Abstract
Background
The births of more than 8 million infants have been enabled globally through assisted reproductive technologies (ARTs), including conventional in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) with either fresh embryo transfer (ET) or frozen embryo transfer (FET). However, the safety issue regarding ARTs has drawn growing attention with accumulating observations of rising health risks, and underlying epigenetic mechanisms are largely uncharacterized.
Methods
In order to clarify epigenetic risks attributable to ARTs, we profiled DNA methylome on 137 umbilical cord blood (UCB) and 158 parental peripheral blood (PPB) samples, histone modifications (H3K4me3, H3K4me1, H3K27me3 and H3K27ac) on 33 UCB samples and transcriptome on 32 UCB samples by reduced representation bisulfite sequencing (RRBS), chromatin immunoprecipitation sequencing (ChIP-seq), and RNA sequencing (RNA-seq), respectively.
Findings
We revealed that H3K4me3 was the most profoundly impacted by ICSI and freeze-thawing operation compared with the other three types of histone modifications. IVF-ET seemed to introduce less disturbance into infant epigenomes than IVF-FET or ICSI-ET did. ARTs also decreased the similarity of DNA methylome within twin pairs, and we confirmed that ART per se would introduce conservative changes locally through removal of parental effect. Importantly, those unique and common alterations induced by different ART procedures were highly enriched in the processes related to nervous system, cardiovascular system and glycolipid metabolism etc., which was in accordance with those findings in previous epidemiology studies and suggested some unexplored health issues, including in the immune system and skeletal system.
Interpretation
Different ART procedures can induce local and functional epigenetic abnormalities, especially for DNA methylation and H3K4me3, providing an epigenetic basis for the potential long-term health risks in ART-conceived offspring.
Funding sources
This study was funded by National Natural Science Foundation of China (81,730,038; 81,521,002), National Key Research and Development Program (2018YFC1004000; 2017YFA0103801; 2017YFA0105001) and Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16020703). Yang Wang was supported by Postdoctoral Fellowship of Peking-Tsinghua Center for Life Science.
Keywords: ART, ICSI, Frozen embryo transfer, DNA methylation, Histone modifications, H3K4me3
Research in context.
Evidence before this study
Accumulating observations of rising health risks in ART-conceived offspring have linked ART treatment with potential epigenetic abnormities in offspring. Previous studies mainly focused on epigenetic influence of ARTs on either limited number of genes or repeat elements. In recent years, several groups have reported the impact of ART on genome-wide DNA methylation using Illumina 450 K or EPIC DNA methylation array. However, the similarities and differences among various ART processes have not been fully elucidated and no study has quantified the parental background biases at genome-wide level. In addition, genome-wide changes of histone modifications in ART-conceived offspring and their regulatory role on gene expression have not been reported so far.
Added value of this study
Overall, we systematically profiled DNA methylomes, four genome-wide histone modifications and transcriptomes of offspring and revealed that distinct ART procedures would cause their own unique effects as well as certain common effects on epigenomes. Our study revealed that ART-induced epigenetic alterations tended to be away from active cis-regulatory elements. We showed the reduced similarity in DNA methylomes within ART-conceived twin pairs. In addition, our study also confirmed that a large percentage of DNA methylome changes in offspring could be explained by their parental background biases. Meanwhile, we reported genome-wide impacts of ARTs on four different histone modifications (H3K4me3, H3K4me1, H3K27me3 and H3K27ac) for the first time. IVF-ET showed almost no disturbance in either type of histone modification. H3K4me3 was the most affected histone modification by ICSI and freeze-thawing procedures.
Implications of all the available evidence
To the best of our knowledge, this is the first time that genome-wide maps of DNA methylation, different histone modifications and gene expression have comprehensively been investigated to clarify the multilayer effects of various ART procedures on epigenomes in offspring. Our study provides an epigenetic basis for potential long-term health risks in ART-conceived offspring that reinforces the need to review different types of ARTs in human. Moreover, H3K4me3 might serve as a sensitive biomarker for assessing the influence of aggressive procedures in ART, such as ICSI and freeze-thawing.
Alt-text: Unlabelled box
1. Introduction
Assisted reproductive technology (ART) has become routine in infertility treatment; indeed, more than eight million ART-conceived infants have been born worldwide [1]. However, conventional in vitro fertilization and fresh embryo transfer (IVF-ET) will introduce extraordinary changes in the environment where oocytes mature and the early embryo develops [2]. Moreover, intracytoplasmic sperm injection (ICSI), which was initially used to address severe male infertility, has replaced IVF as the most commonly used method for ART-mediated fertilization in many countries [3]. This more invasive fertilization procedure introduces additional mechanical damage, bypasses the complicated process of sperm-egg recognition and alters a series of downstream reactions [4]. Embryo cryopreservation enables embryos to be preserved for further transplantation, but both cryogens and freeze-thawing operation may cause damage to embryos [5]. All those unfavorable factors have raised concerns regarding the long-term health of ART-conceived children in recent years [6]. Despite claims to the contrary [7], [8], [9], accumulating evidences have linked ART with potentially increased risks of neurodevelopmental disorders, cardiovascular dysfunction and metabolic abnormality in offspring and even preeclampsia during pregnancy [10], [11], [12].
Epigenetic modifications, such as DNA methylation and histone modifications, play key roles in regulating gene expression and are relatively sensitive to environmental factors [13]. Preimplantation embryos undergo dramatic genome-wide epigenetic reprogramming [14,15] that coincides with the time frame of ART treatments. Thus, ART-associated perturbations may disturb the establishment and maintenance of epigenomic patterns and increase the relevant health risks of ART-conceived children in later life [16]. Notwithstanding, published research on the association between ART and DNA methylation in offspring are limited to either specific genes [17,18] or repeated sequences [19] and selected CpG sites on array [20], [21], [22], [23], [24]. Meanwhile, few studies have parsed the parental inheritance bias where considerable reports have revealed that parental genetic backgrounds, health situations, nutritional conditions and living habits have potential impacts on the epigenomes of neonates [25,26]. In addition, genome-wide changes in histone modifications of ART-conceived infants have not been reported and the effects of each specific ART procedure have not been fully elucidated so far.
Here, we integrated genome-wide maps of DNA methylation, four histone modifications associated with promoter/enhancer function (H3K4me1, H3K4me3, H3K27ac and H3K27me3), and gene expression for nuclear families to investigate the specific multilayer effects of various ART procedures on epigenomes in offspring. We found that various ART treatment would not dramatically disturb the global epigenomes of neonates but subtly induced local and functional changes. Our comprehensive analysis not only accords with the findings in previous epidemiology studies but also reveals unexplored health risks in offspring from epigenomic aspect, and may serve as a valuable resource for researchers on the epigenetic influences of ART procedures.
2. Materials and methods
2.1. Ethics statement
All blood samples were obtained after written informed patient consent and were fully anonymized; each individual was assigned a code in this study. The Reproductive Study Ethics Committee of Peking University Third Hospital provided study approval (approved protocol no.201752–044 in 20,170,620). All relevant ethical regulations were followed.
2.2. Study design
The overall objective of this study was to investigate the specific effects of various ART procedures on epigenomes in offspring, including DNA methylome and different histone modifications (H3K4me1, H3K4me3, H3K27ac and H3K27me3). For this study, nuclear family-based cohorts were employedincluding mothers who planned to give birth at the Third Hospital of Peking University with an accurate initial medical history. None of the infants in any of the families suffered from any birth defects. Families in which the mothers suffered from any of the following diseases were excluded: preeclampsia, hyperthyroidism, hypothyroidism, Hashimoto's disease, hypertension, autoimmune disease, polycystic ovarian syndrome (PCOS), severe metabolic syndrome, intrauterine infection or any other pregnancy complication that might have a strong influence on the health of progeny. Families with triplets or stillbirth were also excluded. A total of 32 nuclear families with singletons and 68 nuclear families with twins were recruited in this study and were divided into two main categories: ART-conceived pregnancy and spontaneously conceived pregnancy. All ART cycles were performed at the Center for Reproductive Medicine, Third Hospital of Peking University. The ART-conceived pregnancy families were further subdivided into four groups based on the type of ART applied: the IVF-ET, IVF-FET, ICSI-ET and ICSI-FET groups. According to sex and fetus number, the subjects in the whole groups were subdivided into five subtypes: male twins, female twins, opposite-sex twins, single boys, and single girls. RRBS were performed on umbilical cord blood (UCB) and parental peripheral blood (PPB) samples of selected nuclear families to evaluate the influence of different types of ARTs on DNA methylome. We also perform ChIP-seq to examine histone modifications (H3K4me1, H3K4me3, H3K27ac, and H3K27me3) and RNA-seq to examine the transcriptomes of selected UCB samples, respectively. All blood samples were obtained after written informed patient consent and were fully anonymized; each individual was assigned a code in this study. The detailed classification and related clinical information are shown in Supplementary Table S1.
2.3. Sample collection and treatment
PPB and UCB were collected on the delivery day. Approximately 3 mL blood samples were drawn from each individual and processed through the following steps as soon as possible to prevent degradation. One of three 300 µL whole-blood aliquots was used for DNA extraction, and the other two were stored at −80 °C for backup; the rest of the whole blood was used for RNA extraction immediately. For families in which histone modifications were detected, approximately 20 mL of blood was collected, and peripheral blood mononuclear cell (PBMC) extraction was performed immediately.
2.4. DNA extraction
Genomic DNA (gDNA) was extracted from stored whole blood samples using the QIAmp DNA Blood Mini Kit (Qiagen Cat# 51,106), according to manufacturer's instruction and stored in −80 °C if not used immediately.
2.5. RNA extraction
Total RNA was prepared from fresh blood by QIAmp RNA Blood Mini Kit (Qiagen Cat# 52,304), according to manufacturer's instruction and stored in −80 °C if not used immediately.
2.6. PBMCs extraction and fixation
PBMCs were isolated by Ficoll density gradient centrifugation (TBD Cat# LDS1075). Then about 10 million PMBCs were fixed with freshly prepared 1% formaldehyde in 1X PBS for 10 min at room temperature. Glycine buffer (125 mmol/L) was added to quench the crosslinking reaction, with further incubation for 5 min at room temperature. After washing once with cold 1X PBS, fixed PBMCs pellet was cryopreserved at −80 °C for subsequence ChIP-seq.
2.7. RRBS
RRBS was performed by following the published protocol with little modification [27]. In brief, approximately 0.3% unmethylated lambda DNA (dam-; dcm-) (Thermo Scientific Cat# SD0021) was spiked into 500 ng of high-quality gDNA, and the mixture was digested by MspI (Thermo Scientific Cat# ER0541). After purification with Agencourt AMPure XP beads (Agencourt Cat# A63881), the digested DNA was end-repaired and tailed with deoxyadenosine using Klenow Fragment exo- (Thermo Scientific Cat# EP0422). Then, NEBNext adapters (NEB Cat# E7335) were ligated overnight with T4 DNA ligase at a high concentration (NEB Cat# M0202M) with subsequent digestion by Uracil-Specific Excision Reagent (USER) enzyme (NEB Cat# M5505L). Bisulfite conversion was conducted with a MethylCode Bisulfite Conversion Kit (Thermo Scientific Cat# MECOV-50). The converted samples were size-selected by excising gel slices containing 160 bp~700 bp DNA (2% TAE gel). After recovery using a gel DNA recovery kit (VISTECH Cat# PC0313), the converted fragments were amplified through PCR with a maximum of 12 cycles with Kapa HiFi U+ Master Mix (Kapa Biosystems Cat# KK2801). Barcodes were also introduced during this process. The purified PCR-amplified products were quantified using Qubit dsDNA high-sensitivity dye in a Qubit 3.0 Fluorometer (Thermo Scientific Cat# Q33216/Q32854). The exact fragment distributions were examined with a Fragment Analyzer™ Automated CE System (Analysis Kit: Cat# DNF-474–0500) following the detection of molar concentration using a Library Quant Kit for Illumina (NEB Cat# E7630L). The final qualified libraries were sequenced using the PE150 strategy on an Illumina X Ten sequencer.
2.8. ChIP-seq
ChIP-seq was performed as previously described [28]. Briefly, isolated nuclei were sonicated into fragments of an average of 200–500 bp using an M220 Focused-ultrasonicator (Covaris) with the following parameters: burst, 200; cycle, 20%; and intensity, 8. After centrifugation (13,000 rpm for 10 min at 4 °C), sheared chromatin in supernatant was precleared by incubating the samples with protein A Dynabeads (Invitrogen Cat# 10002D) at 4 °C for 1 h with rotation. The supernatant, which contained 12.5–50 µg of chromatin and was further diluted into 250 µL, was transferred to low-binding EP tubes and incubated with 1 µg or 2 µg of the following antibodies at 4 °C overnight with rotation: H3K4me3 (Millipore Cat# 07–473, RRID:AB_1977252), H3K27me3 (Abcam Cat# ab6002, RRID:AB_305,237), H3K4me1 (Abcam Cat# ab8895, RRID:AB_306847), and H3K27ac (Abcam Cat# ab4729, RRID:AB_2118291). After being washed with cold RIPA (10 mM pH 7.6 HEPES, 1 mM EDTA, 4 mM LiCl, 1% NP-40, 0.1% N-lauryl sarcosine) and TEN (1 mM EDTA, 10 mmol/L pH 8 Tris, 50 mM NaCl), the immunoprecipitated pellets consisting of chromatin immune complexes and protein A Dynabeads (Invitrogen Cat# 10002D) were recovered by digestion with 20 µg of Proteinase K (Qiagen Cat# 19,131) in 200 µL of elution buffer at 65 °C for over 6 h; a 10% input control sample stored previously was also processed. DNA was purified using a MinElute PCR Purification Kit (Qiagen Cat# 28,006). Then, libraries were constructed using a NEXTflex™ ChIP-Seq Kit (Bioo Scientific Cat# 5143–02) according to the manufacturer's instructions. Briefly, 5~10 ng of DNA was end-repaired. Then, fragments of 180–280 bp were size-selected using Agencourt AMPure XP beads (Beckman Cat# A63881). After being tailed with deoxyadenosine and ligated to barcoded ChIP-seq adapters (Bioo Scientific Cat# 514,124), the DNA fragments were amplified by PCR with 12 cycles. The final qualified libraries for the same sample were pooled together and sequenced as described in the above RRBS protocol. The requirement of data output was 12 G of raw data for the input library and 10 G of raw data on average for each histone modification library.
2.9. RNA-seq
mRNA libraries were constructed using NEBNext® UltraTM II RNA Library Prep Kit for Illumina®(NEB Cat# E7770L/E7775L) following the vendors protocol. Briefly, poly(A) mRNA was purified from approximately 500 ng of input total RNA by NEBNext Magnetic Oligo d(T)25 Beads (NEB Cat# E7490). After fragmentation and subsequently the first strand/Second strand cDNA synthesis, purified double-stranded DNA fragments were filled into blunt ends and dA-tailing. The ligation reaction was performed with further addition of USER Enzyme to cut the hairpin loop structure within NEBNext Adaptor (NEB Cat# E7335). Finally, adaptor-ligated DNA fragments were size-selected using SPRIselect Beads (Beckman Cat# B23317) and the index was introduced in PCR amplification reaction. The quality of purified library was assessed as described above.
2.10. RRBS data analysis
Paired-end 150-nucleotide (nt) RRBS reads were generated with a HiSeq X Ten sequencer. Before read mapping, adapters, low-quality bases in the reads and reads smaller than 50 bases were removed with Trim Galore (version 0.4.1; https://github.com/FelixKrueger/TrimGalore) and Cutadapt [29] (version 1.9.1) to optimize the paired-end alignments. The remaining reads were aligned to the Homo sapiens reference genome (human GRCh38/hg38) using Bismark [30] (version 0.18.1) with the parameter “–bowtie2”. The uniquely mapped reads with less than 2% mismatch were retained for further analyses. Bisulfite conversion efficiency was estimated by aligning the reads to the spiked-in phage λ genome. The sequencing data and bisulfite conversion efficiency, and the number of covered CpG sites were summarized in Supplementary Table S3.
For DNA methylation calling, the 3 nt at both ends of each read were removed to prevent bias in DNA methylation level estimation based on M-bias reports by using the script “bismark_methylation_extractor” in Bismark. Then, the number of reads supporting C (methylated) and supporting T (unmethylated) for each covered CpG site was determined with the Bismark methylation extractor. Only the CpG sites with greater than fivefold read coverage (>=6) were passed to the script “bismark2bedGraph” in Bismark to generate a bedGraph file as well as a coverage file, both of which were sorted by chromosomal position.
There were data of 140 neonates, 79 fathers, and 79 mothers from 30 singleton nuclear families and 55 twin nuclear families (49 twin families with parents, 6 monozygotic twin families without parents). Based on unsupervised clustering of these RRBS samples, 3 neonate samples as outliers were removed. Finally, 137 samples for neonates and 158 samples for parents were used to downstream analysis.
As there were 5 groups of neonate samples, it was necessary that the CpG sites were covered in most of the samples. Therefore, first, only autosomal CpG sites with greater than 5-fold read coverage in at least 18 samples in each neonate group were selected. Second, the overlapping sites among the selected CpG sites from the 5 groups were taken into consideration for further analysis. Therefore, Cs (cytosines) of CHH and CHG context, on sex chromosomes (X and Y), or with low coverage in CpG context, all of them were removed; only autosomal Cs of CpG context covered by at least 90 neonatal samples were used to further analysis. The DNA methylation level of each retained CpG was determined by the ratio of the number of methylated reads to the total number of reads. The DNA methylation levels for different genomic regions, such as tiles, promoters, repetitive regions, imprinting regions and metastable epialleles, were determined by the ratios of the numbers of methylated reads to the total numbers of reads across all the retained CpG sites in the corresponding regions.
2.11. Identification of DMRs
DMRs between any two groups of RRBS samples were identified with the R/Bioconductor package methylKit (version 1.6.1), which uses logistic regression to calculate p-values [31]. The p-values were adjusted to q-values using the Sliding Linear Model (SLIM) method. The parameters used for methylKit included a 200 bp window and a 50 bp step size. Only the 200 bp window with 3 or more kept CpG sites were used to downstream analysis. Each of the 5 groups of newborn twins was divided into 2 subgroups for separation of twin pairs. In comparisons of any two groups (four subgroups) of twins, the DMRs between corresponding subgroups (four comparisons) were generated by using methylKit with a q-value threshold of 0.05 and a mean methylation difference of 5%. Then, to identify DMRs between any two groups of neonates, six comparisons were generated from all neonates (1 comparison), singletons (1 comparison), and twins (4 comparisons). DMRs that appeared in at least four comparisons were selected for newborn twins to improve the confidence. To identify DMRs between any two groups of fathers or mothers, three comparisons were generated from fathers or mothers of all neonates (1 comparison), fathers or mothers of all singletons (1 comparison), and fathers or mothers of all twins (1 comparison). DMRs that appeared in at least two comparisons were selected for fathers or mothers to improve the confidence. Finally, the hyper- and hypo-DMRs between any two groups were merged as the final DMRs if they had a common region of at least 1 bp.
2.12. ChIP-seq data analysis
In order to optimize the paired-end alignments, adapters, low-quality bases in the reads and reads smaller than 50 bases were removed with Trimmomatic (version 0.36) before read mapping [32]. The remaining reads were aligned to the Homo sapiens reference genome (human GRCh38/hg38) using the Burrows-Wheeler Aligner (BWA) (version 0.7.12) with the default parameters in paired-end mode [33]. For duplicated reads that aligned to the same position with the same orientation, only the read with highest mapping quality was retained to prevent potential PCR bias by using picard (version 1.4.2) (http://broadinstitute.github.io/picard/). The nonredundant reads with no more than 2% mismatches that uniquely mapped to the reference genome were retained for further analyses. The sequencing data and the number of peaks were summarized in Supplementary Table S3.
The MACS2 peak caller [34] (version 2.1.0) with the parameter settings “–keep-dup=1, –broad” was used to identify peaks of histone modifications. BAM files were converted to BigWig files and the read numbers normalized to the numbers per 20 million aligned reads with bamCoverage in deepTools [35] (version 3.1.0), and the BigWig files were viewed using the Integrative Genomics Viewer (IGV) [36] (version 2.4.5). In box-violin plots, histone modification signals at 20 bp intervals (ChIP reads minus input reads; retained only if the value was >0) were calculated with DANPOS [37] (version 2.2.2) and plotted using the R package ggplot2 (version 2.2.0) (Wickham H. ggplot2: elegant graphics for data analysis [M]. Springer, 2016.). The genome-wide distributions of ChIP signals were determined by counting the histone modification signals within nonoverlapping 1 kb windows tiled over the human genome. The R/Bioconductor package DiffBind (version 2.2.12) was used [38], which employs DESeq2 (version 1.16.1) or edgeR (version 3.18.1) to identify differential peaks (DPs) with q-values 〈 0.05 and fold changes 〉 2 for H3K4me1, H3K4me3, H3K27ac, and H3K27me3, respectively. The shared outputs of the DESeq2 and edgeR methods were taken as the final DPs. Based on the combined ChIP signal of the four histone modifications, 12 chromatin states were generated to separate the genome into distinct functional regions by using chromHMM [39] (version 1.15).
2.13. RNA-seq data analysis
The 150 nt paired-end RNA-seq reads were preprocessed similarly to the ChIP-seq reads with the same software. Briefly, the retained reads were mapped to the Homo sapiens reference genome (human GRCh38/hg38) using STAR [40] (version 2.5.3a) with the default parameters. The uniquely mapped reads with less than 2% mismatch were passed to StringTie [41] (version 1.3.3b) for transcript assembly, and the transcripts per million (TPM) value was also generated for each gene. Batch effects of TPM values were corrected with the R/Bioconductor package SVA [42] (version 3.22.0). Finally, the DEGs were called with DEseq2 [43] with the criterion of an adjusted p-value<0.1 (the p-values were attained by the Wald test and adjusted by BH method). The sequencing data are summarized in Supplementary Table S3.
2.14. Sample clustering
Hierarchical clustering, principal component analysis (PCA) and multidimensional scaling (MDS) were used for unsupervised clustering of the epigenomes of CTRL and ART samples with the R functions ‘hclust’, ‘prcomp’, and ‘cmdscale’, respectively, in the R base package stats. For hierarchical clustering of binary classification, its accuracy was measured by counting the maximum number of correctly assigned samples and dividing by the total number. K-means clustering were used for unsupervised clustering of DMRs by using R function kcca with parameter “family=kccaFamily (which=NULL, dist=distCor)” in the flexclust (version 1.3.3) package. To calculate distance and similarity matrices, we used the dist and cor functions, respectively, of the stats package in R with the parameter “method=euclidean”.
2.15. Functional enrichment and overlapping analysis of genomic regions
Potential biological functions of genomic regions, such as DMRs, histone modification peaks and differential peaks, are determined by gene ontology (GO) and human phenotype ontology (HPO) analysis. GO enrichment analysis was carried out using the R/Bioconductor package ChIPseeker [44] (version 1.10.3) and Homo sapiens annotation package org.Hs.eg.db (version 3.5.0). The enrichment analysis of human phenotype was performed by Erichr [45,46], and the top 20 terms with the highest p-value (<0.05) were used. Two or more genomic regions were considered as overlapping regions if they had 1-bp common region at least, which was implemented by subcommand mergePeaks with parameter “-d given” in HOMER [47] (version 4.9).
2.16. Statistical analysis
Fisher's exact test was applied for comparing gender and mode of birth while unpaired two-tailed ANOVA was used for comparing other clinical features of parents or progenies (including “Paternal/Maternal age”, “Paternal/Maternal body height”, “Paternal /Maternal body mass”, “Paternal /Maternal BMI”, “Gestational week”, “Birth weight” and “Birth length”) among different conception groups using R package ggstatsplot [48] (version 0.1.1). For violin-box plots, the center dot indicated the average, the significance of differences between two groups was determined by Wilcoxon rank sum test. To compare the distribution of Pearson correlation coefficients, unpaired and two-tailed t-test was performed unless otherwise specified. Hypergeometric test was used to determine the significance of overlap between two groups of genomic regions or genes. The statistical significance was determined using hypergeometric test in gene ontology (GO) terms and KEGG pathway enrichment analysis, with p value adjusted by the multiple test adjustment (Benjamini-Hochberg, BH).
2.17. Role of funding source
The Funders didn't play any roles in study design, samples collection, production, analyzation, and interpretation of data, paper writing, or the decision to submit the paper for publication.
3. Results
3.1. Global epigenomic profiles in ART-conceived neonates
To systematically study the DNA methylomic effects of different types of ARTs on offspring, we performed reduced-representation bisulfite sequencing (RRBS) on 137 umbilical cord blood (UCB) samples and 158 parental peripheral blood (PPB) samples from nuclear families with either singletons or twins (Fig. 1a, Supplementary Fig. S1a, and Supplementary Table S1). The samples were classified into five groups based on the mode of conception: spontaneous (CTRL), IVF-ET, IVF-FET, ICSI-ET and ICSI-FET. There were no significant differences in the clinical features of the neonates among the different groups; in addition, maternal ages were under 35 years old and parental body mass index (BMI) values were comparable in general (Supplementary Table S2). In addition, we performed chromatin immunoprecipitation sequencing (ChIP-seq) on 33 UCB samples to examine histone modifications (H3K4me1, H3K4me3, H3K27ac, and H3K27me3) and RNA sequencing (RNA-seq) on 32 UCB samples to examine the transcriptomes of neonates (Fig. 1a, Supplementary Fig. S1a, and Supplementary Table S1).
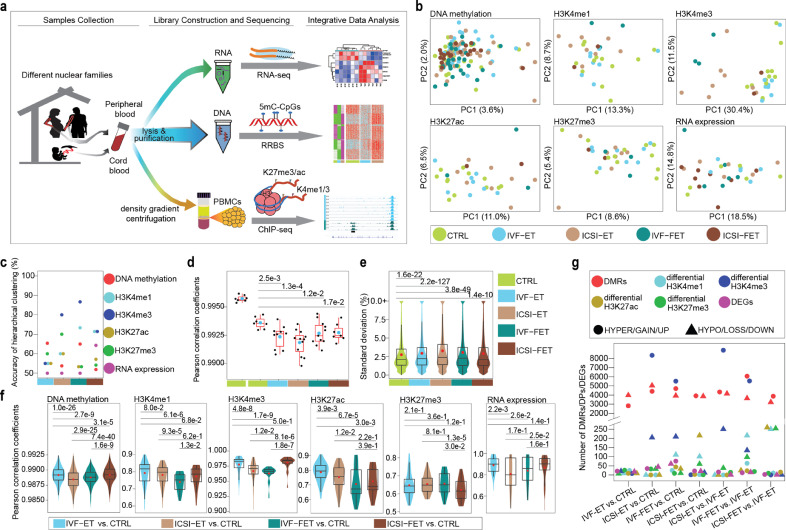
Fig. 1.
Epigenome profiling in ART-conceived neonates. (a) Graphical overview of the study design. (b) Principal component analysis (PCA) for all the five groups (CTRL, IVF-ET, ICSI-ET, IVF-FET, and ICSI-FET) of neonatal samples by using each layer of the reference epigenome and transcriptome. For DNA methylation (sample size: n = 137), only the 100-bp tiles covered all neonatal samples were used; ChIP-seq peaks of all neonatal samples (sample size: n = 33) for each histone modification were pooled together, then the overlapping peaks were merged; genes with TPM=0 in all neonatal samples(sample size: n = 32) are removed. Finally, the number of genomic regions for each epigenomic layer to generate the PCAs: DNA methylation (467,097 100-bp tiles), H3K4me1 (195,570 peaks), H3K4me3 (86,157 peaks), H3K27ac (324,900 peaks), H3K27me3 (257,175 peaks), and RNA expression (26,326 genes). (c) The accuracy of hierarchical clustering for IVF-ET versus CTRL (cyan), ICSI-ET versus CTRL (sienna), IVF-FET versus CTRL (dark cyan), and ICSI-FET versus CTRL (dark sienna) neonatal samples. Samples and genomic regions used were the same as in (b). (d) Box plots for the distribution of the within-twin-pair Pearson correlation coefficients for genome-wide DNA methylation in each group as in (b), respectively. Twin pairs in CTRL group were classified into two subgroups, monozygotic and dizygotic twins. Each black dot represents the Pearson correlation coefficient and the cyan dots are the arithmetic means. The p-value between two groups was determined by unpaired and two-tailed t-test. (e) Violin-box plots showed the distribution of standard deviations of genome-wide DNA methylation for neonatal samples in each group mentioned in (b), respectively. The p-value between two groups was determined by Wilcoxon rank-sum test. (f) For each layer of the reference epigenome and transcriptome, violin-box plots showed the distribution of Pearson correlation coefficients between CTRL and one of the four ART groups. The red dots are the arithmetic means. The p-value between two groups was determined by Wilcoxon rank-sum test. (g) The number of DMRs, DPs of four histone modifications, and DEGs for neonatal samples were shown for seven comparisons: IVF-ET versus CTRL, ICSI-ET versus CTRL, IVF-FET versus CTRL, ICSI-FET versus CTRL, ICSI-ET versus IVF-ET, IVF-FET versus IVF-ET, and ICSI-FET versus IVF-ET.
The results showed that the global DNA methylation levels, histone modifications, and transcriptomes of individual neonates were overall similar among the CTRL and four ART groups (Supplementary Fig. S1, b, c and d). There were also no noticeable intergroup differences for DNA methylation of various functional genomic regions, such as promoters, enhancers and repeats (Supplementary Fig. S1e). Unsupervised clustering of each layer of the reference epigenome showed no obvious subgroups but rather showed broadly distributed patterns among neonates, except for H3K4me3 in the IVF-FET and ICSI-ET groups (Fig. 1b and Supplementary Fig. S2a). These observations were further verified in hierarchical clustering analyses of the CTRL group versus specific ART subtype groups or ARTs as a whole; the accuracies in these analyses were close to a completely random value (50%) for binary classification but were markedly higher with regard to H3K4me3 in the IVF-FET and ICSI-ET groups (Fig. 1c and Supplementary Fig. S2b-h). These results suggested that ART processes do not dramatically disturb the overall epigenomes and transcriptomes of neonates in general but that H3K4me3 might be exceptionally sensitive to disturbance by specific ART procedures.
Correlation analysis for the DNA methylomes of twin pairs showed an overall high correlation coefficient greater than 0.99 (Fig. 1d). However, the coefficient for monozygotic twins was significantly higher than that for dizygotic twins in the CTRL group (Fig. 1d), consistent with previous studies [49,50]. Surprisingly, the correlation coefficients in all four ART subgroups were also relatively lower than in the CTRL group. This phenomenon could also be observed when only same-sex twins were analyzed or in any two non-twin-pair neonates in each group (with exclusion of twin-pair bias) (Figure S3a, S3b, and S3c). Meanwhile, the standard deviations of the DNA methylation levels of all neonates in the different ART groups were slightly but significantly higher than that in CTRL group (Fig. 1d and 1e). Above results implied that ART itself might increase the heterogeneity of DNA methylome of neonates, and reduced the within-pair similarity in ART-conceived twins. Intergroup correlation analysis further revealed that IVF-ET group was the most similar to CTRL group with regard to H3K4me1, H3K4me3, H3K27ac, transcriptomes and DNA methylomes genome-wide or on some special elements (Fig. 1f and Supplementary Fig. S3d), indicating that IVF-ET might have less epigenetic influence on neonates than the other three ART processes. These findings suggested that imperceptible epigenomic disturbances were introduced into ART-conceived neonates and might vary among different ART subtypes.
3.2. Conservative epigenomic changes in ART-conceived offspring
To elucidate the epigenetic effects of ARTs on neonates, we performed pairwise comparisons between groups for DNA methylation, histone modifications, and gene expression. In particular, to identify differentially methylated regions (DMRs), six comparisons were generated using all neonates, only singleton neonates, and four groups of neonates from twin cohorts. DMRs in at least four comparisons were selected for downstream analysis to improve the confidence of the analysis (Supplementary Fig. S3e, see Methods for more details). The numbers of hyper-/hypo-DMRs, gain/loss differential histone modifications (differential peaks, DPs), and up-/downregulated differentially expressed genes (DEGs) among the different comparison groups were shown in Fig. 1g and Supplementary Table S4 (see Supplementary Table S5 to S10 for more details).
Only a few DPs were observed in the comparison between the IVF-ET and CTRL groups for four histone modifications, suggesting that IVF-ET has little impact on histone modification (Fig. 1g and Supplementary Table S4). However, a lot of H3K4me3 DPs were detected in the comparison between the ICSI-ET/IVF-FET and CTRL groups (ICSI-ET: 8352 gain, 205 loss; IVF-FET: 5526 gain, 210 loss), which were consistent with the observation in unsupervised clustering (Fig. 1b, and Supplementary Fig. S2, a, f-g), The difference detected in DMRs was mainly less than 15% and the majority of the increased DPs in the ICSI-ET/IVF-FET groups were in regions with originally weak signals in the CTRL and IVF-ET groups (Supplementary Fig. S4, a and b). All of the DEGs, DMRs and DPs for each ART group versus the CTRL group were further validated at the individual level and broadly distributed on the genome scale (Supplementary Fig. S4, c, d and e, and Supplementary Fig. S5). Together, these results indicated that ART processes potentially caused conservative epigenetic changes distributed widely throughout the genome.
3.3. Parent-derived and ART-derived dna methylomic differences in offspring
To evaluate parental influence on the DNA methylomes of progeny, the DNA methylation levels of DMRs identified in neonates were further analyzed in parents. There were significant differences between the two groups of fathers or mothers (Fig. 2a and Supplementary Fig. S6a; p<0.01, Wilcoxon rank-sum test). In addition, 30.9 - 50.1% of DMRs identified in neonates significantly overlapped with DMRs identified in their corresponding parents (Fig. 2b, Supplementary Fig. S6b, and Supplementary Table S14). To investigate the effects of ARTs per se, the overlapping DMRs were removed and the remaining neonatal DMRs were used as the final neonatal DMRs for downstream analysis (Supplementary Fig. S6c, Supplementary Table S11 and Supplemental Table 15). As expected, no significant differences were observed in the DNA methylation levels of the final neonatal DMRs between the corresponding parental groups (Supplementary Fig. S6, d and e). However, the significant differences persisted in all seven pairwise comparisons when we compared the overall DNA methylation level of final neonatal DMRs in corresponding cord blood samples in different genders respectively (Supplementary Fig. S7). Above results together indicated ART per se was also able to induce conservative DNA methylomic changes, which were affected little by gender difference.
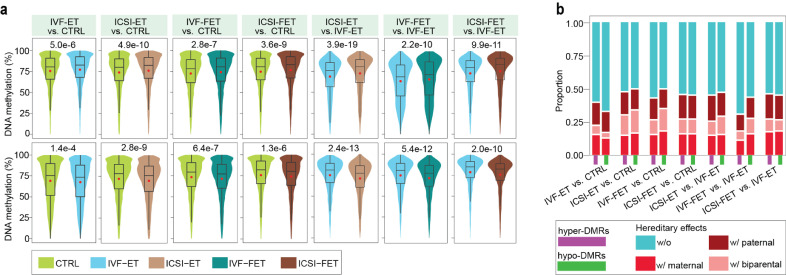
Fig. 2.
Removal of the parental effects in neonatal DMRs. (a) Violin-box plots displayed the distribution of DNA methylation level in paternal samples at hyper- (upper) and hypo-DMRs (lower) of the seven comparisons, IVF-ET versus CTRL, ICSI-ET versus CTRL, IVF-FET versus CTRL, ICSI-FET versus CTRL, ICSI-ET versus IVF-ET, IVF-FET versus IVF-ET, and ICSI-FET versus IVF-ET neonatal samples. The p-value between two groups was determined by Wilcoxon rank-sum test. (b) Bar graph showed the proportion of DMRs with/without hereditary effects in the hyper- or hypo-DMRs of each comparison. Any one of the 14 groups of DMRs were classified into four subgroups, without hereditary effects (w/o), only with paternal hereditary effects (w/paternal, only overlapped with the DMRs of comparison for the corresponding fathers), only with maternal hereditary effects (w/ maternal, only overlapped with the DMRs of comparison for the corresponding mothers), and with biparental hereditary effects (w/ biparental, overlapped with the DMRs of comparisons for the corresponding fathers and mothers).
3.4. Epigenomic alteration of regulatory regions in IVF-ET-conceived offspring
IVF-ET, the most basic process of ART treatment, is generally recommended as a first-line ART therapy for couples with female infertility [51]. A total of 1703 hyper-DMRs and 2658 hypo-DMRs were identified in the IVF-ET group compared with the CTRL group (Supplementary Table S11). Gene Ontology (GO) enrichment analysis showed that the associated genes of DMRs were enriched in a broad range of processes, including processes related to the nervous system, respiratory system and cardiovascular system, etc. (Fig. 3a and Supplementary Table S16). Human Phenotype Ontology (HPO) analysis also indicated that alterations in DNA methylome induced by IVF-ET might lead to abnormal phenotypes in ocular, cardiovascular and skeletal system, tooth morphology and metabolism, etc. (Supplementary Fig. S8a and Supplementary Table S16).
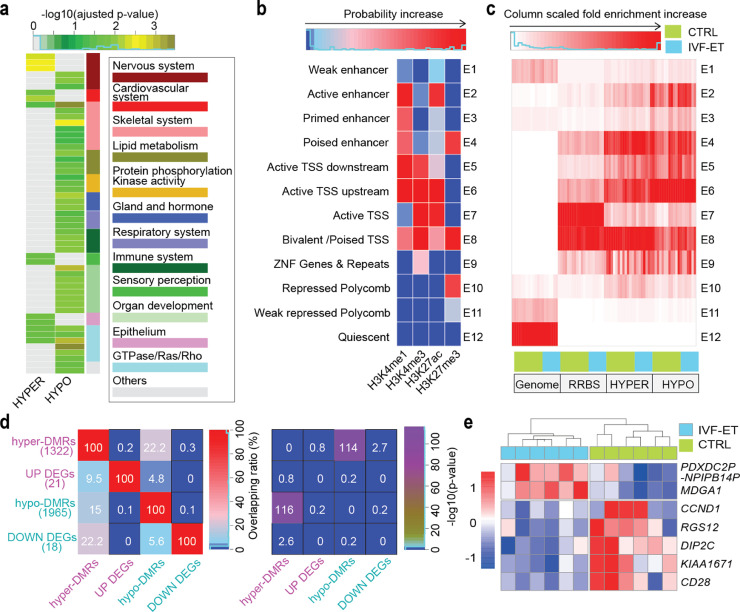
Fig. 3.
IVF effects on the epigenome of the offspring. (a) Gene ontology (biological process) analysis for the hyper- and hypo-DMRs of IVF-ET versus CTRL, all enrichment terms with adjusted p-value < 0.05 were grouped and shown. The p-value was determined by hypergeometric test and adjusted by BH method. (b) Based on the four histone modifications in all neonatal samples, 12 chromatin states were defined by chromHMM. Each row of the heatmap corresponds to a specific chromatin state, and each column corresponds to a different histone mark, H3K4me1, H3K4me3, H3K27me3, or H3K27ac. (c) In each neonatal sample of CTRL and IVF-ET groups, column scaled fold enrichment of the chromatin states at genome-wide regions (Genome), RRBS covered regions (RRBS), hyper- and hypo-DMRs (HYPER and HYPO) was shown. (d) Heatmap showed overlapping ratio among associated genes of DMRs and DEGs for IVF-ET versus CTRL (left), and their statistical significances (right) determined by hypergeometric test. (e) Heatmap showed the expression level of DEGs overlapped with the associated genes of DMRs (PDXDC2P-NPIPB14P, MDGA1, CCND1, RGS12, DIP2C, KIAA1671, and CD28) among neonatal samples in CTRL and IVF-ET.
Given the distinct roles of various genomic regions in regulating gene expression, we performed ChromHMM using four types of histone modifications and identified 12 chromatin states to investigate the specific impacts of IVF-ET on the DNA methylation of functional elements (Fig. 3b). The regions covered in our DNA methylation data were mainly the regions related with transcription start site (TSS) (E7 and E8), consistent with the features of highly enriched CpG regions in RRBS. Notably, a large fraction of DMRs were concentrated on Active enhancer (E2), Poised enhancers (E4), Active TSS upstream sites (E6) and Bivalent/Poised TSSs (E8), but almost depleted from Active TSSs (E7) (Fig. 3c). GO analysis for those DMRs in different chromatin states suggested that those hyper-DMRs in Weak enhancer (E1), Bivalent/Poised TSSs (E8) and ZNF Genes & Repeats (E9) might be associated with the interference on nervous system, while hyper-DMRs in Repressed Polycomb (E10) might account for the influence on cardiovascular system. Meanwhile, hypo-DMRs in Active TSS upstream (E6) and Weak repressed polycomb (E11) might highly correlate with immune system and skeletal system, respectively (Supplementary Fig. S8b and Supplementary Table S16).
The intersection of the DMR associated genes with the DEGs revealed that the majority of the DMRs were not associated with transcriptional changes in their associated genes (Fig. 3d and Supplementary Table S17). For DEGs overlapped with hypo-DMRs associated genes, MDGA1 was upregulated in the IVF-ET group, while RGS12 was downregulated; in the former, the DMRs occurred in promoters, while in the latter they occurred in introns. The expression of genes associated with hyper-DMRs in introns or distal intergenic regions, such as KIAA1671 (introns), DIP2C, CCND1 and CD28, was decreased (Fig. 3e). Polymorphisms/mutations in MDGA1 and DIP2C are associated with mental disease, and dysregulation of KIAA1671, CCND1, and DIP2C has been reported to take part in tumorigenesis; furthermore, RGS12 dysfunction contributes to tumorigenesis as well as pathological cardiac hypertrophy, osteoclast genesis and bone destruction [52], [53], [54], [55], [56], [57]. Taken together, our findings indicated that DNA methylomic changes caused by IVF-ET might affect multiple aspects related with neonates’ health, but mainly kept away from active TSS regions.
3.5. Identification of altered epigenomic profiles associated with ICSI procedure
To investigate the differences between those two ways of fertilization, we first compared ICSI-ET with IVF-ET and identified more than 4500 DMRs and 9000 H3K4me3 DPs (Supplementary Table S4 and Supplementary Table S11; 2365 hyper-DMRs and 2386 hypo-DMRs; 8914 gain DPs and 270 loss DPs). Although there were no overlapped genomic locations, the changes shared common associated genes and GO terms (Supplementary Fig. S9, a, b and c), with only a few genes showing consistent changes in expression with changes in the epigenome (Supplementary Table S18). Taking the differences between the ICSI-ET and CTRL groups into account enabled us to determine the abnormal effects of ICSI per se. Thereafter, the DMRs and H3K4me3 DPs of the ICSI-ET group versus the IVF-ET group were refined, the former based on k-means clustering of DNA methylation levels among the ICSI-ET, IVF-ET and CTRL groups and the latter based on integrated comparison between DPs of ICSI-ET versus CTRL (n = 8557) and DPs of ICSI-ET versus IVF-ET (n = 9184). Six clusters of DMRs (C1~C3 for hyper-DMRs, C4~C6 for hypo-DMRs) and overlapping H3K4me3 DPs (n = 7317) that showed similar change tendencies in the comparison of ICSI-ET versus IVF-ET and ICSI-ET versus CTRL groups were selected for downstream analysis (Fig. 4a and 4b, Supplementary Fig. S9d and Supplementary Table S19). The associated genes of hyper-DMRs and gain DPs were enriched in processes involving the development of skeletal system and Wnt signaling morphogenesis, respectively (Fig. 4c and Supplementary Table S20). Meanwhile, HPO annotation of those DMRs also revealed that the disturbance in those regions might potentially introduce adverse effect on appendage and skeletal system (Supplementary Fig. S9e and Supplementary Table S20). Though there were only 84 loss DPs of H3K4me3, they were highly enriched in GO terms of immune system (Fig. 4c and Supplementary Table S20).
Fig. 4.
Separate epigenetic effects of ICSI. (a) K-means clustering for the hyper- (left) and hypo-DMRs (right) of ICSI-ET versus IVF-ET. In green-purple heatmaps, the hyper-DMRs (hypo-DMRs) were classified into seven (nine) clusters by k-means clustering on row scaled average DNA methylation level. With the same row order, row scaled DNA methylation level in each neonatal sample was also shown in cyan-red heatmaps. The number of DMRs in the selected clusters were also shown. (b) Venn diagram for the overlap between of the gain (upper) and lost (lower) H3K4me3 DPs of ICSI-ET versus CTRL, and ICSI-ET versus IVF-ET neonatal samples. (c) For the selected hyper- (union of cluster C1-C3) and hypo-DMRs (union of cluster C4-C6), gain (7233) and loss (84) H3K4me3 DPs, their gene ontology (biological process) enrichment analysis results were grouped and shown, if its enrichment terms existed (adjusted p-value < 0.05; The p-value was determined by hypergeometric test and adjusted by BH method.). (d) The chromatin states distribution of the selected hyper- and hypo-DMRs and H3K4me3 DPs (7233 GAIN, 84 LOSS) in each neonatal sample of CTRL, IVF-ET, and ICSI-ET. The chromatin states were defined in Fig. 3b.
Like the observation in IVF-ET, the selected DMRs for ICSI also tended to avoid the active TSSs (Fig. 4d). Associated genes of hyper DMRs in ZNF Genes & Repeats (E9) and weak repressed Polycomb (E11) were enriched in the processes of immune and nervous system, respectively. Associated genes of hypo-DMRs in Active TSS (E7) and ZNF Genes & Repeats (E9) were also enriched in the processes of immune and nervous system, respectively (Supplementary Fig. S9f and Supplementary Table S20). As expected, H3K4me3 peaks were mainly enriched in regions around TSSs (E5, E6, E7, E8). However, regions for the selected gain H3K4me3 DPs in CTRL and IVE-ET group were highly enriched in active enhancer (E2) and poised enhancer (E4) and shifted to ZNF Genes & Repeats (E9) in ICSI-ET. For loss H3K4me3 DPs, the situation was reversed (Fig. 4d). GO terms for the selected gain H3K4me3 DPs in E2 and E4 were mainly related to the regulation of leukocytes, cell migration/adhesiveness and the development of several systems (Supplementary Fig. S9f and Supplementary Table S20). These results suggested that ICSI might alter H3K4me3 at enhancers and affect multiple processes. Together, the results regarding both the DNA methylome and H3K4me3 modification raise the question of whether ICSI may have a potential impact on health of offspring, with the skeletal systems and immune system as the representatives.
3.6. Specific epigenomic changes induced by freeze-thawing procedure
We then investigated the differences between the IVF-FET and IVF-ET groups and observed 6251 DMRs and 5683 DPs of H3K4me3 (Supplementary Table S4 and Supplementary Table S11; 4191 hyper-DMRs and 2060 hypo-DMRs; 5548 gain DPs and 135 loss DPs). Compared to the number of DPs in ICSI-ET versus IVF-ET group or CTRL group, the notably increased numbers of H3K4me1, H3K27me3 and H3K27ac DPs suggested that the freeze-thawing procedure might introduce more disturbance of histone modifications than ICSI operation. In addition, the DMRs and DPs (of H3K4me3 and H3K27ac) shared common associated genes and GO terms (Fig. 5a, Supplementary Fig. S10a, Supplementary Table S21 and Supplementary Table S22), implying that freeze-thawing procedure might cause certain functional changes through different epigenetic layers.
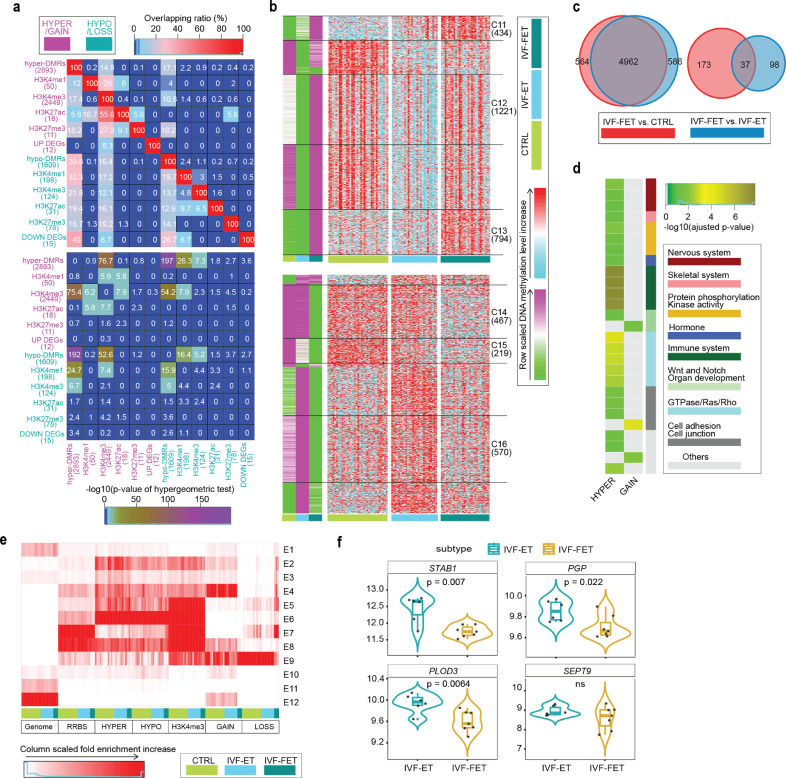
Fig. 5.
Specific epigenetic effects of freezing-thaw procedure. (a) Overlapping ratio among DEGs, associated genes of DMRs and DPs for IVF-FET versus IVF-ET neonatal samples (upper), and their statistical significances (lower) determined by hypergeometric test. (b) K-means clustering for the hyper- (upper) and hypo-DMRs (lower) between IVF-FET versus IVF-ET neonatal samples. In green-purple heatmaps, the hyper-DMRs (hypo-DMRs) were classified into six (seven) clusters by k-means clustering on row scaled average DNA methylation level. With the same row order, row scaled DNA methylation level in each neonatal sample was also shown in cyan-red heatmaps. The number of DMRs in the selected clusters were also shown. (c) Venn diagrams displayed the overlap between the gain (left) and lost (right) H3K4me3 DPs of IVF-FET versus CTRL and that of IVF-FET versus IVF-ET. (d) For the selected hyper- (union of cluster C11-C13) and hypo-DMRs (union of cluster C14-C16), overlapping gain (4962) and loss (37) H3K4me3 DPs, their gene ontology (biological process) enrichment analysis results were grouped and shown, if the enrichment terms existed (adjusted p-value < 0.05; The p-value was determined by hypergeometric test and adjusted by BH method). (e) The chromatin states distribution of the selected hyper- and hypo-DMRs, overlapping 4962 gain and 37 loss H3K4me3 DPs in each neonatal sample of CTRL, IVF-ET, and IVF-FET. The chromatin states were defined in Fig. 3b. (f) Violin-box plots showed the expression level of DEGs (adjusted p-value < 0.1; The p-values were attained by the Wald test and adjusted by BH method) overlapped with DMR associated genes (PGP, PLOD3, STAB1, and SETP9) among neonatal samples in IVF-ET and IVF-FET. The p-value between IVF-ET and IVF-FET was determined by Wilcoxon rank-sum test.
To further explore the specific effects of freeze-thawing procedure per se, we selected six subgroups of DMRs of IVF-FET versus IVF-ET through k-means clustering of scaled DNA methylation levels among the IVF-FET, IVF-ET and CTRL groups (Fig. 5b and Supplementary Table S23; cluster C11~13 in hyper-DMRs and cluster C14~C16 in hypo-DMRs), as well as H3K4me3 DPs (n = 4999) identified by intersection in the comparison of IVF-FET versus IVF-ET (n = 5683) and IVF-FET versus CTRL (n = 5736) (Fig. 5c). The replications of DMRs and DPs for each group were highly comparable (Supplementary Fig. S10b). The associated genes of selected hyper-DMRs were highly enriched in the processes of neutrophil-mediated immunity and the regulation of GTPase/Ras (Fig. 5d). The results of HPO analysis further revealed that the selected DMRs might be related to carcinogenesis (Supplementary Fig. S10c and Supplementary Table S22). The majority of selected hyper/hypo-DMRs were inclined to evade from Active TSS (Fig. 5e), and the associated genes of those DMRs in Active enhancer (E2) and Active TSS upstream (E6) were highly enriched in the processes of immune system, metabolism of purine/carbohydrate, muscle and skeletal system (Supplementary Fig. S10d and Supplementary Table S22). Poised enhancer (E4) was the most overrepresented states for the selected gain H3K4me3 DPs in CTRL/IVF-ET group and became ZNF Genes & Repeats (E9) in IVF-FET. Associated genes for the selected gain H3K4me3 DPs in E2 and E4 were enriched in the processes of various systems (Fig. 5e, Supplementary Fig. S10d and Supplementary Table S22). It was also worth mentioning that the genes PGP, PLOD3, and STAB1 in the IVF-FET group all exhibited decreased expression with hyper-DMRs in their promoters. In addition, the expression level of SEPT9 also showed decreased tendency in the IVF-FET group, with hyper-DMRs detected in the TSS upstream region and hypo-DMRs detected in the TSS downstream region (Fig. 5f and Supplementary Table S21). Considering that the dysregulation of the PGP, PLOD3, STAB1 and SEPT9 genes [58], [59], [60], [61], as well as the GTPase signaling pathway is highly associated with carcinogenesis [62], the above results implied that disturbance in the epigenome introduced by cryopreservation per se might potentially affect the immune system and increase the risk of carcinogenesis later in life.
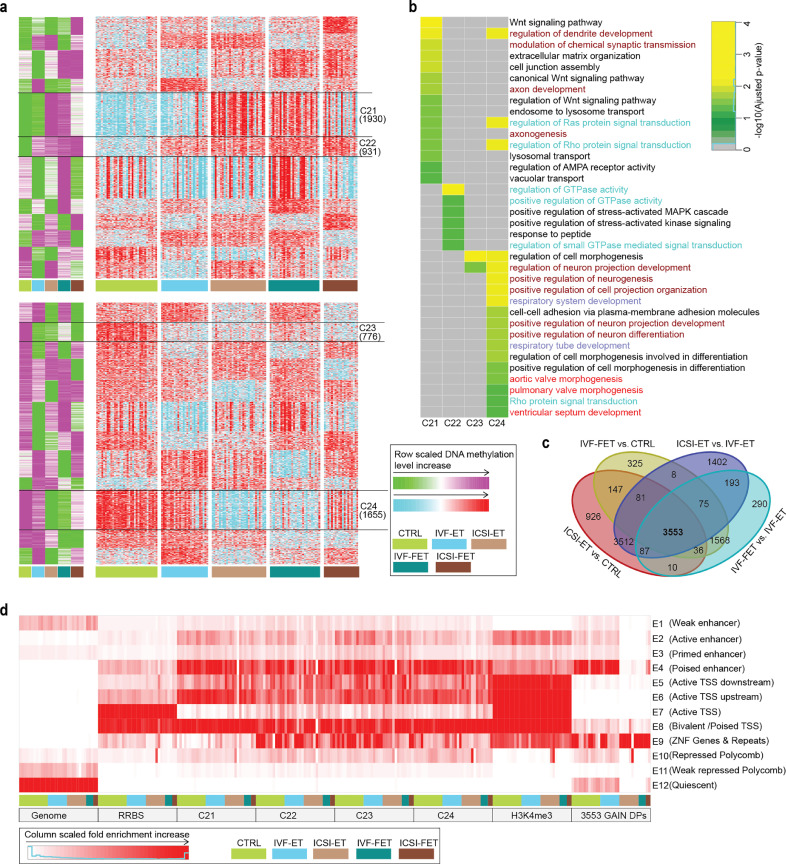
3.7. Common epigenetic effects among different ART processes
Since various ART processes share common procedures, we further analyzed the relationship among DMRs/DPs to investigate common epigenetic effects of different ART treatments. Clearly, the DMRs identified in seven pairwise comparisons were overlapped significantly in genomic location and shared many common GO terms (Supplementary Fig. S11a and S11b, and Supplementary Table S24). Moreover, a considerable number of DMRs showed similar change tendencies in different ART groups compared with CTRL group through k-means clustering using DMRs from all seven groups (Fig. 6a and Supplementary Table S25; C22, n = 931; C23, n = 776). The associated genes of C22 and C23 were highly enriched in the regulation of GTPase activity, stress-activated MAPK cascade, cell morphogenesis and neuron projection development in GO analysis (Fig. 6b and Supplementary Table S24).
Fig. 6.
Common Epigenetic effect of different ARTs. (a) K-means clustering for the all merged hyper- (up) and hypo-DMRs (down) of the seven comparisons. The hyper- and hypo-DMRs of the seven comparisons were merged if they have 1-bp common region at least, respectively. In green-purple heatmaps, DMRs were classified into twelve clusters by k-means clustering on row scaled average DNA methylation level. With the same row order, row scaled DNA methylation level in each neonatal sample was also shown in cyan-red heatmaps. The number of DMRs in the selected clusters were also shown. (b) For the selected clusters of hyper-DMRs (C21, C22) and hypo-DMRs (C23, C24), all enrichment terms of gene ontology (biological process) with adjusted p-value < 0.05 were grouped and shown (The p-values were attained by the hypergeometric test and adjusted by BH method). (c) Venn diagrams displayed the overlap of the gain H3K4me3 DPs for ICSI-ET versus CTRL, IVF-FET versus CTRL, ICSI-ET versus CTRL, and IVF-FET versus IVF-ET. (d) The chromatin states distribution in each neonatal sample for the selected DMRs and overlapping gain H3K4me3 DPs. The chromatin states were defined in Fig. 3b.
Interestingly, clusters C21 and C24 seemed to represent regions where ICSI and freeze-thawing procedure might introduce similar alterations, revealed by the similar change on DNA methylation levels in the ICSI-ET and IVF-FET groups compared with CTRL and IVF-ET groups (Fig. 6a and Supplementary Table S25). These DMRs were related to Wnt signaling pathway, respiratory, cardiovascular and nervous system. Similarly, there were noticeable overlaps between ICSI- and freeze-thawing-specific gain H3K4me3 DPs (Fig. 6c and Supplementary Table S25; n = 3553), which were mainly involved in immune (B lymphocytopenia, Abnormality of B cell number, Neutropenia) and skeletal system by HPO analysis (Supplementary Fig. S10c and Supplementary Table S24). As mentioned previously, those DMRs were also noticeably absent from Active TSS (E7), with the increased enrichment in ZNF Genes & Repeats (E9) for DMRs in clusters C21 and C24 (Fig. 6d). Meanwhile, the difference between ICSI-FET and IVF-ET would be expected as a mixed effect of ICSI and freeze-thawing operation. Indeed, large percentage of those DMRs showed similar changes in ICSI-ET or IVF-FET (C31~C33; C35, C36, C38). However, 13% hyper-DMRs (C34) and 19% hypo-DMRs (C37) were uniquely found in ICSI-FET groups, implying complex interactions between ICSI and freeze-thawing procedure (Supplementary Fig. S11d and Supplementary Table S25). Overall, the above results together suggested that in addition to causing their own unique effects, as mentioned previously, distinct ART procedures also leaded to certain common effects on the epigenomes of offspring.
4. Discussion
Safety concerns regarding ARTs are as old as ARTs themselves [63]. Although several studies suggest that ART may have adverse effects on the long-term health of offspring, the underlying epigenetic mechanisms remain to be elucidated. We systematically explored the influences of fertilization procedures and freeze-thawing operation on descendant genome-wide DNA methylation, histone modifications and gene expression. Recruitment of nuclear families with twins provided us with perfect biological replicates in each family and enabled us to eliminate ART-irrelevant parental impacts as much as possible. Our study illustrates that the epigenomes of neonates conceived by ART are overall similar to those of naturally conceived children, which may partially explain why most ART offspring are generally healthy. However, ART do increase the heterogeneity of the DNA methylome within twin pairs and induce local subtle changes in different epigenetic layers, supporting the theory that ART increases risks of epigenomic abnormality [17], [18], [19]. We also found that a large percentage of the DNA methylomic changes in ART offspring were derived from parents, highlighting the necessity of removing parental bias in assessing the influences of ARTs per se. Moreover, we reported the genome-wide impacts of ARTs on four types of histone modifications in humans for the first time. The epigenomes of IVF-ET conceived infants seemed to be closer to those of naturally conceived infants than those of IVF-FET or ICSI-ET conceived infants and showed almost no disturbance in either type of histone modification. It suggests that ART-inherent procedures, including controlled ovarian hyperstimulation (COH), in vitro culture, in vitro fertilization, etc., may not increase the risks of abnormal histone reprogramming; however, ICSI and freeze-thawing operation may do so. What surprised us was that H3K4me3 was the most profoundly impacted by ICSI and freeze-thawing operations, compared with the other three types of histone modifications (H3K4me1, H3K27me3 and H3K27ac). H3K4me3 signals around some DPs regions seem to form relatively moderate but broad patterns, similar to unclassical domains reported elsewhere [64]. Therefore, H3K4me3 may serve as a sensitive histone modification for assessment of the influences of ARTs though remain to be further verified.
Recently, Boris Novakovic, et al. reported ART-related changes in DNA methylation of blood from ART and non-ART-conceived individuals in the CHART cohort by using the EPIC array [65]. However, there were only 18 DMRs identified in their neonates’ group, which might stem in part from the limited number and special genomic locations of selected CpGs. In fact, our data revealed that the distributions of epigenetic changes tended to be away from active cis-regulatory elements and were not largely associated with transcriptional changes in corresponding genes. Our final neonatal specific DMRs set also contains five neonatal DMRs reported by Boris Novakovic, et al. (as highlighted in Supplementary Table 16). It supported the credibility of epigenetic changes identified in our study and suggested that the epigenetic effects of ART are to a certain extent universal and conserved in different samples.
Functional enrichment of epigenetic changes suggested that epigenetic disorders caused by ICSI might interfere with the processes associated with skeletal system, which has been highlighted in another study about the potential impact of ART on DNA methylome [66]. Our results also implied that freeze-thawing procedure, as well as ICSI, might increase the risks of immune dysfunction in offspring. Though the risks of hospital admission in FET-conceived children and asthma medication in ART-conceived children have been reported to be higher than CTRL conceived children [67,68], direct evidence regarding the effects of ART on the immune system is lacking so far and long-term follow-up studies are required. Previous study has revealed elevated rates of preeclampsia in women who have undergone FET [69]. GTPases, especially Rho kinases, play essential roles in extravillous trophoblast cell (EVT) invasion [70], and limited EVT invasion following poor remodeling of arteries is widely observed in preeclampsia [71]. More interestingly, our results suggested that freeze-thawing operation might cause dysregulation in the GTPase/Ras signaling pathway in offspring. Thus, the epigenetic abnormality we report may also possibly explain the increased risks of preeclampsia in FET compared with fresh ET pregnancies. Our findings emphasize that it is reasonable to inform patients of potential risks associated with ICSI and embryo cryopreservation and that it would be wise to reconsider overutilization of ICSI or routine freezing of all embryos.
Apart from the influences for given ART operations, we also revealed the common effects induced by various ART processes. Further analysis showed that ART-induced DNA methylation changes were enriched in the processes of cardiovascular system and glucolipid metabolism in the comparisons of all four ART groups versus CTRL groups. Since the abnormalities in lipid profiles and higher rates of cardiovascular dysfunction have been reported in ART-achieved children [11,72], it implies that the disturbance on epigenome by ART in offspring may increase the risk of metabolic syndrome [73]. In addition, we identified a considerable number of common DMRs among different ART groups compared with CTRL group. It is noteworthy that the regulation of GTPase activity is also the most overrepresented terms in GO analysis for those DMRs, which is in line with those studies suggesting that pregnancies conceiving by ART is related with the increased risk for certain cancers in offspring and preeclampsia compared with natural pregnancy [74,75].The common DMRs induced by both ICSI and freeze-thawing procedure were enriched in the processes involving in neuron, consistent with the concern that these two kinds of aggressive ART operation might increase the risk of mental disorders in offspring [76]. As suggested by the reports that improved in vitro culture systems for animals will affect the epigenome less than earlier versions [77,78], continuous optimization for ART procedures is urgently needed to simulate the in vivo environment and reduce potential epigenetic abnormalities in offspring.
Our study was mainly focused on the effects of different fertilization methods and freeze-thawing and only involved newborns, lacking postnatal follow-up data on the enrolled populations. However, it is worth noting that the two stable DMRs (CHRNE- and PRSS16- associated) persisting in adults reported previously [65] were both observed in our final neonatal DMRs identified. It suggested that a larger multicenter randomized controlled trial (RCT) along with detection of the epigenetic profiles of offspring in later life would be helpful for elucidation of continued epigenetic changes, their association with the long-term health risks, and exploration of the specific epigenetic impacts of other factors in ART treatment, such as the duration of embryo culture or the composition of culture system. Whole blood samples are used in the current study; whether the alterations in H3K4me3 and DNA methylation are general or only occur in some specific cell types is still waiting to be illuminated. Further studies on other tissues will be helpful to evaluate the inter-tissue universality of ART's epigenetic influence, too.
In conclusion, our results provide an epigenetic basis for the increased long-term health risks in ART offspring. We highlight ART clinical interventions that require particular surveillance. More effort should be expended to optimize current ART systems, and the choice of appropriate procedures requires careful evaluation. Since epigenomic changes might be maintained throughout the human lifespan [79] and can potentially be transmitted to subsequent generations, long-term follow-up and health evaluation of ART offspring are necessary to provide more robust clinical evidence.
Author contributions
W.C., X.M., Y.P. and S.K. wrote the manuscript. W.C. and X.M. collected the study materials and samples and patient data, performed the experiments. Y.Wei., Y.Z., S.T. and W.Z. were helpful for the recruitment of family and sample collection for this study. Y.P. and W.C. developed analysis methods and performed bioinformatic analysis. J.Q., L.Y. and Y.Wang. developed the experimental conception and designs. All the authors read and approved the final version of the manuscript.
Data sharing
Bioinformatics pipelines and scripts used for our analysis are available at https://github.com/CTLife/ART_Epigeome. All the datasets of RRBS, ChIP-seq, and RNA-seq included in this study have been uploaded to NCBI Gene Expression Omnibus (GEO) database, which are available via accession number GSE136849.
Declaration of Interests
All authors declared that they had no competing conflicts of interests to disclose.
Acknowledgments
Acknowledgments
We thank the families who participated in this study. We thank Robert Norman for his advice and discussion about the manuscript.
Funding sources
This study was funded by National Natural Science Foundation of China (81730038; 81521002), National Key Research and Development Program (2018YFC1004000; 2017YFA0103801; 2017YFA0105001) and Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16020703). Y.Wang. was supported by Postdoctoral Fellowship of Peking-Tsinghua Center for Life Science.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103076.
Contributor Information
Yang Wang, Email: yangwang@bjmu.edu.cn.
Liying Yan, Email: yanliyingkind@aliyun.com.
Jie Qiao, Email: jie.qiao@263.net.
Appendix. Supplementary materials
References
- 1.Fauser B.C. Towards the global coverage of a unified registry of IVF outcomes. Reprod. Biomed. Online. 2019;38(2):133–137. doi: 10.1016/j.rbmo.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Winston R.M., Hardy K. Are we ignoring potential dangers of in vitro fertilization and related treatments? Nat Med. 2002;8(10):14–18. doi: 10.1038/ncb-nm-fertilityS14. [DOI] [PubMed] [Google Scholar]
- 3.Esteves S.C., Roque M., Bedoschi G., Haahr T., Humaidan P. Intracytoplasmic sperm injection for male infertility and consequences for offspring. Nat Rev Urol. 2018;15(9):535–562. doi: 10.1038/s41585-018-0051-8. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig M., Schröder A.K., Diedrich K. Impact of intracytoplasmic sperm injection on the activation and fertilization process of oocytes. Reprod. Biomed. Online. 2001;3(3):230–240. doi: 10.1016/s1472-6483(10)62042-1. [DOI] [PubMed] [Google Scholar]
- 5.Kopeika J., Thornhill A., Khalaf Y. The effect of cryopreservation on the genome of gametes and embryos: principles of cryobiology and critical appraisal of the evidence. Hum. Reprod. Update. 2014;21(2):209–227. doi: 10.1093/humupd/dmu063. [DOI] [PubMed] [Google Scholar]
- 6.Fleming T.P., Watkins A.J., Velazquez M.A., Mathers J.C., Prentice A.M., Stephenson J. Origins of lifetime health around the time of conception: causes and consequences. Lancet. 2018;391(10132):1842–1852. doi: 10.1016/S0140-6736(18)30312-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fruchter E., Beck-Fruchter R., Hourvitz A., Weiser M., Goldberg S., Fenchel D. Health and functioning of adolescents conceived by assisted reproductive technology. Fertil. Steril. 2017;107(3):774–780. doi: 10.1016/j.fertnstert.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Kuiper D., Hoek A., la Bastide-Van Gemert S., Seggers J., Mulder D.J., Haadsma M. Cardiovascular health of 9-year-old IVF offspring: no association with ovarian hyperstimulation and the in vitro procedure. Human Reproduction. 2017;32(12):2540–2548. doi: 10.1093/humrep/dex323. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe N., Fujiwara T., Suzuki T., Jwa S.C., Taniguchi K., Yamanobe Y. Is in vitro fertilization associated with preeclampsia? A propensity score matched study. BMC Pregnancy Childbirth. 2014;14(1):69. doi: 10.1186/1471-2393-14-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berntsen S., Söderström-Anttila V., Wennerholm U.-.B., Laivuori H., Loft A., Oldereid N.B. The health of children conceived by ART:‘the chicken or the egg?’. Hum Reprod Update. 2019;25(2):137–158. doi: 10.1093/humupd/dmz001. [DOI] [PubMed] [Google Scholar]
- 11.Scherrer U., Rexhaj E., Allemann Y., Sartori C., Rimoldi S.F. Cardiovascular dysfunction in children conceived by assisted reproductive technologies. Eur. Heart J. 2015;36(25):1583–1589. doi: 10.1093/eurheartj/ehv145. [DOI] [PubMed] [Google Scholar]
- 12.Choux C., Carmignac V., Bruno C., Sagot P., Vaiman D., Fauque P. The placenta: phenotypic and epigenetic modifications induced by Assisted Reproductive Technologies throughout pregnancy. Clin Epigenetics. 2015;7(1):87. doi: 10.1186/s13148-015-0120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feil R., Fraga M.F. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Fan X., Wang R., Lu X., Dang Y.-.L., Wang H. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018;28(8):819–832. doi: 10.1038/s41422-018-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q., Xie W. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol. 2018;28(3):237–253. doi: 10.1016/j.tcb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Gluckman P.D., Hanson M.A., Cooper C., Thornburg K.L. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359(1):61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazaraviciute G., Kauser M., Bhattacharya S., Haggarty P., Bhattacharya S. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum. Reprod. Update. 2014;20(6):840–852. doi: 10.1093/humupd/dmu033. [DOI] [PubMed] [Google Scholar]
- 18.Song S., Ghosh J., Mainigi M., Turan N., Weinerman R., Truongcao M. DNA methylation differences between in vitro-and in vivo-conceived children are associated with ART procedures rather than infertility. Clin Epigenetics. 2015;7(1):41. doi: 10.1186/s13148-015-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh J., Coutifaris C., Sapienza C., Mainigi M. Global DNA methylation levels are altered by modifiable clinical manipulations in assisted reproductive technologies. Clin Epigenetics. 2017;9(1):14. doi: 10.1186/s13148-017-0318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melamed N., Choufani S., Wilkins-Haug L.E., Koren G., Weksberg R.J.E. Comparison of genome-wide and gene-specific DNA methylation between ART and naturally conceived pregnancies. Epigenetics. 2015;10(6):474–483. doi: 10.4161/15592294.2014.988041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estill M.S., Bolnick J.M., Waterland R.A., Bolnick A.D., Diamond M.P., Krawetz S.A.J.F. Assisted reproductive technology alters deoxyribonucleic acid methylation profiles in bloodspots of newborn infants. Fertil. Steril. 2016;106(3) doi: 10.1016/j.fertnstert.2016.05.006. 629-39. e10. [DOI] [PubMed] [Google Scholar]
- 22.El Hajj N., Haertle L., Dittrich M., Denk S., Lehnen H., Hahn T. DNA methylation signatures in cord blood of ICSI children. Human Reproduction. 2017;32(8):1761–1769. doi: 10.1093/humrep/dex209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choufani S., Turinsky A.L., Melamed N., Greenblatt E., Brudno M., Bérard A. Impact of assisted reproduction, infertility, sex and paternal factors on the placental DNA methylome. Hum. Mol. Genet. 2018;28(3):372–385. doi: 10.1093/hmg/ddy321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castillo-Fernandez J.E., Loke Y.J., Bass-Stringer S., Gao F., Xia Y., Wu H. DNA methylation changes at infertility genes in newborn twins conceived by in vitro fertilisation. Genome Med. 2017;9(1):28. doi: 10.1186/s13073-017-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobi E.W., Goeman J.J., Monajemi R., Gu H., Putter H., Zhang Y. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun. 2014;5:5592. doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer T., Trump S., Ishaque N., Thürmann L., Gu L., Bauer M. Environment‐induced epigenetic reprogramming in genomic regulatory elements in smoking mothers and their children. Mol. Syst. Biol. 2016;12(3) doi: 10.15252/msb.20156520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu H., Smith Z.D., Bock C., Boyle P., Gnirke A., Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6(4):468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Li Y., Guo C., Lu Q., Wang W., Jia Z. ISL1 and JMJD3 synergistically control cardiac differentiation of embryonic stem cells. Nucl Acids Res. 2016;44(14):6741–6755. doi: 10.1093/nar/gkw301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 30.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S. deepTools2: a next generation web server for deep-sequencing data analysis. Nucl Acids Res. 2016;44(W1):W160–W1W5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen K., Xi Y., Pan X., Li Z., Kaestner K., Tyler J. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013;23(2):341–351. doi: 10.1101/gr.142067.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst J., Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3(9):e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu G., Wang L.G, He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 45.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucl Acids Res. 2016;44(W1):W90–WW7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patil I., Powell C.… ggstatsplot:“ggplot2” Based Plots with Statistical Details. CRAN. 2018 [Google Scholar]
- 49.Kaminsky Z.A., Tang T., Wang S.-.C., Ptak C., Oh G.H., Wong A.H. DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet. 2009;41(2):240. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 50.Gordon L., Joo J.E., Powell J.E., Ollikainen M., Novakovic B., Li X. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012;22(8):1395–1406. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tournaye H. Male factor infertility and ART. Asian J Androl. 2012;14(1):103–108. doi: 10.1038/aja.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kähler A.K., Djurovic S., Kulle B., Jönsson E.G., Agartz I., Hall H. Association analysis of schizophrenia on 18 genes involved in neuronal migration: MDGA1 as a new susceptibility gene. American J. Medical Genetics Part B: Neuropsychiatric Genetics. 2008;147(7):1089–1100. doi: 10.1002/ajmg.b.30726. [DOI] [PubMed] [Google Scholar]
- 53.Yuan X., Cao J., Liu T., Li Y., Scannapieco F., He X. Regulators of G protein signaling 12 promotes osteoclastogenesis in bone remodeling and pathological bone loss. Cell Death & Differentiation. 2015;22(12):2046–2057. doi: 10.1038/cdd.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang J., Chen L., Yao Y., Tang C., Ding J., Fu C. Pivotal role of regulator of G-protein signaling 12 in pathological cardiac hypertrophy. Hypertension. 2016;67(6):1228–1236. doi: 10.1161/HYPERTENSIONAHA.115.06877. [DOI] [PubMed] [Google Scholar]
- 55.Sathyan K., Nalinakumari K., Abraham T., Kannan SJOo. CCND1 polymorphisms (A870G and C1722G) modulate its protein expression and survival in oral carcinoma. Oral Oncol. 2008;44(7):689–697. doi: 10.1016/j.oraloncology.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Larsson C., Ali M.A., Pandzic T., Lindroth A.M., He L., Sjöblom TJBc. Loss of DIP2C in RKO cells stimulates changes in DNA methylation and epithelial-mesenchymal transition. BMC Cancer. 2017;17(1):1–12. doi: 10.1186/s12885-017-3472-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin H.-.J., Huang Y.-.C., Lin J.-.M., Liao W.-.L., Wu J.-.Y., Chen C.-.H. Novel susceptibility genes associated with diabetic cataract in a Taiwanese population. Ophthalmic Genet. 2013;34(1–2):35–42. doi: 10.3109/13816810.2012.736590. [DOI] [PubMed] [Google Scholar]
- 58.Possik E., Madiraju S.M., Prentki M. Glycerol-3-phosphate phosphatase/PGP: role in intermediary metabolism and target for cardiometabolic diseases. Biochimie. 2017;143:18–28. doi: 10.1016/j.biochi.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Song L., Gong Y., He B. Detection of colorectal cancer by DNA methylation biomarker SEPT9: past, present and future. Biomark Med. 2014;8(5):755–769. doi: 10.2217/bmm.14.8. [DOI] [PubMed] [Google Scholar]
- 60.Baek J.-.H., Yun H.S., Kwon G.T., Lee J., Kim J.-.Y., Jo Y. PLOD3 suppression exerts an anti-tumor effect on human lung cancer cells by modulating the PKC-delta signaling pathway. Cell Death Dis. 2019;10(3):156. doi: 10.1038/s41419-019-1405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rantakari P., Patten D.A., Valtonen J., Karikoski M., Gerke H., Dawes H. Stabilin-1 expression defines a subset of macrophages that mediate tissue homeostasis and prevent fibrosis in chronic liver injury. Proc Natl Acad Sci U S A. 2016;113(33):9298–9303. doi: 10.1073/pnas.1604780113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sahai E., Marshall C.J. RHO–GTPases and cancer. Nat Rev Cancer. 2002;2(2):133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 63.Cohen M.E. The brave new baby and the law: fashioning remedies for the victims of in vitro fertilization. Am J Law Med. 1978;4:319–336. [PubMed] [Google Scholar]
- 64.Chen K., Chen Z., Wu D., Zhang L., Lin X., Su J. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat. Genet. 2015;47(10):1149–1157. doi: 10.1038/ng.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novakovic B., Lewis S., Halliday J., Kennedy J., Burgner D.P., Czajko A. Assisted reproductive technologies are associated with limited epigenetic variation at birth that largely resolves by adulthood. Nat Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-11929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y., Li X.-.Z., Chen S.-.C., Wang L., Tan Y.-.J., Li X.-.C., et al. Comparison of Genome-Wide DNA Methylation Profiles of Fetal Tissues Conceived by In Vitro Fertilization and Natural Conception. (October 16, 2018). Available at SSRN: https://ssrn.com/abstract=3267672 or http://dx.doi.org/10.2139/ssrn.3267672. [DOI] [PMC free article] [PubMed]
- 67.Pelkonen S., Gissler M., Koivurova S., Lehtinen S., Martikainen H., Hartikainen A.-.L. Physical health of singleton children born after frozen embryo transfer using slow freezing: a 3-year follow-up study. Human Reproduction. 2015;30(10):2411–2418. doi: 10.1093/humrep/dev203. [DOI] [PubMed] [Google Scholar]
- 68.Kuiper D.B., Seggers J., Schendelaar P., Haadsma M.L., Roseboom T.J., Heineman M.J. Asthma and asthma medication use among 4-year-old offspring of subfertile couples–association with IVF? Reprod. Biomed. Online. 2015;31(5):711–714. doi: 10.1016/j.rbmo.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 69.Wei D., Liu J.-.Y., Sun Y., Shi Y., Zhang B., Liu J.-.Q. Frozen versus fresh single blastocyst transfer in ovulatory women: a multicentre, randomised controlled trial. The Lancet. 2019;393(10178):1310–1318. doi: 10.1016/S0140-6736(18)32843-5. [DOI] [PubMed] [Google Scholar]
- 70.Saso J., Shields S.-.K., Zuo Y., Chakraborty C. Role of Rho GTPases in human trophoblast migration induced by IGFBP1. Biol Reprod. 2012;86(1):1–9. doi: 10.1095/biolreprod.111.094698. [DOI] [PubMed] [Google Scholar]
- 71.Redman C.W., Sargent I.L. Latest advances in understanding preeclampsia. Science. 2005;308(5728):1592–1594. doi: 10.1126/science.1111726. [DOI] [PubMed] [Google Scholar]
- 72.Guo X.-.Y., Liu X.-.M., Jin L., Wang T.-.T., Ullah K., Sheng J.-.Z. Cardiovascular and metabolic profiles of offspring conceived by assisted reproductive technologies: a systematic review and meta-analysis. Fertil. Steril. 2017;107(3):622–631.e5. doi: 10.1016/j.fertnstert.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 73.Alberti K.G.M., Zimmet P., Shaw J. The metabolic syndrome—A new worldwide definition. The Lancet. 2005;366(9491):1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 74.Chen X.-.K., Wen S.W., Bottomley J., Smith G.N., Leader A., Walker M.C. In vitro fertilization is associated with an increased risk for preeclampsia. Hypertens Pregnancy. 2009;28(1):1–12. doi: 10.1080/10641950802001859. [DOI] [PubMed] [Google Scholar]
- 75.Williams C.L., Bunch K.J., Stiller C.A., Murphy M.F., Botting B.J., Wallace W.H. Cancer risk among children born after assisted conception. New England J. Medicine. 2013;369(19):1819–1827. doi: 10.1056/NEJMoa1301675. [DOI] [PubMed] [Google Scholar]
- 76.Sandin S., Nygren K.-.G., Iliadou A., Hultman C.M., Reichenberg A. Autism and mental retardation among offspring born after in vitro fertilization. JAMA. 2013;310(1):75–84. doi: 10.1001/jama.2013.7222. [DOI] [PubMed] [Google Scholar]
- 77.Canovas S., Ivanova E., Romar R., Garcia-Martinez S., Soriano-Ubeda C., Garcia-Vazquez F.A. DNA methylation and gene expression changes derived from assisted reproductive technologies can be decreased by reproductive fluids. Elife. 2017;6:e23670. doi: 10.7554/eLife.23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan K., An L., Miao K., Ren L., Hou Z., Tao L. Impaired imprinted X chromosome inactivation is responsible for the skewed sex ratio following in vitro fertilization. Proceed. National Academy of Sciences. 2016;113(12):3197–3202. doi: 10.1073/pnas.1523538113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dominguez-Salas P., Moore S.E., Baker M.S., Bergen A.W., Cox S.E., Dyer R.A. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun. 2014;5:3746. doi: 10.1038/ncomms4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.