For bacteria, the preferred form of the essential nutrient phosphate is inorganic monophosphate (Pi), but phosphate can also be extracted from a variety of phosphocompounds. Pathogens, including Staphylococcus aureus, experience Pi limitation within the host, suggesting that the use of alternative phosphate sources is important during infection. However, the alternative phosphate sources that can be used by S. aureus and others remain largely unexplored. We screened a library of phosphorus-containing compounds for the ability to support growth as a phosphate source. S. aureus could use a variety of phosphocompounds, including nucleotides, phosphosugars, and phosphoamino acids. Subsequent genetic analysis determined that a majority of these alternative phosphate sources are first processed extracellularly to liberate Pi, which is then imported through Pi transporters.
KEYWORDS: Staphylococcus aureus, phosphate acquisition, organophosphate, glycerol-3-phosphate, alkaline phosphatase
ABSTRACT
Phosphate is an essential nutrient that Staphylococcus aureus and other pathogens must acquire from the host during infection. While inorganic monophosphate (Pi) is the preferred source of this nutrient, bacteria can also obtain it from phosphate-containing organic molecules. The Pi-responsive regulator PhoPR is necessary for S. aureus to cause infection, suggesting that Pi is not freely available during infection and that this nutrient must be obtained from other sources. However, the organophosphates from which S. aureus can obtain phosphate are unknown. We evaluated the ability of 58 phosphorus-containing molecules to serve as phosphate sources for S. aureus. Forty-six of these compounds, including phosphorylated amino acids, sugars, and nucleotides, supported growth. Among the organophosphate sources was glycerol-3-phosphate (G3P), which is commonly found in the mammalian host. Differing from the model organism Escherichia coli, S. aureus does not import G3P intact to obtain Pi. Instead, S. aureus relies on the phosphatase PhoB to release Pi from G3P, which is subsequently imported by Pi transporters. To determine if this strategy is used by S. aureus to extract phosphate from other phosphate sources, we assessed the ability of PhoB- and Pi transporter-deficient strains to grow on the same library of phosphorus-containing molecules. Sixty percent of the substrates (28/46) relied on the PhoB/Pi transporter pathway, and an additional 10/46 (22%) were PhoB independent but still required Pi transport through the Pi transporters. Cumulatively, these results suggest that in Pi-limited environments, S. aureus preferentially generates Pi from organophosphates and then relies on Pi transporters to import this nutrient.
IMPORTANCE For bacteria, the preferred form of the essential nutrient phosphate is inorganic monophosphate (Pi), but phosphate can also be extracted from a variety of phosphocompounds. Pathogens, including Staphylococcus aureus, experience Pi limitation within the host, suggesting that the use of alternative phosphate sources is important during infection. However, the alternative phosphate sources that can be used by S. aureus and others remain largely unexplored. We screened a library of phosphorus-containing compounds for the ability to support growth as a phosphate source. S. aureus could use a variety of phosphocompounds, including nucleotides, phosphosugars, and phosphoamino acids. Subsequent genetic analysis determined that a majority of these alternative phosphate sources are first processed extracellularly to liberate Pi, which is then imported through Pi transporters.
INTRODUCTION
Phosphate is an essential nutrient that pathogens must acquire from the host during infection. As such, phosphate acquisition and regulatory systems are critical for the virulence of a variety of bacterial pathogens (1–8). The preferred source of phosphate for bacteria is inorganic monophosphate (Pi). Phosphate can also be obtained from a variety of phosphate-containing molecules (9, 10). However, outside of model organisms and closely related microbes, how pathogens cope with the absence of Pi is unknown (9, 11). Capable of infecting a wide variety of tissues, Staphylococcus aureus is a significant threat to human health, and public health organizations have called for new strategies to combat S. aureus (12–15). Understanding the aspects of staphylococcal physiology that are important during infection and how pathogens obtain essential nutrients such as phosphate has the potential to inform the development of antimicrobials.
Both Pi importers and a phosphate-responsive signal transduction system are necessary for S. aureus to cause infection, and altering these processes has been associated with changes in antibiotic sensitivity (16–20). S. aureus possesses three Pi transporters that enable it to acquire Pi in distinct environments (16). Two of these transporters, PitA and NptA, are expressed regardless of Pi availability but function optimally at opposite pHs: PitA functions best at acidic pHs, and NptA functions best at alkaline pHs. The third transporter, PstSCAB, is an ABC-type transporter that is expressed only during Pi limitation, where it is also optimal for Pi acquisition (16). The loss of NptA and either PitA or PstSCAB significantly attenuates the virulence of S. aureus, underscoring the importance of Pi transporters for infection (16). Yet despite its three Pi transporters, S. aureus additionally requires regulatory systems that respond to Pi limitation for infection, indicating that Pi is not freely available within the host (17). This observation suggests that S. aureus must use alternative phosphate sources during infection. However, the organophosphates from which S. aureus can obtain Pi are largely unknown, as are the pathways through which they are acquired.
To maintain Pi homeostasis, many bacteria, including S. aureus, contain a Pi-responsive two-component system named PhoPR (PhoBR in Escherichia coli) that is important for growth during Pi starvation and infection (9, 17, 21, 22). Upon Pi limitation, PhoPR is activated and induces the expression of a set of genes responsible for scavenging phosphate, collectively known as the Pho regulon (23–25). In E. coli and others, the Pho regulon includes the high-affinity PstSCAB system, a secreted, nonspecific alkaline phosphatase, and transporters for alternative organic sources of phosphate, such as glycerol-3-phosphate (G3P) (9, 23–26). Although the Pho regulon has not been comprehensively elucidated in S. aureus, the expression of NptA and PstSCAB is PhoPR dependent (17). Notably, in some tissues, the requirement of PhoPR for staphylococcal infection extends beyond its regulation of NptA and PstSCAB, indicating that Pi transporter-independent Pho regulon members are important for virulence (17).
G3P can be a source of both phosphate and carbon for S. aureus (27, 28). In the context of the host, G3P is found in various host tissues and fluids as well as the cell wall of cocolonizing bacteria (29, 30). S. aureus encodes a glycerophosphodiesterase, GlpQ, that liberates G3P from both wall teichoic acids produced by closely related staphylococci and host-derived phospholipids (27, 28). Of note, the expression of GlpQ increases upon phosphate starvation (28). Together, these data suggest that G3P may be an important source of phosphate for S. aureus within skin-associated bacterial communities and in the context of infection. However, how S. aureus utilizes G3P liberated by GlpQ as a source of phosphate is unknown. In E. coli, two PhoBR-regulated pathways promote the use of G3P as a phosphate source. First, G3P is a substrate for the conserved, periplasmic phosphate starvation-inducible alkaline phosphatase PhoA, which cleaves G3P into glycerol and Pi, releasing Pi for uptake through a Pi importer (9, 10). The homolog of this enzyme in S. aureus is PhoB, which is located on the surface of the bacterium (31). Second, E. coli possesses UgpBAEC, an ABC-type transporter that imports G3P (9, 10, 32, 33). Thus, E. coli can use G3P as a phosphate source in both a Pi transporter-dependent and -independent manner.
Given the evidence that insufficient Pi is available for S. aureus during infection, we set out to identify other phosphocompounds that can serve as phosphate sources for S. aureus and the mechanisms by which they are used. A library of 58 phosphorus-containing compounds was screened for their ability to promote staphylococcal growth in Pi-depleted medium. Forty-six of the compounds supported staphylococcal growth, including a variety of inorganic and organophosphate molecules, among them G3P. Next, the pathway by which S. aureus acquires phosphate from G3P was assessed. These studies revealed that, differing from E. coli, S. aureus does not contain a Ugp system for G3P import. Instead, S. aureus depends solely on the phosphatase PhoB to liberate Pi, which is subsequently imported by Pi transporters. Further analysis revealed that the majority of the alternative phosphate sources are used by S. aureus in a PhoB- and/or Pi transporter-dependent manner. Cumulatively, these findings suggest that S. aureus can obtain Pi from a variety of phosphate sources but that their utilization is dependent on the extracellular release of Pi and its subsequent import by Pi transporters.
RESULTS
S. aureus can utilize a variety of organophosphates as alternative phosphate sources.
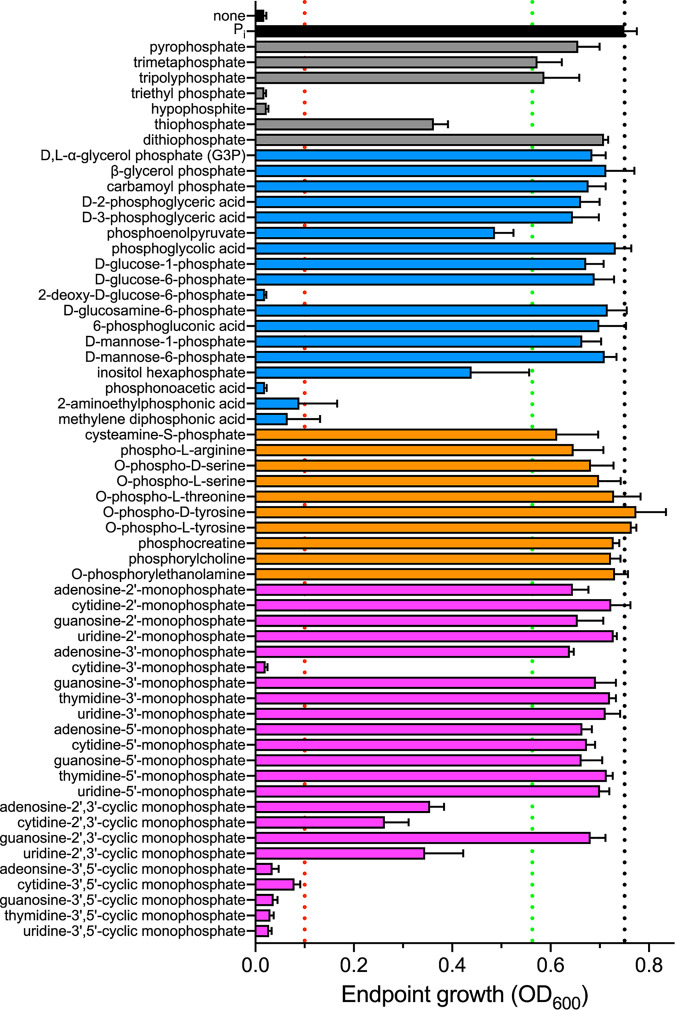
While it is highly likely that S. aureus can use organophosphates as a source of phosphate, the diversity of compounds that it can use is unknown. To address this gap in knowledge, a library of 58 phosphorus-containing compounds was screened for the ability to support the growth of S. aureus in a Pi-limited growth medium (phosphate-free M9-based medium [PFM9]). The library consisted of organic sources of phosphate, including phosphorylated sugars, amino acids, and nucleotides, as well as inorganic sources, such as thiophosphate. The ability of S. aureus to grow on these compounds was compared to that with growth medium supplemented with Pi or no phosphate source (Fig. 1). Twelve (21%) of the compounds did not support the growth of S. aureus as a phosphate source (optical density at 600 nm [OD600] of <0.1), including several cyclic nucleotides and three phosphonate (carbon-phosphorus bond-containing) molecules (Fig. 1, substrates below the red dotted line; Table 1; see Fig. S1 in the supplemental material). Six of the remaining 46 compounds (13%) resulted in intermediate growth of S. aureus, defined as a final growth yield less than 75% of the growth on Pi (Fig. 1, substrates between the red and green dotted lines). The other 40 out of 46 phosphorylated compounds (87%) promoted the growth of S. aureus to within 25% of the yield obtained with Pi, suggesting that these are strong alternative phosphate sources (Fig. 1, substrates above the green dotted line). Cumulatively, these results indicate that S. aureus is capable of using a wide variety of phosphate-containing molecules, including phosphorylated amino acids, nucleotides, and sugars, as phosphate sources.
FIG 1.
S. aureus can use a variety of phosphorylated molecules as phosphate sources. A library of 58 phosphorus-containing compounds, including inorganic molecules (gray), carbohydrates (blue), amino acids (orange), and nucleotides (purple), was screened for the growth of S. aureus in defined, phosphate-depleted medium buffered to pH 7.4. Bacteria were precultured overnight in a Pi-limiting medium. Growth was monitored by assessing optical density at 600 nm (OD600), and endpoint growth after 10 h is reported. The screen was performed in biological triplicate; error bars indicate standard deviations. An OD600 of >0.1 (red dotted line) was used as the threshold to define phosphate sources that support growth. The black dotted line delineates growth on the positive control, Pi. The green dotted line denotes 75% of the growth on the positive control, used as the threshold to define good phosphate sources.
TABLE 1.
PhoB and Pi transporter dependencies of 58 phosphorus-containing compounds screened for growth of S. aureusa
| Source |
|---|
| Nonphosphate sources |
| Triethyl phosphate |
| Hypophosphite |
| Adenosine-3′,5′-cyclic monophosphate |
| Guanosine-3′,5′-cyclic monophosphate |
| 2-Deoxy-d-glucose-6-phosphate |
| Cytidine-3′-monophosphate |
| Cytidine-3′,5′-cyclic monophosphate |
| Uridine-3′,5′-cyclic monophosphate |
| Phosphonoacetic acid |
| 2-Aminoethyl phosphonic acid |
| Methylene diphosphonic acid |
| Thymidine-3′,5′-cyclic monophosphate |
| Pi transporter-dependent, PhoB-independent phosphate sources |
| AMP |
| Guanosine-2′-monophosphate |
| GMP |
| Guanosine-2′,3′-cyclic monophosphate |
| d-Glucose-6-phosphate |
| CMP |
| Uridine-3′-monophosphate |
| UMP |
| Thymidine-3′-monophosphate |
| dTMP |
| PhoB- and Pi transporter-independent phosphate sources |
| Thiophosphate |
| Dithiophosphate |
| Carbamoyl phosphate |
| Guanosine-3′-monophosphate |
| Phosphoglycolic acid |
| Cysteamine S-phosphate |
| Phospho-l-arginine |
| O-Phospho-d-tyrosine |
| PhoB- and Pi transporter-dependent phosphate sources |
| Pyrophosphate |
| Trimetaphosphate |
| Tripolyphosphate |
| Adenosine-2′-monophosphate |
| Adenosine-3′-monophosphate |
| Adenosine-2′,3′-cyclic monophosphate |
| d,l-α-Glycerol phosphate (G3P) |
| β-Glycerol phosphate |
| d-2-Phosphoglyceric acid |
| d-3-Phosphoclyceric acid |
| Phosphoenolpyruvate |
| d-Glucose-1-phosphate |
| d-Glucosamine-6-phosphate |
| 6-Phosphogluconic acid |
| Cytidine-2′-monophosphate |
| Cytidine-2′,3′-cyclic monophosphate |
| d-Mannose-1-phosphate |
| d-Mannose-6-phosphate |
| O-Phospho-d-serine |
| O-Phospho-l-serine |
| O-Phospho-l-threonine |
| Uridine-2′-monophosphate |
| Uridine-2′,3′-cyclic monophosphate |
| O-Phospho-l-tyrosine |
| Phosphocreatine |
| Phosphorylcholine |
| O-Phosphorylethanolamine |
| Inositol hexaphosphate |
Categorization of phosphate sources was based on the endpoint growth of S. aureus PhoB- and/or Pi transporter-deficient strains normalized to wild-type growth on Pi. Nonphosphate sources were those on which wild-type S. aureus did not reach a terminal OD600 of >0.1. PhoB- and Pi transporter-dependent phosphate sources were those in which ΔphoB mutants reached a terminal OD600 of <70% of that of the wild type at either the permissive or nonpermissive pH. Pi transporter-dependent, PhoB-independent substrates were classified as those on which the growth of the ΔpstSCAB ΔpitA mutant was <70% of that of the wild type at the nonpermissive pH compared to growth at the permissive pH. PhoB- and Pi transporter-independent phosphate sources were those in which all mutants grew to ≥70% of the growth of the wild type at both pHs.
The use of G3P as a phosphate source induces the expression of alkaline phosphatase.
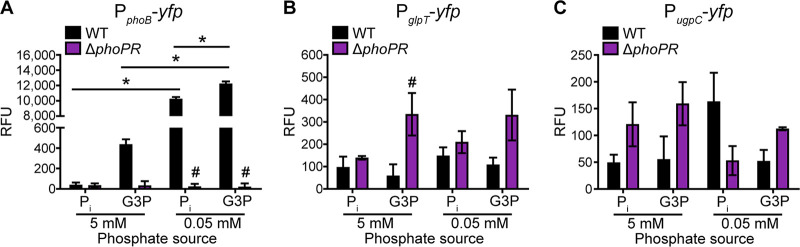
Among the organophosphates on which S. aureus grew robustly was G3P, a phospholipid and bacterial cell wall component which has previously been shown to act as a phosphate source for S. aureus (27). As G3P might represent a significant source of phosphate for S. aureus both during infection and in polymicrobial communities, how phosphate is obtained from this molecule was investigated in more detail. S. aureus contains three genes that may be involved in the use of G3P as a phosphate source: an alkaline phosphatase gene, phoB (NWMN_2526); a G3P/Pi antiporter gene, glpT (NWMN_0329); and a putative high-affinity G3P importer subunit gene, ugpC (NWMN_0151). Of note, while NWMN_0151 is annotated as the ATP-binding subunit UgpC of a Ugp system, it is encoded within a four-gene operon (NWMN_0151 to NWMN_0154) that is predicted to encode an ABC-type maltose importer (34). Initially, the expression of these three loci was assessed using yfp promoter fusions following growth with both excess (5 mM) and limiting (0.05 mM) Pi. The expression of phoB, but not glpT or the ugpC locus, was strongly induced in phosphate-limited medium (Fig. 2). These results indicate that only phoB responds strictly to phosphate availability. However, it was conceivable that glpT or the ugpC locus is induced only when Pi is limiting and that its substrate is present. When 0.05 mM G3P was provided as a phosphate source, phoB, but not glpT or the ugpC locus, was strongly induced relative to when either 5 mM Pi or G3P was provided (Fig. 2). Notably, when a high concentration (5 mM) of G3P was provided, phoB was expressed at a higher level than when a similar concentration of Pi was provided (Fig. 2A). This observation suggests that PhoB is potentially important for the use of G3P as a phosphate source in the absence of Pi. In contrast, the expression of neither glpT nor ugpC increased significantly when 5 mM G3P was the sole phosphate source instead of Pi (Fig. 2B and C). To further evaluate whether these genes are potentially involved in the use of G3P as an alternative phosphate source, their expression was also measured in a ΔphoPR mutant (17). The expression of phoB was entirely ablated in the ΔphoPR mutant, suggesting that PhoB is a member of the PhoPR-controlled phosphate starvation regulon (Fig. 2A). In contrast, the expression of glpT and ugpC did not decrease in the ΔphoPR mutant compared to the wild type (WT) (Fig. 2B and C). Cumulatively, these results suggest that PhoB, but not GlpT or the Ugp system, is responsive to phosphate availability.
FIG 2.
Expression of phoB responds to phosphate starvation and is PhoPR dependent. Shown are data for the expression of phoB (A), glpT (B), and ugpC (C) in wild-type and ΔphoPR S. aureus after 10 h of growth in PFM9 with 0.5% glucose supplemented with excess (5 mM) or limiting (0.05 mM) Pi or G3P as the sole phosphate source. Expression was assessed by measuring fluorescence using the PphoB-yfp, PglpT-yfp, and PugpC-yfp reporter plasmids. Strains were precultured overnight in TSB. *, P < 0.05 for the indicated comparison by two-way analysis of variance (ANOVA) with Sidak’s multiple-comparison test; #, P < 0.05 for the ΔphoPR mutant compared to the wild type in the same growth medium by two-way ANOVA with Tukey’s multiple-comparison test (n = 3). Error bars indicate standard errors of the means (SEM).
glpT and ugpC are induced by the absence of glucose.
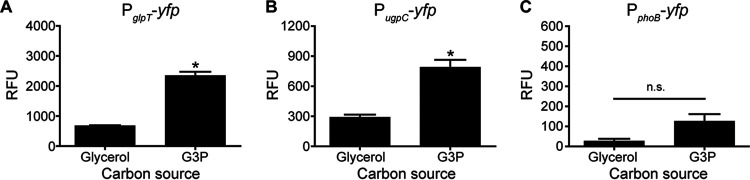
As glpT and ugpC expression was not phosphate responsive, whether they are regulated by carbon source availability was investigated. Their expression was assessed in Pi-replete (5 mM Pi) medium in which glycerol or G3P was provided as the carbon source. Differing from glucose-containing growth medium, when glycerol and G3P were provided as the sole carbon source, glpT and the ugpC locus were expressed at appreciable levels (Fig. 3A and B). The expression levels of glpT and the ugpC locus were ∼3- and 4-fold higher, respectively, in the presence of G3P than in the presence of glycerol (Fig. 3A and B). The expression of phoB was not substantially induced when glycerol or G3P was provided as the sole carbon source (Fig. 3C). As G3P did not induce the expression of glpT or the ugpC locus when glucose was present (Fig. 2B and C), this suggests that their expression is subject to carbon catabolite repression. These results suggest roles for GlpT and the UgpC locus in carbon metabolism rather than phosphate assimilation.
FIG 3.
Expression of glpT and ugpC responds to carbon availability. Shown are data for the expression of glpT (A), ugpC (B), and phoB (C) in wild-type S. aureus after 10 h of growth in PFM9 with 5 mM Pi and 5 mM either glycerol or G3P as the sole carbon source. Expression was assessed by measuring fluorescence using the PglpT-yfp, PugpC-yfp, and PphoB-yfp reporter plasmids. Strains were precultured overnight in TSB. *, P < 0.05 for G3P compared to glycerol by an unpaired t test (n = 3); n.s., not significant. Error bars indicate SEM.
PhoB is important for the use of G3P as a phosphate source.
As phoB is induced when G3P is present as a phosphate source, we next assessed whether PhoB is needed for growth when G3P is present as the sole phosphate source. The ΔphoB mutant grew similarly to the wild type when Pi was the phosphate source, regardless of the concentration (Fig. 4A and B). However, when excess (5 mM) G3P was the phosphate source, the ΔphoB mutant had a strong growth defect (Fig. 4C), suggesting that PhoB is important for utilizing G3P as a phosphate source. With a limiting amount (0.05 mM) of G3P in the medium, the ΔphoB mutant grew similarly to the wild type, presumably because the amount of phosphate that PhoB could liberate from G3P is relatively negligible compared to background growth under these conditions (Fig. 4D). The growth phenotype of the ΔphoB mutant in 5 mM G3P could be complemented by the ectopic expression of phoB (Fig. 4E). The loss of phoB in the community-acquired methicillin-resistant S. aureus (MRSA) strain USA300 JE2 also resulted in a growth defect when the strain was grown on G3P as a phosphate source (Fig. 4F). Cumulatively, these results suggest that PhoB is critical for the ability of S. aureus to use G3P as a Pi source.
FIG 4.
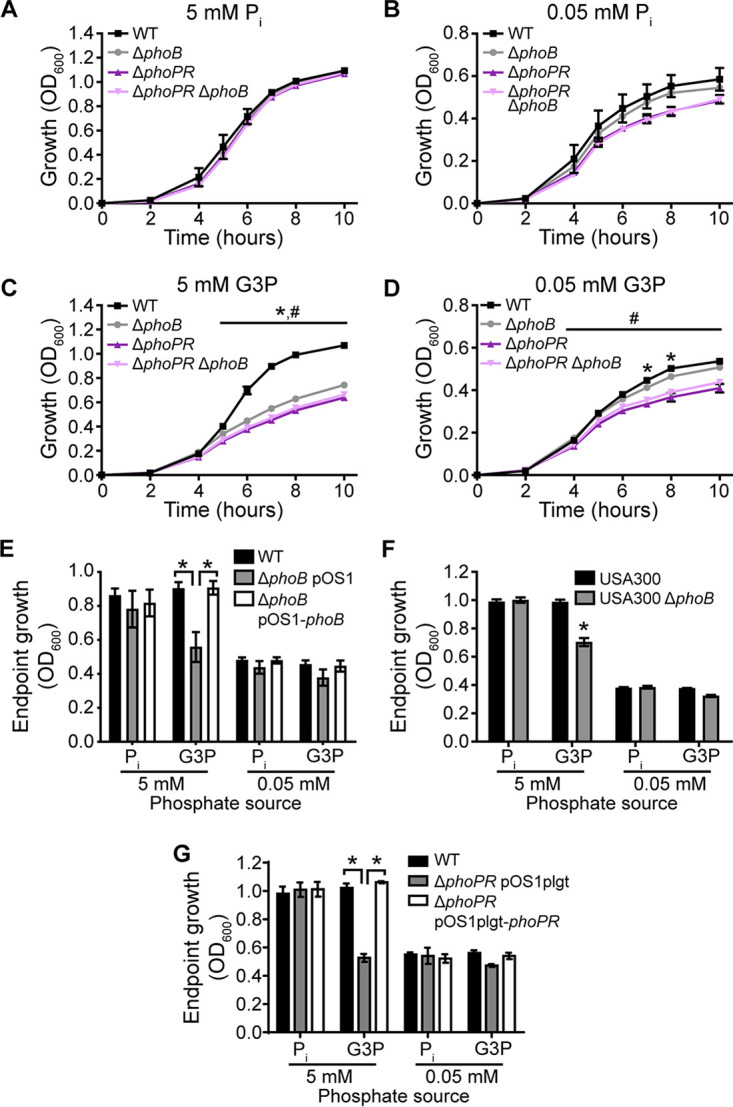
PhoB is important for the use of G3P as a phosphate source. (A to D) Growth of wild-type S. aureus (Newman) and the indicated mutants in PFM9 with 0.5% glucose supplemented with the phosphate source Pi (A and B) or G3P (C and D) in excess (A and C) or limiting (B and D) amounts. *, P < 0.05 for the ΔphoB mutant compared to the wild type; #, P < 0.05 for the ΔphoPR mutant compared to the ΔphoB mutant (by two-way ANOVA with Tukey’s multiple-comparison test) (n = 4). (E) Endpoint growth after 10 h of wild-type S. aureus with an empty vector (pOS1), the ΔphoB mutant with an empty vector, or the ΔphoB mutant with a vector containing phoB in PFM9 with 0.5% glucose and the indicated concentrations of Pi or G3P as the phosphate source. *, P < 0.05 for the indicated comparison by two-way ANOVA with Tukey’s multiple-comparison test (n = 3). (F) Endpoint growth after 10 h of wild-type S. aureus USA300 (JE2) or the ΔphoB mutant in PFM9 with 0.5% glucose supplemented with the indicated concentrations of Pi or G3P as the phosphate source. *, P < 0.05 for the phoB::erm strain compared to the wild type by two-way ANOVA with Sidak’s multiple-comparison test (n = 3). (G) Endpoint growth after 10 h of wild-type S. aureus with an empty vector (pOS1), the ΔphoPR mutant with an empty vector, or the ΔphoPR mutant with a vector containing phoPR in PFM9 with 0.5% glucose and the indicated concentrations of Pi or G3P as the phosphate source. *, P < 0.05 for the indicated comparison by two-way ANOVA with Tukey’s multiple-comparison test (n = 3). For panels A to G, strains were precultured overnight in TSB. Error bars indicate SEM.
PhoPR is necessary to utilize G3P as a phosphate source.
The finding that phoB is induced in a PhoPR-dependent manner when G3P is the sole phosphate source raises the possibility that other components of the Pho regulon could be necessary for the use of G3P as a phosphate source. To test this idea, the growth of a ΔphoPR mutant was compared to that of wild-type S. aureus and a ΔphoB mutant in excess (5 mM)- and limiting (0.05 mM)-phosphate media with either Pi or G3P provided as the sole phosphate source. Consistent with our previous findings (17), the ΔphoPR mutant grew similarly to the wild type in Pi-replete medium (Fig. 4A) but had a growth defect in Pi-limiting medium (Fig. 4B). When 5 mM G3P was provided as a phosphate source, the ΔphoPR mutant had a pronounced growth defect relative to the wild type (Fig. 4C). This defect was reversed by expressing phoPR from a plasmid (Fig. 4G). Notably, the growth defect of the ΔphoPR mutant was slightly but statistically significantly more pronounced than that of the ΔphoB mutant. The growth of the ΔphoPR mutant in the presence of 0.05 mM G3P was similar to that in 0.05 mM Pi but again was slightly worse than that of the ΔphoB mutant (Fig. 4B and D). A ΔphoPR ΔphoB double mutant grew similarly to the ΔphoPR mutant under all conditions tested (Fig. 4A to D). Cumulatively, these results suggest that the induction of PhoB by PhoPR substantially contributes to the growth of S. aureus on G3P but that other components of the Pho regulon, such as those involved in the phosphate-sparing response (17), may also be important.
The use of G3P by S. aureus as a phosphate source is Pi transporter dependent.
The observation that PhoB is necessary for the use of G3P as a phosphate source suggests that Pi would be liberated extracellularly, necessitating the subsequent use of Pi transporters. To test this model, a phenotypically Pi transport-deficient strain was used to determine if S. aureus requires both PhoB and Pi transporters in order to use G3P as a phosphate source. A ΔpstSCAB ΔpitA mutant grown at an acidic pH becomes phenotypically a triple Pi transporter mutant, as the remaining Pi transporter, NptA, functions poorly under this condition (16). We assessed the growth of the wild type and the ΔphoB, ΔpstSCAB ΔpitA, and ΔpstSCAB ΔpitA ΔphoB mutants at permissive and nonpermissive pHs (pH 7.4 and 6.4, respectively) with either Pi or G3P provided as the phosphate source. Consistent with previous results (16), the ΔpstSCAB ΔpitA double mutant grew similarly to the wild type on Pi at pH 7.4 but reached a lower terminal optical density at pH 6.4 (Fig. 5A and B). Similar to the observations in Fig. 4, the ΔphoB mutant grew as well as wild-type bacteria when Pi was provided as a phosphate source but had a severe growth defect when G3P was provided (Fig. 5A and C). The ΔpstSCAB ΔpitA mutant also had a growth defect relative to the wild type at the nonpermissive pH when G3P was provided as the phosphate source (Fig. 5D). The expression of phoB from a plasmid reversed the growth defect of the ΔphoB mutant when G3P was provided as the sole phosphate source in acidic medium (Fig. 5E). Similarly, the ectopic expression of pitA reversed the growth defect of the ΔpstSCAB ΔpitA mutant (Fig. 5F). To preserve the native regulation of pitA, the complementation construct also included the upstream gene pitR. These results indicate that Pi transporters are necessary for S. aureus to use G3P as a phosphate source. As expected, the ΔpstSCAB ΔpitA ΔphoB triple mutant grew worse than the wild type when G3P was the phosphate source or when Pi was provided at the nonpermissive pH (Fig. 5B to D). Unexpectedly, the ΔphoB mutant grew to a higher optical density when G3P was the phosphate source in medium at pH 6.4 than in medium at pH 7.4 (Fig. 5C and D). However, this pathway is Pi transporter dependent, as the increased growth of the ΔphoB mutant is abolished in the ΔpstSCAB ΔpitA ΔphoB strain (Fig. 5D). This observation suggests an increase in the presence of Pi produced independently of PhoB in acidic environments; regardless, Pi transporters would still be necessary to take up the liberated Pi. Cumulatively, these observations establish that PhoB and Pi transporters are necessary for S. aureus to use G3P as a phosphate source.
FIG 5.
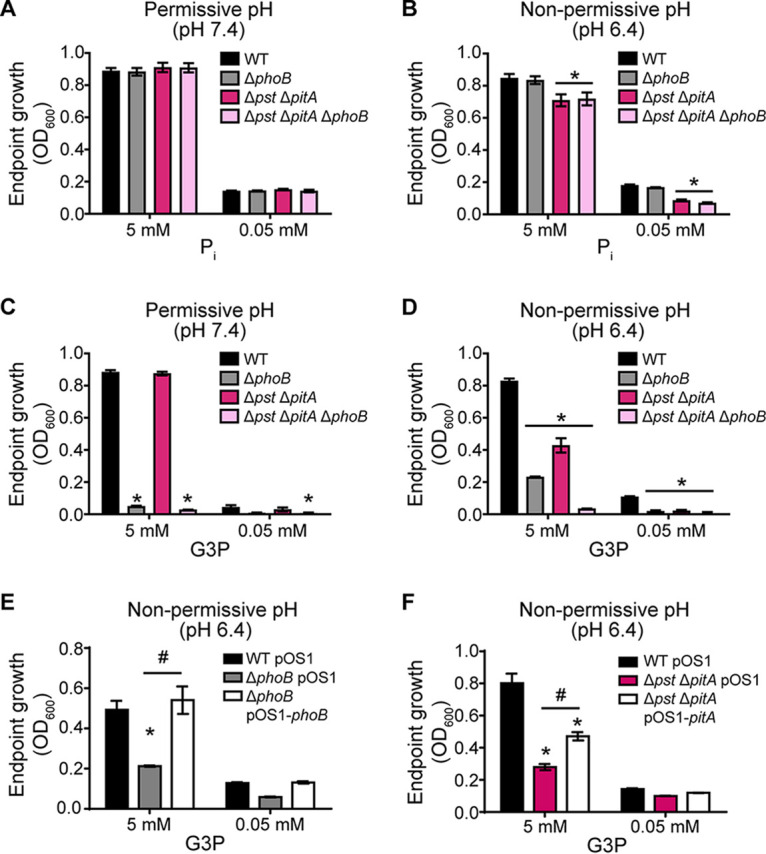
Use of G3P as a phosphate source by S. aureus is PhoB and Pi transporter dependent. Shown are data for endpoint growth after 10 h of wild-type S. aureus and the indicated mutants (Δpst indicates ΔpstSCAB) in PFM9 with 0.5% glucose supplemented with the indicated concentrations of Pi (A and B) or G3P (C to F) as the phosphate source and buffered to pH 7.4 (A and C) or pH 6.4 (B, D, and F). Strains were precultured overnight in a Pi-limiting medium at the permissive pH so that the cells were equivalently phosphate limited at the start of the growth assay. *, P < 0.05 compared to the wild type by two-way ANOVA with Tukey’s multiple-comparison test; #, P < 0.05 for the indicated comparison by two-way ANOVA with Tukey’s multiple-comparison test (n ≥ 3). Error bars indicate SEM.
S. aureus relies on PhoB and Pi transporters to obtain phosphate from most alternative phosphate sources.
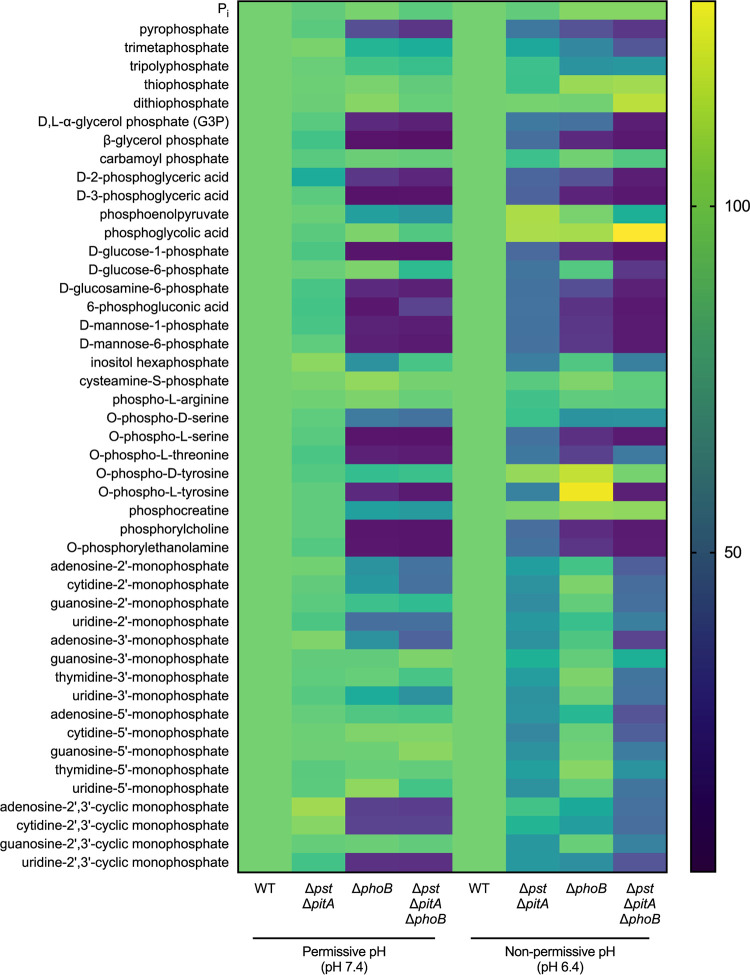
Given the finding that S. aureus relies on PhoB and Pi transporters to use G3P, we investigated if this was a general strategy employed by S. aureus or specific to G3P. To address this question, we evaluated the ability of ΔphoB, ΔpstSCAB ΔpitA, and ΔpstSCAB ΔpitA ΔphoB mutants to grow on the same library of phosphorus-containing compounds that was previously screened with wild-type S. aureus. In addition to screening the library at a pH of 7.4, it was also screened at a pH of 6.4, where the ΔpstSCAB ΔpitA and ΔpstSCAB ΔpitA ΔphoB mutants would have a reduced ability to utilize Pi liberated by PhoB. Changing the pH of the growth medium did not change which phosphorous-containing compounds wild-type S. aureus could use as a phosphate source (Fig. S2).
The ΔphoB, ΔpstSCAB ΔpitA, and ΔpstSCAB ΔpitA ΔphoB mutants were unable to grow on the 12 compounds in the library that were previously found to be unable to support the growth of wild-type S. aureus (Fig. S1 and S3). Based on the endpoint growth yield of the mutants relative to the wild type on the 46 phosphocompounds that can serve as phosphate sources for S. aureus, three classes of compounds were identified: (i) PhoB- and Pi transporter-dependent phosphate sources, (ii) PhoB- and Pi transporter-independent phosphate sources, and (iii) PhoB-independent but Pi transporter-dependent phosphate sources (Fig. 6; Table 1). Consistent with a role in liberating Pi from organophosphates, all PhoB-dependent compounds were also Pi transporter dependent. More than 60% (28/46) of the compounds that S. aureus can use as a phosphate source were PhoB and Pi transporter dependent (Fig. 6; Table 1). Validating the screen, G3P was identified as one of the PhoB- and Pi transporter-dependent substrates (Fig. 6; Table 1). Other substrates in this group included a range of phosphorylated sugars, nucleotides, and amino acids, suggesting that S. aureus relies on its inducible alkaline phosphatase and Pi importers to acquire phosphate from a wide variety of sources. Approximately 17% (8/46) of the compounds could be used independently of PhoB and Pi transporters, suggesting that S. aureus may express dedicated transporters to import these molecules intact. These eight substrates included several phosphorylated amino acids (cysteamine S-phosphate, phospho-l-arginine, and O-phospho-d-tyrosine), one nucleotide (guanosine-3′-monophosphate), and other organophosphate (carbamoyl phosphate and phosphoglycolic acid) and inorganic phosphate (thiophosphate and dithiophosphate) compounds (Fig. 6; Table 1). The remaining 22% (10/46) of the compounds that support staphylococcal growth, nine of which were nucleotides, are PhoB independent but Pi transporter dependent (Fig. 6; Table 1). In total, 38 of the 46 compounds that S. aureus could use required the presence of a functional Pi importer. Cumulatively, the results of the screen suggest that the liberation of Pi and the subsequent import of Pi are the preferred mechanisms for obtaining phosphate by S. aureus.
FIG 6.
S. aureus requires its alkaline phosphatase PhoB and Pi transporters to acquire phosphate from most alternative phosphate sources. Forty-six phosphorus-containing compounds were screened for their ability to support the growth of S. aureus wild-type, ΔphoB, ΔpstSCAB ΔpitA, and ΔpstSCAB ΔpitA ΔphoB strains at pH 7.4 and pH 6.4 (Δpst indicates ΔpstSCAB). Strains were precultured overnight in a Pi-limiting medium at the permissive pH so that the cells were equivalently phosphate limited at the start of the growth assay. Growth was measured by assessing the OD600 for 10 h. For each phosphate source, endpoint growth values were then normalized to the growth of the wild type (WT) within each pH. Percent growth relative to the WT is represented as a heat map. The screen was performed in biological triplicate.
DISCUSSION
Pathogens, including S. aureus, must obtain essential inorganic nutrients from a variety of environments within the host. One of these nutrients, phosphate, can be taken up via dedicated importers for Pi monomers or transported as part of organophosphate compounds (9, 10). While the preferred source of phosphate for bacteria is Pi, the requirement of Pi starvation-responsive two-component systems for the virulence of many pathogens indicates that Pi is not freely available in all host environments (1, 17, 22, 35–38). Given that S. aureus is among the pathogens that experience Pi limitation in vivo, we screened a library of 58 phosphorus-containing compounds and identified 46 phosphate sources for S. aureus, including phosphorylated sugars, nucleotides, and amino acids, as well as several inorganic phosphate sources that can be used to provide phosphate. Subsequent experiments revealed that the majority of these compounds are used as phosphate sources in a PhoB- and Pi transporter-dependent manner. These findings lead to a model in which S. aureus preferentially uses PhoB in combination with Pi transporters to extract phosphate from a variety of organophosphates (Fig. 7).
FIG 7.
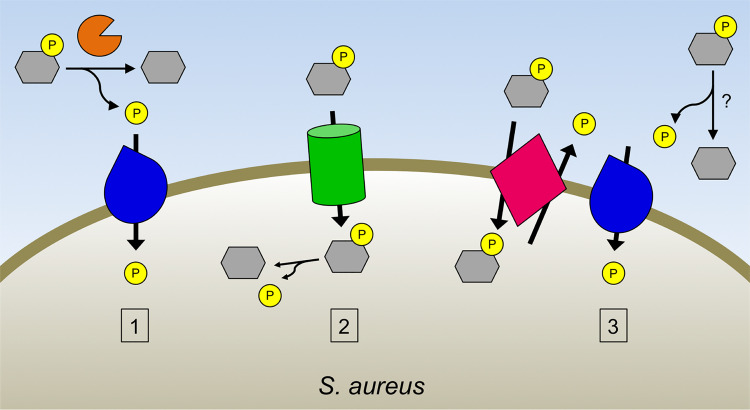
Model of phosphate acquisition from alternative phosphate sources by S. aureus. S. aureus can acquire phosphate from a variety of phosphosubstrates during phosphate starvation through three pathways: the alkaline phosphatase PhoB (orange) cleaves Pi (yellow) from phosphorylated compounds (gray) extracellularly, with the liberated Pi subsequently imported by Pi importers (blue) (pathway 1); phosphorylated molecules can be imported intact through phosphosubstrate importers (green), with Pi subsequently released intracellularly (pathway 2); and extracellular Pi is generated independently of PhoB, such as by phosphosubstrate/Pi antiporters (pink) or by other phosphatases, which is then imported through Pi importers (pathway 3).
Among the phosphate sources for S. aureus identified in the screen was G3P, a compound that has been studied as an alternative phosphate source for E. coli (39). While G3P is known to promote the growth of S. aureus as a phosphate and carbon source (27), the pathways through which it is used for these purposes had not been determined. Here, we reveal that to use G3P as a phosphate source, S. aureus must first process G3P extracellularly with its inducible alkaline phosphatase, PhoB, and then import Pi through Pi transporters. Differing from E. coli, our results indicate that S. aureus lacks a phosphate starvation-regulated G3P importer to transport G3P intact. Based on its expression pattern, the annotated staphylococcal ugpC gene does not appear to encode part of a Ugp-like transporter but instead is likely part of an operon involved in carbohydrate uptake, as previously suggested (34). The Pho regulon of Bacillus subtilis also lacks a Ugp system, but compared to E. coli, which possesses one inducible alkaline phosphatase, B. subtilis contains an expanded repertoire of three phosphatases (24). These observations cumulatively suggest that some organisms, including B. subtilis and S. aureus, may more strongly rely on phosphatases and subsequent Pi import to acquire phosphate. Of note, while both E. coli and S. aureus contain PstSCAB and PitA Pi transporters, S. aureus also possesses an additional Pi importer, NptA, which expands the environments in which it can import Pi (16). E. coli does have an NptA homolog; however, a pstSCAB pitA mutant of E. coli cannot grow on Pi, indicating that E. coli NptA does not function as a Pi importer (39, 40). Therefore, although S. aureus lacks a dedicated transporter for a common organophosphate source, it may compensate for this by an increased biochemical capacity for Pi import.
A screen of phosphate sources revealed that, similar to the pathway through which S. aureus uses G3P as a phosphate source, this pathogen relies on the enzymatic function of PhoB and then its Pi importers to acquire phosphate from most of the phosphocompounds that support growth. Given the importance of PhoB in using alternative phosphate sources, this finding suggests that PhoB may represent one of the PhoPR-regulated factors other than Pi transporters that are important for systemic staphylococcal infection (17). The screen also revealed that 8 compounds, including amino acids, a nucleotide, and other organic and inorganic phosphocompounds, can be phosphate sources independent of Pi transporters, suggesting that S. aureus may express dedicated importers for their uptake. The observation that nucleotides and phosphorylated amino acids can serve as phosphate sources in this manner was unexpected because aside from G3P and a few hexose-phosphates, phosphorylated compounds are not thought to be able to cross the cytoplasmic membrane (9). Continued genetic and biochemical analyses will be required to appreciate how S. aureus obtains phosphate from these molecules.
Interestingly, S. aureus can also use 10 compounds, including 9 nucleotides and hexose-6-phosphate, as phosphate sources in a PhoB-independent yet Pi transporter-dependent manner. We envision S. aureus having two mechanisms for using such substrates: (i) a substrate/Pi antiporter that would release Pi, subsequently allowing for the reuptake of Pi via the Pi transporters and, ultimately, net phosphate acquisition, or (ii) another phosphatase to act on the substrate. Correspondingly, one of the compounds in this category, glucose-6-phosphate, can be imported by S. aureus via the substrate-inducible hexose-6-phosphate/Pi antiporter UhpT (41). However, given that an additional export-and-import step must be taken for a net gain of one phosphate, we anticipate that such pathways of phosphosubstrate utilization are artificial. While nonphosphorylated nucleosides can be transported across the membrane, nucleotide transporters have not been described (42–44). Similar to B. subtilis, Vibrio cholerae encodes three inducible phosphatases, in addition to a PhoB homolog, that promote phosphate acquisition from molecules, including nucleotides (44). The observation that the S. aureus ΔphoB mutant grows better on G3P as a phosphate source at pH 6.4 than at pH 7.4 (Fig. 5B and D) suggests that there may be an additional phosphatase deployed under this condition that can act on G3P. Intriguingly, a secreted tyrosine phosphatase, PtpA, was recently found to be required for the virulence of S. aureus (45). While PtpA interacts with the host protein coronin-1A, presumably to modulate signaling pathways during infection, perhaps Pi liberated by PtpA and potentially other host-targeting phosphatases can also serve as a phosphate source.
Overall, this study provides molecular insight into the range of phosphate sources available to S. aureus. Detailed analysis of the use of G3P as a phosphate source and subsequent screening of potential phosphate sources underscore the importance of PhoB and the Pi acquisition systems in S. aureus. In addition, the work here adds to a growing body of literature that reveals key differences in phosphate acquisition and metabolism in pathogenic organisms such as S. aureus and established models built from the study of nonpathogenic organisms such as E. coli and B. subtilis.
MATERIALS AND METHODS
Bacterial strains and cloning.
S. aureus strain Newman and derivatives were used for all experiments unless otherwise noted. S. aureus was cultivated in tryptic soy broth (TSB) and on tryptic soy agar (TSA) plates and stored in brain heart infusion (BHI) broth containing 30% glycerol at −80°C. E. coli, used for subcloning, was grown in LB and on LB agar plates and stored in LB containing 30% glycerol at −80°C. Both species were grown at 37°C. As needed to maintain plasmids, 100 μg/ml ampicillin or 10 μg/ml chloramphenicol was added to growth media.
The phoB::erm allele was obtained from the Nebraska Transposon Mutant Library (NTML) (46), and the antibiotic resistance cassette was swapped using a protocol provided in the NTML toolkit (47). Mutant alleles were subsequently transduced into the desired background via Phi85 phage (Table 2). For complementation studies, phoB and pitA were cloned into the pOS1 vector (48) under the control of their native promoter using the indicated primers (Tables 3 and 4). For expression studies, the promoters of phoB, glpT, and ugpC were cloned into the pAH5 vector using the indicated primers (Tables 3 and 4) (49). All PCR-generated constructs were verified by sequencing.
TABLE 2.
Strains used in this study
| Genotype or name | Description | Reference |
|---|---|---|
| Newman | Wild-type strain of Staphylococcus aureus | |
| ΔphoB | phoB::erm allele from the NTMLa transduced into Newman | This study |
| ΔphoPR | Clean deletion of phoPR in Newman | 17 |
| ΔphoPR ΔphoB | phoB::erm antibiotic resistance cassette swapped to kan and transduced into ΔphoPR | This study |
| ΔpstSCAB ΔpitA | pitA::erm allele from the NTML transduced into the strain with a clean deletion of pstSCAB | 16 |
| ΔpstSCAB ΔpitA ΔphoB | phoB::erm antibiotic resistance cassette swapped to kan and transduced into the ΔpstSCAB ΔpitA mutant | This study |
| USA300 JE2 | Wild-type strain of Staphylococcus aureus | 46 |
| ΔphoB (USA300 JE2) | phoB::erm mutant from the NTML | 46 |
NTML, Nebraska Transposon Mutant Library (46).
TABLE 3.
Primers used in this study
| Primer name | Sequence |
|---|---|
| phoB comp F | GGCCGAATTCCTATAATCTTCTTCCTTCAATTGAATTATCC |
| phoB comp R | GCGCGGATTCGAACACCTTGTTATAGTGCTCG |
| phoB prom F | CGCGCTGCAGCTATAATCTTCTTCCTTCAATTGAATTATCC |
| phoB prom R | CGCGGGTACCAAGACATCCTCCTGAGTATATGAC |
| pitA comp F | GATTGTACTGAGAGTGCACCATAATTTTTGAAATTAATATCAGTACACTAAAATTATAC |
| pitA comp R | GCTAGCTTGGCTGCAGGTCGACGTTAGAAAAATAAGTTAAGTATATAGAATAGTAAAC |
| glpT prom F | CGCGCTGCAGACTATCCCTCCTATTAGTATAAATTTTATACCAGC |
| glpT prom R | CGCGGGTACCAAAATCCTCCTTAATATGTATTTATATGCATTTTGTG |
| ugpC prom F | CGCGCTGCAGTTATAGAAGGGTGCCCGCAGTC |
| ugpC prom R | CGCGGGTACCGTTATGCCTCCCATACTTTGTTTACAGTTTGATTG |
TABLE 4.
Plasmids used in this study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pOS1 | pOS1 (empty vector) | 48 |
| pOS1-phoB | pOS1 containing the phoB CDS under the control of the native promoter | This study |
| pOS1-pitA | pOS1 containing the pitRA locus under the control of the native promoter | This study |
| pOS1 Plgt | pOS1 (empty vector) with the lgt promoter | 48 |
| pOS1 Plgt-phoPR | pOS1 Plgt containing the phoPR CDS | 17 |
| pAH5::Pempty-yfp | pAH5 with no promoter driving yfp expression | 50 |
| pAH5::PphoB-yfp | pAH5 with the phoB promoter driving yfp expression | This study |
| pAH5::PglpT-yfp | pAH5 with the glpT promoter driving yfp expression | This study |
| pAH5::PugpC-yfp | pAH5 with the ugpC promoter driving yfp expression | This study |
CDS, coding DNA sequence.
Growth medium, growth assays, and expression analysis.
Phosphate-free M9-based medium (PFM9) was used for growth assays and was described previously (16). PFM9 was buffered with 70 mM either HEPES (pH 7.4) or morpholinepropanesulfonic acid (MOPS) (pH 6.4). Pi source stocks were made by mixing NaH2PO4 and Na2HPO4 and adjusting the mixture to the desired pH. G3P (d,l-α-glycerol phosphate, purchased from Sigma [catalog no. 17766]) was resuspended in water at 20 mM and filter sterilized. For phosphate limitation growth assays, bacteria were inoculated into TSB and grown overnight on a roller drum for 16 to 18 h. Where indicated, to prestarve strains for phosphate, bacteria were inoculated into 5 ml PFM9 plus 70 mM HEPES (pH 7.4) supplemented with 0.5% glucose and 500 μM Pi and grown overnight on a roller drum for 16 to 18 h. Cultures grown overnight were inoculated 1:100 into a 96-well round-bottom plate containing 100 μl/well PFM9 containing the indicated phosphate and carbon sources. Plates were incubated at 37°C with shaking at 180 rpm. Bacterial growth was monitored by measuring the optical density at 600 nm (OD600). Expression (RFU [relative fluorescence units]) was determined by measuring fluorescence (excitation/emission wavelengths of 505/535 nm), normalizing the values to the OD600, and then subtracting the RFU of empty vector controls.
Identification of phosphate sources for S. aureus.
Phenotype MicroArray PM4A microplate phosphorus and sulfur source plates (Biolog, Inc., Haywood, CA) were screened to identify compounds that can be used as phosphate sources by S. aureus. For these assays, bacteria were inoculated into 5 ml PFM9 plus 0.5% glucose and 70 mM HEPES (pH 7.4) supplemented with 500 μM Pi and grown on a roller drum for 16 to 18 h. Cultures grown overnight were diluted 1:100 into PFM9 plus 0.5% glucose with either 70 mM HEPES (pH 7.4) or 70 mM MOPS (pH 6.4). The Biolog plates were then inoculated with 100 μl of this mixture. The plates were incubated at 37°C with shaking at 180 rpm. Bacterial growth was monitored by measuring the OD600 for 10 h. The screen was performed in biological triplicate (on three separate days).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Vallee Foundation and National Institutes of Health grants (R01 AI118880 and R21 AI149115) to T.E.K.-F. This work was also supported in part by a James R. Beck graduate research fellowship in microbiology awarded to J.L.K.
The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication. This work does not represent the views of the Vallee Foundation or the National Institutes of Health.
J.L.K., A.J.L.M., K.M.G., J.N.R., and T.E.K.-F. performed the research and analyzed the data. J.L.K., A.J.L.M., J.N.R., and T.E.K.-F. designed the experiments and wrote the paper.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lamarche MG, Wanner BL, Crepin S, Harel J. 2008. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev 32:461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- 2.Valdivia RH, Falkow S. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 3.Burall LS, Harro JM, Li X, Lockatell CV, Himpsl SD, Hebel JR, Johnson DE, Mobley HL. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect Immun 72:2922–2938. doi: 10.1128/iai.72.5.2922-2938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lathem WW, Crosby SD, Miller VL, Goldman WE. 2005. Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc Natl Acad Sci U S A 102:17786–17791. doi: 10.1073/pnas.0506840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin AJ, Miller VL. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol Microbiol 32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 6.Merrell DS, Hava DL, Camilli A. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol 43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 7.Hava DL, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- 8.Peirs P, Lefevre P, Boarbi S, Wang XM, Denis O, Braibant M, Pethe K, Locht C, Huygen K, Content J. 2005. Mycobacterium tuberculosis with disruption in genes encoding the phosphate binding proteins PstS1 and PstS2 is deficient in phosphate uptake and demonstrates reduced in vivo virulence. Infect Immun 73:1898–1902. doi: 10.1128/IAI.73.3.1898-1902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wanner BL. 1996. Phosphorus assimilation and control of the phosphate regulon, p 1357–1381. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 10.Rao NN, Torriani A. 1990. Molecular aspects of phosphate transport in Escherichia coli. Mol Microbiol 4:1083–1090. doi: 10.1111/j.1365-2958.1990.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 11.Devine KM. 2018. Activation of the PhoPR-mediated response to phosphate limitation is regulated by wall teichoic acid metabolism in Bacillus subtilis. Front Microbiol 9:2678. doi: 10.3389/fmicb.2018.02678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core Surveillance MRSA Investigators. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 14.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 15.Balasubramanian D, Harper L, Shopsin B, Torres VJ. 2017. Staphylococcus aureus pathogenesis in diverse host environments. Pathog Dis 75:ftx005. doi: 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelliher JL, Radin JN, Grim KP, Parraga Solorzano PK, Degnan PH, Kehl-Fie TE. 2018. Acquisition of the phosphate transporter NptA enhances Staphylococcus aureus pathogenesis by improving phosphate uptake in divergent environments. Infect Immun 86:e00631-17. doi: 10.1128/IAI.00631-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelliher JL, Radin JN, Kehl-Fie TE. 2018. PhoPR contributes to Staphylococcus aureus growth during phosphate starvation and pathogenesis in an environment-specific manner. Infect Immun 86:e00371-18. doi: 10.1128/IAI.00371-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelliher JL, Brazel EB, Radin JN, Joya ES, Parraga Solorzano PK, Neville SL, McDevitt CA, Kehl-Fie TE. 2020. Disruption of phosphate homeostasis sensitizes Staphylococcus aureus to nutritional immunity. Infect Immun 88:e00102-20. doi: 10.1128/IAI.00102-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mechler L, Bonetti EJ, Reichert S, Flotenmeyer M, Schrenzel J, Bertram R, Francois P, Gotz F. 2016. Daptomycin tolerance in the Staphylococcus aureus pitA6 mutant is due to upregulation of the dlt operon. Antimicrob Agents Chemother 60:2684–2691. doi: 10.1128/AAC.03022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mechler L, Herbig A, Paprotka K, Fraunholz M, Nieselt K, Bertram R. 2015. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 59:5366–5376. doi: 10.1128/AAC.00643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chekabab SM, Harel J, Dozois CM. 2014. Interplay between genetic regulation of phosphate homeostasis and bacterial virulence. Virulence 5:786–793. doi: 10.4161/viru.29307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C, Huang TW, Wen SY, Chang CY, Tsai SF, Wu WF, Chang CH. 2012. Genome-wide PhoB binding and gene expression profiles reveal the hierarchical gene regulatory network of phosphate starvation in Escherichia coli. PLoS One 7:e47314. doi: 10.1371/journal.pone.0047314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allenby NE, O’Connor N, Pragai Z, Ward AC, Wipat A, Harwood CR. 2005. Genome-wide transcriptional analysis of the phosphate starvation stimulon of Bacillus subtilis. J Bacteriol 187:8063–8080. doi: 10.1128/JB.187.23.8063-8080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allenby NE, Laing E, Bucca G, Kierzek AM, Smith CP. 2012. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic Acids Res 40:9543–9556. doi: 10.1093/nar/gks766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torriani A. 1960. Influence of inorganic phosphate in the formation of phosphatases by Escherichia coli. Biochim Biophys Acta 38:460–469. doi: 10.1016/0006-3002(60)91281-6. [DOI] [PubMed] [Google Scholar]
- 27.Jorge AM, Schneider J, Unsleber S, Gohring N, Mayer C, Peschel A. 2017. Utilization of glycerophosphodiesters by Staphylococcus aureus. Mol Microbiol 103:229–241. doi: 10.1111/mmi.13552. [DOI] [PubMed] [Google Scholar]
- 28.Jorge AM, Schneider J, Unsleber S, Xia G, Mayer C, Peschel A. 2018. Staphylococcus aureus counters phosphate limitation by scavenging wall teichoic acids from other staphylococci via the teichoicase GlpQ. J Biol Chem 293:14916–14924. doi: 10.1074/jbc.RA118.004584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A. 2018. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown S, Santa Maria JP Jr, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okabayashi K, Futai M, Mizuno D. 1974. Localization of acid and alkaline phosphatases in Staphylococcus aureus. Jpn J Microbiol 18:287–294. doi: 10.1111/j.1348-0421.1974.tb00812.x. [DOI] [PubMed] [Google Scholar]
- 32.Schweizer H, Argast M, Boos W. 1982. Characteristics of a binding protein-dependent transport system for sn-glycerol-3-phosphate in Escherichia coli that is part of the pho regulon. J Bacteriol 150:1154–1163. doi: 10.1128/JB.150.3.1154-1163.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argast M, Boos W. 1980. Co-regulation in Escherichia coli of a novel transport system for sn-glycerol-3-phosphate and outer membrane protein Ic (e, E) with alkaline phosphatase and phosphate-binding protein. J Bacteriol 143:142–150. doi: 10.1128/JB.143.1.142-150.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitko NP, Grosser MR, Khatri D, Lance TR, Richardson AR. 2016. Expanded glucose import capability affords Staphylococcus aureus optimized glycolytic flux during infection. mBio 7:e00296-16. doi: 10.1128/mBio.00296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakimoto S, Nakayama-Imaohji H, Ichimura M, Morita H, Hirakawa H, Hayashi T, Yasutomo K, Kuwahara T. 2013. PhoB regulates the survival of Bacteroides fragilis in peritoneal abscesses. PLoS One 8:e53829. doi: 10.1371/journal.pone.0053829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Kruger WM, Humphreys S, Ketley JM. 1999. A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology 145:2463–2475. doi: 10.1099/00221287-145-9-2463. [DOI] [PubMed] [Google Scholar]
- 37.Parish T, Smith DA, Roberts G, Betts J, Stoker NG. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149:1423–1435. doi: 10.1099/mic.0.26245-0. [DOI] [PubMed] [Google Scholar]
- 38.Chekabab SM, Jubelin G, Dozois CM, Harel J. 2014. PhoB activates Escherichia coli O157:H7 virulence factors in response to inorganic phosphate limitation. PLoS One 9:e94285. doi: 10.1371/journal.pone.0094285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sprague GF Jr, Bell RM, Cronan JE Jr. 1975. A mutant of Escherichia coli auxotrophic for organic phosphates: evidence for two defects in inorganic phosphate transport. Mol Gen Genet 143:71–77. doi: 10.1007/BF00269422. [DOI] [PubMed] [Google Scholar]
- 40.Motomura K, Hirota R, Ohnaka N, Okada M, Ikeda T, Morohoshi T, Ohtake H, Kuroda A. 2011. Overproduction of YjbB reduces the level of polyphosphate in Escherichia coli: a hypothetical role of YjbB in phosphate export and polyphosphate accumulation. FEMS Microbiol Lett 320:25–32. doi: 10.1111/j.1574-6968.2011.02285.x. [DOI] [PubMed] [Google Scholar]
- 41.Park JY, Kim JW, Moon BY, Lee J, Fortin YJ, Austin FW, Yang SJ, Seo KS. 2015. Characterization of a novel two-component regulatory system, HptRS, the regulator for the hexose phosphate transport system in Staphylococcus aureus. Infect Immun 83:1620–1628. doi: 10.1128/IAI.03109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe K, Tomioka S, Tanimura K, Oku H, Isoi K. 2011. Uptake of AMP, ADP, and ATP in Escherichia coli W. Biosci Biotechnol Biochem 75:7–12. doi: 10.1271/bbb.100063. [DOI] [PubMed] [Google Scholar]
- 43.Gumpenberger T, Vorkapic D, Zingl FG, Pressler K, Lackner S, Seper A, Reidl J, Schild S. 2016. Nucleoside uptake in Vibrio cholerae and its role in the transition fitness from host to environment. Mol Microbiol 99:470–483. doi: 10.1111/mmi.13143. [DOI] [PubMed] [Google Scholar]
- 44.McDonough E, Kamp H, Camilli A. 2016. Vibrio cholerae phosphatases required for the utilization of nucleotides and extracellular DNA as phosphate sources. Mol Microbiol 99:453–469. doi: 10.1111/mmi.13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gannoun-Zaki L, Patzold L, Huc-Brandt S, Baronian G, Elhawy MI, Gaupp R, Martin M, Blanc-Potard AB, Letourneur F, Bischoff M, Molle V. 2018. PtpA, a secreted tyrosine phosphatase from Staphylococcus aureus, contributes to virulence and interacts with coronin-1A during infection. J Biol Chem 293:15569–15580. doi: 10.1074/jbc.RA118.003555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneewind O, Mihaylova-Petkov D, Model P. 1993. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J 12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. 2009. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods 77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia YM, Barwinska-Sendra A, Tarrant E, Skaar EP, Waldron KJ, Kehl-Fie TE. 2017. A superoxide dismutase capable of functioning with iron or manganese promotes the resistance of Staphylococcus aureus to calprotectin and nutritional immunity. PLoS Pathog 13:e1006125. doi: 10.1371/journal.ppat.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.