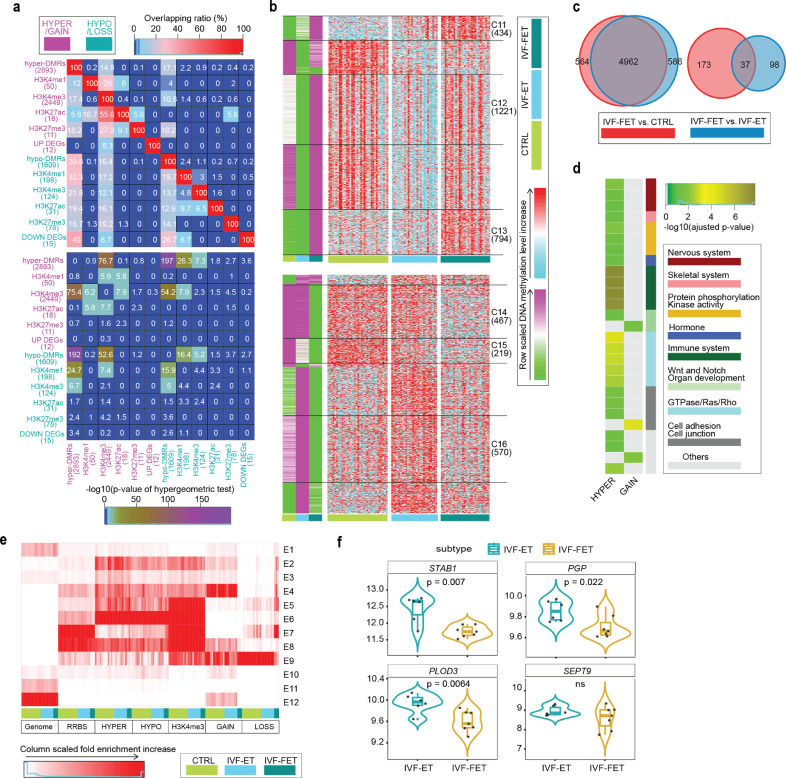
Fig. 5.
Specific epigenetic effects of freezing-thaw procedure. (a) Overlapping ratio among DEGs, associated genes of DMRs and DPs for IVF-FET versus IVF-ET neonatal samples (upper), and their statistical significances (lower) determined by hypergeometric test. (b) K-means clustering for the hyper- (upper) and hypo-DMRs (lower) between IVF-FET versus IVF-ET neonatal samples. In green-purple heatmaps, the hyper-DMRs (hypo-DMRs) were classified into six (seven) clusters by k-means clustering on row scaled average DNA methylation level. With the same row order, row scaled DNA methylation level in each neonatal sample was also shown in cyan-red heatmaps. The number of DMRs in the selected clusters were also shown. (c) Venn diagrams displayed the overlap between the gain (left) and lost (right) H3K4me3 DPs of IVF-FET versus CTRL and that of IVF-FET versus IVF-ET. (d) For the selected hyper- (union of cluster C11-C13) and hypo-DMRs (union of cluster C14-C16), overlapping gain (4962) and loss (37) H3K4me3 DPs, their gene ontology (biological process) enrichment analysis results were grouped and shown, if the enrichment terms existed (adjusted p-value < 0.05; The p-value was determined by hypergeometric test and adjusted by BH method). (e) The chromatin states distribution of the selected hyper- and hypo-DMRs, overlapping 4962 gain and 37 loss H3K4me3 DPs in each neonatal sample of CTRL, IVF-ET, and IVF-FET. The chromatin states were defined in Fig. 3b. (f) Violin-box plots showed the expression level of DEGs (adjusted p-value < 0.1; The p-values were attained by the Wald test and adjusted by BH method) overlapped with DMR associated genes (PGP, PLOD3, STAB1, and SETP9) among neonatal samples in IVF-ET and IVF-FET. The p-value between IVF-ET and IVF-FET was determined by Wilcoxon rank-sum test.