Abstract
Background
Prioritization of breast cancer patients based on the risk of resistance to tamoxifen plays a significant role in personalized therapeutic planning and improving disease course and outcomes.
Methods
In this work, we demonstrate that a genome-wide pathway-centric computational framework elucidates molecular pathways as markers of tamoxifen resistance in ER+ breast cancer patients. In particular, we associated activity levels of molecular pathways with a wide spectrum of response to tamoxifen, which defined markers of tamoxifen resistance in patients with ER+ breast cancer.
Findings
We identified five biological pathways as markers of tamoxifen failure and demonstrated their ability to predict the risk of tamoxifen resistance in two independent patient cohorts (Test cohort1: log-rank p-value = 0.02, adjusted HR = 3.11; Test cohort2: log-rank p-value = 0.01, adjusted HR = 4.24). We have shown that these pathways are not markers of aggressiveness and outperform known markers of tamoxifen response. Furthermore, for adoption into clinic, we derived a list of pathway read-out genes and their associated scoring system, which assigns a risk of tamoxifen resistance for new incoming patients.
Interpretation
We propose that the identified pathways and their read-out genes can be utilized to prioritize patients who would benefit from tamoxifen treatment and patients at risk of tamoxifen resistance that should be offered alternative regimens.
Funding
This work was supported by the Rutgers SHP Dean's research grant, Rutgers start-up funds, Libyan Ministry of Higher Education and Scientific Research, and Katrina Kehlet Graduate Award from The NJ Chapter of the Healthcare Information Management Systems Society.
Keywords: Therapeutic resistance, Pathway markers, Tamoxifen, Breast cancer
Research in context.
Evidence before this study
Treatment resistance plays a central role in disease management and outcomes, especially for patients with oncologic malignancies. Several groups have studied response to tamoxifen in ER+ breast cancer, yet the identification of markers that accurately predict tamoxifen resistance remains limited.
Added value of this study
In this work, we derived a computational framework to predict treatment resistance to tamoxifen in ER+ breast cancer patients based on behavior of their molecular pathways, defined from changes in mRNA expression profiles. Our analysis identified five molecular pathways and their corresponding read-out genes that successfully predict risk to fail tamoxifen, as validated in two independent patient cohorts. These pathways have not been previously reported as associated with risk of tamoxifen resistance and provide an accurate estimate of response to tamoxifen, outperforming previously known resistance signatures.
Implications of all the available evidence
Our study derived a marker panel to identify patients at risk of primary tamoxifen failure. Together with other studies, our work builds a foundation for personalized therapeutic planning for patients with ER+ breast cancer patients.
Alt-text: Unlabelled box
1. Introduction
Despite recent advances in diagnosis, classification, and therapeutic management, breast cancer (BC) remains one of the leading causes of cancer-related death in women worldwide [[1], [2], [3]]. Nearly 70% of all diagnosed cases of breast tumors are estrogen receptor-positive (ER+) [4,5], making treatments that have anti-estrogen effects in the breast cells, such as tamoxifen, the standard-of-care for patients with ER+ breast cancers [4,[6], [7], [8], [9]]. Despite the significant success of tamoxifen administration, nearly 30% of treated patients develop therapeutic resistance, ultimately leading to metastasis and lethality [1,10]. Therefore, prioritization of patients based on the risk of resistance to tamoxifen before treatment administration could play a significant role in personalize therapeutic planning for patients with ER+ breast cancer and builds a foundation to improve disease course and outcomes.
Tamoxifen is a selective estrogen receptor modulator (SERM) and has agonist or antagonist activity depending on the tissue type [11]. In the breast cells, tamoxifen directly binds to the ER, blocking estrogen from attaching to the receptor and thus inhibiting the activity of estrogen-regulated genes and causing the repression of estrogenic effects [4,5,12,13]. However, the emergence of alternative mechanisms of estrogenic stimulation has been shown to cause resistance to tamoxifen. For example, some studies have demonstrated that ER+ breast cancers that overexpress HER2 and EGFR can activate the components of downstream signaling pathways which then stimulate both ER and estrogen receptor co-activator AIB1, and thus induce the estrogen agonistic activity of tamoxifen in breast cancer cells [14,15]. Another study noticed that the increased expression of HER2 signaling can also downregulate progesterone receptor (PR) levels in the ER+ breast tumors, where losing the PR expression serves as a biomarker of hyperactive growth factor signaling, leading to another possible mechanism of tamoxifen resistance [16]. Despite the emerging role of HER2 in tamoxifen resistance, it only accounts for 10% of ER+ breast cancers [12,17], suggesting more complex resistance mechanisms in these cases and presenting a central clinical problem for patients with ER+ breast cancer [4,5,10,12].
In recent years, several groups have developed gene expression signatures of tamoxifen response for ER+ patients, including 10 gene-signature by Men et al. [18], 21 gene-signature by Paik et al. [19] (known as Oncotype DX), and 2 gene-signature by Ma et al. [20] While these signatures provide substantial advances to our understanding of individual genes involved in resistance, they do not yet capture the complex interplay between biological mechanisms that governs tamoxifen resistance. Here, we propose a pathway-centric computational framework to elucidate tamoxifen resistance and demonstrate that it outperforms known gene-based approaches. Advantages of our pathway-based approach lies in (i) its ability to identify a tightly connected cooperative group of genes unified by the same function [[21], [22], [23]]; (ii) studying molecular pathways, rather than individual genes, produces more reliable read-out outputs as they are less susceptible to experimental noise [24]; (iii) pathway-level view enhances our understanding of the biological mechanisms related to disease and treatment response [[25], [26], [27], [28]]; and finally (iv) looking at alterations in biological pathways enhances the likelihood of identifying potential therapeutic targets to preclude or overcome resistance.
In this work, we have established a systematic pathway-centric computational framework to elucidate molecular pathways as markers of tamoxifen resistance in ER+ breast cancer patients, which we call pathER. Through the analysis of pathway activity in each ER+ patient and their association with response to tamoxifen (Training cohort, n = 53), we identified five biological pathways as markers of tamoxifen resistance: Retrograde Neurotrophin Signalling, Loss of NLP from Mitotic Centrosomes, RNA Polymerase III Transcription Initiation from Type 2 Promoter, EIF2 pathway, and Valine, Leucine and Isoleucine Biosynthesis. We have demonstrated the ability of the identified five candidate pathways to predict the risk of tamoxifen resistance in two independent patient cohorts [29] (Test cohort 1, n = 66: log-rank p-value = 0.02, accuracy of leave one out cross-validation (LOOCV) = 85.8%; Test cohort 2, n = 77: log-rank p-value = 0.01, accuracy of LOOCV = 82.5%) and their independence from known covariates, such as age, tumor grade, tumor size, lymph node status, and PR status, as the absence of PR in ER+ tumor can be an indicator of HER2 activation and an aggressive phenotype [16] (Test cohort 1, adjusted hazard ratio = 3.11; Test cohort 2, adjusted hazard ratio = 4.24). Furthermore, we performed stratified Kaplan-Meier survival analyses on the PR+ and PR- patients as well as patients with Ki-67 low and Ki-67 high status and demonstrated the five candidate pathways can predict risk of resistance to tamoxifen in each PR and Ki-67 group with high accuracy. Importantly, as a negative control, we have demonstrated that the identified five candidate pathways did not classify patients simply based on the disease aggressiveness (log-rank p-value = 0.7, hazard ratio = 1.246) and that in fact pathways associated with disease aggressiveness do not overlap with the five candidate pathways. We have compared our method to other computational techniques to tackle treatment response, including Epsi et al. [28] (which utilized extreme-responder analysis, using tails of the treatment response distribution to define a treatment response signature), Zhong et al. [30] (which used Support Vector Machine approach as a base), Yu et al. [31] (which uses random forest approach as a base), and mRNA data alone (without considering molecular pathways) and demonstrated that our method outperforms these techniques in predicting risk of resistance to tamoxifen. Furthermore, we have compared our pathway signature to other known signatures of tamoxifen response [18], [19], [20] and have shown the superiority of our pathway-based approach (adjusted hazard ratio = 3.11, hazard p-value = 0.0278). Finally, to enhance clinical applicability of our finding, we derived five read-out genes (each of which reflects activity changes in a corresponding pathway) and defined their treatment failure scoring system, indicating risk of developing tamoxifen resistance in two patient cohorts (Test cohort 1, adjusted hazard ratio = 3.1; Test cohort 2, adjusted hazard ratio = 6.95). Thus, we propose that the identified five candidate pathways and their read-out genes can potentially be used to prioritize patients who would benefit from tamoxifen treatment as their first-line therapy, and to identify patients at risk of tamoxifen resistance who should be offered an alternative regimen plan.
2. Materials and Methods
2.1. Patient cohorts utilized for study analysis
All gene expression datasets of patients with ER+ breast cancer were obtained from publicly available GEO data repository [32] from multi-institutional multi-PI comprehensive Loi et al. [29] study GSE6532 (Supplementary Fig. 1, Supplementary Table 1): (i) KIT-GSE6532 utilized as a Training cohort; (ii) GUYT-GSE6532, utilized as Test cohort 1; (iii) OXFT-GSE6532, utilized as a Test cohort 2; and (iv) KIU-GSE6532, utilized as a negative control cohort. Training cohort contains patient profiles of primary ER+ breast tumors (n = 57), archived at the Uppsala University Hospital (Uppsala, Sweden), profiled on Affymetrix Human Genome U133A array and Affymetrix Human Genome U133B array. Test cohort 1 contains patient profiles of primary tumors from patients with ER+ breast cancer (n = 70), archived at the Guy's Hospital (London, United Kingdom), profiled on Affymetrix Human Genome U133 Plus 2.0 Array. Test cohort 2 contains patient profiles of primary ER+ breast tumors (n = 77), archived at the John Radcliffe Hospital (Oxford, United Kingdom), profiled on Affymetrix Human Genome U133A, B array. Negative control cohort consists of not-treated patients with ER+ primary breast tumors (n = 51), profiled on Affymetrix Human Genome U133A, B array. All primary tumors samples in Training and Test cohorts were collected through surgery, diagnosed between 1980 and 1995 and received tamoxifen-only treatment for 5 years post-diagnosis as their adjuvant treatment.
2.2. Data normalization and filtering
For each gene expression microarray dataset, a matrix of RMA (Robust Microarray Analysis) normalized signal intensity values was used [29]. Using the most updated annotation file from GEO and the latest Affymetrix annotation files from Thermo Fisher database [33], each probe set ID was annotated to gene ID; thereafter, probe IDs that annotated to different gene IDs or did not annotate to any gene ID were excluded. When multiple probe set IDs were mapped to the same gene, probes with the highest coefficient of variation (CV) were selected [34, 35].
2.3. Breast cancer molecular subtypes
Gene expression classifier of the breast cancer subtypes (PAM50) was applied to assign breast cancer patients to one of the intrinsic molecular subtypes: luminal A, luminal B, HER2-enriched, triple-negative/basal-like, and normal-like [36,37]. The subtype classification of each patient was determined based on the closeness between the average expression profile of 50 genes in each subtype centroid and the corresponding gene expression pattern of patient tumor, where the distances were measured utilizing Spearman's rank correlation [36]. We utilized genefu package in R, intrinsic.cluster.predict function with pam50 [38] to assign subtype membership and eliminate samples with HER2-enriched, triple-negative/basal-like, and normal-like subtypes (i.e., non ER+).
2.4. Single-sample gene set enrichment analysis (ssGSEA)
To estimate pathway enrichment in each ER+ patient, we performed single-sample (i.e., single-patient) pathway enrichment analysis, where standardized gene expression profile for each patient was used as reference and genes from each biological pathway were used as a query in an unweighted (i.e., each gene had the same weight) single-sample gene set enrichment analysis (ssGSEA) [39,40]. For such analysis gene expression values for each gene were transformed into standardized scores (i.e., z-scores) in order to bring the expression level into a common scale across all samples [41,42]. Z-score for each gene in each sample was computed by subtracting the average intensity of this gene across samples from the intensity of this gene in each sample and dividing it by the gene's standard deviation (SD), where mean and standard deviation were estimated for each gene across all samples [41]. In this way, after such z-scoring, each gene's mean is standardized to 0 and standard deviation to 1. Ranked list of z-scores across all genes for a given sample then defines a single-sample (i.e., single-patient) signature, utilized as a reference for pathway enrichment analysis.
To acquire a comprehensive list of pathways, we utilized MSigDB C2 pathway database [43] which includes curated selections of 833 pathways obtained from human gene sets using Reactome [44], KEGG [45], and BioCarta databases. Reactome database is a curated resource that describes fundamental biological processes including cell signaling, metabolism, regulatory, and human diseases with particular focus on signaling and metabolic pathways derived from a wide list of biomedical experiments and literature references [44,[46], [47], [48], [49], [50]]. KEGG database is an integrated resource of genomic, chemical, metabolic, regulatory, signaling, health, disease, drug, and systematic functional data and contains carefully manually curated human pathway maps, which are literature-based [45,47,[50], [51], [52], [53]]. BioCarta collection is considered the major human pathway resources of metabolic and signaling pathways and is based on literature reference annotations [50,54,55].
Each pathway from the C2 collection (i.e., genes from each pathway) was used as a query set for unweighted ssGSEA. The ssGSEA normalized enrichment scores (NESs), and p-values were assessed utilizing 1,000 gene permutations. NES for each of the 833 pathways (i.e., also referred to as pathway activity levels) indicated how much each pathway was enriched/active in each single-sample signature. In particular, the positive NES would indicate a pathways enrichment in the top of the rank-ordered list (i.e., overexpressed part) of the signature (pathway was active) and the negative NES would indicate pathway enrichment in the bottom of the rank ordered list (i.e., underexpressed part) of the signature (pathway was repressed).
2.5. Associating the activity levels of molecular pathways with therapeutic response
The activity levels of each pathway (i.e., NES) were then associated with tamoxifen response, across all patients in a Training cohort, using Cox proportional hazards model [56], adjusted for common covariates, such as age, tumor grade, tumor size, lymph node status, and PR status. For this, we utilized R coxph function from survival package [57]. To establish a robust threshold which should be utilized to select most significantly associated pathways, we evaluated predictive ability of the pathways as a group. For this, we first sorted pathways based on their significance (i.e., p-values) from the Cox proportional hazards analysis, which measured association of pathway activity levels with response to tamoxifen across all samples in the Training cohort. We then started from the most significant pathway (from the Cox analysis) and added the next most significant pathway, one at a time, evaluating their predictive ability as a group. Thus, the evaluated groups of pathways were (i) Pathway 1; (ii) Pathways 1 and 2; (iii) Pathways 1, 2, and 3; etc. until all pathways were utilized. We finally firecorded predicted ability of each group and the cutoff point was determined as the one, where the addition of the next pathway would not benefit an overall predictive ability of the group.
Furthermore, given that many of the 833 pathways exhibit parent-child relationships or are heavily overlapping, to prevent model overfitting we examined the final list of pathways for such relationships and overlaps, and if such situation occurred, we prioritized pathways with higher association with tamoxifen response. For example, from two significantly overlapping pathways, we selected one that has higher hazard ratio of being associated to tamoxifen response, defining a list of final candidate pathways (i.e., five final pathways were selected).
2.6. Clinical validation in independent patient cohorts
For validation studies, the activity levels of the final candidate pathways were used to stratify patients based on the risk of relapse due to treatment resistance in independent Test cohorts. Patient cohorts were subjected to t-distributed Stochastic Neighbor Embedding (t-SNE) clustering [58], using all pairs of high-dimensional (i.e., 5-dimensions in this study) points [59,60] and successfully distinguishing groups of patients that have similar pathway activity levels. Subsequently, k-means clustering [61] was utilized on t-SNE-derived data, as suggested in [59, 60] to obtain two groups of patients with distinct pathway activity patterns, using kmeans function in R [62].
The ability of the activity levels of the final candidate molecular pathways to efficiently distinguish patient clusters was determined through receiver operating characteristics (ROC) analysis [63] on multiple (i.e., multivariable) logistic regression model, where normalized enrichment scores (i.e., NESs) of the final candidate pathways were used as input parameters (i.e., independent/predictor variables) and patient clusters were utilized as a dependent/response variable. ROC curves were evaluated using the area under the curve (AUC) [64], where AUC score of 0.5 indicates a random predictor. The logistic regression analysis was conducted using glm [65] function, and ROC analysis was performed using pROC [66] and ggplot2 packages in R.
Differences in therapeutic response between the patient groups were evaluated through Kaplan-Meier treatment-related survival analysis [67] and Cox proportional hazards model using survival and survminer packages [56] in R. Log-rank p-value was utilized to assess the statistical significance of the Kaplan-Meier survival analysis and Wald p-value and hazard ratio were utilized for multivariable Cox proportional hazards model through survdiff and coxph functions from survival package.
To ensure that predictive ability of the final candidate pathways is non-random, we performed random model (i.e., randomness) analysis. For this, predictive ability of the final (five) candidate pathways was compared to predictive ability of the five pathways selected at random. Such random selection of five pathways was done 1,000 times and log-rank p-value (from Kaplan-Meier survival analysis) was noted for each random run. The nominal p-value was then calculated as the number of times five pathways selected at random reached or outperformed the final five candidate pathways.
To estimate the predictive accuracy of our model and obtain a more accurate indication of how well our finding behaves toward a new incoming patient, we conducted Leave-One-Out Cross-Validation (LOOCV) [68]. In this method, one patient is “excluded/eliminated” and the rest of the patients are utilized for training purposes to the regression model. After that, a removed patient is assumed to be a new incoming patient and is assigned a risk of developing tamoxifen resistance. This process is repeated for each patient within a given dataset. LOOCV was implemented using multiple logistic regression model, where patient clusters membership was used as a response variable and normalized enrichment scores of our candidate pathways were utilized as input parameters. The logistic regression analysis was performed using glm [65] function, and LOOCV analysis was prepared using cv.glm function from boot package in R.
To ensure that the identified pathways were not Training cohort-specific and would not be missed if Training and Test cohorts are switched, we applied our method to the Test cohorts and compared identified pathways using pathway-on-pathway GSEA. Pathways from the Test cohorts ranked by their hazard ratios were used as a reference pathway list and top 100 pathways from the Training cohort ranked by their hazard ratios were used as a query pathway set.
2.7. Comparative analysis to other commonly utilized approaches
To assess the advantages of our approach over other commonly used techniques, we compared its performance to (i) extreme-responder analysis, described in Epsi et al. [28]; (ii) SVM-based method [30]; (iii) PRES random forest-based method [31]; and (iv) expression data alone (without utilizing biological pathways). In each case, we utilized Training cohort for model training and Test cohort 1 for model validation. For methods that require a signature of treatment response, we compared groups of patients with poor and favorable response to tamoxifen in the Training cohort by selecting: patients that experienced events within 1 year of tamoxifen administration (i.e., non-responders, n = 4); and patients that did not experience any relapses for more than 9 years (i.e., responders, n = 4) to define a differential expression signature of tamoxifen response (i.e., through two-sample two-tailed Welch t-test [69] using t.test function in R). For Epsi et al. method, we then subjected the differential expression signature to pathway enrichment analysis, where this signature was used as a reference and groups of genes from each pathway were used as a query gene set, and treated most significant pathways as candidate pathway markers. For SVM and PRES random forest, we subjected the differential expression signature (i.e., based on the proposed significance level) to the model training using Training cohort (here, information about biological pathways was not utilized and pure gene expression was used, as suggested in such methods). The SVM analysis was performed using svm function from e1071 package, and PRES random forest analysis was prepared using train function from caret package in R. For comparison with gene expression alone, the algorithm was applied directly to expression levels of each gene, which were associated with treatment response outcome using adjusted Cox proportional hazards model, in the same way it was applied to biological pathways. Ability of the identified determinants to predict response to tamoxifen was evaluated using Cox proportional hazards model through survival and survminer packages in R.
2.8. Assigning a risk score of treatment failure for each patient
To enhance clinical utility and applicability of our findings, we have defined read-out genes, which serve as representatives for each biological pathway and make good candidates for affordable clinical evaluation. Such read-out genes were defined as those (i) whose expression levels significantly correlated with pathway activity changes, for corresponding pathways; (ii) that were significantly associated with response to tamoxifen.
We then utilized these read out genes (one per pathway) to define a risk to develop resistance to tamoxifen. The risk score was calculated as a weighted sum of the read-out genes expression values, multiplied by their ROC values (which defined their ability to differentiate patients with good and poor response in a Training set) as
where k is a read-out gene, x(k) is expression value of k, and w(k) is a weight (i.e., ROC value) for k. The risk scores were then separated into low/intermediate risk (≤ mean+1SD) and high risk (> mean+1SD) groups, which were further evaluated using Kaplan-Meier survival analysis and Cox proportional hazards model.
2.9. Cancer dependency map by DepMap
To evaluate the association of the read-out genes to tamoxifen sensitivity in human cancer cell lines, we performed cancer dependency map analysis, using DepMap web portal [70], which utilizes PRISM Repurposing [71], CTD2 [72,73], and GDSC databases [74]. Dependency map screens for sensitivity to multiple anti-cancer drugs (including tamoxifen) across various human cancer cell lines. Measures of dose response are obtained using the area under the dose-response curve (AUCs) scores for each drug–cancer cell lines pair where large AUC scores show decreased sensitivity to the drug and small scores show increased sensitivity to the drug. We have utilized mRNA expressions of our identified read-out genes to query this resource, where large AUCs showed poor or no response to tamoxifen and smaller AUCs values showed favorable response.
2.10. Statistical analysis
Statistical analysis was performed using R studio version 3.5.1 for statistical computing. For single-sample (i.e., single-patient) analysis, data were z-scored on individual gene level. For this, the mean and standard deviation were first estimated for each gene across all samples in the dataset. Subsequently, z-score for each gene was defined as the difference between its intensity value and the mean of that gene across the samples and divided by the standard deviation for that gene. The ranked list of z-scores for each gene in a sample then defined single-sample (i.e., single-patient) signature. Pathway activity levels were estimated as Normalized Enrichment Scores (NESs) from the single-sample Gene Set Enrichment Analysis (ssGSEA), where NESs and p-values were estimated using 1,000 gene permutations. Cox proportional hazards model was utilized to associate pathway activity levels with treatment-related relapse-free survival (tRFS). When adjusting for common covariates, multivariable Cox proportional hazards model was utilized and its significance was reported using hazard ratio, hazard p-value, and Wald test. Kaplan-Meier survival analysis was utilized to estimate difference in treatment-related survival between two groups of patients, with log-rank p-value used to indicate significance. All survival analyses were subjected to adjustment for common covariates (e.g., tumor grade, tumor size, lymph node positivity, age, and PR negativity). Patients’ cohorts were obtained from public repositories and all the code was assembles using freely available R packages, as described above, with no restrictions.
2.11. Funding sources
Antonina Mitrofanova has been supported by the Rutgers SHP Dean's research grant and Rutgers start-up funds. Sarra M. Rahem has been supported by the Libyan Ministry of Higher Education and Scientific Research. Nusrat Epsi has been supported by the Katrina Kehlet Graduate Award from The NJ Chapter of the Healthcare Information Management Systems Society (NJHIMSS). The funders had no role in study design, data collection, data analyses, interpretation, or writing of report. The authors have not been paid to write this article by any agency.
3. Results
3.1. Overview
We present a genome-wide pathway-centric computational analysis to identify molecular pathways predictive of risk of resistance to tamoxifen in ER+ breast cancer patients. Our approach has the following steps:
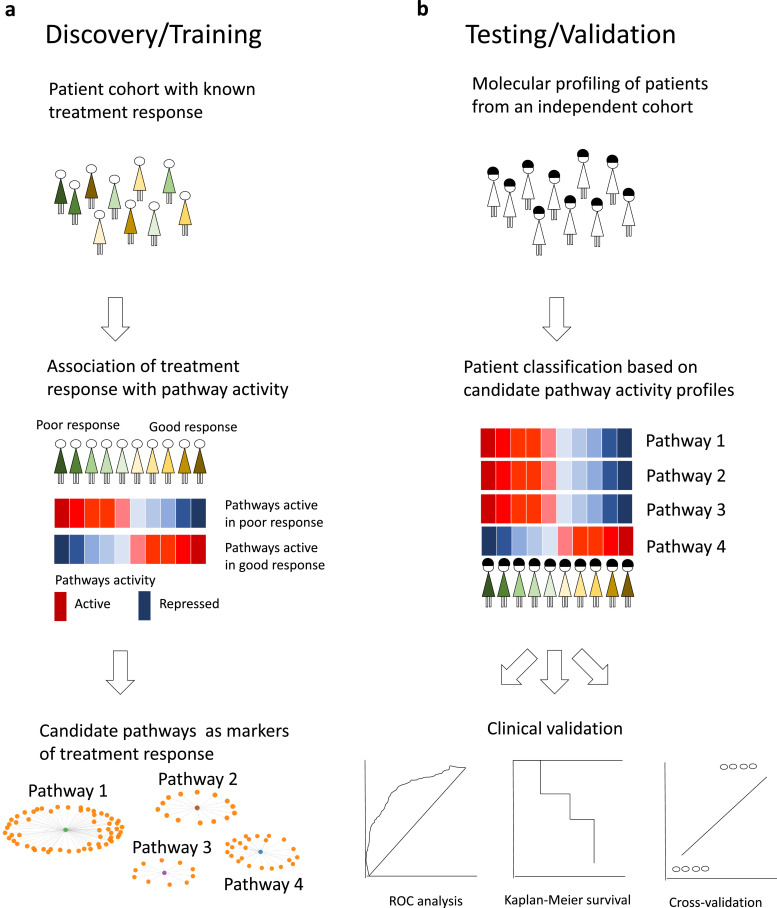
Training phase (Fig. 1a): (i) activity levels of biological pathways are estimated in each ER+ breast cancer patient (across a wide spectrum of responses, present in a clinical setting) that received adjuvant tamoxifen (Supplementary Fig. 1, Supplementary Table 1); (ii) these pathway activity levels are then associated with tamoxifen response across all patients, adjusted for common covariates;
Fig. 1.
Schematic representation of the pathway-centric approach. (a)Training phase: identification of molecular pathways of tamoxifen resistance. (b)Testing phase: clinical validation of identified candidate pathways and multi-modal prediction evaluation.
Testing phase (Fig. 1b): (iii) pathways that are significantly associated with the risk of tamoxifen failure are then subjected to clinical validation analysis in independent patient cohorts (Supplementary Fig. 1, Supplementary Table 1), for their ability to predict tamoxifen resistance for new incoming patients; (iv) finally, ability of the candidate pathways to predict the risk of tamoxifen resistance is compared to known gene signatures of resistance and overall disease aggressiveness, alongside comparison to other methods.
3.2. Training phase: identifying molecular pathways that govern primary tamoxifen resistance
To accurately define therapeutic response to tamoxifen in ER+ breast cancer patients, we carefully selected gene expression profiles for the Training cohort (Loi et al. [29], KIT-GSE6532) of primary ER+ breast tumors collected through surgery, not subjected to any neoadjuvant (i.e., prior to sample collection) treatment, and administered adjuvant (i.e., post-operative) 5-year long tamoxifen administration, with available clinical follow-up data (n = 57) (Supplementary Fig. 1, Supplementary Table 1).
To avoid inconsistencies in BC classification, we subjected patient profiles of the Training cohort to a 50-gene Prediction Analysis of Microarrays panel [36] (PAM50) classification. PAM50 classification categorized BC patients from the Training cohort into the five intrinsic molecular subtypes: luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-enriched, triple-negative/basal-like, and normal-like, known to differ in their clinical outcomes [75,76] and therapy choice [77]. ER+ BC, which is the phenotype of interest in our study, is contained within the luminal A and luminal B subtypes and is excluded from HER2-enriched, triple-negative/basal-like, and normal-like subtypes (Supplementary Table 1). Out of 57 post-operative tamoxifen-treated patients, 4 patients were classified as HER2-enriched, basal-like, or normal-like, and thus were excluded from further analysis.
Our objective was to evaluate tamoxifen response across all 53 patient samples (on the individual-patient level) and associate them with changes in biological pathway activities (Fig. 2a). In order to be able to evaluate each patient sample individually, we scaled (i.e., z-scored, see Materials and Methods) gene expression profiles on individual gene levels so that each gene had mean 0 and standard deviation 1 over all samples in the Training cohort [41]. The list of genes ranked by their z-scores in each sample then defined an individual-patient signature. We then utilized each individual-patient signature to evaluate activity levels of biological pathways using unweighted single-sample Gene Set Enrichment Analysis (ssGSEA) [39,40], where pathways were obtained from widely utilized MSigDB C2 pathway collection (which includes 833 pathways from Reactome [44], KEGG [45], and BioCarta databases) (Supplementary Dataset 1a-1c). For this analysis, each patient signature was used as a reference and each pathway as a query gene set. Activity levels of biological pathways were defined by their enrichment in each patient signature, mathematically represented by the Normalized Enrichment Scores (NES) from the GSEA analysis, where positive NES corresponds to enrichment in the over-expressed part of the signature and negative NES corresponds to enrichment in the under-expressed part of the signature (Fig. 1a, Fig. 2a).
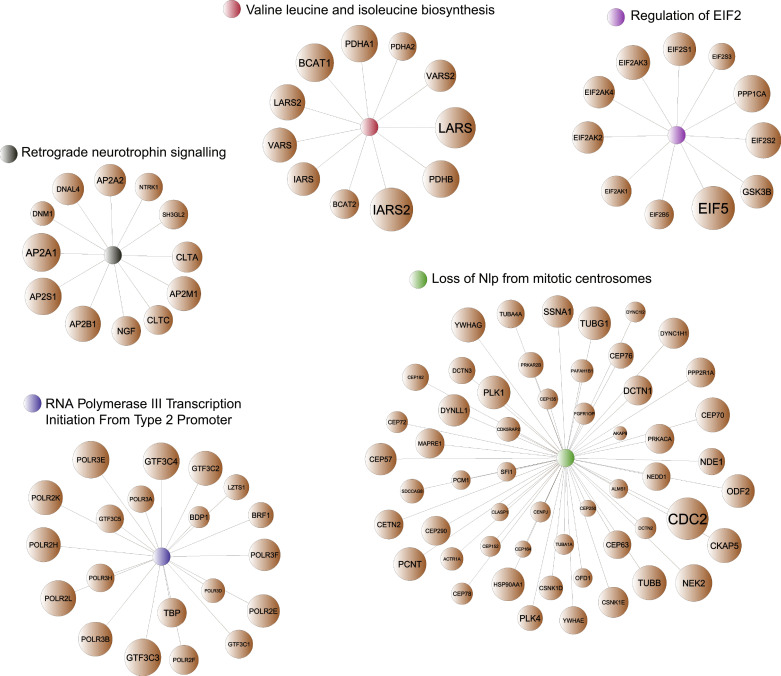
Fig. 3.
Graphical representation of the five candidate pathways and their significantly contributing genes. Network-based representation of the five candidate pathways. Selected genes shown (brown nodes) correspond to genes that contribute to significant enrichment of each pathway in the patient single-sample signatures. Node sizes represent number of times each gene appears in the leading edge in the single-sample pathway enrichment analysis (i.e., indicating significant changes in activity of this pathway across Training cohort).
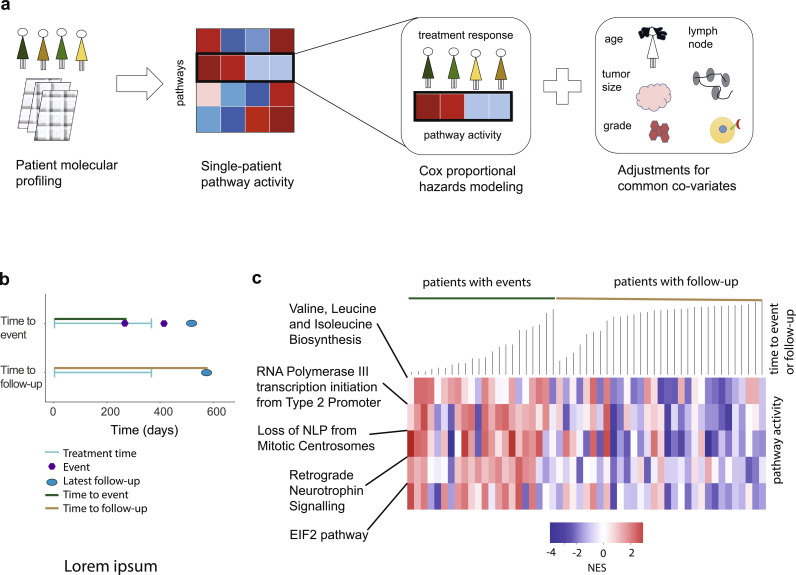
Fig. 2.
Training phase: pathway-centric approach identifies five biological pathways that govern tamoxifen response. (a) Schematic representation of the Testing phase of our approach: (left) patient molecular profiles are collected and analyzed; (middle) pathway activities are estimated in each patient using single-patient pathway enrichment analysis; (right) pathway activities are associated with response to tamoxifen using Cox proportional hazards modeling and are adjusted to common covariates, including age, tumor grade, tumor size (> 2 cm vs ≤ 2 cm), lymph node status, and PR status. (b) Graphical illustration of tamoxifen-related treatment response or follow-up. Time to event (top): time interval between tamoxifen administration and earliest relapse is indicated by green line. Time to follow-up (bottom): time interval between tamoxifen administration and latest follow-up date is indicated by brown line (no tamoxifen-related events observed). (c) Heatmap representation of the pathway activity levels (i.e., NES) and their association with time to tamoxifen-related relapse or follow-up, in the Training cohort. Green line marks the group of patients with tamoxifen-related relapse, sorted from the shortest to the longest time to relapse. Brown line marks the group of patients with follow-up and without disease relapse until the latest follow-up, sorted from the shortest to longest time to follow-up.
Next essential step in our analysis was to associate changes in pathway activity levels to tamoxifen treatment response. In general, we defined treatment-related relapse free survival (tRFS) as the interval between tamoxifen administration (which occurred immediately after surgery) and the earliest relapse (defined as local, regional, or distant metastasis) or the latest follow-up (these patients did not develop an event until their latest follow-up). When a patient had a relapse during or after the therapy administration, time to therapy related relapse was defined from therapy start to the earliest relapse (Fig. 2b, top schematics, green line). When a patient never experienced a relapse, therapy-related relapse-free survival was measured from therapy start to the latest follow-up (Fig. 2b, bottom schematics, brown line). In this dataset, 41.5% of patients experienced tamoxifen-related events (i.e., relapse), making it ideally suited for Training purposes.
To estimate association between the activity levels of the biological pathways and tRFS across a wide spectrum of tamoxifen response (taking into account a heterogeneity of response to tamoxifen, present in a clinical setting), we utilized Cox proportional hazards model [56], ideally suited when time to event or follow-up is available. The Cox proportional hazards model was estimated between each pathway activity level (i.e., NESs, independent/predictor variable) and tamoxifen tRFS (i.e., dependent/response variable) across all 53 patients in the Training cohort (Supplementary Dataset 1c). Furthermore, to account for the effect of other factors, this analysis was adjusted for commonly utilized covariates, as suggested in [78], such as age, tumor grade, tumor size (> 2 cm vs ≤ 2 cm), lymph node status, and PR status (note that decreased PR levels are associated with increased HER2 signaling [16]) (Fig. 2a). Such analysis identified five molecular pathways (Fig. 2c, Supplementary Table 2), most significantly associated with response to tamoxifen (hazard p-value ≤ 0.00075, Supplementary Fig. 2a-b, see Materials and Methods), including Retrograde Neurotrophin Signalling, Loss of NLP from Mitotic Centrosomes, RNA Polymerase III Transcription Initiation from Type 2 Promoter, EIF2 pathway, and Valine Leucine and Isoleucine Biosynthesis, adjusting for parent-child relationships inherent in pathway databases (see Materials and Methods, Fig. 3).
3.3. Testing phase: Clinical validation in independent patient cohorts
The next essential step in our analysis was to evaluate the ability of five candidate pathways to predict treatment response to tamoxifen in independent non-overlapping clinical cohorts. For this, we utilized two patient cohorts for testing/validation purposes: (i) Test cohort 1 [29] (GUYT-GSE6532, n = 70) of primary breast tumors obtained at surgery, from patients that did not receive any neoadjuvant treatment and received only adjuvant tamoxifen, with 28.78% of patients having tamoxifen-related events; and (ii) Test cohort 2 [29] (OXFT-GSE6532, n = 77) of primary breast tumors obtained at surgery, from patients that did not receive any neoadjuvant treatment and received only adjuvant tamoxifen, with 25.97% of patients with tamoxifen-related events (Supplementary Table 1). Both Test cohorts had clinical characteristics, neoadjuvant, and adjuvant conditions comparable to the Training cohort (Supplementary Table 1). Similar to the analysis done on the Training cohort, we performed PAM50 classification on the two Test cohorts, eliminating 4 patients from Test cohort 1 and keeping all patients for Test cohort 2.
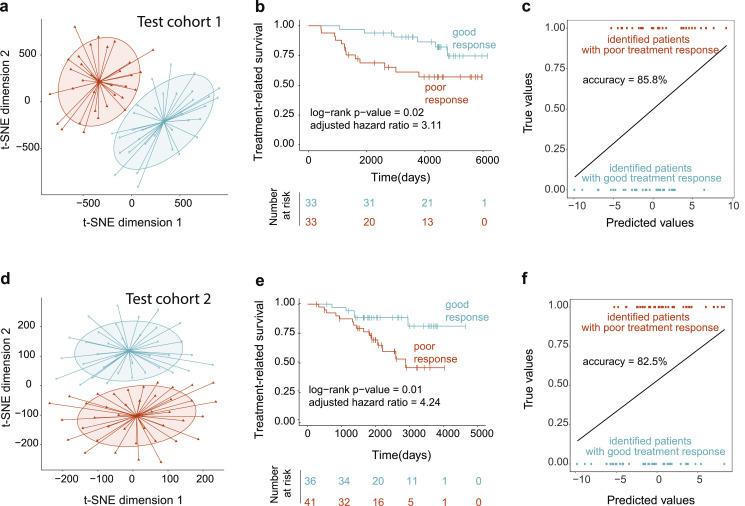
Our main objective was to investigate if activity levels of the five candidate pathways could predict risk of resistance to tamoxifen in two independent Test cohorts. For this, we estimated activity levels for five candidate pathways in each patient in the Test cohorts (similarly to Training cohorts, see Materials and Methods) and subjected patients to t-distributed Stochastic Neighbor Embedding (t-SNE) clustering [58] as suggested in [59] for investigation of samples relationships. T-SNE analysis, which displays five-dimensional dataset in a two-dimensional space, stratified patients into two groups based on their pathway activity levels. The low-dimensional output (i.e., 2-dimensional) of t-SNE were then subjected to the k-means clustering [61] to correctly assign group membership (Fig. 4a for Test cohort 1 and Fig. 4d for Test cohort 2) one group with increased pathways’ activities (orange) and one group with decreased pathways’ activities (turquoise), mimicking the relationship that was observed in the Training cohort (Fig. 2c). We confirmed the strength of group separation through Receiver Operating Characteristic (ROC) analysis [63] using multiple logistic regression model (Supplementary Fig. 3a-b), where normalized enrichment scores of 5 pathways were used as input parameters (i.e., independent/predictor variables) and selected patient groups were utilized as a dependent/response variable. The efficiency of ROC analysis was estimated using area under the curve (AUC) [64], where AUC of 0.5 denotes a random predictor and AUC score of 1 denotes a perfect predictor (i.e., full separation of the patient groups). This analysis confirmed that the activity levels of the five candidate pathways can be effectively used for classifying patients into distinct groups (Test cohort 1, AUC = 0.929; Test cohort 2, AUC = 0.867; Supplementary Fig. 3a-b).
Fig. 4.
The five candidate pathways predict patients at risk of tamoxifen resistance in independent patient cohorts. (a, d) T-SNE and subsequent k-means clustering of Test cohort 1 (a) and Test cohort 2 (d) based on activity levels of the five candidate pathways demonstrates patient separation into two groups: orange group (with overall increased activity levels of the five candidate pathways) and turquoise group (with overall decreased activity levels of the five candidate pathways). (b, e) Kaplan-Meier treatment-related survival analysis comparing two patient groups in Test cohort 1 (b) and in Test cohort 2 (e). Log-rank p-values and adjusted hazard ratios are indicated. (c, f) Leave-one-out cross-validation (LOOCV) correctly identified patients with poor response to tamoxifen (orange) and patients with favorable response to tamoxifen (turquoise) in Test cohort 1 (c) and Test cohort 2 (f). Accuracy values (%) are indicated.
To assess if these patient groups significantly differ in their tamoxifen response, we analyzed therapy-related relapse-free survivals between the groups using Kaplan-Meier survival analysis [67] and adjusted Cox proportional hazards model [56], which demonstrated that the identified patient groups had a significant difference in their response to tamoxifen (Test cohort 1, log-rank p-value = 0.02, Fig. 4b; Test cohort 2, log-rank p-value = 0.01, Fig. 4e). We have also adjusted these analyses for common covariates [78] (i.e., age, tumor grade, tumor size, lymph node status, and PR status), demonstrating that these covariates did not significantly impact the predictive ability of our findings (Test cohort 1, adjusted hazard ratio = 3.11, adjusted hazard p-value = 0.044, 95% confidence interval CI: = 1.03-9.396, Fig. 4b; Test cohort 2, adjusted hazard ratio = 4.24, adjusted hazard p-value = 0.012, CI: 1.3708- 13.120, Fig. 4e).
To ensure that these results are non-random, we compared the ability of the five candidate pathways to predict treatment response to the five pathways selected at random (see Materials and Methods), which confirmed highly non-random ability of the five candidate pathways to predict response to tamoxifen in ER+ breast cancer patients (Test cohort 1, random model p-value = 0.031; Test cohort 2, random model p-value = 0.025, Supplementary Fig. 3c-d).
Furthermore, we evaluated predictive accuracy of our model in the two test cohorts using Leave-One-Out Cross-Validation (LOOCV), which simulates a situation when a new incoming patient needs to be evaluated for her risks of developing resistance to tamoxifen. In particular, in LOOCV, one patient is “removed”, and the model is trained on the remaining patients, followed by the prediction of risk of resistance for the removed patient. The process is repeated for each patient. Using this analysis, we demonstrated the accurate performance of our model in predicting poor and favorable tamoxifen response for new incoming patients (Test cohort 1, accuracy for LOOCV = 85.8%, Fig. 4c; Test cohort 2, accuracy for LOOCV = 82.5%, Fig. 4f). Taken together, these findings indicate that the five candidate pathway signature could successfully predict patients at risk of tamoxifen resistance in independent patient cohorts.
Finally, to ensure that the identified pathways were not only specific for one patient cohort (e.g., Training cohort), we applied our method to both Test cohort 1 and Test cohort 2, and compared identified pathways to those from the Training cohort, which demonstrated their striking similarity (comparison of Test cohort 1 and Training cohort, GSEA positive tail NES = 5.46, p-value <0.001, negative tail NES = -6.45, p-value < 0.001; comparison of Test cohort 2 and Training cohort, GSEA positive tail NES = 5.35, p-value <0.001, negative tail NES = -5.16, p-value < 0.001; Supplementary Fig. 3e-f), indicating that pathways of resistance to tamoxifen in ER+ breast cancer patients are significantly conserved among different cohorts.
3.4. Comprehensive comparison of tamoxifen response and overall disease aggressiveness
A fundamental question in studying therapeutic response lies in its comparison to and differentiation from overall disease aggressiveness. Our comprehensive investigation of this question was four-fold: (i) we identified pathways implicated in disease aggressiveness and compared their overlap with the candidate five pathways of tamoxifen response; (ii) we evaluated if the five candidate pathways can predict breast cancer aggressiveness in an independent (negative control) cohort; (iii) we evaluated the ability of the five candidate pathways to predict tamoxifen response, given different status of PR receptor and Ki-67 proliferation index, which are known indicator of breast cancer aggressiveness; and (iv) we evaluated if known published signatures of disease aggressiveness could predict response to tamoxifen.
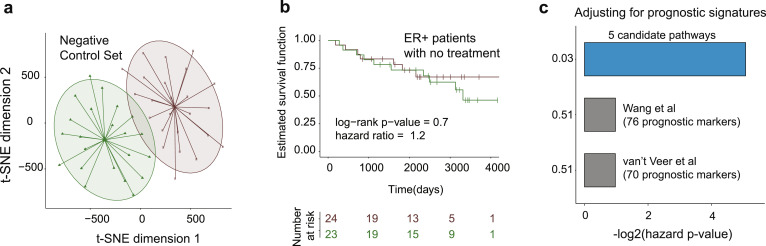
First, to examine if our 5 candidate pathways overlap with pathways implicated in disease aggressiveness, we developed treatment-free prognostic pathway signature using a patient cohort that received surgery only (KIU-GSE6532, n = 51, negative control cohort) [29]. Out of 51 surgery-treated patients, 4 patients were removed, based on the PAM50 classification. We further applied our single-sample pathway-based discovery approach (as in the Training phase) and associated them to the RFS, which identified 3 pathways of aggressiveness (see Materials and Methods) that showed no overlap with the five candidate pathways, signifying that none of our candidates are involved in cancer severity and are indeed specific to tamoxifen response.
Second, we evaluated if the five candidate pathways could separate patients based on overall disease aggressiveness. For this, we evaluated predictive ability of the five candidate pathways on the BC patient cohort that did not receive any treatment after surgery (negative control cohort, as above). We subjected the dataset to the single-sample pathway enrichment analysis (for the five candidate pathways, similarly to Test cohorts analysis). T-SNE clustering based on activity levels of five candidate pathways (Fig. 5a) with subsequent Kaplan-Meier survival analysis (Fig. 5b) on this cohort demonstrated that the five pathways do not separate patients base on their disease aggressiveness (hazard ratio = 1.2, log-rank p-value = 0.7, RFS was considered as a clinical endpoint), but rather specific for tamoxifen response. We have also examined the effect of covariates (i.e., age, tumor grade, tumor size, and PR status), on disease progression in this setting and demonstrated that as expected our candidate pathways remain insignificant, with tumor size significantly contributing to the disease progression (adjusted hazard p-value = 0.0307).
Fig. 5.
Five candidate pathways do not predict and are not affected by overall disease aggressiveness. (a) T-SNE and subsequent k-means clustering based on the activity levels of the five candidate pathways in the negative control cohort. (b) Kaplan-Meier survival analysis on negative control cohort confirms that the five candidate pathways do not predict disease aggressiveness. Log-rank p-value and hazard ratio are indicated. (c) Multivariable Cox proportional hazards model representing analysis for five candidate pathways adjusted for various prognostic signatures in breast cancer, including Wang et al. (76 prognostic markers, with 57 present on U133 Plus 2.0) and van‘t Veer et al. (70 prognostic markers, with 53 present on U133 Plus 2.0). Adjusted hazard p-values are reported.
Third, given that the PR receptor status (which also reflects HER2 signaling) is a known indicator of breast cancer aggressiveness, we performed a stratified Kaplan-Meier analysis on Test cohort 1 (for which this information was available). For this, we divided Test cohort 1 into two groups: one with PR-positive status and one with PR-negative status. We then subjected both groups separately to t-SNE clustering, which have demonstrated that the five candidate pathways separated each group into patient sub-groups with high and low levels of pathway activities. Subsequent Kaplan-Meier survival analysis (Supplementary Fig. 4a and Supplementary Fig. 4b, respectively) showed that these patient-subgroups significantly differ in their response to treatment (group with PR-positive tumors, c-index = 0.698, Supplementary Fig. 4a; group with PR-negative tumors, c-index = 0.769, Supplementary Fig. 4b), demonstrating that our five candidate pathways are able to predict patients at risk of tamoxifen resistance regardless of the PR-status. Similar analysis was performed on patients with different Ki-67 proliferation index (i.e., low levels of Ki-67 corresponding to Luminal A subtype and high levels of Ki-67 corresponding to Luminal B subtype) and demonstrated that our five candidate pathways predict patients at risk of tamoxifen resistance independently of Ki-67 status (group with Luminal A/ Ki-67 low, c-index = 0.657, Supplementary Fig. 4c; group with Luminal B/ Ki-67 high, c-index = 0.658, Supplementary Fig. 4d). Moreover, given that age is an important factor in female hormonal status, we also performed stratified analysis for different age groups (i.e., patient age < 50 years and patient age ≥ 50 years, where 50 was chosen for a threshold as the average menopausal age) and confirmed the predictive ability of our five candidate pathways regardless of the age groups (age < 50 years, c-index = 0.756, Supplementary Fig. 4e; age ≥ 50 years, c-index = 0.616, Supplementary Fig. 4f).
Finally, to demonstrate that the predictive ability of the five candidate pathways is not affected by other known markers of disease aggressiveness, we investigated if commonly known gene-based prognostic signatures can predict tamoxifen response or affect predictive ability of the candidate five pathways. For this, we gathered several known signatures of overall BC aggressiveness (i.e., prognostic signatures), including Wang et al. signature [79] (76 prognostic markers, with 57 present on U133 Plus 2.0) and van‘t Veer et al. signature [80] (70 prognostic markers, with 53 present on U133 Plus 2.0) and subjected them to adjusted multivariable Cox proportional hazards model, alongside the five candidate pathway signature, in the Test cohort 1. This analysis confirmed that the prognostic signatures were not predictive of tamoxifen response and did not impact predictive ability of the five candidate pathways (adjusted hazard p-value = 0.03, Fig. 5c). Taken together, these findings indicate that our five-pathway signature of tamoxifen response is not indicative of overall breast cancer aggressiveness and is indeed specific to response to tamoxifen.
3.5. Comparative analysis to commonly utilized methods and known signatures of tamoxifen response
To evaluate predictive advantages of the five candidate pathways, we took a comprehensive approach and first (i) compared the predictive ability of the five candidate pathways to predictions from other commonly used methods, including approaches based on extreme-responder analysis (i.e., tails of the distribution), support vector machine (SVM), random forest, and mRNA gene expression alone; and second (ii) assessed if the predictive ability of the five candidate pathways outperforms other known signatures of tamoxifen response.
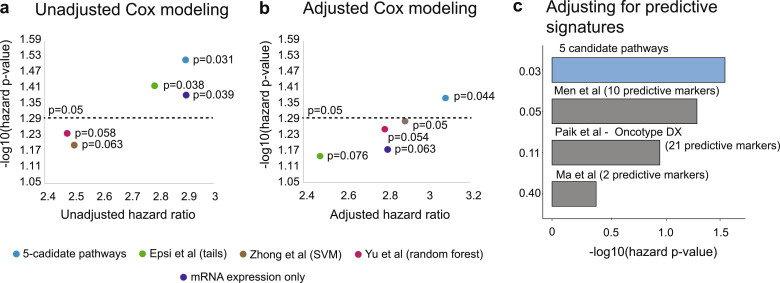
First, we compared predictive ability of the five candidate pathways to predictions from other commonly utilized methods, such as (i) Epsi et al. [28] method, which utilized extreme-responder analysis, using tails of the treatment response distribution to define a treatment response signature; (ii) Zhong et al. [30] method, which used Support Vector Machine approach as a base; (iii) Yu et al. [31] method, also referred to as Personalized REgimen Selection (PRES), which used random forest approach as a base; and (iv) mRNA expression alone (without taking into account information about molecular pathways) (see Materials and Methods). To assure that all methods are comparable to our pathway-centric method, we trained Epsi et al., Zhong et al., Yu et al., and expression-only methods on the Training cohort, with each producing a list of predictions, either pathways or gene lists, depending on the method (112 predictions for Epsi et al.; 5 predictions for Zhong et al.; 3 predictions for Yu et al., and 13 (p-value < 0.001) predictors for mRNA expression alone). We then followed by validating these predictions on the Test cohort 1, similarly to our pathway-centric method. Such analysis demonstrated that the five candidate pathways outperform predictions (either pathways or gene lists, depending on the method) identified by other four methods in their ability to predict the risk of tamoxifen treatment resistance (Fig. 6a: five candidate pathways, hazard ratio = 2.91, hazard p-value = 0.031; Epsi et al., hazard ratio = 2.79, hazard p-value = 0.038; Zhong et al., hazard ratio = 2.53, hazard p-value = 0.063; Yu et al., hazard ratio = 2.48, hazard p-value = 0.058; mRNA expression alone, hazard ratio = 2.93, hazard p-value = 0.039). Furthermore, we adjusted these analyses for the effect of common covariates (similarly to our original training phase), including age, tumor grade, tumor size, lymph node status and PR status and re-confirmed that the five candidate pathways retain their significant predictive ability and outperform the other methods (Fig. 6b: five candidate pathways, adjusted hazard ratio = 3.11, adjusted hazard p-value = 0.044; Epsi et al., adjusted hazard ratio = 2.48, adjusted hazard p-value = 0.076; Zhong et al., adjusted hazard ratio = 2.96, adjusted hazard p-value = 0.05; Yu et al., adjusted hazard ratio = 2.81, adjusted hazard p-value = 0.054; mRNA expression alone, hazard ratio = 2.78, hazard p-value = 0.063).
Fig. 6.
Predictive ability of the five candidate pathways outperforms markers from other methods and known signatures of tamoxifen response. (a, b) Comparison of the predictive ability of the five candidate pathways (blue) to the candidate identified by other approaches, including Epsi et al. extreme-responder analysis (green), Zhong et al. SVM-based method (brown) and Yu et al. PRES random forest-based method (pink), and mRNA expression alone (purple) through unadjusted (a) and adjusted for common covariates (b) Cox proportional hazards model. P-values for unadjusted and adjusted hazard ratios are indicated. (c) Multivariable Cox proportional hazards model representing analysis for the five candidate pathways adjusted for different predictive signatures of tamoxifen response, including Men et al. (10 predictive markers, with 9 present on U133 Plus 2.0), Paik et al. (Oncotype DX, 21 predictive markers), and Ma et al. (2 predictive markers). Adjusted hazard p-values are indicated.
Second, to confirm that the predictive ability of the five candidate pathways outperforms other known signatures in their ability to predict tamoxifen treatment response, we selected known signature of tamoxifen response (i.e., predictive signatures), such as (i) Men et al. [18] (10 predictive markers, with 9 present on U133 Plus 2.0); (ii) Paik et al. [19] (also now as Oncotype DX, 21 predictive markers); and (iii) Ma et al. [20] (2 predictive markers) (Fig. 6c) and used them in adjusted multivariable Cox proportional hazards model, alongside the five candidate pathway signature, utilizing Test cohort 1, as above. This analysis demonstrated that the additional predictive signatures do not significantly affect the ability of the five candidate pathways to predict the risk of tamoxifen resistance (Fig. 6c, adjusted hazards p-value = 0.03). Taken together, these results demonstrate that the five candidate pathway signature can be utilized to predict patients at risk of developing resistance to tamoxifen in a clinical setting and build a foundation for personalized therapeutic advice for patients with ER+ breast cancer.
3.6. Defining risk of tamoxifen resistance using pathway read-out genes
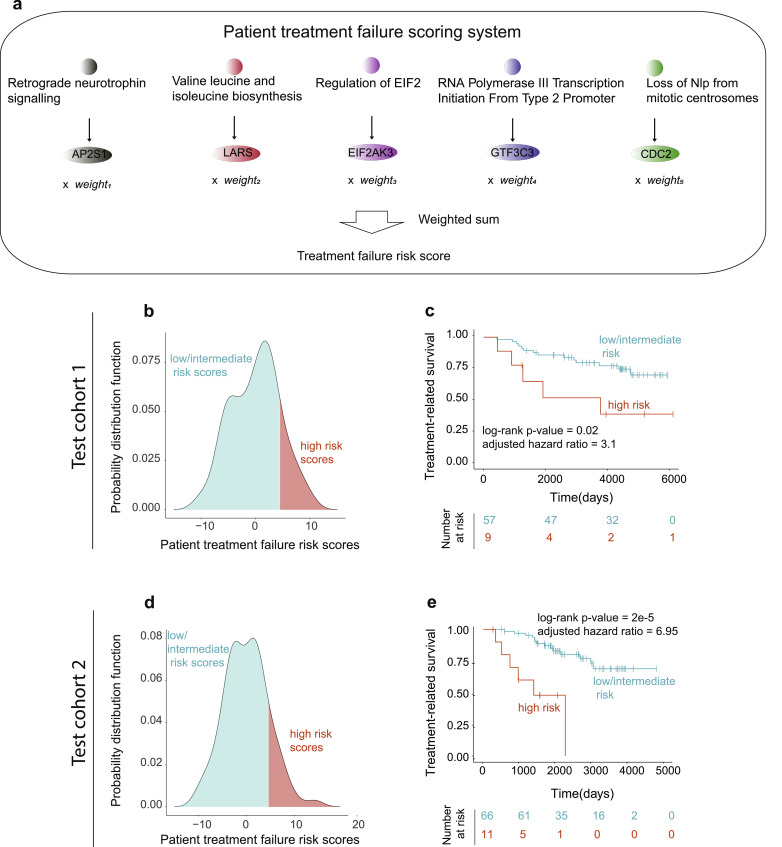
Utilization of pathway activity levels in clinical setting might be hampered by the number of genes in each pathway and need for a full transcriptomic profiling of patients, which might be both time- and cost- sensitive. To address these issues and bring our predictions closer to clinical utilization, we have aimed to identify a so-called “read-out” genes for each pathway. Expression of these genes would (i) accurately reflect pathway activity levels (i.e., estimated through Spearman correlation between gene expression levels and pathway activity levels) and (ii) be significantly associated with treatment response (i.e., through adjusted Cox proportional hazards); thus making them suitable as marker read-outs for tamoxifen resistance. Using such analyses, we identified five read-out genes (one for each pathway, Supplementary Fig. 5) in Training cohort., such as (i) AP2S1 (Retrograde Neurotrophin Signalling pathway), (ii) CDC2 (Loss of NLP from Mitotic Centrosomes pathway), (iii) GTF3C3 (RNA Polymerase III Transcription Initiation from Type 2 Promoter pathway), (iv) EIF2AK3 (EIF2 pathway), and (v) LARS (Valine Leucine and Isoleucine Biosynthesis pathway)
We first evaluated the association of the read-out genes to tamoxifen sensitivity in human cancer cell lines by performing cancer dependency map analysis using DepMap web portal [70], which utilizes PRISM Repurposing [71], CTD2 [72,73], and GDSC databases [74] (see Materials and Methods). We have utilized mRNA expressions levels of the identified read-out genes to query this resource, where large AUCs values showed poor or no response to tamoxifen and smaller AUCs values showed favorable response to tamoxifen. Overall, this analysis showed decreased sensitivity to tamoxifen treatment in different human cancer cell lines (high AUCs), based on the expression levels of the read-out genes in these cell lines, which is consistent with conclusions made in our paper.
To further the utilization of such read-out genes into the clinical setting, we used their expression levels to define a patient risk score to develop tamoxifen resistance. For this, we first performed ROC analysis for each read-out gene in the Training cohort, which reflected each gene's ability to separate patients into good and poor response groups. Ranks of these ROC scores from the Training cohort were then utilized as weights for each read-out gene, so that the risk score of tamoxifen resistance was defined as the weighted sum of expression values for read-out genes (where expression values were multiplied by the weights corresponding to the ranks of ROC values) (Fig. 7a). The risk scores were defined for both Test cohort 1 and Test cohort 2 and risk score distribution defined high risk (>mean+1SD, where mean+1SD for both cohorts were equal to 4.5 score) and low/intermediate risk (≤mean+1SD) patients (Fig 7b, d). We then subjected patient groups with high and low/intermediate risk scores to Kaplan-Meier survival analysis (Fig. 7c, e), which demonstrated that risk scores based on read-out genes are equally effective (compared to activity levels of five candidate pathways) in predicting tamoxifen response in both Test cohort 1 and Test cohort 2, making them suitable candidates for potential clinical integration.
Fig. 7.
Risk scores of tamoxifen resistance identity patients with significant difference in treatment-related survival. (a) Schematic representation of the risk score to fail tamoxifen (i.e., treatment failure score). Top: read-out genes for candidate molecular pathways are assigned; Middle: expression values for the read-out genes are multiplied by their corresponding weights (i.e., ROC values); Bottom: the weighted expression values are then summed and utilized to assign low/intermediate or high risk of failing tamoxifen. (b-e) Validation studies using risk scores in Test cohort 1 (b, c) and Test cohort 2 (d, e). (b) Distribution of risk scores in Test cohort 1, with low/intermediate and high risk patients indicated. (c) Kaplan-Meier survival analysis, comparing low-intermediate and high risk patient groups in Test cohort 1. (d) Distribution of risk scores in Test cohort 2, with low/intermediate and high risk patients indicated. (e) Kaplan-Meier survival analysis, comparing low-intermediate and high risk patient groups in Test cohort 2.
4. Discussion
In this study, we have demonstrated that a pathway-centric genome-side computational approach is able to uncover biological pathways, highly associated with risk of tamoxifen resistance in ER+ breast cancer patients. The important advantage of our approach is that it identifies a tightly connected group of genes - biological pathways - as opposed to individual (possibly distantly connected genes), thus (i) decreasing the chances of experimental noise present in biological experiments; (ii) improving our understanding of the mechanisms implicated in therapeutic resistance; and (iii) increasing the likelihood of identifying a functionally relevant signature, which could be utilized to study mechanisms of primary resistance and their potential therapeutic targeting. Furthermore, these biological pathways have been shown to be highly associated with a wide spectrum of treatment responses across patient cohorts (as opposed to selecting a limited category of patients for analysis), effectively reflecting heterogeneity of response to tamoxifen present in a clinical setting. Even though this work is focused on identifying cases of resistance to tamoxifen, our method can be broadly applicable to other therapeutic interventions and cancer types.
Our computational analysis has identified five molecular pathways implicated in tamoxifen resistance, including (i) Retrograde Neurotrophin Signalling, (ii) Loss of NLP from Mitotic Centrosomes, (iii) RNA Polymerase III Transcription Initiation from Type 2 Promoter, (iv) EIF2 pathway, and (v) Valine Leucine and Isoleucine Biosynthesis. Interestingly, many of these pathways have been shown to be closely related to carcinogenic mechanisms and therapeutic response in various cancers. In particular, the Retrograde Neurotrophin Signalling pathway is implicated in metabolic detoxification, mitosis, clathrin-mediated vesicles development, and enriched with bladder cancer predisposition loci [81]. One of the genes from this pathway, Neurotrophic tyrosine kinase receptor type 1 (NTRK1), is a recognized oncogene frequently altered in various tumor types [82] and its gene fusions have previously been identified in glioblastoma [82], colon cancer [83], papillary thyroid carcinoma [84], and non-small cell lung cancers [85]. Clinical studies of tumor response to NTRK1 fusion-targeted therapy have indicated that this oncogene represents a treatment target in human cancer [86].
Ninein-like protein (NLP) (i.e., also known as NINL) is a part of the Loss of NLP from the Mitotic Centrosomes pathway. The role of human centrosomal NLP expression in breast, lung, ovarian, head and neck cancers has been widely demonstrated [87]. The NLP gene amplification accounts for NLP overexpression in human breast and lung cancer cells [87]. The deregulated expression of NLP in cell models leads to mitotic spindle aberrations, spindle checkpoint defects, chromosomal missegregation, cytokinesis failure, stimulation of chromosomal instability, anchorage-independent growth, and cell malignant transformation [87]. Recently, it has been discovered that NLP co-localizes and interacts with BRCA1 at inter-phasic centrosome and thus the disruptions of BRCA1 function could affect NLP co-localization to centrosomes and induce the genomic instability [88]. Interestingly, it has been reported that the NLP overexpression may also cause breast cancer resistance to paclitaxel chemotherapy [89]. Furthermore, a positive correlation between expression of NLP and PLK1 (i.e., another gene implicated in the Loss of NLP from the Mitotic Centrosomes pathway) has recently been discovered, implicated in chemoresistance, particularly to taxane agents [89] and tumor growth in general, in breast cancer and other cancer types [89,90].
In the Eukaryotic Initiation Factor 2 (eIF2) pathway, phosphorylation of eIF2α has been shown to play a significant role in maintaining normal cellular homeostasis and regulating cell growth [91], with dysregulation of eIF2 signaling pathway stimulating the cancerous tumors transformation [92]. The overexpression of eIF2α has been observed in several cancers, such as gastrointestinal cancer [93] and non-Hodgkin's lymphomas [94] and has been proposed as a potential therapeutic target [95].
Finally, in the Valine Leucine and Isoleucine Biosynthesis pathway, valine, leucine, and isoleucine are important branched-chain amino acids (BCAAs) for normal growth and development [96]. In the BCAA catabolism pathway, the first step is transamination, catalyzed by the branched chain amino acid transferase isozymes BCATs: a mitochondrial (BCATm) and a cytosolic (BCATc) isozyme [[97], [98], [99]]. Mitochondrial BCATm (BCAT2) expression can drive the development of pancreatic ductal adenocarcinoma under the regulation of the mitochondrial malic enzyme 2 [100,101]. Cytosolic BCATc (BCAT1) is overexpressed in glioblastoma [102], nasopharyngeal carcinoma [103], and cancers with elevated c-MYC [103]. It has been recommended to consider BCAT1 as a promising target for glioblastoma and nasopharyngeal carcinoma treatments [102,103].
Furthermore, to enhance clinical applicability of our findings, we defined five read-out genes, one per pathway: (i) AP2S1 (Retrograde Neurotrophin Signalling pathway), (ii) CDC2 (Loss of NLP from Mitotic Centrosomes pathway), (iii) GTF3C3 (RNA Polymerase III Transcription Initiation from Type 2 Promoter pathway), (iv) EIF2AK3 (EIF2 pathway), and (v) LARS (Valine Leucine and Isoleucine Biosynthesis pathway). One of these genes, LARS, has been reported as a potential metabolic onco-target [[104], [105], [106], [107]], where its direct inhibition suppresses cell proliferation via the p21 signaling, leading to apoptosis [107]. Furthermore, CDC2 has been shown to be implicated in tamoxifen response and serves as a positive control in this study. In particular, CDC2 mRNA expression has been shown to be significantly correlated with the poor response to tamoxifen therapy by several groups [[108], [109], [110]] and its inhibition is suggested as a potential therapeutic strategy for tamoxifen-resistant breast tumors [110]. We propose that the identified candidate pathways and their read-out genes should be further investigated for their potential use as targets for solo treatments or in combination with ER-targeting agents for ER+ breast cancer patients at risk of developing resistance to tamoxifen.
One of the limitations of our study is in the limited availability of the epigenomic profiles for our patient cohorts. In fact, DNA and histone methylation has been suggested to be responsible for inactivation of ER [111]. Thus, further examination of the role of epigenomic modulations and their interplay with transcriptomic changes is an invaluable next step for in-depth understanding of molecular mechanisms implicated in hormone therapy resistance.
Furthermore, miRNAs (micro-RNAs) have received substantial attention for their role in regulating pathway functionality [112]. For example, miR-15a/miR-16’s deletion or down-regulation contributes to dysregulated of cell cycle in chronic lymphocytic leukemia [113] and non-small cell lung cancer [114]. Even though miRNA data are not available in our cohorts, we foresee the importance of miRNA analysis for further understanding mechanisms of pathway dysregulation, especially when applies to therapeutic resistance [[115], [116], [117]]. The presence of miRNAs in tumor-derived exosomes has recently been postulated to play important roles in facilitating metastasis, and this work suggests that exosomes containing tumor-derived miRNAs which regulate one of these five pathways may also play a role in the spread of tamoxifen resistance [118].
In addition, availability of single-cell profiles for investigation of therapeutic response has proven to be invaluable [119] in understanding of therapeutic targets for complex diseases, including cancer. Thus, as such profiles become available, we foresee their immediate utilization for elucidation of mechanisms of primary and secondary therapy resistance.
In conclusion, we have demonstrated that a systematic computational pathway-centric method could identify molecular pathways and their read-out genes to predict tamoxifen resistance. We propose that our finding can be ultimately utilized to prioritize and determine (i) cases at higher risk of developing resistance to tamoxifen that should be considered for alternative treatment manipulations (for instance, alternative endocrine therapy, radiation therapy, or chemotherapy etc.) and (ii) cases who would benefit maximally from tamoxifen therapy.
Data availability and sharing
Data utilized for Training and Testing and their clinical characteristics are freely available from GEO repository GSE6532. We have also included data objects (Supplementary Dataset 1a, Supplementary Dataset 1b, and Supplementary Dataset 1c), which will allow a user to easily reproduce the results of our discovery.
Ethics
We have only utilized de-identified patient data from GEO repository in this study, thus no IRB approval for this study was required.
Author contributions
S.R. and A.M. designed the study and wrote the manuscript. S.R performed the computational and statistical analysis. N.E. generated network image, Dependency Map output, and helped generate and format Supplementary Data 1. F.C. provided useful feedback and discussion suggestions. All authors read and approved the manuscript.
Declaration of Competing Interests
The authors declare no competing financial interests. Dr. Mitrofanova and Dr. Rahem filed a U.S. Provisional Patent Application No. 63/045,878 on June 30, 2020.
Acknowledgements
We would like to thank all members of Mitrofanova lab, especially Sukanya Panja, for thoughtful discussions and advice about this work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103047.
Appendix. Supplementary materials
References
- 1.Zhang MH, Man HT, Zhao XD, Dong N, Ma SL. Estrogen receptor-positive breast cancer molecular signatures and therapeutic potentials (Review) Biomed Rep. 2014;2(1):41–52. doi: 10.3892/br.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedraza V, Gomez-Capilla JA, Escaramis G, Gomez C, Torne P, Rivera JM. Gene expression signatures in breast cancer distinguish phenotype characteristics, histologic subtypes, and tumor invasiveness. Cancer. 2010;116(2):486–496. doi: 10.1002/cncr.24805. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Chang M. Tamoxifen resistance in breast cancer. Biomol Ther (Seoul) 2012;20(3):256–267. doi: 10.4062/biomolther.2012.20.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40. doi: 10.1186/s13058-015-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Group EBCTC. Tamoxifen for early breast cancer: an overview of the randomised trials. The Lancet. 1998;351(9114):1451–1467. [PubMed] [Google Scholar]
- 7.Hackshaw A, Roughton M, Forsyth S, Monson K, Reczko K, Sainsbury R. Long-term benefits of 5 years of tamoxifen: 10-year follow-up of a large randomized trial in women at least 50 years of age with early breast cancer. J Clin Oncol. 2011;29(13):1657–1663. doi: 10.1200/JCO.2010.32.2933. [DOI] [PubMed] [Google Scholar]
- 8.Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S. Early Breast Cancer Trialists’ Collaborative G. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378(9793):771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies C, Pan H, Godwin J, Gray R, Arriagada R, Raina V. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. The Lancet. 2013;381(9869):805–816. doi: 10.1016/S0140-6736(12)61963-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genom. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallo MA, Kaufman D., editors. Antagonistic and agonistic effects of tamoxifen: significance in human cancer. Seminars in oncology; 1997. [PubMed] [Google Scholar]
- 12.Fox EM, Arteaga CL, Miller TW. Abrogating endocrine resistance by targeting ERalpha and PI3K in breast cancer. Front Oncol. 2012;2:145. doi: 10.3389/fonc.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osborne CK. Tamoxifen in the treatment of breast cancer. New Engl J Med. 1998;339(22):1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 14.Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96(12):926–935. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 15.Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95(5):353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 16.Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23(30):7721–7735. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Dowsett M, Allred C, Knox J, Quinn E, Salter J, Wale C. Relationship between quantitative estrogen and progesterone receptor expression and human epidermal growth factor receptor 2 (HER-2) status with recurrence in the Arimidex, Tamoxifen, Alone or in Combination trial. J Clin Oncol. 2008;26(7):1059–1065. doi: 10.1200/JCO.2007.12.9437. [DOI] [PubMed] [Google Scholar]
- 18.Men X, Ma J, Wu T, Pu J, Wen S, Shen J. Transcriptome profiling identified differentially expressed genes and pathways associated with tamoxifen resistance in human breast cancer. Oncotarget. 2018;9(3):4074–4089. doi: 10.18632/oncotarget.23694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 20.Ma X-J, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5(6):607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Wang Y, Shen B, Zhang D. Molecular signature of cancer at gene level or pathway level? Case studies of colorectal cancer and prostate cancer microarray data. Comput Math Methods Med. 2013 doi: 10.1155/2013/909525. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Chen J, Li Q, Wang H, Liu G, Jing Q. Identifying novel prostate cancer associated pathways based on integrative microarray data analysis. Comput Biol Chem. 2011;35(3):151–158. doi: 10.1016/j.compbiolchem.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Myers JS, von Lersner AK, Robbins CJ, Sang Q-XA. Differentially expressed genes and signature pathways of human prostate cancer. PloS One. 2015;10(12) doi: 10.1371/journal.pone.0145322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abraham G, Kowalczyk A, Loi S, Haviv I, Zobel J. Prediction of breast cancer prognosis using gene set statistics provides signature stability and biological context. BMC Bioinf. 2010;11(1):277. doi: 10.1186/1471-2105-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci. 2005;102(38):13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E, Chuang H-Y, Kim J-W, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4(11) doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network‐based classification of breast cancer metastasis. Mol Syst Biol. 2007;3(1):140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epsi NJ, Panja S, Pine SR. pathCHEMO, a generalizable computational framework uncovers molecular pathways of chemoresistance in lung adenocarcinoma. 2019;2:334. [DOI] [PMC free article] [PubMed]
- 29.Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Q, Fang J, Huang Z, Yang Y, Lian M, Liu H. A response prediction model for taxane, cisplatin, and 5-fluorouracil chemotherapy in hypopharyngeal carcinoma. Sci Rep. 2018;8(1):12675. doi: 10.1038/s41598-018-31027-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu K, Sang Q-XA, Lung P-Y, Tan W, Lively T, Sheffield C. Personalized chemotherapy selection for breast cancer using gene expression profiles. Sci Rep. 2017;7:43294. doi: 10.1038/srep43294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ThermoFisher Scientific . 2018, May 25. Human genome u133 set - support materials.www.affymetrix.com/support/technical/byproduct.affx?product=hgu133 Available from: [Google Scholar]
- 34.Negi SK, Guda C. Global gene expression profiling of healthy human brain and its application in studying neurological disorders. Sci Rep. 2017;7(1):897. doi: 10.1038/s41598-017-00952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnatkevic Iute A, Fulcher BD, Fornito A. A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage. 2019;189:353–367. doi: 10.1016/j.neuroimage.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chia SK, Bramwell VH, Tu D, Shepherd LE, Jiang S, Vickery T. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res. 2012;18(16):4465–4472. doi: 10.1158/1078-0432.CCR-12-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haibe-Kains B, Schroeder M, Bontempi G, Sotiriou C, Quackenbush J. genefu: Relevant functions for gene expression analysis, especially in breast cancer. R/Bioconductor version. Development. 2011;(212) [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5(2):73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee E, Chuang HY, Kim JW, Ideker T, Lee D. Inferring pathway activity toward precise disease classification. PLoS Comput Biol. 2008;4(11) doi: 10.1371/journal.pcbi.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liberzon A. Molecular signatures database (MSigDB) 3.0. Bioinforma. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37(suppl_1):D619–DD22. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–DD62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(D1):D472–D4D7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang R, Grishagin I, Wang Y, Zhao T, Greene J, Obenauer JC. The NCATS BioPlanet–an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front Pharmacol. 2019;10:445. doi: 10.3389/fphar.2019.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jupe S, Akkerman JW, Soranzo N, Ouwehand WH. Reactome-a curated knowledgebase of biological pathways: megakaryocytes and platelets. J Thromb Haemost JTH. 2012;10(11):2399. doi: 10.1111/j.1538-7836.2012.04930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauer-Mehren A, Furlong LI, Sanz F. Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Mol Syst Biol. 2009;5(1):290. doi: 10.1038/msb.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T. PID: the pathway interaction database. Nucleic Acids Res. 2009;37(suppl_1):D674–D6D9. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M, Goto S. KEGG kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007;36(suppl_1):D480–D4D4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsui IF, Chari R, Buys TP, Lam WL. Public databases and software for the pathway analysis of cancer genomes. Cancer Inf. 2007;3 [PMC free article] [PubMed] [Google Scholar]
- 54.Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8(3):1–13. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galperin MY. The molecular biology database collection: 2004 update. Nucleic Acids Res. 2004;32(suppl_1):D3–D22. doi: 10.1093/nar/gkh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox DR. Regression models and life-tables. Journal of the Royal Statistical Society: Series B (Methodological). 1972;34(2):187-202.
- 57.Therneau TM, Grambsch PM. Modeling survival data: extending the Cox model: Springer Science & Business Media; 2013.
- 58.Maaten Lvd, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9(Nov):2579–2605. [Google Scholar]
- 59.Taskesen E, Reinders MJ. 2D representation of transcriptomes by t-SNE exposes relatedness between human tissues. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0149853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mwangi B, Soares JC, Hasan KM. Visualization and unsupervised predictive clustering of high-dimensional multimodal neuroimaging data. J Neurosci Methods. 2014;236:19–25. doi: 10.1016/j.jneumeth.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 61.Hartigan JA, Wong MA. Algorithm AS 136: A k-means clustering algorithm. J R Stat Soc Ser C (Appl Stat) 1979;28(1):100–108. [Google Scholar]
- 62.stat. K-means clustering 2019, Mar 21 [Available from: https://stat.ethz.ch/R-manual/R-devel/library/stats/html/kmeans.html.
- 63.Hajian-Tilaki K. Receiver Operating Characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4(2):627–635. [PMC free article] [PubMed] [Google Scholar]
- 64.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 65.Zeileis A, Kleiber C, Jackman S. Regression models for count data in R. J statistical software. 2008;27(8):1–25. [Google Scholar]
- 66.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1(4):274–278. doi: 10.4103/0974-7788.76794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mosteller F, Tukey JW. Data analysis, including statistics. Handb Soc Psychol. 1968;2:80–203. [Google Scholar]
- 69.Welch BL. The generalization of student’s problems when several different population variances are involved. Biometrika. 1947;34(1/2):28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 70.depmap portal. Explore the cancer dependency map 2019 [Available from:https://depmap.org/portal/.
- 71.Corsello SM, Nagari RT, Spangler RD, Rossen J, Kocak M, Bryan JG. Non-oncology drugs are a source of previously unappreciated anti-cancer activity. bioRxiv. 2019 [Google Scholar]
- 72.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5(11):1210–1223. doi: 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem biology. 2016;12(2):109–116. doi: 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166(3):740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rouzier R, Pusztai L, Delaloge S, Gonzalez-Angulo AM, Andre F, Hess KR. Nomograms to predict pathologic complete response and metastasis-free survival after preoperative chemotherapy for breast cancer. J Clin Oncol. 2005;23(33):8331–8339. doi: 10.1200/JCO.2005.01.2898. [DOI] [PubMed] [Google Scholar]
- 78.Cianfrocca M, Goldstein LJ. Prognostic and predictive factors in early-stage breast cancer. Oncologist. 2004;9(6):606–616. doi: 10.1634/theoncologist.9-6-606. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 80.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 81.Menashe I, Figueroa JD, Garcia-Closas M, Chatterjee N, Malats N, Picornell A. Large-scale pathway-based analysis of bladder cancer genome-wide association data from five studies of European background. PloS one. 2012;7(1):e29396. doi: 10.1371/journal.pone.0029396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J, Lee Y, Cho H-J, Lee Y-E, An J, Cho G-H. NTRK1 fusion in glioblastoma multiforme. PLoS One. 2014;9(3):e91940. doi: 10.1371/journal.pone.0091940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martin-Zanca D, Hughes SH, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319(6056):743. doi: 10.1038/319743a0. [DOI] [PubMed] [Google Scholar]
- 84.Greco A, Pierotti M, Bongarzone I, Pagliardini S, Lanzi C, Della GP. TRK-T1 is a novel oncogene formed by the fusion of TPR and TRK genes in human papillary thyroid carcinomas. Oncogene. 1992;7(2):237–242. [PubMed] [Google Scholar]
- 85.Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19(11):1469. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5(1):25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li J, Zhan Q. The role of centrosomal Nlp in the control of mitotic progression and tumourigenesis. Br J cancer. 2011;104(10):1523. doi: 10.1038/bjc.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jin S, Gao H, Mazzacurati L, Wang Y, Fan W, Chen Q. BRCA1 interaction of centrosomal protein Nlp is required for successful mitotic progression. J Biol Chem. 2009;284(34):22970–22977. doi: 10.1074/jbc.M109.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao W, Song Y, Xu B, Zhan Q. Overexpression of centrosomal protein Nlp confers breast carcinoma resistance to paclitaxel. Cancer Biol Ther. 2012;13(3):156–163. doi: 10.4161/cbt.13.3.18697. [DOI] [PubMed] [Google Scholar]
- 90.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6(4):321. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 91.Burwick N, Aktas BH. The eIF2-alpha kinase HRI: A potential target beyond the red blood cell. Expert Opin Therap Targets. 2017;21(12):1171–1177. doi: 10.1080/14728222.2017.1397133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Donze O, Jagus R, Koromilas A, Hershey J, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14(15):3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lobo MV, Martín ME, Pérez MI, Alonso FJM, Redondo C, Álvarez MI. Levels, phosphorylation status and cellular localization of translational factor eIF2 in gastrointestinal carcinomas. Histochem J. 2000;32(3):139–150. doi: 10.1023/a:1004091122351. [DOI] [PubMed] [Google Scholar]
- 94.Wang S, Rosenwald IB, Hutzler MJ, Pihan GA, Savas L, Chen J-J. Expression of the eukaryotic translation initiation factors 4E and 2α in non-Hodgkin's lymphomas. Am J Pathol. 1999;155(1):247–255. doi: 10.1016/s0002-9440(10)65118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burwick N, Zhang MY, de la Puente P, Azab AK, Hyun TS, Ruiz-Gutierrez M. The eIF2-alpha kinase HRI is a novel therapeutic target in multiple myeloma. Leukemia Res. 2017;55:23–32. doi: 10.1016/j.leukres.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hutson SM, Sweatt AJ, LaNoue KF. Branched-chain amino acid metabolism: implications for establishing safe intakes. The J Nutr. 2005;135(6):1557S–1564S. doi: 10.1093/jn/135.6.1557S. [DOI] [PubMed] [Google Scholar]
- 97.Hutson SM, Fenstermacher D, Mahar C. Role of mitochondrial transamination in branched chain amino acid metabolism. J Biol Chem. 1988;263(8):3618–3625. [PubMed] [Google Scholar]
- 98.Wallin R, Hall TR, Hutson SM. Purification of branched chain aminotransferase from rat heart mitochondria. J Biol Chem. 1990;265(11):6019–6024. [PubMed] [Google Scholar]
- 99.Hall T, Wallin R, Reinhart G, Hutson S. Branched chain aminotransferase isoenzymes. Purification and characterization of the rat brain isoenzyme. J Biol Chem. 1993;268(5):3092–3098. [PubMed] [Google Scholar]
- 100.Mayers JR, Torrence ME, Danai LV, Papagiannakopoulos T, Davidson SM, Bauer MR. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353(6304):1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dey P, Baddour J, Muller F, Wu CC, Wang H, Liao W-T. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature. 2017;542(7639):119. doi: 10.1038/nature21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tönjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19(7):901. doi: 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou W, Feng X, Ren C, Jiang X, Liu W, Huang W. Over-expression of BCAT1, a c-Myc target gene, induces cell proliferation, migration and invasion in nasopharyngeal carcinoma. Mol Cancer. 2013;12(1):53. doi: 10.1186/1476-4598-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci. 2008;105(32):11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genom Hum Genet. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- 106.Rock FL, Mao W, Yaremchuk A, Tukalo M, Crépin T, Zhou H. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007;316(5832):1759–1761. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 107.Gao G, Yao Y, Li K, Mashausi DS, Li D, Negi H. A human leucyl-tRNA synthetase as an anticancer target. OncoTargets Ther. 2015;8:2933. doi: 10.2147/OTT.S88873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jansen M, Reijm E, Sieuwerts A, Ruigrok-Ritstier K, Look M, Rodriguez-Gonzalez F. High miR-26a and low CDC2 levels associate with decreased EZH2 expression and with favorable outcome on tamoxifen in metastatic breast cancer. Breast Cancer Res Treat. 2012;133(3):937–947. doi: 10.1007/s10549-011-1877-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Egeland NG, Lunde S, Jonsdottir K, Lende TH, Cronin-Fenton D, Gilje B. The role of microRNAs as predictors of response to tamoxifen treatment in breast cancer patients. Int J Mol Sci. 2015;16(10):24243–24275. doi: 10.3390/ijms161024243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnson N, Bentley J, Wang L, Newell D, Robson C, Shapiro G. Pre-clinical evaluation of cyclin-dependent kinase 2 and 1 inhibition in anti-estrogen-sensitive and resistant breast cancer cells. Br J Cancer. 2010;102(2):342–350. doi: 10.1038/sj.bjc.6605479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sharma D, Blum J, Yang X, Beaulieu N, Macleod AR, Davidson NE. Release of methyl CpG binding proteins and histone deacetylase 1 from the estrogen receptor α (ER) promoter upon reactivation in ER-negative human breast cancer cells. Mol Endocrinol. 2005;19(7):1740–1751. doi: 10.1210/me.2004-0011. [DOI] [PubMed] [Google Scholar]
- 112.Jin S, Zeng X, Fang J, Lin J, Chan SY, Erzurum SC. A network-based approach to uncover microRNA-mediated disease comorbidities and potential pathobiological implications. npj Syst Biol Appl. 2019;5(1):41. doi: 10.1038/s41540-019-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Braga TV, Evangelista FCG, Gomes LC, Araujo S, Carvalho MDG, Sabino AP. Evaluation of MiR-15a and MiR-16-1 as prognostic biomarkers in chronic lymphocytic leukemia. Biomed Pharmacother = Biomedecine & pharmacotherapie. 2017;92:864–869. doi: 10.1016/j.biopha.2017.05.144. [DOI] [PubMed] [Google Scholar]
- 114.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69(13):5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 115.Cava C, Colaprico A, Bertoli G, Bontempi G, Mauri G, Castiglioni I. How interacting pathways are regulated by miRNAs in breast cancer subtypes. BMC Bioinf. 2016;17(12):348. doi: 10.1186/s12859-016-1196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cimino D, De Pitta C, Orso F, Zampini M, Casara S, Penna E. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. The FASEB J. 2013;27(3):1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17(1):147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gawel DR, Serra-Musach J, Lilja S, Aagesen J, Arenas A, Asking B. A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Med. 2019;11(1):47. doi: 10.1186/s13073-019-0657-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data utilized for Training and Testing and their clinical characteristics are freely available from GEO repository GSE6532. We have also included data objects (Supplementary Dataset 1a, Supplementary Dataset 1b, and Supplementary Dataset 1c), which will allow a user to easily reproduce the results of our discovery.