Abstract
Objectives
COVID-19 has been arguably the most important public health concern worldwide in 2020, and efforts are now escalating to suppress or eliminate its spread. In this study we undertook a meta-analysis to estimate the global and regional seroprevalence rates in humans of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and to assess whether seroprevalence is associated with geographical, climatic and/or sociodemographic factors.
Methods
We systematically reviewed PubMed, Scopus, Embase, medRxiv and bioRxiv databases for preprints or peer-reviewed articles (up to 14 August 2020). Study eligibility criteria were population-based studies describing the prevalence of anti-SARS-CoV-2 (IgG and/or IgM) serum antibodies. Participants were people from different socioeconomic and ethnic backgrounds (from the general population), whose prior COVID-19 status was unknown and who were tested for the presence of anti-SARS-CoV-2 serum antibodies. We used a random-effects model to estimate pooled seroprevalence, and then extrapolated the findings to the global population (for 2020). Subgroup and meta-regression analyses explored potential sources of heterogeneity in the data, and relationships between seroprevalence and sociodemographic, geographical and/or climatic factors.
Results
In total, 47 studies involving 399 265 people from 23 countries met the inclusion criteria. Heterogeneity (I2 = 99.4%, p < 0.001) was seen among studies; SARS-CoV-2 seroprevalence in the general population varied from 0.37% to 22.1%, with a pooled estimate of 3.38% (95%CI 3.05–3.72%; 15 879/399 265). On a regional level, seroprevalence varied from 1.45% (0.95–1.94%, South America) to 5.27% (3.97–6.57%, Northern Europe), although some variation appeared to relate to the serological assay used. The findings suggested an association of seroprevalence with income levels, human development indices, geographic latitudes and/or climate. Extrapolating to the 2020 world population, we estimated that 263.5 million individuals had been exposed or infected at the time of this study.
Conclusions
This study showed that SARS-CoV-2 seroprevalence varied markedly among geographic regions, as might be expected early in a pandemic. Longitudinal surveys to continually monitor seroprevalence around the globe will be critical to support prevention and control efforts, and might indicate levels of endemic stability or instability in particular countries and regions.
Keywords: COVID-19, General population, Global seroprevalence, Meta-analysis, SARS-CoV-2, Serum antibodies (IgG and/or IgM), Subgroup analyses
Introduction
Coronavirus 19 disease (COVID-19)—a severe, acute respiratory syndrome caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—was first identified in Wuhan, China, in December 2019 [1,2], and spread within months to most nations of the world [3]. By 16th August 2020, this pandemic disease was affecting people in 213 countries and territories, with about 21 million confirmed cases and around 800,000 deaths reported globally [4]. The diagnosis and management of COVID-19 are based on the detection of SARS-CoV-2 in nasopharyngeal swabs from patients presenting with clinical signs (including fever, dry cough and/or shortness of breath), or in suspected cases, by reverse transcription polymerase chain reaction (RT-PCR) [5,6]. Since the manifestation of SARS-CoV-2 infection ranges from asymptomatic to fatal, the surveillance of confirmed COVID-19 cases might not be representative for a particular community [7,8]. Although RT-PCR is currently recognized as ‘reference standard’ for the diagnosis of SARS-CoV-2 infection [5], a significant number of asymptomatic or subclinically infected individuals is likely to remain undetected. Therefore, it is plausible or likely that the actual number of people exposed to, or infected with, is underestimated [[7], [8], [9]]. Serological screening represents a critical adjunct to PCR-based detection/diagnosis and is a key tool to evaluate the cumulative prevalence of SARS-CoV-2 infection, and to monitor seroconversion [10] and seroreversion [11,12] in individuals and a community; such screening is useful to gain insight into the dynamics of specific antibody responses during and after the spread of the virus and, if undertaken routinely, to inform health authorities, politicians and policy-makers about seroprevalence at any given stage during an epidemic [13,14]. The prevalence of specific serum antibodies (IgG and/or IgM) against SARS-CoV-2 can provide a sound indication of exposure to SARS-CoV-2 in a population [7,9]. Due to an apparent persistence of antibodies to SARS-CoV-2 (particularly IgG) after viral clearance [7], it is expected that serological monitoring and surveillance provide relevant datasets to estimate the cumulative prevalence of SARS-CoV-2 infection/exposure in a population [7,15], and may even indicate the immune status of individuals or populations [8,9].
Several commercial and in-house immunoassays are being used for the detection of IgG and/or IgM serum antibodies to SARS-CoV-2; these are mainly enzyme-linked immunosorbent assays (ELISAs), chemiluminescence immunoassays (CLIAs) or lateral flow assays (LFIAs) [16,17]. The diagnostic specificity and sensitivity of these methods vary and depend on the use of recombinant or purified protein antigens—e.g. spike (S), envelope (E), membrane (M), nucleocapsid (N) or receptor binding domain (RBD) proteins—and the rigor of assay optimization [18,19].
Since April 2020, sero-epidemiological studies have been reported from a number of countries most affected by COVID-19, including Brazil, China, France, Germany, Iran, Italy, Spain, England and the USA [9,[20], [21], [22], [23], [24], [25], [26], [27]]. As the pandemic spreads, it is crucial that a rapid and thorough analysis be undertaken to estimate global seroprevalence at a moment in time. In this study, 6 months after the commencement of the pandemic, we undertook a meta-analysis to estimate the global and regional seroprevalences of SARS-CoV-2 in people of the general population (whose prior COVID-19 status was unknown), and assessed whether geographical, climatic and sociodemographic factors impact on seroprevalence.
Methods
Search strategy and selection criteria
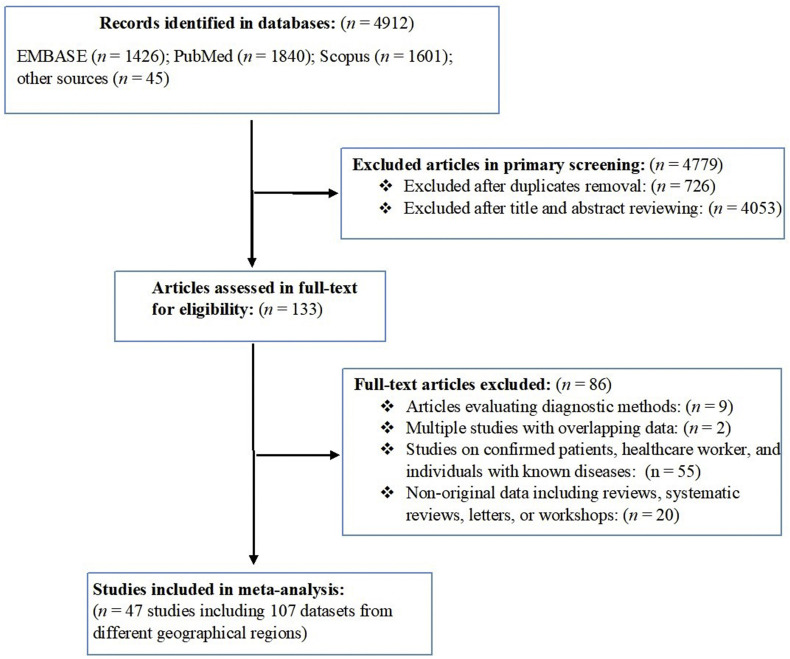
This study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (cf. Fig. 1 ). We performed a systematic literature search in the databases PubMed, Scopus, Embase, medRxiv and bioRxiv in August 2020 using the following terms: “SARS-CoV-2”, “COVID-19”, “coronavirus”, “antibody”, “ELISA”, “seroprevalence” and “population”, without language or geographical restriction (Supplementary Material Fig. S1). Additional related articles were retrieved manually from Google Scholar and critically evaluated. All articles were imported to Endnote software X8 (Thompson and Reuters, Philadelphia, USA), and duplicates were removed. Two independent reviewers (AR, MS) studied all titles and abstracts for eligibility. Included were all peer-reviewed population-based studies, preprints, and research reports which reported the prevalence of anti-SARS-CoV-2 serum antibodies in the ‘general population’ (i.e. randomly selected people of different ages, occupations, educational and ethnic backgrounds, and socioeconomic status, living in a defined geographical region, whose prior COVID-19 status was unknown). Articles were excluded if they (a) involved suspected, confirmed or hospitalized COVID-19 patients, (b) were performed in at-risk populations (e.g. healthcare workers) or individuals with known diseases (e.g. cancer or dialysis patients), (c) recorded prevalence based on clinical manifestation, computed tomography scan or PCR, (d) were comparative studies of diagnostic methods, (e) used datasets that overlapped with those of other articles, (f) were case reports or case studies, or (g) were editorials, commentaries, reviews or systematic reviews.
Fig. 1.
Search strategy and study selection process, indicating numbers of studies (and associated datasets) excluded or included.
Extraction of data and quality evaluation
After the screening of published articles for eligibility, relevant data and information from each eligible study were entered into a specific form in Microsoft Excel (version 2016; Microsoft Corporation, Redmond, USA). Two co-authors (AR and MNS) independently collated data from all eligible studies, and two (MS and SE) independently evaluated these data. Any inconsistencies were discussed and a consensus decision was made. The following items were obtained from each study (if described): primary author; publication year; country; city; study design and period; type of serological methods used; sensitivity and specificity of diagnostic methods; number of people screened; the number of people seropositive for SARS-CoV-2 antibodies; and data regarding age, sex and ethnicity.
All geographical areas (i.e. cities and countries) investigated were classified according to ‘Sustainable Development Goal’ (SDG) regions or subregions defined by the United Nations [28]. For individual countries, we recorded information on the total numbers of confirmed cases and deaths (up to 15th August 2020) reported by the World Health Organization (WHO) [29], World Bank's income category [30], gross national income per capita [31], and the human development index (HDI) [32]. Furthermore, we recorded total global, regional and national populations (both sexes combined) in 2020, estimated by the United Nations [33]. If sample size(s) and the numbers of seropositive people were specified in studies, we extracted and critically appraised data for separate geographic regions. We also recorded latitude, longitude, mean relative humidity, and mean environmental temperature in geographic regions/subregions during the study period using the database timeanddate.com (weblink: https://www.timeanddate.com). The quality of studies included in the meta-analysis was assessed using the Joanna Briggs Institute (JBI) Prevalence Critical Appraisal Tool [34]. Individual articles were assessed as to whether they adequately described the following: sample collection, recruitment method, subjects and the setting, number of subjects, information on subjects, results, reliability of results, statistical analysis method(s), subpopulation analysis and confounder adjustment (‘yes’ or ‘no’ answer). For each study, the number of ‘yes’ answers to these ten criteria was counted; the higher the number of ‘yes’ answers, the lower the risk of bias in a study.
Meta-analysis
All analyses were carried out using Stata statistical software (v.13 Stata Corp., College Station, TX, USA). To conservatively estimate the pooled seroprevalence of SARS-CoV-2 in the general population, we used a DerSimonian and Laird random-effects model (REM) [35]. For this purpose, first, we estimated the seroprevalence in individual countries by synthesizing the seroprevalence rates of all studies from the same country, and then we calculated the seroprevalences of SARS-CoV-2 for the WHO-defined regions (if studies were available for at least two countries) by synthesizing the data for countries within the same SDG region. We calculated the pooled seroprevalence rates at a 95% confidence interval (CI) using the ‘metaprop’ command in Stata software. We estimated heterogeneity using the I 2 statistic; an I 2 >75% and p < 0.05 were considered to represent substantial heterogeneity [36]. To estimate the number of people exposed to SARS-CoV-2, we extrapolated seroprevalence estimates to the total human population (in 2020) living in a country and a region according to the UN Population Division [28].
To explore possible sources of heterogeneity and also effects of sociodemographic, geographical and climatic parameters on SARS-CoV-2 seroprevalence, we undertook several subgroup analyses by REM as well as random effects meta-regression ecological analyses using the ‘metareg’ command in Stata [37]. These analyses were performed considering the following: SDG regions; serological method used; age, sex and ethnicity of people; country income level, country HDI; latitude, longitude; mean environmental temperature; mean relative humidity; and time during the pandemic. To assess the effect of these variables on seroprevalence, we carried out random effects meta-regression analyses using the ‘metareg’ command in Stata [37]. Further meta-regression analyses were performed to assess whether seroprevalence was associated with the total number of confirmed cases or deaths in individual countries. As publication bias is not relevant for prevalence studies [38], it was not assessed. Results were considered as statistically significant if p < 0.1.
Results
Study characteristics
Our search of electronic databases identified a total of 4912 articles; following the removal of duplicate articles and a critical appraisal of article titles and abstracts, 133 potentially relevant articles were identified for full-text evaluation (Fig. 1). After applying the eligibility criteria, 47 articles were included in the quantitative synthesis; these 47 eligible articles contained 107 datasets representing 399 265 people from 23 countries in six SDG regions. Of these datasets, 74 were from Europe and Northern America, 17 from Latin America and the Caribbean, 13 from Eastern and South-Eastern Asia, one from Central and Southern Asia, one from North Africa and Western Asia, and one from Sub-Saharan Africa. We did not identify a published study from Oceania. Information on the studies included is provided in the Supplementary Material Table S1. Most articles included (44 studies) had a low risk of bias (score: 7–10/10), and only three studies had a moderate risk (6/10) of bias (Table 2 ).
Table 2.
Prevalence of serum antibodies to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the general population according to a priori defined subgroups
| Variable/subgroups | Number of datasets | Number of people screened (total) | Number of seropositive people | Pooled seroprevalence % (95%CI) |
|---|---|---|---|---|
| Gender | ||||
| Male | 29 | 145 368 | 6186 | 5.33 (4.35–6.31) |
| Female | 29 | 151 790 | 6958 | 5.05 (4.06–6.04) |
| Age | ||||
| ≤19 | 11 | 18 333 | 535 | 2.28 (1.01–3.56) |
| 20–49 | 15 | 96 109 | 4268 | 3.22 (1.90–4.55) |
| 50–64 | 15 | 75 589 | 3769 | 2.98 (1.59–4.36) |
| ≥65 | 12 | 41 421 | 1634 | 2.57 (1.39–3.76) |
| Type of population | ||||
| General | 68 | 227 428 | 6483 | 2.43 (2.16–2.70) |
| General adult | 18 | 169 016 | 9201 | 5.31 (4.12–6.50) |
| General children | 2 | 1821 | 162 | 8.76 (7.46–10.06) |
| Serological method | ||||
| LFIA | 58 | 224 922 | 10 023 | 3.95 (3.17-4.74) |
| ELISA | 23 | 38 159 | 1417 | 3.53 (2.65–4.40) |
| CLIA | 15 | 80 435 | 1907 | 2.73 (2.03–3.42) |
| Virus neutralisation assay | 10 | 40 648 | 645 | 1.32 (0.90–1.74) |
| Microsphere immunoassay | 1 | 15 101 | 1887 | 12.50 (11.97–13.03) |
| Type of procedure | ||||
| Commercial kit | 83 | 334 334 | 13 870 | 3.33 (2.95–3.71) |
| In-house | 24 | 64 931 | 2009 | 3.63 (2.79–4.48) |
| Race/ethnicity | ||||
| White, non-Hispanic | 7 | 114 544 | 5662 | 3.76 (1.43–6.08) |
| Black, non-Hispanic | 7 | 7287 | 649 | 9.96 (2.95–16.97) |
| Brown/Hispanic | 7 | 14 347 | 1016 | 8.76 (0.01–18.65) |
| Multiple race/Asian/other/unknown | 7 | 8139 | 709 | 5.78 (1.76–9.79) |
LFIA, lateral flow immunoassay; ELISA, enzyme-linked immunosorbent assay; CLIA, chemiluminescence immunoassay.
SARS-CoV-2 seroprevalence
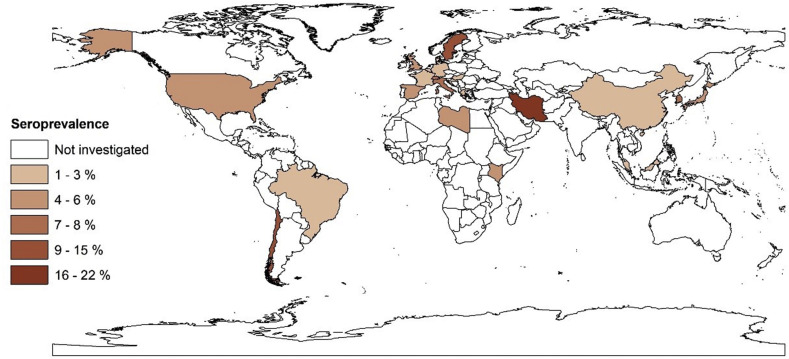
Analysis of the 107 datasets selected from the 47 articles showed that 15 879 people from a general population of 399 265 had specific serum antibodies to SARS-CoV-2, indicating a pooled seroprevalence of 3.38% (95%CI 3.05–3.72%). Significant heterogeneity (I 2 = 99.4%, p < 0.001) was seen among studies. An extrapolation to the global population (2020) indicated that ~263.5 million (range: 237 741 369 to 289 966 523) people had been exposed to SARS-CoV-2 (14th July 2020). More details on the overall and regional SARS-CoV-2 seroprevalences and burdens are given in Table 1. According to SDG subregions (for which two or more countries were represented), seroprevalences were: 5.27% (3.97–6.57%) in Northern Europe; 4.41% (2.20–6.61%) in Southern Europe; 4.41% (3.03–5.79%) in Northern America; 3.17% (1.96–4.38%) in Western Europe; 2.02% (1.56–2.49%) in Eastern Asia; and 1.45% (0.95–1.94%) in South America. Countries with the highest seroprevalences were Iran (22.1%), Sweden (15.02%), Chile (10.7%), Switzerland (7.9%), Italy (7.27%), South Korea (7.5%), Spain (5.0%) and the USA (4.4%). Fig. 2 shows the SARS-CoV-2 seroprevalence estimates for individual countries.
Table 1.
Global, regional and national pooled prevalence of serum antibodies to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the general population (results from 47 studies containing 107 datasets performed in 23 countries)
| WHO regions/country | Number datasets | Number of people screened (total) | Number of seropositive people | Pooled seroprevalence % (95%CI) |
Estimated global or country's population (2020) | Estimated number of people exposed to SARS-CoV-2 (95%CI) |
|---|---|---|---|---|---|---|
| Global | 107 | 399 265 | 15 879 | 3.38 (3.05–3.72) | 7 794 799 000 | 263 565 606 (237 741 369–289 966 523) |
| Europe and northern America | 74 | 272 265 | 13 109 | 4.21 (3.52–4.90) | 1 116 506 000 | 47 004 902 (39 301 011–54 708 794) |
| Northern America | 22 | 51 544 | 3146 | 4.41 (3.03–5.79) | 368 870 000 | 16 267 167 (11 176 761–21 357 573) |
| United states | 22 | 51 544 | 3146 | 4.41 (3.03–5.79) | 331 003 000 | 14 597 232 (10 029 390–19 165 073) |
| Western Europe | 13 | 16 933 | 658 | 3.17 (1.96–4.38) | 196 146 000 | 6 217 828 (3 844 461–8 591 194) |
| Belgium | 2 | 7391 | 293 | 3.46 (3.04–3.88) | 11 590 000 | 401 014 (352 336–449 692) |
| France | 5 | 1198 | 30 | 2.19 (1.20–3.18) | 65 274 000 | 1 429 500 (783 288–2 075 713) |
| Germany | 4 | 3806 | 81 | 2.23 (0.79–3.67) | 83 784 000 | 1 868 388 (661 893–3 074 872) |
| Switzerland | 1 | 2766 | 219 | 7.92 (6.94–8.99) | 8 655 000 | 685 476 (600,657–778,084) |
| Luxembourg | 1 | 1862 | 35 | 1.88 (1.31–2.60) | 626 000 | 11 768 000 (8 200–16 276) |
| Southern Europe | 26 | 71 478 | 3242 | 4.41 (2.20–6.61) | 152 215 000 | 6 712 681 (3 348 730–10 061 411) |
| Croatia | 2 | 1494 | 19 | 1.05 (0.56–1.60) | 4 105 000 | 43 102 (22 988–65 680) |
| Italy | 4 | 2323 | 145 | 7.27 (2.48–11.9) | 60 462 000 | 4 395 587 (1 499 457–7 249 393) |
| Spain | 19 | 61 075 | 3054 | 5.01 (4.83–5.18) | 46 755 000 | 2 342 425 (2 258 266–2 421 909) |
| Greece | 1 | 6586 | 24 | 0.36 (0.23–0.54) | 10 423 000 | 37 522 (23 972–56 284) |
| Eastern Europe | 1 | 10 474 | 69 | 0.66 (0.51–0.83) | 293 013 000 | 1 933 885 (1 494 366–2 432 007) |
| Hungary | 1 | 10 474 | 69 | 0.66 (0.51–0.83) | 9 660 000 | 63 756 (49 266–80 178) |
| Northern Europe | 12 | 121 836 | 5994 | 5.27 (3.97-6.57) | 106 261 000 | 5 599 954 (4 218 561–6 981 347) |
| England | 9 | 99 908 | 5544 | 5.65 (4.61–6.69) | 67 886 000 | 3 835 559 (3 129 544–4 541 573) |
| Denmark | 2 | 21 715 | 418 | 1.77 (1.60–1.95) | 5 792 000 | 102 518 (92 672–112 944) |
| Sweden | 1 | 213 | 32 | 15.0 (10.5–20.5) | 10 099 000 | 1 516 869 (1 061 405–2 074 334) |
| Eastern and south-eastern Asia | 13 | 89 648 | 1855 | 2.02 (1.56–2.49) | 2 346 709 000 | 47 403 521 (36 608 660- 58 433 054) |
| Eastern Asia | 12 | 88 832 | 1852 | 2.02 (1.56–2.49) | 1 678 090 000 | 33 897 418 (26 178 204- 41 784 441) |
| China | 8 | 86 416 | 1756 | 1.63 (1.13–2.13) | 1 439 324 000 | 23 460 981 (16 264 361 – 30 657 601) |
| Japan | 3 | 2218 | 81 | 3.62 (2.84–4.39) | 126 476 000 | 4 578 431 (3 591 918 – 5 552 296) |
| South-Korea | 1 | 198 | 15 | 7.58 (4.30–12.2) | 51 269 000 | 3 886 190 (2 204 567 – 6 249 691) |
| South-Eastern Asia | 1 | 816 | 3 | 0.37 (0.08-1.07) | 668 620 000 | 2 473 894 (534 896–7 154 234) |
| Malaysia | 1 | 816 | 3 | 0.37 (0.08–1.07) | 32 366 000 | 119 754 (25 893–346 316) |
| Latin America and the Caribbean | 17 | 33 596 | 618 | 1.45 (0.95–1.94) | 653 962 000 | 9 482 449 (6 212 639–12 686 862) |
| South America | 17 | 33 596 | 618 | 1.45 (0.95-1.94) | 430 760 000 | 6 246 020 (4 092 220–8 356 744) |
| Brazil | 15 | 32 352 | 479 | 0.96 (0.52–1.40) | 212 559 000 | 2 040 566 (1 105 306- 2 975 826) |
| Chile | 2 | 1244 | 139 | 10.78 (9.1–12.5) | 19 116 000 | 2 060 704 (1 731 909 -2 389 500) |
| Sub-Saharan Africa | 1 | 3098 | 174 | 5.62 (4.83-6.49) | 1 094 366 000 | 61 503 369 (52 857 878–71 024 353) |
| Kenya | 1 | 3098 | 174 | 5.62 (4.83–6.49) | 53 771 000 | 3 021 930 (2 597 139–3 489 738) |
| Central and southern Asia | 1 | 528 | 117 | 22.16 (18.7–26.0) | 2 014 709 000 | 446 459 514(376 549 112- 522 816 985) |
| Iran | 1 | 528 | 117 | 22.16 (18.7–26.0) | 83 993 000 | 18 612 848 (15 698 291–21 796 183) |
| Northern Africa and western Asia | 1 | 130 | 6 | 4.62 (1.71–9.78) | 525 869 000 | 24 295 147(8 992 359- 51 429 988) |
| Libya | 1 | 130 | 6 | 4.62 (1.71–9.78) | 6 871 000 | 317 440 (117 494 – 671 983) |
Fig. 2.
Estimated seroprevalence rates of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the general human population in different countries using the geographic information system (GIS).
Seroprevalence according to sex, age and population
Of the 47 studies included, 29 reported separate, pooled seroprevalences for males and females. Of 145 368 males and 151 790 females, 6186 males (5.33%, 4.35–6.31%) and 6958 females (5.05%, 4.06–6.04) had specific serum antibodies against SARS-CoV-2. Fifteen studies reported pooled seroprevalences for different age groups; subgroup analyses revealed pooled seroprevalences of 2.28% (1.01–3.56%), 3.22% (1.90–4.55%), 2.98% (1.59–4.36%) and 2.57% (1.39–3.76%) in people aged ≤19, 20–49, 50–64 and ≥ 65 years, respectively (Table 2).
Of the 47 studies, 36 tested people of all age groups, whereas nine and two studies tested only adults and children, respectively (Table 2). Subgroup analysis revealed pooled seroprevalences of 2.43% (2.16–2.70%) in people of all ages, 5.31% (4.12–6.50%) in adults only, and 8.76% (7.46–10.06%) in children only (Table 2).
Seroprevalence in relation to serological assay used
Of 47 studies, 18 utilized rapid LFIAs to detect specific serum antibodies against SARS-CoV-2, 11 used ELISA, 13 used CLIAs, four studies employed a virus neutralization assay, and one used a microsphere immunoassay. Thirty-seven studies used commercial kits and ten employed in-house serological methods. Subgroup analyses, conducted considering the type of serological method employed, revealed pooled seroprevalences of 3.95% (3.17–4.74%), 3.53% (2.65–4.40%), 2.73% (2.03–3.42%) and 1.32% (0.90–1.74%) using LFIA, ELISA, CLIA and neutralization assays, respectively. One study in the USA, which used a microsphere immunoassay, indicated a seroprevalence of 12.5% (11.97–13.03%). Subgroup analysis revealed pooled seroprevalence rates of 3.33% (2.95–3.71%) using commercial assays and 3.63% (2.79–4.48%) employing in-house assays (Table 2).
Seroprevalence in relation to ethnicity
Seven studies (five from the USA, one from England and one from Brazil) had datasets that were stratified according to ethnicity. Subgroup analysis revealed pooled seroprevalences of 3.76% (1.43–6.08%), 9.96% (2.95–16.97%), 8.76% (0.01–18.65%) and 5.78% (1.76–9.79%) in people of white, black, Hispanic and other ethnic backgrounds (Asian/other), respectively (Table 2). In the USA, subgroup analysis revealed pooled seroprevalences of 4.11% (1.45–6.78%), 10.83% (4.81–16.85%), 12.79% (2.33–27.91%) and 5.86% (1.12–10.60%) in people of white/non-Hispanic, black/non-Hispanic, Hispanic and other backgrounds (Asian/other), respectively.
Relationship between seroprevalence and sociodemographic variables
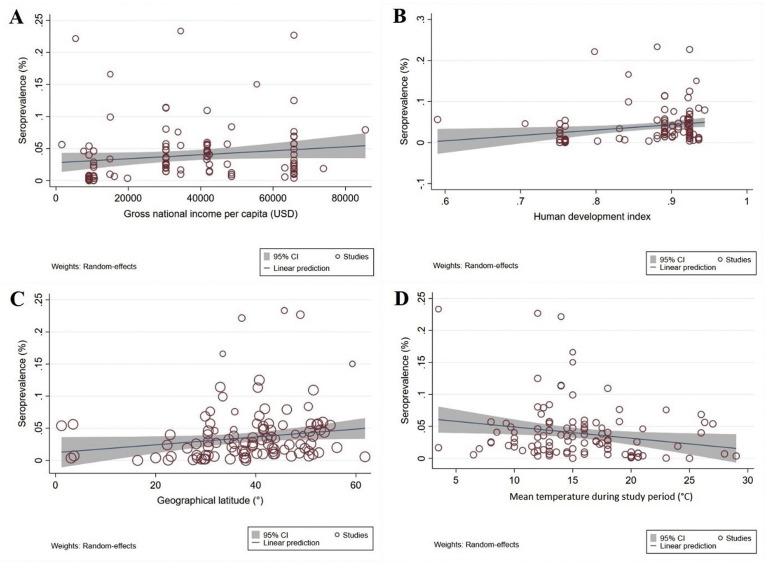
Thirty-five studies represented countries with high income and very high HDI levels; 11 represented countries with upper-middle income levels and high HDIs, and one country had lower-middle income and medium HDI levels. No study was from a low-income or low-HDI country. Subgroup analysis (Table 3 ), according to income and HDI level, revealed higher seroprevalences in countries with high income (4.44%, 3.77–5.1%) and very high HDI levels (4.37%, 3.71–5.02%) than in countries with upper-middle income (1.31%, 1.02–1.59%) and high HDI levels (1.35%, 1.06–1.64%). Random-effects meta-regression analyses showed a significant, increased trend in seroprevalence with higher income levels (coefficient, C = 3.10e-07; p = 0.09) and HDI levels (C = 0.131; p = 0.01) (Figs. 3 A,B).
Table 3.
Prevalence of serum antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the general population based on subgroups according to different sociodemographic geographic parameters and time during, calculated using a random effects model
| Parameters/subgroups | Number of datasets | Number of people screened (total) | Number of seropositive people | Pooled seroprevalence % (95%CI) |
|---|---|---|---|---|
| Income | ||||
| Lower middle | 1 | 3098 | 174 | 5.62 (4.83–6.49) |
| Upper middle | 26 | 120 242 | 2361 | 1.31 (1.02–1.59) |
| High | 80 | 275 925 | 13 344 | 4.44 (3.77–5.1) |
| Human development index | ||||
| Medium | 1 | 3 098 | 174 | 5.62 (4.83-6.49) |
| High | 26 | 119 426 | 2358 | 1.35 (1.06–1.64) |
| Very high | 80 | 276 741 | 13 347 | 4.37 (3.71–5.02) |
| Latitude | ||||
| 0–20° | 5 | 18 007 | 496 | 2.99 (0.71–5.28) |
| 20–40° | 49 | 160 890 | 4034 | 2.29 (2.03–2.56) |
| 40–60° | 53 | 220 368 | 11 349 | 4.68 (3.92–5.43) |
| Longitude | ||||
| 0–30° | 53 | 220 851 | 9969 | 4.15 (3.49–4.82) |
| 30–60° | 16 | 35 478 | 769 | 1.76 (1.18–2.34) |
| 60–90° | 10 | 27 927 | 2435 | 6.36 (3.07–9.66) |
| 90–120° | 18 | 102 378 | 2497 | 2.80 (2.37–3.22) |
| ≥120 | 10 | 12 631 | 209 | 1.63 (1.01–2.25) |
| Relative humidity (%) | ||||
| <60 | 15 | 62 692 | 3763 | 5.84 (3.81–7.87) |
| 60–79 | 76 | 306 057 | 11 159 | 3.41 (2.96–3.85) |
| ≥80 | 16 | 30 516 | 957 | 2.77 (2.01–3.55) |
| Mean temperature (°C) | ||||
| <7 | 4 | 1765 | 102 | 7.87 (1.54–14.20) |
| 7.1–13 | 36 | 111 683 | 5351 | 4.27 (3.23–5.32) |
| 13.1–19 | 43 | 232 763 | 9332 | 4.16 (3.53–4.78) |
| 19.1–25 | 18 | 29 550 | 303 | 0.85 (0.60–1.11) |
| 25.1–30 | 6 | 23 504 | 791 | 3.79 (1.75–5.84) |
| The time from the beginning of the pandemic (days) | ||||
| <15 | 5 | 8222 | 129 | 3.54 (1.84–5.23) |
| 16–30 | 2 | 1943 | 69 | 3.05 (2.29–3.81) |
| 31–45 | 12 | 68 370 | 1470 | 2.51 (2.05–2.97) |
| 46–60 | 24 | 49 663 | 1009 | 1.76 (1.28–2.24) |
| 60–75 | 15 | 22 452 | 614 | 2.59 (1.83–3.34) |
| 76–90 | 27 | 105 317 | 5419 | 3.97 (2.98–4.96) |
| 90–105 | 8 | 36 945 | 1189 | 3.80 (2.22–5.39) |
| 106–120 | 2 | 3557 | 389 | 8.63 (7.72–9.54) |
| 121–135 | 2 | 411 | 47 | 10.34 (7.42–13.26) |
| 136–150 | 9 | 99 908 | 5544 | 5.65 (4.61–6.69) |
Fig. 3.
Ecological random effects meta-regression analyses of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) seroprevalence in the general population in relation to: (A) a country's income level (a statistically significant upward trend in seroprevalence in countries with higher income levels); (B) human development index (HDI) (a statistically significant upward trend in seroprevalence in higher HDI countries); (C) geographical latitude (a statistically significant upward trend in seroprevalence with increasing geographical latitude); and (D) the mean temperature during study implementation (a statistically significant downward trend in seroprevalence with increasing mean temperature).
Relationship between seroprevalence and geographical location, climate or time
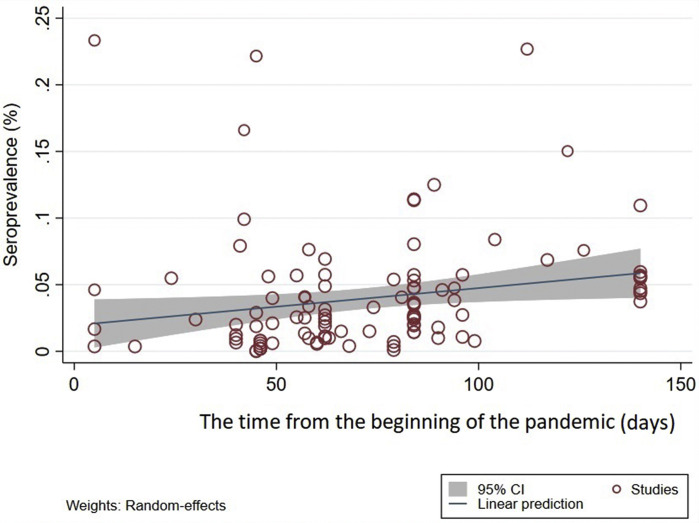
At geographical latitudes of 0–20°, 20-40° and 40-60°, seroprevalences were 2.99% (0.71–5.28%), 2.29% (2.03–2.56%) and 4.68% (3.92–5.43%), respectively; the highest and lowest seroprevalences were at longitudes 60–90° (6.36%, 3.07–9.66%) and ≥120° (1.63%, 1.01–2.25%). In relation to climate, seroprevalences were 5.48% (3.81–7.87%), 3.41% (2.96–3.85%) and 2.77% (2.01–3.55%) in regions with mean relative humidities of <60%, 60–80%, and >80%, respectively. Subgroup analysis indicated that the highest and lowest seroprevalence rates occurred in climes with average environmental temperatures of <7°C (7.87%, 1.54–14.20%) and 19–25°C (0.85%, 0.60–1.11%), respectively (Table 3). There was a significant (C = 0.0007, p = 0.03), increasing trend in seroprevalence with increasing geographical latitude (Fig. 3C), and a non-significant (C = –0.00008, p = 0.316) decreasing trend with geographical longitude (Fig. S2A). Furthermore, there was a significant (C = –0.0017, p = 0.02), decreasing seroprevalence trend with increasing average environmental temperature (Fig. 3D) and a non-significant (C = –0.0006, p = 0.12), decreasing trend with increasing relative humidity (Supplementary Material Fig. S2B). Another subgroup analysis was conducted to explore SARS-CoV-2 seroprevalence over time from the start of the pandemic to the time of sampling/testing in individual studies. The results indicated that seroprevalence in a country was lowest at the beginning of a COVID-19 epidemic, higher at 70 days, and highest 4 months after the start of such an epidemic (p = 0.001, Table 3). Random-effects meta-regression analysis showed a significant increasing trend in seroprevalence over time (C = 0.002, p = 0.02, Fig. 4 ).
Fig. 4.
Random effects meta-regression analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) seroprevalence in the general human population in relation to time from the start of the pandemic to the time of sampling/testing in individual studies (articles) included in the present review. A statistically significant upward trend in seroprevalence is seen over time (C = 0.002, p 0.02).
Association between seroprevalence and confirmed COVID-19 cases (i.e. disease) and death
Meta-regression analyses revealed a non-significant increasing trend in the number of confirmed cases (C = 0.0002, p = 0.921) and of deaths (C = 0.0001, p = 0.640) with increasing seroprevalences (Supplementary Material Fig. S3A,B).
Discussion
Currently, COVID-19 is the number one public health concern worldwide. Here we provide a comprehensive appraisal of SARS-CoV-2 seroprevalence in the ‘general’ human population from continents from which peer-reviewed investigations have been published (up to 14th August 2020), and excluding studies of high-risk patient groups to avoid a overestimation of seroprevalence. The meta-analysis revealed a pooled SARS-CoV-2 seroprevalence of 3.38% (95%CI 3.05–3.72%) relating to ~264 million individuals worldwide at the time of drafting this manuscript. Our findings are in accord with the World Health Organization (WHO) report prediction that 2–3% of the global population might have been infected by the end of the first epidemic wave [39]. Thus, our findings suggest that (at the time of this study) ~97% of the world's population was susceptible to SARS-CoV-2 and COVID-19.
Overall seroprevalence varied markedly among countries and regions, which may be attributable to many factors, including chance variation, cultural practices, political decision-making, policies, mitigation efforts, health infrastructure and prevention/control measures and/or the effectiveness of the implementation of such measures [40,41]. Subgroup analysis suggested higher seroprevalences rates in countries with higher income levels and HDIs. Due to a lack of data for many disadvantaged countries, findings need to be interpreted with caution, but possible explanations might include increased urbanization and population density, higher levels of social interaction and intensity of international travel. Morever, our analysis did not extend to a time when COVID-19 will accelerate in the Southern Hemisphere, especially in Africa and South America, or in South Asia.
Due to variability in the data and the lack of detailed reporting, the present findings should be interpreted with some caution. We did see a lower seroprevalence in white people than in other ethnic minority groups, which is in accord with previous studies [[42], [43], [44], [45]] reporting that minority groups are being disproportionately impacted by COVID-19. According to the Centers for Disease Control and Prevention (CDC) [46], factors suggested to contribute to this disproportionate impact include discrimination in health care, housing, education and finances; communication and language barriers; cultural differences between patients and healthcare providers; lack of health insurance; increased employment of ethnic minority groups in essential work settings such as healthcare facilities, farms, factories, grocery stores, and public transportation; and living in more crowded families or conditions. In this analysis we did not attempt to distinguish prevalence rates within different regions of the USA or other nations. Doing so might have revealed increased seroprevalence in lower-income areas due to the factors mentioned above. Indeed, it was noted that the ‘blue marble health’ concept of poverty-related diseases amongst the poor living in high-income nations might apply to COVID-19, just as it does to neglected tropical diseases, tuberculosis and other poverty-related conditions [47,48].
Regarding the serological tests, our analysis indicated some variation in diagnostic sensitivity (detecting IgG and/or IgM) among the serological assays used in published studies. A recent investigation indicated sensitivity and specificity of 85% and 99% for both antibody isotypes [49]. This review showed that similar seroprevalences were established by ELISA (3.9%) and LFIA (3.5%), while lower seroprevalence rates were obtained using CLIA (2.7%) and virus neutralization (1.3%) assays. Two recent meta-analyses [50,51] showed that sensitivities were consistently lower for the LFIA (66–80%) assay compared with ELISA (84–93%) and CLIA (90–97%), while a specificity of >95% was calculated for all methods. Variation in sensitivity could be attributable to differences in the antigens used (i.e. recombinant or purified protein), the antibody conjugate employed, and cut-off set for an assay [50,52]. A Cochrane review indicated that the combination of the detection of IgG and IgM achieved a sensitivity of 30.1% 1–7 days, 72.2% for 8–14 days, 91.4% for 15–21 days after the onset of symptoms [53]. In the present study [22,25,54,55], four studies used virus neutralization to detect serum antibodies to SARS-CoV-2, all exhibiting a sensitivity and specificity of >98%. Neutralization assays are more time-consuming to perform (3–5 days) and are carried out in biosafety level 3 (BSL-3) laboratories [56]; therefore, these assays might be less suited for routine use. One study [57] used a microsphere immunoassay to detect serum antibodies to SARS-CoV-2 and indicated a high seroprevalence (12.5%). Although this method has been approved by the Food and Drug Administration (FDA), results might be interpreted with caution, given that only one study has been published to date.
The present analyses indicate an increasing trend for seroprevalence at higher latitudes and lower mean environmental temperatures and relative humidities. This trend seems consistent with some previous laboratory, epidemiological and mathematical modelling studies [[58], [59], [60], [61]], showing that environmental temperature and humidity play key roles in the survival and transmission of seasonal respiratory viruses. The highest seroprevalences were estimated for latitudes between 40°N and 56°N, in accordance with a previous study [61], indicating a substantial community spread of SARS-CoV-2 up to March 2020 in areas located in a narrow band in the 30°N and 50°N ‘corridor’. The finding of higher seroprevalence rates in areas with a low mean relative humidity and temperature accords with recent epidemiological [61] and laboratory [62] studies of coronavirus survival. Both temperature and humidity are known to be critical factors in determining survival and community transmission of SARS-CoV, MERS-CoV and influenza [61,63,64]. Processes or mechanisms proposed to be linked to cold temperatures and low humidity include airborne droplet stabilization, increased viral replication in the nasopharyngeal mucosa or respiratory epithelium, and/or reduced local innate immune responses, as evidenced for other respiratory viruses [58,61,[65], [66], [67]]. However, much more research is needed to explore these proposals and the effects of geographical locations and/or climate factors on SARS-CoV-2 seroprevalence and COVID-19 prevalence.
As the present study represents a first ‘snapshot’ of SARS-CoV-2 seroprevalence based on a critical evaluation of published information, it has a number of limitations. First, there is a lack of peer-reviewed, population-based studies from many countries across the globe at an early phase of the pandemic, and some studies included here lacked data on sex and age of subjects tested. We hope that these limitations can be addressed over the coming months and years, so that longitudinal investigations will provide future estimates that will be more representative of the situation worldwide, and so that, eventually, conclusions might be reached regarding endemic stability and instability in particular countries and regions. Second, different serological methods/assays (with varying sensitivities and specificities) were employed in different studies, which will have some effect on our global estimate, although subgroup analyses were undertaken to assess a potential effect of the serological methods used. Third, pooled analyses showed significant heterogeneity. As such heterogeneity was expected in meta-analyses of global prevalence estimates [[68], [69], [70]], we explored possible sources of heterogeneity, including geographic region and diagnostic methods. However, we did not find the source of this heterogeneity.
This study reinforces the major global health threat posed by SARS-CoV-2 infection and its very rapid spread, with the global seroprevalence rising to 3.38% only months after the commencement of the pandemic. This prevalance suggests, though, that ~96% of the world's population are still susceptible to infection, which is alarming. This means that many countries could still face multiple surges in cases, overwhelming medical systems. We have seen in many locations that hospital beds, intensive care units (ICUs) and ventilators have reached capacity. For instance, early on in New York City the USA had to send mercy ships to handle the surge in need. Therefore, countries should have plans and medical resources in place for future, unexpected waves of COVID-19.
There are indications from some countries that mortality rates for COVID-19 are higher than those officially reported [[71], [72], [73]]. Hence, until a vaccine or vaccines is/are available, the focus needs to be on education and prevention and strict quarantine measures. Presently, masks and safe physical distancing are our key means of reducing exposure, infections, disease and deaths. A global meta-analysis [74] showed that applying physical distancing of ≥1 m and usage of personal protective equipment (PPE, including face mask, eye and body protection) results in a major reduction in transmission/infection risk. However, the lack of preparedness in many countries to control a rapidly spreading, highly virulent and pathogenic virus, combined with limited or no biosecurity strategies/policies on how to deal with pandemics in populations, meant that such simple measures were not introduced initially. Our study calls for routine surveys to monitor temporal changes in seroprevalence in a location. In the context of epidemics and pandemics, such surveys might be conducted on a monthly or 2-weekly basis to allow authorities to assess the spread of the virus and exposure levels in populations. A global plan is required to monitor SARS-CoV-2 seroprevalence to assist prevention and control efforts. We aim to continue to follow the global seroprevalence situation over time, and to report on trends and changes.
Author contributions
AR, RBG and PJH conceived the study. AR, MS, MNS and SE conducted the searches and collected data. AR, MS, SMR and ML analysed and interpreted the datasets. AR, AHM, RBG and PJH drafted and edited the manuscript. All authors commented on or edited drafts and approved the final version of the manuscript.
Transparency declaration
The authors declare no conflict of interest. This study was supported by the Health Research Institute at the Babol University of Medical Sciences, Babol, Iran (IR.MUBABOL.REC.1399.304). RBG's research programme is supported by the National Health and Medical Research Council (NHMRC) of Australia, the Australian Research Council (ARC), Yourgene Health Singapore, and Melbourne Water.
Acknowledgements
Sincere thanks to Constantine E. Gasser for critical reading of the manuscript and comments.
Editor: J. Rodriguez-Baño
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2020.10.020.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hick J.L., Biddinger P.D. Novel coronavirus and old lessons—preparing the health system for the pandemic. N Engl J Med. 2020;382:e55. doi: 10.1056/NEJMp2005118. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization (WHO) Coronavirus disease 2019 (COVID-19) Situation Report—209. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200816-covid-19-sitrep-209.pdf?sfvrsn=5dde1ca2_2 Available from:
- 5.Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58:e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X., Sun J., Nie S., Li H., Kong Y., Liang M. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med. 2020;26:1193–1195. doi: 10.1038/s41591-020-0949-6. [DOI] [PubMed] [Google Scholar]
- 8.Stringhini S., Wisniak A., Piumatti G., Azman A.S., Lauer S.A., Baysson H. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet. 2020;396:313–319. doi: 10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolff F., Dahma H., Duterme C., Van den Wijngaert S., Vandenberg O., Cotton F. Monitoring antibody response following SARS-CoV-2 infection: Diagnostic efficiency of four automated immunoassays. Diagn Microbiol Infect Dis. 2020;98:115140. doi: 10.1016/j.diagmicrobio.2020.115140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosado J., Cockram C., Merkling S.H., Demeret C., Meola A., Kerneis S. Serological signatures of SARS-CoV-2 infection: Implications for antibody-based diagnostics. medRxiv. 2020 doi: 10.1101/2020.05.07.20093963. published online May 11; (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang A.T., Garcia-Carreras B., Hitchings M.D., Yang B., Katzelnick L.C., Rattigan S.M. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv. 2020 doi: 10.1101/2020.04.14.20065771. published online April 20; (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization Global surveillance for COVID-19 disease caused by human infection with the 2019 novel coronavirus. https://www.who.int/publications-detail/global-surveillance-for-human-infection-with-novel-coronavirus-(2019-ncov) Available from:
- 14.Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. N Engl J Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 15.Thomas S.N., Altawallbeh G., Zaun C., Pape K., Peters J.M., Titcombe P.J. Initial determination of COVID-19 seroprevalence among outpatients and healthcare workers in Minnesota using a novel SARS-CoV-2 total antibody ELISA. 2020. Available at SSRN: [DOI] [PMC free article] [PubMed]
- 16.Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., André E. Diagnostic performance of 7 rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect. 2020;26:1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isabel M., Damien G., Benoit K., Hafid D., Soleimani R., Vincenzo C. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti-SARS-CoV-2 antibodies. J Clin Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peiris J., Guan Y., Yuen K. Severe acute respiratory syndrome. Nat Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallal P., Hartwig F., Horta B., Victora G.D., Silveira M., Struchiner C. Remarkable variability in SARS-CoV-2 antibodies across Brazilian regions: nationwide serological household survey in 27 states. medRxiv. 2020 doi: 10.1101/2020.05.30.20117531. published online May 30; (preprint) [DOI] [Google Scholar]
- 21.Xu X., Sun J., Nie S., Li H., Kong Y., Liang M. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med. 2020;26:1193–1195. doi: 10.1038/s41591-020-0949-6. [DOI] [PubMed] [Google Scholar]
- 22.Gallian P., Pastorino B., Morel P., Chiaroni J., Ninove L., de Lamballerie X. Lower prevalence of antibodies neutralizing SARS-CoV-2 in group O French blood donors. Antivir Res. 2020;181:104880. doi: 10.1016/j.antiviral.2020.104880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer B., Knabbe C., Vollmer T. SARS-CoV-2 IgG seroprevalence in blood donors located in three different federal states, Germany, March to June 2020. Eur Surveill. 2020;25:2001285. doi: 10.2807/1560-7917.ES.2020.25.28.2001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shakiba M., Nazari S.S.H., Mehrabian F., Rezvani S.M., Ghasempour Z., Heidarzadeh A. Seroprevalence of COVID-19 virus infection in Guilan province, Iran. medRxiv. 2020 doi: 10.1101/2020.04.26.20079244. published online May 1; (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Percivalle E., Cambiè G., Cassaniti I., Nepita E.V., Maserati R., Ferrari A. Prevalence of SARS-CoV-2 specific neutralising antibodies in blood donors from the Lodi Red Zone in Lombardy, Italy, as at 06 April 2020. Eur Surveill. 2020;25:2001031. doi: 10.2807/1560-7917.ES.2020.25.24.2001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward H., Atchison C., Whitaker M., Ainslie K., Elliott J., Okell L. Antibody prevalence for SARS-CoV-2 following the peak of the pandemic in England: REACT2 study in 100,000 adults. medRxiv. 2020 doi: 10.1101/2020.08.12.20173690. published online August 21; (preprint) [DOI] [Google Scholar]
- 27.Havers F.P., Reed C., Lim T., Montgomery J.M., Klena J.D., Hall A.J. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23–May 12, 2020. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.4130. published online July 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The sustainable development goals (SDGs) report 2019. Regional groupings. https://unstats.un.org/sdgs/report/2019/regional-groups/ Available from:
- 29.World Health Organization (WHO) Coronavirus disease (COVID-19), Situation Report–198. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200805-covid-19-sitrep-198.pdf?sfvrsn=f99d1754_2 Available from:
- 30.World Bank Group database. World Bank Country and Lending Groups https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups Available from:
- 31.World Bank Group database Gross national income per capita 2019. https://databank.worldbank.org/data/download/GNIPC.pdf Available from:
- 32.United Nations Development Program http://hdr.undp.org/en/composite/HDI Available from:
- 33.United Nations Total population (both sexes combined) by region, subregion and country, annually for 1950–2100 (thousands) https://population.un.org/wpp/Download/Standard/Population/ Available from:
- 34.Munn Z., Moola S., Riitano D., Lisy K. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int J Health Policy Manag. 2014;3:123–128. doi: 10.15171/ijhpm.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 36.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 37.Harbord R.M., Higgins J.P. Meta-regression in Stata. Stata J. 2008;8:493–519. [Google Scholar]
- 38.Hunter J.P., Saratzis A., Sutton A.J., Boucher R.H., Sayers R.D., Bown M.J. In meta-analyses of proportion studies, funnel plots were found to be an inaccurate method of assessing publication bias. J Clin Epidemiol. 2014;67:897–903. doi: 10.1016/j.jclinepi.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 39.World Health Organization (WHO) WHO; Geneva: 20 Apr 2020. Coronavirus disease (COVID-2019) press briefing 20 April 2020.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/media-resources/press-briefings# Available from: [Google Scholar]
- 40.Miller A., Reandelar M.J., Fasciglione K., Roumenova V., Li Y., Otazu G.H. Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID-19: an epidemiological study. medRxiv. 2020 doi: 10.1101/2020.03.24.20042937. published online March 28; (preprint) [DOI] [Google Scholar]
- 41.Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stokes E.K., Zambrano L.D., Anderson K.N., Marder E.P., Raz K.M., Felix S.E.B. Coronavirus Disease 2019 Case Surveillance—United States, January 22–May 30, 2020. Morb Mortal Wkly Rep. 2020;69:759. doi: 10.15585/mmwr.mm6924e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Killerby M.E., Link-Gelles R., Haight S.C., Schrodt C.A., England L., Gomes D.J. Characteristics associated with hospitalization among patients with COVID-19—Metropolitan Atlanta, Georgia, March–April 2020. Morb Mortal Wkly Rep. 2020;69:790. doi: 10.15585/mmwr.mm6925e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price-Haywood E.G., Burton J., Fort D., Seoane L. Hospitalization and mortality among black patients and white patients with Covid-19. N Engl J Med. 2020;382:2534–2543. doi: 10.1056/NEJMsa2011686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millett G.A., Jones A.T., Benkeser D., Baral S., Mercer L., Beyrer C. Assessing differential impacts of COVID-19 on Black communities. Ann Epidemiol. 2020;47:37–44. doi: 10.1016/j.annepidem.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention (CDC) Health Equity Considerations and Racial and Ethnic Minority Groups. https://www.cdc.gov/coronavirus/2019-ncov/community/health-equity/race-ethnicity.html Available from:
- 47.Hotez P.J. 2020. Poverty and the Impact of COVID-19: The Blue-Marble Health Approach. [Google Scholar]
- 48.Hotez P.J., Bottazzi M.E., Singh S.K., Brindley P.J., Kamhawi S. Will COVID-19 become the next neglected tropical disease? PLoS Negl Trop Dis. 2020;14 doi: 10.1371/journal.pntd.0008271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caini S., Bellerba F., Corso F., Díaz-Basabe A., Natoli G., Paget J. Meta-analysis of diagnostic performance of serological tests for SARS-CoV-2 antibodies up to 25 April 2020 and public health implications. Eur Surveill. 2020;25:2000980. doi: 10.2807/1560-7917.ES.2020.25.23.2000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kontou P.I., Braliou G.G., Dimou N.L., Nikolopoulos G., Bagos P.G. Antibody tests in detecting SARS-CoV-2 infection: a meta-analysis. Diagnostics. 2020;10:319. doi: 10.3390/diagnostics10050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bastos M.L., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.-P., Johnston J.C. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee C.Y.-P., Lin R.T., Renia L., Ng L.F. Serological approaches for COVID-19: epidemiologic perspective on surveillance and control. Front Immunol. 2020;11:879. doi: 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deeks J.J., Dinnes J., Takwoingi Y., Davenport C., Spijker R., Taylor-Phillips S. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;25:CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang L., Hou W., Zhao L., Zhang Y., Wang Y., Wu L. The prevalence of antibodies to SARS-CoV-2 among blood donors in China. medRxiv. 2020 doi: 10.1101/2020.07.13.20153106. published online July 14; (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sam I.-C., Chong Y.M., Tan C.W., Chan Y.F. Low post-pandemic wave SARS-CoV-2 seroprevalence in Kuala Lumpur and Selangor, Malaysia. J Med Virol. 2020 doi: 10.1002/jmv.26426. published online August 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.8259. published online May 6. [DOI] [PubMed] [Google Scholar]
- 57.Rosenberg E.S., Tesoriero J.M., Rosenthal E.M., Chung R., Barranco M.A., Styer L.M. Cumulative incidence and diagnosis of SARS-CoV-2 infection in New York. medRxiv. 2020 doi: 10.1101/2020.05.25.20113050. published online May 29; (preprint) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowen A.C., Mubareka S., Steel J., Palese P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007;3:e151. doi: 10.1371/journal.ppat.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barreca A.I., Shimshack J.P. Absolute humidity, temperature, and influenza mortality: 30 years of county-level evidence from the United States. Am J Epidemiol. 2012;176:S114–S122. doi: 10.1093/aje/kws259. [DOI] [PubMed] [Google Scholar]
- 60.Żuk T., Rakowski F., Radomski J.P. Probabilistic model of influenza virus transmissibility at various temperature and humidity conditions. Comput Biol Chem. 2009;33:339–343. doi: 10.1016/j.compbiolchem.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 61.Sajadi M.M., Habibzadeh P., Vintzileos A., Shokouhi S., Miralles-Wilhelm F., Amoroso A. Temperature, humidity, and latitude analysis to estimate potential spread and seasonality of coronavirus disease 2019 (COVID-19) JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casanova L.M., Jeon S., Rutala W.A., Weber D.J., Sobsey M.D. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl Environ Microbiol. 2010;76:2712–2717. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Otter J., Donskey C., Yezli S., Douthwaite S., Goldenberg S., Weber D. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: the possible role of dry surface contamination. J Hosp Infect. 2016;92:235–250. doi: 10.1016/j.jhin.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Doremalen N., Bushmaker T., Munster V. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Eur Surveill. 2013;18:20590. doi: 10.2807/1560-7917.es2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- 65.Yang W., Marr L.C. Dynamics of airborne influenza A viruses indoors and dependence on humidity. PloS One. 2011;6 doi: 10.1371/journal.pone.0021481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaffer F., Soergel M., Straube D. Survival of airborne influenza virus: effects of propagating host, relative humidity, and composition of spray fluids. Arch Virol. 1976;51:263–273. doi: 10.1007/BF01317930. [DOI] [PubMed] [Google Scholar]
- 67.Kudo E., Song E., Yockey L.J., Rakib T., Wong P.W., Homer R.J. Low ambient humidity impairs barrier function and innate resistance against influenza infection. Proc Natl Acad Sci. 2019;116:10905–10910. doi: 10.1073/pnas.1902840116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Veroniki A.A., Jackson D., Viechtbauer W., Bender R., Bowden J., Knapp G. Methods to estimate the between-study variance and its uncertainty in meta-analysis. Res Synth Methods. 2016;7:55–79. doi: 10.1002/jrsm.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwatra G., Cunnington M.C., Merrall E., Adrian P.V., Ip M., Klugman K.P. Prevalence of maternal colonisation with group B streptococcus: a systematic review and meta-analysis. Lancet Infect Dis. 2016;16:1076–1084. doi: 10.1016/S1473-3099(16)30055-X. [DOI] [PubMed] [Google Scholar]
- 70.Platt L., Easterbrook P., Gower E., McDonald B., Sabin K., McGowan C. Prevalence and burden of HCV co-infection in people living with HIV: a global systematic review and meta-analysis. Lancet Infect Dis. 2016;16:797–808. doi: 10.1016/S1473-3099(15)00485-5. [DOI] [PubMed] [Google Scholar]
- 71.Burn-Murdoch J., Romei V., Giles C. Financial Times; April 2020. Global coronavirus death toll could be 60% higher than reported.https://www.ft.com/content/6bd88b7d-3386-4543-b2e9-0d5c6fac846c Available from: [Google Scholar]
- 72.Modi C., Boehm V., Ferraro S., Stein G., Seljak U. How deadly is COVID-19? A rigorous analysis of excess mortality and age-dependent fatality rates in Italy. medRxiv. 2020 doi: 10.1101/2020.04.15.20067074. published online May 14; (preprint) [DOI] [Google Scholar]
- 73.Baud D., Qi X., Nielsen-Saines K., Musso D., Pomar L., Favre G. Real estimates of mortality following COVID-19 infection. Lancet Infect Dis. 2020;20:773. doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chu D.K., Akl E.A., Duda S., Solo K., Yaacoub S., Schünemann H.J. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.