PIN+] was transferred by cytoduction from a single donor (BY4742) (SI Appendix, Table S1) (57). The resulting [PIN+] recipient strains were transformed with centromeric plasmid p1520 (SI Appendix, Table S2) (30, 31) carrying the [PSI+]-suppressible nonsense allele ura3-14 (58), and the Sup35 prion-forming domain (NM) driven by the GAL1 promoter. Strains were grown 2 d in glucose or galactose, and cells were plated on −Ura plates to select Ura+ ([PSI+]) clones. To check that these Ura+ clones are [PSI+], they were then tested for GuHCl curability by transient growth on 1 mM or 5 mM GuHCl.
For spontaneous [PSI+] generation, the frequency of [PSI+] clones in ssb1/2∆, zuo1∆, or ssz1∆ was increased 19-, 16-, or 13-fold, respectively, compared to the WT (Table 1). For [PSI+] generation induced by Sup35NM overexpression, ssb1/2∆, zuo1∆, and ssz1∆ strain showed 40-, 37-, and 51-fold increased [PSI+] clones compared with the WT strain (Table 1).
Table 1.
De novo generation of [PSI+] variants is elevated in ssb-rac deficient cells
| Total Ura+/106 cells | ||
| Host | Spontaneous [PSI+] | Induced [PSI+] |
| WT | 0.36 ± 0.1 | 43 ± 6 |
| ssb1Δssb2Δ | 7.2 ± 0.5 | 1,740 ± 200 |
| zuo1Δ | 6.0 ± 0.7 | 1,600 ± 160 |
| Ssz1Δ | 4.5 ± 0.4 | 2,200 ± 230 |
Strains MS327, MS515, MS527, and MS510 (top to bottom) carry the same [PIN+]. For spontaneous [PSI+], cells were grown 2 d in 2% glucose liquid culture at 30 °C, and 107 yeast cells were spread on standard SC plates without uracil. For induced [PSI+] formation, strains carrying p1520 with SUP35NM driven by a galactose-inducible promoter were grown 2 d in 2% galactose/2% raffinose medium at 30 °C, and 105 yeast cells were spread on SC plates without uracil. The average number of colonies formed after 5 d of incubation at 30 °C is shown (the data from at least three independent experiments were combined). Numbers represent Ura+ colonies with [PSI+] that were confirmed by GuHCl curability. Ura+ colonies ± SD is shown.
Normal Levels of Ssb1p, Zuo1p, or Ssz1p can Cure Most [PSI+] Variants Isolated in Absence of Each Component.
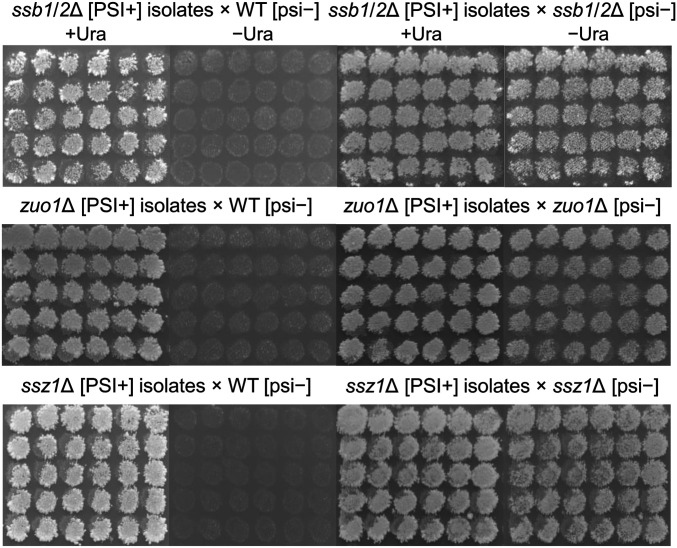
Altered levels or alteration of Sis1, Hsp104, Ssa, Ssb, or Sup35 proteins can destabilize [PSI+] (8, 23, 59–63), but the levels of these and Ydj1 are unaltered in ssz1, zuo1, or ssb1/2 mutants (43, 44, 64). We tested for Ssb1/2p, Zuo1p, or Ssz1p as prion-curing factors without protein overproduction. GuHCl-curable Ura+ [PSI+] clones (from Table 1) were crossed with the isogenic [psi−] WT strain MS173 to complement the deletion in each strain. For ssb1Δssb2Δ [PSI+], 60 of 72 isolates gave Ura− diploids on mating with WT. For zuo1Δ [PSI+] and ssz1Δ [PSI+], 32 of 50 and 46 of 60 isolates, respectively, similarly produced Ura− diploids with the WT [psi−] strain. Among the isolates from each mutant strain that gave Ura− diploids, 30 were mated with either the WT [psi−] strain MS173 or a ssb1/2Δ [psi−] strain MS520, zuo1Δ [psi−] strain MS528, and ssz1Δ [psi−] strain MS514. All of the diploids formed with the WT (heterozygotes) were Ura−, but the diploids with the same deletion mutant [psi−] strains were Ura+ (Fig. 1). Among 30 isolates in each case, 12 isolates were randomly selected and tested for [PSI+] stability in the haploid, heterozygotes, or homozygotes by subcloning on YPAD medium. Both the haploid and diploid subclones were then replica-plated to –Uracil media. All [PSI+] prion variants were more stable in the original haploid and homozygous diploid than in the complemented heterozygous diploid (Table 2).
Fig. 1.
Most [PSI+] prion variants isolated in ssb1/2Δ, zuo1∆, and ssz1∆ strains are lost in the presence of the WT allele of SSB1, ZUO1, and SSZ1, respectively (see text). Such [PSI+] isolates in each deletion strain MS515, MS527, and MS510 were mated for 2 d on YPAD with either isogenic WT MS173 or each deletion strain MS520, MS528, or MS514 and replica-plated to minimal media with and without uracil. The presence of p1520 (pCEN LEU2 ura3-14) in all strains enables scoring [PSI+]. Diploids formed with WT are almost all Ura− as a result of elimination of [PSI+] (Left). Diploids formed with deletion strains are Ura+ indicating stable maintenance of [PSI+] (Right).
Table 2.
[PSI+] isolates are more stable in ssb-racΔ strains than in the ssb-racΔ/+ diploid strains
| Isolate | Ura+/total subclones | ||||||||
| [PSI+sbs] in | [PSI+zos] in | [PSI+szs] in | |||||||
| ssb1/2Δ | ssb1/2Δ/+ | ssb1/2Δ/− | zuo1Δ | zuo1Δ/+ | zuo1Δ/− | ssz1Δ | ssz1Δ/+ | ssz1Δ/− | |
| 1 | 30/30 | 1/20 | 20/20 | 28/30 | 3/20 | 18/20 | 30/30 | 1/20 | 18/20 |
| 2 | 30/30 | 2/20 | 20/20 | 30/30 | 0/20 | 19/20 | 30/30 | 0/20 | 19/20 |
| 3 | 25/30 | 1/20 | 20/20 | 22/30 | 1/20 | 19/20 | 29/30 | 0/20 | 20/20 |
| 4 | 30/30 | 0/20 | 18/20 | 24/30 | 1/20 | 19/20 | 30/30 | 0/20 | 20/20 |
| 5 | 30/30 | 2/20 | 17/20 | 28/30 | 1/20 | 20/20 | 30/30 | 0/20 | 20/20 |
| 6 | 30/30 | 1/20 | 18/20 | 28/30 | 1/20 | 19/20 | 30/30 | 0/20 | 20/20 |
| 7 | 26/30 | 0/20 | 19/20 | 27/30 | 2/20 | 19/20 | 30/30 | 2/20 | 20/20 |
| 8 | 30/30 | 3/20 | 20/20 | 26/30 | 2/20 | 20/20 | 27/30 | 0/20 | 19/20 |
| 9 | 30/30 | 0/20 | 20/20 | 21/30 | 1/20 | 18/20 | 25/30 | 7/20 | 18/20 |
| 10 | 27/30 | 1/20 | 19/20 | 24/30 | 1/20 | 20/20 | 27/30 | 6/20 | 18/20 |
| 11 | 30/30 | 2/20 | 20/20 | 29/30 | 2/20 | 18/20 | 27/30 | 4/20 | 18/20 |
| 12 | 30/30 | 1/20 | 19/20 | 30/30 | 1/20 | 19/20 | 30/30 | 2/20 | 19/20 |
| Total | 348/360 | 14/240 | 232/240 | 317/360 | 16/240 | 228/240 | 349/360 | 22/240 | 229/240 |
| % Ura+ | 96.7% | 5.8% | 96.7% | 88.1% | 6.7% | 95.0% | 96.9% | 9.2% | 95.4% |
Twelve [PSI+] isolates in each haploid strain were obtained and were either subcloned on YPAD medium or were mated with isogenic WT strain MS173 and the each of diploids formed were subcloned on YPAD medium. Each haploid strain, heterozygous- and homozygous-diploids were replica-plated to −Ura plates to test the stability of [PSI+]. Each prion variant was more stable in its original host (haploids) or homozygous diploids than in the complemented (heterozygous) diploids.
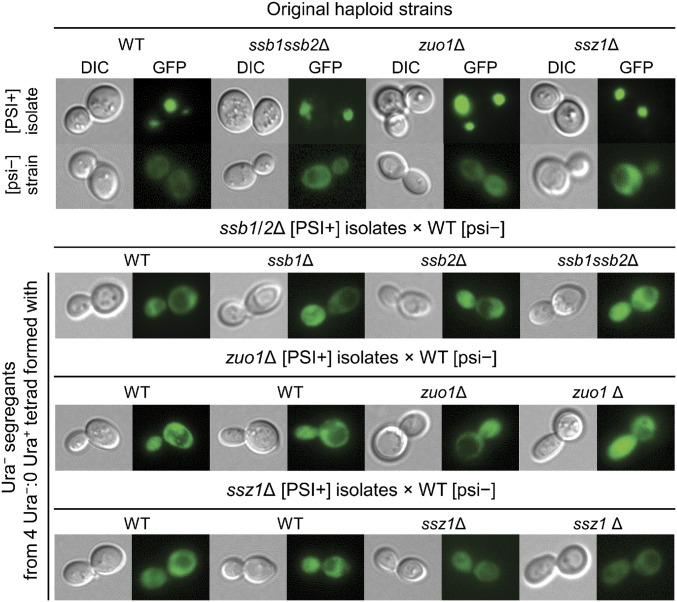
Meiotic segregation of each Ura− heterozygote showed mostly 4 Ura−: 0 Ura+ segregation, 20 of 24 tetrads for ssb1ssb2Δ/+, 8 of 12 tetrads for zuo1/+, and 21 of 26 tetrads for ssz1/+, respectively. Only a few mutant segregants showed a Ura+ or very weak Ura+ phenotype, but the majority of mutant segregants were Ura−: 75 to 100% (average 92.6%) for ssb1/2Δ segregants, 33 to 100% (average 68.2%) for zuo1Δ segregants, 50 to 100% (average 83.7%) for ssz1Δ segregants. This shows that [PSI+] is gradually lost in the heterozygous diploids. Although ssb1/2Δ, ssz1Δ, and zuo1Δ mutants have a slight nonsense-suppression phenotype (43, 65), the meiotic analysis shows that the Ura− phenotype of the heterozygous diploids is due to loss of [PSI+], not simply an effect on translation termination (these are, after all, ribosomal proteins). However, if, after mating with the isogenic WT [psi−] strain MS173, the diploids were immediately sporulated before loss of [PSI+] variants could happen in the diploids, segregations were two Ura− SSZ1: two Ura+ ssz1Δ (20 of 21 tetrads) or two Ura− ZUO1: two Ura+ zuo1Δ (14 of 15 tetrads). In case of ssb1ssb2Δ/+, all of the 22 Ura+ spore clones were ssb1Δ ssb2Δ. This confirms that it is the ssb or zuo1 or ssz1 mutation that allows propagation of these [PSI+] variants. The combined results from meiotic analysis show that each of these [PSI+] prion variants is eliminated by normal levels of Ssb1/2p, Zuo1p, or Ssz1p.
Based on their loss of [PSI+] on restoration of normal levels of Ssb1/2p, Zuo1p, or Ssz1p, these variants (all guanidine-curable and mitotically stable in the mutant) are denoted [PSI+sbs] for Ssb1/2p-sensitive, [PSI+zos] for Zuo1p-sensitive, and [PSI+szs] for Ssz1p-sensitive, respectively. To confirm loss of [PSI+] variants in all different Ura− segregants that showed 4 Ura−: 0 Ura+ segregation, formation of aggregate structures was examined. To detect prion aggregates, Sup35NM-GFP was transiently overexpressed in Ura− segregants and [PSI+] or [psi−] strains with the same backgrounds. Like the WT [psi−][PIN+] or mutant [psi−][PIN+] strain, most cells of each Ura−segregant examined showed a diffuse GFP signal (Fig. 2). Single or multiple foci were rarely detected in in the Ura− segregants, but were detected in many cells of different [PSI+]-carrying strains (Fig. 2). In addition, to double-check loss of [PSI+sbs], [PSI+zos], or [PSI+szs] in WT strains, cytoduction was conducted using each [PSI+] variant-carrying mutant strain as a donor and WT [psi−] or mutant [psi−] strain as a recipient. The majority of WT recipients in each case showed a Ura− phenotype (SI Appendix, Table S3). To verify that these Ura− phenotypes are due to loss of [PSI+] in the WT cytoductants, reverse-cytoductions were performed using Ura− WT cytoductants as donors and each mutant [psi−] strain as a recipient. Almost all reverse-cytoductants still showed a Ura− phenotype in each mutant recipient strain. For unknown reasons, too few cytoductants were obtained in the combination of mutant donor and mutant recipient, to enable analysis of such matings. Once again loss of different [PSI+] variants in WT strains was confirmed by examining the formation of fluorescent structures. As expected, mainly a diffuse GFP signal was observed in each Ura− WT cytoductant like the control WT [psi−] strain (SI Appendix, Fig. S1). Taken together, these combined results demonstrated that many [PSI+] variants isolated in each mutant strain are indeed lost in the presence of normal levels of the corresponding SSB, ZUO1, or SSZ1 in WT strains.
Fig. 2.
[PSI+] prion variants are lost in heterozygous diploids formed with an isogenic WT strain. (Top) Original haploids of [PSI+] strains (WT MS224, [PSI+] isolates in each deletion strain MS515, MS527, and MS510: left to right), and [psi−] strains (MS327, MS520, MS528, and MS514: left to right), and Ura− segregants (Top to Bottom) were transformed with a 2-µ plasmid pH770/pM18 encoding Sup35NM-GFP controlled by the GAL1 promoter. Each Ura− segregant was obtained from a 4 Ura−: 0 Ura+ tetrad of the indicated heterozygous diploid, and its genotype was examined by replica-plating. The genotype of cells examined is shown above the image. After GAL induction for 16 h in 2% (wt/vol) raffinose, 2% (wt/vol) galactose minimal medium, Sup35NM-GFP aggregates or diffused GFP signal were observed using fluorescence confocal microscopy (magnification 1500×).
To further confirm that Ssb1, Zuo1p, or Ssz1p function in eliminating the [PSI+] prion variant in each mutant strain, pRS313 (CEN vector) or pRS313 with the corresponding gene (pM76 = pRS313-SSB1, pM75 = pRS313-ZUO1, pM78 = pRS313-SSZ1) controlled by its own promoter, were transformed into each [PSI+] isolate, respectively. Transformants were selected in media containing uracil and then replica-plated to plates lacking uracil to analyze the stability of [PSI+] variants. Most of the transformants carrying pSSB1, pZUO1, or pSSZ1 were unable to grow on media lacking uracil, indicating that each [PSI+] prion variant was eliminated by the restored normal levels of Ssb1, Zuo1p, or Ssz1p (Table 3). From Ura− transformants, 50 subclones for each case that had lost pSSB1, pZUO1, or pSSZ1 were found to remain Ura− in 86%, 92%, or 100% of cases. The maintenance of Ura− phenotype in each subclone after plasmid loss indicates that [PSI+] prion variants are indeed eliminated by each plasmid, not that its phenotype is merely disguised (Table 3).
Table 3.
Restored normal level of components of ribosome-associated chaperone can eliminate each of [PSI+] variants
| Isolate | ssb1/2Δ [PSI+sbs] transformants (Ura+/total transformants) | zuo1Δ [PSI+zos] transformants (Ura+/total transformants) | ssz1Δ [PSI+szs] transformants (Ura+/total transformants) | |||
| Vector | pSSB1 | Vector | pZUO1 | Vector | pSSZ1 | |
| 1 | 49/50 | 16/50 | 49/50 | 25/50 | 48/50 | 5/30 |
| 2 | 48/50 | 18/50 | 46/50 | 16/50 | 47/50 | 6/30 |
| 3 | 47/50 | 17/50 | 48/50 | 30/50 | 48/50 | 5/30 |
| 4 | 47/50 | 16/30 | 49/50 | 27/50 | 48/50 | 6/30 |
| 5 | 48/50 | 14/40 | 50/50 | 14/50 | 48/50 | 7/30 |
| 6 | 49/50 | 18/45 | 48/50 | 31/50 | 50/50 | 15/50 |
| 7 | 26/26 | 4/30 | 26/27 | 7/50 | 50/50 | 9/40 |
| 8 | 23/24 | 14/50 | 29/29 | 7/50 | 46/50 | 5/50 |
| 9 | 28/29 | 16/50 | 32/34 | 32/50 | 45/50 | 7/40 |
| 10 | 27/28 | 14/50 | 52/57 | 14/50 | 44/50 | 5/31 |
| 11 | 24/28 | 9/50 | 38/44 | 18/50 | 36/40 | 8/30 |
| 12 | 28/30 | 3/50 | 32/33 | 6/50 | 34/40 | 11/50 |
| Total | 444/465 | 159/545 | 499/524 | 227/600 | 544/580 | 81/441 |
| % Ura+ | 95.48% | 29.17% | 95.23% | 37.8% | 93.79% | 20.18% |
Twelve strains carrying [PSI+sbs], [PSI+zos], or [PSI+szs] were transformed with the CEN plasmid pRS313 or the same plasmid carrying SSB1, ZUO1, or SSZ1, respectively, under their native promoters (pM76 = pSSB1, pM75 = pZUO1, pM78 = pSSZ1). Transformants were selected in the presence of uracil and were replica-plated to a plate lacking uracil to test the stability of [PSI+]. Subclones of Ura− transformants that had lost pM76, pM75, or pM78 were tested for uracil auxotrophy by replica-plating; 86–100% of such subclones in each case showed Ura− phenotype, indicating that the [PSI+] variants had been largely eliminated by pM76, pM75, and pM78.
Ssb1/2p-Sensitive [PSI+sbs] Variants Differ from Zuo1p-Sensitive or Ssz1p-Sensitive [PSI+] Variants in Propagation Ability.
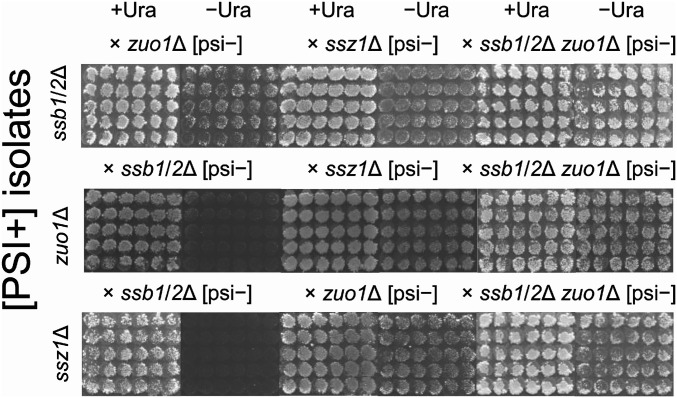
Yeast strains lacking any one or all three of the components of the ribosome-bound tripartite Ssb1/2p-RAC system show similar phenotypes, such as slow growth, hypersensitivity to several translation-inhibitors, and sensitivity to high salt or low temperature (32–34, 36–38). Thereby, all three appear to be functionally equivalent in protecting newly synthesized nascent polypeptide chains from misfolding or aggregation (38–40). First, we tried to test whether each [PSI+] prion variant can propagate in each other deletion strain even in presence of the originally missing protein, for example, infecting [PSI+sbs] into a zuo1Δ [psi−] or ssz1Δ [psi−] recipient strain. However, like our result described above and in SI Appendix, Table S3, we were unable to obtain enough cytoductants to analyze the combination of mutant donor and mutant recipient, and this was not caused by [PSI+]. We then examined the stability of each [PSI+] variant in different backgrounds using doubly heterozygous diploids. The ssb1/2Δ [PSI+sbs] isolates were mated with zuo1Δ [psi−], ssz1Δ [psi−], or ssb1/2Δzuo1Δ [psi−] strains, and each were subcloned on YPAD and further tested for [PSI+] by assaying uracil auxotrophy. Almost all diploids formed with ssb1/2Δzuo1Δ were Ura+ (96 of 100) like ssb1/2Δ/− homozygotes (Fig. 1 and Table 2), but not the diploids with zuo1Δ [psi−] or ssz1Δ [psi−]. The latter diploids grew on −Ura media, but only slowly (Fig. 3), and were mixtures of Ura+ (35of 100 and 11 of 100, respectively) and Ura− phenotype (SI Appendix, Table S4). The diploids formed with either zuo1Δ [PSI+zos] or ssz1Δ [PSI+szs] and ssb1/2Δ [psi−] were nearly all Ura− (Fig. 3 and SI Appendix, Table S4), like diploids with the WT (Fig. 1 and Table 2), while the diploids with a strain lacking the other partner of RAC were faithfully Ura+ (Fig. 3 and SI Appendix, Table S4). Propagation abilities of [PSI+sbs] and [PSI+zos or szs] in identical doubly heterozygous diploids (ssb1ssb2Δ/+ zuo1Δ/+ [or ssz1Δ/+]) are different. Note that the same two parent strains were used in these crosses; in one case, [PSI+sbs] was generated in the ssb1ssb2Δ parent, while in the other cross, [PSI+zos] (or [PSI+szs]) was generated in the other parent. The nuclear genotypes of the diploids were the same; only the [PSI+] prion variant was different. The behavior of [PSI+zos] and [PSI+szs] were exactly similar. These results indicate that [PSI+] isolates in the ssb1/2∆ strain and those in zuo1∆ or ssz1∆ strains have distinct characteristics, even though these three molecular chaperones are part of the same complex on the ribosomes and their mutants show similar characteristics for cellular functions.
Fig. 3.
[PSI+sbs], [PSI+zos], and [PSI+szs] prion variants are differentiated by their propagation ability in doubly heterozygous diploids. [PSI+sbs], [PSI+zos], and [PSI+szs] isolates were mated on YPAD with ssb1/2Δ [psi−] MS520, zuo1Δ [psi−] MS528, ssz1Δ [psi−] MS514, or ssb1/2Δzuo1Δ [psi−] MS560. Doubly heterozygous diploids were replica-plated to minimal media with and without uracil, supplemented with adenine and histidine for diploid selection.
The Effects of Deletion of SSB1/2, ZUO1, SSZ1, or of [PSI+] on the Ribosome Association and Stability of Chaperones.
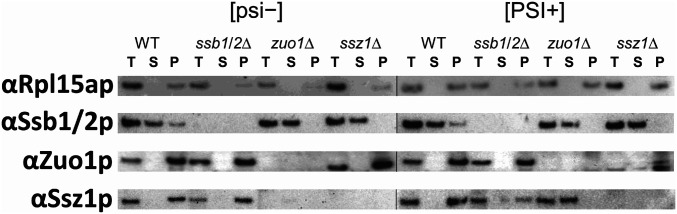
The deletion of a RAC component gene leads to the release of at least a portion of Ssb1/2p from the ribosome to the cytosol (44). This seems to affect prion generation and propagation. To understand the mechanisms of inhibition of prion generation or propagation by Ssb1/2p or RAC, the levels and soluble fraction of Ssb1/2p, Zuo1p, or Ssz1p was analyzed by SDS/PAGE and immunoblotting with antibodies. Total extracts (T) of each strain with/without [PSI+] were prepared and soluble (S) and pellet (P) fractions were separated using ultracentrifugation (Material and Methods).
In WT [psi−] or WT [PSI+] cells, Ssb1/2p was found in both soluble and pellet fraction, with a lower amount in the pellet (Fig. 4). However, in either zuo1∆ or ssz1∆ cells, Ssb1/2p was found only in the soluble fraction, not in the pellet fraction (below the detection level), in both [psi−] and [PSI+] cells (Fig. 4). This confirms again that ribosome-associated Ssb1/2p is released from ribosomes in either zuo1∆ or ssz1∆ cells (of our strain background) (44). As a ribosome-associated protein, both Zuo1p and Ssz1p in WT cells or ssb1/2∆ cells are detected only in the pellet fraction like ribosomal protein Rpl15a (Fig. 4). Zuo1p (expected size 49 kDa) was present, but slightly decreased in size in ssz1∆ [psi−] cells, and decreased in amount and appearing as two bands in ssz1∆ [PSI+] cells (Fig. 4 and SI Appendix, Fig. S2A). Ssz1p was completely destabilized in zuo1∆ [psi−] cells, but not destabilized in zuo1∆ cells with any of several [PSI+] variants (Fig. 4 and SI Appendix, Fig. S2B). This undegraded Ssz1p remained in the soluble fraction, not in the pellet, indicating that Ssz1p was released from ribosome, but not destabilized. These Western blots indicate that the stability of Zuo1p (both in [psi−] and [PSI+]) or Ssz1p (only in [psi−]) is severely decreased by ssz1∆ and zuo1∆, respectively. However, in [PSI+] cells, the stability of Ssz1p is not changed by zuo1∆, but the ribosome binding of Ssz1p is severely impaired.
Fig. 4.
The fractionation of cell extracts from WT, ssb1/2Δ, zuo1Δ, and ssz1Δ with and without [PSI+]. Total cell extracts (T) from each strain were separated into a postribosomal supernatant (S) and a ribosomal pellet (P) using ultracentrifugation. Corresponding amounts of T, S, and P fraction were separated by SDS/PAGE, one gel for [psi−] strain extracts and one for [PSI+] extracts, and further analyzed by Western blot, with the use of antibodies specific to Rpl15a, Ssb1/2p, Zuo1p, and Ssz1p, respectively. For better detection of Zuo1p from pellet fractions, both S and P fractions were overloaded by two-fold compared to the T fraction.
Taking these data together, we find that the location of Ssb1/2p, not its stability, is altered by absence of Zuo1p or Ssz1p. However, both the fraction and the stability of each RAC protein is altered by absence of its partner, and not by ssb1/2∆. This can suggest that the difference between [PSI+sbs] and [PSI+zos or szs] has resulted from different cellular environments based on the location and stability of Ssb1/2p, Zuo1p, or Ssz1p.
[PSI+] Variant Sensitivity to Overproduced Ribosome-Associated Chaperones.
Overproduced Ssb1p is known to antagonize propagation of weak [PSI+] variants in WT cells without changing Hsp104 levels (44, 60, 64, 66). In both zuo1∆ and ssz1∆, the spontaneous loss of weak and strong [PSI+] variants are increased (44). To test whether each of our [PSI+] is destabilized by excess Ssb1p, two plasmids carrying SSB1 driven by its own promoter, one single copy (pRS313) and the other high copy (pRS423), were transformed into each [PSI+] variant-carrying strain. Transformants were then replica-plated on media with and without uracil.
Of [PSI+sbs] colonies transformed with the single-copy pRS313-SSB1, 77% lost the prion (as in Table 3), as did 97% of those transformed with the high-copy pRS423-SSB1 (SI Appendix, Table S5), indicating again that the [PSI+sbs] variant is sensitive to Ssb. The meiotic crosses mentioned above support the same conclusion. [PSI+zos] variants were not affected by an extra copy of SSB1 on a CEN plasmid, but were slightly destabilized when Ssb1p was overproduced from pRS423-SSB1. Like [PSI+zos] variants, [PSI+szs] variants are not affected by an extra copy of normally expressed SSB1. However, about half of [PSI+szs] transformants carrying pRS423-SSB1 showed a Ura− phenotype, indicating that the [PSI+szs] variant is somewhat sensitive to overproduced Ssb1p (SI Appendix, Table S5).
Previously, either overproduced Ssb1p or Zuo1p was found to partially rescue the growth and antibiotic sensitivity phenotypes of a deletion of SSZ1 (38). As mentioned above, high-copy SSB1 destabilizes [PSI+szs] in an ssz1∆ strain (SI Appendix, Tables S5 and S6). As expected, overproduced Ssz1p efficiently cures [PSI+szs] variants (SI Appendix, Table S6). Interestingly, overproduced Zuo1p also cures [PSI+szs] partly; about 30% of transformants were Ura−, indicating that the prion curing function of Ssz1p is also partially rescued by its overproduced partners. Although overproduction of these chaperones does not actually occur in normal cells while our variant is being cured, these results indicate that the prion curing ability of Ssz1p does not require those of Ssb1p and Zuo1p, but all three are needed for their common cellular function (38).
Is the [PIN+] Prion Variant Altered in the Absence of Ssb or RAC?
Deletion of Hsp90 and related cochaperone genes leads to alteration of [PIN+] prion variants, changing their efficiency of [PSI+] induction, Rnq1-GFP aggregate formation, and dominance of prion variant, even after the deleted gene is restored (67). We tested whether [PSI+] inducibility of [PIN+] was altered by each deletion. [PIN+] in each deletion strain and the WT strain (MS327) were transferred into same background, a [psi−][pin−] WT recipient strain (MS173), produced by transient GuHCl treatment. These WT strains were grown in galactose, and cells were plated on −Ura plates to score [PSI+] clones.
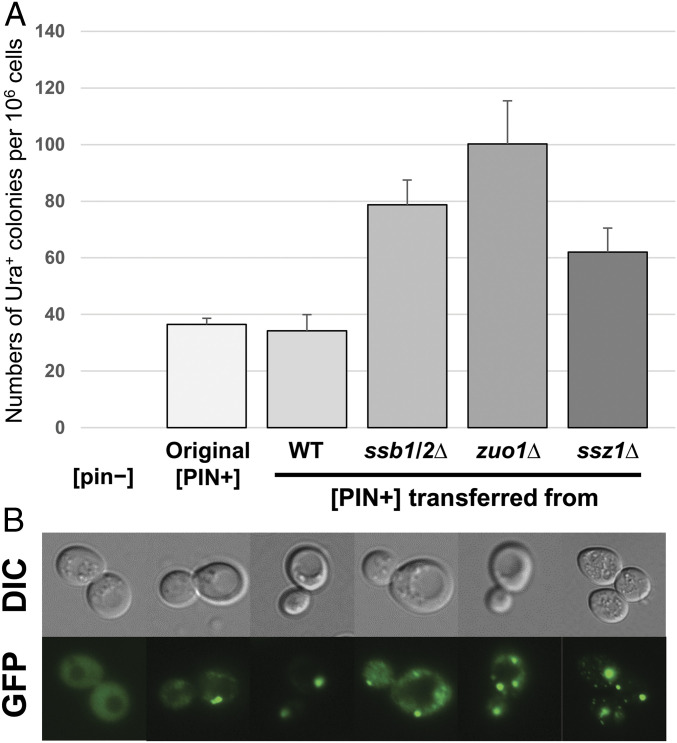
The frequency of Ura+ guanidine-curable clones induced by Sup35NM overproduction per 106 cells in WT [PIN+]ssb1/2∆, WT [PIN+]zuo1∆, or WT [PIN+]ssz1∆ was increased about 2-, 2.5-, or 1.5-fold, respectively, compared to BY4742 (WT [PIN+]original) or WT [PIN+] (Fig. 5A). To examine formation of fluorescence structures, Rnq1-GFP was transiently overexpressed in the WT strain with different sources of [PIN+] in Fig. 5A. In the WT [pin−] strain, all of cells tested for this study showed diffused GFP signal (Fig. 5B and Table 4). In WT [PIN+]original or WT [PIN+]WT strain, the frequency of diffuse signal, single foci, and multiple foci were about 35%, 47%, and 17%, respectively (Fig. 5B and SI Appendix, Table S5). WT [PIN+]ssb1/2∆ cells had slightly different content of single and multiple foci, 42.6% and 22.9%, respectively (Table 4). [PIN+]zuo1∆ or [PIN+]ssz1∆ carrying cells showed very similar frequency consisting of ∼19% of diffuse, 41% of single foci, and 40% of multiple foci (Fig. 5B and Table 4). The combined results indicate that deletion of SSB1/2, ZUO1, or SSZ1 leads to slight alteration of [PIN+] prion in [PSI+] induction and Rnq1-GFP aggregate formation. However, these modest changes do not account for the dramatic increases in [PSI+] generation frequency in these mutants.
Fig. 5.
Alteration of [PIN+] prion by ssb1/2Δ, zuo1Δ, and ssz1Δ. (A) The efficiency of [PSI+] induction was slightly elevated in a WT strain carrying [PIN+] from ssb1/2Δ, zuo1Δ, or ssz1Δ. Into each mutant strain (MS515, MS527, MS510) and the WT (MS327), all made [pin−], the [PIN+] from from BY4742 (=“Original [PIN+]”) was introduced by cytoduction. The mutants showed high [PSI+] generation (Table 1). Then the [PIN+] in each mutant and WT strain was transferred back into MS173 (previously made [pin-] by guanidine curing) by cytoduction. The “[pin−]” strain is MS173 cured of [PIN+]. In these strains, all with WT genome but [PIN+] from different sources, [PSI+] was induced by overproduction of Sup35NM in 2% (wt/vol) raffinose, 2% (wt/vol) galactose minimal medium. Cells were plated on −Ura plates, and arising Ura+ ([PSI+]) clones were tested for GuHCl curability using transient growth on plates containing 5 mM guanidine. (B) The formation of Rnq1-GFP aggregates in different [PIN+]-carrying strains. BY4742 [pin−], BY4742 [PIN+], MS562, MS563, MS564, and MS565 were transformed with pM60 (pCEN LEU2) expressing ADH1 promoted Rnq1-GFP. Transformants were directly used for fluorescence confocal microscopy observation (magnification 1500×).
Table 4.
The frequency of cells with different numbers of Rnq1-GFP foci
| Strain | GFP signals | ||
| Diffused, % | Single foci, % | Multiple foci, % | |
| WT [pin−] | 100 | 0 | 0 |
| WT [PIN+]original | 35.6 | 47.1 | 17.3 |
| WT [PIN+]WT | 35.0 | 47.4 | 17.6 |
| WT [PIN+]ssb1/2∆ | 34.5 | 42.6 | 22.9 |
| WT [PIN+]zuo1∆ | 19.5 | 40.8 | 39.7 |
| WT [PIN+]ssz1∆ | 18.9 | 41.9 | 39.2 |
Strains MS317, MS562-MS565 (top to bottom), transformed with plasmids encoding RNQ1−GFP were grown on SD-Leu. The frequencies of cells with different number of aggregates were scored in 100 to 150 cells. Superscripts in the strain column indicate different sources of [PIN+].
Induced [URE3] Generation Is Not Elevated in ssz1∆ Cells.
In BY241, the Ure2p-regulated DAL5 promoter drives ADE2, so that [URE3] cells are Ade+, but [ure-o] cells are Ade−. The frequency of [URE3] induced by overproduction of Ure2 N (amino acids 1 to 65) was investigated in WT [ure-o] and ssz1∆ [ure-o] strains. The average of Ade+ clones in WT and ssz1∆ strains was 846 ± 163 and 940 ± 43 per 105 cells, respectively. These Ade+ [URE3] clones were then tested for GuHCl curability by transient growth on 4 mM GuHCl for WT or 1.5 mM GuHCl for ssz1∆. Thirty of 32 Ade+ clones in WT and 28 of 32 Ade+ clones in the mutant were GuHCl curable, indicating that they carried a [URE3]. Eleven randomly selected [URE3] isolates, each from WT or ssz1∆ strains, were tested for [URE3] stability in the haploid or heterozygous diploid by subcloning on YPAD media and replica-plating to −Ade plates. All [URE3] isolates in ssz1∆ strains were mostly stable in both the original ssz1∆ haploid and in the heterozygous diploid (SI Appendix, Fig. S3 and Table S7), behaving the same as variants isolated in the WT host. This result was confirmed by transforming ssz1∆ or WT [URE3] isolates with a single-copy plasmid carrying SSZ1 expressed by its own promoter (pM89). These transformants were no more likely to lose [URE3] than those transformed with the vector whether the host was ssz1∆ or WT (SI Appendix, Table S8).
Discussion
Ssb1/2p, Zuo1p, and Ssz1p were previously shown to repress prion generation (43–45), and we have now shown that the majority of the prions arising in their absence are cured by normal levels of the respective protein; these latter prions are the focus of this work. We confirm previous reports that an increased frequency of “insensitive” [PSI+] variants also occurs in the mutants, but we find that the majority of [PSI+] variants arising are cured in the normal environment. We show by genetic analyses and cell biological analysis that curing of the [PSI+] prion variants by normal levels of the WT allele is occurring, not simply an effect on the phenotype. Thus Ssb1/2p, Zuo1p, and Ssz1p are components of an antiprion system. We show that [PSI+sbs] variants differ from [PSI+szs] or [PSI+zos] variants based on consistently different stability in the identical doubly heterozygous sbs1/2Δ/+ ssz1Δ/+ (or zuo1Δ/+) host. In investigating the mechanisms involved in the elevated generation of the new prion variants and their curing, we find that Ssz1p is very unstable in the absence of Zuo1p, and Zuo1p is somewhat unstable in the absence of Ssz1p. Interestingly, in the presence of [PSI+], Ssz1p is stable in the zuo1Δ strain, but soluble- , no longer ribosome-bound.
Why Is Prion Generation Elevated in ssb1/2 and RAC Mutants?
A large part of the increase in [PSI+] frequency was the appearance of the novel curable variants that could not propagate in the WT strain. The most straight-forward explanation for the appearance of these variants as well as increased incidence of variants not cured in the WT is that Ssb1/2 and RAC chaperones are important for proper folding of nascent proteins, and that improperly folded Sup35p is preferred or required for the growth of the [PSI+] amyloid filaments (43). This mechanism might suggest that other prions would be affected similarly, but we find that [URE3] generation is not increased in an ssz1∆ strain and that ssz1∆ [URE3] isolates are not cured by restored Ssz1p, suggesting that alternative explanations should be considered.
One known effect of deletion of SSB1/2, ZUO1, or SSZ1 is slight nonsense suppression (43, 65), too weak to explain the elevated [PSI+] appearance (43), and our genetic and cytologic analysis shows that the [PSI+sbs], [PSI+szs], and [PSI+zos] prions are indeed lost in the WT, not just phenotypically masked. But the existence of this nonsense suppression suggests some functional interaction of the Ssb and RAC components with the Sup35/Sup45 termination apparatus, through the ribosome structure. Thus, it is possible that it is an effect on the mature Sup35p (rather than only nascent Sup35p) that is responsible for elevated [PSI+] formation. We previously explained the antiprion action of Upf proteins by their specific interactions with Sup35p (31). Similarly, direct interactions of Ssb1/2 with Sup35p have been described previously (64), and the [PSI+] specificity of ribosome associated chaperones might be a result of a direct interaction of these components with mature Sup35p.
Deficiency of specific chaperones (Hsc82, Aha1p, Cpr6p, Cpr7p, Sba1p, Tah1p, and Sse1p) was reported to alter [PIN+] variant phenotypes, namely, [PSI+]-inducibility, Rnq1-GFP aggregate formation and variant dominance (67). In that study, the [PIN+] prion transferred to a WT strain conferred the elevated or reduced [PSI+] inducing activity of the mutant, indicating that the [PIN+] prion had been altered in the mutant strain. In our study, [PSI+] inducing efficiency of the [PIN+] prion that had experienced the ssb1/2, ssz1, or zuo1 deletion strain was increased by only about twofold in each case when transferred to the WT (Fig. 5A). However, compared to the >30-fold effect of the deletion itself on [PSI+] induction, it is not likely that alteration of [PIN+] by each deletion is important in determining the inducing efficiency. The small differences in inducing activity, and the modest variation of the pattern of Rnq1-GFP foci in each case suggests that [PIN+]zuo1∆, [PIN+]ssz1∆, and [PIN+]ssb1/2∆ are essentially the same as their common parent (Fig. 5B and Table 4).
What Controls Generation and Curing of [PSI+sbs] vs. [PSI+szs] or [PSI+zos]?
The existence of prion variants, with different biological properties, results from different amyloid structures of a single prion protein (68–70). The architecture of amyloid-based yeast prions (71–73) led to an explanation of how a protein could template its own conformation and the stable propagation of many distinct prion variants from one single protein sequence (74, 75). [PSI+] prion variants previously isolated in ssb1/2∆ were not cured by restored SSB1 on a CEN plasmid (43). [PSI+] isolates in either zuo1∆ or ssz1∆ were more often unstable in their own mutant strain (44). These previous studies used the [PSI+]-suppressible ade1-14 nonsense allele in a different strain background. We confirm the existence of variants not cured in a WT cell but, unlike previous studies, find a majority of variants that are cured in a normal WT strain. This difference may be due to the different strain backgrounds and reporters used.
[PSI+sbs] variants differ from both [PSI+szs] and [PSI+zos] prion variants, because the latter two are rapidly lost in doubly heterozygous ssb1,2Δ/+ ssz1Δ (or zuo1Δ)/+ diploids, while [PSI+sbs] propagates and is only slowly lost from the very same diploids (Fig. 3 and SI Appendix, Tables S3 and S4). Beyond the prion variant differences, one would like to understand what component is causing prion loss in each case. This prion variant difference must result from the conditions under which variants were isolated. As shown in Fig. 4 (Fig. 4, Left: [psi−]), [PSI+zos] and [PSI+szs] were generated under the condition with no ribosomal Ssb1/2p, but with soluble Ssb1/2p, indicating that both variants are resistant to the soluble (cytosolic) Ssb1/2p. [PSI+sbs] must be inhibited by ribosome-bound or soluble Ssb1/2p, because Ssz1p and Zuo1p are ribosome-bound even without Ssb1/2p. While ssb1/2Δ [PSI+sbs] × WT produces rapid curing, ssb1/2Δ [PSI+sbs] × ssz1Δ (or zuo1Δ) show slower curing, even though the latter diploids have more soluble Ssb1/2p than the former, being partially deficient (haplo-insufficient) in Ssz1p (or Zuo1p). We infer that it is ribosome-bound Ssb1/2p that inhibits propagation of [PSI+sbs]. Because ssz1Δ (or zuo1Δ) strains lack ribosome bound Ssb1/2, but have elevated soluble Ssb1/2p, [PSI+szs] and [PSI+zos] may be inhibited by ribosome-bound Ssb1/2 or by Ssz1p (or Zuo1p). Just because [PSI+sbs] differs from [PSI+szs] and [PSI+zos], we speculate that it is not ribosomal Ssb1/2p, but rather Ssz1p (or Zuo1p) itself that blocks [PSI+szs] and [PSI+zos].
Deletion of SSB1/2, ZUO1, SSZ1, or all show the same cellular phenotypes, and the same stimulation of [PSI+] formation, suggesting a common function (32–34, 36–38, 44). Here we found that RAC deletions lead to the generation of substantially different prion variants than ssb1/2Δ strains. Most recently, RAC was found to bind to translating nascent peptide chains at the ribosomal tunnel exit like ribosomal Ssb1/2p (76). Ribosome-associated Zuo1p, Ssz1p, and Ssb1/2p sequentially contact growing nascent chains of minimum length 40, 45, and 50 residues, respectively, and hand over chains to the next chaperone in a relay for cotranslational de novo protein folding. Accordingly, the absence of different components may differently affect the site-specific contact with the translating nascent chain, inducing distinct misfolding of Sup35p and thus different arrays of prion variants. Replacing the missing chaperone should prevent the occurrence of a specifically misfolded Sup35p needed for a specific amyloid structure, thus explaining the curing of [PSI+sbs], [PSI+szs], and [PSI+zos] by limiting the growth of the amyloid fibers. Inhibition of Sup35 NM fibrilization by purified RAC and Ssb1p in vitro was previously reported, suggesting that nascent Sup35 polypeptides are likely shielded from possible prion conformations upon emerging from the ribosome (77).
In earlier work, Ssb1/2p was suggested to prevent formation of the misfolded intermediates for Sup35p prion aggregates and stimulate degradation of these intermediates, but not final [PSI+] prion aggregates (43). Especially, ribosomal bound Ssb1/2p was also reported to repress formation of [PSI+] prion aggregates, and the relocation of Ssb1/2p from ribosome to cytosol enhanced prion formation, but antagonized prion propagation by competing with cytosolic Ssa for binding to prion aggregates (44–46). In this work we studied a different class of prion variants, not detected in the earlier work, and so the roles of Ssb1/2p and RAC in preventing generation and in curing these prions could be different.
Why Does [PSI+] Stabilize Ssz1p in zuo1Δ Cells?
Zuo1p is directly bound to the ribosome and Ssz1p is bound to the ribosome through Zuo1p (41). Ssb1/2p relies for its ribosome association on Zuo1p and Ssz1p. We find that in zuo1Δ [psi−] cells Ssz1p is degraded, but not in [PSI+] cells. Although stable in [PSI+] cells, Ssz1p is not ribosome-associated, so it is unlikely to interact with the normal ribosome-bound Sup35p, or sense the frequent absence of Sup35p on ribosomes in [PSI+] cells. It is possible that the [PSI+] prion leads to the partial inactivation of some E3 ligase or autophagy factor that would otherwise lead to degradation of Ssz1p in zuo1Δ.
This study confirms that prions are more abundant and more differentiated than was previous thought (reviewed in ref. 20). Yeast has evolved a wide (and widening) array of systems to prevent prion formation, to cure most prion variants, and to limit the damage caused by those variants that survive those measures. It is hoped that the variety of antiprion systems found in yeast will stimulate a similar search in mammals, and provide tools useful in treatment of the common human amyloidoses, increasingly found to have infectious aspects.
Materials and Methods
Nomenclature.
Yeast prions are shown in brackets to indicate they are nonchromosomal genes (e.g., [PSI+], [URE3], or [PIN+]). Specific types of prion variants are indicated within the brackets (e.g., [PSI+sbs]). Strains of origin of [PIN+] prion are indicated with a superscript (e.g., “WT [PIN+]zuo1∆” for [PIN+] transferred from a zuo1∆ strain into the WT strain).
Strains and Media.
Strains used in this study are listed in SI Appendix, Table S1. Gene disruption mutants were generated by PCR-amplifying yeast genomic DNA of the corresponding strains from the Saccharomyces cerevisiae knockout collection or mating with isogenic strains from the knockout collection, and further confirmed using PCR-amplification (78). Media used were as described by Sherman (79). Induction of GAL1-promoted genes was conducted using galactose-raffinose–containing media as previously described (31). The ade1-14 allele carrying parental strains were derivatives of strains used for previous studies (31). Scoring [PSI+] with the suppressible ura3-14 mutation and inducing prion formation routinely used p1520 (CEN LEU2 ura3-14 GAL1promoter-SUP35NM). For all of the [PSI+] scoring experiments, p1520 was maintained in strains/colonies without leucine to avoid loss of this plasmid.
Plasmids.
Plasmids used in this study are listed in SI Appendix, Table S2. SSB1 with 357 bp and ZUO1 with 500 bp upstream of the ORF were amplified and ligated into pRS313 (CEN HIS3) cut with BamHI and XhoI forming pM76 and pM75, respectively. SSZ1 with 390 bp upstream of the ORF was amplified and ligated into pRS313 cut with BamHI and SalI forming pM78. The same strategy was used for generating pRS423 (2µ HIS3)-based SSB1, ZUO1, and SSZ1 plasmids, forming pM85, pM87, and pM86, respectively. An ADH1-promoted Rnq1p-GFP expressing plasmid was generated previously (18).
Cytoduction.
In mating kar1 mutants fail in nuclear fusion so daughter cells have a mixture of the parental cytoplasms. Cytoduction donors are ρ+, recipients are ρo from ethidium exposure, both isogenic to the knockout bank strains, BY4741 and BY4742, both carrying p1520 (CEN LEU2 ura3-14) bearing PGAL1-SUP35NM (30). In effect, cytoplasm is transferred from donors (with induced [PSI+] variants) and recipients. After mating, mixtures were streaked on media selecting against the donor. Colonies were replica-plated to YPG, media selective for diploids and media lacking uracil. Clones growing on YPG but not on diploid selection media were cytoductants. Ura+ clones propagated [PSI+]. When donor and recipient strains are both ssb-rac∆, resulting cytoductants rarely appeared making this type of analysis impractical.
Antibodies.
Antibodies to Ssb1/2p, Zuo1p, and Ssz1p were generous gifts from Elizabeth A. Craig, University of Wisconsin–Madison, Madison, WI. Rpl15p antibody was purchased from Bio-Rad.
Supplementary Material
Acknowledgments
We especially thank Thomas Ziegelhoffer (University of Wisconsin–Madison) for detailed guidance and kind advice about protein analysis. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016954117/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Wickner R. B., [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Stansfield I. et al., The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14, 4365–4373 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frolova L. et al., A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372, 701–703 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Cox B. S., PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20, 505–521 (1965). [Google Scholar]
- 5.King C.-Y. et al., Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. U.S.A. 94, 6618–6622 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D., In vitro propagation of the prion-like state of yeast Sup35 protein. Science 277, 381–383 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Glover J. R. et al., Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Jung G., Jones G., Wegrzyn R. D., Masison D. C., A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics 156, 559–570 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King C.-Y., Diaz-Avalos R., Protein-only transmission of three yeast prion strains. Nature 428, 319–323 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S., Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Lacroute F., Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 106, 519–522 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masison D. C., Maddelein M.-L., Wickner R. B., The prion model for [URE3] of yeast: Spontaneous generation and requirements for propagation. Proc. Natl. Acad. Sci. U.S.A. 94, 12503–12508 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edskes H. K., Gray V. T., Wickner R. B., The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. U.S.A. 96, 1498–1503 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor K. L., Cheng N., Williams R. W., Steven A. C., Wickner R. B., Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283, 1339–1343 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Brachmann A., Baxa U., Wickner R. B., Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 24, 3082–3092 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper T. G., Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol. Rev. 26, 223–238 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGlinchey R. P., Kryndushkin D., Wickner R. B., Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. U.S.A. 108, 5337–5341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayashiki T., Kurtzman C. P., Edskes H. K., Wickner R. B., Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. U.S.A. 102, 10575–10580 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly A. C., Shewmaker F. P., Kryndushkin D., Wickner R. B., Sex, prions, and plasmids in yeast. Proc. Natl. Acad. Sci. U.S.A. 109, E2683–E2690 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickner R. B., Son M., Edskes H. K., Prion variants of yeast are numerous, mutable, and segregate on growth, affecting prion pathogenesis, transmission barriers and sensitivity to anti-prioin systems. Viruses 11, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wickner R. B. et al., Yeast prions: Proteins templating conformation and an anti-prion system. PLoS Pathog. 11, e1004584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickner R. B. et al., Anti-prion systems in yeast and inositol polyphosphates. Biochemistry 57, 1285–1292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W., Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880–884 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Moriyama H., Edskes H. K., Wickner R. B., [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20, 8916–8922 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung G. C., Masison D. C., N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 173, 611–620 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorkovskiy A., Reidy M., Masison D. C., Wickner R. B., Hsp104 disaggregase at normal levels cures many [ PSI+] prion variants in a process promoted by Sti1p, Hsp90, and Sis1p. Proc. Natl. Acad. Sci. U.S.A. 114, E4193–E4202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryndushkin D. S., Shewmaker F., Wickner R. B., Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 27, 2725–2735 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanneganti V., Kama R., Gerst J. E., Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol. Biol. Cell 22, 1648–1663 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickner R. B., Bezsonov E., Bateman D. A., Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc. Natl. Acad. Sci. U.S.A. 111, E2711–E2720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickner R. B., Kelly A. C., Bezsonov E. E., Edskes H. K., [PSI+] prion propagation is controlled by inositol polyphosphates. Proc. Natl. Acad. Sci. U.S.A. 114, E8402–E8410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Son M., Wickner R. B., Nonsense-mediated mRNA decay factors cure most [PSI+] prion variants. Proc. Natl. Acad. Sci. U.S.A. 115, E1184–E1193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson R. J., Ziegelhoffer T., Nicolet C., Werner-Washburne M., Craig E. A., The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71, 97–105 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Pfund C. et al., The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17, 3981–3989 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan W. et al., Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17, 4809–4817 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallstrom T. C., Katzmann D. J., Torres R. J., Sharp W. J., Moye-Rowley W. S., Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 1147–1155 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gautschi M. et al., RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. U.S.A. 98, 3762–3767 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hundley H. et al., The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. U.S.A. 99, 4203–4208 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gautschi M., Mun A., Ross S., Rospert S., A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. U.S.A. 99, 4209–4214 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bukau B., Deuerling E., Pfund C., Craig E. A., Getting newly synthesized proteins into shape. Cell 101, 119–122 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Hartl F. U., Hayer-Hartl M., Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295, 1852–1858 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Sinning I., Rospert S., Two chaperones locked in an embrace: Structure and function of the ribosome-associated complex RAC. Nat. Struct. Mol. Biol. 24, 611–619 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Deuerling E., Gamerdinger M., Kreft S. G., Chaperone interactions at the ribosome. Cold Spring Harb. Perspect. Biol. 11, a033977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chernoff Y. O., Newnam G. P., Kumar J., Allen K., Zink A. D., Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol. Cell. Biol. 19, 8103–8112 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiktev D. A., Melomed M. M., Lu C. D., Newnam G. P., Chernoff Y. O., Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol. 96, 621–632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amor A. J. et al., The ribosome-associated complex antagonizes prion formation in yeast. Prion 9, 144–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chernoff Y. O., Kiktev D. A., Dual role of ribosome-associated chaperones in prion formation and propagation. Curr. Genet. 62, 677–685 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Matveenko A. G., Barbitoff Y. A., Jay-Garcia L. M., Chernoff Y. O., Zhouravleva G. A., Differential effects of chaperones on yeast prions: CURrent view. Curr. Genet. 64, 317–325 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., Liebman S. W., Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147, 507–519 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sondheimer N., Lindquist S., Rnq1: An epigenetic modifier of protein function in yeast. Mol. Cell 5, 163–172 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W., Prions affect the appearance of other prions: The story of [PIN(+)]. Cell 106, 171–182 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Li X., Rayman J. B., Kandel E. R., Derkatch I. L., Functional role of Tia1/Pub1 and Sup35 prion domains: Directing protein synthesis machinery to the tubulin cytoskeleton. Mol. Cell 55, 305–318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bradley M. E., Edskes H. K., Hong J. Y., Wickner R. B., Liebman S. W., Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. U.S.A. 99 (suppl. 4), 16392–16399 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuite M. F., Mundy C. R., Cox B. S., Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics 98, 691–711 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung G., Masison D. C., Guanidine hydrochloride inhibits Hsp104 activity in vivo: A possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43, 7–10 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Ferreira P. C., Ness F., Edwards S. R., Cox B. S., Tuite M. F., The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40, 1357–1369 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Jung G., Jones G., Masison D. C., Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. U.S.A. 99, 9936–9941 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manogaran A. L., Fajardo V. M., Reid R. J. D., Rothstein R., Liebman S. W., Most, but not all, yeast strains in the deletion library contain the [PIN(+)] prion. Yeast 27, 159–166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manogaran A. L., Kirkland K. T., Liebman S. W., An engineered nonsense URA3 allele provides a versatile system to detect the presence, absence and appearance of the [PSI+] prion in Saccharomyces cerevisiae. Yeast 23, 141–147 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higurashi T., Hines J. K., Sahi C., Aron R., Craig E. A., Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. U.S.A. 105, 16596–16601 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chacinska A. et al., Ssb1 chaperone is a [PSI+] prion-curing factor. Curr. Genet. 39, 62–67 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Doel S. M., McCready S. J., Nierras C. R., Cox B. S., The dominant PNM2- mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137, 659–670 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirkland P. A., Reidy M., Masison D. C., Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 188, 565–577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reidy M. et al., Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet. 10, e1004720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allen K. D. et al., Hsp70 chaperones as modulators of prion life cycle: Novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169, 1227–1242 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rakwalska M., Rospert S., The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 9186–9197 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kushnirov V. V., Kochneva-Pervukhova N. V., Chechenova M. B., Frolova N. S., Ter-Avanesyan M. D., Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 19, 324–331 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lancaster D. L., Dobson C. M., Rachubinski R. A., Chaperone proteins select and maintain [PIN+] prion conformations in Saccharomyces cerevisiae. J. Biol. Chem. 288, 1266–1276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W., Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanaka M., Collins S. R., Toyama B. H., Weissman J. S., The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Dergalev A. A., Alexandrov A. I., Ivannikov R. I., Ter-Avanesyan M. D., Kushnirov V. V., Yeast Sup35 prion structure: Two types, four parts, many variants. Int. J. Mol. Sci. 20, 2633 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shewmaker F., Wickner R. B., Tycko R., Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. U.S.A. 103, 19754–19759 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baxa U. et al., Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry 46, 13149–13162 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Gorkovskiy A., Thurber K. R., Tycko R., Wickner R. B., Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc. Natl. Acad. Sci. U.S.A. 111, E4615–E4622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wickner R. B., Edskes H. K., Shewmaker F., Nakayashiki T., Prions of fungi: Inherited structures and biological roles. Nat. Rev. Microbiol. 5, 611–618 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wickner R. B. et al., Yeast prions: Structure, biology, and prion-handling systems. Microbiol. Mol. Biol. Rev. 79, 1–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y. et al., The ribosome-associated complex RAC serves in a relay that directs nascent chains to Ssb. Nat. Commun. 11, 1504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shorter J., Lindquist S., Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 27, 2712–2724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winzeler E. A. et al., Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Sherman F., “Getting started with yeast” in Guide to yeast genetics and molecular biology, Guthrie C., Fink G. R., Eds. (Academic Press, San Diego, 1991), Vol. 194, pp. 3–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.