Significance
ABCB1 is a membrane transport protein that protects cells from toxic compounds. At the same time, it limits the uptake of orally administered drugs and contributes to multidrug resistance in cancer cells. Small-molecule inhibitors of ABCB1 could alleviate these negative effects, but their development requires detailed insight into how such compounds interfere with ABCB1-catalyzed drug extrusion. Our study shows how ABCB1 inhibitors bind in pairs and can block the function of the transporter by interacting with structural features that are important for its transport function. Our results therefore provide insight into the mechanism of ABCB1 and will be valuable for the development of more effective inhibitors.
Keywords: ABC transporter, ABCB1, P-glycoprotein, single-particle cryoelectron microscopy, structure
Abstract
ABCB1 detoxifies cells by exporting diverse xenobiotic compounds, thereby limiting drug disposition and contributing to multidrug resistance in cancer cells. Multiple small-molecule inhibitors and inhibitory antibodies have been developed for therapeutic applications, but the structural basis of their activity is insufficiently understood. We determined cryo-EM structures of nanodisc-reconstituted, human ABCB1 in complex with the Fab fragment of the inhibitory, monoclonal antibody MRK16 and bound to a substrate (the antitumor drug vincristine) or to the potent inhibitors elacridar, tariquidar, or zosuquidar. We found that inhibitors bound in pairs, with one molecule lodged in the central drug-binding pocket and a second extending into a phenylalanine-rich cavity that we termed the “access tunnel.” This finding explains how inhibitors can act as substrates at low concentration, but interfere with the early steps of the peristaltic extrusion mechanism at higher concentration. Our structural data will also help the development of more potent and selective ABCB1 inhibitors.
ABCB1, also known as P-glycoprotein or MDR1, is an ATP-binding cassette (ABC) transporter expressed at several blood–organ barriers, where it extrudes a wide spectrum of xenobiotic compounds outside the cell, thereby protecting tissues from toxic substances (1, 2). This is pharmacologically important because it reduces uptake of certain orally administered drugs and limits the delivery of therapeutics into the brain across the blood–brain barrier, where ABCB1 is highly expressed. Due to its function, ABCB1 has a significant impact on the absorption, distribution, metabolism, and excretion (ADME) of drugs in the human body (3). The protein is also overexpressed in some tumors, where its ability to translocate diverse compounds contributes to multidrug resistance (MDR) (4). Among the therapeutic compounds transported by ABCB1 are the vinca alkaloids vinblastine and vincristine that block beta-tubulin polymerization during cell division and have been used in antitumor therapies (5, 6). In cancer cells that overexpress ABCB1, vinblastine and vincristine uptake is strongly reduced (5, 7).
As the contribution of ABCB1 to MDR became established, efforts were directed at developing inhibitors. The rationale was that preventing ABCB1-associated extrusion of chemotherapeutic drugs would increase their efficacy. Several inhibitory compounds were developed and showed promise in cellular assays; however, most did not pass clinical trials due to insufficient selectivity, unsatisfactory efficacy, or excessive toxicity (8). This also applies to the third-generation inhibitors elacridar, tariquidar, and zosuquidar, which are more potent than their precursors and had shown potential during preclinical evaluation (9–12). Although these setbacks have reduced interest in pursuing ABCB1 inhibitors as a general strategy for overcoming MDR, there is still an interest in understanding ABCB1 inhibition for special applications, including the improvement of oral uptake of drugs. In parallel, high-affinity inhibitors have been instrumental in investigating the mechanism of ABCB1. In mouse studies and in studies using porcine kidney epithelial cells overexpressing ABCB1, it was shown that when present at low concentrations, both tariquidar and elacridar can be transported and therefore act as substrates (13), suggesting that there may not be a clear-cut distinction between substrates and inhibitors.
In addition to small-molecule compounds, monoclonal antibodies were developed to modulate or inhibit ABCB1 (14). The best known are MRK16, generated against leukemia cells resistant to adriamycin (15), and UIC2, generated from BALB/c mice immunized with BALB/c 3T3-1000 cells that overexpress ABCB1 (14). While neither antibody was successful in clinical applications, both have since been used for functional studies as they can modulate the function of ABCB1 in vitro (14, 16, 17). Whereas UIC2 efficiently blocks transport of all investigated ABCB1 substrates, MRK16 interferes with the transport of bulky and large substrates such as vincristine and actinomycin D, but not with that of small and flat molecules such as doxorubicin (14).
Due to the clinical significance of the protein, the structure and mechanism of ABCB1 have been extensively investigated. ABCB1 is a monomeric ABC transporter with two pseudosymmetric halves, each containing a transmembrane domain (TMD) and a nucleotide-binding domain (NBD). High-resolution structures of a multitude of related bacterial and archaeal ABC exporters as well as ABCB1 homologs of mouse, worm, and red algae were determined by X-ray crystallography (18–20). However, only through the use of single-particle cryoelectron microscopy (cryo-EM) was it possible to determine structures of the human protein. A structure of taxol-bound human ABCB1 revealed a drug-binding pocket at the center of the membrane, surrounded by 12 transmembrane (TM) helices (21). The structure of a catalytically inactive variant (ABCB1-EQ) revealed a conformation featuring a closed NBD dimer containing trapped ATP and a collapsed translocation pathway between the TMDs (22). This was interpreted to represent a posttransport state.
ABCB1 is thought to operate by ATP-driven alternating access. By harnessing energy from binding and hydrolyzing ATP, the TMD conformations cycle between inward facing, occluded, outward facing, and collapsed (21, 23, 24). The reaction cycle can be interrupted by unspecific inhibitors such as vanadate, which binds to the conserved ATPase sites and traps a posthydrolytic state. In contrast, most small-molecule inhibitors are expected to bind to the TMDs and compete with substrates for binding to the drug-binding pocket. At present, there are no structures of fully human ABCB1 bound to small-molecule inhibitors, but a recent structure of human–mouse chimeric ABCB1 revealed that two zosuquidar molecules can bind in the central drug-binding cavity (25). Cyclic peptide inhibitors were previously visualized bound to mouse ABCB1 by X-ray crystallography (18, 26).
The development of more specific ABCB1 inhibitors will tremendously benefit from high-resolution structural insight. In addition, we hypothesized that elucidating how human ABCB1 interacts with substrate drugs and inhibitors can reveal how mechanical forces facilitate drug extrusion. To accomplish this, we determined five cryo-EM structures of drug-free and small-molecule-bound human ABCB1.
Results and Discussion
Functional Characterization of ABCB1.
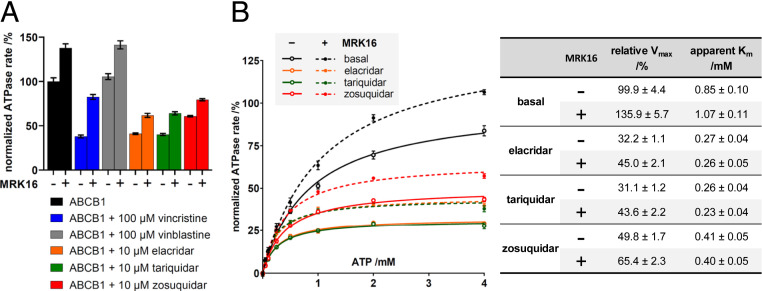
Human wild-type ABCB1 was expressed in HEK293 cells and increased the cells’ resistance to vinblastine when compared to uninduced cells (SI Appendix, Fig. S1A). The observed level of resistance to vinblastine was similar to that reported for ABCB1-expressing MDCK cells (27). Upon addition of verapamil, an inhibitor of ABCB1, the resistance of the overexpressing HEK293 cells to vinblastine was reduced (SI Appendix, Fig. S1A), demonstrating that the overexpressed protein was functionally competent. To investigate the activity of purified ABCB1, the transporter was reconstituted into liposomes and nanodiscs containing a mixture of brain polar lipid extract (BPL) and cholesterol. We observed distinct basal ATPase rates: in liposomes, the hydrolysis rate was ∼200 nmol(Pi)·min−1·mg−1, whereas in nanodiscs, we observed a higher ATPase rate of ∼650 nmol(Pi)·min−1·mg−1 (SI Appendix, Fig. S1B). High basal levels of ATPase rates are consistent with the previously reported ATPase activity of ABCB1 in nanodiscs containing BPL and cholesterol (21), but higher than the activity in detergent micelles or lipid bilayers without cholesterol (28). Upon addition of verapamil (SI Appendix, Fig. S1B), distinct levels of stimulation of ATPase rates were observed in proteoliposomes and nanodiscs, both of which were lower than reported in other studies (28, 29). However, a consistent maximum ATPase rate of ∼750 nmol(Pi)·min−1·mg−1 was observed in both nanodiscs and liposomes. In proteoliposomes, the EC50 value of verapamil-stimulated ATPase activity was 24 µM, which is in agreement with that observed for membrane vesicles containing overexpressed P-glycoprotein (30) or ABCB1 reconstituted in brain phosphatidylethanolamine (31).
While proteoliposomes are more suitable for functional studies of transporters, they are generally not useful for high-resolution structure determination of proteins by single-particle cryo-EM. In contrast, nanodiscs have been successfully used for this purpose over the past years. In addition to our structural studies, we therefore characterized nanodisc-reconstituted ABCB1 by determining the modulation of its ATPase activity by drugs (Fig. 1A). We found that addition of elacridar, tariquidar, or zosuquidar reduced the basal ATPase rate by ∼50 to 60%. In contrast, addition of verapamil, taxol, or vinblastine showed either a slight stimulation (taxol 10% and verapamil 15%) or no measurable effect (vinblastine). Intriguingly, the substrate vincristine did not alter the ATPase rate at concentrations below 1 μM, but strongly reduced the activity at higher concentrations (SI Appendix, Fig. S1C). For our structural studies, we focused on drugs that showed a pronounced effect on ABCB1 function at the chosen conditions.
Fig. 1.
Functional characterization of human ABCB1. (A) Normalized ATPase activity of nanodisc-reconstituted ABCB1 in the absence (black bars) or presence of different drugs, and in the absence (−) or presence (+) of the Fab fragment of antibody MRK16. The concentration of ATP was 2 mM throughout. n = 3, error bars represent SDs. (B) Normalized ATPase activity of ABCB1 as a function of ATP concentration in the absence or presence of inhibitors, and in the absence (−) or presence (+) of the MRK16-Fab. The points were plotted with nonlinear regression of the Michaelis–Menten equation. Relative Vmax and apparent Km are displayed in the table. n = 3, error bars represent SDs.
Because we used the inhibitory antibody fragment MRK16-Fab to achieve sufficiently high resolution in our structural studies, we investigated the effect of MRK16-Fab on nanodisc-reconstituted ABCB1. We found that MRK16-Fab stimulated the ATPase activity by 30 to 40%. This effect was also observed in the presence of vinblastine, elacridar, tariquidar, and zosuquidar and was even more pronounced (100% increase) in the presence of vincristine (Fig. 1). However, MRK16-Fab did not alter the apparent affinity for vincristine (SI Appendix, Fig. S1D), nor did it change the Km for ATP even in the presence of high-affinity inhibitors (Fig. 1B). These findings suggest that our nanodisc-reconstituted ABCB1 sample, in complex with MRK16-Fab, is well suited for structural studies, since it contains lipids mimicking the membrane environment and cholesterol molecules that stabilize the protein (32).
Drug-Free and Drug-Bound Structures of the ABCB1–MRK16-Fab Complex.
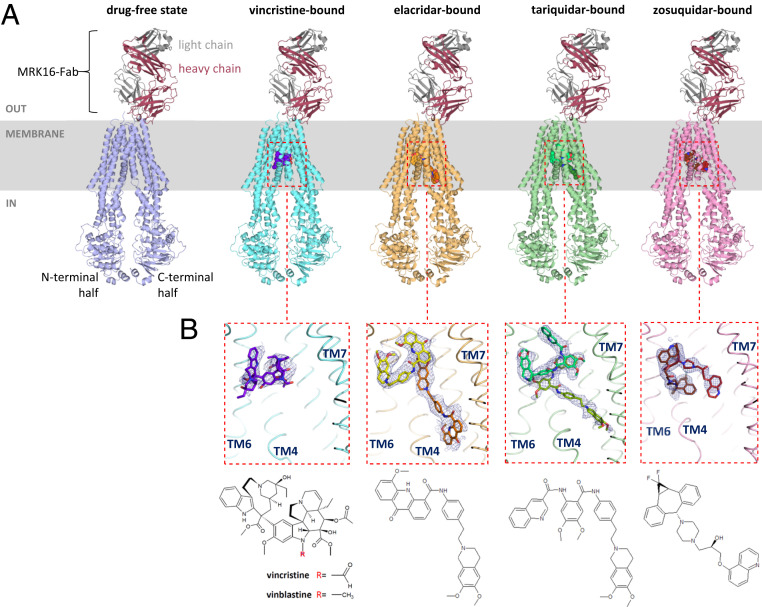
We formed five ABCB1–MRK16-Fab complexes in the absence of any drug or in the presence of vincristine, elacridar, tariquidar, or zosuquidar and used single-particle cryo-EM for high-resolution structure determination. The use of MRK16-Fab increased the mass of the particles and helped break the pseudo twofold symmetry, allowing for high-resolution structure determination. All complexes were resolved to resolutions higher than 4 Å (SI Appendix, Figs. S2–S6). In the drug-free state, resolved to 3.9-Å resolution, a nanobody specific to one of the NBDs was also present, which was added with the goal to aid with particle alignment. However, due to the high flexibility of the NBDs (relative to the TMDs), the density of the NBD-attached nanobody was not resolved. Therefore, the nanobody was not used for the subsequent drug-bound structures. Elacridar- and tariquidar-bound ABCB1–MRK16-Fab were determined at 3.6-Å resolution, and zosuquidar-bound ABCB1–MRK16-Fab was determined at 3.5-Å resolution. The highest resolution was observed for the vincristine-bound state of ABCB1–MRK16-Fab (3.2 Å). The transmembrane regions, including the binding pocket and the variable domain of MRK16-Fab, were consistently well resolved, allowing for de novo model building (Fig. 2A). In contrast, the density for the NBDs was of lower resolution and previous structures (21) were used for model building.
Fig. 2.
Overview of ABCB1–MRK16-Fab structures. (A) Ribbon diagrams of drug-free and drug-bound states of ABCB1–MRK16-Fab structures. The drug molecules are represented in sphere representation. (B) Close-up view of the binding pocket of vincristine- and inhibitor-bound structures, with adjacent TM helices shown as backbone splines and labeled. The thresholds of the EM density maps were adjusted such that the level of density covering the surrounding TM helices is comparable. Drug molecules are shown as sticks and their corresponding chemical structures are shown underneath.
All five structures captured an occluded conformation of ABCB1, which is characterized by transmembrane helices TM4 and TM10 kinked toward the pseudosymmetric axis of the transporter, forming a gate at the entrance of the drug-binding pocket. The only major difference among the structures is in and around the central drug-binding cavity, where we observed either no interpretable density besides noise (drug-free state), density consistent with a single molecule of vincristine, or density corresponding to two molecules of either elacridar, tariquidar, or zosuquidar. In contrast to the human–mouse chimeric ABCB1–UIC2-Fab data (25), the inward-open conformation (with straight TM4 and TM10) was not observed in our datasets. This suggests that binding of MRK16-Fab may shift the conformational equilibrium of lipid-embedded ABCB1 from inward open to occluded. This is in agreement with the observed stimulation of ATPase activity by MRK16-Fab, since the formation of the occluded conformation and the concomitant kinking of TM4 and TM10 decrease the distance between the NBDs.
The observed EM density suggested one main orientation of bound vincristine (Fig. 2 B, Left), but alternative orientations are possible. Vincristine is a globular molecule that is bound in a location similar to that of taxol in the previously reported ABCB1–UIC2-Fab structure (21). We identified 13 residues within a distance of 3.5 Å of the bound vincristine, the majority of which are aromatic side chains (SI Appendix, Fig. S7). Intriguingly, the formyl moiety attached to the indole nitrogen of vincristine (Fig. 2B), a feature that distinguishes the molecule from vinblastine, is located in close proximity to the carboxamide group of the Q990 side chain, suggesting that there might be strong contact (see below). Another view (SI Appendix, Fig. S7B) suggests some flexibility of the bound vincristine molecule, even though the density allowed confident fitting of the scaffold. While there are mostly insignificant structural differences when compared to the drug-free state, TM12 of vincristine-bound ABCB1–MRK16-Fab revealed structural changes in response to drug binding (SI Appendix, Fig. S7D). In the occluded state, a kink at position P996 separates TM12 into two alpha-helices. The segment F971–P996 lines the binding pocket, but in the vincristine-bound structure, the Ca positions of residues V981–F983 are slightly displaced, allowing the side chain of F983 to shift closer to the vincristine molecule and interact with its indolyl ring. These observations indicate that TM12 contributes to the plasticity of the binding pocket.
In contrast to the substrate-bound structure, two inhibitor molecules are bound in the central cavity of ABCB1 (Fig. 2 B, Middle and SI Appendix, Fig. S8). For elacridar and tariquidar, one of these adopts a globular, U-shaped conformation that is fully inside the binding pocket of ABCB1. The second molecule adopts a distinct, l-shaped conformation, with the acridonyl/dimethoxytetrahydroisoquinoline ring systems located between the central cavity and the cytoplasmic gate region formed by the kinks in TM4 and TM10. These parts of the l-shaped elacridar and tariquidar molecules are not well defined in the EM density maps, suggesting that they are somewhat flexible. The interactions of the drugs with ABCB1 in these regions are also weaker and less specific, as exemplified by the side chain of W232 for which the EM density suggests two distinct rotamers. The l-shaped and U-shaped tariquidar conformations match the conformers predicted by computational analyses (33). In the zosuquidar-bound complex, we also observed two inhibitor molecules (Fig. 2 B, Right) similar in conformation and position to those previously observed in the human–mouse chimeric ABCB1–UIC2-Fab complex (Protein Data Bank [PDB]: 6qee) (25) (SI Appendix, Fig. S9). Contrary to another human multidrug-resistant transporter, ABCG2, which was reported to have a preference for flat compounds (34), our structures suggest that ABCB1 preferentially binds globular compounds (or compounds that can adopt a globular conformation). These molecules often contain aromatic ring systems, which we observed are likely to be positioned in the “top” part of the cavity, close to residues Y953 and F983 (SI Appendix, Figs. S7–S9).
Our results indicate two distinct modes of how ABCB1 might be inhibited by small molecules: The first involves binding a molecule that fully occupies and strongly interacts with the central pocket, as observed for vincristine, a substrate that inhibits the ATPase activity of ABCB1 when present at high concentrations. While we do not expect vincristine to sterically block the transporter, it is possible that its formyl moiety and the amide group of Q990 may transiently form a covalent bond based on the intermediate state of the aldehyde–amide condensation reaction (35). Full condensation would require two amide groups interacting with the aldehyde group, but in ABCB1, where only one glutamine side chain is available, we expect that a reversible, intermediate step of the reaction occurs. This interaction might alter the architecture of the binding pocket and thus prevent ATP hydrolysis, resulting in a lowered ATPase activity rate and hence, the inhibitory effect of vincristine at high concentration.
Another way to inhibit the transporter is the tight binding of a second inhibitor molecule that extends beyond the central cavity and binds additional areas of ABCB1, as observed mainly in our elacridar- and tariquidar-bound structures. Previous research suggested that elacridar and tariquidar can be transported by ABCB1 when present at very low concentration (13). Given our structural data, we conclude that at low (nanomolar) concentrations, these molecules can be transported only if a single molecule occupies the binding pocket. At higher (micromolar) concentrations, the equilibrium is shifted and two molecules are bound. The U-shaped, cavity-bound molecule may be thought of as a substrate, whereas the second, l-shaped molecule can be thought of as a noncompetitive inhibitor. However, due to insufficient sensitivity, this cannot be experimentally explored using an ATPase assay. Given that the ATPase activity is reduced only to 40 to 50% in the presence of elacridar, zosuquidar, or tariquidar (SI Appendix, Fig. S1C), the NBDs of ABCB1 are evidently still flexible enough to form the closed sandwich dimer required for ATP hydrolysis to occur. This could explain how inhibitors can uncouple ATPase activity from the transport and relatively high ATPase rates are observed (36) even though transport is inhibited.
Binding and Inhibition by MRK16-Fab.
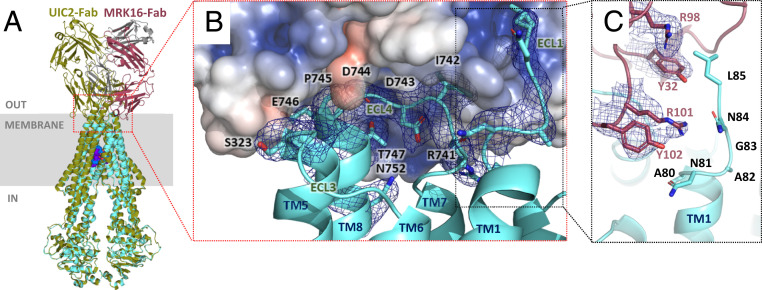
Although a crystal structure of the isolated MRK16-Fab was reported (37), it was not established how MRK16 binds to ABCB1. Our structures reveal a conformational epitope located on the exterior of ABCB1. While UIC2 also recognizes an external ABCB1 epitope (25), MRK16 binds differently (Fig. 3A). The interface is also distinct from previous predictions (16, 38) and involves mostly polar and charged residues from ECL3 and ECL4 (Fig. 3 A and B). The total buried surface area amounts to ∼808 Å2 and contains residues 741 to 747 of ABCB1 (ECL4 residues) as well as Q750 and N751 from TM8, and S323 and G324 from ECL3. MRK16 also contacts the external region of TM1 (residues 81 to 85), which no longer has a helical structure. The two complementarity-determining regions (CDRs) of the light chain of MRK16 (CDR1 and CDR3) and all three CDRs of the heavy chain participate in binding (SI Appendix, Fig. S10). A key contact is formed by R101 and Y102 of the heavy chain, which engage in a cation–pi interaction (Fig. 3C), through which they appear to prevent TM1 of ABCB1 from forming an alpha-helix beyond residue N81.
Fig. 3.
ABCB1–MRK16-Fab interface. (A) Ribbon diagram of vincristine-bound ABCB1–MRK16-Fab complex (ABCB1 colored cyan) superimposed with the taxol-bound ABCB1–UIC2-Fab structure [PDB: 6qex (21), ABCB1 colored olive]. (B) Close-up of the ABCB1–MRK16 interface. ABCB1 is shown as a cyan ribbon except for residues in contact with MRK16, which are shown as sticks. The EM density of the ABCB1 residues in contact with MRK16 is shown as a blue mesh. MRK16-Fab is shown as a transparent electrostatic surface (blue, positive charges; red, negative charges). Transmembrane helices, extracellular loops, and ABCB1 residues are labeled. (C) Close-up of TM1 of ABCB1 at the interface with MRK16 (colored as in A). EM density (blue mesh) is shown for MRK16 residues in contact with ABCB1 residues of TM1. Note that the arginine residue R101 causes an unwinding of TM1 of ABCB1.
Both UIC2 and MRK16 inhibit ABCB1 by preventing substrate release at the external side of the membrane, but without interfering with ATPase activity or disrupting drug binding. Substrate release can be visualized based on the outward-open conformation of the bacterial Sav1966 transporter, showing separation of TM helices and loops into two “wings” (39). For ABCB1, extracellular loops ECL1, ECL5, and ECL6 form one wing, whereas ECL2, ECL3, and ECL4 form the other. UIC2 binds to ECL1, ECL3, and ECL4, clamping the wings together and thus preventing the transporter from converting to an outward-facing conformation and releasing the substrate (25). MRK16, on the other hand, has only minor contact with ECL1 and might thus interfere less with the conversion of ABCB1 to outward open. If the interaction with ECL1 is broken, the wings of the transporter will be able to separate, even if only partially. This could facilitate extrusion of small or flat molecules such as doxorubicin (544 Da), while inhibiting the transport of bulky and larger molecules, such as vincristine (825 Da) or actinomycin D (1,255 Da), fully consistent with previous biochemical studies (14).
Access Tunnel to the Binding Pocket.
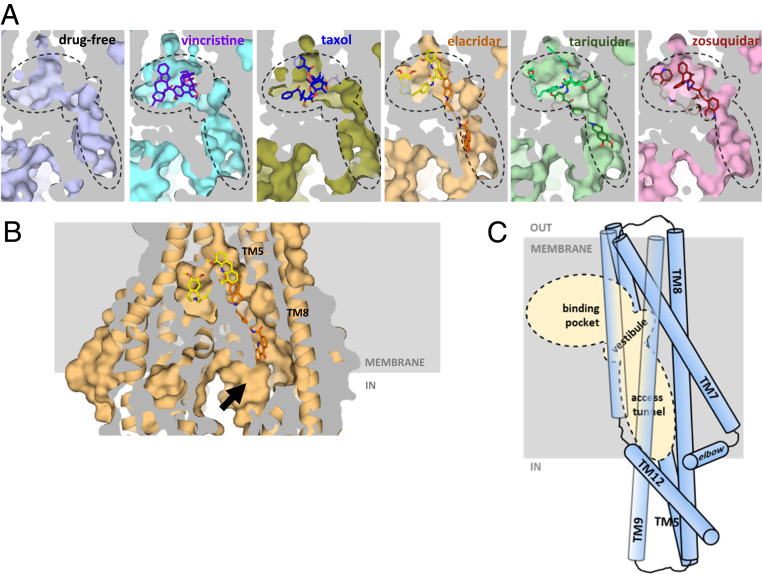
A closer inspection of the three inhibitor-bound structures revealed a cavity within ABCB1 that extends from the central drug-binding pocket to the cytoplasmic gate formed by the kinks in TM4 and TM10 (Fig. 4A). We named this cavity “access tunnel” because it can provide access for small solutes from the cytoplasm to the central pocket even when ABCB1 adopts an occluded conformation (Fig. 4B). Where the access tunnel and the central drug-binding pocket meet, a vestibule is formed (Fig. 4). Our structures show that substrate molecules (vincristine and taxol) and one of the two high-affinity inhibitor molecules are fully enclosed within the central binding pocket, while the second molecule of the inhibitors is partially located in the vestibule (zosuquidar) or even reaches into the access tunnel (tariquidar and elacridar). It is worth noting that the access tunnel is not located on the central pseudosymmetry axis of ABCB1, but is offset and closer to the C-terminal half of the transporter. It is surrounded by five transmembrane helices (TM5, TM7, TM8, TM9, and TM12) and its surface contains several phenylalanine residues that interact with the bound inhibitors. The side chains of F343, F994, and F239 interact with the l-shaped elacridar (SI Appendix, Fig. S8A), while F303 and F994 interact with zosuquidar in the vestibule (SI Appendix, Fig. S9C). Some of the conserved residues in the access tunnel were previously investigated by mutagenesis and were shown to affect substrate transport (29, 40). Intriguingly, there are three residues in the binding pocket (Y307, Q725, and V982) whose mutagenesis did not significantly alter substrate transport but did interfere with inhibition by tariquidar (41).
Fig. 4.
Access tunnel and vestibule in the occluded conformation of ABCB1. (A) Surface representation of ABCB1 structures presented in this study and taxol-bound ABCB1 (PDB: 6qex) (21). The view is parallel to the membrane and the section shown covers the central drug-binding pocket and the cavities (vestibule and access tunnel). These cavities are indicated with dashed lines. (B) Surface and ribbon representation of elacridar-bound ABCB1–MRK16-Fab structure showing the general architecture of the central cavity in the occluded state. The access tunnel extends toward the cytoplasmic side of the membrane allowing water molecules to enter the cavity (black arrow). (C) Schematic of the drug-binding pocket, vestibule, and access tunnel within ABCB1. The transmembrane helices that surround the access tunnel are shown as cylinders. The dashed area corresponds to the areas shown in A.
In the occluded conformation, the access tunnel is too narrow to facilitate the passage of the substrate or inhibitor molecules investigated here. We therefore speculate that a more inward-open conformation shown earlier (21) or possibly even more inward-facing states allow substrates or inhibitors to access the drug-binding pocket, which may occur directly from the inner leaflet of the membrane as proposed earlier (42–45). We further speculate that as long as one or more noncovalently bound molecules fit fully within the binding pocket of ABCB1, subsequent transport is possible. However, if a molecule (or pairs of molecules) protrude into the vestibule or the access tunnel, the transport reaction cannot proceed. We conclude that the access tunnel of human ABCB1 is a key feature and likely the site where small molecules selectively modulate the transporter.
Recent electron paramagnetic resonance (EPR) data showed that upon adding ATP, zosuquidar stabilizes human ABCB1 in a distinct high energy state, more so than tariquidar (46). These data are in line with our observation that zosuquidar protrudes only to the vestibule and therefore allows for a more noticeable rearrangement of the TM helices than what is seen for the tariquidar-bound ABCB1, where tariquidar is also present in the access tunnel. Further research is needed to confirm the correlation between the EPR and the cryo-EM results.
Implications for the Transport Mechanism.
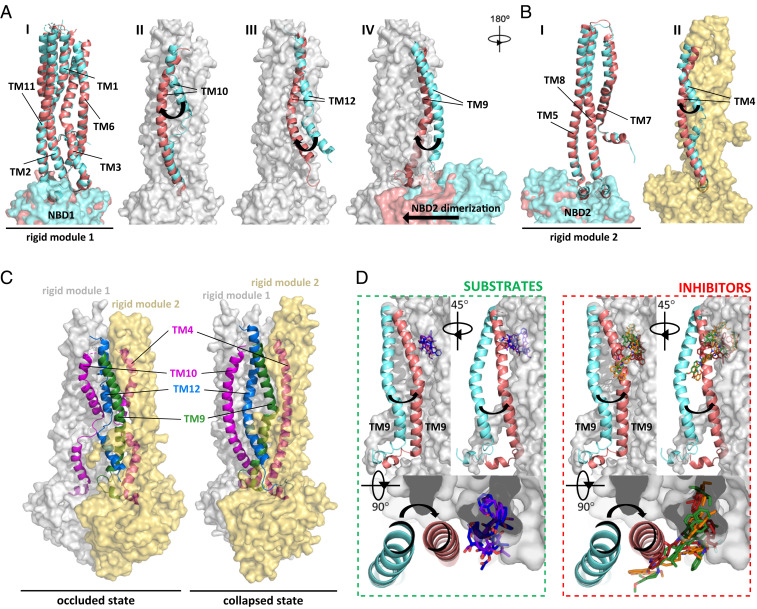
In addition to revealing the mechanism of ABCB1 inhibition by high-affinity inhibitors and MRK16, our results provide insight into how peristaltic forces may contribute to the extrusion of substrates upon ATP binding and hydrolysis. Transport mechanisms invoking peristalsis were previously proposed for other ABC transporters, including the bacterial vitamin B12 transporter BtuCD-F and the human multidrug transporter ABCG2 (47, 48). By superimposing our structures with all available ABCB1 structures, we identified two modules within the TMDs that move as rigid entities as the transporter cycles between inward-open, occluded, and collapsed conformation (Fig. 5 A–C). The first module consists of TM1, TM2, TM3, TM6, TM11, and NBD1 (Fig. 5A, module shown as a gray surface), while the second module consists of TM5, TM7, TM8, and NBD2 (Fig. 5B, module shown as a yellow surface). While these modules move as rigid bodies, the remaining four TM helices (TM4, TM9, TM10, and TM12) undergo major rearrangements. The key changes between occluded and collapsed conformations can be visualized by showing the rigid modules as surfaces and the four mobile TM helices as ribbons (Fig. 5C). As the NBDs move to form a closed dimer in response to ATP binding, the rigid modules move closer to the vertical pseudosymmetry axis, which likely triggers conformational changes in the four mobile TM helices. TM4 and TM10 thus change from strongly kinked to a straighter conformation, whereas TM12 forms an α-helix along its full length and TM9 changes its position and degree of bending. TM12 appears to have a key role in pushing bound substrates out of the drug-binding pocket, as it occupies the space available to substrates in the occluded conformation. The importance of TM12 for drug transport was previously acknowledged (49, 50). We propose here that TM9 has an equally important but distinct role. In the collapsed conformation, TM9 obstructs the access tunnel and vestibule (Fig. 5D, superposition as in panel A) and can only fully shift when a substrate (vincristine, taxol) is bound in the central cavity. By contrast, in the presence of inhibitors (elacridar, tariquidar, and zosuquidar) this shift is not possible because TM9 and the molecule(s) would clash with the second (l-shaped) inhibitor molecule. We therefore propose that TM9 of ABCB1 assumes the role of an “initiator” of the peristaltic extrusion mechanism. The mechanism that we propose here might not be limited only to ABCB1, but could also apply to the other B-subfamily ABC transporters that rely on a peristaltic mechanism.
Fig. 5.
Rigid modules and mobile TM helices facilitate the ABCB1 transport mechanism. (A and B) Superposition of vincristine-bound ABCB1 in an occluded conformation (cyan, this study) and collapsed conformation of ATP-bound ABCB1-EQ mutant (pink, PDB: 6c0v) (22) identified two rigid modules shown as gray and yellow surface, respectively, and four TM domains with distinct conformations. (A) Superposition of five TM domains and NBD1 that define rigid module 1, with no rearrangement seen between the occluded and collapsed conformations (subpanel I). In subpanels II to IV, rigid module 1 is shown as a gray surface and the distinct conformations of TM10, TM12, and TM9 are shown individually in the two states. Subpanel IV also shows the large conformational changes caused by the dimerization of the NBDs. (B) Superposition of three TM domains and NBD2 that define rigid module 2, with no rearrangement seen between the occluded and collapsed conformations (subpanel I). In subpanel II, rigid module 2 is shown as a yellow surface and the distinct conformations of TM4 are shown. (C) Comparison of occluded and collapsed states, with the rigid modules represented as transparent surfaces colored as in A and B and mobile TM domains shown as ribbons and labeled. The view is as in A. (D) Role of TM9: Following the superposition of structures, rigid module 1 is shown as a gray surface, TM9 is shown as a ribbon, and substrates (vincristine, taxol) or inhibitors (elacridar, tariquidar, zosuquidar) are shown as sticks. This shows that the shift of TM9 is prevented by bound inhibitors but possible in the presence of substrates.
Conclusions
Our structural data provide insight into the distinct interactions of ABCB1 with inhibitors. We conclude that molecules that only occupy the central cavity behave like substrates, even if they belong to the inhibitor family (e.g., tariquidar at low concentrations). In contrast to substrates, inhibitors can bind in pairs and occupy not only the drug-binding pocket but extend into the vestibule and/or access tunnel. Our structural analysis identifies TM9 as an important player in helping ABCB1 distinguish substrates from inhibitors. When a substrate is bound, TM9 is able to initiate the peristaltic extrusion mechanism by moving into the space provided by the access tunnel. A tightly bound inhibitor in the access tunnel will prevent this motion. Our study also reveals the ABCB1–MRK16 interface and rationalizes how this antibody can inhibit drug transport, while simultaneously increasing ATPase activity. Finally, our structures provide a solid basis toward a rational design and development of improved ABCB1 modulators and inhibitors.
Methods
Protein Expression and Purification.
Wild-type human ABCB1 expression and purification was performed as previously described (21, 25) with minor modifications. Briefly, the protein (GenBank ID AAA59576.1) was expressed as a fusion construct with the C-terminal 3C protease cleavage site followed by the eYFP-rho-1D4 tag from a stable cell line generated with the tetracycline-inducible Flp-In T-REx system (Thermo Fisher Scientific) following the company’s guidelines. Cells were adapted, grown, and maintained in EX-CELL 293 (Sigma) medium supplemented with 2% fetal bovine serum (FBS, Thermo Fisher Scientific), 100 units/mL/100 µg/mL penicillin/streptomycin and 6 mM l-Glutamine at 37 °C under humidified conditions with 6% CO2.
The induction was done with a final 0.5 to 1.0 µg/mL tetracycline concentration for 48 h under the same conditions. Cells were harvested and stored at −80 °C. After thawing, the pellet was dounce homogenized in suspension buffer (25 mM Hepes, pH 7.4, 150 mM NaCl, 20% glycerol) supplemented with a protease inhibitor mixture (Roche cOmplete) and DNaseI (Roche) before the detergents n-Dodecyl-β-D-maltopyranoside (DDM), octaethlyene glycol dodecyl ether (C12E8) and Cholesteryl Hemisuccinate (CHS) were added to final concentrations of 0.4% DDM/0.1% C12E8/0.1% CHS (wt/vol). The extraction was performed for 2 h at 4 °C, followed by centrifugation at 100,000 × g for 30 min. The supernatant was applied to equilibrated Sepharose-coupled rho-1D4 antibody (University of British Columbia) resin (1D4 resin). The immobilization was performed overnight at 4 °C. The resin was washed four times with 10 column volumes (CVs) of running buffer (25 mM Hepes, pH 7.4, 150 mM NaCl, 20% glycerol, 0.01%/0.01%/0.004% DDM/C12E8/CHS) followed by elution with washing buffer supplemented with 0.5 mg/mL 1D4 peptide (GenScript) for a minimum of 5 h at 4 °C. Elution was collected, concentrated with Amicon Ultra centrifugal filters, and passed through desalting PD-10 column (GE Healthcare) equilibrated with running buffer 2 (25 mM Hepes, pH 7.4, 150 mM NaCl, 10% glycerol, 0.4%/0.1%/0.1% DDM/C12E8/CHS) to decrease concentration of glycerol prior to the nanodisc reconstitution. The final sample was measured at absorbance of 280 nm, using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) to determine concentration, and validated by SDS/PAGE gel.
Cytotoxicity Assay.
The wild-type ABCB1 stable cell line was grown and maintained in adherent cultures in Dulbecco's Modified Eagle Medium (DMEM, Thermo Fisher Scientific) supplemented with 10% FBS and 100 units/mL/100 µg/mL penicillin/streptomycin at 37 °C under humidified conditions with 5% CO2. For induction, the media were exchanged into DMEM supplemented with 2% FBS and 100 units/mL/100 µg/mL penicillin/streptomycin prior to addition of tetracycline to a final 0.5 µg/mL. The induction was done for 15 h under the same conditions. Induced and uninduced cells were plated at a density of ∼5,000 cells per well on a 96-well plate. After 2 h, when cells attached, the medium was exchanged with a fresh one containing either vinblastine sulfate (Fluka) or R(+)-verapamil monohydrochloride hydrate (Sigma) at various concentrations. Cells in the media containing vinblastine were incubated for another 24 to 48 h before cell viability was measured using Cell Proliferation Reagent WST-1 (Roche). Cells in the media containing verapamil were incubated for 3 h and vinblastine at three different concentrations (0.1 μM, 0.5 μM, and 1.0 μM) was directly added to the media, followed by 24 to 48 h incubation and viability measurement. The experiments were performed in triplicates. Data were calculated by subtracting Bmin values and fit to a sigmoidal dose–response curve in GraphPad Prism 9 and normalized to a Bmax-Bmin value. Data from the second experiment were normalized to a Bmax-Bmin value calculated from the plots at the lowest and the highest vinblastine concentrations, respectively.
Nanodisc Reconstitution.
A mixture of Brain Polar Extract lipids (Avanti Polar) and cholesterol (Avanti Polar) (4:1 wt/wt) was solubilized in 1.5%/0.3% DDM/CHS followed by mixing with the protein. Next, membrane scaffold protein MSP1D1 was added to the mixture. The stoichiometry of the mixture was kept at a molar ratio of 1:10:350 (protein to MSP to lipids). For the preparation of the protein sample for cryo-EM (in a complex with MRK16 Fab), the mixture was supplemented with MRK16 Fab in a molar ratio of 1:3 ABCB1:MRK16; for the preparation of the protein sample for functional experiments that step was skipped. The mixture was diluted with HBS (25 mM Hepes, pH 7.4, 150 mM NaCl) to reach the final glycerol concentration of 4%, followed by a 25-min incubation at room temperature. Finally, prewashed Bio-Beads SM-2 (Bio-Rad) were added at 0.8 g/mL concentration and incubated for 2 h at 4 °C. The eluted mixture was transferred to 1D4 resin equilibrated with HBS for another immobilization and to ensure separation from empty nanodiscs. The mixture was incubated overnight at 4 °C. Resin was washed four times with 10CV of HBS and elution was done by incubation with HBS supplemented with 3C protease (1:10 wt/wt protease:protein) at 4 °C for 2 h. The elution was concentrated and the final purification step was performed by size exclusion chromatography (SEC) with HBS as the running buffer.
Liposome Reconstitution.
Brain Polar Extract lipids and cholesterol (Avanti Polar) mixture (4:1 wt/wt) were used for proteoliposome preparation as previously described (51). Briefly, the mixture was extruded 11 times through a 1.0-μm polycarbonate filter (Whatman) and destabilized with 0.17% (vol/vol) Triton X-100. Detergent-solubilized ABCB1 was mixed with destabilized lipids at a 100:1 (wt/wt) lipid:protein ratio, incubated, and reconstituted by adding prewashed Bio-Beads SM-2 (Bio-Rad). Proteoliposomes were spun and resuspended in HBS to a final lipid concentration of 10 mg/mL. The reconstitution efficiency was determined by the Schaffner and Weissmann method (52).
Antibody Expression, Purification, and Fragmentation.
Human-specific MRK16 antibodies were expressed from hybridoma cells, cultured in Wheaton CeLLine Bioreactors following the producer’s recommendation, similar to the previously published UIC2 antibody method (25), followed by Protein G (GenScript) affinity purification and fragmentation using Fab Preparation Kit protocol (Thermo Fisher). The purified Fab fragment was validated by SEC, SDS/PAGE (both in native and reducing conditions), and by binding to the ABCB1 protein.
ATPase Activity Assay.
ATPase activity assays of nanodisc- and liposome-reconstituted ABCB1 were performed following a protocol that relies on detecting how much inorganic phosphate is released (53). Reconstituted ABCB1 sample was used at a concentration in the range of 0.01 to 0.05 mg/mL of protein (nanodiscs) or 2.5 mg/mL of lipids (proteoliposomes). MRK16-Fab was used in molar excess of at least threefold to ensure saturation. Vincristine sulfate (Fluka), vinblastine sulfate (Fluka), taxol (Sigma), elacridar hydrochloride (MedKoo), tariquidar (MedKoo), zosuquidar hydrochloride (Sigma), and R(+)-verapamil monohydrochloride hydrate (Sigma) were prepared as stock solutions in dimethyl sulfoxide (DMSO) at a concentration in the range of 10 to 50 mM, protected from oxidation and diluted accordingly. To analyze the stimulatory or inhibitory effects of each drug, we performed ATPase assays at varying concentrations of the respective drug in the presence of 2 mM ATP and 10 mM MgCl2 at 37 °C. To analyze the change in affinity for ATP, we performed ATPase assays in the absence and in the presence of elacridar, tariquidar, zosuquidar, and with or without Fab, at varying ATP concentrations (0 to 4 mM), 10 mM MgCl2 at 37 °C. The selected inhibitors were used at a saturating amount (in excess of the respective IC50 values), i.e., 10 µM each. ATPase rates were determined using linear regression in GraphPad Prism 8, and normalized to the basal ATPase rate. The means are represented along with the SEs of the three technical replicates. Apparent Km and Vmax were determined from the fit to the Michaelis–Menten model of the corresponding ATPase rates, which for comparison were normalized according to the basal Vmax value.
EM Sample Preparation.
Nanodisc-reconstituted ABCB1–MRK16-Fab complex was purified and concentrated up to 0.3 to 0.5 mg/mL. For the drug-bound sample, a compound solution in DMSO was added to a final concentration of 100 µM (vincristine), 20 µM (elacridar or tariquidar), or 10 µM (zosuquidar), but without exceeding the final DMSO concentration of 1%. In the drug-free state, a nanobody specific to NBD was added to aid with particle alignment, but due to the flexibility of the NBDs, the density of the nanobody was not resolved, and thus not built in the final model. Samples were applied to glow discharged Quantifoil R1.2/1.3 carbon/copper grids and plunge frozen in liquid ethane using Vitrobot Mark IV (FEI) at 4 °C and 100% humidity.
EM Data Acquisition and Processing.
Drug-bound samples were imaged on a Titan Krios 300-kV microscope (Thermo Fisher Scientific) equipped with a Gatan K3 camera and a Gatan BioQuantum 1967 energy filter and the drug-free state sample was imaged on a Titan Krios 300-kV microscope (Thermo Fisher Scientific) equipped with a Gatan K2-Summit camera and a Gatan Quantum-LS (GIF) energy filter. Automated data acquisition was performed with SerialEM (54) for the vincristine-bound state and with EPU (Thermo Fisher Scientific) for all other samples. Data were collected in superresolution mode at the nominal magnification of 130,000 (vincristine bound, elacridar bound, and tariquidar bound) and 165,000 (drug-free state, zosuquidar bound). Image stacks contained 40 frames with an average dose of 1.6 electrons/Å2 per frame. The defocus was set in the range of −0.6 µm to −2.4 µm with a 0.2-µm step.
The data processing pipelines are presented in SI Appendix, Figs. S2–S6 and Table S1 for each structure. We describe in detail the EM data processing for one structure, since the same pattern applies to all datasets. For vincristine-bound ABCB1–MRK16-Fab data were collected in two sessions yielding 7,701 and 5,791 multiframe micrographs that were imported into Relion 3 (55), motion corrected, dose weighted, and binned by 2 to a pixel size of 0.69 Å/pix (56) (SI Appendix, Fig. S3). The contrast transfer function (CTF) was estimated with gctf (57). From a total of 12,469 micrographs, 2,823,122 particles were autopicked based on a previously generated three-dimensional (3D) template, followed by extraction with threefold binned pixel size of 2.07 Å/pix. After two-dimensional (2D) classification, 1,467,190 particles were selected for 3D classification. The best classes were selected and at this step, particles were combined from the two data collections yielding 811,631 particles. Particles were reextracted (unbinned to 0.69 Å/pix) and 3D refined, yielding a map at 4.1 Å. Another 3D classification was performed, without alignment and with a mask without the nanodisc and NBD regions. Particles from the best class (the highest resolution, the best class with visible TM4 and TM10, 160,170 particles) were 3D refined, followed by CTF refinement and Bayesian polishing. The final 3D refinement, followed by postprocessing (B-factor sharpening) was performed with applying sequential, soft masks: a full mask (yielding a map 3.4 Å), a mask without nanodisc density (3.3 Å), a mask without nanodisc and NBDs (3.2 Å), and a mask without nanodisc and constant domain of the Fab resulting in a final map at 3.20-Å resolution. The final calibrated pixel size was experimentally defined as 0.66 Å/pix and the B-factor adjusted to −75 Å2. Local resolution was calculated in Relion 3.1. The data processing for the apo state ABCB1–MRK16-Fab complex is shown in SI Appendix, Fig. S2, elacridar-bound ABCB1–MRK16-Fab complex in SI Appendix, Fig. S4, tariquidar-bound ABCB1–MRK16-Fab complex in SI Appendix, Fig. S5, and zosuquidar-bound ABCB1–MRK16-Fab complex in SI Appendix, Fig. S6.
Model Building and Refinement.
The final postprocessed EM maps were used for model building in Coot (58). All maps showed good resolvability in the transmembrane region of the transporter and variable domain of the antibody that allowed the building of these regions de novo based on the protein sequence from already available structures: taxol-bound ABCB1 (PDB: 6qex) and crystal structure of MRK16–Fab-fragment (PDB: 1bln). The NBD regions and the constant domain of the Fab region were modeled by rigid-body fitting of these domains from the same PDB structures. Vincristine, elacridar, and tariquidar molecules, and their geometry, were generated from SMILE codes using eLBOW (59). The zosuquidar atom coordinates and their geometry were copied from the previous model of zosuquidar-bound human–mouse chimeric ABCB1–UIC2-Fab (PDB: 6fn1). The vincristine molecule was built in similar conformation to the vinblastine molecule bound to tubulin (60). Coordinate refinement was performed in Phenix (61) and the model validations, using MolProbity (62), are presented in SI Appendix, Table S1.
Figure Preparation.
Calculations and graph preparation were performed in GraphPad Prism 9 (GraphPad software). Images of the EM maps were prepared in UCSF Chimera (63) and images of the models were prepared in PyMOL (the PyMOL Molecular Graphics System, Version 2.3 Schrödinger). The ligand schematic view was prepared using LigPlot+ (64).
Supplementary Material
Acknowledgments
We thank Miroslav Peterek of the Scientific Center for Optical and Electron Microscopy (ScopeM) at ETH Zürich for technical support. We also acknowledge Nina Tremp for help with antibody expression and purification. This research was supported by grants from the Swiss Cancer League (to K.P.L.), the Swiss National Science Foundation (SNF) through National Centers of Competence in Research Structural Biology and TransCure, as well as SNF grant 310030_189111 to K.P.L. Further support was provided by the Japan Society for the Promotion of Science, Ministry of Education, Culture, Sports, Science and Technology (SPS-MEXT, grant JP17H06327 to N.F.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010264117/-/DCSupplemental.
Data Availability.
Data associated with this research have been deposited in the Electron Microscopy Data Bank (EMDB) (EMD-11666, EMD-11667, EMD-11670, EMD-11671, EMD-11672). Coordinates for deposited models are available at the Protein Data Bank with IDs: 7A65 (drug-free ABCB1–MRK16-Fab), 7A69 (vincristine-bound ABCB1–MRK16-Fab), 7A6C (elacridar-bound ABCB1–MRK16-Fab), 7A6E (tariquidar-bound ABCB1–MRK16-Fab), and 7A6F (zosuquidar-bound ABCB1–MRK16-Fab).
References
- 1.Fromm M. F., Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol. Sci. 25, 423–429 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Wishart D. S. et al., Drugbank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giacomini K. M. et al.; International Transporter Consortium , Membrane transporters in drug development. Nat. Rev. Drug Discov. 9, 215–236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robey R. W. et al., Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 18, 452–464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martino E. et al., Vinca alkaloids and analogues as anti-cancer agents: Looking back, peering ahead. Bioorg. Med. Chem. Lett. 28, 2816–2826 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Zhou X., Xu Z., Li A., Zhang Z., Xu S., Double-sides sticking mechanism of vinblastine interacting with α,β-tubulin to get activity against cancer cells. J. Biomol. Struct. Dyn. 37, 4080–4091 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Wang R. C. et al., Sensitivity of docetaxel-resistant MCF-7 breast cancer cells to microtubule-destabilizing agents including vinca alkaloids and colchicine-site binding agents. PLoS One 12, e0182400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowley E., McDevitt C. A., Callaghan R., . “Generating inhibitors of P-Glycoprotein: Where to, now?” in Methods in Molecular Biology, Zhou J., Ed. (Humana Press, 2010), pp. 405–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robert J., Jarry C., Multidrug resistance reversal agents. J. Med. Chem. 46, 4805–4817 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Hyafil F., Vergely C., Du Vignaud P., Grand-Perret T., In vitro and in vivo reversal of multidrug resistance by GF120918, an acridonecarboxamide derivative. Cancer Res. 53, 4595–4602 (1993). [PubMed] [Google Scholar]
- 11.Roe M. et al., Reversal of P-glycoprotein mediated multidrug resistance by novel anthranilamide derivatives. Bioorg. Med. Chem. Lett. 9, 595–600 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Cripe L. D. et al., Zosuquidar, a novel modulator of P-glycoprotein, does not improve the outcome of older patients with newly diagnosed acute myeloid leukemia: A randomized, placebo-controlled trial of the eastern cooperative oncology group 3999. Blood 116, 4077–4085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bankstahl J. P. et al., Tariquidar and elacridar are dose-dependently transported by P-glycoprotein and bcrp at the blood-brain barrier: A small-animal positron emission tomography and in vitro study. Drug Metab. Dispos. 41, 754–762 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Mechetner E. B., Roninson I. B., Efficient inhibition of P-glycoprotein-mediated multidrug resistance with a monoclonal antibody. Proc. Natl. Acad. Sci. U.S.A. 89, 5824–5828 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamada H., Tsuruo T., Functional role for the 170- to 180-kDa glycoprotein specific to drug-resistant tumor cells as revealed by monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A. 83, 7785–7789 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie T. K., Kwon H., Atkins W. M., Conformational analysis of human ATP-binding cassette transporter ABCB1 in lipid nanodiscs and inhibition by the antibodies MRK16 and UIC2. J. Biol. Chem. 286, 39489–39496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuruo T., Hamada H., Sato S., Heike Y., Inhibition of multidrug-resistant human tumor growth in athymic mice by anti-P-glycoprotein monoclonal antibodies. Jpn. J. Cancer Res. 80, 627–631 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aller S. G. et al., Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323, 1718–1722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin M. S., Oldham M. L., Zhang Q., Chen J., Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kodan A. et al., Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog. Proc. Natl. Acad. Sci. U.S.A. 111, 4049–4054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alam A., Kowal J., Broude E., Roninson I., Locher K. P., Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y., Chen J., Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 359, 915–919 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 24.van Wonderen J. H. et al., The central cavity of ABCB1 undergoes alternating access during ATP hydrolysis. FEBS J. 281, 2190–2201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alam A. et al., Structure of a zosuquidar and UIC2-bound human-mouse chimeric ABCB1. Proc. Natl. Acad. Sci. U.S.A. 115, E1973–E1982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szewczyk P. et al., Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein. Acta Crystallogr. D Biol. Crystallogr. 71, 732–741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen H., Lee F. Y., Gan J., Ixabepilone, a novel microtubule-targeting agent for breast cancer, is a substrate for P-glycoprotein (P-gp/MDR1/ABCB1) but not breast cancer resistance protein (BCRP/ABCG2). J. Pharmacol. Exp. Ther. 337, 423–432 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Nandigama K., Lusvarghi S., Shukla S., Ambudkar S. V., Large-scale purification of functional human P-glycoprotein (ABCB1). Protein Expr. Purif. 159, 60–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vahedi S., Chufan E. E., Ambudkar S. V., Global alteration of the drug-binding pocket of human P-glycoprotein (ABCB1) by substitution of fifteen conserved residues reveals a negative correlation between substrate size and transport efficiency. Biochem. Pharmacol. 143, 53–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lusvarghi S., Ambudkar S. V., ATP-dependent thermostabilization of human P-glycoprotein (ABCB1) is blocked by modulators. Biochem. J. 476, 3737–3750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loo T. W., Clarke D. M., Identification of residues in the drug-binding domain of human P-glycoprotein. Analysis of transmembrane segment 11 by cysteine-scanning mutagenesis and inhibition by dibromobimane. J. Biol. Chem. 274, 35388–35392 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Pollock N. L. et al., Improving the stability and function of purified ABCB1 and ABCA4: The influence of membrane lipids. Biochim. Biophys. Acta 1838, 134–147 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Labrie P. et al., A comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA) of anthranilamide derivatives that are multidrug resistance modulators. J. Med. Chem. 49, 7646–7660 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Jackson S. M. et al., Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat. Struct. Mol. Biol. 25, 333–340 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Noyes W. A., Forman D. B., Aldehyde—amide condensation. I. Reactions between aldehydes and acetamide. J. Am. Chem. Soc. 55, 3493–3494 (1933). [Google Scholar]
- 36.Sauna Z. E., Ambudkar S. V., Characterization of the catalytic cycle of ATP hydrolysis by human P-glycoprotein. The two ATP hydrolysis events in a single catalytic cycle are kinetically similar but affect different functional outcomes. J. Biol. Chem. 276, 11653–11661 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Vasudevan S., Tsuruo T., Rose D. R., Mode of binding of anti-P-glycoprotein antibody MRK-16 to its antigen. A crystallographic and molecular modeling study. J. Biol. Chem. 273, 25413–25419 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Vahedi S. et al., Mapping discontinuous epitopes for MRK-16, UIC2 and 4E3 antibodies to extracellular loops 1 and 4 of human P-glycoprotein. Sci. Rep. 8, 12716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawson R. J. P., Locher K. P., Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Sajid A., Lusvarghi S., Chufan E. E., Ambudkar S. V., Evidence for the critical role of transmembrane helices 1 and 7 in substrate transport by human P-glycoprotein (ABCB1). PLoS One 13, e0204693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chufan E. E. et al., Multiple transport-active binding sites are available for a single substrate on human P-glycoprotein (ABCB1). PLoS One 8, e82463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callaghan R., Providing a molecular mechanism for P-glycoprotein; why would I bother? Biochem. Soc. Trans. 43, 995–1002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raviv Y., Pollard H. B., Bruggemann E. P., Pastan I., Gottesman M. M., Photosensitized labeling of a functional multidrug transporter in living drug-resistant tumor cells. J. Biol. Chem. 265, 3975–3980 (1990). [PubMed] [Google Scholar]
- 44.Gottesman M. M., Ambudkar S. V., Xia D., Structure of a multidrug transporter. Nat. Biotechnol. 27, 546–547 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharom F. J., Complex interplay between the P-glycoprotein multidrug efflux pump and the membrane: Its role in modulating protein function. Front. Oncol. 4, 41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dastvan R., Mishra S., Peskova Y. B., Nakamoto R. K., Mchaourab H. S., Mechanism of allosteric modulation of P-glycoprotein by transport substrates and inhibitors. Science 364, 689–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korkhov V. M., Mireku S. A., Locher K. P., Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature 490, 367–372 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Manolaridis I. et al., Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature 563, 426–430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crowley E., O’Mara M. L., Kerr I. D., Callaghan R., Transmembrane helix 12 plays a pivotal role in coupling energy provision and drug binding in ABCB1. FEBS J. 277, 3974–3985 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Crowley E. et al., Transmembrane helix 12 modulates progression of the ATP catalytic cycle in ABCB1. Biochemistry 48, 6249–6258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geertsma E. R., Nik Mahmood N. A. B., Schuurman-Wolters G. K., Poolman B., Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256–266 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Schaffner W., Weissmann C., A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56, 502–514 (1973). [DOI] [PubMed] [Google Scholar]
- 53.Chifflet S., Torriglia A., Chiesa R., Tolosa S., A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: Application to lens ATPases. Anal. Biochem. 168, 1–4 (1988). [DOI] [PubMed] [Google Scholar]
- 54.Schorb M., Haberbosch I., Hagen W. J. H., Schwab Y., Mastronarde D. N., Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zivanov J. et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, 1–22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng S. Q. et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moriarty N. W., Grosse-Kunstleve R. W., Adams P. D., Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waight A. B. et al., Structural basis of microtubule destabilization by potent auristatin anti-mitotics. PLoS One 11, e0160890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams P. D. et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams C. J. et al., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen E. F. et al., UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Laskowski R. A., Swindells M. B., LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this research have been deposited in the Electron Microscopy Data Bank (EMDB) (EMD-11666, EMD-11667, EMD-11670, EMD-11671, EMD-11672). Coordinates for deposited models are available at the Protein Data Bank with IDs: 7A65 (drug-free ABCB1–MRK16-Fab), 7A69 (vincristine-bound ABCB1–MRK16-Fab), 7A6C (elacridar-bound ABCB1–MRK16-Fab), 7A6E (tariquidar-bound ABCB1–MRK16-Fab), and 7A6F (zosuquidar-bound ABCB1–MRK16-Fab).