Significance
SARS-CoV-2 is an emerging pathogen that has already had catastrophic consequences on the health and well-being of people worldwide. As a zoonotic virus, the implications for animal populations are largely unknown. This manuscript describes a pilot study in which domestic cats and dogs were assessed for their susceptibility to infection. While neither species developed clinical disease in this study, cats shed infectious virus for up to 5 d and infected naive cats via direct contact, while dogs do not appear to shed virus. Cats that were reinfected with SARS-CoV-2 mounted an effective immune response and did not become reinfected. These studies have important implications for animal health and suggest that cats may be a good model for vaccine development.
Keywords: cats, SARS-CoV-2, experimental infection, transmission
Abstract
The pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has reached nearly every country in the world with extraordinary person-to-person transmission. The most likely original source of the virus was spillover from an animal reservoir and subsequent adaptation to humans sometime during the winter of 2019 in Wuhan Province, China. Because of its genetic similarity to SARS-CoV-1, it is probable that this novel virus has a similar host range and receptor specificity. Due to concern for human–pet transmission, we investigated the susceptibility of domestic cats and dogs to infection and potential for infected cats to transmit to naive cats. We report that cats are highly susceptible to infection, with a prolonged period of oral and nasal viral shedding that is not accompanied by clinical signs, and are capable of direct contact transmission to other cats. These studies confirm that cats are susceptible to productive SARS-CoV-2 infection, but are unlikely to develop clinical disease. Further, we document that cats developed a robust neutralizing antibody response that prevented reinfection following a second viral challenge. Conversely, we found that dogs do not shed virus following infection but do seroconvert and mount an antiviral neutralizing antibody response. There is currently no evidence that cats or dogs play a significant role in human infection; however, reverse zoonosis is possible if infected owners expose their domestic pets to the virus during acute infection. Resistance to reinfection holds promise that a vaccine strategy may protect cats and, by extension, humans.
The coronavirus disease (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), originated in the Wuhan province of China in late 2019 and within 4 mo spread to nearly every country in the world. Sequence analysis and epidemiological investigations suggest that the virus was of animal origin, possibly bat, and was potentially first introduced into the human population via an intermediate animal host in the Huanan seafood market in Wuhan, China (1, 2). The virus quickly adapted to humans, and human-to-human transmission became the almost immediate source of subsequent infections, with direct contact and aerosol droplets as the primary routes of infection (3). Early indications suggested that SARS-CoV-2, much like SARS-CoV-1, infects host cells by binding to the angiotensin-converting enzyme 2 (ACE2), a receptor that is expressed in many animal species, although notably not in mice or rats (4). Thus, while humans are almost certainly the sole source of infection to other humans, multiple early studies suggest other animals are susceptible to infection as well (5–7).
The first report of reverse zoonosis, or transmission from human to animal, was reported from Hong Kong, where a COVID-19 patient’s dog tested PCR-positive for SARS-CoV-2 multiple times (5). In following weeks, other instances of domestic pets becoming infected following exposure to humans were documented, including another dog in Hong Kong and a cat with clinical disease in Belgium (6). Serologic studies so far have failed to identify domestic dogs and cats as a primary source of human infection (8). Importantly, a survey of veterinary students with confirmed COVID-19 infection was unable to identify SARS-CoV-2 antibodies in their pets (9). Despite the low probability of pet-to-human or human-to-pet transmission, it remains important to clarify what role, if any, that domestic pets play in SARS-CoV-2 transmission.
The first published study involving cat experimental infections showed that cats could become infected by SARS-CoV-2 and potentially transmit virus to other cats via aerosols, as determined by PCR-positive fecal samples from cats in cages in the same room as directly infected cats. This study also described pathology and mortality in juvenile cats euthanized at 3 and 7 days post infection (DPI) (7). Additional communications described viral shedding and direct contact transmission in cats as well as seroconversion in cats exposed to infected humans (10, 11). The experiments described herein expand upon existing work by providing shedding kinetics in cats over time, assessing virus neutralization, seroconversion, assessing pathology, and exploring transmission. This is the first report of protective immunity against SARS-CoV-2 in cats following repeated exposure. These studies indicate that cats may serve as a suitable animal model for studying SARS-CoV-2 infection and for furthering the development of vaccines and therapeutics for use in both animals and humans. We also confirm an earlier report that dogs do not replicate virus in the upper respiratory tract (7), but document evidence of antiviral neutralizing activity in postexposure canine sera. The role of cats in zoonotic transmission remains an open question, but relatively short duration of shedding and resistance to reinfection suggests risk of this is very low, particularly when cats are kept indoors and thus have limited contact with humans or other susceptible animals.
Materials and Methods
Virus.
SARS-CoV-2 virus strain WA1/2020WY96 was obtained from BEI Resources, passaged twice in Vero E6 cells and stocks frozen at −80 °C in Dulbecco’s Modified Eagle Medium (DMEM) with 5% fetal bovine serum and antibiotics. Virus stock was titrated on Vero E6 cells using standard double-overlay plaque assay (12), and plaques were counted 72 h post infection to determine plaque-forming units (pfu) per mL.
Animals.
Seven adult (1 male, 6 female, 5–8 y old) cats were obtained from a closed breeding colony held at Colorado State University in a pathogen-free environment in an Association for Assessment and Accreditation of Laboratory Animal Care International accredited animal facility. Cats were screened negative for feline enteric coronavirus antibody prior to transfer. Three dogs (female, 5–6 y old) were obtained from Ridglan Farms (Blue Mounds, WI). Cats and dogs were transferred to the Animal Disease Laboratory, an Animal Biosafety Level-3 facility at Colorado State University, group-housed, and fed dry/wet food mix with access to water ad libitum. Animals were allowed several days to acclimate before temperature-sensing microchips (Lifechips, Destron-Fearing) were inserted s.c. in the dorsum. Baseline weights, body temperatures, clinical evaluation, and oral swabs were obtained prior to inoculation. All animals were in apparent good health at the onset of the study.
Virus Challenge.
Cats were lightly anesthetized with 30–50 mg s.c. ketamine hydrochloride (Zetamine), and dogs were sedated with 1–3 mg xylazine prior to SARS-CoV-2 inoculation (day 0). Virus diluted in phosphate buffered saline (PBS) was administered to both species via pipette into the nares (500 µL/nare) for a total volume of 1 mL; animals were observed until fully recovered from anesthesia. Virus back-titration was performed on E6 cells immediately following inoculation, confirming that cats received 3.0E5 pfu and dogs received 1.4E5 pfu.
Sampling.
Cat cohort 1 (n = 3).
Oropharyngeal swabs were collected on 1–5, 7, 10, and 14 DPI using a polyester-tipped swab applicator. Swabs were placed in BA-1 medium (Tris-buffered MEM containing 1% bovine serum albumin) supplemented with gentamicin, amphotericin B, and penicillin/streptomycin. Nasal flushes were performed on 1, 3, 5, 7, 10, and 14 DPI by instilling 1 mL BA-1 dropwise into the nares of awake or lightly anesthetized cats and collecting nasal discharge into a sterile Petri dish by allowing the wash fluid to be sneezed out or dripped onto the dish. Blood (5 mL into serum separator tubes) was collected prior to inoculation and on 7, 14, 21, 28, 35, and 42 DPI. At 28 DPI, cats were reinoculated with 3.0E5 pfu of homologous virus. Oronasal sample collection was performed 1, 3, 5, 7, 10, and 14 d after reinoculation (days 29, 31, 33, 35, 38, and 42 post initial inoculation), at which time cats were euthanized and tissues collected for histopathology.
Cat cohort 2 (n = 4).
Two of the four cats were lightly anesthetized and challenged with SARS-CoV-2 as for cohort 1. Forty-eight hours post infection, two naive cats were introduced into the room with the infected cats and sampled on the same schedule as before. The two directly challenged cats were euthanized on 5 DPI and the following tissues collected for virus isolation and histopathology: nasal turbinates, trachea, esophagus, mediastinal lymph node, lung, liver, spleen, kidney, small intestine, uterus, and olfactory bulb. Tissues were collected into BA-1 frozen at −80 °C and homogenized prior to plaque assay. Additional tissues collected for histopathology included heart, colon, pancreas, hemilung lobe, and mesenteric lymph nodes. Thoracic radiographs were also obtained for these two cats prechallenge and just prior to euthanasia. The remaining two cats were euthanized at 30 DPI and necropsied; these cats will be referred to as contact cats hereafter.
Dogs (n = 3).
Dogs were sampled at the same frequency and using the same methods as cats in cohort 1 for 42 DPI. Dogs were not rechallenged.
Clinical Observations.
Body temperatures were recorded daily at approximately the same time each morning for the duration of the study using the thermal microchips. Cats and dogs were observed twice daily for the first 7 d post challenge and at least once daily for the duration of the study. Body weights were obtained weekly. Thoracic radiographs (3-view) were taken prechallenge and at 5 DPI just prior to euthanasia for the experimentally inoculated cats in cohort 2 and reviewed by several veterinarians. Clinical evaluation included temperament and assessment for presence of any clinical signs of disease including ocular discharge, nasal discharge, ptyalism, coughing/sneezing, dyspnea, diarrhea, lethargy, anorexia, moribund. None of the animals exhibited clinical signs of disease characterized by any of these symptoms at any time during the study.
Viral Assays.
Virus isolation was performed on all oral swab, nasal flush, and 5-DPI tissue samples by double-overlay plaque assay on Vero E6 cells as previously described (12). Briefly, 6-well plates with confluent monolayers of cells were washed once with PBS and inoculated with 100 μL of serial 10-fold dilutions of samples, incubated for 1 h at 37 °C, and overlaid with a 0.5% agarose in MEM containing 2% fetal bovine serum and antibiotics/antifungal agents. A second overlay with neutral red dye was added at 48 h, and plaques were counted at 72 h. Viral titers were reported as the log10 pfu per mL.
Plaque reduction neutralization assays (PRNT) were performed as previously described (13). Serum was heat-inactivated for 30 min at 56 °C, and twofold dilutions prepared in BA-1 starting at a 1:5 dilution were aliquoted onto 96-well plates. An equal volume of virus was added to the serum dilutions and incubated for 1 h at 37 °C. Following incubation, serum–virus mixtures were plated onto Vero E6 plates as described for virus isolation assays. Antibody titers were recorded as the reciprocal of the highest dilution in which >90% of virus was neutralized.
ELISA.
Serum samples from cats were heat-inactivated and tested by plaque assay to verify samples were noninfectious prior to conducting ELISA analysis. Positive control antibodies to the receptor-binding domain (RBD) and full-length spike protein were human mAb CR3022 antibody (Absolute Antibody) and human IgG whole molecule (Jackson Immuno Research). Positive control for the nucleocapsid ELISA was SARS-CoV nucleoprotein rabbit monoclonal antibody (Sino Biological, Inc.). Negative controls were reagent-grade human sera (compared to mAb CR3022). Nonpooled cat sera from two specific pathogen-free, naive experimental animals and five field isolate bioarchived samples obtained prior to 2019 were used as negative controls for assay validation (14, 15). ELISA protocols were adapted from protocols for SARS-CoV-2 ELISA described by Amanat et al. (16). ELISA plates (Thermo) were coated at 2 µg/mL with spike glycoprotein RBD from SARS-CoV-2, WuHan-Hu-1 recombinant from HEK293T cells (BEI), or spike glycoprotein (Stabilized) from SARS-CoV-2, Wuhan-Hu-1, recombinant from Baculovirus (BEI). SARS-CoV-2 Spike RBB-His and 2019-nCoV spike protein S1+S2 ectodomain (ECD) (Sino Biological) were used on plates used to test dog seroreactivity. SARS-CoV-2 nucleocapsid protein was a gift of Brian Geiss (Colorado State University, Fort Collins, CO). Prior to running experimental cat sera, the assay was optimized using positive and negative control sera described above. Samples and controls were diluted 1:50 in ELISA diluent (1X PBS, tween, milk powder) and run in duplicate. Human sera controls were developed using anti-human IgG horse radish peroxidase (HRP) (Thermo), cat sera was developed using anti-cat IgG HRP (Thermo) or anti-cat IgM (Novus Biologicals), dog sera was developed using anti-dog IgG HRP (Sigma), and rabbit monoclonal antibody (mAb) SARS-CoV nucleocapsid protein (NP) was detected by anti-rabbit IgG HRP (Thermo). Secondary antibodies were diluted 1:3,000, and SigmaFast o-phenylenediamine dihydrochloride (OPD) was prepared in water for injection and added to wells. Plates were read at 490 nm using a Multiskan Spectrum spectrophotometer (Thermo Fisher). The mean of negative control sera OD490 plus three times the SD of the negative control readings were used to determine cutoff values for each plate.
qRT-PCR.
Plaques were picked from culture plates from each cat to confirm SARS-CoV-2 viral shedding. RNA extractions were performed per the manufacturer’s instructions using Qiagen QiaAmp Viral RNA mini kits. RT-PCR was performed as recommended using the E_Sarbeco primer probe sequence as described by Corman and colleagues (17) and the SuperScript III Platinum One-Step qRT-PCR system (Invitrogen), with the following modification; the initial reverse transcription was at 50 °C. Standard curves were obtained by serial dilution of stock viral RNA from the original WA1/2020WY96 SARS-CoV-2 isolate.
Histopathology.
Tissues from cats were fixed in 10% buffered formalin for 12 d and transferred to 70% ethanol prior to sectioning for hematoxylin and eosin (H&E) staining. Slides were read by a board-certified veterinary pathologist.
Results
Clinical Disease.
None of the cats in either cohort displayed any clinical signs of disease, and all remained afebrile (temperature < 39.5 °C) throughout the study. Body weights were maintained over time. No evidence of lung involvement or any other radiographically detectable abnormalities were noted (images not shown). Similarly, dogs inoculated with SARS-CoV-2 remained clinically normal and afebrile.
Viral Shedding.
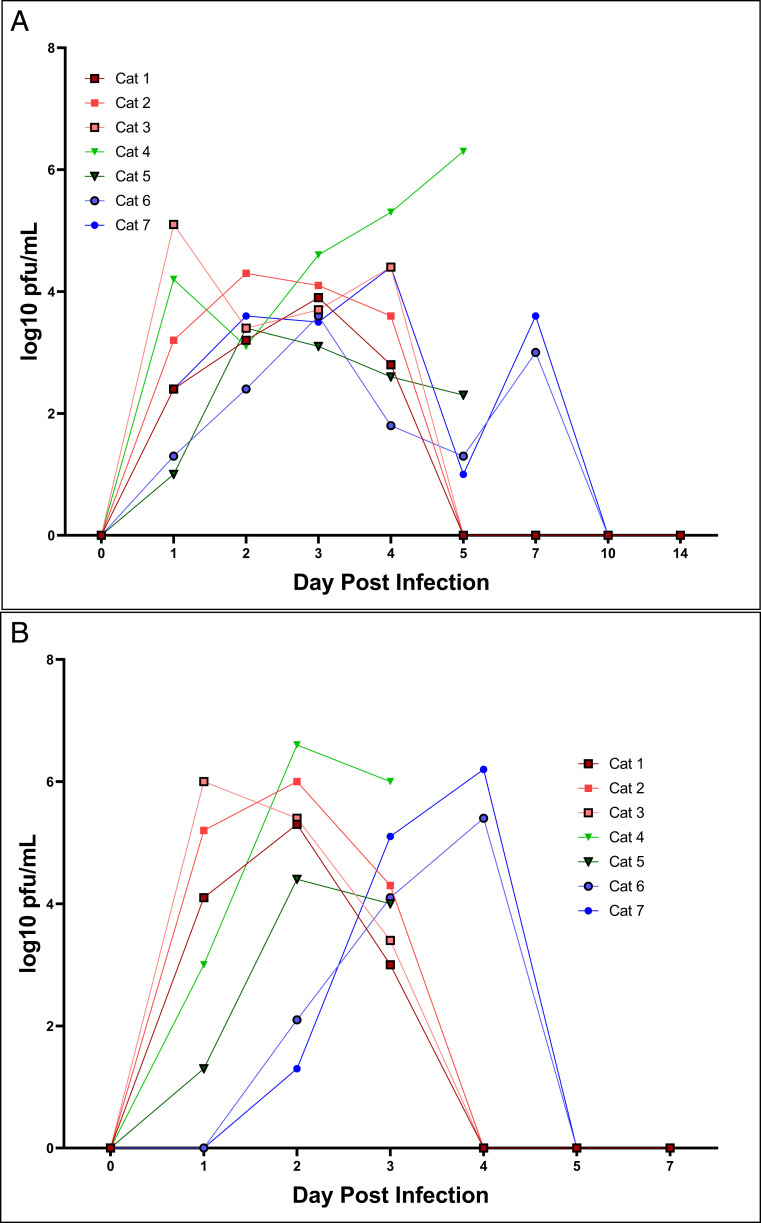
In cohort 1, all three cats shed virus both orally and nasally for up to 5 DPI, with peak titers achieved from nasal shedding at day 3. Nasal titers were ∼1 log higher than oral swabs collected at the same time (Fig. 1). There was some variability in titer over the course of infection that is likely attributable to sample collection (i.e., number of sneezes), but overall the data demonstrate clear presence of infectious virus in both the nasal cavity and the oropharynx for multiple days post infection. In cohort 2, the two directly inoculated cats shed virus for 5 DPI both orally and nasally, with a similar pattern to cohort 1. The contact cats, however, shed infectious virus orally by 24 h post exposure, and the duration of shedding was prolonged compared to the inoculated cats, with peak shedding occurring at 7 d post exposure (Fig. 1). Virus was isolated from trachea, nasal turbinates, and esophagus from cats in cohort 2 necropsied on day 5. Infectious virus was not found in the lung or other organs of either cat. Viral shedding was not detected by plaque assay from any of the dogs at any point post infection.
Fig. 1.
Inoculation and exposure with SARS-CoV-2 leads to nasal and oropharnygeal shedding in cats. SARS-CoV-2 virus is detected by plaque assay from (A) nasal and (B) oropharyngeal secretions of cats 1–5 DPI. Viral titers expressed as log10 pfu/mL. Cats 1, 2, and 3 represent cohort 1. Cats 4, 5, 6, and 7 represent cohort 2. Cats 4 and 5 were euthanized on 5 DPI. Cats 6 and 7 were introduced to the infected cats in cohort 2 on 2 DPI.
Pathology.
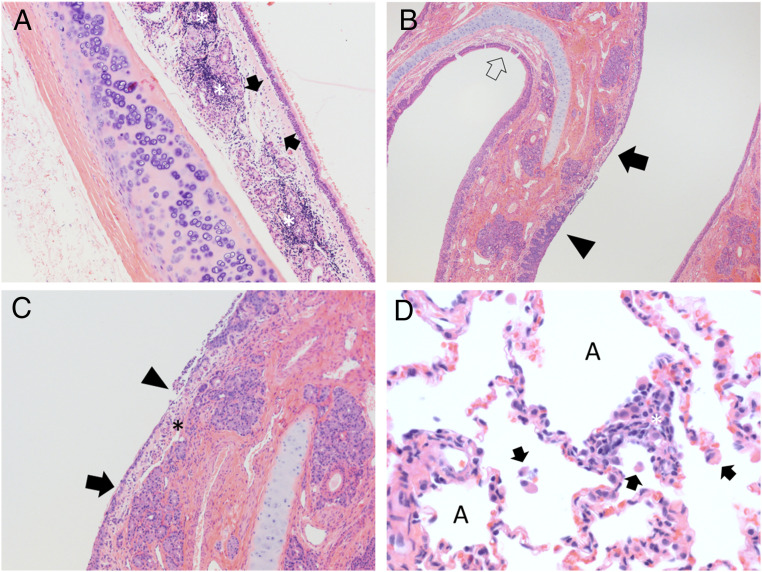
Gross lesions were not observed in any of the necropsied cats or dogs. Histologically, in both cats sacrificed at 5 DPI from cohort 2, moderate ulcerative, suppurative lymphoplasmacytic rhinitis was observed in the nasal turbinates along with mild lymphoplasmacytic tracheitis (Fig. 2 A–C). These cats also had minimal alveolar histiocytosis with edema. Both cats from cohort 2 which were introduced at 2 DPI and sacrificed at 28 DPI had moderate lymphoplasmacytic rhinitis with rare fibroplasia. All three cats from cohort 1 sacrificed 42 DPI had mild lung changes, including mild interstitial lymphocytic pneumonia with peribronchiolar and perivascular lymphocytic cuffing and alveolar histiocytosis (Fig. 2D). Two of these cats also had minimal tracheitis and mild rhinitis, but largely the lesions in the upper respiratory tract appear decreased in comparison to the early timepoint cats, while lung pathology was more evident in these animals compared to those sacrificed during acute infection.
Fig. 2.
SARS-CoV-2 exposure results in acute upper respiratory inflammation and mild lung infiltrates during later courses of infection. (A) Cat 4, cohort 2, trachea 5 DPI. The submucosa is expanded by edema (arrows) and abundant lymphocytic inflammatory infiltrates (asterisks) which dissect and disrupt submucosal glands. H&E stain, 100× magnification. (B) Cat 5, cohort 2, nasal turbinates, 5 DPI. Normal thickness respiratory mucosa is present in the section (open arrow). Nasal respiratory epithelium ranges from hyperplastic (filled black arrow) to ulcerated (arrowhead). The submucosa in regions of ulceration is edematous and infiltrated by scattered neutrophils and mononuclear cells. H&E stain, 40× magnification. (C) Cat 5, cohort 2, nasal turbinates, 5 DPI. Nasal respiratory epithelium ranges from attenuated (arrow) to ulcerated (arrowhead) with overlying remnant cellular debris. The submucosa (asterisk) in regions of ulceration is edematous and infiltrated by scattered neutrophils and mononuclear cells. H&E stain, 100× magnification. (D) Cat 1, cohort 1, lung, 42 DPI. Alveolar spaces (“A”) contain scattered mononuclear cells (arrows). The alveolar wall is expanded by mixtures of mononuclear cells and occasional neutrophils (asterisk). H&E stain, 400× magnification.
Seroconversion.
All animals were seronegative against SARS-CoV-2 at the time of infection (<50% viral neutralization at 1:10 serum dilution). Cats in both cohort 1 and the direct contact cats developed neutralizing activity as measured by PRNT as early as 7 DPI. Neutralizing titers in all cats reached or exceeded 1:2,560 by 14 DPI and either maintained or increased in titer between 28 and 42 DPI. Cats reinoculated at 28 DPI displayed a moderate increase in PRNT titer in the 14 d following exposure (Table 1). Dogs developed neutralizing antibodies by 14 DPI and peaked at 21 DPI with titers between 1:40 and 1:80 (Table 1).
Table 1.
Antibody titers (PRNT90) for cats and dogs infected with SARS-CoV-2. NT, not tested
| Animal | Preinfection (%) | 7 DPI | 14 DPI | 21 DPI | 28 DPI | 42 DPI |
| Cat 1 (cohort 1) | <50 | 320 | 5,120 | 2,560 | 2,560 | 10,240 |
| Cat 2 (cohort 1) | <50 | 80 | 5,120 | 2,560 | 2,560 | 5,120 |
| Cat 3 (cohort 1) | <50 | 80 | 2,560 | 2,560 | 1,280 | 5,120 |
| Cat 4 (cohort 2) | <50 | — | — | — | — | — |
| Cat 5 (cohort 2) | <50 | — | — | — | — | — |
| Cat 6 (cohort 2) | <50 | NT | 2,560 | 5,120 | 5,120 | — |
| Cat 7 (cohort 2) | <50 | NT | 2,560 | 1,280 | 10,240 | — |
| Dog 1 | <50 | <10 | 10 | 40 | 40 | 80 |
| Dog 2 | <50 | 10 | 80 | 20 | 20 | 40 |
| Dog 3 | <50 | <10 | 20 | 40 | 20 | 20 |
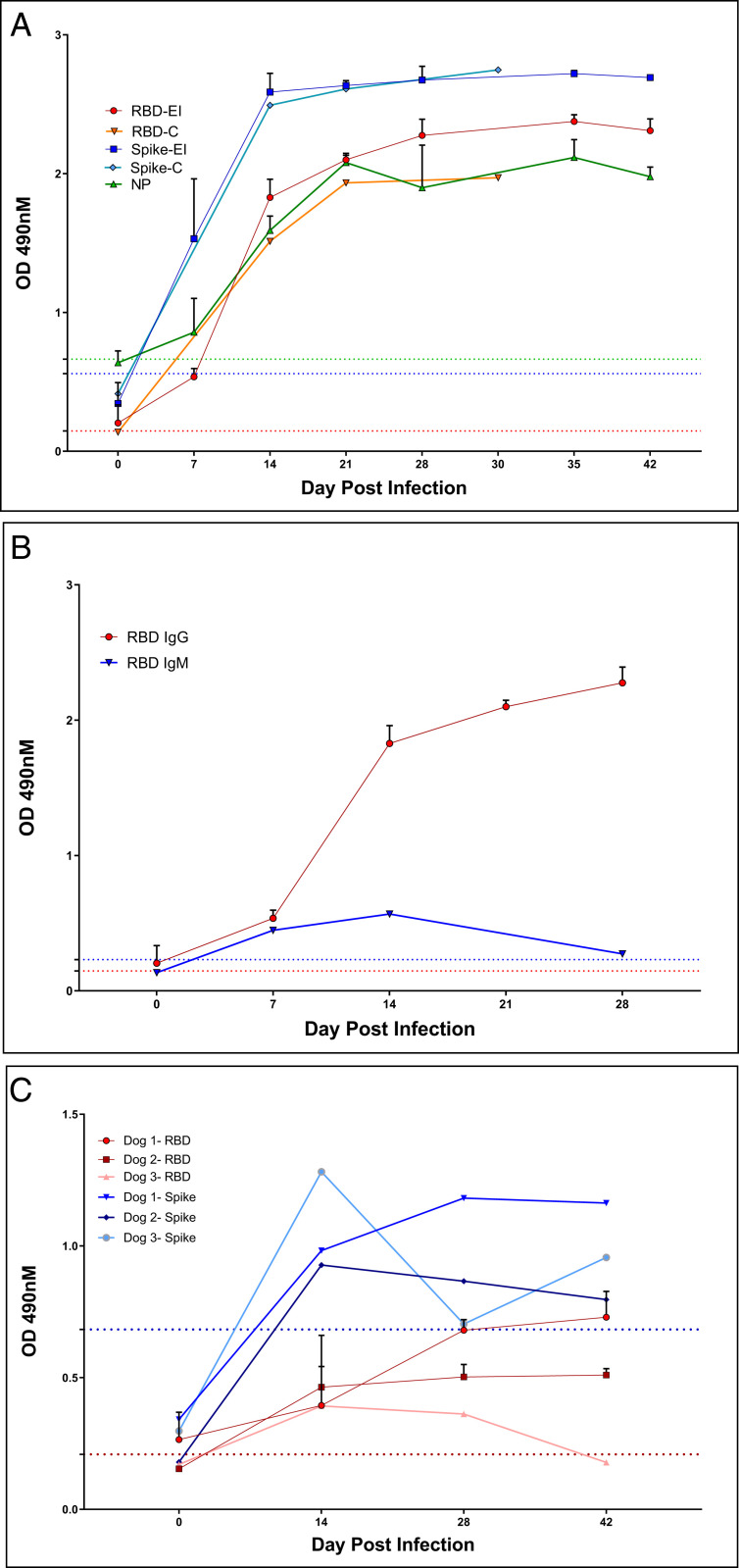
IgG antibody responses exceeding OD490 cutoff values were detected at 7 DPI against both the complete spike glycoprotein and RBD in all experimentally inoculated cats, and seroconversion against NP was detected in 2 of 3 cats at this time. By day 14 all five cats had optical density (OD) values that neared the upper limit of detection in the spike ELISA; RBD and NP OD saturation was obtained by day 21 and did not increase following reexposure (Fig. 3A). Rates of seroconversion and absorbance levels were similar between contact cats and experimentally infected cats. Seroconversion to spike protein was most rapid and robust, and the specificity of response to RBD exceeded that of NP. Seroconverted cat OD values for all three antigens exceeded absorbances of specific pathogen free (SPF) or field domestic cats, and background was highest for NP. IgM antibodies against RBD were detected at days 7 and 14 but not at day 28. IgG responses were much more robust than IgM (Fig. 3B). Dogs seroconverted against RBD and spike antigens starting day 14, but OD values were significantly lower than for cats and varied over time (Fig. 3C).
Fig. 3.
Cats and dogs infected with SARS-CoV-2 rapidly develop antibodies against viral antigens. (A) Sera from cats with intranasal inoculation of SARS-CoV-2 (n = 3, ‘EI’) or exposed to inoculated cats (n = 2, ‘C’) were evaluated for seroreactivity to RBD, Spike, or NP for 30–42 d post exposure. IgG reactivity to Spike and RBD was evident at day 7, and all animals had clearly seroconverted by day 14. (B) IgM against RBD was transiently detected at low levels relative to IgG on days 7 and 14 post exposure in cats (experimentally inoculated animals, n = 3). Bars represent 1 SE of the mean. Dogs infected with SARS-CoV-2 seroconvert versus Spike and RBD antigen with lower reactivity than cats (C). Sera tested on days indicated. IgG reactivity was evident by day 14 but plateaued and/or waned by day 42. Dashed lines indicate cut off values for seropositive diagnosis. Colors correspond to RBD (red), Spike (blue), or Nucleocapsid (green) ELISAs.
Reinfection.
Rechallenged cats in cohort 1 were sampled for oral and nasal shedding for 7 d post exposure by viral isolation, and shedding was not detected by plaque assay in any cat at any timepoint following rechallenge.
Discussion
The COVID-19 pandemic has affected virtually every country in the world and is the most significant outbreak of an emerging zoonotic pathogen in the current century. The SARS-CoV-2 virus is one of three emergent zoonotic coronaviruses capable of causing significant disease in humans in the last two decades, following SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) (18). The overall trend of disease emergence favors viral spillover from animals to humans, and land use and wildlife encroachment are just two of the factors contributing to this phenomenon (19). The continued presence of live animal markets provides optimal conditions for emergence of zoonoses (20). As with SARS-CoV-1 and MERS-CoV, SARS-CoV-2 is of probable bat origin based on phylogenetic analysis (2), but unlike its predecessors, SARS-CoV-2 has rapidly evolved for highly efficient human-to-human transmission (21). While animals, including domestic animals and pets, are frequently implicated as the source of emerging pathogens, reverse zoonosis of SARS-CoV-2 is more probable, as human cases are far more prevalent than domestic animals and there is no evidence to date of infected cats or dogs transmitting SARS-CoV-2 to humans. Similar results were seen with SARS-CoV-1, where domestic cats exposed to the virus by infected humans became infected, and cats experimentally infected shed virus for several days (22, 23). There have been several cases of pets becoming infected by SARS-CoV-2 following exposure to infected humans in New York, Hong Kong, Belgium, Germany, Spain, France, and Russia (5, 24–29). Other animal exposures from infected humans include farmed mink, which display respiratory signs, gastrointestinal signs, and even sudden death following infection (30). In several of these cases, including nondomestic felids at the Bronx Zoo and pet cats in New York and Europe, animals displayed signs of respiratory disease and/or conjunctivitis. None of the cats or dogs in this study exhibited any clinical signs of disease, but individual animal health status, age, and comorbidities may be responsible for this variability. Two other studies assessing experimental infection in cats have reported variation in respiratory sign; thus, further studies relating to clinical disease expression in cats are warranted (7, 10). Pathological changes in cats suggest that mild subclinical disease in otherwise healthy animals can occur. This is not altogether different from human infections, where the majority of cases are relatively mild but more severe disease tends to occur in older patients with significant comorbidities (31). In a recent serosurvey of cats in Wuhan, China, nearly 14.7% of sampled animals were seropositive for SARS-CoV-2 by RBD ELISA, suggesting that the cat population in areas with high human transmission is also likely to be exposed to the virus (11). Considering that the number of human infections has reached the millions and yet only a handful of animals have tested PCR-positive, it seems unlikely that domestic pets are a significant source of infection or are at serious risk for developing severe disease. Importantly, infected cats shed for no more than 5 d following exposure, suggesting that cats, if exposed to infected humans, will develop and clear infection rapidly. In comparison, humans typically have an incubation period of ∼5 d and can shed virus for more than 3 wk (32, 33). Thus, if symptomatic humans follow appropriate quarantine procedures and stay home with their pets, there is minimal risk of a potentially exposed cat infecting another human. Infected pet cats should not be allowed to roam freely outdoors to prevent potential risk of spreading infection to other outdoor cats or wildlife. More research into the susceptibility of wildlife species and potential for establishment of infection in outdoor cat populations is necessary to identify risk factors and mitigation strategies to prevent establishment of reservoir infections in feral cats or other wildlife.
The development of animal models for studying SARS-CoV-2 is an important step in research methodologies. Rhesus macaques, hamsters, and ferrets are all suitable models for replicating asymptomatic or mildly clinical disease and, while not often used as a traditional animal model, this work demonstrates that cats may serve as an alternative model (34–36). The cats in this study developed subclinical pathological changes in the upper respiratory tract early in the course of infection with more lower respiratory tract pathology later following viral clearance, which suggests that, while subclinical, viral infection of cats is not completely benign and may make their utility as an animal model more relevant to mild human disease. Additionally, the relatively high-titer viral shedding produced by cats and the rapidity of transmission may make them an ideal model for simulation of aerosols. As such, cat models may be quite instrumental for understanding the shed/spread kinetics of SARS-CoV-2. Perhaps most importantly, cats develop significant neutralizing antibody titers and are resistant to reinfection, although the duration of immunity is not currently known. This could prove a useful measurement for subsequent vaccine trials for both human and animal vaccine candidates.
Acknowledgments
We thank Todd Bass and the histology laboratory at Colorado State University for preparation of tissue cassettes and slides for histopathology and Dr. Brian Geiss for providing the SARS-CoV-2 nucleocapsid protein. This work was funded by the Animal Models Core, Colorado State University. SARS-Related Coronavirus 2, Isolate USA-WA1/2020 (NR-52281) was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), NIH. The following reagents were produced under HHSN272201400008C and obtained through BEI Resources, NIAID, NIH: Spike Glycoprotein RBD from SARS-Related Coronavirus 2, Wuhan-Hu-1, Recombinant from HEK293 cells, NR-52306, and Spike Glycoprotein (Stabilized) from SARS-Related Coronavirus 2, Wuhan-Hu-1, Recombinant from Baculovirus, NR-52396.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Bogoch I. et al., Pneumonia of unknown aetiology in Wuhan, China: Potential for international spread via commercial air travel. J. Travel Med. 27, taaa008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P. et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q. et al., Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 382, 1199–1207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan Y., Shang J., Graham R., Baric R. S., Li F., Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94, e00127-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sit T. H. C. et al., Infection of dogs with SARS-CoV-2. Nature, 10.1038/s41586-020-2334-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chini M., Coronavirus: Belgian woman infected her cat [Internet]. The Brussels Times, 01 April 2020. https://www.brusselstimes.com/all-news/belgium-all-news/103003/coronavirus-belgian-woman-infected-her-cat/. Accessed 30 May 2020.
- 7.Shi J. et al., Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 368, 1016–1020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng J. et al., Serological survey of SARS-CoV-2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transbound. Emerg. Dis. 67, 1745–1749 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Temmam Sarah et al., Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Health, 10.1016/j.onehlt.2020.100164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halfmann P. J. et al., Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 383, 592–594 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q. et al., SARS-CoV-2 neutralizing serum antibodies in cats: A serological investigation. Microbiology, 10.1101/2020.04.01.021196 (2020). [DOI] [Google Scholar]
- 12.Kropinski A. M., Mazzocco A., Waddell T. E., Lingohr E., Johnson R. P., “Enumeration of bacteriophages by double agar overlay plaque assay” in Bacteriophages, Clokie M. R. J., Kropinski A. M., Eds. (Methods in Molecular Biology, Humana Press, Totowa, NJ, 2009), Vol. 501, pp. 69–76. [DOI] [PubMed] [Google Scholar]
- 13.Perera R. A. et al., Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill. 25, 2000421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carver S. et al., Pathogen exposure varies widely among sympatric populations of wild and domestic felids across the United States. Ecol. Appl. 26, 367–381 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Sprague W. S. et al., Prior puma lentivirus infection modifies early immune responses and attenuates feline immunodeficiency virus infection in cats. Viruses 10, 210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amanat F. et al., A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 26, 1033–1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corman V. M. et al., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 2000045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guarner J., Three emerging coronaviruses in two decades. Am. J. Clin. Pathol. 153, 420–421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olival K. J. et al., Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L.-F., Eaton B. T., “Bats, civets, and the emergence of SARS” in Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-Species Transmission, Childs J. E., Mackenzie J. S., Richt J. A., Eds. (Current Topics in Microbiology and Immunology, Springer, 2007), Vol. 315, pp. 325–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan J. F.-W. et al., A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 395, 514–523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Brand J. M. A. et al., Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet. Pathol. 45, 551–562 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Martina B. E. E. et al., Virology: SARS virus infection of cats and ferrets. Nature 425, 915 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chini M., COVID-19 update (58): Belgium, animal, cat, clinical case. The Brussels Times, 27 March 2020. archive no. 20200327.7151215.
- 25.Government of Hong Kong , COVID-19 update (70): China (Hong Kong) animal, cat, pets & stock. Press release 0200402.7173286 (31 March 2020).
- 26.Davidson M., COVID-19 update (124): USA (NY) animal, cat, lion, OIE, 20200423.7259119 (22 April 2020).
- 27.Volz A, COVID-19 update (181): Germany (BY), France (AC), cat, OIE animal case definition, 20200513.7332909 (13 May 2020).
- 28.Vlasov N., COVID-19 update (212): Russia (Moskva) animal, cat, OIE, 20200526.7379578 (26 May 2020).
- 29.Matthews S., Chalmers V., COVID-19 update (334): Animals, Netherlands, mink, spread, UK, cat, first case, OIE identification number 20200727.7617582 (27 July 2020).
- 30.Clinical and pathological findings in SARS-CoV-2 disease outbreaks in farmed mink (Neovison vison). Vet. Pathol. 57, 653–657 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Nikolich-Zugich J. et al., SARS-CoV-2 and COVID-19 in older adults: What we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience 42, 505–514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauer S. A. et al., The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: Estimation and application. Ann. Intern. Med. 172, 577–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noh J. Y. et al., Asymptomatic infection and atypical manifestations of COVID-19: Comparison of viral shedding duration. J. Infect., 10.1016/j.jinf.2020.05.035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y.-I. et al., Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe 27, 704–709.e2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munster V. J. et al., Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature, 10.1038/s41586-020-2324-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan J. F.-W. et al., Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis., ciaa325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and SI Appendix.