Significance
Balanced rearrangements involving 11q23/KMT2A are among the most frequent chromosomal abnormalities in acute myeloid leukemia (AML). We analyzed the mutational status of 81 genes, clinical features and outcomes of patients with recurring 11q23/KMT2A rearrangements. We found that mutations in genes of the RAS pathway were most frequent, and that there were differences in mutation patterns among patients with different 11q23/KMT2A rearrangements. Outcomes of patients age <60 y with t(9;11)(p22;q23)/KMT2A-MLLT3, currently classified in the intermediate-risk group of the 2017 European LeukemiaNet classification, were superior to outcomes of intermediate-risk patients without t(9;11). Older patients with t(9;11) and patients with other 11q23/KMT2A rearrangements had poor outcomes. Our study improves understanding of the mutational landscape and clinical implications for AML patients with 11q23/KMT2A rearrangements.
Keywords: acute myeloid leukemia, KMT2A, next-generation sequencing, gene mutations, clinical outcome
Abstract
Balanced rearrangements involving the KMT2A gene, located at 11q23, are among the most frequent chromosome aberrations in acute myeloid leukemia (AML). Because of numerous fusion partners, the mutational landscape and prognostic impact of specific 11q23/KMT2A rearrangements are not fully understood. We analyzed clinical features of 172 adults with AML and recurrent 11q23/KMT2A rearrangements, 141 of whom had outcome data available. We compared outcomes of these patients with outcomes of 1,097 patients without an 11q23/KMT2A rearrangement categorized according to the 2017 European LeukemiaNet (ELN) classification. Using targeted next-generation sequencing, we investigated the mutational status of 81 leukemia/cancer-associated genes in 96 patients with 11q23/KMT2A rearrangements with material for molecular studies available. Patients with 11q23/KMT2A rearrangements had a low number of additional gene mutations (median, 1; range 0 to 6), which involved the RAS pathway (KRAS, NRAS, and PTPN11) in 32% of patients. KRAS mutations occurred more often in patients with t(6;11)(q27;q23)/KMT2A-AFDN compared with patients with the other 11q23/KMT2A subsets. Specific gene mutations were too infrequent in patients with specific 11q23/KMT2A rearrangements to assess their associations with outcomes. We demonstrate that younger (age <60 y) patients with t(9;11)(p22;q23)/KMT2A-MLLT3 had better outcomes than patients with other 11q23/KMT2A rearrangements and those without 11q23/KMT2A rearrangements classified in the 2017 ELN intermediate-risk group. Conversely, outcomes of older patients (age ≥60 y) with t(9;11)(p22;q23) were poor and comparable to those of the ELN adverse-risk group patients. Our study shows that patients with an 11q23/KMT2A rearrangement have distinct mutational patterns and outcomes depending on the fusion partner.
In acute myeloid leukemia (AML), recurring cytogenetic abnormalities are frequently present, provide important prognostic information, and guide therapeutic decisions (1–5). Recurrent, balanced rearrangements involving the lysine methyltransferase 2a (KMT2A) gene (also known as the MLL gene), which is located at 11q23 and encodes a histone H3 lysine 4 methyltransferase, occur in ∼3% to 7% of adult patients with de novo AML (3–13). Balanced chromosome rearrangements involving 11q23 and KMT2A are very heterogeneous, with at least 77 different 11q23/KMT2A fusion partners reported in AML patients (6). Most of the rearrangements result in fusion proteins, which can deregulate the transcriptional program of cells, lead to specific gene expression patterns that include high expression of HOXA genes and thereby contribute to leukemogenesis (14–16).
Among the recurrent 11q23/KMT2A rearrangements in AML, the most common is t(9;11)(p22;q23) [hereinafter referred to as t(9;11)], which results in a fusion of KMT2A with the MLLT3 gene (previously known as AF9). AML with this translocation is recognized as a specific disease entity in the 2016 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia (17). Other translocations common in AML include t(6;11)(q27;q23) [t(6;11) hereinafter] resulting in a fusion with AFDN (previously known as MLLT4 and AF6); t(11;19)(q23;p13.1), resulting in a fusion with ELL; and t(11;19)(q23;p13.3), resulting in a fusion with MLLT1 (also termed ENL) (3, 4, 6, 7, 10, 18).
Previous studies have suggested that patient prognosis is associated with the 11q23/KMT2A fusion partner. For example, patients with t(9;11) had better outcomes compared with patients with other 11q23/KMT2A rearrangements in several studies of both adult (7, 9, 10) and pediatric AML (19), although some other studies (12, 13) did not confirm these results. In addition, in a large series of children with AML, t(1;11)(q21;q23) was associated with favorable clinical outcomes and t(6;11)(q27;q23), t(10;11)(p12;q23), and t(10;11)(p11.2;q23) were associated with unfavorable clinical outcomes independent of other prognostic factors (20). The 2017 European LeukemiaNet (ELN) risk stratification schema includes AML patients with t(9;11) in the intermediate-risk group, whereas patients with all other 11q23/KMT2A balanced rearrangements [t(v;11)(v;q23) hereinafter] are included in the adverse-risk group, regardless of age (1). Further evaluation of the clinical parameters and outcomes of the AML patients with specific recurrent 11q23/KMT2A rearrangements potentially may contribute to refining of the current ELN classification.
Along with cytogenetics, gene mutations could potentially improve risk stratification of AML patients (1, 2). Previous studies of mutations in AML patients with specific balanced 11q23/KMT2A rearrangements have reported a predominance of RAS mutations in patients with 11q23/KMT2A rearrangements (21–23). Whether the presence of additional gene mutations influences the clinical outcomes of AML patients with 11q23/KMT2A rearrangements is currently unclear.
To address this question, we analyzed pretreatment characteristics, clinical outcomes and mutational data of 81 leukemia/cancer-associated genes (24) in AML patients with well-defined 11q23/KMT2A rearrangements, who were similarly treated on Cancer and Leukemia Group B (CALGB)/Alliance for Clinical Trials in Oncology (Alliance) protocols.
Results
Frequency and Pretreatment Characteristics of Patients with Specific 11q23/KMT2A Rearrangements.
Among 172 patients with 11q23/KMT2A rearrangements, the most frequent abnormality was t(9;11)/KMT2A-MLLT3, detected in 75 patients (44%), followed by t(6;11)/KMT2A-AFDN in 29 patients (17%), t(11;19)(q23;p13.1)/KMT2A-ELL in 20 (12%), t/ins(10;11)(p13;q23)/KMT2A-MLLT10 in 14 (8%), t(11;19)(q23;p13.3)/KMT2A-MLLT1 in 11 (6%), and t(11;17)(q23;q25)/KMT2A-SEPTIN9 in 5 (3%). An additional 18 patients had less common translocations, each of which occurred in only one or two patients. Detailed cytogenetic data of these patients, who constitute a subset referred to hereafter as the “other 11q23/KMT2A translocations,” are provided in SI Appendix, Table S1. In addition, details of secondary chromosome abnormalities detected in patients with the most frequently recurring 11q23/KMT2A rearrangements are provided in SI Appendix, Table S2.
The pretreatment characteristics of AML patients with 11q23/KMT2A rearrangements are shown in SI Appendix, Table S3. The median age of the entire patient cohort was 43 y (range, 17–84 y), and 80% of the patients were younger than 60 y at diagnosis. The age distribution among patient groups with specific 11q23/KMT2A rearrangements did not differ significantly (P = 0.65). There were differences in the percentages of blood (P = 0.001) and bone marrow (BM) (P < 0.001) blasts among patient groups with different 11q23/KMT2A rearrangements. Patients with t(11;19)(q23;p13.1) had the lowest blast percentages in BM (median, 55%) and blood (median, 11%), whereas patients with t/ins(10;11) had the highest blast percentages in BM (median, 89%) and blood (median, 77%) (SI Appendix, Table S3).
Mutational Landscape of AML Patients with 11q23/KMT2A Rearrangements.
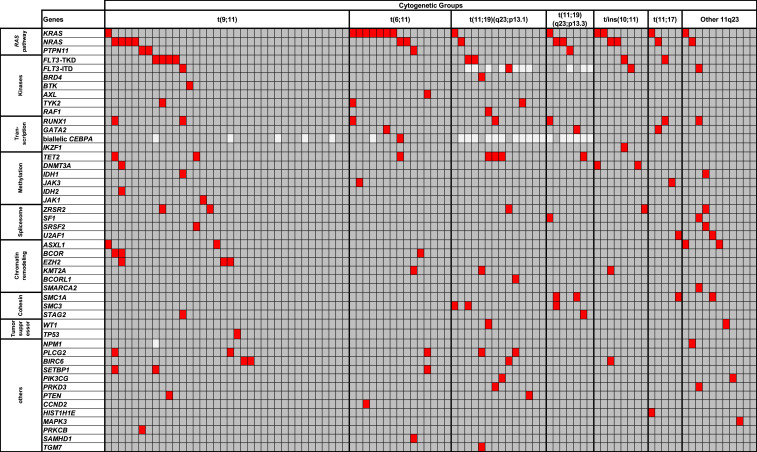
Ninety-six patients had material available for molecular analysis using our targeted sequencing panel. We identified 134 gene mutations in these 96 patients (median, 1 mutation per patient; range, 0 to 6). In line with previous reports (21–23), the most frequently mutated genes were KRAS (in 14 of 96 patients; 15%) and NRAS (in 13 patients; 14%). Mutations in KRAS and NRAS were mutually exclusive in our cohort, as were PTPN11 mutations, found in four patients (4%) (Fig. 1). Altogether, mutations in the genes composing the RAS pathway (KRAS, NRAS, and PTPN11) were detected in 32% of patients with 11q23/KMT2A rearrangements. Beside RAS pathway mutations, only tyrosine kinase domain mutations in the FLT3 gene (FLT3-TKD; 8%) and mutations in the RUNX1 (7%), TET2 (7%), PLCG2 (5%), and ZRSR2 (5%) genes were found with a frequency of at least 5%. Other recurrent gene mutations that are relatively common in AML patients without 11q23/KMT2A rearrangements, such as internal tandem duplications of the FLT3 gene (FLT3-ITD), mutations in the DNMT3A and NPM1 genes, and biallelic mutations of CEBPA (1), were infrequent in patients with 11q23/KMT2A rearrangements, detected in 4%, 3%, 1%, and 1% of the patients, respectively.
Fig. 1.
Oncoprint of mutations detected in 96 patients with AML harboring recurrent rearrangements involving 11q23/KMT2A. Each column represents one patient, and each row represents a single gene. Red, mutated; dark gray, wild-type; light gray, mutation status not determined. The frequencies of these gene mutations in patients with each recurrent 11q23/KMT2A rearrangement are provided in SI Appendix, Table S4. No mutation was detected in any of the following genes analyzed: AKT1, ARAF, ATM, BCL2, BRAF, BRINP3, CBL, CCND1, CSNK1A1, CTNNB1, ETV6, FBXW7, GATA1, GSK3B, HNRNPK, IKZF3, IL7R, JAK2, KIT, KLHL6, MAPK1, MED12, MYD88, NOTCH1, PHF6, PIK3CD, PLEKHG5, RAD21, SF3A1, SF3B1, SYK, U2AF2, XPO1, and ZMYM3.
We next compared patient subsets with different 11q23/KMT2A rearrangements to identify molecular similarities and differences. The distribution of mutations in these subsets is shown in Fig. 1, and these mutations are listed in SI Appendix, Table S4. All analyzed patient subsets had a low number of mutations in common. Whereas each patient with a t/ins(10;11) or t(11;17) had at least one mutation, 36% of patients with t(9;11), 20% of those with t(6;11), and 20% of those with other 11q23/KMT2A translocations did not harbor mutations in any of the genes that we sequenced (Fig. 1).
As mentioned above, the most frequently mutated genes were KRAS and NRAS, and they were mutated in at least one patient in each of the 11q23/KMT2A subsets. The distribution of NRAS mutations was not significantly different among the subsets (P = 0.67). On the other hand, KRAS mutations occurred with a significantly higher frequency in patients with t(6;11) compared with patients with all other 11q23/KMT2A rearrangements combined (47% vs. 9%; P = 0.001).
Variant Allele Fractions of Detected Mutations and the Proportion of Cells with the 11q23/KMT2A Rearrangement.
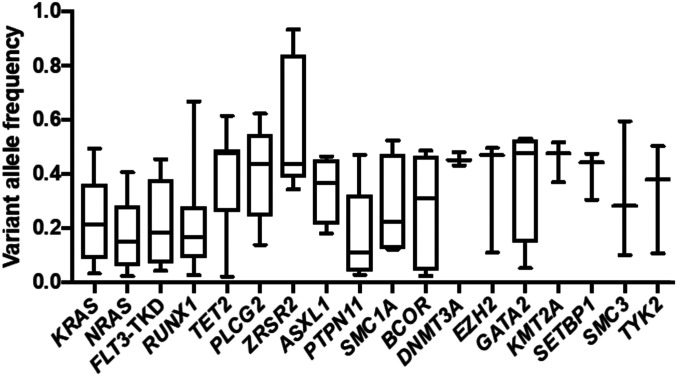
Next, we determined variant allele fractions (VAFs) for the 18 most frequent gene mutations assessed using targeted amplicon sequencing (Fig. 2). Similar to previous studies (21, 25), here the detected gene mutations had a broad range of VAFs. Higher VAFs were found for mutations in such genes as ZRSR2, PLCG2, GATA2, TET2, ASXL1, and DNMT3A, the last three of which are known to be recurrently mutated in age-associated clonal hematopoiesis of indeterminate potential (26, 27). On the other hand, mutations in the KRAS, NRAS, PTPN11, and RUNX1 genes or the FLT3-TKD had lower VAFs, indicating that these mutations may be subclonal and likely occurred later in leukemogenesis. In contrast, when we analyzed the percentages of metaphase cells containing the rearrangements involving 11q23/KMT2A in G-banded preparations, we found that in a vast majority of the patients (165 of 172; 96%), these rearrangements were found in >50% of the analyzed metaphase cells (median, 100%; range, 17 to 100%). Likewise, in the 34 patients for whom interphase fluorescence in situ hybridization data were available, 31 (91%) harbored rearrangements of the KMT2A gene in >50% of the analyzed nuclei (median, 86.5%; range, 10 to 100%). These data suggest that the 11q23/KMT2A rearrangement is an important and possibly early event in leukemogenesis.
Fig. 2.
VAFs for the 18 genes most frequently mutated in patients with AML and 11q23/KMT2A rearrangements.
Treatment Outcomes of Patients with 11q23/KMT2A Rearrangements.
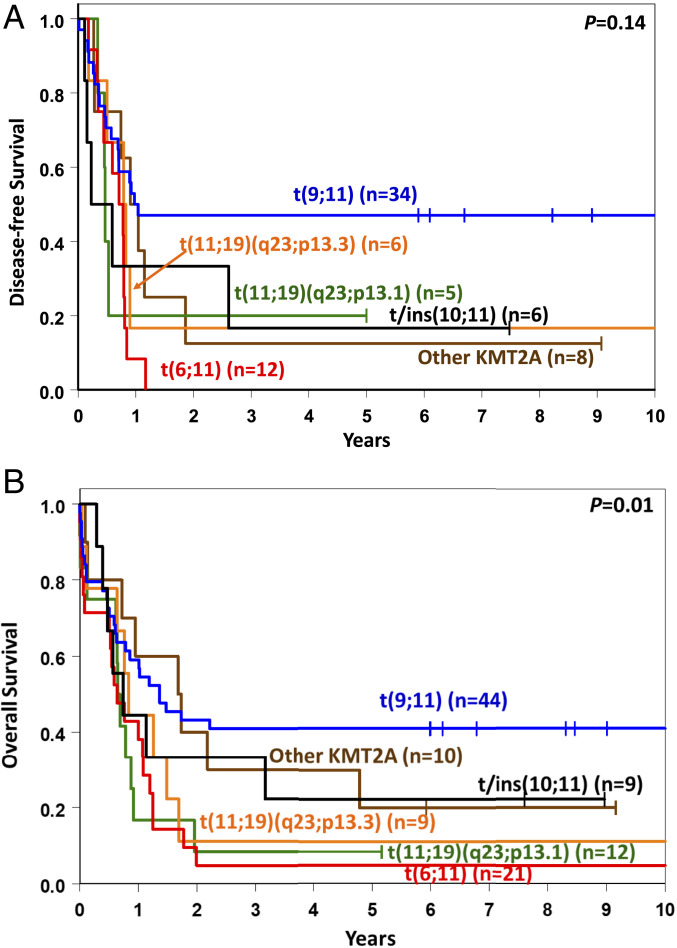
Among 108 younger adult patients (age <60 y) for whom outcome data were available, those with different 11q23/KMT2A rearrangements had varying complete remission (CR) rates, with the lowest CR rate of 42% in patients with t(11;19)(q23;p13.1), a 57% rate in patients with t(6;11), and the highest rates, 77% and 80%, in patients with t(9;11) and patients with other 11q23/KMT2A translocations, respectively (Table 1). However, a high CR rate translated into a relatively good outcome only for younger patients with t(9;11), with a 3-y disease-free (DFS) rate of 47% and an overall survival (OS) rate of 41%. Patients with t(6;11) had the shortest survival, with no patients surviving 3 y disease-free and only one patient (5%) alive at 3 y. The OS of patients in the remaining 11q23/KMT2A groups was also rather poor, with 3-y OS rates of 8% for patients with t(11;19)(q23;p13.1), 11% for those with t(11;19)(q23;p13.3), 30% for those with other 11q23/KMT2A translocations, and 33% for those with t/ins(10;11) (Table 1 and Fig. 3). Consequently, because of the similarity in outcomes of patients with 11q23/KMT2A rearrangements other than t(9;11), we combined these patients into one group, denoted t(v;11)(v;q23). As expected, patients with t(v;11)(v;q23) had a shorter DFS (3-y rate: 13% vs. 47%, P = 0.007) and OS (3-y rate: 16% vs. 41%, P = 0.004) than patients with t(9;11) (SI Appendix, Table S5 and Fig. S1).
Table 1.
Outcomes of younger adult patients (age <60 y) with AML and 11q23/KMT2A rearrangements
| Rearrangements involving 11q23/KMT2A* | CR, % | DFS | OS | ||
| Median, y | % Disease-free at 3 y (95% CI) | Median, y | % Alive at 3 y (95% CI) | ||
| t(9;11)(p22;q23) (n = 44) | 77 | 1.0 | 47 (30–63) | 1.4 | 41 (26–55) |
| t(6;11)(q27;q23) (n = 21) | 57 | 0.7 | 0 | 0.6 | 5 (0–20) |
| t(11;19)(q23;p13.1) (n = 12) | 42 | 0.5† | ‒ | 0.7 | 8 (1–31) |
| t(11;19)(q23;p13.3) (n = 9) | 67 | 0.8‡ | ‒ | 0.8 | 11 (1–39) |
| t/ins (10;11)(p13;q23) (n = 9) | 67 | 0.4§ | ‒ | 0.7 | 33 (8–62) |
| Other t(11q23) (n = 10) | 80 | 1.0 | 13 (1–42) | 1.7 | 30 (7–58) |
| All patients (n = 108) | 68 | 0.8 | 29 (19–39) | 0.9 | 26 (18–34) |
CI, confidence interval.
The group of patients with t(11;17) is not listed in the table because only three patients were available for outcome analyses. These patients are included in the “all patients” group.
The five patients who achieved CR had the following DFS: 0.3, 0.5, 0.5, 0.5, and 5.0+ (+ indicates that the patient has not died and DFS is determined using last follow-up date) years.
The six patients who achieved CR had the following DFS: 0.2, 0.5, 0.8, 0.8, 0.9, and 11.1+ years.
The six patients who achieved CR had the following DFS: 0.1, 0.2, 0.2, 0.6, 2.6 and 7.5+ years.
Fig. 3.
Clinical outcomes of younger adult patients (age <60 y) with AML harboring the most frequent recurring rearrangements involving 11q23/KMT2A. (A) DFS. (B) OS.
In addition, since several previous studies have shown that mutational patterns can refine risk stratification of AML patients (1, 2, 17, 25, 28), we assessed the impact of the presence of one or more additional mutations vs. no mutation on the outcomes of younger patients with t(9;11) and those with all remaining 11q23/KMT2A rearrangements combined into a single group. As shown in SI Appendix, Fig. S2 A and B, younger patients with t(9;11) who harbored at least one additional mutation tended to have a longer DFS (P = 0.07) than patients without any mutation in addition to t(9;11); however, there was no significant difference in OS between these patient groups. The mutations found in patients with t(9;11) were heterogeneous; among 15 different mutations, only mutations in the NRAS gene were detected in three patients, and FLT3-TKD and mutations in the PLCG2, PTPN11, TET2, and ZRSR2 genes were detected in two patients each. In contrast, the presence of additional mutations had no significant impact on the DFS or OS of patients with t(v;11)(v;q23) (SI Appendix, Fig. S2 C and D). Notably, the numbers of patients in both of the foregoing comparisons were small.
Among 33 older patients (age ≥60 y) in our cohort for whom outcome data were available, 16 harbored t(9;11) and 17 had other rearrangements involving 11q23/KMT2A. Because of relatively small patient numbers, we were only able to compare outcomes of patients with t(9;11) and all those with t(v;11)(v;q23) combined. We found that the former tended to have better rates of CR (75% vs. 41%; P = 0.08), but not of DFS (3-y rate, 0% vs. 29%; P = 0.12) or OS (3-y rate, 6% vs. 12%; P = 0.92) (SI Appendix, Fig. S3).
Outcomes of Patients with de novo 11q23/KMT2A-Rearranged AML with Respect to the 2017 ELN Classification.
According to the 2017 ELN recommendations (1), patients with t(9;11) are classified in the intermediate-risk group regardless of age or any accompanying gene mutations, whereas patients with t(v;11)(v;q23) are included in the adverse-risk group. We compared clinical outcomes of patients with t(9;11) and, separately, of those with t(v;11)(v;q23) with outcomes of 1,097 similarly treated AML patients without 11q23/KMT2A rearrangements who were classified into the 2017 ELN favorable-, intermediate-, or adverse-risk groups (1).
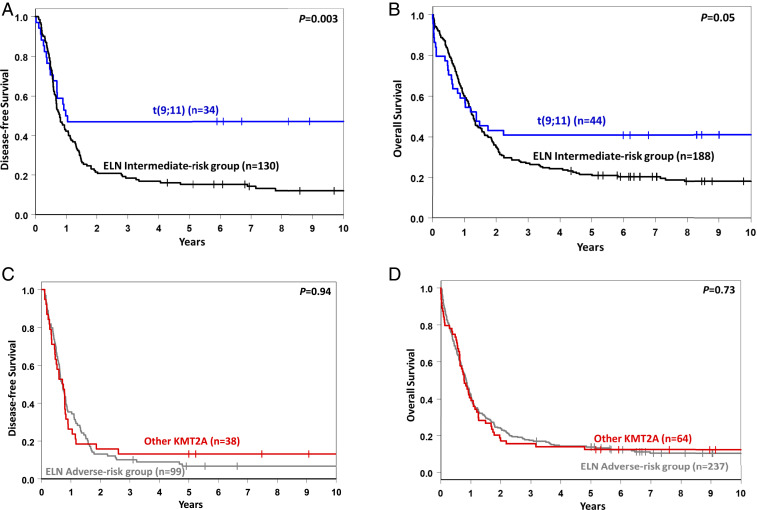
We compared the baseline clinical characteristics and treatment outcome of 44 younger patients with t(9;11) with those of 188 younger patients classified in the ELN intermediate-risk group who did not have t(9;11). The former had lower white blood cell counts (median, 11.9 × 109/L vs. 28.8 × 109/L; P = 0.01), a higher percentage of BM blasts (median, 87% vs. 73%; P < 0.001), and more frequent extramedullary involvement (43% vs. 23%; P = 0.01) than other patients in the ELN intermediate-risk group (SI Appendix, Table S6). Concerning outcome, although CR rates did not differ significantly (77% vs. 70%; P = 0.36), patients with t(9;11) had longer DFS (3-y rate, 47% vs. 19%; P = 0.003; Fig. 4A) and OS (3-y rate, 41% vs. 27%; P = 0.05) than non-t(9;11) ELN intermediate-risk patients (Fig. 4B and SI Appendix, Table S7).
Fig. 4.
Outcomes of younger adult patients (age <60 y) with AML. (A and B) DFS (A) and OS (B) of patients with t(9;11) vs. other patients classified in the 2017 ELN intermediate-risk group. (C and D) DFS (C) and OS (D) of patients with 11q23/KMT2A rearrangements other than t(9;11) [i.e., t(v;11)(v;q23)] vs. other patients classified in the 2017 ELN adverse-risk group.
We then examined how the improved outcomes of 44 younger patients with t(9;11) compared with the outcomes of younger patients classified in the 2017 ELN favorable-risk group (n = 449). A comparison of pretreatment clinical characteristics showed that patients with t(9;11) had lower white blood cell counts (median, 11.9 × 109/L vs. 25.0 × 109/L; P = 0.03), and higher percentages of BM blasts (median, 87% vs. 63%; P < 0.001) than other patients in the 2017 ELN favorable-risk group (SI Appendix, Table S8). With regard to outcome, we found that younger patients with t(9;11) tended to have a lower CR rates than patients classified in the 2017 ELN favorable-risk group (77% vs. 88%; P = 0.06), as well as a shorter OS (3-y rate, 41% vs. 61%; P = 0.03). DFS did not differ significantly between the two groups (3-y rate, 47% vs. 52%; P = 0.83) (SI Appendix, Table S9 and Fig. S4).
Patients with t(v;11)(v;q23) are classified in the 2017 ELN adverse-risk group. We compared pretreatment characteristics and outcomes of 63 younger patients with t(v;11)(v;q23) with those of 237 patients classified in the ELN adverse-risk group who did not carry t(v;11)(v;q23). The former were younger (median age, 38 y vs. 49 y; P < 0.001) and had higher percentages of BM blasts (median, 78% vs. 65%; P < 0.001), higher hemoglobin levels (median, 9.5 g/dL vs. 9.0 g/dL; P = 0.05), and a higher frequency of extramedullary disease (34% vs. 19%; P = 0.02; SI Appendix, Table S10). The CR rate of patients with t(v;11)(v;q23)/KMT2A-rearranged AML was higher than that of the other ELN adverse-risk patients (60% vs. 42%; P = 0.01). However, there were no significant between-group differences in either DFS (3-y rate, 13% vs. 10%; P = 0.91; Fig. 4C) or OS (3-y rate, 16% vs. 17%; P = 0.73; Fig. 4D and SI Appendix, Table S11).
Since DFS and OS did not differ significantly between older patients with t(9;11) and those with t(v;11)(v;q23), we compared the outcomes of 16 older patients with t(9;11) with those of 223 older non–11q23/KMT2A-rearranged patients classified in the 2017 ELN adverse-risk group. As shown in SI Appendix, Table S12, there was no statistically significant difference in either DFS (3-y rate, 17% vs. 3%; P = 0.33) or OS (3-y rate, 6% vs. 4%; P = 0.56) between the older patients with t(9;11) and older patients classified in the 2017 ELN adverse-risk group, even though the former group had a higher CR rate (75% vs. 29%; P < 0.001).
Discussion
Our data on a relatively large cohort of AML patients with t(9;11) and other recurrent balanced rearrangements involving 11q23/KMT2A reveal both similarities and differences among AML patients with specific 11q23/KMT2A translocations. Translocation t(9;11) was the most common, followed by t(6;11) and t(11;19)(q23;p13.1). This is consistent with previous studies, although several of those studies combined patients with t(11;19)(q23;p13.1) and patients with t(11;19)(q23;p13.3) into one subset, making assessment of the incidence of each type of t(11;19) difficult (3, 8, 9). Both the data presented here and in our previous report (29) emphasize the fact that t(11;19)(q23;p13.1) and t(11;19)(q23;p13.3) represent two distinct rearrangements associated with unique clinical and molecular features and thus should not be considered to represent a single rearrangement.
Two previous studies used next-generation sequencing to analyze AML patients with several separate recurrent 11q23/KMT2A rearrangements (21, 22). Grossmann et al. (22) examined the mutational status of 15 genes in 85 patients representing five different 11q23/KMT2A subsets and found a high incidence (∼50%) of mutations in genes constituting the RAS signaling pathway: NRAS, 22%; KRAS, 20%; PTPN11, 3%; and BRAF, 2.5%. Similar results were reported by Lavallée et al. (21), who used a whole transcriptome approach to investigate 31 patients belonging to five different 11q23/KMT2A subgroups. Our analysis of the molecular landscape of 96 de novo AML patients with 11q23/KMT2A rearrangements not only substantiates the high frequency of RAS signaling pathway mutations (although we did not detect any mutations in the BRAF gene), but also reveal an uneven distribution of specific RAS mutations among the subgroups with different 11q23/KMT2A rearrangements. Whereas the NRAS mutations occurred with a similar frequency among these subgroups, we observed—in contrast to the aforementioned studies (21, 22)—the highest incidence of KRAS mutations in patients with t(6;11), 47% of whom carried this mutation, as opposed to only 9% of patients with other 11q23/KMT2A rearrangements, including only 3% of patients with t(9;11). Importantly, the VAFs of mutations in all RAS signaling pathway genes were low, which is consistent with the literature data indicating that RAS mutations are likely subclonal and acquired after the formation of other genetic abnormalities, such as the 11q23/KMT2A rearrangements (21, 22). Indeed, our observation that in most patients, 11q23/KMT2A rearrangements were found in either all or the majority of metaphase cells analyzed supports the view that acquisition of the 11q23/KMT2A rearrangement represents an early, disease-initiating event during leukemogenesis (30). Nevertheless, even though the mutations in the RAS pathway genes and in the FLT3 gene are subclonal and possibly acquired later, they can accelerate the onset of leukemia and promote disease progression and clonal expansion of KMT2A-MLLT3-positive cells, as demonstrated by a recent study using a retroviral mouse AML model (31). Further studies are needed to determine the exact roles played by other infrequent but recurring mutations that we identified in our cohort of 11q23/KMT2A-rearranged patients.
The prognostic impact of the 11q23/KMT2A rearrangements in AML has been studied for >30 y, but these studies have produced somewhat conflicting results (3–5, 7, 9–13, 18–20, 32, 33). Our initial report (7) demonstrating better outcomes in adults with t(9;11) than in patients with 11q23/KMT2A-balanced abnormalities involving other partner chromosomes was confirmed by a study from the German AML Intergroup in a large cohort of younger patients (up to 60 y) that also included patients with secondary and therapy-related AML (10). Moreover, Chen et al. (9) found improved outcomes of patients with t(9;11) in a cohort comprising younger and older patients. However, our present study demonstrates that the prognostic significance of t(9;11) is dependent on patient age. The younger patients with de novo AML and t(9;11) have had significantly better outcomes than patients with all other 11q23/KMT2A rearrangements combined. In contrast, the outcomes of older patients with t(9;11) were very poor and similar to the outcomes of patients with t(v;11)(v;q23), confirming our earlier observations (28).
In the current 2017 ELN classification, the presence of t(9;11) is a criterion for classifying all patients with this translocation in the intermediate-risk group, irrespective of patient age or the potential coexistence of rare, concurrent adverse-risk gene mutations (1). Our data show that AML patients age <60 y with t(9;11) have significantly better DFS and OS than other AML patients classified in the ELN intermediate-risk group. However, even though the DFS of these younger patients with t(9;11) was not significantly different from that of patients categorized in the 2017 ELN favorable-risk group, the OS of the former group was shorter and CR rates tended to be lower than those of the latter group. We note that we only analyzed patients with de novo AML, which is important not only because the frequency of 11q23/KMT2A rearrangements is increased in patients with therapy-related AML, especially after treatment with topoisomerase II inhibitors (34), but also because the clinical outcome of patients with therapy-related AML with t(9;11) is very poor (35).
The clinical outcome of de novo AML patients age ≥60 y with t(9;11) was also very poor. In fact, their survival was comparable to that of older patients classified in the 2017 ELN adverse-risk group, both those who harbored 11q23/KMT2A rearrangements other than t(9;11) and those with other adverse-risk cytogenetic and molecular abnormalities. However, the relatively small number of older patients with t(9;11) that we studied (n = 16) makes confirmation of our results necessary.
Our comparison of outcomes of younger patients with 11q23/KMT2A rearrangements other than t(9;11) with those of non–11q23/KMT2A-rearranged patients classified in the 2017 ELN adverse-risk group revealed that the higher CR rates of patients with t(v;11)(v;q23) did not translate into a benefit in DFS or OS. This indicates the need for allogeneic stem cell transplantation (allo-SCT) and/or novel therapeutic approaches for these patients.
In summary, our analysis of the mutational landscape of patients with 11q23/KMT2A rearrangements revealed a relatively low number of mutations, the most common of which involve the RAS pathway. With regard to outcome, patients with t(v;11)(v;q23) had short survival regardless of age, but there was an age difference for patients with t(9;11). The younger patients had relatively favorable outcomes, whereas the older patients with t(9;11) had very poor outcomes.
Methods
Patients and Treatment.
We investigated 172 adult patients diagnosed with de novo AML and a balanced 11q23/KMT2A rearrangement for whom clinical pretreatment data were available. For outcome analyses, patients who underwent allo-SCT in first CR were excluded. This was done because most CALGB treatment protocols did not allow performing allo-SCT in first CR on the study, and patients undergoing allo-SCT had to be taken off the protocol, resulting in either absent or incomplete follow-up data. Hence, we analyzed the outcomes of 141 adult patients with a balanced 11q23/KMT2A rearrangement. We also analyzed outcomes of 1,097 adults with de novo AML who had no 11q23/KMT2A rearrangement and were classified by the 2017 ELN risk groups as a comparison cohort. Because of differences in the treatment protocols, outcomes of younger adult patients (age <60 y) were analyzed separately from those of older patients (age ≥60 y). Almost all patients (98.9%) received similar cytarabine/daunorubicin-based induction chemotherapy on the CALGB/Alliance trials, and most younger patients were enrolled onto protocols with consolidation consisting of high-dose chemotherapy and autologous SCT (36–45); 1.1% of the patients were treated with decitabine with or without bortezomib (46). Details of the treatment protocols are provided in SI Appendix. In addition, 96 of the 172 adult patients with AML diagnosed with a balanced 11q23/KMT2A had material available for molecular studies and were analyzed using targeted amplicon sequencing (24). All patients provided written informed consent, and study protocols were approved by the Institutional Review Board at each participating center (listed in SI Appendix) in accordance with the Declaration of Helsinki.
Cytogenetic and Molecular Analyses.
All patients were enrolled on companion protocols CALGB 8461 (cytogenetic studies), CALGB 9665 (leukemia tissue bank), and CALGB 20202 (molecular studies). Cytogenetic analyses of pretreatment BM and/or blood samples were performed by institutional laboratories approved by the CALGB/Alliance using unstimulated short-term (24- or 48-h) cultures. Cytogenetic results were confirmed by central karyotype review (47).
Viable cryopreserved BM or blood cells were stored for future analyses before starting treatment. Mononuclear cells from BM or blood were enriched by Ficoll-Hypaque gradient and cryopreserved in liquid nitrogen until thawed at 37 °C for analysis. DNA extractions were performed using the Qiagen DNeasy Blood and Tissue Kit. The mutational status of 80 protein-coding genes was determined centrally at The Ohio State University by targeted amplicon sequencing using the Illumina MiSeq platform as described previously (24). All variants that occurred with VAFs of <0.10 or were sequenced to a depth of <15 reads were defined as not mutated. In addition, variants were excluded when they occurred only in one read direction if sequenced in both directions, if the region contained many variants with low quality scores, or if they occurred in all analyzed samples including run controls. Samples with high background noise were also excluded from analysis. Samples were considered nonevaluable for a specific gene if ≥85% of the amplicons covering the target regions within the coding sequence of the gene were sequenced to a depth of <15 reads. Testing for the presence or absence of FLT3-ITD was performed as described previously (48). In addition to the 80 genes analyzed using the targeted amplicon sequencing panel, testing for CEBPA mutations was performed with Sanger sequencing as described previously (49); thus, mutational status was assessed in a total of 81 genes. In accordance with the revised WHO classification of myeloid neoplasms and acute leukemias (17), only patients with biallelic CEBPA mutations were considered CEBPA-mutated.
Statistical Analyses.
Clinical endpoints were defined as described previously (1) and in SI Appendix. Basic clinical characteristics were compared using Fisher’s exact test for categorical variables and Wilcoxon’s rank-sum test for continuous variables (50). Estimated probabilities of DFS and OS were calculated using the Kaplan–Meier method, and differences between survival distributions were evaluated using the log-rank test (50). All analyses were performed by the Alliance Statistics and Data Center on a database locked on January 10, 2019, using SAS 9.4 and TIBCO Spotfire S+ 8.2.
Supplementary Material
Acknowledgments
We thank the patients who participated in the clinical trials and their families supporting them; Donna Bucci, Christopher Manring, and the CALGB/Alliance Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center for sample processing and storage services; and Lisa J. Sterling for data management. This work was supported in part by the National Cancer Institute (Grants CA233338, CA101140, CA140158, CA180861, CA196171, CA016058, CA180821, CA180882, and CA077658, and Grant R35 CA198183, to J.C.B.), the Leukemia Clinical Research Foundation, the Warren D. Brown Foundation, the Pelotonia Fellowship Program (to A.-K.E.), and by allocation of computing resources from The Ohio Supercomputer Center. Support to Alliance for Clinical Trials in Oncology and Alliance Foundation Trials programs is listed at https://acknowledgments.alliancefound.org. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interest statement: J.S.B. serves as a consultant/advisory board member for AbbVie, AstraZeneca, and KITE Pharma. The other authors declare no potential conflicts of interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014732117/-/DCSupplemental.
Data Availability.
All study data are included in main text and SI Appendix.
References
- 1.Döhner H. et al., Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Döhner H., Weisdorf D. J., Bloomfield C. D., Acute myeloid leukemia. N. Engl. J. Med. 373, 1136–1152 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Byrd J. C. et al., Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461). Blood 100, 4325–4336 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D. et al.; National Cancer Research Institute Adult Leukaemia Working Group , Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 116, 354–365 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Grimwade D., Mrózek K., Diagnostic and prognostic value of cytogenetics in acute myeloid leukemia. Hematol. Oncol. Clin. North Am. 25, 1135–1161, vii (2011). [DOI] [PubMed] [Google Scholar]
- 6.Meyer C. et al., The MLL recombinome of acute leukemias in 2017. Leukemia 32, 273–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mrózek K. et al., Adult patients with de novo acute myeloid leukemia and t(9;11)(p22;q23) have a superior outcome to patients with other translocations involving band 11q23: A Cancer and Leukemia Group B study. Blood 90, 4532–4538 (1997). [PubMed] [Google Scholar]
- 8.Cox M. C. et al., Chromosomal aberration of the 11q23 locus in acute leukemia and frequency of MLL gene translocation: Results in 378 adult patients. Am. J. Clin. Pathol. 122, 298–306 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Chen Y. et al., Prognostic significance of 11q23 aberrations in adult acute myeloid leukemia and the role of allogeneic stem cell transplantation. Leukemia 27, 836–842 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krauter J. et al., Prognostic factors in adult patients up to 60 years old with acute myeloid leukemia and translocations of chromosome band 11q23: Individual patient data-based meta-analysis of the German Acute Myeloid Leukemia Intergroup. J. Clin. Oncol. 27, 3000–3006 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Mrózek K., Heerema N. A., Bloomfield C. D., Cytogenetics in acute leukemia. Blood Rev. 18, 115–136 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Schoch C. et al., AML with 11q23/MLL abnormalities as defined by the WHO classification: Incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood 102, 2395–2402 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Harrison C. J. et al., Cytogenetics of childhood acute myeloid leukemia: United Kingdom Medical Research Council treatment trials AML 10 and 12. J. Clin. Oncol. 28, 2674–2681 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Armstrong S. A. et al., MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30, 41–47 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Mullighan C. G. et al., Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia 21, 2000–2009 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Valk P. J. M. et al., Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 350, 1617–1628 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Arber D. A. et al., The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127, 2391–2405 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Blum W. et al., Adult de novo acute myeloid leukemia with t(6;11)(q27;q23): Results from Cancer and Leukemia Group B study 8461 and review of the literature. Cancer 101, 1420–1427 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Climent J. A., Espinosa R. 3rd, Thirman M. J., Le Beau M. M., Rowley J. D., Abnormalities of chromosome band 11q23 and the MLL gene in pediatric myelomonocytic and monoblastic leukemias. Identification of the t(9;11) as an indicator of long survival. J. Pediatr. Hematol. Oncol. 17, 277–283 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Balgobind B. V. et al., Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: Results of an international retrospective study. Blood 114, 2489–2496 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavallée V.-P. et al., The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat. Genet. 47, 1030–1037 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Grossmann V. et al., High incidence of RAS signalling pathway mutations in MLL-rearranged acute myeloid leukemia. Leukemia 27, 1933–1936 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Sandhöfer N. et al., Dual PI3K/mTOR inhibition shows antileukemic activity in MLL-rearranged acute myeloid leukemia. Leukemia 29, 828–838 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Eisfeld A.-K. et al., The mutational oncoprint of recurrent cytogenetic abnormalities in adult patients with de novo acute myeloid leukemia. Leukemia 31, 2211–2218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metzeler K. H. et al.; AMLCG Study Group , Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 128, 686–698 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Rothenberg-Thurley M. et al., Persistence of pre-leukemic clones during first remission and risk of relapse in acute myeloid leukemia. Leukemia 32, 1598–1608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal S. et al., Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mrózek K. et al., Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J. Clin. Oncol. 30, 4515–4523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhatnagar B. et al., Clinical features and gene- and microRNA-expression patterns in adult acute leukemia patients with t(11;19)(q23;p13.1) and t(11;19)(q23;p13.3). Leukemia 30, 1586–1589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ley T. J. et al.; Cancer Genome Atlas Research Network , Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyrenius-Wittsten A. et al., De novo activating mutations drive clonal evolution and enhance clonal fitness in KMT2A-rearranged leukemia. Nat. Commun. 9, 1770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arthur D. C. et al., The clinical significance of karyotype in acute myelogenous leukemia. Cancer Genet. Cytogenet. 40, 203–216 (1989). [DOI] [PubMed] [Google Scholar]
- 33.Grimwade D. et al.; The Medical Research Council Adult and Children’s Leukaemia Working Parties , The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. Blood 92, 2322–2333 (1998). [PubMed] [Google Scholar]
- 34.Gill Super H. J. et al., Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood 82, 3705–3711 (1993). [PubMed] [Google Scholar]
- 35.Bloomfield C. D. et al., 11q23 balanced chromosome aberrations in treatment-related myelodysplastic syndromes and acute leukemia: Report from an International Workshop. Genes Chromosomes Cancer 33, 362–378 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Mayer R. J. et al.; Cancer and Leukemia Group B , Intensive postremission chemotherapy in adults with acute myeloid leukemia. N. Engl. J. Med. 331, 896–903 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Moore J. O. et al., Sequential multiagent chemotherapy is not superior to high-dose cytarabine alone as postremission intensification therapy for acute myeloid leukemia in adults under 60 years of age: Cancer and Leukemia Group B Study 9222. Blood 105, 3420–3427 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolitz J. E. et al., Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: Final induction results of Cancer and Leukemia Group B study 9621. J. Clin. Oncol. 22, 4290–4301 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Marcucci G. et al., A phase III randomized trial of intensive induction and consolidation chemotherapy ± oblimersen, a pro-apoptotic Bcl-2 antisense oligonucleotide in untreated acute myeloid leukemia patients >60 years old. J. Clin. Oncol. 25, 7012 (2007). [Google Scholar]
- 40.Attar E. C. et al., Bortezomib added to daunorubicin and cytarabine during induction therapy and to intermediate-dose cytarabine for consolidation in patients with previously untreated acute myeloid leukemia age 60 to 75 years: CALGB (Alliance) study 10502. J. Clin. Oncol. 31, 923–929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blum W. et al.; Alliance for Clinical Trials in Oncology , Maintenance therapy with decitabine in younger adults with acute myeloid leukemia in first remission: A phase 2 Cancer and Leukemia Group B study (CALGB 10503). Leukemia 31, 34–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stone R. M. et al.; Cancer and Leukemia Group B , Granulocyte-macrophage colony-stimulating factor after initial chemotherapy for elderly patients with primary acute myelogenous leukemia. N. Engl. J. Med. 332, 1671–1677 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Kolitz J. E. et al.; Cancer and Leukemia Group B , P-glycoprotein inhibition using valspodar (PSC-833) does not improve outcomes for patients younger than age 60 years with newly diagnosed acute myeloid leukemia: Cancer and Leukemia Group B study 19808. Blood 116, 1413–1421 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee E. J. et al., Parallel phase I studies of daunorubicin given with cytarabine and etoposide with or without the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age or older with acute myeloid leukemia: Results of Cancer and Leukemia Group B study 9420. J. Clin. Oncol. 17, 2831–2839 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Baer M. R. et al., Low-dose interleukin-2 immunotherapy does not improve outcome of patients age 60 years and older with acute myeloid leukemia in first complete remission: Cancer and Leukemia Group B study 9720. J. Clin. Oncol. 26, 4934–4939 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roboz G. J. et al., A randomized trial of 10 days of decitabine alone or with bortezomib in previously untreated older patients with acute myeloid leukemia: CALGB 11002 (Alliance). Blood Adv. 2, 3608–3617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mrózek K. et al., Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int. J. Oncol. 33, 239–244 (2008). [PMC free article] [PubMed] [Google Scholar]
- 48.Whitman S. P. et al., Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: A Cancer and Leukemia Group B study. Cancer Res. 61, 7233–7239 (2001). [PubMed] [Google Scholar]
- 49.Marcucci G. et al., Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J. Clin. Oncol. 26, 5078–5087 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vittinghoff E., Glidden D. V., Shiboski S. C., McCulloch C. E., Regression Methods in Biostatistics: Linear, Logistic, Survival and Repeated- Measures Models, (Springer, 2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in main text and SI Appendix.