Abstract
Currently, miR-21 and CXCR4 are being extensively investigated as two key regulators in glioma malignancy. In this study, we investigated the combined effects of these two factors on glioma progression. Herein, the expression of miR-21 and CXCR4 was increased in tumor tissues and cell lines. Inhibition of miR-21, CXCR4, and miR-21 and CXCR4 together all reduced the migration, invasiveness, proliferation, and enhanced apoptosis in glioma cells, as well as reduced tumor volume and mass in xenograft model. The inhibition effect was strongest in double-targeted knockdown of miR-21 and CXCR4 group, whose downstream pathways involved in AKT axis and ERK axis activation. In conclusion, our findings reported that double-targeted knockdown of miR-21 and CXCR4 could more effectively inhibit the proliferation, migration, invasion, and growth of transplanted tumor and promote cell apoptosis, which were involved in the PI3K/AKT and Raf/MEK/ERK signaling pathways.
1. Introduction
Glioma is the most common and feared type of tumor, with an annual incidence of 1 per 10,000 population [1]. Currently, treatments for malignant glioma are mainly surgical resection, followed by chemotherapy or radiotherapy [2]. However, these comprehensive treatments are not very effective, and the median survival of patients afflicted with malignant glioma is less than 1 year, due to the aggressive proliferation and insidious invasion of cells [3, 4]. Thus, recent studies are focusing on the factors that can effectively control the invasive and proliferative capacity of malignant glioma cells to develop targeted salvage treatments to improve patients' life quality.
miR-21 is an oncomiR which attracts many attentions, because it takes part in almost all steps of tumor growth and metastasis [5]. Evidence shows that miR-21 is overexpressed in gliomas and glioma cells compared to normal tissues, and its expression level is positively correlated with glioma grade [5, 6]. Elevated miR-21 expression leads to increased glioma cell proliferation, invasion and also chemo-resistance, which indicates poor prognosis and tumor recurrence of glioma patients [7–10]. On the other hand, downregulation of miR-21 leads to repression of antiapoptotic capacity, reduction of migratory, and invasion, as well as increase of chemical-induced death in glioma cells [8, 11–13]. All of these make miR-21 not only a potential glioma marker for diagnosis and prognosis but a target for novel therapeutic intervention.
Another factor that draws great attention for playing a crucial role in malignant glioma biological regulation is C-X-C Chemokine Receptor 4 (CXCR4), a transmembrane G-protein-coupled receptor. Statistics shows that CXCR4 level is elevated in malignant glioma when compared to normal cells derived from the same tumor [14]. Particularly, its overexpression is concentrated in invading regions and associated with increased glioma tumor grade and malignancy [1, 15]. Numerous studies suggest that the overexpression of CXCR4 facilitates proliferation, angiogenesis, invasion, and metastasis, as well as chemotherapy and radiotherapy resistance of glioma in several glioma cell lines and mouse models [16–21]. Administration of either CXCR4 neutralizing antibody or CXCR4 siRNA impairs the enhanced glioma malignancy (proliferation, angiogenesis, invasion, metastasis, and postchemotherapy and postradiotherapy recurrence) and increases median survival in vivo glioma models [17–20, 22–27]. Thus, the potential of CXCR4 as an antitumor target has been a popular research topic.
The two moleculars miR-21 and CXCR4 as a single target, respectively, have drawn wide attention of researches. However, several studies have shown that in gene therapy of cancers, combination of multigene therapy tends to be more effective than single-gene therapy strategy, which may provide a promising treatment in cancer. Herein, we investigated the combined effects of miR-21 and CXCR4 by lentiviral-mediated gene recombinant technology on glioma malignancy. Double-targeted knockdown of miR-21 and CXCR4 was performed in glioma cell lines (U87 and U251), and its effects on glioma malignant progression were evaluated in cells and xenograft mouse models. Knockdown of either miR-21 or CXCR4 decreased glioma proliferation, invasion, and migration and enhanced apoptosis. Double-targeted knockdown generated a significantly enhanced inhibition effect on tumorigenicity even compared to single-targeted knockdown of miR-21 or CXCR4 alone. Also, we found that decrease of tumor progression was related to the PI3K/AKT and Raf/MEK/ERK pathways which are responsible for glioma growth, invasiveness, chemo- and radiotherapy resistance, and recurrence [11, 13]. Thus, this study might also pave ways for further research on glioma pathology and physiology.
2. Material and Methods
2.1. Samples
A total of 25 patients were recruited in this study. All patients were diagnosed with malignant glioma, and diagnosis was confirmed by both clinical symptoms and the histology consensus from at least two neuropathologists at the Department of Oncology in the First Affiliated Hospital of Zhengzhou University. As recommended in the 2016 WHO Classification of Tumours of the Central Nervous System, all tumor samples were histologically and pathologically diagnosed; among them, 18 cases were WHO grade IV, and 7 cases were WHO grades II-III. The surgical removed tumor tissues, and adjacent peritumoral brain tissues (between 1 cm and 2 cm from the tumor border) were immediately snap frozen using liquid nitrogen and then stored at -80°C fridge for further qRT-PCR and western blot analysis. Written informed consents for the pathological analyses and experiments were obtained from all of the donors before their surgery. All samples were handled and processed in compliance to the institutional ethical and legal standards, which was approved by the Research Ethics Committee of the First Affiliated Hospital of Zhengzhou University.
2.2. Cell Lines and Culture
Human glioma cell lines U87 and U251, normal glial cell HAc, and embryonic kidney cell HEK-293T were obtained from the American Type Culture Collection (ATCC, USA). All cells were cultured in Dulbecco's modified Eagle medium (DMEM)/high glucose (Hyclone, Logan, UT) supplemented with 10% FBS (Invitrogen, CarIsbad, CA, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) and maintained in humidified incubator at 37°C with 5% CO2 and 95% of air.
2.3. qRT-PCR Analysis
Total RNA of the cultured glioma cells (U87 and U251), HAc cells or tissues samples was extracted with the TRIzol reagents (Invitrogen) in accordance with the manufacturer's instruction. For detecting miR-21 and CXCR4 mRNA, cDNA was first amplified by reverse transcription using the PrimeSCript RT reagent kit (Takara Bio, Inc, Otsu, Japan). Then, the relative expressions were measured with the TaqMan microRNA Assay kit (Applied Biosystems, Foster City, CA, USA) and SYBR-Green PCR Master Mix Kit (Takara) on an ABI7500 system (Applied Biosystem), respectively. The primer sequences used in this study were as followings: CXCR4: sense: 5′-GAAACCCTCAGCGTCTCAGT-3′, antisense: 5′-AGTAGTGGGCTAAGGGCACA-3′; miR-21: sense: 5′-GCGTGTCGGGTAGCTTATCAGAC-3′, antisense from TaqMan microRNA Assay kit; GAPDH: sense: 5′-GGGAAACTGTGGCGTGAT-3′, antisense: 5′-GAGTGGGTGTCGCTGTTGA-3′; and U6 sense: 5′-CTCGCTTCGGCAGCACATATACT-3′, antisense: 5′-ACGCTTCACGAATTTGCGTGTC-3′. All experiments were performed in triplicate. The relative CXCR4 and miR-21 levels were obtained from comparing to the expression level of GAPDH and U6 snRNA, respectively. The relative expression levels were calculated using 2-ΔΔCt method.
2.4. Western Blot Assay
The cultured cells and collected tissues samples were first washed twice with ice-cold PBS and then lysed in RIPA buffer supplemented with protease and phosphatase inhibitor cocktails. 30 μg of protein was separated by 10% SDS-PAGE and then electrophoretically transferred onto a PVDF membrane (Millipore, Bedford, MA). The blots were incubated in 1 × PBS containing 5% skimmed milk powder for 1 hr at room temperature to block the nonspecific binding. Following washing (TBST, 3×, 10 min each), the blots were incubated with primary antibodies overnight at 4°C. After another wash, the blots were exposed to horseradish peroxidase-conjugated secondary antibody for 2 hr at room temperature and visualized using the enhanced chemiluminescence detection kit (Sigma). The following primary antibodies were used in this study: CXCR4 (Abcam, Cambridge, MA, USA), β-actin (Cell Signaling Technology, Danvers, MA, USA), AKT (Cell Signaling Technology), phospho-AKT (Cell Signaling Technology), ERK1/2 (Cell Signaling Technology), and phospho-ERK1/2 (Cell Signaling Technology). The protein levels were digitalized using ImageJ 1.48 version (NIH, Bethesda, Maryland) (Java 1.8.9_66), and relative protein levels were obtained by comparing the protein level to the level of β-actin.
2.5. Construction of the Recombinant Lentiviral Vectors
Negative control and anti-miR-21 sequences were cloned into lentiviral vector (pLent-hU6-EF1-GFP-Puro) to generate the negative control recombinant plasmid (pLenti-Lv-NC) and miR-21 knockdown recombinant plasmid (pLenti-anti-miR-21), respectively. sh-CXCR4 was cloned into lentiviral vector pLenti-hU6-GFP-Puro (Shanghai sheng-gong) to generate the CXCR4 knockdown recombinant plasmid (pLenti-sh-CXCR4). The double-targeted knockdown plasmid (pLenti-anti-miR-21 + sh-CXCR4) was obtained by connected sh-CXCR4 fragment with pLenti-anti-miR-21 plasmid using Ready-to-Use Seamless Cloning Kit (Shanghai sheng-gong).
Before the lentiviruses packing, HEK-293T cells were cultured in 6-well plates until reach 80% confluence. Then, cells were cotransfected with the recombinant lentiviral vectors (Lv-NC, anti-miR-21, sh-CXCR4, and anti-miR-21 + sh-CXCR4) and two auxiliary packaging plasmids (pCMV-Δ8.2 and pCMV-VSV-G). 72 hours after the transfection, the lentivirus particles were collected by centrifugating at 25000 rpm for 1.5 h at 4°C. All lentiviral constructs were inserted a puromycin resistance sequence for drug screen.
2.6. Lentiviral Infection of Glioma Cells (U87 and U251)
Before the infection, glioma cells (U87 and U251) were routinely cultured and seeded on 6-well plates at a density of 5 × 104 cells/well. When cell density reached 50% confluence, cells were infected with lentivirus particles (MOI = 20) in presence of 8 μg/ml polybrene (GeneCopoeia, Guangzhou, China) for 24 hr. Then, the culture was replaced by fresh culture medium containing 2% FBS and cultured for another 72 hr. The effectiveness of lentiviral particles that expressing anti-miR-21, sh-CXCR4, or anti-miR-21 + sh-CXCR4 was examined by qRT-PCR and western blot as described above.
2.7. MTT Assay
Logarithmically growing glioma cells (U87 and U251) with different treatments were plated into 96-well culture clusters (Costar, Cambridge, MA, USA), at a density of 2000 cells/well and cultured for different time points (0, 24, 48, 72 hr). Cell viability was assessed by adding 10 μl MTT (0.5 mg/ml) to each well and incubated for 4 hr. After removing the cell medium, 100 μl DMSO was added, and cell proliferation was measured at 490 nm wavelength. Measurements of cell viability for each treatment were done in triplicates from three independent experiments. Treatments included (1) anti-miR-21, (2) sh-CXCR4, (3) anti-miR-21 + sh-CXCR4, and (4) Lv-NC.
2.8. Flow Cytometry Analysis
Annexin V-PE/7-AAD Apoptosis detection Kit I (BD Bioscience, USA) was employed to determine the apoptosis of transfected glioma cells (U87 and U251). Cells were harvested after transfected with lentiviruses, washed twice with PBS, and then resuspended in 400 μl binding buffer. After the resuspension, cells were incubated with 5 μl Annexin V-PE and 10 μl 7-AAD solutions at room temperature for 10 min in the dark. The analysis was performed by flow cytometry (FACSCalibur; Becton Dickinson). For each sample, 10,000 cells were analyzed. All of the experiments were performed in triplicate. Transfection treatments include (1) anti-miR-21, (2) sh-CXCR4, (3) anti-miR-21 + sh-CXCR4, and (4) Lv-NC.
2.9. Transwell Assay
The invasion of glioma cells (U87 and U251) was assayed using BioCoat Matrigel invasion chambers (BD Bioscience) with poly carbonic membrane (6.5 mm in diameter, 8 μM pore size). Before the start of the assay, the chambers were inserted into 24-well culture plates and pretreated with serum-free DMEM medium at 37°C for 30 min. In the transwell assay, the transfected cells were resuspended in 300 μl serum-free DMEM at a density of 5 × 104 cells/ml and added to the upper chamber. 500 μl DMEM supplemented with 20% FBS was added into the lower chamber of each well. The plates were incubated in humidified incubator (37°C, 5% CO2) for 20-24 hr. After the incubation, nonmigrated cells in the upper chamber were removed mechanically with a cotton swap, while the invaded cells in the lower side of the membranes were fixed with 5% glutaraldehyde for 15 min at 4°C, washed with PBS twice, and stained with 1% crystal violet for 15 min. Pictures were photographed at 200x magnification, and cell numbers were counted from at least five randomly selected fields.
2.10. Scratch Assay
Transfected glioma cells (U87 and U251) were suspended and plated in 6-well plates until they reach 70-80% confluence. At the initial time, a scratch was made on the cell monolayer using a sterile 200 μl micropipette tip. Pictures were photographed at different time points (0, 24 hr), with 40x magnification.
2.11. Generation of Tumor Xenografts and In Vivo Treatments
All the animal procedures were approved by Ethics of Animal Experiments of the First Affiliate Hospital of Zhengzhou University, and all the experiments were performed in strict accordance with the institutional guidelines. BALB/C nude mice weighting 21.2 ± 3 g and being 5 weeks old were housed in sterile cages under standard conditions (23 ± 2°C, 45-55% humidity, 12 hr light duration). The mice were allowed for free access to water and acclimatized for one week before the experiments. The mice were randomly divided into 4 groups. Immediately prior to the xenograft experiment, the transfected (Lv-NC, anti-miR-21, sh-CXCR4, and anti-miR-21 + sh-CXCR4) cells were suspended to single-cell in sterile saline. The transfected U87 cells were injected into the left axillary subcutaneous location of BALB/C nude mice. The xenograft procedure was designed according to the published ones [28, 29]. In brief, immediately prior to the xenograft experiment, the transfected cells were suspended to single-cell in sterile saline. Following the surgery, mice were monitored for tumor growth every 5 days. The formula V (volume) = (long diameter × short diameter2)/2 was used to calculate the tumor volume. Mice were weighted and sacrificed at 4 weeks postxenograft; then, tumors were collected and measured. Immediately after the measuring, xenografts were stored at -80°C for further qRT-PCR and western blot analyses.
2.12. Statistical Analysis
Statistical analysis was performed with SPSS 19.0 with an overall significance level of P < 0.05. Differences between the groups were computed by two-tailed unpaired Student t-test.
3. Results
3.1. miR-21 and CXCR4 Expressions Were Increased in Malignant Glioma Tissues and Cells
In order to determine the effect of double-targeted knockdown of miR-21 and CXCR4 on malignant glioma tumorigenicity, we first confirmed the dysregulation of these two factors in malignant glioma tissues. qRT-PCR was employed to assess the expression of miR-21 and CXCR4 at genetic level. Both miR-21 and CXCR4 average expressions were significantly upregulated in glioma tissues compared to paracancerous tissues (Figures 1(a)–1(c)). Further, we examined the expression of miR-21 and CXCR4 in two invasive-prone glioma cell lines (U87 and U251) in comparison to a normal glial cell line (HAc). The expression levels of miR-21 and CXCR4 were both significantly increased in U251 and U87 cells (Figures 1(d)–1(f)). The results suggested that miR-21 and CXCR4 might play important roles in mediating malignant glioma aggressiveness, as well as the tumorigenicity.
Figure 1.
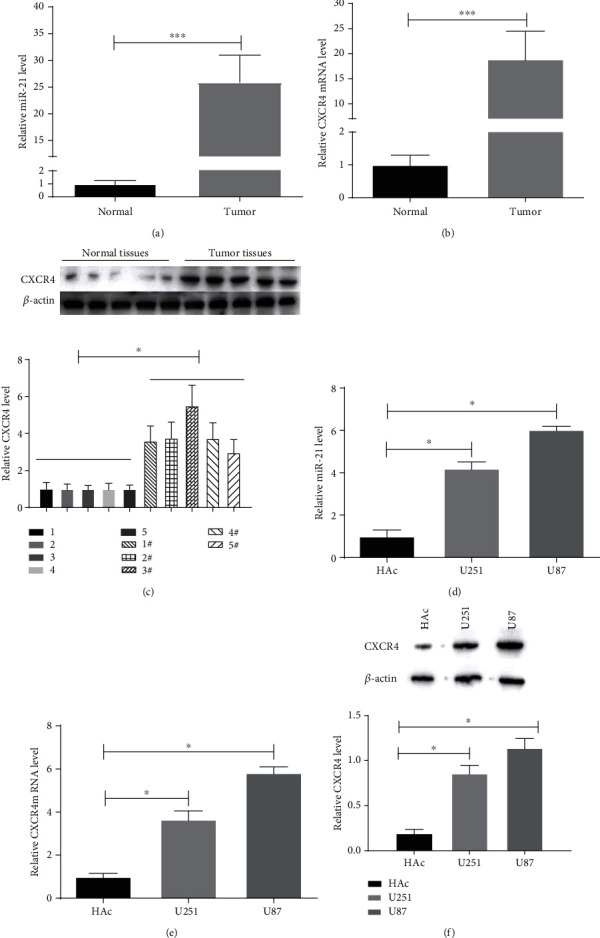
miR-21 and CXCR4 levels in malignant glioma tissues and cells. (a, b) The expression of miR-21 and CXCR4 was detected by qRT-PCR in glioma and normal adjacent tissues samples. (c) The levels of CXCR4 were determined by western blot in tumor and normal tissues samples. (d, e) The expression of miR-21 and CXCR4 was measured by qRT-PCR in glioma cells (U251 and U87) and control cells (HAc). (f) CXCR4 protein levels were tested by western blot. ∗P < 0.05.
3.2. The Construction of Stable Anti-miR-21, sh-CXCR4, and Anti-miR-21 + sh-CXCR4 Glioma Cells
We constructed miR-21 knockdown (anti-miR-21), CXCR4 knockdown (sh-CXCR4), and miR-21 and CXCR4 double-targeted knockdown (anti-miR-21 + sh-CXCR4) U87 and U251 cells to investigate the anti-miR-21 + sh-CXCR4 generated effects on glioma tumorgenicity. qRT-PCR was used to detect the efficiency of miR-21 knockdown at genetic level, and data showed that the knockdown efficiency reached more than 70% in both U87 and U251 cells (Figure 2(a)). The efficiency of sh-CXCR4 was measured by qRT-PCR and western blot, and the results showed that CXCR4 expression level in U251 and U87 cells was significantly repressed compared to the negative control and the mock group (Figures 2(b)–2(d)). Moreover, the construction of anti-miR-21 + sh-CXCR4 was successful with the efficiency of miR-21 and CXCR4 inhibition in U87 and U251 cells (Figures 2(e)–2(g)).
Figure 2.
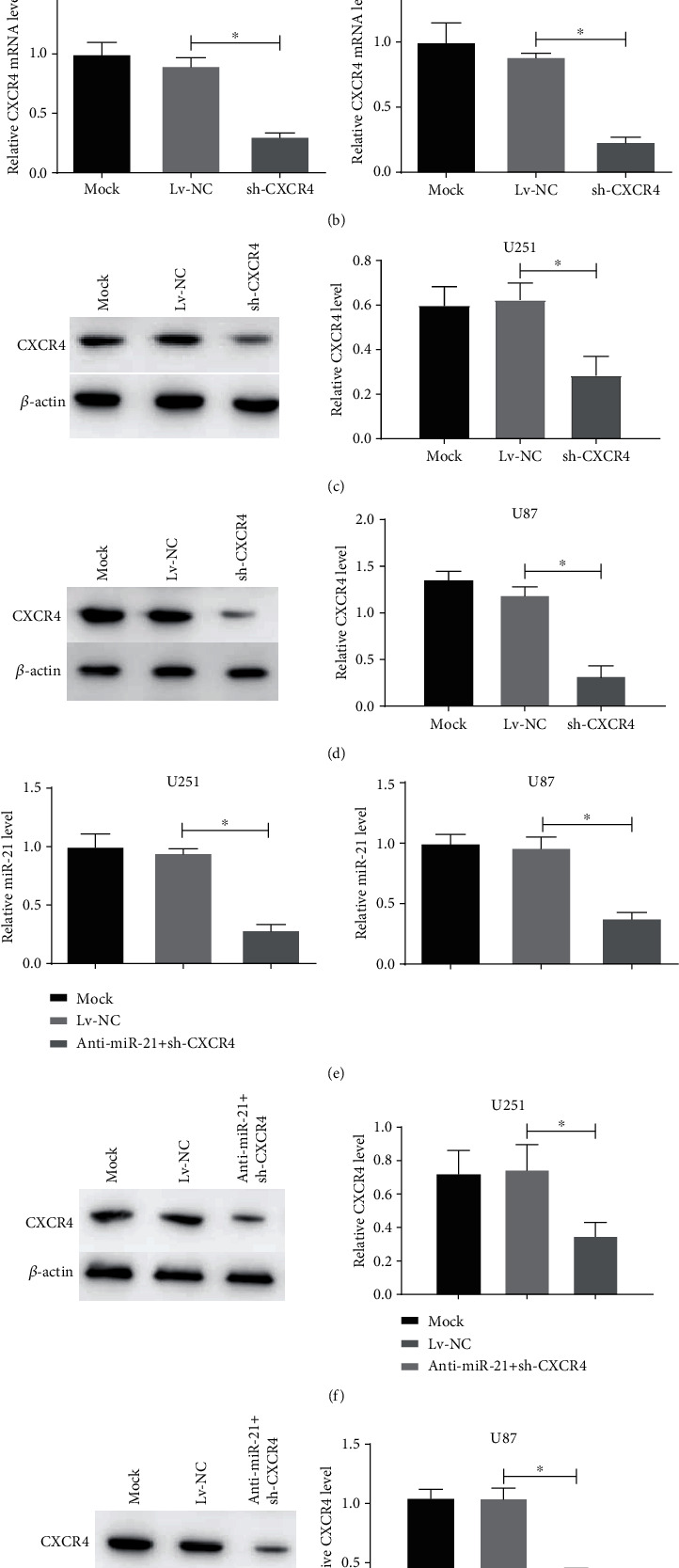
miR-21 and CXCR4 levels of stable glioma cells transfected with anti-miR-21, sh-CXCR4, or anti-miR-21 + sh-CXCR4. (a, b) miR-21 and CXCR4 levels in the transfected U251 and U87 cells were measured using qRT-PCR. (c, d) CXCR4 protein levels in the transfected U251 and U87 cells were measured using western blot. (e) miR-21 and CXCR4 levels in the transfected U251 and U87 cells were measured using qRT-PCR. (f, g) CXCR4 protein levels in the transfected U251 and U87 cells were measured using western blot. ∗P < 0.05.
3.3. Double-Targeted Knockdown of miR-21 and CXCR4 Inhibited Proliferation and Enhanced Apoptosis of Glioma Cells
To explore the roles of miR-21 and CXCR4 in sustaining aggressive proliferation and antiapoptosis in glioma cells, functional experiments were performed. MTT assay was performed on the transfected U87 and U251 cells for the evaluation of cell proliferation at different time points (0, 24, 48, 72 hr). In comparison to the negative control group, anti-miR-21 or sh-CXCR4 alone was able to significantly decrease cell proliferation of U87 and U251 (Figures 3(a) and 3(b)). Interestingly, in the anti-miR-21 + sh-CXCR4 group, the inhibition efficiency was significantly higher than that in the anti-miR-21 or sh-CXCR4 alone group. Next, flow cytometry analysis was used to assess the apoptosis in these transfected cells. Statistically, more apoptotic cells were found in both the anti-miR-21 and sh-CXCR4 groups, and as we expected, the anti-miR-21 + sh-CXCR4 group generated the most apoptotic cells (Figures 3(c) and 3(d)). Thus, our results demonstrated that anti-miR-21 + sh-CXCR4 had an enhanced effect on both suppressing glioma cell proliferation and proapoptosis.
Figure 3.
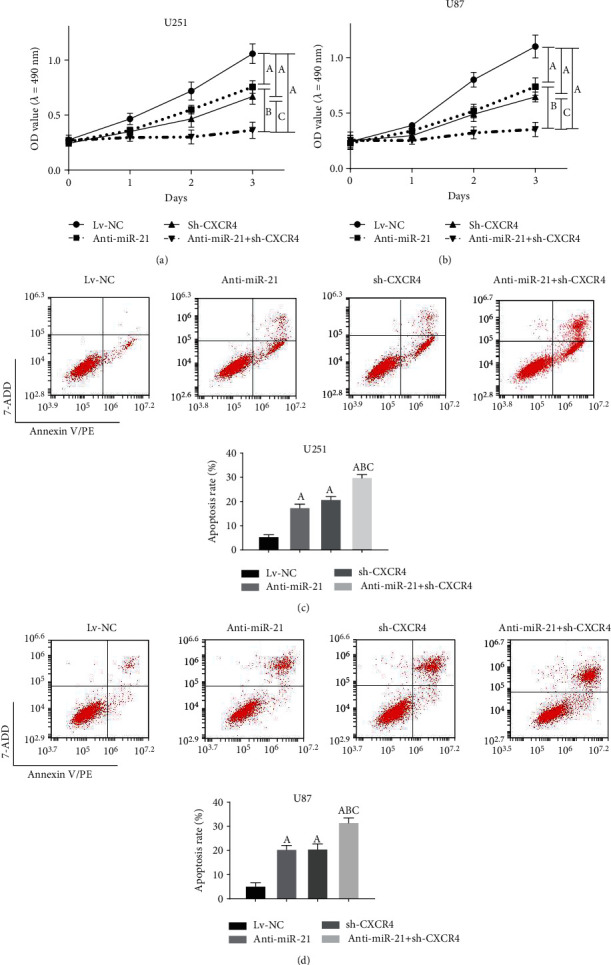
Double-targeted knockdown of miR-21 and CXCR4 inhibited proliferation of glioma cells. (a, b) MTT assays were employed to assess the proliferation of transfected U251 and U87 cells. (c, d) Flow cytometry was used to analyze the apoptosis in transfected U251 and U87 cells. P < 0.05, (A) vs. Lv-NC group; (B) vs. anti-miR-21 group; (C) vs. sh-CXCR4 group.
3.4. Double-Targeted Knockdown of miR-21 and CXCR4 Inhibited Invasion and Migration of Glioma Cells
Transwell assay was performed to evaluate the regulation of miR-21 and CXCR4 on glioma cell invasiveness. Anti-miR-21 or sh-CXCR4 alone was able to significantly limit the invasiveness of glioma cells (Figures 4(a) and 4(b)). Herein, our data presented that anti-miR-21 + sh-CXCR4 significantly inhibited the cell invasiveness compared to single-targeted knockdowns (anti-miR-21 and sh-CXCR4) (Figures 4(a) and 4(b)). Anti-miR-21 or sh-CXCR4 alone significantly reduced migration in both U87 and U251 cells, while anti-miR-21 + sh-CXCR4 suppressed the cell migration compared to single-targeted knockdowns in both cell lines (Figures 4(c) and 4(d)). This revealed the potent repressive effects of anti-miR-21 + sh-CXCR4 on invasion and migration capabilities of glioma cells.
Figure 4.
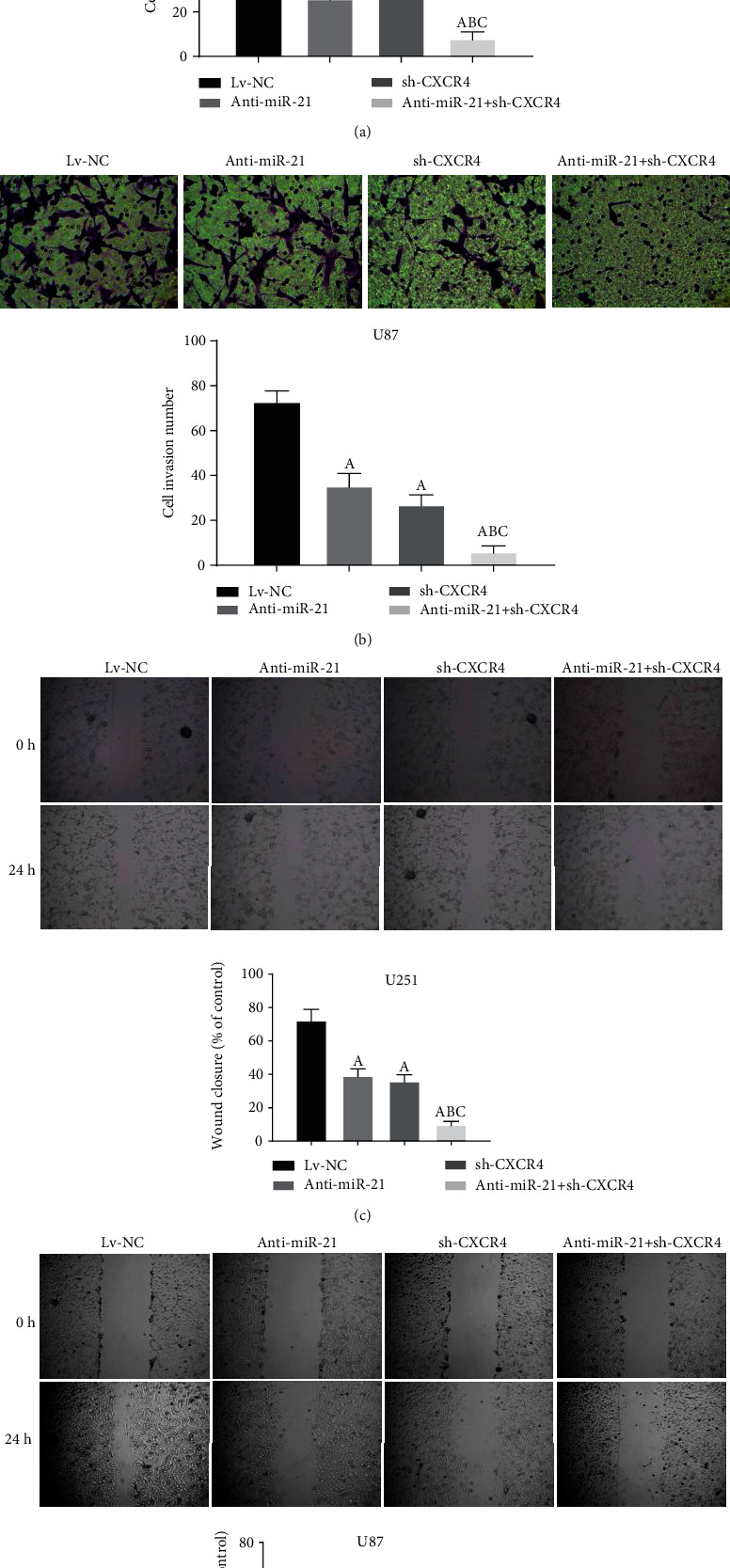
Double-targeted knockdown of miR-21 and CXCR4 inhibited invasion and migration of glioma cells. (a, b) Transwell assay was performed on the transfected U251 and U87 cells. Magnification, 200x. (c, d) Scratch assays were performed to detect migration of transfected cells. Magnification, 40x. P < 0.05, (A) vs. Lv-NC group; (B) vs. anti-miR-21 group; (C) vs. sh-CXCR4 group.
3.5. Double-Targeted Knockdown of miR-21 and CXCR4 Slowed Tumor Growth in Glioma Xenograft Mouse Model
To better illustrate the roles of miR-21 and CXCR4 together in tumor growth of glioma, we employed a U87 xenograft mouse model. No obvious difference between the body weight of mice from different treatment groups (Lv-NC, anti-miR-21, sh-CXCR4, and anti-miR-21 + sh-CXCR4, Figure 5(a)). We measured tumor volume every 5 days. The growth of glioma tumor was significantly suppressed in anti-miR-21 or sh-CXCR4 treatment alone groups compared to negative control, while the anti-miR-21 + sh-CXCR4 treatment obviously reduced the tumor growth compared to the anti-miR-21 or sh-CXCR4 group (Figures 5(b) and 5(c)). Furthermore, the miR-21 and CXCR4 levels in xenografts were detected. And further, the results had confirmed that the remarkable effect on tumor growth suppression was contributed by inhibition of miR-21 and CXCR4 (Figures 5(d)–5(f)). Thus, our data reported that anti-miR-21 + sh-CXCR4 could diminish growth of glioma xenograft in vivo.
Figure 5.
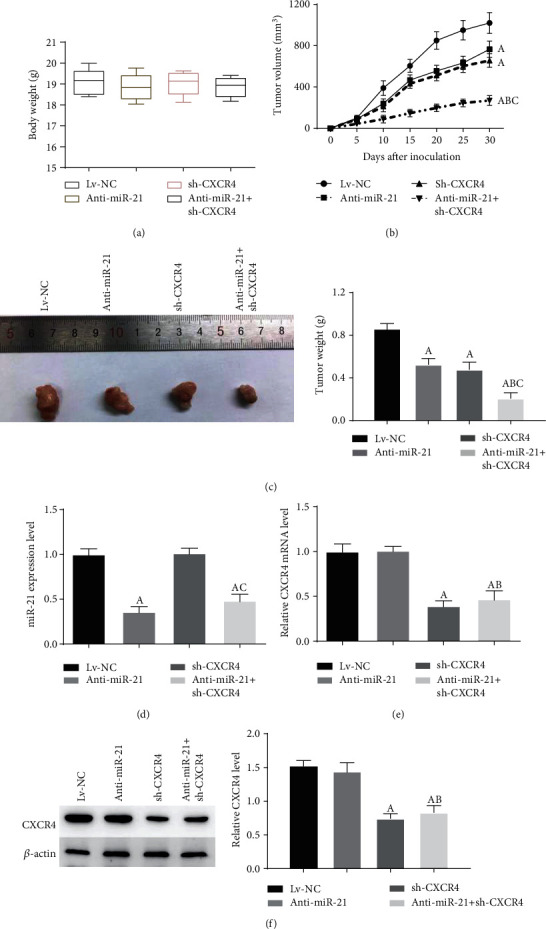
Double-targeted knockdown of miR-21 and CXCR4 inhibited glioma xenograft growth. (a) Mice were weighted at 30 day postinoculation. (b) Tumor volumes were monitored for 30 days. (c) Tumors were dissected and weighted. (d, e) miR-21 and CXCR4 levels in xenograft tumor tissues were measured using qRT-PCR. (f) CXCR4 protein levels were tested using western blot. P < 0.05, (A) vs. Lv-NC group; (B) vs. anti-miR-21 group; (C) vs. sh-CXCR4 group.
3.6. Double-Targeted Knockdown of miR-21 and CXCR4 Inhibited Malignant Glioma Progression by Suppressing of the PI3K/AKT and Raf/MEK/ERK Pathways
We further investigated the potential downstream pathways that might be responsible for the repressive effect of anti-miR-21 + sh-CXCR4 on tumor progression. In this study, we focused on two pathways, (1) PI3K/AKT and (2) Raf/MEK/ERK, which both play a vital role in glioma cell fate and are aberrant in the miR-21 and/or CXCR4 overexpression environment [11, 30–34]. The results from western blot analysis presented no statistically difference in AKT, ERK1/2 among different treatment groups both in glioma cells and xenografts (Figures 6(a)–6(c)). However, anti-miR-21, sh-CXCR4, and anti-miR-21 + sh-CXCR4 all showed a significantly suppressive effect on p-AKT and p-ERK1/2, with anti-miR-21 + sh-CXCR4 group presenting the strongest inhibition effect in both cell lines and xenografts. These results demonstrated a potent suppressive effect of anti-miR-21 + sh-CXCR4 on activation of AKT axis and ERK axis. These results suggested that double-targeted knockdown of miR-21 and CXCR4 inhibited malignant glioma progression by suppressing of the PI3K/AKT and Raf/MEK/ERK pathways.
Figure 6.
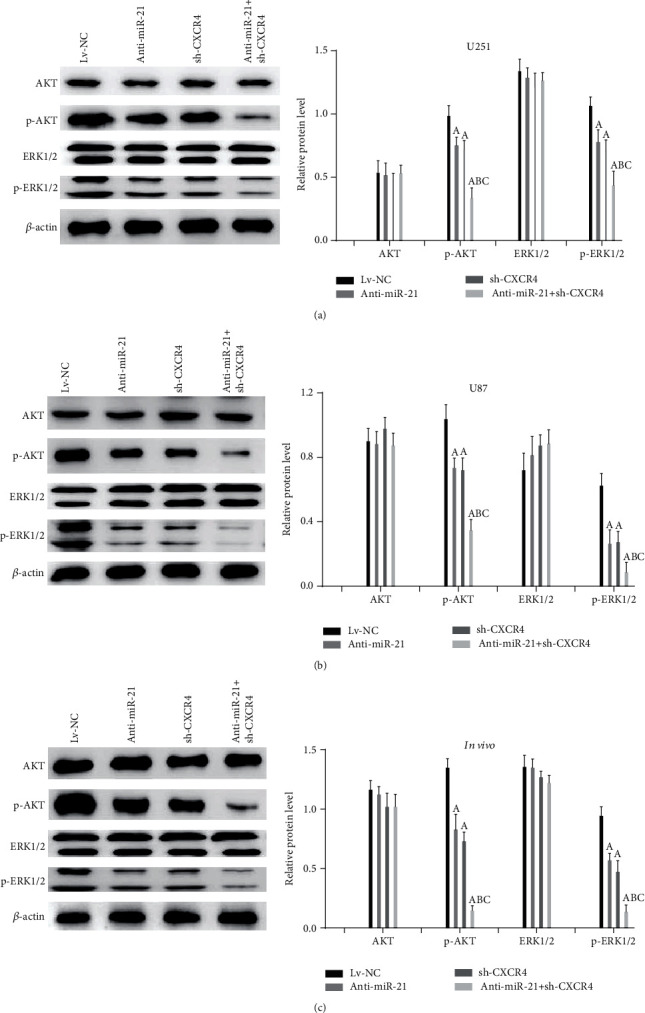
Double-targeted knockdown of miR-21 and CXCR4 inhibited the PI3K/AKT and Raf/MEK/ERK pathways in glioma xenograft tissues and cells. (a–c) Related proteins were tested using western blot. P < 0.05, (A) vs. Lv-NC group; (B) vs. anti-miR-21 group; (C) vs. sh-CXCR4 group.
4. Discussion
Glioma is the most common primary malignant brain tumor. Several novel therapies have been developed for the treatment of malignant glioma, including gene therapy, targeted molecular therapy, immunotherapy, and stem therapy. Among them, gene therapy presents great efficiency in suppressing glioma progression [34]. Particularly, the combination of multigene therapy tends to be more effective than the single-gene one [35]. It is reported that patients undergoing surgery and tumor resection between 1 cm and 2 cm from the tumor and have absence of tumor cells in this area had better survival than patients who had the presence of tumor cells [36]. In addition, studies show that there are some shared gene expression and molecular pathways in glioblastoma tissues and peritumoral regions, and there are differences between short-term and long-term survivors [37, 38]. In our study, we confirmed that the miR-21 and CXCR4 expressions were significantly upregulated in glioma tissues compared to normal tissues and cells, which suggested that miR-21 and CXCR4 might play important roles in mediating malignant glioma aggressiveness, as well as the tumorigenicity.
miR-21 is the first miRNA reported involved in human malignant glioma regulation [9]. Its expression has been reported positively correlated with clinical grade of glioma malignancy [5, 10]. Also, the vital roles of miR-21 in glioma initiation, maintenance, and survival have been proved by numerous studies [5, 6, 10, 12, 39]. Studies show that miR-21 downregulation represses cell proliferation and invasiveness, enhances apoptosis, and sensitizes resistant to chemo- and radiotherapy [5, 13, 39, 40]. In our study, we confirmed the increased miR-21 in malignant glioma tissues and glioma cells as well as the regulation of it in glioma progression.
CXCR4 is another factor that plays a vital role in glioma pathology (i.e., neoplastic transformation, malignant tumor progression, infiltration, metastasis, angiogenesis, and vasculogeneis) [21, 41, 42]. Our data confirmed that the CXCR4 expression was upregulated in malignant glioma tissues and glioma cells in comparison to normal tissues or glial cells. The potential of CXCR4 as a promising therapeutic target for malignant glioma treatment has been extensively investigated, and several targeting drugs have already been undergoing clinical trials [21]. However, most research topics are focused on employing CXCR4 antagonist as a single treatment or in complement with radio- or chemotherapy [21]. In our present study, we reported that double-targeted knockdown of miR-21 and CXCR4 presented a better suppressive effect on tumor progression when compared to single-targeted knockdown of miR-21 or CXCR4 alone. Anti-miR-21 + sh-CXCR4 almost abrogated the proliferation, cell invasion, and migration in migration-prone glioma cells. Also, anti-miR-21 + sh-CXCR4 repressed xenograft tumor growth.
Both PI3K/AKT and Raf/MEK/ERK are common and crucial signaling for cancer development [43]. Activated ATK and ERK phosphorylate and activated downstream proteins involved in regulating cell survival, proliferation, and metabolic pathways [44, 45]. Studies demonstrate that dysregulation of both pathways are observed in various cancers, and the inhibition of both pathways would be promising strategy for cancer therapy [44]. A published study has shown that CXCR4-mediated activation of downstream proproliferative signaling through the Raf/MEK/Erk or PI3K/Akt pathways is modulated by PTEN status in cells [46]. Many studies have reported that miR-21 regulates cell proliferation and apoptosis via targeting the PTEN/PI3K/AKT pathway in cancer [47–49]. miR-21 overexpression has been reported to activate the Raf/MEK/ERk pathway in vestibular schwannomas cells [50]. In addition, a previous report has shown that ERK1/2 activation is detected in glioblastoma multiforme and in peritumor tissues, which suggests that the ERK1/2 pathway may participate in GBM growth, tumor invasion, and progression [51]. A study has shown that there are some shared molecular pathways in glioblastoma tissues and peritumoral regions [37]. According to the above studies, both miR-21 and CXCR4 are involved in positively regulating the Akt and MAPK pathways. However, direct inhibition of these pathways is unpractical given the complexity of their crosstalk regulatory mechanisms [52].
A previous study has shown that in the gene therapy of cancer, multigene silencing was better than single gene silencing in regulating cell functions, which may provide a promising treatment in cancer [53]. Interestingly, our results presented that double-targeted knockdown of miR-21 and CXCR4 powerfully suppressed PI3K/AKT and Raf/MEK/ERK activation, which might have significant therapeutic values and offer some insights for further investigation of the molecular mechanism in glioma pathology. Since the overexpression of miR-21 and/or CXCR4 is also observed in many other cancers, such as ovarian, gastric, colonic, pancreatic, breast, and prostate cancers [9, 54], our study could also shed some light on the efficacious treatment development for them.
5. Conclusion
In summary, compared to knockdown of miR-21 or CXCR4, double-targeted knockdown of miR-21 and CXCR4 could more effectively inhibit the proliferation, migration, invasion, and growth of transplanted tumor and promote cell apoptosis, which were involved in the PI3K/AKT and Raf/MEK/ERK signaling pathways. Therefore, double-targeted miR-21 and CXCR4 may be potential and promising therapeutic targets for malignant glioma salvage treatment.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Yang X. J., Long H. A., Sheng-ping Y. U. Study on invasion and migration of malignant glioma. Chinese Journal of Contemporary Neurology & Neurosurgery. 2018;18(1):p. 36. [Google Scholar]
- 2.Sottoriva A., Spiteri I., Piccirillo S. G. M., et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences of United States of America. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skalsky R. L., Cullen B. R. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 2011;6(9, article e24248) doi: 10.1371/journal.pone.0024248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Põlajeva J., Swartling F. J., Jiang Y., et al. miRNA-21 is developmentally regulated in mouse brain and is co-expressed with SOX2 in glioma. BMC Cancer. 2012;12(1) doi: 10.1186/1471-2407-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lei B. X., Liu Z. H., Li Z. J., Li C., Deng Y. F. miR-21 induces cell proliferation and suppresses the chemosensitivity in glioblastoma cells via downregulation of FOXO1. International Journal of Clinical and Experimental Medicine. 2014;7(8):2060–2066. [PMC free article] [PubMed] [Google Scholar]
- 6.Tao T., Wang Y., Luo H., et al. Involvement of FOS-mediated miR-181b/miR-21 signalling in the progression of malignant gliomas. European Journal of Cancer. 2013;49(14):3055–3063. doi: 10.1016/j.ejca.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Kwak H. J., Kim Y. J., Chun K. R., et al. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene. 2011;30(21):2433–2442. doi: 10.1038/onc.2010.620. [DOI] [PubMed] [Google Scholar]
- 8.Gabriely G., Wurdinger T., Kesari S., et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Molecular and Cellular Biology. 2008;28(17):5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melnik B. C. MiR-21: an environmental driver of malignant melanoma? Journal of Translational Medicine. 2015;13(1) doi: 10.1186/s12967-015-0570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermansen S. K., Nielsen B. S., Aaberg-Jessen C., Kristensen B. W. miR-21 is linked to glioma angiogenesis: a co-localization study. Journal of Histochemistry & Cytochemistry. 2015;64(2):138–148. doi: 10.1369/0022155415623515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao Y.-Y., Zhang T.-L., Wu L.-X., et al. AKT axis, miR-21, and RECK play pivotal roles in dihydroartemisinin killing malignant glioma cells. International Journal of Molecular Sciences. 2017;18(2):p. 350. doi: 10.3390/ijms18020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo G., Luo W., Sun X., et al. MicroRNA-21 promotes migration and invasion of glioma cells via activation of Sox2 and β-catenin signaling. Molecular Medicine Reports. 2017;15(1):187–193. doi: 10.3892/mmr.2016.5971. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Gwak H.-S., Kim T. H., Jo G. H., et al. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012;7(10):p. e47449. doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C., Cui G. H., Liu F., Wu Q. L., Chen Y. The role of stromal cell derived factor-1/CXCR4 biological axis in tumor metastasis of non-Hodgkin lymphoma. Zhonghua yi xue za zhi. 2007;87(10):695–697. [PubMed] [Google Scholar]
- 15.Chatterjee S., Azad B. B., Nimmagadda S. The Intricate Role of CXCR4 in Cancer. Advances in Cancer Research. 2014;124:31–82. doi: 10.1016/b978-0-12-411638-2.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bajetto A., Barbieri F., Dorcaratto A., et al. Expression of CXC chemokine receptors 1–5 and their ligands in human glioma tissues: role of CXCR4 and SDF1 in glioma cell proliferation and migration. Neurochemistry International. 2006;49(5):423–432. doi: 10.1016/j.neuint.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Redjal N., Chan J. A., Segal R. A., Kung A. L. CXCR4 inhibition synergizes with cytotoxic chemotherapy in gliomas. Clinical Cancer Research. 2006;12(22):6765–6771. doi: 10.1158/1078-0432.CCR-06-1372. [DOI] [PubMed] [Google Scholar]
- 18.Rubin J. B., Kung A. L., Klein R. S., et al. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proceedings of the National Academy of Sciences of the United States of America. 2011;100(23):13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercurio L., Ajmone-Cat M. A., Cecchetti S., et al. Targeting CXCR4 by a selective peptide antagonist modulates tumor microenvironment and microglia reactivity in a human glioblastoma model. Journal of Experimental & Clinical Cancer Research. 2016;35(1) doi: 10.1186/s13046-016-0326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagner J.-P., Sarfraz Y., Ortenzi V., et al. Multifaceted C-X-C chemokine receptor 4 (CXCR4) inhibition interferes with anti–vascular endothelial growth factor therapy–induced glioma dissemination. The American Journal of Pathology. 2017;187(9):2080–2094. doi: 10.1016/j.ajpath.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckert F., Schilbach K., Klumpp L., et al. Potential role of CXCR4 targeting in the context of radiotherapy and immunotherapy of cancer. Frontiers in Immunology. 2018;9 doi: 10.3389/fimmu.2018.03018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercurio L., Cecchetti S., Ricci A., et al. Phosphatidylcholine-specific phospholipase C inhibition down- regulates CXCR4 expression and interferes with proliferation, invasion and glycolysis in glioma cells. PLoS One. 2017;12(4, article e0176108) doi: 10.1371/journal.pone.0176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Q., Wu H., Wang H., et al. N-terminal polypeptide derived from vMIP-II exerts its antitumor activity by inhibiting the CXCR4 pathway in human glioma. International Journal of Oncology. 2017;50(4):1160–1174. doi: 10.3892/ijo.2017.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yadav V. N., Zamler D., Baker G. J., et al. CXCR4 increases in-vivo glioma perivascular invasion, and reduces radiation induced apoptosis: a genetic knockdown study. Oncotarget. 2016;7(50):83701–83719. doi: 10.18632/oncotarget.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi W.-T., Yang Y., Xu Y., An J. Targeting chemokine receptor CXCR4 for treatment of HIV-1 infection, tumor progression, and metastasis. Current Topics in Medicinal Chemistry. 2014;14(13):1574–1589. doi: 10.2174/1568026614666140827143541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Xie Y., Oupický D. Potential of CXCR4/CXCL12 chemokine axis in cancer drug delivery. Current Pharmacology Reports. 2016;2(1):1–10. doi: 10.1007/s40495-015-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esencay M., Newcomb E. W., Zagzag D. HGF upregulates CXCR4 expression in gliomas via NF-κB: implications for glioma cell migration. Journal of Neuro-Oncology. 2010;99(1):33–40. doi: 10.1007/s11060-010-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin E. Y., Cooper D. D., Abbott K. L., et al. Neural precursor-derived pleiotrophin mediates subventricular zone invasion by glioma. Cell. 2017;170(5):845–859.e19. doi: 10.1016/j.cell.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Li Q., Lin L., et al. Targeting the Notch1 oncogene by miR-139-5p inhibits glioma metastasis and epithelial-mesenchymal transition (EMT) BMC Neurology. 2018;18(1):p. 133. doi: 10.1186/s12883-018-1139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ching C. B., Hansel D. E. Expanding therapeutic targets in bladder cancer: the PI3K/Akt/mTOR pathway. Laboratory Investigation. 2010;90(10):1406–1414. doi: 10.1038/labinvest.2010.133. [DOI] [PubMed] [Google Scholar]
- 31.Du W., Pang C., Xue Y., Zhang Q., Wei X. Dihydroartemisinin inhibits the Raf/ERK/MEK and PI3K/AKT pathways in glioma cells. Oncology Letters. 2015;10(5):3266–3270. doi: 10.3892/ol.2015.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D., Zuo D., Luan C., et al. Glioma cell proliferation controlled by ERK activity-dependent surface expression of PDGFRA. PLoS One. 2014;9(1, article e87281) doi: 10.1371/journal.pone.0087281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo G., Yao W., Zhang Q., Bo Y. Oleanolic acid suppresses migration and invasion of malignant glioma cells by inactivating MAPK/ERK signaling pathway. PLoS One. 2013;8(8, article e72079) doi: 10.1371/journal.pone.0072079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y., Wu Z., Wu T., et al. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death & Disease. 2016;7(3, article e2123) doi: 10.1038/cddis.2015.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J., Jin C., Hao S., Luo G., Fu D. Pancreatic cancer: gene therapy approaches and gene delivery systems. Expert Opinion on Biological Therapy. 2010;10(1):73–88. doi: 10.1517/14712590903321454. [DOI] [PubMed] [Google Scholar]
- 36.Mangiola A., de Bonis P., Maira G., et al. Invasive tumor cells and prognosis in a selected population of patients with glioblastoma multiforme. Cancer. 2008;113(4):841–846. doi: 10.1002/cncr.23624. [DOI] [PubMed] [Google Scholar]
- 37.Fazi B., Felsani A., Grassi L., et al. The transcriptome and miRNome profiling of glioblastoma tissues and peritumoral regions highlights molecular pathways shared by tumors and surrounding areas and reveals differences between short-term and long-term survivors. Oncotarget. 2015;6(26):22526–22552. doi: 10.18632/oncotarget.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangiola A., Saulnier N., de Bonis P., et al. Gene expression profile of glioblastoma peritumoral tissue: an ex vivo study. PLoS One. 2013;8(3, article e57145) doi: 10.1371/journal.pone.0057145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeh W.-L., Lin H.-Y., Huang C.-Y., et al. Migration-prone glioma cells show curcumin resistance associated with enhanced expression of miR-21 and invasion/anti-apoptosis-related proteins. Oncotarget. 2015;6(35):37770–37781. doi: 10.18632/oncotarget.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P., Liu Y.-h., Yao Y.-l., et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cellular Signalling. 2015;27(2):275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Dutt P., et al. Stromal cell-derived factor-1α and stem cell factor/kit ligand share signaling pathways in hemopoietic progenitors: a potential mechanism for cooperative induction of chemotaxis. The Journal of Immunology. 1998;161(7):3652–3658. [PubMed] [Google Scholar]
- 42.Ono A., Freed E. O. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. Journal of Virology. 1999;73(5):4136–4144. doi: 10.1128/JVI.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pillozzi S., Masselli M., de Lorenzo E., et al. Chemotherapy resistance in acute lymphoblastic leukemia requires hERG1 channels and is overcome by hERG1 blockers. Blood. 2011;117(3):902–914. doi: 10.1182/blood-2010-01-262691. [DOI] [PubMed] [Google Scholar]
- 44.Dueppers P., Jankowiak S., Schelzig H., Wagenhäuser M. U., Oberhuber A. Spontaneous rupture of an isolated iliac artery dissection in a young man because of cystic medial degeneration Erdheim–Gsell. Annals of Vascular Surgery. 2015;29(3):596.e11–596.e13. doi: 10.1016/j.avsg.2014.10.041. [DOI] [PubMed] [Google Scholar]
- 45.Redman E. K., Brookes P. S., Karcz M. K. Role of p90(RSK) in regulating the Crabtree effect: implications for cancer. Biochemical Society Transactions. 2013;41(1):124–126. doi: 10.1042/BST20120277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Begley L. A., Kasina S., Shah R. B., Macoska J. A. Signaling mechanisms coupled to CXCL12/CXCR4-mediated cellular proliferation are PTEN-dependent. American Journal of Clinical and Experimental Urology. 2015;3(2):91–99. [PMC free article] [PubMed] [Google Scholar]
- 47.Hao X. J., Xu C. Z., Wang J. T., et al. miR-21 promotes proliferation and inhibits apoptosis of hepatic stellate cells through targeting PTEN/PI3K/AKT pathway. Journal of Receptor and Signal Transduction Research. 2018;38(5-6):455–461. doi: 10.1080/10799893.2019.1585452. [DOI] [PubMed] [Google Scholar]
- 48.Liu H. Y., Zhang Y. Y., Zhu B. L., et al. miR-21 regulates the proliferation and apoptosis of ovarian cancer cells through PTEN/PI3K/AKT. European Review for Medical and Pharmacological Sciences. 2019;23(10):4149–4155. doi: 10.26355/eurrev_201905_17917. [DOI] [PubMed] [Google Scholar]
- 49.Shi B., Deng W., Long X., et al. miR-21 increases c-kit+cardiac stem cell proliferationin vitrothrough PTEN/PI3K/Akt signaling. PeerJ. 2017;5, article e2859 doi: 10.7717/peerj.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang P., Sun D., Jiang F. Ailanthone promotes human vestibular schwannoma cell apoptosis and autophagy by downregulation of miR-21. Oncology Research. 2018;26(6):941–948. doi: 10.3727/096504018X15149775533331. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Lama G., Mangiola A., Anile C., et al. Activated ERK1/2 expression in glioblastoma multiforme and in peritumor tissue. International Journal of Oncology. 2007;30(6):1333–1342. [PubMed] [Google Scholar]
- 52.Ghayad S., Cohen P. Inhibitors of the PI3K/Akt/mTOR pathway: new hope for breast cancer patients. Recent Patents on Anti-Cancer Drug Discovery. 2010;5(1):29–57. doi: 10.2174/157489210789702208. [DOI] [PubMed] [Google Scholar]
- 53.Song Y., Dong M. M., Yang H. F. Effect of silencing VEGF, c-myc, survivin and hTERT by multiple shRNA expression vectors on the xenografted human nasopharyngeal carcinoma in nude mice. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2010;45(11):935–940. [PubMed] [Google Scholar]
- 54.Richardson P. J. CXCR4 and glioblastoma. Anti-Cancer Agents in Medicinal Chemistry. 2016;16(1):59–74. doi: 10.2174/1871520615666150824153032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.


