Significance
Of the skeleton, teeth preserve well in the fossil record and have the strongest genetic signature and thus great potential for reconstructing evolutionary processes involved in human origins. The enamel–dentine junction (EDJ) is especially important, because it is more frequently preserved and somewhat decoupled from dietary function. Researchers have used the EDJ to test hypotheses of human evolution, but this trait has not yet been comprehensively studied across human populations. This study is the largest investigation of EDJ morphology to date, spanning 161 archaeological individuals from six continents. These data demonstrate that the EDJ evolves neutrally, and biomedical imaging of EDJ morphology can therefore be used as a noninvasive method to investigate ancient population movement and human genetic structure.
Keywords: dentition, phenetic variation, genetic structure, neutral evolution, geometric morphometrics
Abstract
Teeth have been studied for decades and continue to reveal information relevant to human evolution. Studies have shown that many traits of the outer enamel surface evolve neutrally and can be used to infer human population structure. However, many of these traits are unavailable in archaeological and fossil individuals due to processes of wear and taphonomy. Enamel–dentine junction (EDJ) morphology, the shape of the junction between the enamel and the dentine within a tooth, captures important information about tooth development and vertebrate evolution and is informative because it is subject to less wear and thus preserves more anatomy in worn or damaged specimens, particularly in mammals with relatively thick enamel like hominids. This study looks at the molar EDJ across a large sample of human populations. We assessed EDJ morphological variation in a sample of late Holocene modern humans (n = 161) from archaeological populations using μ-CT biomedical imaging and geometric morphometric analyses. Global variation in human EDJ morphology was compared to the statistical expectations of neutral evolution and “Out of Africa” dispersal modeling of trait evolution. Significant correlations between phenetic variation and neutral genetic variation indicate that EDJ morphology has evolved neutrally in humans. While EDJ morphology reflects population history, its global distribution does not follow expectations of the Out of Africa dispersal model. This study increases our knowledge of human dental variation and contributes to our understanding of dental development more broadly, with important applications to the investigation of population history and human genetic structure.
Teeth have been studied for decades and continue to reveal information relevant to human evolution. Many parts of the primate dentition are heritable and vary significantly inter- and intraspecifically, with variation resulting from both neutral (e.g., drift and migration) and adaptive processes (1–6). A large body of work has also demonstrated that neutral genetic variation in modern humans is patterned geographically with significant correlations between genetic variation and phenotypic variation of neutrally evolving traits (7–16).
Neutrally evolving phenotypic traits, or traits that are free from adaptive pressures, provide considerable insight into ancient human population structure and population movement in archaic and fossil communities (8, 11–14, 16). There is increasing interest in which traits of the human skeleton capture neutral genetic evolution and can be used in place of, or in combination with, more destructive DNA analyses, thereby allowing for increased sampling that preserves and protects fragile and rare specimens including holotypes and irreplaceable archaeological samples (7). The dentition is an ideal functional anatomical unit in which to investigate adaptive and neutral genetic processes, as teeth develop before eruption and function and are thus subject to fewer environmental perturbations, and dental morphology is significantly heritable (3, 17–20). To date, several studies have investigated neutral evolution of external dental morphologies (11, 21–24), but global variation in the internal morphology of the teeth remains understudied. Assessing whether dental morphology evolves neutrally in humans and other primates is of particular interest to paleoanthropologists, as neutral traits offer a way to characterize population movement in the absence of readily available genetic data, as is the case in the fossil record. This offers the opportunity to test hypotheses of range expansions, such as those proposed by the “Out of Africa” model. On the other hand, if dental traits are not evolving neutrally, they are instead likely the result of adaptive processes and therefore provide crucial insight into human origins and the evolution of human biological variation.
The outer enamel surface (OES) of the dentition has been well-studied, with research contributing to our understanding of dental development [through studies using a phenotype–genotype back approach (3)], diet, attrition, ontogeny, and morphological evolution (25–33). However, the external morphology of the dentition is subject to extensive wear processes during an individual’s lifespan. This is particularly notable in archaic and fossil populations, as well as contemporary human populations with a nonindustrialized, processed diet, limiting the utility of these morphologies in many studies (34–37). With advances in scanning technologies alleviating the need for destructive analyses (38, 39), greater attention has now been placed on the internal morphology of teeth (34–36, 40–42).
The enamel–dentine junction (EDJ) is the boundary between the enamel crown of the tooth and the underlying supportive tissues, and it captures important information about tooth development and evolution (36, 39, 42–45). The morphology of the EDJ (SI Appendix, Fig. S1) is informative, because it is not subject to the same extent of wear as the OES and thus preserves more morphology in worn specimens, particularly in mammals with relatively thick enamel like hominids (34–37). Additionally, the EDJ is formed early in development, marking the shape of the tooth at the beginning of mineralization prior to eruption, and is thus less subject to environmental and developmental perturbation (37, 39). The early development of the EDJ suggests that its topography is largely genetically determined, making it an excellent trait for tests of variation and population movement in the archaeological and fossil records.
Paleoanthropologists are particularly interested in the EDJ, because it is often the most well-preserved tooth anatomy in the fossil record (34–36). Morphological variation of the EDJ has been used extensively to investigate taxonomic relationships in the hominoid fossil record (34, 35, 42, 44, 46–48) but has not yet been comprehensively explored in modern humans. Many studies suggest that variation is taxonomically patterned (34, 39, 43). However, other studies report that considerable variation in traits of the EDJ precludes their use in taxonomic assessment (36, 37). Investigations of morphological concordance between the EDJ and OES have reported a strong phenotypic relationship between discrete traits of these anatomical layers in primates (48–50), especially for Pan (43), but others argue that anthropoids have relatively low levels of concordance compared to other euarchontoglirans (51). Overall, research suggests that traits of the EDJ and OES can be differentially expressed, and homologous accessory cusps at the OES may have different developmental origins and underlying EDJ morphology (36, 44, 45). Additionally, enamel deposition and thickness vary across the tooth surface and interspecifically (52–57), and quantitative genetic analyses of enamel thickness in primates indicate a complex genetic archetype that is additive and independent of sex and tooth size (58). There is also considerable metameric variation in EDJ morphology, and humans frequently express greater variation along the tooth row of a single individual than between individuals (39, 44). Metameric variation in EDJ morphology increases along the molar row, consistent with studies on other dental traits in primates (25, 40, 41, 47, 59). Given this reported variation, it has become essential to understand the extent of disparity in EDJ morphology before this well-preserved phenotype can be utilized in archaeological and paleontological investigations of taxonomy and evolution.
This study is of particular importance to investigation of human origins using the fossil record because it: 1) Uses a methodology that takes principles and theories from evolutionary genetics and applies them to morphological data in a manner that is directly applicable to the study of fossils; and, 2) the EDJ, the morphological trait assessed here, is widely preserved in the fossil record. Using a combination of traditional and modern geometric morphometric methods, this study undertakes an investigation of global variation in the internal dental anatomy (EDJ) of 161 late Holocene modern humans spanning six continents (Fig. 1).
Fig. 1.
Global variation in EDJ surface anatomy as represented by exemplary individuals from each continental sample. See Table 1 for continental sample sizes.
We used Holocene individuals to limit the variation introduced from more recent and wide-spread interregional and intercontinental movement (60). This dataset constitutes one of the most comprehensive investigations of human EDJ morphology to date. Our data capture the three-dimensional (3D) surface anatomy of the EDJ. We focused on the left permanent first mandibular molar and tested two hypotheses: 1) Global patterns of EDJ morphology vary according to expectations of neutral evolution, and 2) global patterns of human EDJ morphology vary according to the expectations of the Out of Africa dispersal hypothesis.
Global Patterns of EDJ Morphology Vary According to Expectations of Neutral Evolution.
We first investigate the mode of evolution of human dental morphology using EDJ topography. Our hypothesis is that phenetic distance calculated from EDJ variation of the first molar is a proxy of genetic distance. This invokes the classic model of phenetic variance being the result of additive genetic variance—that is, linear genotype to phenotype (G:P) mapping (15)—and tests whether the EDJ has evolved neutrally (where genetic drift and migration are the causative mechanisms of variation) by determining whether global phenotypic variation has a linear relationship to global differentiation in neutral genetic markers.
Furthermore, under “neutral-like” trait evolution and human dispersal, an isolation-by-distance pattern can be expected, in which phenotypic distance increases with increasing geographic distance between populations (16, 61–64). Using μ-CT biomedical imaging to characterize internal anatomies, particularly those that had previously been disregarded or assumed to be useful only in destructive DNA analyses, is a powerful method for investigating neutral genetic variation and population movement, as was recently shown using the inner ear bony labyrinth (7). Research focused on global variation of external dental morphology has reported neutral evolution of those traits (21–24). This study builds from this previous work and predicts a similar result for the underlying EDJ morphology.
Global Patterns of Human EDJ Morphology Vary According to the Expectations of the Out of Africa Dispersal Hypothesis.
The second hypothesis is that under the recent African origin (RAO) Out of Africa dispersal model, phenetic distance should increase with increasing geographic distance from the African source population, while within-population variation should decrease (15, 65). The RAO dispersal, or serial founder, model makes predictions about gene flow following recent expansion of Homo sapiens out of Africa since the last glacial period (∼100,000 yBP) that occurred independently of earlier expansions by Homo erectus and Neanderthals (15). One of the main predictions of this dispersal model is that the original populations should retain higher genetic (or phenotypic) variance than populations on the expansion front, because the expanding populations are the product of a series of founder effects (also known as “genetic surfing”) (66–68). Our study tests this hypothesis through regression of both within-population and between-population phenetic distances against geographic and genetic distance from Africa.
Results
Test of Neutral Evolution.
A Procrustes superimposition of 7 fixed and 95 sliding landmarks of the EDJ of left permanent mandibular first molars demonstrates that populations that are genetically more similar have more similar EDJ morphology, as captured by 3D EDJ surface anatomy (Fig. 2 and SI Appendix, Figs. S1 and S2).
Fig. 2.
Linear regression of pairwise kinship coefficients (n = 15) calculated from neutral genetic data (genetic Rij) and continental mean shape of Procrustes coordinates (phenetic Rij; r = 0.56, P = 0.03). The 95% confidence interval is plotted in gray. See SI Appendix, Table S2 for values.
Using the Rij method, Rathmann et al. (21) previously demonstrated that phenetic variation of external dental morphology is correlated with neutral genetic variation in humans. This method was applied to the EDJ morphology of the human dentition by calculating a pairwise Rij-matrix using population mean shape generated from aligned Procrustes coordinates and comparing phenetic variation to neutral genetic variation [Rathmann’s genetic Rij-matrix (21)]. Consistent with findings by Rathmann et al. (21) for the external morphology of the dentition, there is a significant correlation between mean pairwise shape differences (phenetic variation) of the EDJ and neutral genetic variation between populations (Fig. 2) (r = 0.56, P = 0.03). Population pairs that include Australia fall closest to the best-fit line and drive the regression (SI Appendix, Fig. S3). Kinship coefficients from comparison of northern and southern Africa are greater than zero for both phenetic and genetic R-matrices, indicating that these populations are most similar. The distance between two populations in neutral allele frequencies is randomly distributed due to drift. Correlation between neutral genetic variation and phenetic variation thus provides evidence that the phenotype of interest likely also evolved under drift in humans, as selection would have imposed a nonrandom pattern (21).
Out of Africa Dispersal Model.
Variation in EDJ surface anatomy is significantly correlated with neutral genetic variation between populations but deviates from the expectations of an RAO Out of Africa dispersal model. Geographic distance from southern Africa is significantly correlated with allele-sharing distance from southern Africa (r = 0.94, P = 0.02), as to be expected. However, within-population variation in EDJ morphology does not decrease with increasing geographic distance from southern Africa (SI Appendix, Fig. S4) (r = 0.26, P = 0.62), contradicting the predictions of the classic Out of Africa model of trait evolution. Greatest within-population phenetic variation in this sample is among the Asian rather than African populations. Additionally, phenetic distance from southern Africa (measured as Procrustes distance) is not correlated with allele-sharing distance from southern Africa (SI Appendix, Fig. S5) (r = 0.03, P = 0.97). However, there are no significant differences between pairwise Procrustes distances (Kruskal–Wallis rank sum test, P = 0.4159), evidence that variation is greatest between individuals rather than between continental populations. These results may be influenced by unequal sample sizes, and increased sampling in future studies will likely contribute to our understanding of global variation in EDJ morphology.
Morphological Variation.
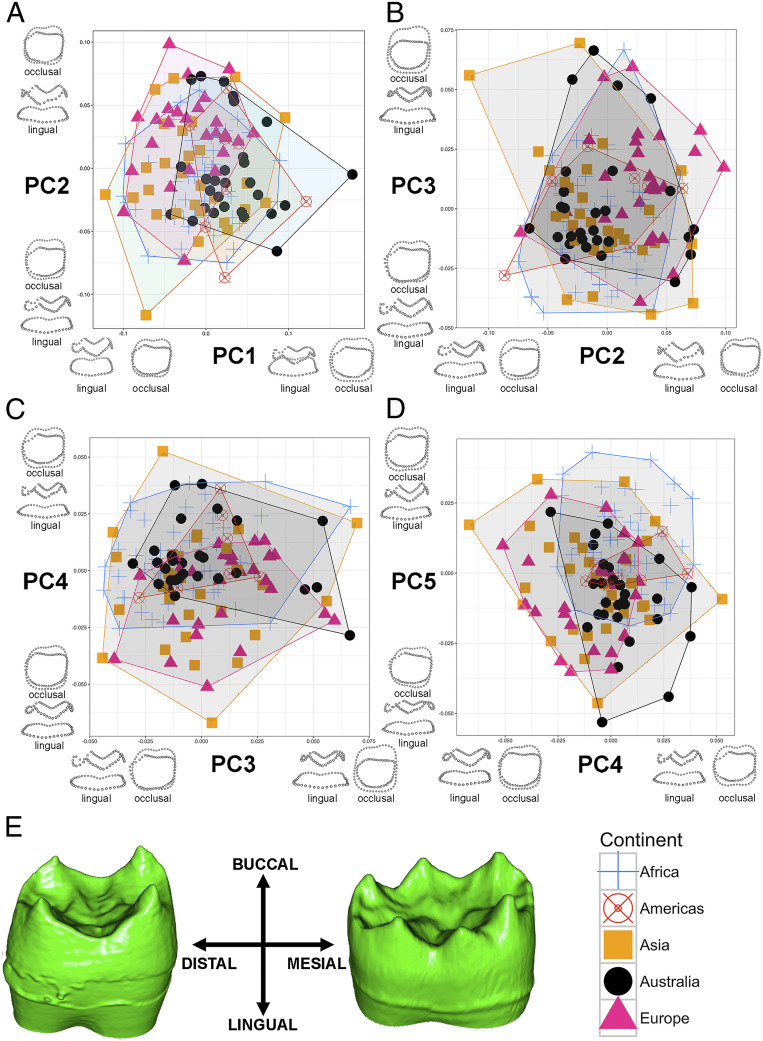
Patterns of EDJ surface anatomy vary significantly across continental populations (Fig. 3) (Procrustes ANOVA, P = 0.001).
Fig. 3.
Comparison of EDJ morphology across continents as captured by PCs derived from Procrustes superimposition of 3D data. Extremes of EDJ shape are shown along the axes. (A) PC1 plotted against PC2, (B) PC2 plotted against PC3, (C) PC3 plotted against PC4, (D) PC4 plotted against PC5. See figure for legend. PC1 comprises 30.2% of the variation, PC2 comprises 19.1% of the variation, PC3 comprises 8.0% of the variation, PC4 comprises 5.2% of the variation, and PC5 comprises 4.2% of the variation. North and South America were combined for analyses due to small sample sizes. Sample sizes: Africa (n = 51), Americas (n = 8), Asia (n = 30), Australia (n = 28), Europe (n = 27). (E) Variation in EDJ morphology of the left mandibular first molar, exemplified by individuals from Europe (Left) and northern Africa (Right). Note the lack of dentine horn associated with the hypoconulid in the European individual. This figure is available without convex hulls as SI Appendix, Fig. S6.
Quantitative and visual analyses demonstrate significant variation in EDJ morphology across continental populations (see SI Appendix for more details). The dominant morphological difference between continental populations is in positioning of the dentine horns that underlie the cusps, particularly the distal cusps, the hypoconid and entoconid, exemplified by Europeans and potentially associated with variable hypoconulid presence and expression (Fig. 3). Variation in EDJ surface morphology, and positioning of the dentine horns relative to one another and to the cervical line, drives the significant results seen in this study.
However, overall, there is greater morphological variation within rather than across continental populations. The greatest morphological variation in human EDJ morphology is in overall cervical height (captured by principal component 1, PC1): That is, the distance from the inferior intersection between cusps to the cervical line (SI Appendix, Fig. S2). PC1 is significantly correlated with centroid size of EDJ morphology, and PC1 captures variation in EDJ cervical height, suggesting that cervical height is likely associated with overall tooth size and therefore body size (P < 0.001) (69–75). Additionally, multivariate regression of common allometric component scores against log centroid size demonstrates a significant allometric relationship between EDJ size and shape (P < 2.2e-16, r = 0.99). In the largest teeth, the dentine crown outline is larger relative to the cervical outline. As crown size is significantly correlated with body size in primates (76); the relationship between centroid size and the relative size of EDJ dentine crown to cervical outline further highlights the effects of allometry on EDJ morphology in humans.
Discussion
From the results of this study, it can be inferred that variation in EDJ morphology is significantly correlated with neutral genetic variation in humans and evolves neutrally, with variation resulting from genetic drift and migration rather than adaptive processes. Based on the nonrandom distribution of EDJ morphology that is correlated with variation in neutral allele frequencies, the evidence presented here supports the hypothesis that EDJ morphology is heritable, or more similar in more closely related individuals. The geographic structure of neutrally evolving traits reflects population movement on a global scale (16, 61–64). As a neutrally evolving trait, EDJ also reflects population movement, as evidenced by significant correlations between genetic and phenetic distances in this study. Patterns of EDJ variation may be useful for interpreting the archaeological record, as more variation likely reflects greater genetic disparity, and samples can be obtained without requiring destructive genetic testing.
Despite evidence for heritability, EDJ morphology does not follow the predictions of an RAO, serial founder, Out of Africa model of dispersal. This may be a result of reversible and repeatable evolution of trait morphology through time. Alternatively, the finding that EDJ morphology does not follow the classic Out of Africa model’s expectations of decreasing variation may not be particularly surprising, as anthropological genetics research continues to demonstrate a high degree of admixture in past human populations (77–80). There is growing support for a model of multiple range expansions out of Africa over the last 100,000 y, although this hypothesis also predicts decreased diversity with increasing distance from Africa (81, 82). The findings here may be explained by increased admixture outside of Africa, with increased diversity potentially resulting from genetic input from extinct hominid groups (60). Additionally, unequal sample sizes may be contributing to the finding of increased variance outside of Africa, as might successive range expansion signatures or predominantly random selective differences. Humans have a complex population history and range. Greater sampling of continental regions, particularly Asia and the Americas, and additional tests of selection and adaptation, will likely further refine our understanding of EDJ variation and patterns of its distribution on a global scale.
There is a strong allometric effect of tooth size on EDJ shape in this study, and there are significant differences in EDJ surface anatomy across continental populations. Tooth size, which is significantly correlated with body size, plays a strong role in dental variation in humans, affecting not only tooth area but also tooth height and cusp orientation (83–87). Given the strong allometric effect of tooth size on EDJ shape, future work focusing on the relationship between body size and dental variation will continue to be illuminating for our understanding of dental development and global patterns of dental variation in modern and archaeological human populations.
Several factors may underlie EDJ variation, including geographic dispersal, neutral and nonneutral evolutionary processes, and allometry. This study clearly demonstrates the role of neutral genetic variation in shaping global patterns of EDJ surface anatomy in humans. But the impact of other factors on EDJ shape, including selection, adaptation, and developmental constraints, remains to be thoroughly quantified in future studies. Thus, future work focusing on these factors, through studies of asymmetry and metameric variation for example, will help illuminate whether EDJ morphology has been under selection in modern humans. This study substantially increases our knowledge of modern human dental variation and contributes to our understanding of dental development more broadly, with taxonomic implications for the hominid fossil record. Additionally, we suggest that EDJ surface anatomy is an excellent trait in which to pursue genotype–phenotype mapping. As sampling EDJ surface anatomy is a noninvasive procedure, this study emphasizes the utility of this trait for investigating large-scale population history and genetic structure in archaeological and fossil populations.
Materials and Methods
High-definition μ-CT biomedical imaging and geometric morphometrics were used to assess EDJ morphological variation in a sample of late Holocene modern humans (n = 161) from archaeological populations (Table 1 and SI Appendix, Table S1). Of the permanent molars, the first molar is least affected by postnatal environmental factors, because the shape of the EDJ is established before secretion of the enamel matrix (17, 31, 88). Given that environmental effects on the topographical morphology of the EDJ of the first molar (M1) are minimal, and based on previous work on external dental anatomy (21, 22), the present study tests several expectations for human M1 EDJ variation. Only individuals with fully developed permanent molar dentition were included in the study. All individuals were μ-CT–scanned using a Nikon industrial CT in Zürich, Switzerland, and Cambridge, United Kingdom. All CT scans were performed using a 225-kV rotating target with average beam energy 210 kV, average beam current 360 µA, average power 80 W, and average isotropic voxel size 0.049 mm3 (or 49 µm3). The 3D scan reconstructions were completed using Nikon CT Pro-3D software. The data volumes were median-filtered to facilitate threshold segmentation and produce 3D models of the EDJ for each individual using software packages VG Studio Pro and Avizo.
Table 1.
Sample sizes of late Holocene individuals included in this study
| Continental area | Region | n | Average dispersal distance, Dd* |
| Africa | Northern Africa | 42 | 6.2 |
| Southern Africa | 9 | 1.95 | |
| Americas | North America | 5 | 23.47 |
| South America | 3 | 43.7 | |
| Asia | Northern Asia | 2 | 19.5 |
| Southern Asia | 10 | 19.35 | |
| Western Asia | 18 | 15.9 | |
| Australia | Australia | 28 | 28.4 |
| Europe | Europe | 27 | 12.08 |
| Unknown | Unknown | 17 | NA |
| Total | 161 |
See SI Appendix, Table S1 for included populations, ID numbers for included individuals, and geological age. n is sample size. Unknown individuals were not included in regional or continental comparisons.
Average dispersal distance in 1,000 km.
The 3D mesh cleaning was performed using Meshmixer 3.4.35 (Autodesk, 2017), and the 3D surface data were then imported into FoRM-IT (Fossil Reconstruction and Morphometry Interactive Toolkit) (89) to quantify EDJ shape using a total of k = 102 3D landmarks (fixed landmarks kf = 7, sliding curve semilandmarks kc = 95) (SI Appendix, Fig. S2).
A Procrustes superimposition of the 7 fixed and 95 sliding landmarks was performed focusing exclusively on the left permanent mandibular first molars. From the Procrustes superimposition, PC scores and Procrustes distances were exported for further analyses. All statistical analyses were conducted in R v.3.2.3 (90). All data and code for this study are available on GitHub (https://github.com/teslamonson/EDJ) (91).
Although this study focuses on nonallometric shape variation in EDJ morphology, a cursory investigation of allometry was completed in the sample with Procrustes regression using the geomorph package (function procD.allometry) (92) and through regression of the common allometric component against log centroid size using the morpho package (function CAC) (93). The common allometric component was assessed across the sample as a whole.
Through quantification of Procrustes-aligned coordinates and principal components scores, variation in EDJ morphology was compared across populations using Procrustes ANOVA with the procD.lm function in the geomorph package (92), as well as through principal component analyses (prcomp) visualized using ggplot2 (94). Pairwise Procrustes distances were statistically compared using a Kruskal–Wallis rank sum test. Samples from North and South America were combined for analyses due to low sample sizes in these regions. All other comparisons were generated at the continental level. Individuals with unknown affiliation (n = 17) were excluded from continental and regional comparisons. More details about the methods used in this study are available in SI Appendix.
Test of Neutral Evolution.
Previous studies have demonstrated that geographical patterns of phenotypic variation reflect population history when that phenotype has been evolving neutrally or without pressures from strong directional selection (15). To evaluate EDJ morphological variation and expectations of neutral evolution, we calculated a phenetic R-matrix using pairwise comparisons of continental mean shape variation generated from Procrustes coordinates. The phenetic R-matrix was statistically compared to a genetic R-matrix previously calculated by Rathmann et al. (21) using bivariate regression and statistical correlation. Positive values indicate continental populations that are more similar to one another. Protocols for calculating the R-matrices are further outlined in SI Appendix.
Tests of the Out of Africa Hypothesis.
Previous work has repeatedly demonstrated the use of geographic distance as a proxy for neutral genetic evolution of certain phenotypes (16, 64). A dispersal distance for each human population was calculated using longitude and latitude and fixing Johannesburg, South Africa, as the point of dispersal (Table 1). Johannesburg is a standard location of origin in sub-Saharan Africa for studies investigating human dispersal. These data were evaluated with an agent-based reproduction-and-diffusion approach. Distances are in kilometers; they are generally larger than great-circle-segment distances but correlate well with them.
Another prediction of neutral evolution in humans is decreasing phenotypic diversity with increasing distance from southern Africa, a result of the impact of bottlenecks and founder effects during episodes of human dispersal (15). To test for neutral evolution using this proxy, the coefficient of variation (CV) was calculated for each continental population using the Procrustes-aligned coordinates. The CVs were used to represent within-population phenetic variation and compared to geographic distance across continental populations (SI Appendix, Table S3). We also statistically compared phenetic distance from southern Africa to population-mean genetic distances calculated from previously published allele-sharing distances between populations (21). Phenetic distance between populations was calculated as group-mean Procrustes distances from southern Africa.
Supplementary Material
Acknowledgments
We thank Marta Mirazón Lahr, Trisha Biers, Madeleine Geiger, and Ket Smithson at the Cambridge Duckworth Collection, and the Department of Anthropology at the University of Zurich for assistance with μ-CT scanning and for facilitating access to museum collections; and May Berenbaum, P. David Polly, Andrew Weitz, Tim White, and two anonymous reviewers for their helpful comments and feedback on earlier versions of this paper. This work was funded by the Swiss National Science Foundation, SNF Grant #CR32I3_166053.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. G.T.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008037117/-/DCSupplemental.
Data Availability.
All data and code for this study are available on GitHub (https://github.com/teslamonson/EDJ) (91).
References
- 1.Hanihara T., Ishida H., Metric dental variation of major human populations. Am. J. Phys. Anthropol. 128, 287–298 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Hlusko L. J., Sage R. D., Mahaney M. C., Modularity in the mammalian dentition: Mice and monkeys share a common dental genetic architecture. J. Exp. Zoolog. B Mol. Dev. Evol. 316, 21–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hlusko L. J., Schmitt C. A., Monson T. A., Brasil M. F., Mahaney M. C., The integration of quantitative genetics, paleontology, and neontology reveals genetic underpinnings of primate dental evolution. Proc. Natl. Acad. Sci. U.S.A. 113, 9262–9267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jheon A. H., Seidel K., Biehs B., Klein O. D., From molecules to mastication: The development and evolution of teeth. Wiley Interdiscip. Rev. Dev. Biol. 2, 165–182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joganic J. L. et al., Additive genetic variation in the craniofacial skeleton of baboons (genus Papio) and its relationship to body and cranial size. Am. J. Phys. Anthropol. 165, 269–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monson T. A. et al., Evidence of strong stabilizing effects on the evolution of boreoeutherian (Mammalia) dental proportions. Ecol. Evol. 9, 7597–7612 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponce de León M. S. et al., Human bony labyrinth is an indicator of population history and dispersal from Africa. Proc. Natl. Acad. Sci. U.S.A. 115, 4128–4133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betti L., Manica A., Human variation in the shape of the birth canal is significant and geographically structured. Proc. Biol. Sci. 285, 20181807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betti L., Balloux F., Amos W., Hanihara T., Manica A., Distance from Africa, not climate, explains within-population phenotypic diversity in humans. Proc. Biol. Sci. 276, 809–814 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betti L., von Cramon-Taubadel N., Manica A., Lycett S. J., Global geometric morphometric analyses of the human pelvis reveal substantial neutral population history effects, even across sexes. PLoS One 8, e55909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanihara T., Morphological variation of major human populations based on nonmetric dental traits. Am. J. Phys. Anthropol. 136, 169–182 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Harvati K., Weaver T. D., Human cranial anatomy and the differential preservation of population history and climate signatures. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 288, 1225–1233 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagani L. et al., Genomic analyses inform on migration events during the peopling of Eurasia. Nature 538, 238–242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prugnolle F., Manica A., Balloux F., Geography predicts neutral genetic diversity of human populations. Curr. Biol. 15, R159–R160 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Relethford J. H., Global patterns of isolation by distance based on genetic and morphological data. Hum. Biol. 76, 499–513 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Bei M., Molecular genetics of tooth development. Curr. Opin. Genet. Dev. 19, 504–510 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillson S., Dental Anthropology, (Cambridge University Press, 1996). [Google Scholar]
- 19.Hughes T. E., Townsend G. C., “Twin and family studies of human dental crown morphology: Genetic, epigenetic, and environmental determinants of the modern human dentition” in Anthropological Perspectives on Tooth Morphology: Genetics, Evolution, Variation, Scott G., Irish J., Eds. (Cambridge University Press, 2013), pp. 31–68. [Google Scholar]
- 20.Townsend G. C., Brown T., Heritability of permanent tooth size. Am. J. Phys. Anthropol. 49, 497–504 (1978). [DOI] [PubMed] [Google Scholar]
- 21.Rathmann H. et al., Reconstructing human population history from dental phenotypes. Sci. Rep. 7, 12495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathmann H., Reyes-Centeno H., Testing the utility of dental morphological trait combinations for inferring human neutral genetic variation. Proc. Natl. Acad. Sci. U.S.A. 117, 10769–10777 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polly P. D., On morphological clocks and paleophylogeography: Towards a timescale for Sorex hybrid zones. Genetica 112-113, 339–357 (2001). [PubMed] [Google Scholar]
- 24.Polly P. D., Paleophylogeography of Sorex araneus: Molar shape as a morphological marker for fossil shrews. Mammalia 68, 233–243 (2003). [Google Scholar]
- 25.Monson T. A., Hlusko L. J., Identification of a derived dental trait in the papionini relative to other Old World monkeys. Am. J. Phys. Anthropol. 155, 422–429 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Monson T. A., Hlusko L. J., The evolution of dental eruption sequence in artiodactyls. J. Mamm. Evol. 25, 15–26 (2018). [Google Scholar]
- 27.Monson T. A., Hlusko L. J., Breaking the rules: Phylogeny, not life history, explains dental eruption sequence in primates. Am. J. Phys. Anthropol. 167, 217–233 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Scott G. R., Turner C. G. II, Townsend G. C., Martinón-Torres M., The Anthropology of Modern Human Teeth: Dental Morphology and its Variation in Recent and Fossil Homo Sapiens, (Cambridge University Press, 2018), vol. 79. [Google Scholar]
- 29.Smith B. H., Dental development as a measure of life history in primates. Evolution 43, 683–688 (1989). [DOI] [PubMed] [Google Scholar]
- 30.Smith T. M., The Tales Teeth Tell: Development, Evolution, Behavior, (MIT Press, 2018). [Google Scholar]
- 31.Thesleff I., From understanding tooth development to bioengineering of teeth. Eur. J. Oral Sci. 126, 67–71 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Ungar P. S., “Tooth form and function: Insights into adaptation through the analysis of dental microwear” in Comparative Dental Morphology, Koppe T., Meyer G., Alt K. W., Eds. (Karger Publishers, 2009), vol. 13, pp. 38–43. [DOI] [PubMed] [Google Scholar]
- 33.Ungar P. S., Evolution’s Bite: A Story of Teeth, Diet, and Human Origins, (Princeton University Press, 2017). [Google Scholar]
- 34.Martin R. M. G., Hublin J. J., Gunz P., Skinner M. M., The morphology of the enamel-dentine junction in Neanderthal molars: Gross morphology, non-metric traits, and temporal trends. J. Hum. Evol. 103, 20–44 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Skinner M. M., Wood B. A., Hublin J. J., Protostylid expression at the enamel-dentine junction and enamel surface of mandibular molars of Paranthropus robustus and Australopithecus africanus. J. Hum. Evol. 56, 76–85 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Ortiz A., Bailey S. E., Hublin J. J., Skinner M. M., Homology, homoplasy and cusp variability at the enamel-dentine junction of hominoid molars. J. Anat. 231, 585–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith T. M., Olejniczak A. J., Reid D. J., Ferrell R. J., Hublin J. J., Modern human molar enamel thickness and enamel-dentine junction shape. Arch. Oral Biol. 51, 974–995 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Macchiarelli R., Bayle P., Bondioli L., Mazurier A., Zanolli C., “From outer to inner structural morphology in dental anthropology: Integration of the third dimension in the visualization and quantitative analysis of fossil remains” in Anthropological Perspectives on Tooth Morphology: Genetics, Evolution, Variation, Scott G., Irish J., Eds. (Cambridge University Press, 2013), pp. 250–277. [Google Scholar]
- 39.Olejniczak A. J. et al., Morphology of the enamel-dentine junction in sections of anthropoid primate maxillary molars. J. Hum. Evol. 53, 292–301 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Morita W., Morphological comparison of the enamel–dentine junction and outer enamel surface of molars using a micro-computed tomography technique. J. Or. Biosci. 58, 95–99 (2016). [Google Scholar]
- 41.Morita W., Morimoto N., Kono R. T., Suwa G., Metameric variation of upper molars in hominoids and its implications for the diversification of molar morphogenesis. J. Hum. Evol. 138, 102706 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Schwartz G. T., Thackeray J. F., Reid C., van Reenan J. F., Enamel thickness and the topography of the enamel-dentine junction in South African Plio-Pleistocene hominids with special reference to the Carabelli trait. J. Hum. Evol. 35, 523–542 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Skinner M. M., Gunz P., Wood B. A., Boesch C., Hublin J. J., Discrimination of extant Pan species and subspecies using the enamel-dentine junction morphology of lower molars. Am. J. Phys. Anthropol. 140, 234–243 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Braga J. et al., The enamel-dentine junction in the postcanine dentition of Australopithecus africanus: Intra-individual metameric and antimeric variation. J. Anat. 216, 62–79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita W. et al., Patterns of morphological variation in enamel-dentin junction and outer enamel surface of human molars. J. Anat. 224, 669–680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinón-Torres M. et al., Talonid crests expression at the enamel–dentine junction of hominin lower permanent and deciduous molars. C. R. Palevol 13, 223–234 (2014). [Google Scholar]
- 47.Pan L. et al., Intra-individual metameric variation expressed at the enamel-dentine junction of lower post-canine dentition of South African fossil hominins and modern humans. Am. J. Phys. Anthropol. 163, 806–815 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Skinner M. M. et al., Dental trait expression at the enamel-dentine junction of lower molars in extant and fossil hominoids. J. Hum. Evol. 54, 173–186 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Guy F., Lazzari V., Gilissen E., Thiery G., To what extent is primate second molar enamel occlusal morphology shaped by the enamel-dentine junction? PLoS One 10, e0138802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skinner M. M. et al., Brief communication: Contributions of enamel-dentine junction shape and enamel deposition to primate molar crown complexity. Am. J. Phys. Anthropol. 142, 157–163 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Selig K. R., López-Torres S., Sargis E. J., Silcox M. T., First 3D dental topographic analysis of the enamel-dentine junction in non-primate euarchontans: Contribution of the enamel-dentine junction to molar morphology. J. Mamm. Evol. 26, 587–598 (2019). [Google Scholar]
- 52.Jernvall J., Thesleff I., Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 92, 19–29 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Kono R. T., Molar enamel thickness and distribution patterns in extant great apes and humans: New insights based on a 3-dimensional whole crown perspective. Anthropol. Sci. 112, 121–146 (2004). [Google Scholar]
- 54.Lucas P., Constantino P., Wood B., Lawn B., Dental enamel as a dietary indicator in mammals. Bioessays 30, 374–385 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Macho G. A., Berner M. E., Enamel thickness of human maxillary molars reconsidered. Am. J. Phys. Anthropol. 92, 189–200 (1993). [DOI] [PubMed] [Google Scholar]
- 56.Martin L., Significance of enamel thickness in hominoid evolution. Nature 314, 260–263 (1985). [DOI] [PubMed] [Google Scholar]
- 57.Schwartz G. T., Taxonomic and functional aspects of the patterning of enamel thickness distribution in extant large-bodied hominoids. Am. J. Phys. Anthropol. 111, 221–244 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Hlusko L. J., Suwa G., Kono R. T., Mahaney M. C., Genetics and the evolution of primate enamel thickness: A baboon model. Am. J. Phys. Anthropol. 124, 223–233 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Hlusko L. J., Expression types for two cercopithecoid dental traits (interconulus and interconulid) and their variation in a modern baboon population. Int. J. Primatol. 23, 1309–1318 (2002). [Google Scholar]
- 60.Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E., Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16, 359–371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crow J. F., Kimura M., An Introduction to Population Genetics Theory, (Burgess, 1970). [Google Scholar]
- 62.Malécot G., The Mathematics of Heredity, (Freeman, 1991). [Google Scholar]
- 63.Morton N. E., “Isolation by distance” in Genetic Structure of Populations, Morton N. E., Ed. (University of Hawaii Press, 1973), pp. 76–77. [Google Scholar]
- 64.Ramachandran S. et al., Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. U.S.A. 102, 15942–15947 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stringer C. B., Andrews P., Genetic and fossil evidence for the origin of modern humans. Science 239, 1263–1268 (1988). [DOI] [PubMed] [Google Scholar]
- 66.Excoffier L., Foll M., Petit R. J., Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 40, 481–501 (2009). [Google Scholar]
- 67.Ibrahim K. M., Nichols R. A., Hewitt G. M., Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity 77, 282–291 (1996). [Google Scholar]
- 68.Slatkin M., Excoffier L., Serial founder effects during range expansion: A spatial analog of genetic drift. Genetics 191, 171–181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caumul R., Polly P. D., Phylogenetic and environmental components of morphological variation: Skull, mandible, and molar shape in marmots (Marmota, Rodentia). Evolution 59, 2460–2472 (2005). [PubMed] [Google Scholar]
- 70.Creighton G. K., Static allometry of mammalian teeth and the correlation of tooth size and body size in contemporary mammals. J. Zool. 191, 435–443 (1980). [Google Scholar]
- 71.Gingerich P. D., Correlation of tooth size and body size in living hominoid primates, with a note on relative brain size in Aegyptopithecus and Proconsul. Am. J. Phys. Anthropol. 47, 395–398 (1977). [DOI] [PubMed] [Google Scholar]
- 72.Gingerich P. D., Schoeninger M. J., Patterns of tooth size variability in the dentition of primates. Am. J. Phys. Anthropol. 51, 457–465 (1979). [DOI] [PubMed] [Google Scholar]
- 73.Gingerich P. D., Smith B. H., Rosenberg K., Allometric scaling in the dentition of primates and prediction of body weight from tooth size in fossils. Am. J. Phys. Anthropol. 58, 81–100 (1982). [DOI] [PubMed] [Google Scholar]
- 74.Goldstein S., Post D., Melnick D., An analysis of cercopithecoid odontometrics. I. The scaling of the maxillary dentition. Am. J. Phys. Anthropol. 49, 517–532 (1978). [DOI] [PubMed] [Google Scholar]
- 75.Gould S. J., On the scaling of tooth size in mammals. Am. Zool. 15, 351–362 (1975). [Google Scholar]
- 76.Hlusko L. J., Lease L. R., Mahaney M. C., Evolution of genetically correlated traits: Tooth size and body size in baboons. Am. J. Phys. Anthropol. 131, 420–427 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Haber M. et al., Chad genetic diversity reveals an African history marked by multiple Holocene Eurasian migrations. Am. J. Hum. Genet. 99, 1316–1324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammer M. F., Woerner A. E., Mendez F. L., Watkins J. C., Wall J. D., Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. U.S.A. 108, 15123–15128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Llorente M. G. et al., Ancient Ethiopian genome reveals extensive Eurasian admixture throughout the African continent. Science 350, 820–822 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Slatkin M., Racimo F., Ancient DNA and human history. Proc. Natl. Acad. Sci. U.S.A. 113, 6380–6387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Groucutt H. S., Petraglia M. D., The prehistory of the Arabian peninsula: Deserts, dispersals, and demography. Evol. Anthropol. 21, 113–125 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Gunz P. et al., Early modern human diversity suggests subdivided population structure and a complex out-of-Africa scenario. Proc. Natl. Acad. Sci. U.S.A. 106, 6094–6098 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biggerstaff R. H., Cusp size sexual dimorphism, and the heritability of maxillary molar cusp size in twins. J. Dent. Res. 55, 189–195 (1976). [DOI] [PubMed] [Google Scholar]
- 84.Hunter J. P., Guatelli-Steinberg D., Weston T. C., Durner R., Betsinger T. K., Model of tooth morphogenesis predicts Carabelli cusp expression, size, and symmetry in humans. PLoS One 5, e11844 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koh C. et al., Genetic integration of molar cusp size variation in baboons. Am. J. Phys. Anthropol. 142, 246–260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kondo S., Townsend G. C., Associations between Carabelli trait and cusp areas in human permanent maxillary first molars. Am. J. Phys. Anthropol. 129, 196–203 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Wolpoff M. H., “Tooth size–body size scaling in a human population” in Size and Scaling in Primate Biology, Jungers W., Ed. (Springer, 1985), pp. 273–318. [Google Scholar]
- 88.Butler P. M., Relative growth within the human first upper permanent molar during the prenatal period. Arch. Oral Biol. 12, 983–992 (1967). [DOI] [PubMed] [Google Scholar]
- 89.Zollikofer C. P. E., de León M. S. P., Tools for rapid prototyping in the biosciences. IEEE Comput. Graph. Appl. 15, 48–55 (1995). [Google Scholar]
- 90.R Core Team , R: A language and environment for statistical computing, (R Foundation for Statistical Computing, Vienna, 2016). [Google Scholar]
- 91.Monson T. A., Fecker D., Scherrer M., EDJ. GitHub. https://github.com/teslamonson/EDJ. Deposited 30 July 2020.
- 92.Adams D., Collyer M., Kaliontzopoulou A., Geomorph: Software for geometric morphometric analyses. R package version 3.1.1, (R Foundation for Statistical Computing, Vienna, 2019). [Google Scholar]
- 93.Schlager S., “Morpho and Rvcg—Shape analysis in R” in Statistical Shape and Deformation Analysis, Zheng G., Li S., Szekely G., Eds. (Academic Press, 2017), pp. 217–256. [Google Scholar]
- 94.Wickham H., ggplot2: Elegant Graphics for Data Analysis, (Springer-Verlag, 2016). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and code for this study are available on GitHub (https://github.com/teslamonson/EDJ) (91).