The transcription of C. difficile toxins TcdA and TcdB is directed by the sigma factor TcdR. TcdC has been proposed to be an anti-sigma factor. The activity of TcdC has been mapped to its C terminus, and the N terminus serves as the membrane anchor. Acting as an anti-sigma factor requires a cytoplasmic localization of the C terminus of TcdC. Using cysteine accessibility analysis and a HiBiT-based system, we show that the TcdC C terminus is located extracellularly, which is incompatible with its role as anti-sigma factor. Furthermore, mutating a cysteine residue at position 51 resulted in the release of TcdC from the bacteria. The codon-optimized version of the HiBiT (HiBiTopt) extracellular detection system is a valuable tool for topology determination of membrane proteins, increasing the range of systems available to tackle important aspects of C. difficile development.
KEYWORDS: tcdC, toxins, membrane, Clostridioides difficile, toxin regulation, HiBiTopt, Clostridium difficile
ABSTRACT
Clostridioides difficile is an anaerobic Gram-positive bacterium that can produce the large clostridial toxins toxin A and toxin B, encoded within the pathogenicity locus (PaLoc). The PaLoc also encodes the sigma factor TcdR, which positively regulates toxin gene expression, and TcdC, which is a putative negative regulator of toxin expression. TcdC is proposed to be an anti-sigma factor; however, several studies failed to show an association between the tcdC genotype and toxin production. Consequently, the TcdC function is not yet fully understood. Previous studies have characterized TcdC as a membrane-associated protein with the ability to bind G-quadruplex structures. The binding to the DNA secondary structures is mediated through the oligonucleotide/oligosaccharide binding fold (OB-fold) domain present at the C terminus of the protein. This domain was previously also proposed to be responsible for the inhibitory effect on toxin gene expression, implicating a cytoplasmic localization of the OB-fold. In this study, we aimed to obtain topological information on the C terminus of TcdC and demonstrate that the C terminus of TcdC is located extracellularly. In addition, we show that the membrane association of TcdC is dependent on a membrane-proximal cysteine residue and that mutating this residue results in the release of TcdC from the bacterial cell. The extracellular location of TcdC is not compatible with the direct binding of the OB-fold domain to intracellular nucleic acid or protein targets and suggests a mechanism of action that is different from that of the characterized anti-sigma factors.
IMPORTANCE The transcription of C. difficile toxins TcdA and TcdB is directed by the sigma factor TcdR. TcdC has been proposed to be an anti-sigma factor. The activity of TcdC has been mapped to its C terminus, and the N terminus serves as the membrane anchor. Acting as an anti-sigma factor requires a cytoplasmic localization of the C terminus of TcdC. Using cysteine accessibility analysis and a HiBiT-based system, we show that the TcdC C terminus is located extracellularly, which is incompatible with its role as anti-sigma factor. Furthermore, mutating a cysteine residue at position 51 resulted in the release of TcdC from the bacteria. The codon-optimized version of the HiBiT (HiBiTopt) extracellular detection system is a valuable tool for topology determination of membrane proteins, increasing the range of systems available to tackle important aspects of C. difficile development.
INTRODUCTION
Clostridioides difficile (Clostridium difficile) (1) is an opportunistic pathogen that can cause disease in individuals with dysbiosis of the gut microbiota (2). The Clostridium difficile infection (CDI) incidence has increased worldwide, and CDI leads to a broad spectrum of symptoms, from mild diarrhea to toxic megacolon and even death (3).
Several factors contribute to the progression and the severity of CDI (2, 4). C. difficile is a Gram-positive anaerobic bacterium that has the ability to form spores, which allows for dissemination and colonization (2). The main virulence factors are the large clostridial toxins, which induce damage to the epithelial cells and which lead to an inflammatory response that underlies the symptoms of CDI (2, 3, 5).
C. difficile strains have been found to produce up to three toxins: toxin A (TcdA), toxin B (TcdB), and binary toxin (CDT) (5, 6). Toxins A and B are encoded by the genes tcdA and tcdB, respectively, located on a 19.6-kb chromosomal region termed the pathogenicity locus (PaLoc) (7). TcdA and TcdB are glucosyltransferases. and once they are translocated to the cytosol of the intestinal epithelial cells, they start a cascade of events that can eventually lead to cell death (2, 5). CDT, encoded by the cdtA and cdtB genes, is an ADP-ribosylating toxin that acts on the actin cytoskeleton (8).
The PaLoc contains at least 3 additional genes that appear to be involved in the regulation of the expression or function of the large clostridial toxins: tcdE, tcdR, and tcdC (5, 6). TcdE is a putative holin-like protein, thought to be involved in toxin secretion; however, its exact role is still unclear (9). TcdR is an RNA polymerase sigma factor that acts as the positive transcriptional regulator of tcdA, tcdB, and tcdE and that also positively regulates its own expression. A direct interaction between TcdR and RNA polymerase allows the recognition of the target promoters and activates expression (5, 10). Expression of tcdR and, consequently, tcdA and tcdB is influenced by different stimuli, such as temperature, nutrient availability, and medium composition (11–14).
Analysis of gene transcription by quantitative PCR has shown that while the expression of tcdA, tcdB, tcdE, and tcdR is low during exponential phase, the expression of these genes strongly increases upon entering stationary phase (15). In contrast, tcdC was found to be highly expressed during exponential phase but to have decreased expression in stationary phase (15). Similar profiles were shown at the protein level, where the levels of TcdC were higher in the exponential growth phase (16). Together, these data suggest that TcdC may act as a negative regulator of toxin transcription. However, several other studies did not find a decrease in tcdC transcription in stationary phase but, rather, showed a constant expression level during the stationary growth phase (13, 17, 18).
Likewise, the association between toxin expression and tcdC gene variants is the subject of debate. Increased virulence in epidemic strains was thought to be caused by deletions and frameshift mutations in tcdC, leading to a severely truncated nonfunctional protein and, presumably, higher toxin titers as a consequence (19, 20). In support of this, it was shown that the introduction of a plasmid-based copy of the wild-type tcdC gene in strain M7404 (PCR ribotype 027, carrying a truncated tcdC) resulted in decreased virulence in hamsters (20). However, mutations in the tcdC gene of clinical isolates did not predict the activity of toxins A and B (18, 21). Moreover, several studies failed to observe a relation between toxin gene expression and the tcdC genotype. Restoration of chromosomal tcdC of outbreak strain R20291 (PCR ribotype 027) to the wild type did not result in altered toxin expression (22), and toxin expression in C. difficile 630Δerm and an isogenic tcdC ClosTron mutant showed no significant differences in toxin levels (17).
Previous studies have characterized the domain structure of TcdC (16, 23). TcdC is a 26-kDa dimeric protein that contains an N-terminal transmembrane region (residues 30 to 50) that allows it to anchor to the cell membrane, a coiled-coil dimerization domain, and a C-terminal functional domain (10, 23). Using surface plasmon resonance experiments, purified full-length TcdC was shown to interact with Escherichia coli core RNA polymerase and prevented the formation of the active holoenzyme TcdR-RNA polymerase (10). Overexpression of C. difficile tcdC in the heterologous host Clostridium perfringens results in repression of TcdR-driven transcription from the tcdA promoter, and the C-terminal domain of TcdC was sufficient for this activity (10). However, it is not clear if TcdR and TcdC are in close proximity inside the bacterial cell.
Due to a lack of structural characterization of TcdC homologues, computational analysis was used to build a structural model of the C-terminal domain of TcdC. This modeling suggested that the domain adopts a dimeric, single-stranded DNA (ssDNA) oligonucleotide/oligosaccharide binding fold (OB-fold) (23). TcdC is capable of binding to ssDNA G-quadruplexes in vitro, but considering the paucity of these structures in the genome sequence of C. difficile, G-quadruplexes might mimic an alternative TcdC binding partner (23).
It is clear that further studies are required to understand TcdC binding partners and their function in transcriptional repression. The prevailing model is that TcdC functions as an anti-sigma factor whose activity depends on the cytosolic localization of the C-terminal OB-fold domain. However, no topological information on the C-terminal domain has been demonstrated to date.
In this study, we aimed to determine whether the C-terminal domain of TcdC is cytosolic or surface exposed and evaluated a codon-optimized version of the HiBiT (HiBiTopt) extracellular detection system as a valuable addition to the molecular tools to study C. difficile. We found that the C-terminal domain of TcdC is located extracellularly and show the value of the HiBiTopt system for topology studies of C. difficile proteins. In addition, we show that the membrane association of TcdC is dependent on a membrane-proximal cysteine residue and that mutating this residue results in the release of TcdC from the bacterial cell.
RESULTS
In silico prediction of TcdC topology suggests an extracellular location of the C-terminal domain.
To analyze the topology of C. difficile TcdC (CD0664 from C. difficile 630), we first analyzed the protein sequence (UniProt accession no. Q189K7) using three different prediction algorithms: TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0) (24), TOPCONS 2.0 (http://topcons.cbr.su.se/) (25), and SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/) (26).
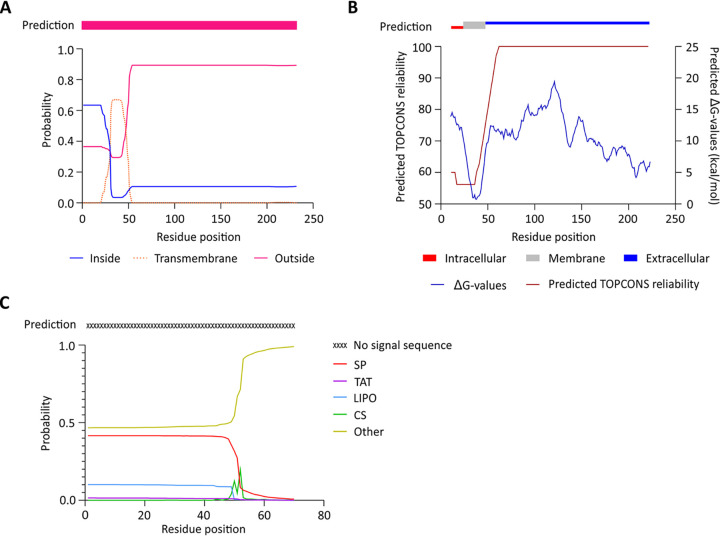
TMHMM 2.0 (24) predicts a transmembrane helix of about 16 residues (residues 31 to 46) with moderate probability. Residues 1 to 13 are predicted to be inside of the cell (probability, 0.63), whereas the C-terminal region (the coiled-coil and OB-fold domains) is predicted to be outside of the cell (probability > 0.8) (Fig. 1A). The consensus of the TMHMM 2.0 1-best algorithm predicts TcdC to be extracellular (Fig. 1A, pink bar). TOPCONS 2.0 (25), which identifies regions with a low free energy difference (ΔG), similarly suggests the presence of a transmembrane helix. The TOPCONS 2.0 consensus prediction is an intracellular N-terminal domain (residues 1 to 26) (Fig. 1B, red bar), a transmembrane helix (residues 27 to 46) (Fig. 1B, gray bar), and an extracellular C-terminal region that encompasses the dimerization and OB-fold domains (residues 47 to 232) (Fig. 1B, blue bar).
FIG 1.
Prediction of a transmembrane helix in TcdC. (A) Output from the prediction by TMHMM 2.0 software (24) through the 1-best algorithm (pink bar) and probability plot for the area inside the cell, the transmembrane region, and the area outside the cell. (B) Prediction by TOPCONS software (25), with consensus in residues 1 to 26 inside the cell, a transmembrane helix (residues 26 to 46), and residues 47 to 232 on the outside of the cell. The TOPCONS reliability score and predicted ΔG values for each residue are shown. (C) Output from the SignalP 5.0 (26) web server for the TcdC amino acid sequence. The probabilities of signal peptide presence from the systems Sec (SP), TAT, and lipoprotein (LIPO) are shown. The predicted cleavage site score (CS) and no signal sequence probability (Other) are depicted.
We also investigated the presence of a potential signal peptide in the TcdC amino acid sequence through the use of SignalP 5.0 (26). However, no known signal peptide was identified, suggesting that TcdC remains tethered to the membrane (Fig. 1C).
Though the reliability of the predictions is relatively low, both TMHMM and TOPCONS support the presence of the transmembrane helix (Fig. 1), consistent with previous observations (10, 16). Strikingly, both methods suggest that the TcdC C terminus is located outside of the cell. As this would be incompatible with a role for the OB-fold domain in sequestering TcdR or repression of TcdR-mediated transcriptional activation, we set out to obtain topological information on the C-terminal domain in C. difficile.
TcdC is accessible for extracellular cysteine labeling.
We analyzed whether cysteines natively present in TcdC are exposed to the extracellular environment in a manner similar to that used for the substituted cysteine accessibility method (SCAM). SCAM subjects the cysteine residues present in the protein of interest to chemical modification with the thiol-specific probe N-(3-maleimidylpropionyl) biocytin (MPB), which has a low level of membrane permeation. The probe forms a stable, nonhydrolyzable bond with the thiol group of a cysteine residue, resulting in the biotinylation of the protein. At low concentrations of MPB, exclusively extracytoplasmic (surface-exposed) cysteines are labeled, providing topological information about the labeled protein (27). A typical SCAM experiment relies on the immunoprecipitation of protein (using antibody specific for the protein of interest), detection of the immunoprecipitated protein (using a second antibody directed at a tag on the protein of interest), and verification of labeling with the MPB (using antibiotin antibodies).
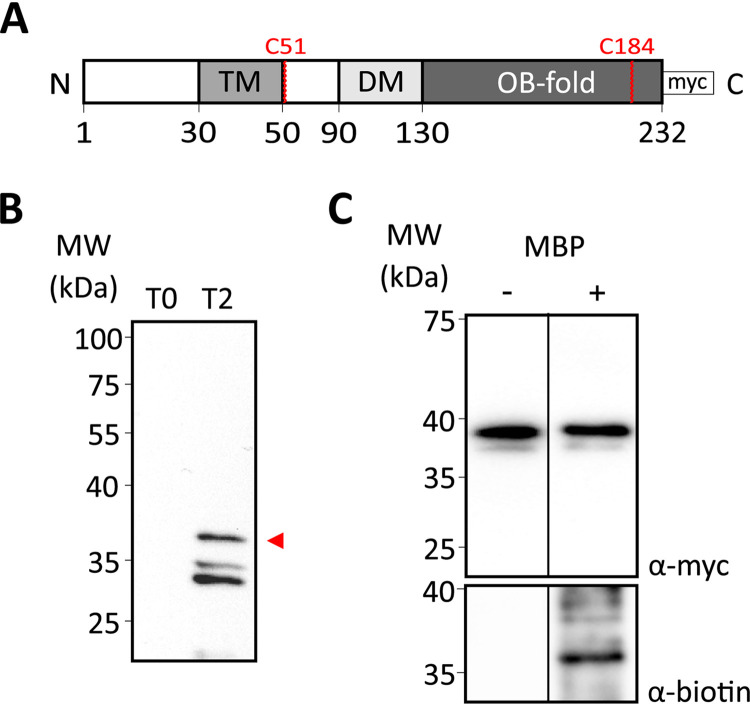
We introduced a C-terminally 3× Myc-tagged TcdC expression construct (TcdC-3×Myc) (Fig. 2A) with an otherwise native protein sequence under the control of the inducible promoter Ptet (28), which can be precipitated with anti-TcdC antibody and detected using anti-Myc antibodies. We affinity purified a previously generated TcdC antibody (17) and verified its specificity on C. difficile lysates by immunoblotting. The TcdC-3×Myc construct was induced, and samples were analyzed before and after induction. A band migrating at the approximate molecular weight of TcdC (37 kDa) was observed only in the induced samples (Fig. 2B, arrowhead), suggesting that the native levels of TcdC under noninducing conditions were below our limit of detection in this assay. Though the predicted molecular weight of the TcdC-3×Myc protein is 26 kDa, the observed molecular weight has been reported to be 37 kDa (10, 23), due to unknown reasons. Several bands with a lower-than-expected molecular weight were detected (Fig. 2B). The fact that these were present only when TcdC-3×Myc was induced (Fig. 2B) suggests possible alternative forms of the protein. Nevertheless, the apparent specificity of the anti-TcdC antibody allowed further analysis.
FIG 2.
Mapping the location of the TcdC C terminus with cysteine accessibility analysis. (A) Schematic representation of the C-terminally 3× Myc-tagged TcdC construct used for the cysteine accessibility analysis. The different domains of TcdC are represented: the transmembrane domain (TM), the dimerization domain (DM), and the OB-fold. The 3× Myc tag and the cysteines residues present on TcdC are represented. (B) Western blot analysis of anti-TcdC antibody specificity in C. difficile 630Δerm lysates harboring pLDJ1 (Ptet-tcdC-3×myc) before (lane T0) and after (lane T2 [time point 2]) induction with 200-ng/ml anhydrotetracycline for 2 h. Full-length TcdC is indicated with an arrowhead. (C) Cysteine labeling analysis of the TcdC-3×Myc construct. The strain harboring the C-terminally 3× Myc-tagged TcdC construct (38 kDa) was induced for 2 h. Samples were collected and either not treated with MPB (lane −) or treated with 1 mM MPB (lane +). Samples were immunoprecipitated and immunoblotted with anti-Myc for TcdC-3×Myc protein detection (top) and antibiotin for biotinylated protein detection (bottom). Cysteine biotinylation of TcdC-3×Myc was observed. MW, molecular weight.
TcdC contains 2 endogenous cysteines: one at position 51, right after the predicted transmembrane domain (residues 30 to 50), and another one at position 184, located in the predicted OB-fold (Fig. 2A). To evaluate the cysteine labeling on the native protein, we assessed the biotinylation of TcdC-3×Myc (Fig. 2C). The signal in the antibiotin Western blot suggests that one or both of the native cysteine residues in TcdC-3×Myc is accessible for labeling by MPB (Fig. 2C). We could not perform a full SCAM analysis (27), as mutation of the native cysteines that is necessary for such an analysis (see further below) resulted in the inconsistent and low-level expression of cell-associated TcdC.
Nevertheless, as only a single membrane-spanning region is predicted, this result suggests that the TcdC C terminus, where both cysteines are present, is located extracellularly.
The HiBiTopt assay for C. difficile confirms the extracellular location of the TcdC C terminus.
We sought to confirm the results of the cysteine accessibility assay in an independent experiment. Previously, we have successfully used luciferase reporter assays to assess promoter activity and in vivo protein-protein interactions in C. difficile (29, 30). Here, we extended the luciferase toolbox for C. difficile by validating an adaptation of the Nano-Glo HiBiT extracellular detection system (Promega) (31).
Similar to the SmBit tag, the HiBiT tag is a small 11-amino-acid peptide that binds to a larger subunit, called LgBiT, to reconstitute a functional luciferase (30–32). However, in contrast to SmBit, HiBiT has been engineered for a high affinity for the LgBiT subunit (32). Due to its molecular weight (19 kDa), extracellularly added LgBiT cannot enter the cell. Thus, a luminescent signal in the presence of the substrate furimazine is observed only if the HiBiT subunit is accessible from the extracellular environment (31).
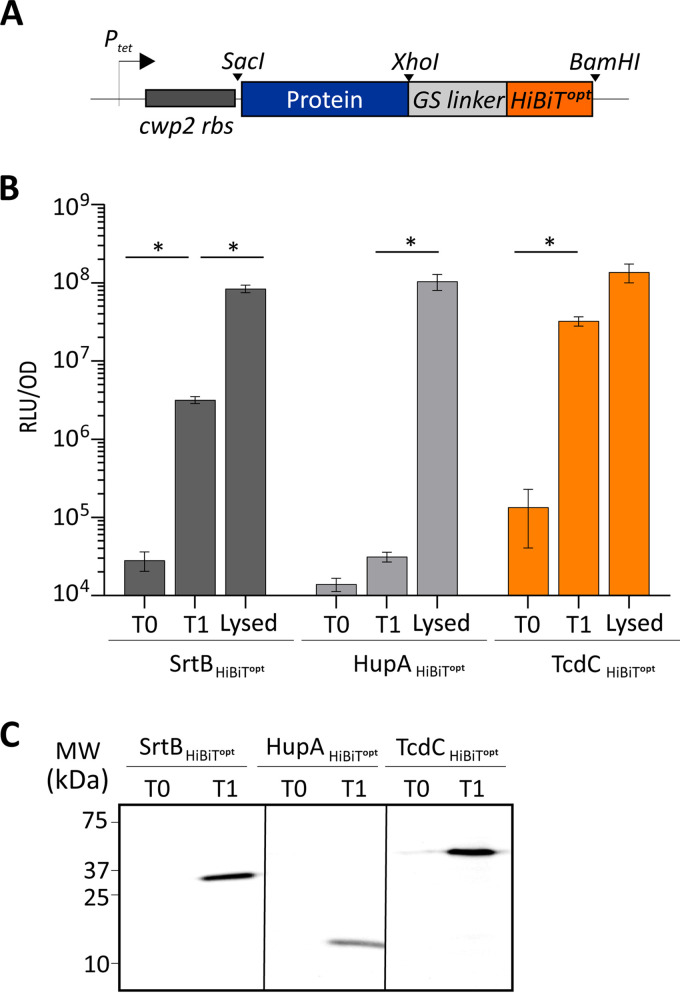
To apply this system for detection of the C. difficile protein topology, we constructed several controls carrying codon-optimized C-terminal HiBiT (HiBiTopt) tags, as schematically represented in Fig. 3A. As a positive control for the detection of extracellular proteins, the sortase B (SrtB) protein (SrtB-HiBiTopt) was selected. Sortases are membrane-anchored enzymes which catalyze the cleavage and transpeptidation of specific substrates and thereby facilitate their attachment to the cell wall (33). The genome of C. difficile strain 630 (and also its derivative, strain 630Δerm) has a single sortase, SrtB, present at the C. difficile cell wall (34, 35). The localization of SrtB and its substrates at the C. difficile cell surface makes SrtB a suitable candidate for the extracellular detection of the reconstituted luminescent signal. As a negative control, the HupA protein (HupA-HiBiTopt) was used. This protein is a cytosolic DNA-binding protein that is not secreted to the extracellular environment and that thus should not be accessible to the LgBiT subunit (30). All the constructs were placed in a modular vector under the control of the anhydrotetracycline (ATc)-inducible promoter Ptet (28). As observed in other bioluminescence assays (29, 30), a background signal from noninduced cells (Fig. 3B, T0 [time zero]) which was comparable to that of a medium-only control (17,872.8 ± 4,397.7 relative light units [RLU]/optical density unit [OD]) (data not shown) was detected. As expected, expression of the positive control, SrtB-HiBiTopt, led to a 2-log increase in the luminescence signal after 45 min of induction (time point 1 [T1]; 3.2 × 106 ± 2.5 × 105 RLU/OD) (Fig. 3B, T1). No significant increase in the luminescent signal was detected in the cells expressing the negative control, HupA-HiBiTopt, confirming that LgBiT does not enter C. difficile cells. The lack of a signal was not due to poor induction, as a clear signal for both SrtB-HiBiTopt and HupA-HiBiTopt was visible in the lysates of the induced samples at the expected molecular weights of 26 kDa and 12 kDa, respectively (Fig. 3C). To confirm that the obtained luciferase signals were derived from cells with an intact cell envelope, we performed lysis on the same samples. Indeed, after lysis of the cells, a clear increase of the luciferase signals was observed for both SrtB-HiBiTopt and HupA-HiBiTopt (Fig. 3B, Lysed). A modest, but significant, 1-log increase was also observed for SrtB-HiBiTopt (8.4 × 107 ± 9.5 × 106 RLU/OD) (Fig. 3B, Lysed). We attributed this increase to enhanced accessibility in SrtB-HiBiTopt for the substrate due to the lysis. However, for cells expressing HupA-HiBiTopt, a significant 4-log increase in the luciferase signal was observed (1.1 × 108 ± 2.4 × 107 RLU/OD) (Fig. 3B, Lysed). These results confirm that LgBiT does not enter C. difficile cells and that the HiBiTopt system, as employed, is suitable for determining the subcellular localization of the C-terminal domain of C. difficile proteins.
FIG 3.
Detection of C-terminal HiBiTopt tags. (A) Representation of the HiBiTopt modular cassette. The protein of interest fused at the C terminus to HiBiTopt through the GS linker is indicated. The positions of the restriction sites used (SacI, XhoI, and BamHI) are marked, and the cwp2 ribosomal binding site (rbs) is represented. (B) The proteins of interest were C-terminally fused to a HiBiT protein tag, and their expression was induced with 50-ng/ml ATc for 45 min. Optical density-normalized luciferase activity (RLU/OD) right before induction (T0), after 45 min of induction (T1), and after subsequent lysis of T1 samples (Lysed) is shown. HiBiTopt-tagged sortase and HupA proteins were used as extracellular and intracellular controls, respectively. TcdC-HiBiTopt-associated luciferase activity is also displayed. The averages for biological quadruplicate measurements are shown, with error bars indicating the standard deviation from the mean. *, P < 0.001 by two-way ANOVA. (C) Blot detection of HiBiTopt-tagged proteins resolved on a 12% SDS-PAGE gel. Sample volumes were normalized for the optical density of the cultures from which they were derived. Expression of HiBiTopt-fused proteins was observed at 0 min (T0) and 45 min (T1) after induction.
Next, HiBiTopt was fused to the C terminus of TcdC (TcdC-HiBiTopt). We observed a 3-log increase in the luminescence signal after 45 min of induction (3.2 × 107 ± 3.0 × 106 RLU/OD at T1) (Fig. 3B). No further increase was detected when the cells were lysed (1.4 × 108 ± 3.7 × 107 RLU/OD) (Fig. 3B, Lysed), supporting an extracellular location of the TcdC C terminus. Through blotting of bacterial lysates and subsequent measurement of the luminescence on the blot (see Materials and Methods), the expression of TcdC-HiBiTopt was confirmed by detection of a clear signal at the expected molecular weight of approximately 39 kDa (Fig. 3C). We observed a low-level signal for the noninduced TcdC-HiBiTopt expression construct, both in the luciferase assay (Fig. 3B, T0) and in the detection of the tagged protein (Fig. 3C, T0), which was not observed for SrtB or HupA. As all proteins are expressed from the same promoter, this possibly indicates the more efficient translation and, thus, higher levels of expression of TcdC-HiBiTopt under noninducing conditions. Alternatively, the differences in luciferase detection levels may be explained by protein stability or the accessibility of the HiBiTopt fusion proteins for the LgBiT subunit, which in turn is affected by the structure and the exact localization of the proteins.
The HiBiTopt experiments indicated that the C terminus of TcdC is located in the extracellular environment (like SrtB is) and not in the intracellular environment (like HupA is).
The cysteine residue at position 51 is important for the membrane association of TcdC.
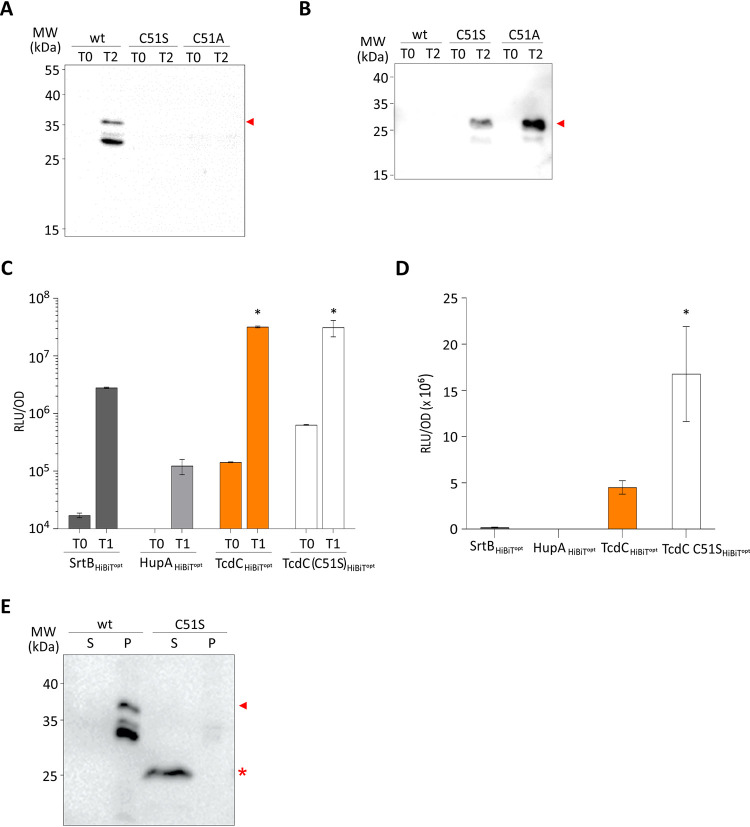
While assessing the extracellular accessibility of the cysteines in TcdC, we planned to perform a classic SCAM analysis that would allow us to determine which one of the extracellular cysteine residues (or both residues) was labeled and could potentially also be used to confirm the subcellular localization of the N terminus of the TcdC protein. To this end, we constructed several mutants of TcdC-3×Myc. During the experiments we observed inconsistent and low-level expression of cell-associated TcdC when the cysteine residue at position 51 was mutated into a serine residue (Fig. 4A). Likewise, mutation to alanine yielded very low levels of TcdC in C. difficile cell lysates, indicating that this effect is not specific to the serine substitution (Fig. 4A). We reasoned that the expression levels of these TcdC mutants were unlikely to differ as a result of a single amino acid change, and therefore, we analyzed the culture supernatant by immunoblotting using anti-TcdC antibodies to assess whether TcdC was released from the cells. Supernatants of cells, pelleted at 2 h postinduction, contained significant amounts of TcdC when cysteine 51 was mutated (Fig. 4B). In contrast, TcdC was cell associated in constructs expressing wild-type TcdC, which contained the cysteine 51 residue. Remarkably, the TcdC present in the supernatant was significantly smaller than the cell-associated TcdC (Fig. 4E), suggesting that TcdC may be the subject of a proteolytic event.
FIG 4.
TcdC C51S affects membrane localization. (A) Western blot analysis, with anti-TcdC, of C. difficile 630Δerm cell lysates harboring pLDJ1 (Ptet-tcdC-3×myc), pLDJ2 [Ptet-tcdC(C51S)-3×myc], and pJC129 [Ptet-tcdC(C51A)-3×myc] before (T0) and after (T2) induction with 200-ng/ml ATc for 2 h. TcdC is indicated with an arrowhead. (B) Western blot analysis, with anti-TcdC, of C. difficile 630Δerm culture supernatants harboring pLDJ1 (Ptet-tcdC-3×myc), pLDJ2 [Ptet-tcdC(C51S)-3×myc], and pJC129 [Ptet-tcdC(C51A)-3×myc] before (T0) and after (T2) induction with 200-ng/ml ATc for 2 h. The secreted/released TcdC is indicated with an arrowhead. (C) Proteins of interest were C-terminally fused to a HiBiT protein tag, and their expression was induced with 50-ng/ml ATc for 45 min. The optical density-normalized luciferase activity (RLU/OD) of the culture (cells plus medium) is shown right before induction (T0) and after 45 min of induction (T1). HiBiTopt-tagged sortase and HupA proteins were used as surface-exposed and intracellular controls, respectively. Luciferase activity with TcdC-HiBiTopt and TcdC(C51S)-HiBiTopt is also displayed. The averages for biological triplicate measurements are shown, with error bars indicating the standard deviation from the mean. *, P < 0.001 by two-way ANOVA. (D) Observed luciferase activity (RLU) in supernatants only from the cells for which the results are shown in panel C. (E) Comparison of cell-associated and cell-released TcdC. The same samples for which the results are shown in panels A and B were run next to each other for a fair comparison of the sizes of the proteins. Cell-associated TcdC is indicated with an arrowhead, and cell-released TcdC is indicated with an asterisk. wt, wild type; S, supernatant; P, pellet.
To confirm these results, we also mutated the cysteine 51 residue in the TcdC-HiBiTopt into a serine residue, monitored the luciferase activity in the culture supernatant of cells, and compared it to that of wild-type TcdC-HiBiTopt. The luciferase activity in the supernatants of cells expressing TcdC(C51S)-HiBiTopt was approximately 4 fold higher than that of cells expressing TcdC-HiBiTopt (Fig. 4D), indicating that TcdC(C51S) was released from the bacterial cells to a greater extent than wild-type TcdC. On the other hand, the total signals (cells and medium together) for TcdC and TcdC(C51S) were equal, showing that the difference in the supernatant was not due to increased expression of TcdC(C51S) (Fig. 4C).
DISCUSSION
The importance of TcdC for the regulation of toxin expression is highly controversial. Though the prevailing model suggests that TcdC is an anti-sigma factor with a role as a negative regulator of toxin transcription (19, 20), several other studies have found no clear relationship between TcdC expression and the toxin levels (17, 18, 21, 22). Previous biochemical analyses of TcdC revealed that it is membrane associated and that the TcdC C terminus comprises a dimerization domain and a domain with a predicted OB-fold important for transcriptional repression (10). However, the localization of the C terminus of TcdC has not been studied to date, and this was addressed in the present study.
In silico analyses of the TcdC amino acid sequence using TMHMM 2.0 (24), TOPCONS 2.0 (25), and SignalP 5.0 (26) suggested the presence of a transmembrane helix and predicted no high-probability cleavage for a secretion signal for any of the canonical secretion pathways (Fig. 1). The prediction of a transmembrane domain is consistent with the findings of previously described biochemical assays demonstrating the association of TcdC with the C. difficile membrane (16, 23). The analysis did not reach a consensus on the localization of the N terminus, due to low reliability scores and differences obtained with the prediction methods (Fig. 1). Nevertheless, both TMHMM 2.0 and TOPCONS 2.0 suggest that the C terminus of TcdC (residues 50 to 232) (Fig. 1) is located on the outside of the cell.
For a preliminary analysis on the C terminus accessibility, we took advantage of the 2 cysteine residues that are natively present after the transmembrane helix of TcdC (Fig. 2A). Biotin labeling of TcdC-3×Myc was detected, suggesting that one or both of the native cysteine residues are accessible for labeling by MPB (27) and are therefore located in the extracellular environment (Fig. 5).
FIG 5.
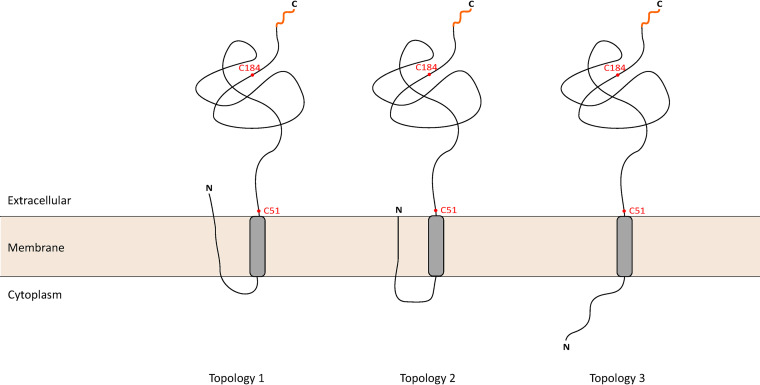
TcdC topology models. TcdC is located in the cell membrane with an extracellular C-terminal region. The localization of the 50-amino-acid N-terminal domain of TcdC is unknown. In topology model 1, the N terminus can cross the cell membrane, exposing the N-terminal domain to the extracellular environment. Another possibility, topology model 2, has the N terminus present in the cell membrane, where it is not accessible from the extracellular or intracellular milieu. Finally, in topology model 3, the N terminus is present in the intracellular environment of the cell. The cysteine residues used for the cysteine accessibility analysis (red dots) and the C-terminal location of the HiBiTopt tag (orange line) are indicated.
Our result was confirmed in independent experiments using the HiBiTopt system, which, to our knowledge, was applied here for the first time in C. difficile. These experiments confirm that the C terminus of TcdC is exposed on the cell surface. Unfortunately, it was not possible to apply the HiBiTopt system to determine the localization of the N terminus of TcdC. Fusions of HiBiTopt at the N terminus of proteins with an established intracellular localization resulted in a high extracellular luciferase signal (data not shown). We attribute this to possible N-terminal processing, which could lead to the release and secretion of the small tag. Further optimization of the HiBiTopt system is essential before the system can be used to assess the localization of both protein termini.
We found that TcdC is released from the cell when a membrane-proximal cysteine residue is mutated in the protein (Fig. 4). As mentioned earlier, TcdC in the culture supernatant appeared to have a lower molecular weight than cell-associated TcdC, suggesting a possible cleavage event. In silico analysis using SignalP suggested that both TcdC(C51S) and TcdC(C51A) are good substrates for signal peptidase (likelihoods of 0.6671 and 0.6901, with probabilities that the cleavage site is between S52 and E53 being 0.5323 and 0.4482, respectively) (see Fig. S2 in the supplemental material), whereas TcdC is not (likelihood of 0.4169; no score for probability was given). As the signal peptidase is located in the extracellular milieu, this provides an additional indication of the extracellular localization of the TcdC C-terminal OB-fold. Alternatively, the decreased size of TcdC(C51S) could be the result of the lack of a cysteine-specific posttranslational modification. Such a modification could also be responsible for the higher-than-expected molecular weight of wild-type TcdC in immunoblotting experiments. Glycosylation is one of the most common posttranslational modifications found in several bacteria, particularly at the cell surface (36), and the higher-molecular-weight TcdC bands could represent glycosylated forms of the protein. Targeted mass spectrometry-based proteomics might shed light on the nature of these modifications, if this is the case. Although our data cannot discriminate between these two hypotheses at this point, we favor the hypothesis that TcdC is cleaved when C51 is not present, as this is clearly supported by the SignalP predictions.
When analyzing the possible release of TcdC mutants, we were unable to detect TcdC by Western blotting in the supernatant of cells expressing TcdC. In contrast, when using the HiBiTopt-tagged TcdC, we were able to detect some luciferase activity in the supernatant, indicating the release of TcdC (Fig. 4D). We can only speculate about this discrepancy. First of all, we cannot compare the sensitivity of the two assays. The fact that we could not detect TcdC in the Western blot may have been due to the limited sensitivity of the assay. Second, the overexpression of TcdC may lead to some release that can be measured with the highly sensitive HiBiTopt-based assay. However, since no significant activity was measured in the supernatants of cells overexpressing sortase-HiBiTopt, the spontaneous release of protein due to overexpression per se does not seem to be a reasonable explanation for the luciferase activity in the supernatant of cells overexpressing TcdC.
When predicting the cleavage of TcdC through SignalP, a low score was given to another possible cleavage system (other than Sec/SPI, TAT/SPI, or Sec/SPII). It is possible that part of TcdC is indeed cleaved by another protease, but we cannot speculate about the nature of this protease. Mutating the membrane-proximal cysteine to serine or alanine converts TcdC into a likely substrate for the efficient Sec/SPI system, possibly explaining the increased signal on the Western blot and in the HiBiTopt-based assay.
Our results show an extracellular localization of the C terminus of TcdC, but we could not fully explore the topology of TcdC. Prediction of the TcdC N terminus was not unambiguous (Fig. 1), and the experimental approaches were unable to conclusively demonstrate the localization of the N terminus of TcdC due to technical limitations. The protein may therefore adopt different orientations, as represented in Fig. 5. The N terminus may be located extracellularly (topology 1), may be embedded in the cell membrane (topology 2), or may be exposed in the intracellular environment (topology 3). Thus, a careful further characterization of the TcdC topology by alternative means is required. Recently, the fluorescence activating and absorption shifting tag (FAST) has been used to label proteins and assess protein topology in bacteria (37). The use of nonpermeant substrates allows the localization of exposed proteins on the surface in Gram-negative and Gram-positive organisms (37). FAST can be used in anaerobic environments and was previously used in C. difficile (38), which makes it a promising candidate to explore the N-terminal location of TcdC.
Our finding that the TcdC C terminus is extracellular challenges the prevailing model of TcdC as an anti-sigma factor. Anti-sigma factors generally sequester their cognate sigma factors away from RNA polymerase (RNAP) using substantial cytoplasmic domains (39). The small N-terminal sequence, which may or may not be intracellular (Fig. 5), is not likely to fulfill this function. Our experiments clearly place the C-terminal domain that previously was postulated to be responsible for transcriptional repression (10) in a different environment than TcdR and RNAP. One has to wonder whether the OB-fold would ever be in contact with these proteins, as was demonstrated in experiments using purified proteins (outside the context of a C. difficile cell) and in heterologous expression systems (10). Experiments that show the repression of TcdR-mediated transcription by coexpression of TcdC were carried out in a heterologous background (10). In this study, it is unclear what directed tcdR transcription, and the observed inhibition of TcdR-mediated transcription by TcdC may be explained by an indirectly lowered level of tcdR transcription. In addition, the inhibitory effect of TcdC was also seen when TcdC was expressed without its N-terminal hydrophobic part, which led to the conclusion that the C-terminal domain is responsible for the inhibitory effect. However, removal of a membrane-binding domain likely results in the mislocalization of the protein compared to its localization in the wild-type situation and does not represent a physiological setup. The fact that this truncated TcdC inhibits TcdR-mediated transcription argues for an aspecific effect of TcdC expression. Lastly, biochemical evidence based on surface plasmon resonance may have been influenced by the fact that full-length TcdC, including the hydrophobic stretch held responsible for the TcdC membrane association, was used. Regardless, it should be noted that an extracellular location of TcdC does not exclude the possibility of its function as a negative regulator of toxin production but, if so, suggests that it does so through an indirect mechanism.
Our data rather imply the binding of the TcdC OB-fold to an extracellular ligand. Bacterial OB-fold proteins have been identified in bacterial genomes and can bind a wide variety of molecules (39, 40). Thus, TcdC might bind extracellular oligonucleotides and/or oligosaccharides. It has previously been shown that the TcdC OB-fold is able to bind G-quadruplex structures, but the physiological relevance of this binding has yet to be determined, and it is conceivable that G-quadruplex binding mimics the binding of its natural substrate, as proposed earlier (23). In the extracellular environment, TcdC might bind oligosaccharides or extracellular DNA (eDNA) (41–43). In Staphylococcus aureus, staphylococcal superantigen-like protein 10 (SSL10) binds to human IgG1 Fc primarily by its N-terminal OB-fold domain and can play a role during S. aureus infections (44). In Salmonella enterica serovar Typhimurium, the small periplasmic protein YdeI contains an OB-fold domain and contributes to the resistance to antimicrobial peptides by interaction with the OmpF porin (45). The VisP protein, a protein from the bacterial oligonucleotide/oligosaccharide-binding fold family also present in S. Typhimurium, binds to the peptidoglycan sugars and also to the inner membrane protein LpxO, mediating resistance and pathogenesis in S. Typhimurium (46). To identify the TcdC ligand(s), a cross-linking and subsequent mass spectrometry based-method with tagged TcdC could be used. In addition, structural studies of TcdC and its ligands could show how the OB-fold of TcdC has evolved and to what extent TcdC contributes to downregulation of the large C. difficile toxins.
In summary, we developed and applied for the first time a modular vector that allows the C-terminal tagging of proteins with a HiBiTopt tag, which extends our existing luciferase tool kit (29, 30). This system offers a useful method for the determination of the topology of C-terminal domains of membrane proteins in cells grown under anaerobic conditions without complex processing of samples. Our study indicates an extracellular localization of the C terminus of TcdC, which is incompatible with its proposed function as an anti-sigma factor. Further studies are required to elucidate the role of TcdC in C. difficile development and toxin regulation.
MATERIALS AND METHODS
Topology prediction.
To determine the topology of the C. difficile TcdC protein (UniProt accession no. Q189K7), the amino acid sequence was analyzed by two computer programs for transmembrane and topology assessment: TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0) (24), with an extensive output format with graphics, and TOPCON 2.0 (http://topcons.cbr.su.se/) (25). The TcdC sequence was analyzed with the SignalP 5.0 program for signal peptide prediction, with a long output for Gram-positive organisms (http://www.cbs.dtu.dk/services/SignalP/) (26). All the results were visualized with GraphPad Prism (version 8.1.2) software.
Strains and growth conditions.
E. coli strains were grown aerobically at 37°C in Luria-Bertani broth (LB; Affymetrix) supplemented with chloramphenicol at 15 μg/ml or 50-μg/ml kanamycin, when required. Plasmids (Table 1) were maintained in E. coli strain DH5α and transformed using standard procedures (47). E. coli CA434 (48) was used for plasmid conjugation with C. difficile strain 630Δerm (34, 49).
TABLE 1.
Plasmids used in this study
| Name | Relevant featuresa | Source or reference |
|---|---|---|
| pCR2.1-TOPO | TA vector; pMB1 oriR km amp | Thermo Fisher |
| pCR2.1TcdC | pCR2.1-TOPO with tcdC km amp | This study |
| pRPF185 | tetR Ptet-gusA catP | 28 |
| pLDJ1 | tetR Ptet-tcdC-3×myc catP | This study |
| pLDJ2 | tetR Ptet-tcdC(C51S)-3×myc catP | This study |
| pAF302 | tetR Ptet-hupA-hiBiTopt catP | This study (Addgene accession no. 137752) |
| pAP233 | tetR Ptet-srtB-hiBiTopt catP | This study (Addgene accession no. 137753) |
| pJC111 | tetR Ptet-tcdC-hiBiTopt catP | This study |
| pJC127 | tetR Ptet-tcdC(C51S)-hiBiTopt catP | This study |
| pJC129 | tetR Ptet-tcdC(C51A)-3×myc catP | This study |
amp, ampicillin resistance cassette; km, kanamycin resistance cassette; catP, chloramphenicol resistance cassette.
C. difficile strains were grown anaerobically in brain heart infusion (BHI) broth (Oxoid) with 0.5% (wt/vol) yeast extract (Sigma-Aldrich) supplemented with Clostridium difficile selective supplement (CDSS; Oxoid) and 15-μg/ml thiamphenicol, when necessary, at 37°C in a Don Whitley VA-1000 workstation or a Baker Ruskin Concept 1000 workstation with an atmosphere of 10% H2, 10% CO2, and 80% N2. All strains are described in Table 2.
TABLE 2.
C. difficile strains used in this study
| Name | Relevant genotype or phenotypea | Origin or references |
|---|---|---|
| 630Δerm | C. difficile 630Δerm Erms | 34, 49 |
| LDJ1 | 630Δerm/pLDJ1 Thiar | This study |
| LDJ2 | 630Δerm/pLDJ2 Thiar | This study |
| AP239 | 630Δerm/pAP233 Thiar | This study |
| JC267 | 630Δerm/pJC111 Thiar | This study |
| JC271 | 630Δerm/pAF302 Thiar | This study |
| JC324 | 630Δerm/pJC127 Thiar | This study |
| JC326 | 630Δerm/pJC129 Thiar | This study |
Erms, erythromycin sensitive; Thiar, thiamphenicol resistant.
The growth was monitored by use of the optical density reading at 600 nm (OD600).
Strain construction.
All oligonucleotides from this study are listed in Table 3. All PCRs were performed on C. difficile 630Δerm genomic DNA (34), unless indicated otherwise. All expression vectors were based on pRPF185 (28). All DNA sequences in the recombinant plasmids were verified by Sanger sequencing of the region of the plasmid encompassing the inserted fragment and the full anhydrotetracycline-inducible promoter.
TABLE 3.
Oligonucleotides used in this study
| Name | Sequence (5′→3′)a |
|---|---|
| oDB0071 | CTGAGCTCCTGCAGTAAAGGAGAAAATTTTATGTTTTCTAAAAAAAATGAT |
| oDB0072 | TAGGATCCGGTTAATTAATTTTCTCTACAGCT |
| oCDTcdCmyc3 | TAGGATCCTTATAAATCTTCTTCACTTATTAATTTTTGTTCTAAATCTTCTTCACTTATTAATTTT |
| oCD_SortaseF | GTCTGAGCTCCTGCAGTAAAGGAGAAAATTTTATGTTGAAAAAATTATATAGAATAG |
| oCD_SortaseR | CCCTCGAGAAATCAATCTACCATGAATCAC |
| oTcdCRev | AAACTCGAGAATTAATTTTCTCTACAGCTATCCCTGG |
| CDTcdCC51SF | CAATATATCCTCACCAGCTAGTTCTGAAGACCATGAGGAG |
| CDTcdCC51SR | CTCCTCATGGTCTTCAGAACTAGCTGGTGAGGATATATTG |
| oJC424 | CAATATATCCTCACCAGCTGCTTCTGAAGACCATGAGGAG |
| oJC425 | CTCCTCATGGTCTTCAGAAGCAGCTGGTGAGGATATATTG |
The restriction enzyme sites used are underlined.
Construction of tcdC-3×myc for cysteine accessibility analysis.
To construct the expression constructs for tcdC-3×myc, the tcdC gene (CD0664 from C. difficile 630 [GenBank accession no. WP_011860905.1]) was amplified by PCR using primers oDB0071 and oDB0072 from C. difficile chromosomal DNA. The PCR product was subsequently cloned into pCR2.1TOPO (Invitrogen), according to the manufacturer’s instructions, to yield the vector pCR2.1TcdC (Table 1). The TcdC fragment was amplified from pCR2.1TcdC with primers oDB0071 and oCDTcdCmyc3, which allows the addition of a 3× Myc tag at the C terminus. The resulting PCR fragment was digested with SacI and BamHI and ligated into pRPF185 that had been digested with the same enzymes, to yield vector pLDJ1 (Table 1), placing tcdC under the control of the anhydrotetracycline-inducible promoter (Ptet).
To mutate the cysteine at position 51 in TcdC, we used the oligonucleotides CDTcdCC51SF and CDTcdCC51SR for the C51S mutation and the oligonucleotides oJC424 and oJC425 for the C51A mutation in a QuikChange reaction, using pCR2.1-TcdC as a template. The mutated TcdC sequences were subsequently subcloned into pRPF185 as described above for wild-type tcdC, yielding plasmids pLDJ2 and pJC129.
Construction of HiBiTopt fusions.
The hupA gene (CD3496 from C. difficile 630 [GenBank accession no. NC_009089.1]) fused at the 3′ end to the hiBiTopt codon-optimized coding sequence (HupA-HiBiTopt) was synthesized by Integrated DNA Technologies, Inc. (IDT). The synthesized fragment (the full sequence is provided in the supplemental material) was digested with BamHI and SacI and cloned into pRPF185 that had been digested with same enzymes, yielding vector pAF302 (Table 1), which can be requested through Addgene (Addgene accession no. 137752).
The srtB gene (CD2718 from C. difficile 630 [GenBank accession no. WP_009903304.1]) was amplified from C. difficile genomic DNA with primers oCD-SortaseF/oCD-SortaseR, digested with XhoI and SacI, and placed into similarly digested pAF302, yielding vector pAP233 (Table 1), which can be requested through Addgene (Addgene accession no. 137753).
The tcdC gene was amplified by PCR with primer set oDB0071/oTcdCRev from C. difficile genomic DNA. The PCR fragment was digested with XhoI and SacI and cloned into similarly digested pAF302, yielding vector pJC111 (Table 1). To generate the equivalent C51S construct, tcdC(C51S) was amplified with oDB0071 and oTcdCRev from pLDJ2 and cloned after SacI-XhoI digestion into similarly digested pJC111 to yield pJC127.
Affinity purification of anti-TcdC polyclonal antibodies.
Polyclonal antibodies against TcdC were raised in rabbits using the peptide CQLARTPDDYKYKKV (17). To reduce background signals, serum from the final bleed from the immunized rabbits was subjected to affinity purification. Recombinant soluble 10×His-TcdCΔN50 (lacking the N-terminal 50 amino acids) (23) was blotted onto a polyvinylidene difluoride (PVDF) membrane. The blot was stained with Ponceau S solution (0.2% [wt/vol] Ponceau S, 1% acetic acid) for 5 min. Subsequently, the blot was washed with Tris-buffered saline (TBS) with Tween 20 (TBST; 500 mM NaCl, 20 mM Tris [pH 7.4], 0.05% [vol/vol] Tween 20) until the TcdC band was visible. The band was cut out of the blot, and the piece of membrane was washed with Ponceau destaining solution (phosphate-buffered saline [PBS], 0.1% NaOH) for 1 min. Subsequently, the blot piece was washed twice with TBST for 5 min each time. The membrane was soaked in acidic glycine buffer (100 mM glycine, pH 2.5) for 5 min and washed twice in TBST for 5 min each time. The blot was blocked in 10% milk powder (in TBST) for 1 h at room temperature. After washing it twice with TBST for 5 min each time, the blot was incubated with 10 ml of 5×-diluted serum (in TBS) overnight at 4°C. Afterwards, the diluted serum was removed and the blot was washed 3 times with TBST for 5 min each time and 2 times with PBS for 5 min each time. The blot was incubated with 1 ml of acidic glycine buffer for 10 min to elute the antibodies. The eluted antibody solution was immediately neutralized by adding 1 M Tris-HCl, pH 8.0. After addition of sodium azide (5 mM) and bovine serum albumin (BSA; 1 mg/ml), the affinity-purified antibodies were stored at 4°C until use in experiments.
Cysteine accessibility analysis.
To perform the cysteine accessibility analysis, 25-ml C. difficile cultures were induced with 200 ng/ml ATc at an OD600 of 0.3 for 2 h. The culture was centrifuged for 10 min at 2,800 × g, and the pellet was frozen at −20°C until needed. Subsequently, the pellet was resuspended in 600 μl GTE buffer (50 mM glucose, 20 mM Tris-HCl, 1 mM EDTA, pH 7.5) supplemented with cOmplete protease inhibitor cocktail (CPIC; Roche Applied Science) and divided into aliquots of 100 μl. For cysteine labeling, 1 mM MPB was added and the samples were incubated for 15 min on ice. The reactions were quenched by the addition of 73 mM β-mercaptoethanol. Samples were washed twice in GTE buffer-CPIC and centrifuged at 2,800 × g. The pellets were resuspended in 100 μl solubilization buffer (50 mm Tris-HCl [pH 8.1], 2% SDS, 1 mM EDTA) with mixing for 5 min and sonication (5- to 10-s pulses 2 times). To remove unspecific binding, 400 μl 0.2% PBS, Triton X-100, CIPC was added to the sample together with 30 μl of a 50% protein A–Sepharose CL-4B (Amersham) slurry in PBS supplemented with 1% BSA that had previously been equilibrated in PBS–1% BSA. After overnight incubation at 4°C, the protein A-Sepharose beads were removed by gentle centrifugation.
For immunoprecipitation, 50 μl of the 50% protein A–Sepharose CL-4B (Amersham) slurry was added to each sample with the affinity-purified polyclonal rabbit anti-TcdC antibody (1:200) and incubated at 4°C with gentle mixing for 2 h. The slurry was pelleted (4,000 × g) and washed 2 times with TENT buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris, 0.5% Triton X-100, pH 7.5), 1% BSA, 0.5 M NaCl; 2 times with TENT buffer, 1% BSA, 0.25 M NaCl; and 2 times with TENT buffer. The pellet was resuspended in 50 μl SDS loading buffer (250 mM Tris-Cl [pH 6.8], 10% SDS, 10% β-mercaptoethanol, 50% glycerol, 0.1% bromophenol blue) and incubated at 50°C for 5 min. The samples were spun down prior to SDS-PAGE analysis.
Immunoblotting and detection.
Proteins were separated on a 12% SDS-PAGE gel and transferred onto PVDF membranes (Amersham), according to the manufacturers’ instructions. The membranes were probed with monoclonal mouse anti-Myc (1:1,500; Invitrogen) or mouse antibiotin (1:1,000) antibodies in PBST. After washing the blots with PBST, a secondary goat anti-mouse antibody conjugated with horseradish peroxidase (1:1,000; Dako) was used. The bands were visualized using Clarity enhanced chemiluminescence Western blotting substrates (Bio-Rad) on an Alliance Q9 Advanced machine (UVItec).
HiBiTopt assay.
C. difficile cells were induced with 50-ng/ml ATc at an OD600 of 0.3 to 0.4 for 45 min. A 1-ml sample was collected and centrifuged (4,000 × g) for SDS-PAGE analysis and for luminescent detection of HiBiTopt-tagged proteins on a blot. Before and after centrifugation, 50-μl samples were collected for analysis.
To measure luciferase activity, the pellets where resuspended in 1 ml PBS and a 50-μl sample was taken for further luciferase detection. The samples were centrifuged (20,000 × g) for 10 min, and the pellets were incubated in 950 μl lysis buffer (10 mM Tris-HCl [pH 7.5], 10 mM EDTA, 0.1 mg/ml lysozyme, CPIC) for 1 h at 37°C. A sample of 50 μl was taken for further luciferase detection.
Samples were incubated with 50 μl of a Nano-Glo HiBiT extracellular detection system, a mixture of the NanoLuc LgBiT protein, and luciferase substrate in buffer (Promega) for 5 min in a 96-well white F-bottom plate. Luciferase activity was measured on a GloMax Multi+ instrument (Promega) with a 0.5-s integration time. All luciferase measurements were taken immediately after sampling. The data were normalized to the OD600 of the culture that the samples were derived from, and statistical analysis was performed by two-way analysis of variance (ANOVA) with Prism (version 7) software (GraphPad Inc., La Jolla, CA).
For luminescent detection of HiBiTopt-tagged proteins on a blot, total protein was resolved on a 12% SDS-PAGE gel and transferred onto PVDF membranes (Amersham). The membranes were washed with TBST and incubated with 200-fold-diluted LgBiT protein in TBST (Promega) for 1 h at room temperature. Nano-Glo luciferase assay substrate (Promega) diluted 500-fold was added, and the mixture was incubated for 5 min at room temperature with gentle shaking. The membranes were analyzed using an Alliance Q9 Advanced machine (UVItec).
Images were prepared for publication in CorelDRAW Graphics Suite X7 software.
Supplementary Material
ACKNOWLEDGMENTS
Work in the group of W.K.S. is supported by a Vidi fellowship (fellowship 864.10.003) of The Netherlands Organization for Scientific Research (NWO) and a Gisela Thier Fellowship from the Leiden University Medical Center.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. 2016. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe 40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. 2016. Clostridium difficile infection. Nat Rev Dis Primers 2:16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czepiel J, Dróżdż M, Pituch H, Kuijper EJ, Perucki W, Mielimonka A, Goldman S, Wultańska D, Garlicki A, Biesiada G. 2019. Clostridium difficile infection: review. Eur J Clin Microbiol Infect Dis 38:1211–1221. doi: 10.1007/s10096-019-03539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaffler H, Breitruck A. 2018. Clostridium difficile—from colonization to infection. Front Microbiol 9:646. doi: 10.3389/fmicb.2018.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandrasekaran R, Lacy DB. 2017. The role of toxins in Clostridium difficile infection. FEMS Microbiol Rev 41:723–750. doi: 10.1093/femsre/fux048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rupnik M, Dupuy B, Fairweather NF, Gerding DN, Johnson S, Just I, Lyerly DM, Popoff MR, Rood JI, Sonenshein AL, Thelestam M, Wren BW, Wilkins TD, Eichel-Streiber C. 2005. Revised nomenclature of Clostridium difficile toxins and associated genes. J Med Microbiol 54(Pt 2):113–117. doi: 10.1099/jmm.0.45810-0. [DOI] [PubMed] [Google Scholar]
- 7.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH, Peto TE, Walker AS, Riley TV, Crook DW, Didelot X. 2014. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol 6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aktories K, Papatheodorou P, Schwan C. 2018. Binary Clostridium difficile toxin (CDT)—a virulence factor disturbing the cytoskeleton. Anaerobe 53:21–29. doi: 10.1016/j.anaerobe.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Olling A, Seehase S, Minton NP, Tatge H, Schroter S, Kohlscheen S, Pich A, Just I, Gerhard R. 2012. Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb Pathog 52:92–100. doi: 10.1016/j.micpath.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Matamouros S, England P, Dupuy B. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol 64:1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 11.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson S, Dupuy B, Mukherjee K, Norin E, Burman LG, Akerlund T. 2003. Expression of Clostridium difficile toxins A and B and their sigma factor TcdD is controlled by temperature. Infect Immun 71:1784–1793. doi: 10.1128/iai.71.4.1784-1793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol 66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 14.Karlsson S, Burman LG, Akerlund T. 2008. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology (Reading) 154:3430–3436. doi: 10.1099/mic.0.2008/019778-0. [DOI] [PubMed] [Google Scholar]
- 15.Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, von Eichel-Streiber C. 1997. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur J Biochem 244:735–742. doi: 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 16.Govind R, Vediyappan G, Rolfe RD, Fralick JA. 2006. Evidence that Clostridium difficile TcdC is a membrane-associated protein. J Bacteriol 188:3716–3720. doi: 10.1128/JB.188.10.3716-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakker D, Smits WK, Kuijper EJ, Corver J. 2012. TcdC does not significantly repress toxin expression in Clostridium difficile 630DeltaErm. PLoS One 7:e43247. doi: 10.1371/journal.pone.0043247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, Gerding DN, Vedantam G. 2010. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol 192:4904–4911. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 20.Carter GP, Douce GR, Govind R, Howarth PM, Mackin KE, Spencer J, Buckley AM, Antunes A, Kotsanas D, Jenkin GA, Dupuy B, Rood JI, Lyras D. 2011. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog 7:e1002317. doi: 10.1371/journal.ppat.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray R, Boyd D, Levett PN, Mulvey MR, Alfa MJ. 2009. Truncation in the tcdC region of the Clostridium difficile PathLoc of clinical isolates does not predict increased biological activity of toxin B or toxin A. BMC Infect Dis 9:103. doi: 10.1186/1471-2334-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Leeuwen HC, Bakker D, Steindel P, Kuijper EJ, Corver J. 2013. Clostridium difficile TcdC protein binds four-stranded G-quadruplex structures. Nucleic Acids Res 41:2382–2393. doi: 10.1093/nar/gks1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182. [PubMed] [Google Scholar]
- 25.Tsirigos KD, Peters C, Shu N, Kall L, Elofsson A. 2015. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 27.Bogdanov M, Zhang W, Xie J, Dowhan W. 2005. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): application to lipid-specific membrane protein topogenesis. Methods 36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem 286:27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira Paiva AM, Friggen AH, Hossein-Javaheri S, Smits WK. 2016. The signal sequence of the abundant extracellular metalloprotease PPEP-1 can be used to secrete synthetic reporter proteins in Clostridium difficile. ACS Synth Biol 5:1376–1382. doi: 10.1021/acssynbio.6b00104. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira Paiva AM, Friggen AH, Qin L, Douwes R, Dame RT, Smits WK. 2019. The bacterial chromatin protein HupA can remodel DNA and associates with the nucleoid in Clostridium difficile. J Mol Biol 431:653–672. doi: 10.1016/j.jmb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Eggers C, Binkowski B, Riching K, Mahan S, Alves J, Schwinn M, Butler B, Daniels D, Zegzouti H, Machleidt T, Wood K, Fan F. 2018. Monitoring protein dynamics at endogenous levels with a luminescent peptide tag. Promega Corporation, Madison, WI. [Google Scholar]
- 32.Schwinn MK, Machleidt T, Zimmerman K, Eggers CT, Dixon AS, Hurst R, Hall MP, Encell LP, Binkowski BF, Wood KV. 2018. CRISPR-mediated tagging of endogenous proteins with a luminescent peptide. ACS Chem Biol 13:467–474. doi: 10.1021/acschembio.7b00549. [DOI] [PubMed] [Google Scholar]
- 33.Jacobitz AW, Kattke MD, Wereszczynski J, Clubb RT. 2017. Sortase transpeptidases: structural biology and catalytic mechanism. Adv Protein Chem Struct Biol 109:223–264. doi: 10.1016/bs.apcsb.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Eijk E, Anvar SY, Browne HP, Leung WY, Frank J, Schmitz AM, Roberts AP, Smits WK. 2015. Complete genome sequence of the Clostridium difficile laboratory strain 630Deltaerm reveals differences from strain 630, including translocation of the mobile element CTn5. BMC Genomics 16:31. doi: 10.1186/s12864-015-1252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers CJ, Roberts AK, Shone CC, Acharya KR. 2015. Structure and function of a Clostridium difficile sortase enzyme. Sci Rep 5:9449. doi: 10.1038/srep09449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaffer C, Messner P. 2017. Emerging facets of prokaryotic glycosylation. FEMS Microbiol Rev 41:49–91. doi: 10.1093/femsre/fuw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chekli Y, Peron-Cane C, Dell'Arciprete D, Allemand J-F, Li C, Ghigo J-M, Gautier A, Lebreton A, Desprat N, Beloin C. 2020. Visualizing the dynamics of exported bacterial proteins with the chemogenetic fluorescent reporter FAST. bioRxiv doi: 10.1101/2020.05.12.090142. [DOI] [PMC free article] [PubMed]
- 38.Streett HE, Kalis KM, Papoutsakis ET. 2019. A strongly fluorescing anaerobic reporter and protein-tagging system for Clostridium organisms based on the fluorescence-activating and absorption-shifting tag protein (FAST). Appl Environ Microbiol 85:e00622-19. doi: 10.1128/AEM.00622-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ginalski K, Kinch L, Rychlewski L, Grishin NV. 2004. BOF: a novel family of bacterial OB-fold proteins. FEBS Lett 567:297–301. doi: 10.1016/j.febslet.2004.04.086. [DOI] [PubMed] [Google Scholar]
- 40.Arcus V. 2002. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr Opin Struct Biol 12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- 41.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ðapa T, Dapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M. 2013. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol 195:545–555. doi: 10.1128/JB.01980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubois T, Tremblay YDN, Hamiot A, Martin-Verstraete I, Deschamps J, Monot M, Briandet R, Dupuy B. 2019. A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. NPJ Biofilms Microbiomes 5:14. doi: 10.1038/s41522-019-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel D, Wines BD, Langley RJ, Fraser JD. 2010. Specificity of staphylococcal superantigen-like protein 10 toward the human IgG1 Fc domain. J Immunol 184:6283–6292. doi: 10.4049/jimmunol.0903311. [DOI] [PubMed] [Google Scholar]
- 45.Pilonieta MC, Erickson KD, Ernst RK, Detweiler CS. 2009. A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J Bacteriol 191:7243–7252. doi: 10.1128/JB.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreira CG, Herrera CM, Needham BD, Parker CT, Libby SJ, Fang FC, Trent MS, Sperandio V. 2013. Virulence and stress-related periplasmic protein (VisP) in bacterial/host associations. Proc Natl Acad Sci U S A 110:1470–1475. doi: 10.1073/pnas.1215416110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 48.Purdy D, O'Keeffe TAT, Elmore M, Herbert M, McLeod A, Bokori-Brown M, Ostrowski A, Minton NP. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol Microbiol 46:439–452. doi: 10.1046/j.1365-2958.2002.03134.x. [DOI] [PubMed] [Google Scholar]
- 49.Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J Med Microbiol 54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.