Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is an increasingly prevalent pathogen. We studied the prevalence of MRSA and its association with vaginitis during pregnancy. Bacteriological investigations of high vaginal swabs of 350 healthy pregnant women attending antenatal clinics were carried out. Staphylococci were isolated from high vaginal swabs of 135 of the women. The staphylococcal isolates were resistant to multiple antibiotics. The PCR amplification of DNA of 20 selected isolates yielded six possessing the mecA gene and 13 the blaZ gene. MRSA possessing both the mecA and blaZ genes were isolated from subjects who reported vaginal discharge and itching.
Keywords: Ado-Ekiti, mecA and blaZ genes, Methicillin-resistant Staphylococcus aureus, pregnancy, vaginitis
Introduction
Staphylococcus aureus is a commensal and an opportunistic pathogen which often exists as part of the normal flora on human skin and mucosal surfaces. S. aureus resistant to methicillin has been reported worldwide; it confers to the organism resistance to all penicillinase-resistant penicillins and cephalosporins [1,2]. The presence of methicillin-resistant S. aureus (MRSA) in pregnant women contributes to the development of life-threatening infections of skin and soft tissues, as well as risks of perinatal transmission to the newborn [3]. MRSA has been reported to colonize the vagina in 14% to 22% of pregnant women. Mothers who are colonized with S. aureus during their third trimester of pregnancy or at the time of delivery are more likely to have infants who also carry the organism [4].
MRSA infections can be divided into hospital-associated (HA) infections and community-associated (CA) infections. They differ not only in respect to their clinical features and molecular biology but also to their antibiotic susceptibility and treatment. Methicillin resistance has occurred in S. aureus by mutation of a penicillin-binding protein, a chromosome-encoded protein [5]. HA-MRSA strains carry a relatively large staphylococcal chromosomal cassette mec (SCCmec) belonging to type I, II or III. These cassettes all contain the signature mecA gene responsible for resistance to many classes of non–β-lactam antibiotics. In contrast, CA-MRSA isolates carry smaller staphylococcal SCCmec elements and have type IV or type V SCCmec. These smaller elements also carry the mecA gene; they are resistant to few non–β-lactam classes of antimicrobials and frequently carry Panton-Valentine leukocidin genes [6].
Penicillin resistance in S. aureus manifests predominantly via the production of β-lactamase encoded by the blaZ gene. However, the blaZ gene is plasmid mediated, unlike the chromosome-mediated mecA gene [7]. This study aimed to evaluate the prevalence of MRSA in pregnant women and to study the contribution of the mecA and blaZ genes to vaginitis.
Materials and methods
Study area and population
High vaginal swabs (HVS) were collected aseptically with the aid of sterile plastic speculums from 350 pregnant women attending the antenatal clinic of Ekiti State University Teaching Hospital, Ado-Ekiti, Nigeria, between April 2017 and March 2018. All samples were transported from the hospital, in leak-proof, watertight containers held under cold conditions, to the Microbiology Laboratory of Afe Babalola University, Ado-Ekiti.
Bacteriologic investigations
The vaginal swabs were streaked on freshly prepared sterile Mannitol salt agar (Oxoid, Basingstoke, UK) and incubated aerobically at 37°C for 24 hours. The bacterial isolates from the Mannitol salt agar were subcultured on freshly prepared nutrient agar plates to obtain a pure culture. The bacterial cultures obtained were Gram stained and microscopically observed for Gram-positive cocci in clusters.
The bacterial isolates were characterized using standard biochemical tests as described by Barrow and Feltham [8]. The following biochemical tests were carried out on the isolates: growth at 10% and 15% sodium chloride; growth at 37°C and 45°C; coagulase, haemolysis, methyl red, Voges-Proskauer, starch hydrolysis, casein hydrolysis, citrate utilization, nitrate reduction, H2S production; production of deoxyribonucleases, alkaline phosphatase, catalase, oxidase, urease; fermentation of sugars which include glucose (with or without gas production), galactose, lactose, sucrose, maltose, mannitol, arabinose, cellobiose, melezitose, raffinose, xylitol and xylose; and novobiocin susceptibility. The bacterial isolates were identified on the basis of their cultural, morphologic and biochemical characteristics using the online resource Gideon Informatics [9], with reference to Schleifer and Bell [10].
Antibiotic susceptibility
The staphylococcal isolates were tested for antibiotic susceptibility using the Kirby-Bauer disc-diffusion method on Müller-Hinton agar as described by the Clinical and Laboratory Standards Institute (CLSI) [11]. The test was carried out by spreading inoculums of 0.5 McFarland suspension of the organism on freshly prepared Müller-Hinton agar, followed by placing standard antibiotic discs (Oxoid) on the plates. The plates were incubated aerobically at 37°C for 24 hours, after which the zones of inhibition were measured and interpreted as described by CLSI [11]. The antibiotics used were cloxacillin (5 μg), amoxicillin/clavulanic acid (30 μg), ampicillin/cloxacillin (10 μg), cefoxitin (10 μg), cefuroxime (30 μg), ceftriaxone (30 μg), ofloxacin (5 μg), pefloxacin (10 μg), ciprofloxacin (30 μg), gentamycin (10 μg), streptomycin (30 μg), trimethoprim/sulfamethoxazole (30 μg), erythromycin (15 μg), clindamycin (10 μg), imipenem (10 μg) and meropenem (10 μg). Resistance to cefoxitin was considered as methicillin resistance [11].
PCR amplification of isolated DNA
The DNA of overnight broth cultures of the staphylococci isolates, incubated at 37°C for 18 to 24 hours, was extracted using a Quick-DNA universal kit (Zymo Research, Irvine, CA, USA). PCR for detection of mecA genes was carried out using the primer pair mecA 1 and mecA 2: 5′-AAA ATC GAT GGT AAA GGT TGG C-3′, 5′-AGT TCT GCA GTA CCG GAT TTG C3-′ [12]; with forward: 5′-GCTTTAAAAGAACTTATTGAGGCTTCA-3′ and reverse: 5′-CCACCGATYTCKTTTATAATTT-3′ used for the blaZ gene [7]. Amplification cycles for mecA were carried out using a thermal cycler, considering an initial denaturation of 94°C for 5 minutes, followed by 40 cycles of denaturing at 94°C for 30 seconds, annealing at 55°C for 30 seconds and extension at 72°C for 1 minute with a final extension of 72°C for 5 minutes. The blaZ amplification was carried out using the thermal cycler, considering an initial denaturation of 94°C for 5 minutes followed by 35 cycles of denaturing at 94°C for 30 seconds, annealing at 55°C for 30 seconds and extension at 72°C for 30 seconds, with a final extension of 72°C for 5 minutes followed by a holding step at 4°C. The amplification products were evaluated by 2% agarose gel electrophoresis followed by ethidium bromide (0.5 mg/mL) staining, visualized on a UV transilluminator and documented using molecular weight markers of 100 bp.
Statistical analysis
Model selection log linear analysis was used to determine the association of sociodemographic variables with the frequency of Staphylococcus isolation using categorical data. All statistical analyses were carried out by SPSS 20.0 for Windows software (IBM, Armonk, NY, USA). A value of p ≤ 0.05 was considered statistically significant.
Results
The study was carried out between April 2017 and March 2018, among 350 apparently healthy pregnant women aged 19 to 43 with mean ± SD of 31.24 ± 4.42 years attending the antenatal clinic of Ekiti State University Teaching Hospital, Ado-Ekiti.
Staphylococcus species were isolated from HVS of 135 (38.6%) of the women. These 135 staphylococcal isolates were made up of S. aureus (56.1%), S. saprophyticus (22.7%), S. haemolyticus (4.5%), S. petrasii (4.5%), S. scheiferi (2.3%), S. epidermidis (6.1%), S. carnosus (1.5%), S. hominis (3.0%), S. warneri (1.5%) and S. hyicus (2.3%). Staphylococcus aureus prevalence in HVS among the entire population of pregnant women studied was 21.2%.
Staphylococcus isolation increased significantly with advancement in age (p = 0.014), with older women (>40 years) yielding 100% Staphylococcus isolation. Isolation of Staphylococcus was not found to be significantly associated with education. Staphylococcus isolation increases significantly with parity, vaginal itching and discharge (p = 0.002). Staphylococcus isolation was not found to be significantly associated with gestation, although women in their second trimester of pregnancy (40.8%) had higher frequency of Staphylococcus isolation compared to women in their third trimester (37.2%) and first trimester (38.4%). Women with history of urinary tract infection, frequent receipt of antibiotics, intra-uterine devices (IUDs) for abortion recorded higher frequencies of Staphylococcus isolation than women without such conditions (Table 1).
Table 1.
Demographic data and distribution of frequency of Staphylococcus isolation from high vaginal swabs of pregnant women
| Characteristic | Variable | No. of samples |
Staphylococcus isolation |
p | |
|---|---|---|---|---|---|
| No | Yes | ||||
| Age | ≤30 years | 148 | 99 (66.9) | 49 (33.1) | 0.014∗ |
| 31–40 years | 194 | 115 (59.3) | 78 (40.2) | ||
| ≥40 years | 8 | 0 (0) | 8 (100) | ||
| Education | No education | 2 | 0 | 2 (100) | 0.395 |
| Primary | 2 | 2 (100) | 0 | ||
| Secondary | 16 | 12 (75) | 4 (25) | ||
| Postsecondary | 330 | 200 (60.6) | 130 (39.4) | ||
| Parity | 0 | 124 | 78 (62.9) | 46 (37.1) | 0.002∗ |
| 1 | 109 | 84 (77.1) | 25 (22.9) | ||
| 2 | 90 | 43 (47.8) | 47 (52.2) | ||
| 3 | 23 | 6 (26.1) | 17 (73.9) | ||
| 4 | 4 | 2 (50) | 2 (50) | ||
| Gestation | First trimester | 43 | 29 (67.4) | 16 (37.2) | 0.954 |
| Second trimester | 49 | 29 (59.2) | 20 (40.8) | ||
| Third trimester | 258 | 159 (61.6) | 99 (38.4) | ||
| Itching | No | 276 | 175 (63.4) | 101 (36.6) | 0.039∗ |
| Yes | 74 | 39 (52.7) | 35 (47.3) | ||
| Discharge | No | 204 | 134 (65.7) | 70 (34.3) | 0.050∗ |
| Yes | 146 | 80 (54.8) | 66 (45.2) | ||
| Smoking | No | 350 | 214 (61.1) | 136 (38.9) | |
| Yes | 0 | 0 | 0 | ||
| HIV status | Negative | 342 | 210 (61.4) | 132 (38.6) | 0.697 |
| Positive | 8 | 4 (50) | 4 (50) | ||
| Antibiotic therapy received | No | 245 | 152 (62.0) | 93 (38.0) | 0.747 |
| Yes | 105 | 62 (59.0) | 43 (41.0) | ||
| UTI | No | 317 | 196 (61.8) | 121 (38.2) | 0.673 |
| Yes | 33 | 19 (57.6) | 14 (42.4) | ||
| UTI treatment | No | 315 | 194 (61.6) | 121 (38.4) | 0.928 |
| Yes | 35 | 21 (60.0) | 14 (40.0) | ||
| Previous IUDs exposure | No | 280 | 175 (62.5) | 105 (37.5) | 0.802 |
| Yes | 70 | 39 (55.7) | 31 (44.3) | ||
| Abortion | No | 264 | 167 (63.3) | 97 (36.7) | 0.328 |
| Yes | 86 | 47 (54.7) | 39 (45.3) | ||
Data are presented as n (%). UTI, urinary tract infection. IUDs, intra-uterine devices.
Statistically significant.
The staphylococcal isolates were resistant to multiple drugs, giving a cluster of resistance to four to 13 antibiotics. The organisms showed high resistance to all the penicillins used in the study, with only amoxicillin/clavulanate having 10.77% susceptibility. Only ceftriaxone gave high susceptibility (70.37%) among the cephalosporins (cephems) tested. Moderate susceptibility values were obtained for clindamycin (a lincosamide) and erythromycin (a macrolide). Out of all the aminoglycosides tested, streptomycin had high susceptibility (89.81%), while high resistance was found against gentamycin. High susceptibility values were obtained for the carbapenems and the fluoroquinolones tested (Table 2).
Table 2.
Susceptibility pattern of Staphylococcus isolates to antimicrobial agents
| Antibiotic group | Antibiotic | Susceptibility (%) |
|---|---|---|
| Penicillin | Amoxicillin/clavulanic acid | 10.77 |
| Amoxicillin/cloxacillin | 0 | |
| Cloxacillin | 0 | |
| Cephalosporin | Ceftin | 5.56 |
| Cefoxitin | 32.31 | |
| Ceftriaxone | 70.37 | |
| Carbapenem | Imipenem | 91.54 |
| Meropenem | 66.15 | |
| Aminoglycoside | Gentamycin | 39.23 |
| Streptomycin | 89.81 | |
| Fluoroquinolone | Ciprofloxacin | 81.54 |
| Ofloxacin | 81.54 | |
| Pefloxacin | 76.85 | |
| Folate pathway inhibitor | Trimethoprim/sulfamethoxazole | 42.59 |
| Lincosamide | Clindamycin | 66.15 |
| Macrolide | Erythromycin | 59.23 |
Staphylococcal isolates were classified as methicillin resistant or sensitive on the basis of resistance to cefoxitin as follows: MRSA, 50 (37.0%); methicillin-sensitive Staphylococcus aureus, 20 (17.0%); methicillin-resistant coagulase-negative Staphylococcus (MRCoNS), 41 (30.4%); and methicillin-sensitive coagulase-negative Staphylococcus (MSCoNS), 21 (15.6%). The MRSA prevalence among the entire cohort of women studied was 14.3%.
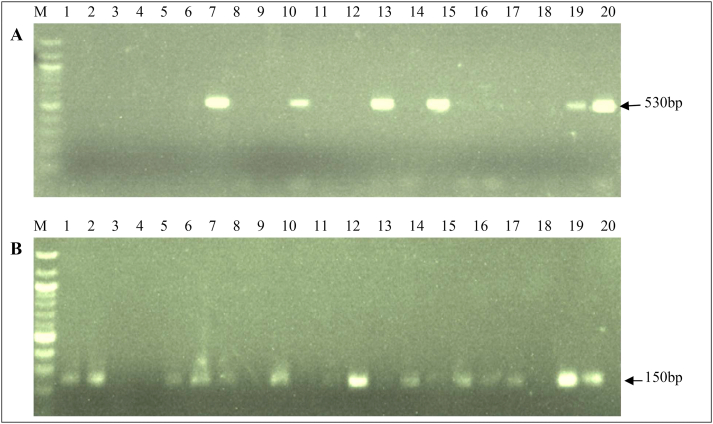
The PCR amplification of DNA extracts of 20 Staphylococcus species isolates, made up of 11 MRSA, two methicillin-susceptible S. aureus (MSSA), five MRCoNS and two MSCoNS, tested for the presence of mecA and blaZ genes, showed that six MRSA isolates possessed the mecA gene, while none of the MSSA, MRCoNS or MSCoNS isolates possessed the mecA gene, a 30% prevalence. The blaZ gene was detected in 13 (65%) of the staphylococcal isolates, made up of eight MRSA, two MSSA and three MRCoNS; neither of the two MSCoNS isolates contained the blaZ gene (Fig. 1). The six MRSA carrying both mecA and blaZ genes were isolated from women experiencing vaginal discharge and itching.
Fig. 1.
PCR amplification of mecA (A) and blaZ (B) genes.
Discussion
Ten different Staphylococcus species were identified from the HVS of the pregnant women in the present study, namely S. aureus, S. saprophyticus, S. haemolyticus, S. petrasii, S. scheiferi, S. epidermidis, S. carnosus, S. hominis, S. warneri, S. hyicus. Staphylococcus aureus was found to be the most prevalent, followed by S. saprophyticus and S. epidermidis. Stanley et al. [13] had previously identified S. aureus as the most common among vaginal staphylococcal isolates in Port-Harcourt, Nigeria.
The staphylococcal isolated from HVS in this study showed resistance to multiple drugs, with high resistance rates for β-lactam drugs and moderate resistance to erythromycin and clindamycin. In a similar study in Hungary by Gajdács and Urbán [14], the resistance rates of Staphylococcus aureus isolated from vaginal samples were below 20% in all the antibiotics tested. They observed highest resistance against erythromycin (11%) and clindamycin (10.85%).
The prevalence of MRSA in this study was 14.3% in pregnant women in Ado-Ekiti (South-West Nigeria), which is at variance with a previous study on MRSA in Ado-Ekiti in South-West Nigeria [15], where a MRSA prevalence rate of 19.2% was reported. Also, our study is at variance with earlier reports from other parts of Nigeria. A prevalence of 12.5% was reported in Maiduguri (Northern Nigeria) [16], 15.1% in Owerri (South-East Nigeria) [17] and 25% in Ekpoma (South-South Nigeria) [18]. However, Gajdács and Urbán [14], in a 10-year retrospective study carried out in Hungary, reported very low prevalence of MRSA, as 97.79% of the Staphylococcus aureus isolated from vaginal samples were susceptible to cefoxitin.
The MRSA isolates showed high resistance to all β-lactam antibiotics, which may be due to the presence of the chromosomal mecA gene, which specifies the production of an abnormal penicillin binding protein, which in turn has low affinity for binding β-lactams [5]. We found that 65% of staphylococci possessed the blaZ gene, which may also account for the high resistance to all β-lactam antibiotics. On the basis of the findings in this study, imipenem, streptomycin, ciprofloxacin, ofloxacin and pefloxacin are the drugs of choice for S. aureus infections in pregnant women.
Resistance of Staphylococcus species isolated from clinical samples to multiple antibiotics has been previously reported in our study area (Ado-Ekiti) [15,19,20]. Multidrug resistance is spreading rapidly among the microbial population and may be traced to the indiscriminate use of antibiotics along with poor hygiene and inadequate infection control, all of which are prevalent in Nigeria and other developing countries [5,21].
The occurrence of MRSA found in association with vaginitis among apparently healthy pregnant women, coupled with its resistance to many frequently prescribed antibiotics, requires increased vigilance in bacteriologic investigations of vaginal infections. There is a need for fast, efficient diagnostic techniques for MRSA for the effective management of its associated infections. A potential novel diagnostic method has been reported by Ábrók et al. [22], who described a combination of MALDI-TOF MS and PBP2’ latex agglutination assay for rapid MRSA detection. This method was found to have positive and negative predictive values of 100% and 99% respectively and can report MRSA colonization 18 to 24 hours after sample collection.
Conflict of interest
None declared.
References
- 1.Gittens-St Hilaire M.V., Chase E., Alleyne D. Prevalence, molecular characteristics and antimicrobial susceptibility patterns of MRSA in hospitalized and nonhospitalized patients in Barbardos. New Microbe. New Infect. 2020;35:100659. doi: 10.1016/j.nmni.2020.100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gajdács M. The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics (Basel) 2019;8:52. doi: 10.3390/antibiotics8020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigos M., Prosniewska M., Seraficka A., Wasiela M. Vagina is not a significant reservoir of methicillin-resistant Staphylococcus aureus in pregnancy. Arch Perinatal Med. 2014;20:108–112. [Google Scholar]
- 4.Jimenez-Teuque N., Tedeschi S., Saye E.J., Mckenna B.D., Langdon W., Wright J.P., al at. Relationship between maternal and neonatal Staphylococcus aureus colonization. Pediatrics. 2012;129:e1252–e1259. doi: 10.1542/peds.2011-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakhundi S., Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31 doi: 10.1128/CMR.00020-18. e00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michael Z.D., Robert S.D. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira L.A., Harnett G.B., Hodge M.M., Cattell J.A., Speersa D.J. Real-time PCR assay for detection of blaZ genes in Staphylococcus aureus clinical isolates. J Clin Microbiol. 2014;52:1259–1261. doi: 10.1128/JCM.03413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrow G.I., Feltham R.K.A. Cambridge University Press; Cambridge: 1993. Cowan and Steel’s manual for the identification of medical bacteria. [Google Scholar]
- 9.Gideon Informatics. Gideon-microbiology-identify bacteria, 1994 -2020. Available at: https://www.gideononline.com/.
- 10.Schleifer K., Bell J.A. Family VIII. Staphylococcaceae fam. nov. In: Vos P.A., Garrity G.M., Jones D., Krieg N.R., Ludwig W., Rainey F.A., editors. Bergey’s manual of systematic bacteriology. 2nd ed. 3—the Firmicutes. Springer; New York: 2015. pp. 392–433. [Google Scholar]
- 11.Clinical and Laboratory Standards Institutes (CLSI) CLSI supplement M100. 28th ed. Clinical and Laboratory Standards Institute; Wayne, PA: 2018. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 12.Strommenger B., Kettlitz C., Werner G., Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 2003;41:4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanley C.N., Ugboma H.A.A., Ibezim E.C., Attama A.A. Prevalence and antibiotic susceptibility pattern of Staphylococcus aureus and other staphylococcal infections in pregnant women attending antenatal clinic in a tertiary hospital in Port Harcourt, Nigeria. J Infect Dis Ther. 2013;2:1–6. [Google Scholar]
- 14.Gajdács M., Urbán E. Epidemiology and resistance trends of Staphylococcus aureus isolated from vaginal samples: a 10-year retrospective study in Hungary. Acta Dermatovenerol. 2019;28:143–147. [PubMed] [Google Scholar]
- 15.Olowe O.A., Kukoyi O.O., Taiwo S.S., Ojurongbe O., Opaleye O.O., Bolaji O.S. Phenotypic and molecular characteristics of methicillin resistant Staphylococcus aureus isolates from Ekiti State, Nigeria. Infect Drug Res. 2013;6:87–92. doi: 10.2147/IDR.S48809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okon K.O., Shittu A.O., Usman H., Adamu N., Balogun S.T., Adesina O.O. Epidemiology and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus aureus recovered from tertiary hospitals in North-Eastern Nigeria. J Med Med Sci. 2013;4:214–220. [Google Scholar]
- 17.Akelere O.J., Anyadoh-Nwadike O.S., Nwadike O.P. Prevalence and antibiogram of multi-drug resistant Staphylococcus aureus among pregnant women attending ante-natal clinics in Owerri, Imo State, Nigeria. Asian J Med Sci. 2013;4:9–14. [Google Scholar]
- 18.Ikheloa J., Inyang N.J., Adelekun A., Agwu E. Molecular analysis of methicillin resistance and beta-lactamase production by clinical isolates of Staphylococcus aureus in Ekpoma, Edo State, Nigeria. Spec Bact Pathogens J. 2015;1:16–22. [Google Scholar]
- 19.Okiki P.A., Obagaye O.C. Prevalence of multi-drug resistant bacteria associated with diarrhoea among infants in Ado-Ekiti, Nigeria. J Biol Agric Healthc. 2015;5:124–132. [Google Scholar]
- 20.Okiki P.A., Idowu R.A., Idris O.O., Osibote I.A., Sobajo O.A. Susceptibility of MDR bacteria associated with respiratory tract infection to methanolic extract of Garcinia kola Heckel (bitter kola) Adv Biol Res. 2015;6:424–435. [Google Scholar]
- 21.Olayinka B.O., Olonitola O.S., Olayinka A.T., Raji B. Antibiotic susceptibility pattern and multiple antibiotic resistance index of Staphylococcus aureus isolates in Zaria, Nigeria. J Trop Biosci. 2004;4:51–54. [Google Scholar]
- 22.Ábrók M., Lázár A., Szécsényi M., Deák J., Urbán E. Combination of MALDI-TOF MS and PBP2′ latex agglutination assay for rapid MRSA detection. J Microbiol Methods. 2018;144:122–124. doi: 10.1016/j.mimet.2017.11.021. [DOI] [PubMed] [Google Scholar]