Significance
Maintaining a functional cell requires the collaboration of a large number of biomolecules that regulate key biochemical processes. The p97 protein is essential in this effort, interacting with adaptor molecules that target it to different regions of the cell for specific functions. Here, we study the structural dynamics of a complex formed between p97 and the p47 adaptor that is critical for membrane assembly. Our data establish that the p47–p97 interaction is partly regulated by regions of p47 that are intrinsically disordered, interacting with p97 in a manner that depends on whether p97 is adenosine triphosphate- or adenosine diphosphate-loaded. Our combined biophysical measurements enable the calculation of structural models of p47–p97, highlighting important interactions within the complex.
Keywords: p97/VCP, p47, methyl-TROSY, intrinsically disordered, structural dynamics
Abstract
VCP/p97, an enzyme critical to proteostasis, is regulated through interactions with protein adaptors targeting it to specific cellular tasks. One such adaptor, p47, forms a complex with p97 to direct lipid membrane remodeling. Here, we use NMR and other biophysical methods to study the structural dynamics of p47 and p47–p97 complexes. Disordered regions in p47 are shown to be critical in directing intra-p47 and p47–p97 interactions via a pair of previously unidentified linear motifs. One of these, an SHP domain, regulates p47 binding to p97 in a manner that depends on the nucleotide state of p97. NMR and electron cryomicroscopy data have been used as restraints in molecular dynamics trajectories to develop structural ensembles for p47–p97 complexes in adenosine diphosphate (ADP)- and adenosine triphosphate (ATP)-bound conformations, highlighting differences in interactions in the two states. Our study establishes the importance of intrinsically disordered regions in p47 for the formation of functional p47–p97 complexes.
Valosin-containing protein (VCP), or p97, is a highly conserved and abundant cytosolic enzyme in the AAA+ superfamily (ATPases associated with diverse cellular activities) that plays an indispensable role in protein homeostasis through its involvement in a range of diverse processes. These include, among others, proteasomal (1–3) and lysosomal degradation (4–6), membrane fusion (7–9), cell cycle regulation (9–11), and apoptosis (12). p97 consists of an amino (N)-terminal domain (NTD) and two tandem ATPase domains (D1 and D2), connected to each other via linkers that are of functional importance (13). Detailed structural studies reveal an enzyme organized as a homohexamer, with D1 and D2 forming two coaxially stacked rings and the NTDs localized to the periphery of the D1 ring (14–16). Upon nucleotide hydrolysis, the NTDs undergo large movements, from above the plane of the D1 ring in the adenosine triphosphate (ATP)-bound state (the “up” conformation) to coplanar with D1 in the adenosine diphosphate (ADP) state (the “down” conformation) (Fig. 1A) (14–16). The up/down conformational equilibrium plays an important regulatory role in the interactions of p97 with various adaptor proteins (17–20), becoming dysregulated upon the introduction of single-site mutations that give rise to a series of diseases collectively referred to as multisystem proteinopathy type 1 (MSP1), in which the lysosomal-degradation pathway becomes impaired (4, 18, 21). At least 30 adaptors have been shown to interact with p97 (22–24), assisting both in the recruitment of the ATPase to designated subcellular locations and in the targeting of p97 to specific substrates. Most of the adaptors bind to the NTD of p97 via a conserved UBX domain (25), or linear peptide motifs such as VIM (26), VBM (27), and SHP/BS1 (28), while some interact with the carboxy (C)-terminal tail of p97 via the PUB (29) and PUL (30) domains.
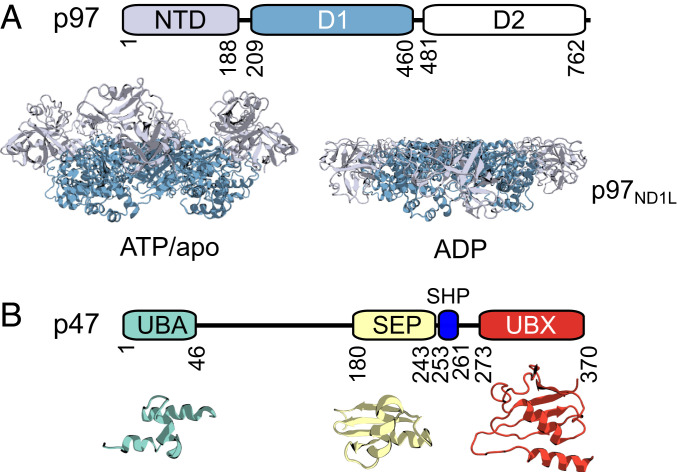
Fig. 1.
Domain architectures of p97 and p47. (A) Domain architecture of a p97 monomer and cartoon representations of hexameric p97ND1L in the ATP/apo (Left) (derived from PDB ID code 5FTN; ref. 14) or ADP (Right) (PDB ID code 5FTK; ref. 14) states. NTDs adopt distinct conformations depending on the nucleotide state of D1. (B) Domain architecture of p47, with structures of the UBA (turquoise) (PDB ID code 1V92; ref. 38), SEP (yellow) (PDB ID code 1VAZ; ref. 38), and UBX (red) (PDB ID code 1S3S; ref. 41) folded domains shown in cartoon depictions below.
The first identified p97 cofactor was p47, isolated as part of a stable p97–p47 complex (31). p47 plays an essential role in directing p97 function to the remodeling of cellular membranes (31–34), including post-mitotic reassembly of the Golgi apparatus (7, 31, 35) through an interaction with the monoubiquitinated SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein syntaxin 5 (36, 37). p47 is comprised of three independently folded motifs, including 1) ubiquitin-associated (UBA); 2) Shp1, eyc, and p47 (SEP); and 3) ubiquitin regulatory X (UBX) domains; the structures of which have been elucidated by solution NMR spectroscopy and X-ray crystallography (38–41) (Fig. 1B). The N-terminal UBA domain, adopting a canonical three-helix bundle (38), is proposed to directly interact with ubiquitinated syntaxin 5 (37, 42), while the C-terminal UBX domain is a ubiquitin-like module that binds to NTDs of p97 (40, 41). The middle SEP domain, which is found in several other p97 cofactors, including p37, UBXD4, and UBXD5, adopts a ββαβ-fold (38, 39). Although its biological function remains unknown, it may be involved in recruiting substrate complexes to p97 for disassembly (43). The three domains in p47 are connected via long, flexible linkers of ∼130 (UBA-SEP) and 30 (SEP-UBX) residues. Full-length p47 is thought to exist as a high-affinity trimer in solution (20, 31, 36, 38, 40, 44–46), with the trimerization mediated by the SEP domain (44), as the SEP domain has been reported to self-associate (38), or via the region between the SEP and UBX domains (38).
Two conserved structural features in p47 have been identified that contribute to a bipartite interaction with p97. These features include a highly conserved loop in the UBX domain that inserts into a hydrophobic groove between the subdomains of a p97 NTD (40, 41) and a short linear SHP motif between SEP and UBX domains (Fig. 1B) that binds to the C-terminal NTD subdomain (28, 46). Although a fragment-based approach has been used to study interactions between p47 and p97 (28, 38, 40, 41, 46), and low-resolution electron cryomicroscopy (cryo-EM) maps of a p47–p97 complex have been presented (44, 47), there remain large gaps in our understanding of the structural details of the intact assembly. For example, it has been proposed that the stoichiometry of the p47–p97 complex is one p47 trimer per p97 hexamer (31, 44), although other data suggest a binding stoichiometry of six p47 monomers per p97 hexamer (47). The role of the linkers in p47 remains unclear—do they simply connect the structured domains that are responsible for function, or do these flexible regions support binding through interactions with distal regions on p47 or with domains of p97? Further, how do the changes to the orientations of the p97 NTDs that accompany ATP hydrolysis affect interactions within the p47–p97 complex or within each bound p47 adaptor? Part of the difficulty in addressing these questions arises from the fact that p47 is an inherently flexible molecule, challenging studies by X-ray crystallography and cryo-EM. Although solution NMR spectroscopy approaches are powerful for studies of dynamic complexes, most of the focus to date has been on relatively small systems. The development of methods for studies of complexes with molecular masses of many hundreds of kilodaltons (48), as is the case for p47–p97, provides an opportunity to explore structural dynamics in a range of important molecules with unprecedented detail (49, 50).
Herein, we present a solution NMR study characterizing the dynamic complex formed between p47 and a 320-kDa p97 construct consisting of residues 1 to 480 of the 762-residue full-length p97 protomer and comprising the NTD, D1, and the linker connecting D1 and D2 (referred to as p97ND1L hereafter) (18, 51, 52). The NMR data have been supplemented by results from other biophysical methods, including cryo-EM, isothermal titration calorimetry (ITC), dynamic light scattering (DLS), small angle X-ray scattering (SAXS), analytical ultracentrifugation (AUC), and molecular dynamics (MD) simulations. In order to provide a structural basis for interpreting measurements on p47–p97ND1L, we first focus on p47. We present strong evidence that p47 is a monomer at concentrations as high as hundreds of micromolar, using a series of different biophysical and biochemical experiments. The linker connecting UBA with SEP is shown to play an active role in stabilizing both intra- and intermolecular interactions. Notably, contacts between p47 linker residues S114 to G130 and the SEP domain in both p47 and the p47–p97ND1L complex are observed, and an SHP motif, localized to the UBA-SEP linker, is identified that binds to p97 NTDs only when they assume the up conformation corresponding to either ATP-bound or apo states. Restraints from the NMR and cryo-EM data are used to calculate models of the p47–p97 complex using a metainference MD approach that takes into account the inherent dynamics of the complex. Our results emphasize that intrinsically disordered regions (IDRs) in p47 contribute to interactions with p97 in a manner dependent on the up/down NTD equilibrium in the ATPase, and, in general, highlight the important roles that IDRs can play in regulating the function of molecular machines. Moreover, they provide an additional framework to understand how MSP1 disease causing mutations can lead to deleterious p97 function.
Results
NMR Studies of p47, Both Free and in Complex with p97ND1L.
As a first step toward characterizing p47 intramolecular and p47–p97ND1L intermolecular interactions, we obtained backbone, sidechain 13Cβ, as well as Ile, Leu, Val, and Met methyl-group chemical-shift assignments of the full-length p47 protein (370 residues). These assignments were generated by using standard transverse relaxation-optimized spectroscopy (TROSY)-based triple-resonance experiments recorded on a [U-2H,15N,13C]p47 sample for backbone and 13Cβ shifts (53) and, in the case of methyl groups, by correlating methyl 13C and 1H shifts with amide resonances via total correlation spectroscopy-based magnetization transfer, using smaller fragments of [U-15N,13C]p47 that included residues M1 to R174, G170 to Q266, G170 to T370, or E291 to T370. This “divide-and-conquer approach” takes advantage of the modular nature of p47, as established by previous studies (38–40, 54) and illustrated in Fig. 1B. In this way, 90% of the backbone resonances and 14/14 Ile (δ1), 53/56 Leu (δ1,δ2), 18/46 Val (γ1,γ2), and 4/4 Met (ε) chemical shifts were assigned, with severe overlap of Val methyl groups limiting their assignment.
The backbone 13Cα and 13Cβ chemical shifts of full-length p47 were used to generate residue-specific secondary-structure propensity (SSP) scores (55) (SI Appendix, Fig. S1A). Consistent with previous structural studies of p47 (38, 40), we observed regions of contiguous secondary structure between residues M1 and G46, V180 and F243, and P273 and T370, corresponding to the UBA, SEP, and UBX domains, respectively (Fig. 1B and SI Appendix, Fig. S1A). Notably, residues P273 to I287 have SSP scores that indicate α-helical secondary structure, in agreement with an X-ray structure of a complex consisting of p47 UBX and p97 NTD and D1 domains (41). This α-helix, however, is absent in the p47 UBX NMR structure (40) due to the shorter construct used, which only spans residues K282 to T370. Additional contiguous secondary structure, based on our measured chemical shifts, was not observed for the remainder of the p47 polypeptide. The SSP data, together with the distribution of cross-peaks in both 15N-1H heteronuclear single-quantum coherence (HSQC) (SI Appendix, Fig. S1B) and 13C-1H heteronuclear multiple-quantum coherence (HMQC) (SI Appendix, Fig. S1C) correlation spectra, with many resonances localized to regions predicted for unfolded structure, strongly support the schematic of Fig. 1B showing p47 comprised of folded domains connected by long IDRs.
Because of the large molecular mass of the p47–p97ND1L complex, in excess of 350 kDa, we have chosen to perform most of our studies using highly deuterated, methyl-protonated proteins, exploiting a methyl-TROSY effect (48) that leads to significant improvements in spectral sensitivity and resolution in applications involving high-molecular weight protein systems (49, 50). Thus, we have prepared highly deuterated p47 samples with methyl labeling at Ile(δ1), Leu(δ1,δ2), Val(γ1,γ2), and Met(ε) positions, referred to in what follows as ILVM-labeling. Note that for Leu and Val, only one of the two isopropyl methyl groups is 13CH3-labeled (the other is 12CD3-labeled) in a nonstereospecific manner, as discussed previously (56). In order to improve the spectral quality further, we have chosen to work with p97ND1L (320 kDa), as opposed to the full-length p97 complex (540 kDa). The p97ND1L truncation has proven to be an excellent model system for studies of the dynamics of the NTD in relation to the D1 ring and how these change depending on the nucleotide state of D1, with the NTD up (ATP)/down (ADP) equilibrium illustrated in Fig. 1A observed for both truncated and full-length constructs (14, 15). Notably, in the apo forms of both p97ND1L (52) and full-length p97, the NTDs adopt the up conformation as well (SI Appendix, Fig. S1D). Perturbations to the up/down equilibrium from MSP1 disease mutations are reproduced in both full-length and the ND1L constructs (18). In addition, we have previously shown, through studies of an ILVM-labeled p97ND1L complex, that the expected interactions of the p97 NTDs with p47 UBX (40, 41, 46) are observed (18). In order to further test the appropriateness of the p97ND1L construct in studies involving p47 here, we have used methyl-TROSY based NMR to ensure that the known interactions from the side of p47 are also found. As expected, significant chemical-shift perturbations (CSPs) of p47 ILVM-methyl resonances are obtained upon addition of perdeuterated p97ND1L (added in fivefold excess; SI Appendix, Fig. S2A), with residues located on the UBX–NTD binding interface affected (SI Appendix, Fig. S2B). A second interaction site on p47 involves the SHP motif (Fig. 1B) (46). A lack of resolved methyl probes on the SHP motif prevented a similar NMR analysis to establish binding of this module to p97ND1L. We have, therefore, introduced a Met residue into SHP and observed spectral changes supporting the expected interaction with the p97ND1L construct as well (SI Appendix, Fig. S2C). Comparative ITC measurements, quantifying the interaction between p97ND1L and either wild-type (WT) p47 or p47 F253A, in which a residue making critical contacts with the NTD of p97 is mutated (46), further establish the expected SHP interactions with the ND1L construct (SI Appendix, Fig. S2D and Table S1A). Finally, ITC measurements quantifying the binding of p47 to p97ND1L are in good agreement with previous results using full-length p97 (44), with similar fitted apparent binding stoichiometries (three p47 monomers:p97 hexamer), ΔH (−30 kcal/mol), and Kd (0.2 to 0.5 μM) values obtained in both cases. Taken together, our results indicate that the p47–p97ND1L complex recapitulates the known structural details of the p47–p97 interaction, as expected since binding occurs via contacts between p47 and the ND1 ring of p97 (44). In all experiments reported here, we have used the apo form of p97 as a surrogate for the ATP-bound state. Under the conditions of our experiments, ATP would be rapidly hydrolyzed. Slowly hydrolyzing mimics of ATP, such as ATPγS, in concert with Walker B mutants that further reduce rates of hydrolysis (18, 57), could be used; however, even in these cases, nucleotide lifetimes are not more than several days, and we prefer to work with the WT sequence.
p47 Is a Monomer in Solution.
The consensus in the literature is that p47 forms a stable trimer, mediated by intermolecular contacts between the central SEP domain of each of the p47 protomers (31, 38, 44, 45). We sought to examine the structural details of this interaction with the intent of producing a model of the trimeric p47 species. However, we were unable to detect any significant concentration-dependent CSPs for p47 across a wide range of sample concentrations spanning 4 to 800 μM (SI Appendix, Fig. S3A), nor were we able to observe intermolecular nuclear Overhauser effects (NOEs) between SEP domains using a p47 sample prepared by mixing initially unfolded [U-2H, Ile(δ1),Met-13CH3]- and [U-2H,15N, Leu,Val-13CH3,12CD3]-labeled proteins that were subsequently refolded (SI Appendix). Next, we conducted pull-down assays where His6-tagged p47 and untagged p471–290 (Fig. 2A) were mixed in a 1:1 molar ratio under denaturing conditions, followed by refolding (Fig. 2B). Assuming that p47 is a stable trimer and that each of the two classes of p47 protomer is equally likely to be incorporated into trimers, three-quarters of the molecules are expected to contain mixtures of p47 and p471–290. However, mixed trimers were not detected, a result that is consistent with monomeric but not oligomeric p47 structures.
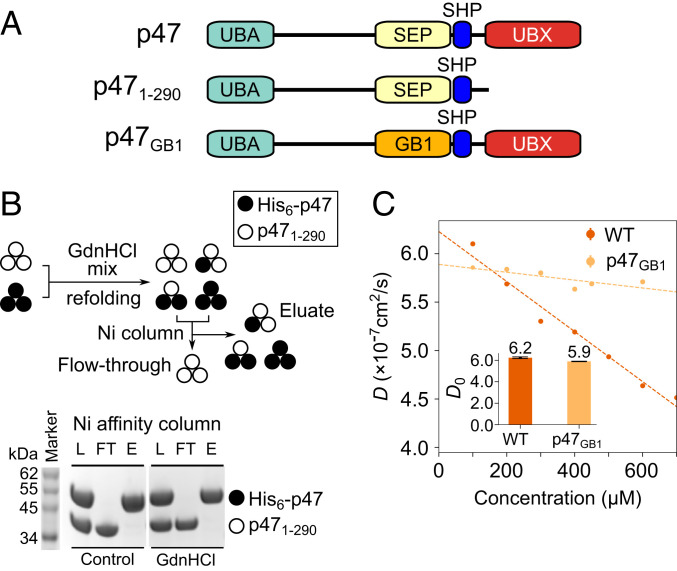
Fig. 2.
p47 is a monomer in solution. (A) Cartoon representation of p47 constructs used in studies illustrated in the figure. (B, Top) Cartoon depiction of GdnHCl mixing/refolding experiment involving His6–p47 and p471–290, followed by purification via Ni affinity chromatography. Assuming p47 is a stable trimer, both His6–p47 and p471–290 monomers are expected in the Ni eluate. (B, Bottom) After mixing His6–p47 and p471–290 in a 1:1 molar ratio, the sample is either applied to the Ni affinity column immediately (left three lanes: Control, instant mixing) or unfolded in a buffer containing 6 M GdnHCl, followed by refolding before application to a Ni-affinity column (right three lanes: GdnHCl mixing). Proteins from Ni loading (L), flow-through (FT), and eluate (E) are read out by sodium dodecyl sulphate–polyacrylamide gel electrophoresis. (C) Translational diffusion rates of WT p47 (dark orange) and p47GB1 (light orange) as a function of protein concentration, obtained by dynamic light scattering at 25 °C. Domain organization of p47GB1 is depicted in A. Inset shows translational diffusion rates of WT p47 and p47GB1 extrapolated to infinitely dilute protein concentration.
These observations raise the question as to whether p47 is in fact a stable trimer in solution. To resolve this issue, we conducted dynamic light scattering (DLS) experiments on two of the samples highlighted in Fig. 2A, under conditions equivalent to those used for NMR experiments and spanning a broad range of p47 concentrations (Fig. 2C). We observed a linear concentration-dependent decrease in measured translational-diffusion coefficients (D) for WT p47 from 6.0 × 10−7 to 4.6 × 10−7 cm2/s for protein concentrations, c, varying from 100 to 700 μM, respectively. The DLS data were then extrapolated back to 0 μM sample concentration (D0) using the relation (58). For WT p47, we obtained D0 and κ parameters of 6.2 ± 0.1 × 10−7 cm2/s and −4.1 ± 0.3 × 10−4 μM−1, respectively (Fig. 2C). Next, we generated a chimeric p47 construct in which the p47 SEP domain, suggested to comprise the putative p47 trimer interface (38, 44), was replaced with GB1, a small, structured, and monomeric domain of approximately the same size as the SEP moiety. This construct thus preserves the overall polypeptide length and relative molecular mass of WT p47, while removing the presumed trimerization domain. Substitution of the SEP domain with GB1 resulted in no significant change in D0 (Fig. 2 C, Inset), suggesting that the overall intrinsic hydrodynamic properties (and oligomeric state, by extension) of WT p47 and p47GB1 are similar. Therefore, either both WT p47 and p47GB1 are capable of trimerizing by a SEP-independent mechanism, or both p47 and p47GB1 are monomeric. Interestingly, the concentration dependence of D observed for p47, as assessed by the parameter κ (−4.1 ± 0.3 × 10−4 μM−1), is different from that measured for p47GB1 (−6.9 ± 1.4 × 10−5 μM−1), perhaps reflecting very weak intermolecular interactions mediated by the SEP domain rather than the formation of stable trimers (see Discussion).
A combined analysis of sedimentation coefficients, s, derived from AUC (SI Appendix, Fig. S3B) and D0 obtained from DLS (Fig. 2C) strongly supports a monomeric p47 structure. The measurement of both D0 and s allows a straightforward calculation of the molecular mass, M, of the particle in question that is shape-independent, via , where , ρ, and T are the partial specific volume of the particle (inverse density), the density of the solvent, and the absolute temperature, respectively (59). A value of M = 42.8 ± 1.0 kDa is obtained, which is in good agreement with the theoretical molecular mass of a single p47 polypeptide chain (40.8 kDa), indicating a monomeric p47 structure.
As a final confirmation that p47 is a monomer, we have performed SAXS experiments, from which the molecular mass of particles in a monodisperse sample can be determined to reasonable accuracy regardless of the particle shape and concentration (60). We acquired scattering profiles for WT p47 (13 mg/mL, 320 μM) as well as for the NTD of p97 (p97NTD) (3 mg/mL, 125 μM) as a control (SI Appendix, Fig. S3C) and obtained molecular masses of 40.6 ± 1.5 and 24.2 ± 0.8 kDa, respectively, that are in close agreement with the predicted monomeric molecular masses of these proteins (24.0 kDa for p97NTD). Thus, p47 is monomeric under the conditions of our measurements.
Charcterizing a Previously Unobserved Intramolecular p47 Interaction.
The schematic of p47 in Fig. 1B, with structured UBA, SEP, and UBX domains connected by linkers drawn as straight lines, conveys the impression that this adaptor can be well described in terms of a “beads on a string” model. However, it is increasingly appreciated that unstructured domains often play important roles in intermolecular recognition or in establishing intramolecular contacts (61, 62). With this in mind, we asked whether the p47 linkers perform functions beyond merely connecting structured domains in this molecule. To answer this question, we performed NMR-binding experiments on fragments that included residues 1 to 174 (p471–174), comprising the UBA domain and the linker region connecting UBA and SEP domains), and residues 171 to 370 (p47171–370), which include SEP, SHP, and UBX domains (Fig. 3A). Upon addition of threefold excess unlabeled p47171–370, we observed CSPs and intensity changes for backbone amide and methyl cross-peaks in spectra recorded of [U-15N,13C]p471–174 (Fig. 3B), with changes in intensity plotted as ΔI/I0 = (I0 − I)/I0, where I and I0 are intensities in the presence and absence of added ligand. ΔI/I0 values of 0 and 1 correspond, therefore, to no change or complete loss of intensity, respectively. Major perturbations are focused on linker residues S114 to G130 (Fig. 3C), providing strong evidence of their association with the p47171–370 fragment.
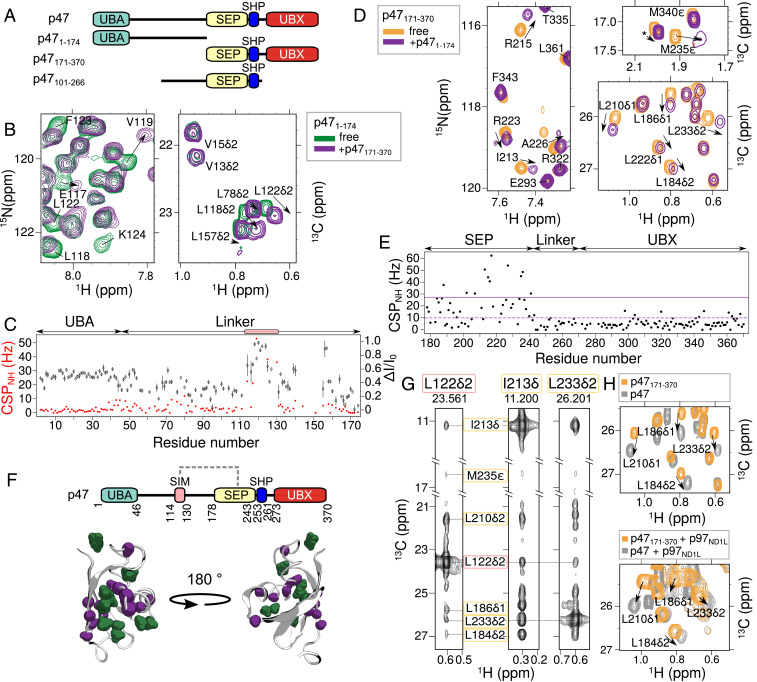
Fig. 3.
The unstructured linker of p47 interacts with the SEP domain. (A) Domain architecture of p47 constructs used in studies illustrated in this figure. (B) Selected regions of 15N-1H HSQC (Left) and 13C-1H constant time (CT)-HSQC (Right) spectra of [U-13C,15N]p471–174 showing perturbations (chemical shift changes and line broadening) upon addition of threefold excess unlabeled p47171–370. (C) CSPs (red spheres) and changes in intensities (black diamond), quantified as ΔI/I0 = (I0 − I)/I0, where I is the intensity of a resonance in the 15N-1H HSQC spectrum of p471–174 upon addition of p47171–370, and I0 is the corresponding intensity in the absence of added ligand. Peaks derived from a continuous set of residues, S114 to G130, show perturbations and are highlighted by the pink box. (D) Selected regions of 15N-1H HSQC (Left) and 13C-1H CT-HSQC (Right) spectra of [U-13C,15N]p47171–370 upon addition of threefold excess unlabeled p471–174. (E) CSPs from 15N-1H HSQC spectra of p47171–370 resulting from the addition of p471–174. Average CSP value and CSP 1σ above average are indicated with dashed and solid lines, respectively. (F, Top) Domain architecture of p47 depicting interaction between the SEP domain and a SIM on the linker (dashed gray line). (F, Bottom) Cartoon representation of the SEP domain (PDB ID code 1VAZ; ref. 38). Residues with CSPs 1σ above the average in the 15N-1H HSQC and 13C-1H CT-HSQC spectra are colored purple and green, respectively. (G) NOEs connecting L122δ2 located in the SIM sequence (pink box) and methyl groups of residues in the SEP domain (yellow box). The NOE dataset was recorded using a [U-2H,ILVM-13CH3]p47101–266 sample at 18.8 T (25 °C). (H) Overlay of 13C-1H HMQC spectra of [U-2H,ILVM-13CH3]p47171–370 (orange) and [U-2H,ILVM-13CH3]-labeled full-length p47 (gray) in the absence (25 °C) (Top) or presence (40 °C) (Bottom) of [U-2H]p97ND1L added to a molar ratio of [p47]([p47171–370]):[p97ND1L] of 3:6 (monomer:monomer). Data were recorded at 18.8 T.
To determine which residues in p47171–370 mediate this interaction, a reciprocal NMR-binding experiment was performed in which excess unlabeled p471–174 was added to [U-15N,13C]p47171–370. Significant CSPs are observed for SEP domain residues L184, K185, Q202, A207, I213, R215, G216, E217, V218, H227, D234, M235, Q236, and D237 (Fig. 3 D–F), indicating that the interaction connects the linker with the SEP domain. We refer to linker residues S114 to G130, therefore, as an SEP-interaction motif (SIM). The strength of the intermolecular SIM–SEP interaction was quantified through an NMR-titration experiment in which [U-2H]p471–174 was titrated into ILVM–p47171–370. A two-state binding model was fit to titration profiles generated from CSPs of the methyl cross-peaks (SI Appendix, Fig. S4A), from which Kd =101 ± 6 μM was obtained. Although the measured affinity is relatively low, it is expected to be considerably higher in the context of an intramolecular association in the full-length protein where the entropic penalty for binding would be reduced relative to the case for bimolecular association. Direct evidence for SIM–SEP contacts has been obtained by recording methyl–methyl NOEs in an ILVM–p47 sample, p47101–266, which includes both the SIM sequence and SEP domain, as indicated in Fig. 3G and SI Appendix, Fig. S4B, where connections between L118, L122 (SIM) and L184, L186, L210, I213, L233, and M235 of the SEP domain are highlighted. CSPs due to the SIM–SEP interaction were also observed in a p47–p97ND1L complex, establishing that these contacts are not disrupted upon binding of p47 to p97 (Fig. 3H).
A Previously Unidentified p47 SHP Motif Leads to a Tripartite p47–p97ND1Lapo-Binding Mechanism.
There are conflicting reports in the literature suggesting that p47 binds p97 in a molar ratio of either 3:6 or 6:6 (monomer:protomer) (31, 44, 47). The most direct experimental evidence, based on ITC, supports a 3:6 model for the interaction of full-length p47 and p97 (44). Curiously, however, in the same study, it was also found that p47171–370 binds p97 with a 6:6 stoichiometry. Given our observations that p47 is monomeric, it is difficult to rationalize the differences between the two constructs without invoking a mechanism by which residues M1 to G170 interfere with binding of more than three ligands. One scenario might be the presence of an additional p97-binding site on p47, within M1 to G170, so that a single p47 molecule would interact simultaneously with adjacent protomers of p97, hence favoring a 3:6 interaction. To test whether an additional binding site exists on p47, we acquired 15N-1H and 13C-1H correlation spectra of [U-15N,13C]p471–174 in the absence and presence of threefold-excess unlabeled p97NTD (Fig. 4 A and B). Calculated changes in peak intensities, ΔI/I0, are plotted in Fig. 4 C, Bottom. The largest ΔI/I0 values were localized to p47 residues K146 to R172; of the 17 well-resolved cross-peaks derived from residues in this region, 14 were broadened beyond detection upon addition of p97NTD. Notably, the sequence of amino acids extending from residues K146 to A159 aligns with that of the canonical p47 SHP motif, as well as with SHP motifs in analogous p47 adaptor proteins (Fig. 4 C, Top).
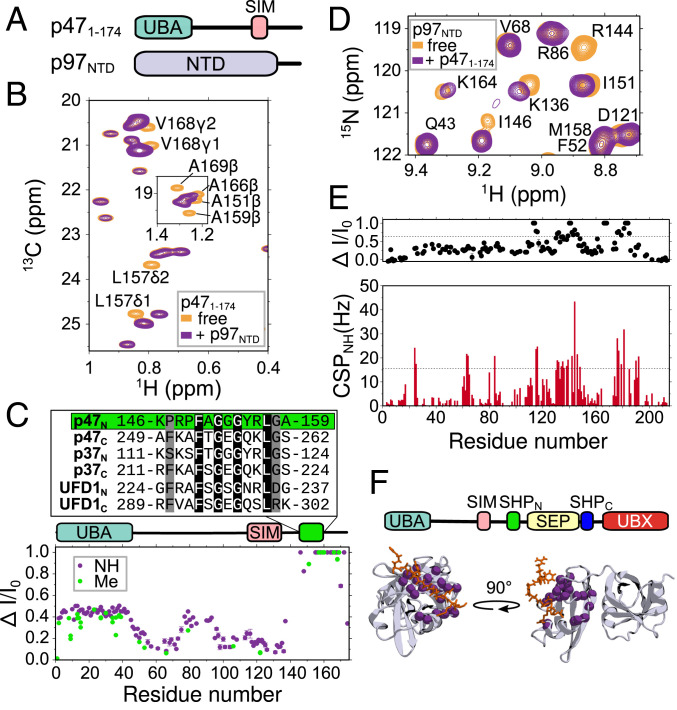
Fig. 4.
p47 interacts with p97NTD through a previously unidentified SHP motif. (A) Domain architecture of p471–174 and p97NTD constructs studied in this figure. (B) 13C-1H CT-HSQC spectra of [U-15N,13C]p471–174 illustrating site-specific cross-peak intensity changes upon addition of threefold excess unlabeled p97NTD. (C, Bottom) Changes in intensity ratios (ΔI/I0) vs. residue for both backbone amide (purple) and methyl (green) cross-peaks of p471–174 upon addition of p97NTD. Domain architecture is illustrated above the plot. Residues K146 to A159 (indicated by the green box) are aligned with SHP motif sequences found in p47, p37 (UniProt (73) accession no. [AC] Q14CS0), and UFD1 (UniProt AC Q92890). (D) 15N-1H correlation spectra of [U-2H,13C,15N]p97NTD illustrating site-specific cross-peak intensity changes and CSPs upon addition of threefold-excess [U-2H]p471–174 that are quantified in E, where dashed lines indicate ΔI/Io values (Top) or CSPs (Bottom) that exceed the average by 1σ. (F, Top) Domain architecture of p47 depicting the central SEP domain flanked by two SHP motifs. (F, Bottom) X-ray structure (PDB ID code 5C1B; ref. 46) of human p97NTD (gray) in complex with a SHP peptide from UFD1 (orange). Backbone nitrogen atoms of p97NTD assigned to cross-peaks with intensity changes, and CSPs above 1σ are indicated with magenta spheres.
We next sought to confirm that the molecular contacts stabilizing the p471–174–p97NTD interaction are those expected for a typical SHP–p97NTD complex (28, 46, 63, 64) by focusing on regions in the NTD that are known to be affected by binding of the canonical SHP motif. Therefore, we acquired 15N-1H correlation spectra of [U-2H,15N,13C]p97NTD in the absence or presence of threefold-molar excess [U-2H]p471–174. We observed site-specific intensity differences upon mixing p97NTD and p471–174 (Fig. 4D), as before, although in this instance, CSPs were also obtained (Fig. 4 D and E). The residues whose amide cross-peaks were perturbed through binding (i.e., those that display both intensity and chemical-shift changes greater than 1 SD above the mean) were mapped onto the structure of p97NTD in complex with an analogous SHP peptide (Protein Data Bank [PDB] ID code 5C1B (46)), making it clear that binding of p471–174 is localized to the SHP-binding motif within the NTD (Fig. 4 F, Bottom). These data, thus, provide strong evidence that the p47 1 to 174 region harbors a previously unidentified SHP motif and further imply that p47 can bind p97 through three distinct binding modules, including a UBX domain and two SHP motifs, designated as SHPN and SHPC, respectively, in what follows (Fig. 4 F, Top).
Given that UBX domains and SHP motifs have different interaction sites on p97NTD (46), binding of either is not mutually exclusive. On the other hand, two SHP motifs would be expected to compete for the same binding site on p97NTD, raising the question of the relative affinity and binding order of the p47 SHPN and SHPC motifs in the context of p97. For instance, it might be anticipated that the proximity of SHPC to UBX (Fig. 1B) would increase its effective binding affinity relative to SHPN so that SHPC would out-compete SHPN for a given SHP-binding site in p97. To address this point, we titrated [U-2H]p97NTD into a solution of [U-2H,15N,13C]p47 and monitored the effects of p97NTD binding on the p47 spectrum (SI Appendix, Fig. S5), focusing on cross-peaks from key residues that are sensitive to the interaction. At an equimolar ratio, CSPs and intensity differences were observed for peaks derived from residues within UBX and SHPC, while cross-peaks from residues in SHPN were largely unaffected. Upon further addition of p97NTD to a two-fold molar equivalent (i.e., two NTDs for each p47), many p47 SHPN cross-peaks were displaced from their initial positions. These data demonstrate a clear binding order for the p47–p97NTD interaction, in which p47 SHPC and UBX motifs bind first to a given NTD, followed by SHPN to a second NTD. Moreover, these data support binding of UBX and SHPC to the same NTD, as the saturation of both UBX- and SHPC-binding sites occurs in concert at equimolar concentrations of p47 and p97NTD.
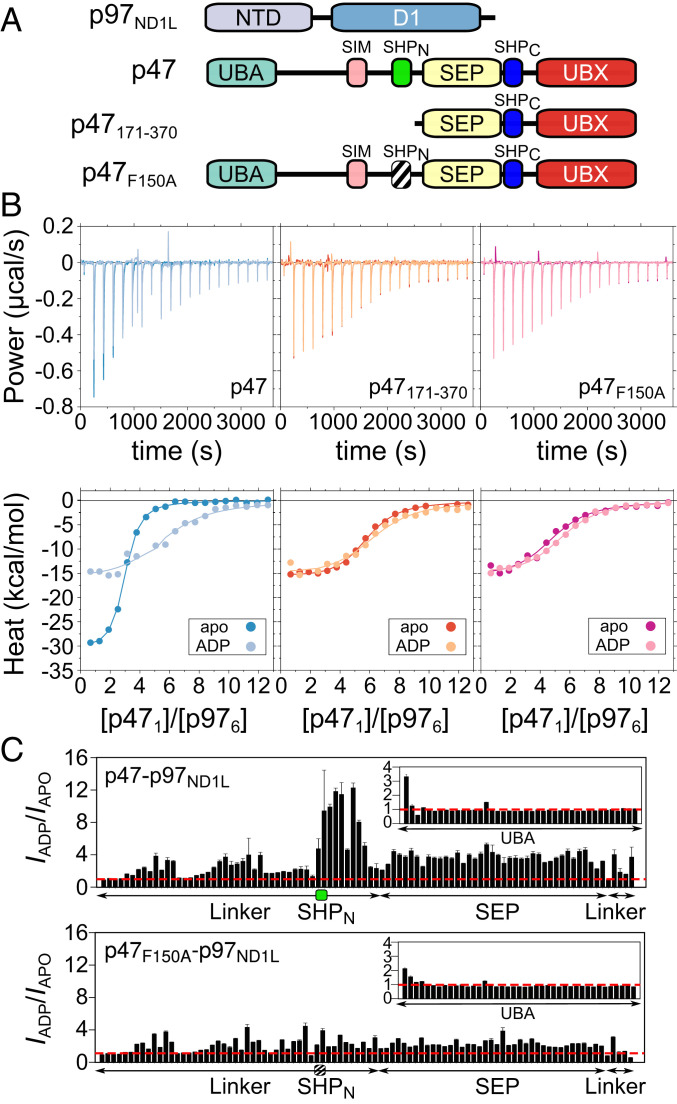
The binding of a pair of p97NTD molecules to a single p47 chain suggests that a single p47 molecule could span two p97 protomers through interactions involving SHPN, SHPC, and UBX motifs. We were interested in testing this idea further and, in particular, in establishing whether the p97 nucleotide state, and therefore the up/down NTD equilibrium, might influence the binding mechanism. To this end, a series of ITC experiments was performed in which full-length p47 and p47171–370 (i.e., no SHPN domain; Fig. 5A) were titrated into solutions of p97ND1L in the absence of nucleotide (p97ND1Lapo, corresponding to the up NTD state; SI Appendix, Fig. S1D) or in the presence of 2 mM ADP (p97ND1LADP; down NTD state) (Fig. 5 B, Left and Middle). In all cases, the data were initially fit to a simple two-state N-equivalent sites binding model (SI Appendix, Materials and Methods and Table S1A), parameterized by an equilibrium dissociation constant (Kd), an enthalpy of binding (ΔH), and a putative binding stoichiometry (N). For the p47–p97ND1Lapo titration, Kd = 0.23 ± 0.16 μM, ΔH = −30.8 ± 1.9 kcal/mol, and N = 2.7 ± 0.3 were obtained. By contrast, the Kd for the p47–p97ND1LADP complex (1.33 ± 0.28 μM) is approximately sixfold larger, and significantly different ΔH and N values were measured (−16.6 ± 1.0 kcal/mol and 5.5 ± 0.4), indicating that the affinity, enthalpy and apparent stoichiometry of p47–p97ND1L binding are dependent on the up/down NTD equilibrium. The differences in binding thermodynamics between apo and ADP states are less apparent for p47171–370–p97ND1L, with Kd = 0.55 ± 0.14 μM (apo) and 0.97 ± 0.27 μM (ADP), ΔH = −15.9 ± 0.4 and −15.5 ± 1.4 kcal/mol, and N = 5.5 ± 0.2 and 6.1 ± 0.3, respectively. These results are in agreement with previous findings from Beuron et al., in which p47 and p47171–370 bound to p97 with apparent 3:6 and 6:6 stoichiometries, respectively (44). To test if this difference is a direct result of whether SHPN binds, we introduced a point mutation at position 150 of p47, resulting in a phenylalanine to alanine substitution (p47F150A). F150 in SHPN is homologous to F253 in SHPC, a residue that is critical for the SHPC–p97 interaction (46). The F150A substitution eliminated the SHPN interaction, as evidenced by similar binding parameters for p47F150A with either p97ND1Lapo or p97ND1LADP, with respective Kd values of 1.01 ± 0.29 and 0.88 ± 0.17 μM, ΔH values of −16.0 ± 1.3 and −15.4 ± 0.6 kcal/mol, and N values of 4.7 ± 0.3 and 5.3 ± 0.2 (Fig. 5 B, Right). Note also that the ΔH values for binding of p47F150A and p47171–370 to p97 are similar (Fig. 5 B, Middle and Right), as would be expected since both p47 constructs do not allow for the establishment of SHPN contacts. Thus, the ITC data lend support to the binding model suggested above in which each p47 ligand interacts with p97ND1L in either a bipartite or tripartite manner, depending on the status of the NTD up/down equilibrium (i.e., nucleotide state), with UBX+SHPC motifs from a single ligand most likely binding to a single p97 protomer, while SHPN can bind to an adjacent protomer in the NTD up (but not down) state. In addition, the calorimetry data are consistent with binding of each p47 ligand to only a single protomer of p97ND1LADP via UBX+SHPC interactions only.
Fig. 5.
Binding of p47 to p97ND1Lapo can occur via a three-prong mechanism and via a two-prong interaction for p97ND1LADP. (A) Domain architectures for p97 and p47 constructs used in the ITC and NMR studies presented in this figure. (B) Experimental ITC isotherms (Top) and integrated heats (Bottom) for titrations of WT p47 (Left), p47171–370 (Middle), and p47F150A (Right) into solutions of p97ND1Lapo or p97ND1LADP. Solid lines represent best fits to a two-state N-equivalent site model, as described in SI Appendix, Materials and Methods. (C) Intensity ratios of amide cross-peaks in spectra recorded of 3:6 complexes of [U-2H,15N]p47–[U-2H]p97ND1L (Top) or [U-2H,15N]p47F150A–[U-2H]p97ND1L (Bottom) in ADP vs. apo states (IADP/Iapo). The red dashed line in each plot indicates an intensity ratio of 1.0, the green box (Top) delineates residues in the SHPN motif, and the black striped box denotes a defunct SHPN motif (F150A). Cross-peaks derived from residues located on SHPC and UBX broaden beyond detection in 15N-1H spectra, and thus intensities for these regions are not shown. Data were recorded at 18.8 T (40 °C).
To test our model that the differential SHPN binding is dependent on the p97 nucleotide state, we performed a series of NMR experiments and quantified peak intensities from 15N-1H TROSY-HSQC datasets recorded on samples of [U-2H,15N]p47 or [U-2H,15N]p47F150A mixed at 3:6 molar ratios (monomer p47:protomer p97) with either [U-2H]p97ND1Lapo or [U-2H]p97ND1LADP. Calculated cross-peak intensity ratios, IADP/IAPO, for p47–p97ND1L and for p47F150A–p97ND1L as a function of residue are plotted in Fig. 5C. Noting that peak intensities increase as local backbone motions become less restricted, the simplest interpretation for IADP/IAPO ratios greater than 1 is that local p47 motions are larger in the ADP state relative to the apo conformation, while values less than 1.0 imply the opposite. Large IADP/IAPO ratios are measured for residues in WT p47 that comprise the SHPN motif and the region immediately C-terminal to it (Fig. 5 C, Top), suggesting that SHPN is significantly more mobile in the ADP state relative to the apo state, as would be expected if SHPN could bind to a p97 NTD in the apo state but not in the ADP conformation. In contrast, similarly large IADP/IAPO enhancements in the vicinity of SHPN are absent in p47F150A (Fig. 5 C, Bottom), indicating that the mobilities of the p47F150A SHPN motif and surrounding residues are not significantly different in the two states. These results (see also SI Appendix, Fig. S6) are consistent with unbound F150A SHPN domains in both apo and ADP states of the p47F150A–p97ND1L complex.
What Really Is the p47:p97 Binding Stoichiometry?
As described above, a stoichiometry parameter, N, is obtained from fits of ITC profiles (Fig. 5B) and for the simple case where each of k ligands binds in the same way to a receptor a value of N = k would be measured. Interpreted in this manner, N = 3 implies a 3:6 binding stoichiometry for the p97 NTD up conformation, while N = 6 measured for the NTD down state indicates that six p47 molecules bind a single p97 hexamer in this case. However, when both tripartite and bipartite binding interactions can occur, such as for p47–p97ND1Lapo, a model based on a single binding interaction such as that used to interpret the ITC data in Fig. 5B is overly simplistic and the extracted parameters must, therefore, be interpreted cautiously (SI Appendix, Fig. S7). Indeed, one would expect that any given p97ND1Lapo hexamer would have a distribution of p47 ligands bound in tri- and bipartite modes and that addition of further ligands would ultimately give rise to a fully bound (6:6) state for p47–p97ND1Lapo, as for p47–p97ND1LADP.
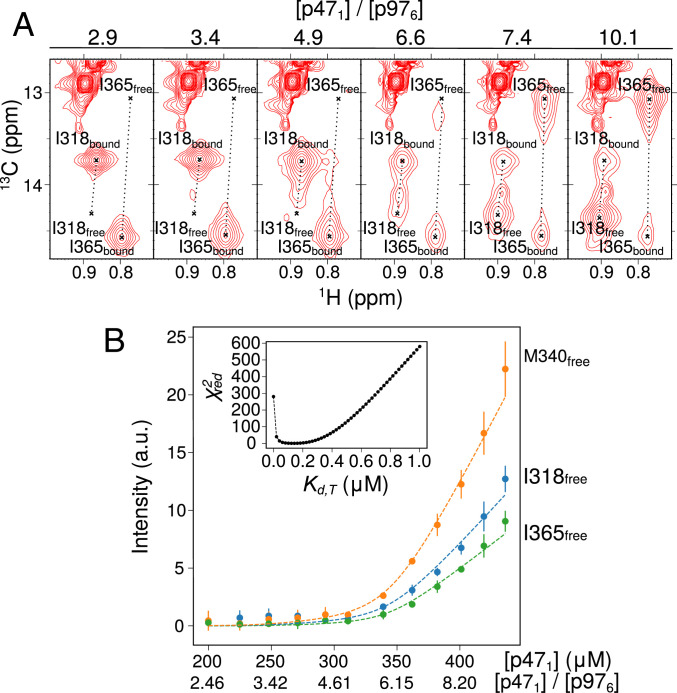
In order to establish whether these additional binding events occur, we first performed NMR experiments in which three equivalents of [U-2H, Ile(δ1),Met-13CH3]p47171–370 were added to a 3:6 p47–p97ND1Lapo complex that was formed from [U-2H, Leu,Val-13CH3,12CD3]p47 and [U-2H]p97ND1Lapo, to determine whether the presumed unoccupied sites on p97 could become bound. In strong support of additional binding interactions, CSPs of Ile and Met methyl groups of residues that report on UBX–NTD interactions were observed with no CSPs for Leu and Val methyl groups, consistent with additional binding of p47171–370 to vacant sites on p97, while the full-length, bound p47 remains in a p97-attached state (SI Appendix, Fig. S8 A–C). We next performed a titration experiment in which full-length ILVM–p47 was added to a 3:6 p47–p97ND1Lapo complex formed from ILVM–p47 and [U-2H]p97ND1Lapo. Each step of the titration was performed by adding a volume V0 of ILVM-p47 after first removing an equivalent amount of the solution so that the sample size remained the same throughout the course of the titration, as described previously (65). Selected regions of 13C-1H HMQC spectra are presented in Fig. 6A and SI Appendix, Fig. S8D, highlighting residues I318, I365, and M340 whose chemical shifts report on UBX–NTD interactions. Notably, cross-peaks from unbound p47 were not observed until [p47]:[p97ND1Lapo] exceeded 6:6 (Fig. 6A and SI Appendix, Fig. S8D), as would be expected if six p47 monomers can interact with a p97 hexamer in the NTD up state. Fig. 6B plots the intensities of methyl groups from I318, I365, and M340 derived from unbound p47 as a function of the [p47]/[p976] ratio. These were fit to a more complex model (SI Appendix, Analysis of NMR Data, Fig. S9, and Table S2) in which the p47 ligands can bind p97ND1Lapo via either bipartite (UBX+SHPC) or tripartite (UBX+SHPC+SHPN) interactions, with dissociation constants of Kd,B and Kd,T, respectively. We prefer to fit the NMR-titration data by setting Kd,B = 0.55 μM, the value obtained from ITC for the bipartite binding of p47171–370 to p97ND1Lapo; a value of Kd,T = 0.15 ± 0.02 μM is obtained, with reduced χ2 values as a function of Kd,T shown in Fig. 6 B, Inset.
Fig. 6.
p47 binds to p97ND1Lapo to form a 6:6 complex. (A) Selective regions of 13C-1H HMQC spectra of [U-2H,IM-13CH3]p47 as a function of [p471]/[U-2H-p97ND1Lapo], highlighting UBX domain residues I318 and I365 of p47, with crosshairs indicating the peak positions in the free and bound states. (B) Intensities of cross-peaks derived from methyl groups of I365 (green), I318 (blue), and M340 (orange) in the p47 free state, upon addition of [U-2H,IM-13CH3]p47 into a preformed solution of [U-2H, IM-13CH3]p47 and [U-2H]p97ND1Lapo ([p47]:[p97]∼2.5:6). Dashed lines represent best fits to a model that takes into account both bipartite (UBX+SHPC) and tripartite (UBX+SHPC+SHPN) p47 binding, as described in SI Appendix, Materials and Methods. The titration data were fit with Kd,B = 0.55 μM (obtained from ITC for the bipartite binding of p47171–370 to p97ND1Lapo) to yield Kd,T = 0.15 ± 0.02 μM. Inset shows reduced χ2 values as a function of Kd,T. Data were recorded at 23.5 T (40 °C). a.u., arbitrary units.
We next revisited the analysis of the ITC data for the titration of p47 into p97ND1Lapo using the more extensive model that takes into account both bi- and tripartite p47 binding. A simultaneous fit of the titration involving full-length p47 and p47171–370 was carried out, with extracted values for Kd,B and Kd,T of 0.52 ± 0.15 and 0.14 ± 0.10 μM, respectively, along with ΔHB = −15.8 ± 0.7 kcal/mol and ΔHT = −31.8 ± 2.1 kcal/mol (SI Appendix, Fig. S10 and Table S1B). Notably, the value of Kd,T extracted in this manner is in excellent agreement with 0.15 ± 0.02 μM obtained by NMR.
The fact that p47 is monomeric and can associate with p97 with up to six bound molecules raises the question of what the p97 occupancy is at the expected cellular concentrations of ligand and receptor. Using mass spectrometry, Aebersold and coworkers have estimated the concentrations of over 7000 proteins produced in the human osteosarcoma cell line U2OS, including p47 and p97 (66). Assuming the total concentrations of p97 and p47 are 0.0955 μM (p97 hexamer) and 0.118 μM, respectively, and using binding constants obtained in this study, it is possible to calculate the relative populations of p97apo and p97ADP complexes with different numbers of bound p47 ligands (zero to six). As p97 has diverse roles in the cell that will require it to bind to a large number of different adaptors, we have performed calculations by assuming that different fractions, p, of p97 are available for binding p47. These are indicated by bar graph plots (SI Appendix, Fig. S11) that are calculated for a total available p97 concentration of 0.0955 μM p ([p47] = 0.118 μM). Notably, the populations of p97apo complexes with more than three bound p47 ligands are extremely small over the complete range of p values examined, while the fractional populations of unbound p97apo molecules range between 1.5 and 20%, and the fraction of p97apo molecules with one to three bound ligands lies between 80 and 97%. At physiological ligand/receptor concentrations, the majority of bound p47 molecules (≥79%) are engaged with p97apo in a tripartite fashion, with UBX+SHPC and SHPN bound to neighboring NTDs. Similar calculations for p97ADP indicate a significantly higher fraction of the completely unbound p97 form (60 to 68%) than for the apo state, with singly ligated p97ADP more probable than the doubly ligated complex.
A Structural Ensemble for the p47–p97 Complex.
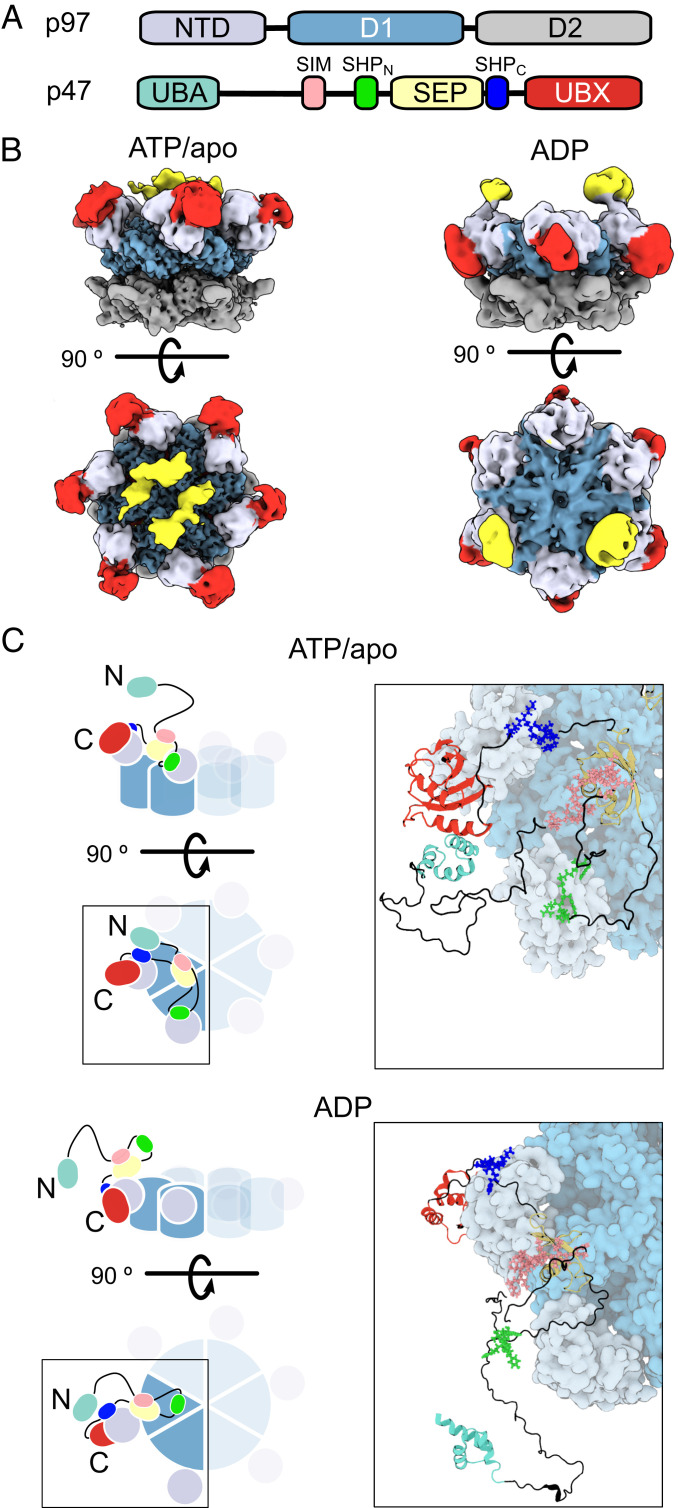
The solution NMR and ITC data provide valuable information that can be used as structural restraints for the determination of p47–p97 structural ensembles based on the interactions that have been identified and described above. Prior to the development of such ensembles, we obtained cryo-EM maps of p47–p97ATP and p47–p97ADP complexes prepared from mixing p97 with p47 (domain architecture in Fig. 7A) in a 6:6 ratio (ATP) or with excess p47, followed by gel filtration (ADP) (Fig. 7B). As expected for a highly dynamic complex, the density for much of p47 was poor, so that only low-resolution features could be identified that, nevertheless, proved valuable in ensemble building. It is noteworthy that we observed a range of p47–p97 configurations from zero to six molecules of bound p47, with no preference for 3:6 complexes (SI Appendix, Fig. S12). Although only 6:6 complexes would be expected at the concentrations of p47 and p97 used, the blotting of EM grids during specimen preparation, or interaction of particles with the air–water interface, appears to lead to removal of p47. Fig. 7 C, Left shows cartoon representations of the p97ND1ATP/apo (Top) and p97ND1ADP (Bottom) complexes, highlighting a single bound p47 ligand. Focusing initially on the p47–p97ATP/apo complex and starting from the C terminus of p47, it is seen that both the UBX domain (red) and the SHPC motif (blue) bind to a single p97 NTD (gray). Continuing to trace the sequence toward the N terminus leads to the SEP domain (yellow), which is localized to the center of p97ND1ATP/apo, as indicated by the cryo-EM map (Fig. 7B). The second SHP motif, SHPN (green), follows the SEP domain and is bound to the NTD of an adjacent p97 protomer. In order to accommodate the interaction between the SIM sequence (pink), that follows SHPN, and the SEP domain, the linker must reverse direction as indicated in the figure. The position of the UBA domain (pale green) with respect to p97 is currently undefined. A similar model can be constructed for p47 bound to p97ND1ADP; however, in this case, SHPN does not bind to p97, as indicated in the schematic. Also shown in Fig. 7 C, Right are representative atomistic models of p47–p97ND1ATP/apo and p47–p97ND1ADP based on metainference MD simulations (67) restrained by experimentally derived NOEs, CSPs, line broadening of NMR resonances, SAXS measurements (SI Appendix, Fig. S13), and cryo-EM maps, as well as by distance restraints to ensure that the interactions between p47 UBX+SHP domains and p97 NTDs are consistent with those established by X-ray crystallography studies (see SI Appendix, Table S3 for a list of experimental restraints). We have assumed only one of the many possible configurations in our modeling in which there is a 3:6 p47:p97ND1 stoichiometry (SI Appendix, Fig. S14) and, further, that both SHPC and SHPN domains are bound in the p47–p97ND1ATP/apo model. As SI Appendix, Fig. S11 highlights, a distribution of bound ligands is expected with both bi- and tripartite interactions in the p97ND1LATP/apo case. Further established in SI Appendix, Fig. S11 is the fact that at physiological ligand and receptor concentrations a 3:6 interaction is at the upper end of what would be expected.
Fig. 7.
Structural models of p47–p97ATP/apo and p47–p97ADP. (A) Domain architectures of p97 and p47, with the color scheme used in B and C. (B) Cryo-EM maps of p47–p97ATP and p47–p97ADP complexes prepared from mixing p47 and p97 in a 6:6 ratio (p47–p97ATP) or by adding a large excess of p47 to p97apo, separated via SEC, followed by addition of ADP (p47–p97ADP). (C) Representations of the p47–p97ND1 complexes in ATP/apo (Top) or ADP-bound (Bottom) states. Cartoon illustrations on the left highlight possible tripartite and bipartite binding configurations for the respective nucleotide state. Right shows representative atomic models derived from the structural ensembles obtained from metainference MD simulations.
Discussion
Many biological processes are controlled by molecular complexes, with interactions both within and between the components of the complex playing an important role in regulating function. A case in point is the p97 protein homeostasis enzyme, whose interactions with a wide range of adaptor proteins direct it to a plethora of different activities in the cell. One such adaptor is p47, and its association with p97 is critical for regulating Golgi reassembly during the cell cycle (7, 31, 35). As a first step toward a detailed understanding of the molecular basis of p47–p97 function, we present a biophysical study of the structural dynamics of p47 and of p47–p97, which complements our previous efforts focusing on p97 (18, 51, 52). Our work clarifies misconceptions in the literature concerning p47 trimerization, identifies motifs that are involved in interactions both within p47 and within the p47–p97 complex, highlights that p47–p97 intermolecular interactions are dependent on the bound nucleotide state of p97, and leads to structural ensembles to model p47–p97ATP/apo and p47–p97ADP complexes. More generally, our study establishes the important role that IDRs can play in forming biomolecular complexes. It also emphasizes the importance of using multiple, orthogonal biophysical techniques to properly characterize molecular interactions, especially those involving multiple binding steps and large regions of disorder, as the discussion below will make clear.
Extensive DLS, AUC, SAXS, and biochemical measurements establish that over a range of concentrations, extending from 100 to 700 μM, p47 is monomeric (Fig. 2 and SI Appendix, Fig. S3A). This conclusion is in contrast to previous biophysical studies that have been interpreted as providing support that p47 forms a stable, high-affinity trimer (31, 44, 45). A seminal paper describing the discovery of p47 as a p97 adaptor protein claims that p47 is trimeric based on its retention volume on a gel-filtration column in comparison with standard samples of known molecular weight (31). In more recent years, we have understood that although this approach is valid for spherical particles, notwithstanding any nonspecific interactions with the gel-filtration media, the presence of IDRs, such as in the case of p47, renders the assumption of a spherical shape invalid. Unusually high mobility on gel-filtration columns has, for example, been noted for the intrinsically disordered, monomeric protein α-synuclein (68) that elutes with a molecular mass four times higher than expected for a spherical particle of 14 kDa. To this point, it is often difficult to establish the oligomeric state of a protein complex based exclusively on a single hydrodynamic measurement when the shape of the complex is unknown. For example, our measured D0 = 6.2 × 10−7 cm2/s for p47 in aqueous solution at 25 °C corresponds to a Stokes radius of 3.96 nm. The corresponding molecular mass for a spherical particle of this size is calculated to be ∼215 kDa, significantly larger than the mass of even a p47 trimer (∼122 kDa). Notably, from the experimentally measured sedimentation coefficient of 2.9 S (SI Appendix, Fig. S3B), a molecular mass of ∼19 kDa is calculated for p47, assuming a spherical particle. Clearly, in the analysis of both diffusion and AUC measurements, the underlying assumption of a sphere leads to errors in the calculated molecular mass. However, by combining D0 (DLS) and sedimentation values (AUC), an estimate of molecular mass can be made that is independent of polypeptide shape; a value of 42.8 ± 1.0 kDa is obtained for p47, in excellent agreement with the molecular mass of 40.6 ± 1.5 kDa from SAXS measurements (SI Appendix, Fig. S3C) and that calculated on the basis of a monomeric p47 structure (40.8 kDa).
Early observations led to conflicting p47–p97 binding models supporting either 3:6 or 6:6 stoichiometries (31, 44, 47). On the basis of ITC measurements, Beuron et al. (44) concluded that the binding stoichiometry of full-length p47–p97 is 3:6 and established a 6:6 binding stoichiometry for the p47171–370–p97 complex. Our study confirms these observations when a simple binding analysis is performed in which each p47 ligand is assumed to bind identically to a single p97 NTD but shows that this conclusion is incorrect, as the binding model used is flawed. Rather, as p47 is monomeric, complexes can be bound with any number of ligands (up to six), and in the case of p97apo/ATP (all NTDs up), but not p97ADP (NTDs down), a set of p47–p97 interactions involving a previously unidentified p47 SHP motif, SHPN, allows for a three-pronged binding mechanism in which UBX and SHPC motifs from a single p47 ligand bind to the NTD of one p97 protomer, while the remaining SHPN motif of the ligand can bind to an NTD from an adjacent protomer. Thus, p47 binding to p97apo/ATP can be either bipartite or tripartite, while binding to p97ADP is bipartite. A binding model is presented, taking the multiplicity of interactions into account, that explains both the ITC and NMR titration data and consistent binding constants are obtained from ITC and NMR titrations, providing confidence in the binding model used. Simulations of ITC isotherms using this binding model followed by fits to the simple N-equivalent sites model that is pervasive in the literature show that extracted stoichiometry values, N, can be variable depending on the affinities and enthalpies that are used and, thus, establish that N does not reflect the number of ligands that are bound (SI Appendix, Fig. S7). Indeed, N ∼ 3 is obtained for the case where affinities for bipartite and tripartite binding are identically high but where ΔHB = −15 kcal/mol (or less) and ΔHT = −32 kcal/mol. Care must be taken, therefore, in the interpretation of ITC derived parameters, as, not unexpectedly, the parameters can be model dependent.
It is of interest to evaluate how many ligands might be bound to a p97 receptor at cellular concentrations. Using p47 binding constants obtained from the present study, along with estimates for protein concentrations in a human cell line (66), we have calculated the relative populations of p97apo and p97ADP complexes with different numbers of bound ligands. Notably, over all concentration ranges examined, the population of p97 molecules bound with more than three ligands is negligibly small for both p47–p97apo and p47–p97ADP (SI Appendix, Fig. S11), and the most populated bound states are those with one (p97ADP) or one to two (p97apo) ligands.
Restraints based on NMR, X-ray crystallography studies defining the interactions between UBX or SHP domains (p47) and NTDs (p97), and cryo-EM data have been used to calculate structural ensembles of p47–p97ND1ATP/apo and p47–p97ND1ADP complexes (Fig. 7 C, Right and SI Appendix, Fig. S14) using a metainference MD strategy that is particularly well suited for studies of dynamic complexes using combinations of experimental data of various origins as restraints (67). In addition to the SHPN–NTD interaction that can occur when the p97 NTDs are in the up state, a further IDR-based contact is highlighted that involves a previously unidentified SIM sequence. This linear polypeptide sequence forms intramolecular contacts with a distinct surface on the SEP domain (Fig. 3), in both isolated p47 and when p47 is complexed to p97 in either the up or down NTD states. The SIM–SEP interaction may lead to transient intermolecular p47–p47 contacts at high protein concentrations, consistent with the differences in concentration dependencies of the diffusion coefficients, D, between the p47 and p47GB1 constructs (Fig. 2C), although it is unclear if such a weak intermolecular interaction is biologically relevant. The intramolecular SIM-SEP interaction, on the other hand, may restrict the conformational space available to p47 UBA domains that bind ubiquitinated substrates and, hence, facilitate their delivery to p97. Alternatively, the SIM may play a more direct role in p97 substrate processing. Several studies have demonstrated that the SIM-binding interfaces on SEP domains from p47 and its analog p37, which also contains a SIM sequence, can mediate heterotypic protein–protein interactions (39, 43). Indeed, it has been shown that the SEP domains of both p47 and p37 directly bind a p97 substrate, protein phosphatase 1 inhibitor 3 (I3), and that this interaction is necessary for extraction of I3 from an inactivated protein phosphatase 1 complex by p97. The molecular details of how these interactions facilitate substrate unfolding will have to await studies involving complexes that include bound substrate molecules.
The results presented here, and previously (18), provide a framework to understand how MSP1 disease-causing mutations can lead to deleterious p97 function. We have shown that these mutations shift NTD conformations toward the up state in p97ADP, resulting in a weakening of the p97ADP–UBXD1 adaptor interaction by decreasing contacts involving one of the two prongs that are used to establish a stable complex (18). However, by shifting the NTD orientations in p97ADP, the mutations will lead to a stronger affinity for p47 than would be the case for the WT protein, as binding involving UBX and both SHP domains becomes possible, decreasing the concentration of p97 that is available for other interactions.
The important role of IDRs and intrinsically disordered proteins (IDPs) in mediating highly regulated, multivalent intermolecular assemblies that are found in a diverse range of cell-signaling and -regulatory processes is becoming increasingly clear. Notable examples include the CDK inhibitor Sic1, which binds to the ubiquitin ligase Cdc4 in a phosphorylation-dependent manner in order to regulate progression of the cell cycle (69, 70), and p27, which regulates eukaryotic cell division through interactions with cyclin-dependent kinase complexes (71). IDPs also play critical roles in the assembly and regulation of macromolecular machines. For instance, many ribosomal proteins contain long, disordered tails that are hypothesized to aid in proper folding of the ribosomal RNA (72). Our study adds to a growing list of the important roles of structural disorder in regulating the function of large complexes and further emphasizes that an understanding of their structural dynamics will require a concerted effort involving methods, such as crystallography and cryo-EM, that focus on the relatively static portions of the complex, and solution NMR, that can extend the window of investigation to regions that are dynamic.
Materials and Methods
Details of DNA constructs, protein expression and purification, experiments including NMR, ITC, SAXS, DLS, AUC, cryo-EM, and MD simulations, along with data fitting, are provided in SI Appendix. Sample conditions used in all experiments are given in SI Appendix, Table S4.
Supplementary Material
Acknowledgments
Z.A.R., R.H., and E.W. were supported by a scholarship (Z.A.R.) and a postdoctoral fellowship (R.H.) from the Canadian Institutes of Health Research (CIHR), and by a Churchill Scholarship (E.W.). L.E.K. and J.L.R. were supported by the Canada Research Chairs program. This research was funded by CIHR Grants FDN-503573 (to L.E.K.), PJT-162186 (to J.L.R.) and supported by the Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering, NIH. Titan Krios cryo-EM data were collected at the Toronto High-Resolution High-Throughput cryo-EM facility, supported by the Canadian Foundation for Innovation and Ontario Research Fund. MD simulations were performed using the Marcopolo computing server at the University of Cambridge and the ARCHER UK National Supercomputing Service.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013920117/-/DCSupplemental.
Data Availability.
All relevant data are included in the article and in SI Appendix.
References
- 1.Shcherbik N., Haines D. S., Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol. Cell 25, 385–397 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richly H. et al., A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120, 73–84 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Verma R., Oania R., Fang R., Smith G. T., Deshaies R. J., Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol. Cell 41, 82–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritz D. et al., Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 13, 1116–1123 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zehner M. et al., Mannose receptor polyubiquitination regulates endosomal recruitment of p97 and cytosolic antigen translocation for cross-presentation. Proc. Natl. Acad. Sci. U.S.A. 108, 9933–9938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramanathan H. N., Ye Y., The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res. 22, 346–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabouille C., Levine T. P., Peters J. M., Warren G., An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell 82, 905–914 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Latterich M., Fröhlich K. U., Schekman R., Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82, 885–893 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Ramadan K. et al., Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450, 1258–1262 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Cao K., Nakajima R., Meyer H. H., Zheng Y., The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115, 355–367 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Parisi E., Yahya G., Flores A., Aldea M., Cdc48/p97 segregase is modulated by cyclin-dependent kinase to determine cyclin fate during G1 progression. EMBO J. 37, e98724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madeo F., Fröhlich E., Fröhlich K. U., A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139, 729–734 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang W. K., Xia D., Role of the D1-D2 linker of human VCP/p97 in the asymmetry and ATPase activity of the D1-domain. Sci. Rep. 6, 20037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee S. et al., 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science 351, 871–875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang W. K. et al., A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 29, 2217–2229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuller J. M., Beck F., Lössl P., Heck A. J. R., Förster F., Nucleotide-dependent conformational changes of the AAA+ ATPase p97 revisited. FEBS Lett. 590, 595–604 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Bodnar N. O., Rapoport T. A., Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169, 722–735.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuetz A. K., Kay L. E., A Dynamic molecular basis for malfunction in disease mutants of p97/VCP. eLife 5, e20143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao M. V., Williams D. R., Cocklin S., Loll P. J., Interaction between the AAA+ ATPase p97 and its cofactor ataxin3 in health and disease: Nucleotide-induced conformational changes regulate cofactor binding. J. Biol. Chem. 292, 18392–18407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulfer S. L., Chou T. F., Arkin M. R., p97 disease mutations modulate nucleotide-induced conformation to alter protein-protein interactions. ACS Chem. Biol. 11, 2112–2116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weihl C. C., Dalal S., Pestronk A., Hanson P. I., Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum. Mol. Genet. 15, 189–199 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Meyer H., Weihl C. C., The VCP/p97 system at a glance: Connecting cellular function to disease pathogenesis. J. Cell Sci. 127, 3877–3883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye Y., Tang W. K., Zhang T., Xia D., A mighty “protein extractor” of the cell: Structure and function of the p97/CDC48 ATPase. Front. Mol. Biosci. 4, 39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchberger A., Schindelin H., Hänzelmann P., Control of p97 function by cofactor binding. FEBS Lett. 589, 2578–2589 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Schuberth C., Buchberger A., UBX domain proteins: Major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 65, 2360–2371 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballar P., Shen Y., Yang H., Fang S., The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J. Biol. Chem. 281, 35359–35368 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Boeddrich A. et al., An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 25, 1547–1558 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruderer R. M., Brasseur C., Meyer H. H., The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J. Biol. Chem. 279, 49609–49616 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Allen M. D., Buchberger A., Bycroft M., The PUB domain functions as a p97 binding module in human peptide N-glycanase. J. Biol. Chem. 281, 25502–25508 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Zhao G., Li G., Schindelin H., Lennarz W. J., An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation. Proc. Natl. Acad. Sci. U.S.A. 106, 16197–16202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo H. et al., p47 is a cofactor for p97-mediated membrane fusion. Nature 388, 75–78 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Roy L. et al., Role of p97 and syntaxin 5 in the assembly of transitional endoplasmic reticulum. Mol. Biol. Cell 11, 2529–2542 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hetzer M. et al., Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 3, 1086–1091 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Uchiyama K., Kondo H., p97/p47-mediated biogenesis of Golgi and ER. J. Biochem. 137, 115–119 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Meyer H. H., Golgi reassembly after mitosis: The AAA family meets the ubiquitin family. Biochim. Biophys. Acta 1744, 108–119 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Rabouille C. et al., Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell 92, 603–610 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Huang S., Tang D., Wang Y., Monoubiquitination of syntaxin 5 regulates golgi membrane dynamics during the cell cycle. Dev. Cell 38, 73–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan X. et al., Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97. EMBO J. 23, 1463–1473 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soukenik M. et al., The SEP domain of p47 acts as a reversible competitive inhibitor of cathepsin L. FEBS Lett. 576, 358–362 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Yuan X. et al., Solution structure and interaction surface of the C-terminal domain from p47: A major p97-cofactor involved in SNARE disassembly. J. Mol. Biol. 311, 255–263 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Dreveny I. et al., Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. EMBO J. 23, 1030–1039 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer H. H., Wang Y., Warren G., Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 21, 5645–5652 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weith M. et al., Ubiquitin-independent disassembly by a p97 AAA-ATPase complex drives PP1 holoenzyme formation. Mol. Cell 72, 766–777.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Beuron F. et al., Conformational changes in the AAA ATPase p97-p47 adaptor complex. EMBO J. 25, 1967–1976 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X. et al., Altered cofactor regulation with disease-associated p97/VCP mutations. Proc. Natl. Acad. Sci. U.S.A. 112, E1705–E1714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hänzelmann P., Schindelin H., Characterization of an additional binding surface on the p97 N-terminal domain involved in bipartite cofactor interactions. Structure 24, 140–147 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Rouiller I., Butel V. M., Latterich M., Milligan R. A., Wilson-Kubalek E. M., A major conformational change in p97 AAA ATPase upon ATP binding. Mol. Cell 6, 1485–1490 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Tugarinov V., Hwang P. M., Ollerenshaw J. E., Kay L. E., Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 125, 10420–10428 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Rosenzweig R., Kay L. E., Solution NMR spectroscopy provides an avenue for the study of functionally dynamic molecular machines: The example of protein disaggregation. J. Am. Chem. Soc. 138, 1466–1477 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Jiang Y., Kalodimos C. G., NMR studies of large proteins. J. Mol. Biol. 429, 2667–2676 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Schütz A. K., Rennella E., Kay L. E., Exploiting conformational plasticity in the AAA+ protein VCP/p97 to modify function. Proc. Natl. Acad. Sci. U.S.A. 114, E6822–E6829 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang R., Ripstein Z. A., Rubinstein J. L., Kay L. E., Cooperative subunit dynamics modulate p97 function. Proc. Natl. Acad. Sci. U.S.A. 116, 158–167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sattler M., Schleucher J., Griesinger C., Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 (1999). [Google Scholar]
- 54.Yuan X. et al., “Letter to the Editor: Complete backbone resonance assignments of p47: The 41kDa adaptor protein of the AAA ATPase p97” in J. Biomol. NMR, (2004), Vol. 28, pp. 309–310. [DOI] [PubMed] [Google Scholar]
- 55.Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D., Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Sci. 15, 2795–2804 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tugarinov V., Kay L. E., An isotope labeling strategy for methyl TROSY spectroscopy. J. Biomol. NMR 28, 165–172 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Wendler P., Ciniawsky S., Kock M., Kube S., Structure and function of the AAA+ nucleotide binding pocket. Biochim. Biophys. Acta 1823, 2–14 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Harding S. E., Johnson P., The concentration-dependence of macromolecular parameters. Biochem. J. 231, 543–547 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svedberg T., Nichols J. B., The application of the oil turbine type of ultracentrifuge to the study of the stability region of carbon monoxide-hemoglobin. J. Am. Chem. Soc. 49, 2920–2934 (1927). [Google Scholar]
- 60.Rambo R. P., Tainer J. A., Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fung H. Y. J., Birol M., Rhoades E., IDPs in macromolecular complexes: The roles of multivalent interactions in diverse assemblies. Curr. Opin. Struct. Biol. 49, 36–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le L. T. M. et al., Structural details of Ufd1 binding to p97 and their functional implications in ER-associated degradation. PLoS One 11, e0163394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim J. J. et al., Structural insights into the interaction of human p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease. FEBS Lett. 590, 4402–4413 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Sekhar A., Nagesh J., Rosenzweig R., Kay L. E., Conformational heterogeneity in the Hsp70 chaperone-substrate ensemble identified from analysis of NMR-detected titration data. Protein Sci. 26, 2207–2220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beck M. et al., The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonomi M., Camilloni C., Cavalli A., Vendruscolo M., Metainference: A Bayesian inference method for heterogeneous systems. Sci. Adv. 2, e1501177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weinreb P. H., Zhen W., Poon A. W., Conway K. A., Lansbury P. T. Jr., NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry 35, 13709–13715 (1996). [DOI] [PubMed] [Google Scholar]
- 69.Mittag T. et al., Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 17772–17777 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nash P. et al., Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Tsytlonok M. et al., Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation. Nat. Commun. 10, 1676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng Z. et al., A creature with a hundred waggly tails: Intrinsically disordered proteins in the ribosome. Cell. Mol. Life Sci. 71, 1477–1504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.The UniProt Consortium , UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research 47, D506–D515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the article and in SI Appendix.