Abstract
Background
Verigene Blood-Culture Gram-Negative is a rapid diagnostic test (RDT) that detects gram-negatives (GNs) and resistance within hours from gram stain. The majority of the data support the use of RDTs with antimicrobial stewardship (AMS) intervention in gram-positive bloodstream infection (BSI). Less is known about GN BSI.
Methods
This was a retrospective quasi-experimental (nonrandomized) study of adult patients with RDT-target GN BSI comparing patients pre-RDT/AMS vs post-RDT/pre-AMS vs post-RDT/AMS. Optimal therapy was defined as appropriate coverage with the narrowest spectrum, accounting for source and co-infecting organisms. Time to optimal therapy was analyzed using Kaplan-Meier and multivariable Cox proportional hazards regression.
Results
Eight-hundred thirty-two patients were included; 237 pre-RDT/AMS vs 308 post-RDT/pre-AMS vs 237 post-RDT/AMS, respectively. The proportion of patients on optimal antibiotic therapy increased with each intervention (66.5% vs 78.9% vs 83.2%; P < .0001). Time to optimal therapy (interquartile range) decreased with introduction of RDT: 47 (7.9–67.7) hours vs 24.9 (12.4–55.2) hours vs 26.5 (10.3–66.5) hours (P = .09). Using multivariable modeling, infectious diseases (ID) consult was an effect modifier. Within the ID consult stratum, controlling for source and ICU stay, compared with the pre-RDT/AMS group, both post-RDT/pre-AMS (adjusted hazard ratio [aHR], 1.34; 95% CI, 1.04–1.72) and post-RDT/AMS (aHR, 1.28; 95% CI, 1.01–1.64), improved time to optimal therapy. This effect was not seen in the stratum without ID consult.
Conclusions
With the introduction of RDT and AMS, both proportion and time to optimal antibiotic therapy improved, especially among those with an existing ID consult. This study highlights the beneficial role of RDTs in GN BSI.
Keywords: antimicrobial stewardhip, gram-negative bloodstream infection, rapid diagnostic testing
Bloodstream infections (BSIs) are a leading cause of health care–related morbidity and mortality [1]. Infections caused by gram-negative (GN) bacteria pose a particularly serious threat, with drug-resistant GNs accounting for ~1700 infections and >600 deaths annually in the United States [2]. Reducing time to in vitro active antibiotic therapy is paramount to improve outcomes, as delays are associated with increased mortality [3–8]. Conversely, antibiotic therapy that is unnecessarily broad contributes to the development of antibiotic resistance [9].
Antimicrobial stewardship (AMS) programs aim to optimize antibiotic therapy for the individual patient while limiting unnecessary antibiotic use in the overall population [10, 11]. An increasingly common AMS intervention is use of rapid diagnostic tests (RDTs), which can decrease time to identification of organisms and key antibiotic resistance mechanisms [12–15]. When used in conjunction with AMS, RDTs lead to improved clinical outcomes in BSIs [14, 16–20]. The benefits of incorporating RDTs into AMS activities and routine clinical practice have been established, but there is limited evaluation focused on GN BSI. Although all acute care hospitals are required to have some form of AMS, very few have the resources necessary to actively and consistently review RDT results in a timely manner [21, 22]. Therefore, the objective of this study was to compare time to optimal therapy and clinical outcomes in GN BSI with a step-wise introduction of RDT followed by RDT with AMS intervention.
METHODS
Study Design and Setting
This was a single-center retrospective quasi-experimental (nonrandomized) study of adult patients aged 18–89 years with GN BSI treated at the University of Maryland Medical Center (UMMC) between September 1, 2014, and October 31, 2018. UMMC is a tertiary care academic hospital with 6 infectious diseases (ID) consult services, 1 medical ID team, and 1.6 full-time equivalent ID/AMS pharmacists. Infectious diseases consults are not required for GN BSI.
Patient Consent Statement
Before initiation, the study was approved by the University of Maryland Baltimore Institutional Review Board with a waiver of informed consent.
Study Population
Patients must have had at least 1 positive blood culture with a GN organism routinely identified by study RDT during hospitalization and received at least 48 hours of antibiotic therapy with GN activity. Patients were included during their first admission with qualifying GN BSI during the study period. This included patients with polymicrobial BSI. Patients were excluded if they expired within 48 hours of blood culture draw. Additionally, patients were excluded if they did not have complete data on antibiotic exposure and other variables needed to assess the outcome of optimal antibiotic therapy including information on phenotypic antibiotic susceptibility, patient allergy history, and concurrent infectious organisms and/or sites of infection.
Microbiology Techniques and Intervention
Routine blood culture testing at UMMC Clinical Microbiology Laboratory consisted of collection in BacTAlert blood culture bottles and initial organism detection through the BacTAlert 3D automated system (bioMérieux, Durham, NC, USA). Once the presence of organisms was confirmed, gram stain was performed. These tests were completed 24 hours a day, 7 days a week, with routine critical callback procedures to ordering providers for all gram stain results. Next, organism identification and automated susceptibility testing were completed with VITEK 2 (bioMérieux, Durham, NC, USA) with antibiotic breakpoints established through the Clinical Laboratory Standards Institute and the European Committee on Antimicrobial Susceptibility Testing [23, 24].
Rapid diagnostic testing with Verigene Blood-Culture Gram-Negative (BC-GN, Luminex Corporation, Austin, TX, USA) was implemented at UMMC September 1, 2015. Verigene BC-GN is a microarray RDT that detects 8 key organisms and 6 genetic resistance determinants within 2.5 hours from gram stain [25]. If gram stain resulted in GN rods, Verigene BC-GN was performed on at least 1 blood culture bottle from the available positive bottle set(s) 24 hours a day, 7 days a week, with additional routine critical callback procedures to ordering providers for all positive RDT results.
Antimicrobial Stewardship Intervention
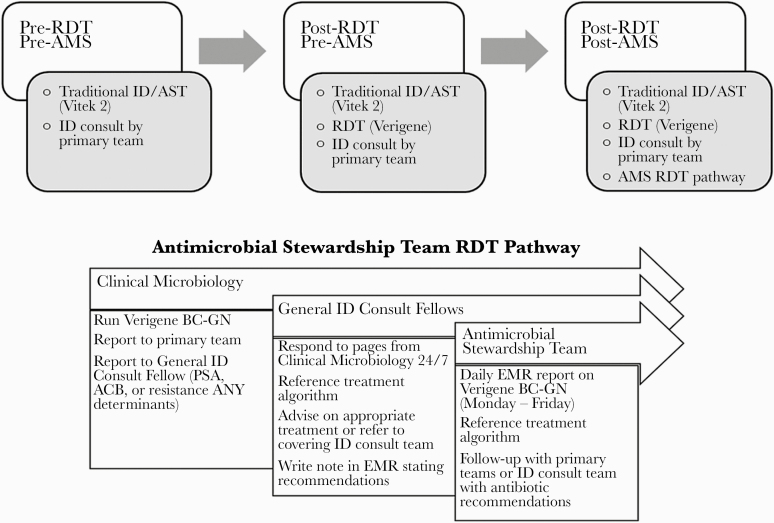
An RDT-based hospital-specific treatment algorithm was developed and validated using local susceptibility data and was implemented to guide clinical practice in March 2017 [26]. After approval of the treatment algorithm, the AMS team worked to create a GN RDT Treatment Pathway to further optimize use of RDT in GN BSI. Due to lack of real-time clinical decision support, the ID Fellow on General ID Consults was contacted with all results for Acinetobacter spp., Pseudomonas aeruginosa, or any resistance marker (eg, CTX-M, KPC) by the laboratory. This occurred 24 hours a day, 7 days a week. During regular working hours, the ID Fellow would either triage the result to the ID consult team that was already seeing the patient, or, if ID was not consulted on the patient, the ID fellow would reach out to the primary team with their interpretation and recommendations. After regular working hours and on the weekend, the ID fellow would reach out to the primary team with their interpretation and recommendations. All other Verigene BC-GN results were reviewed at least daily (Monday to Friday) by a member of the AMS team to review for appropriateness and potential for antibiotic de-escalation (Figure 1). Of note, before implementation of this pathway, AMS did not have baseline activities specifically directed toward interventions for GN BSI or RDT in BSI.
Figure 1.
Schematic of University of Maryland Medical Center Gram-negative Rapid Diagnostic Treatment Pathway with roles and responsibilities of inter-disciplinary team members. Abbreviations: AMS, antimicrobial stewardship; AST, automated susceptibility testing; EMR, electronic medical record; ID, infectious diseases; RDT, rapid diagnostic test.
Study Definitions
The primary exposure was availability of RDT results from Verigene BC-GN, with and without AMS intervention. Thus, we compared 3 groups. The pre-RDT/pre-AMS group consisted of those patients with at least 1 positive blood culture meeting inclusion criteria between September 2014 and August 2015, and the 2 post-RDT groups included post-RDT/pre-AMS intervention between September 2015 and February 2017 and post-RDT/post-AMS intervention between March 2017 and October 2018.
The primary outcome was time to optimal antibiotics, defined as time from blood culture draw to first dose of optimal antibiotic, in hours. Optimal was defined as antibiotics that demonstrated in vitro activity but were also not overly broad in spectrum and accounted for patient allergy history, concurrent infecting organisms, and site(s) of infection, as determined by an ID/AMS pharmacist in conjunction with the RDT-based hospital-specific treatment algorithm. Secondary outcomes included proportion of patients in each group placed on optimal therapy, proportion placed on in vitro active therapy, time to in vitro active therapy, and antibiotic escalation and de-escalation. Clinical outcomes included length of stay (LOS) in days, post-BSI LOS (days of inpatient admission after clearance of blood cultures), ICU LOS, and patient discharge disposition.
Covariates included age, sex, and Charlson Comorbidity Index (CCI). Patients were categorized as immune-suppressed if their primary service was Oncology or Transplant. History of drug resistance was evaluated by presence of diagnostic or surveillance cultures positive for third-generation cephalosporin- or carbapenem-resistant GNs within 1 year of admission. History of beta-lactam or fluoroquinolone antibiotic exposure was determined for the 90 days before admission [27]. ID consult was defined as an ID consult team that completed a patient evaluation within 24 hours of blood draw culture. This was separate from the ID Fellow review that occurred through the GN RDT Treatment Pathway. Source of BSI was categorized as 1 of the following: respiratory, bone/joint, skin and soft tissue, urinary, intra-abdominal infection, endovascular, and unknown/unclear. Simple imputation was used to account for any missing variables, as complete case analysis would decrease the pre-implementation sample.
Additional Molecular Analysis
To further confirm the presence of genetic resistance detected by Verigene BC-GN and better understand the clinical impact of resistant determinants not detected, additional molecular analysis through multiplex polymerase chain reaction (PCR) was completed for Enterobacterales isolates. All available clinical GN isolates from blood cultures were prospectively stored and archived at −80°C. Available Enterobacterales demonstrating phenotypic resistance to advanced-generation cephalosporins, piperacillin-tazobactam, or carbapenems by VITEK 2 (bioMérieux, Durham, NC, USA) or identified to have resistance determinants with Verigene BC-GN were subcultured. PCR consisted of confirmation of detected genetic resistance determinants as well as detection of resistance determinants not included in the Verigene BC-GN panel (blaIMP, blaVIM, blaNDM, blaCTX-M, blaTEM, blaSHV, and blaKPC). The PCR primers used are described elsewhere (Supplementary Table 1) [28–32].
Statistical Methods
Descriptive statistics included frequencies, percentages, means with SDs, or medians with interquartile ranges (IQRs), as applicable. Bivariate analysis of baseline demographics and clinical characteristics between groups was completed using the chi-square or Fisher exact test, as applicable, for nominal variables and analysis of variance or Kuskal Wallis for continuous variables, as applicable. A P value <.05 was considered statistically significant. Modified Bonferroni tests were used to adjust for multiple comparisons, as applicable.
The primary outcome, time to optimal antibiotic therapy, was assessed using Kaplan-Meier survival analysis with a log-rank test. Patients who did not receive optimal therapy were censored on date of death or discharge. Interrupted time series with negative binomial regression was conducted to assess trends in time to optimal therapy. Crude associations between study covariates and the primary outcome were evaluated through a series of univariable Cox proportional hazards regression models with a reference group of pre-RDT/pre-AMS. Potential effect measure modification of the association between exposure group and time to optimal therapy by the presence of ID consult was measured using an interaction term.
To assess for potential confounding, variables were individually entered in the Cox proportional hazards regression models that contained the primary exposure category (RDT and/or AMS). Variables were selected based on a priori biological plausibility or statistical association with the primary outcome (P < .1). Candidate variables determined a priori included exposure group (pre-RDT/AMS vs post-RDT/pre-AMS vs post-RDT/AMS) and ICU at time of GN BSI. Variables with a >10% change in hazard ratio for the association between exposure and optimal antibiotic therapy were considered confounding variables to be entered into the full multivariable model. Variables remained in the model if they remained statistically significant (P < .05) or improved model precision. The proportional hazards assumption was evaluated through assessment of Martingale residuals and the supremum test.
Correlation between Vergiene BC-GN and additional PCR testing was through Pearson’s correlation statistic with Fisher’s Z transformation for 95% CIs. All analyses were completed using SAS, version 9.4 (SAS Institute Inc., Cary, NC, USA).
RESULTS
A total of 832 patients met inclusion; 237 in the pre-RDT/AMS group, 308 in the post-RDT/pre-AMS group, and 237 in the post-RDT/AMS group. All patients had sufficient data to determine whether antibiotic therapy was in vitro active and/or optimal. Infectious Diseases (ID) consult within 24 hours of blood culture draw was missing from 71 patient charts, 70 (98.6%) from the pre-RDT/AMS group. Overall, the mean age of patients (SD) was 55.7 (16) years, the median CCI (IQR) was 2 (1–4), and the most common sources of GN BSI included urinary (240, 28.9%), intra-abdominal (174, 20.9%), and unknown/unclear (150, 18.1%). Empiric antibiotic therapy was in vitro active in 825 (99.3%) patients, and 683 (76.9%) received optimal therapy at some point during the treatment of their GN BSI.
Baseline characteristics were similar between all 3 groups (Table 1). Most patients were in the ICU at time of blood culture draw. Patients in both the post-RDT/pre-AMS and post-RDT/AMS groups were more likely to have a history of infection and/or colonization with a resistant gram-negative organism and antibiotic exposure in the 90 days prior compared with pre-RDT/AMS. Additionally, patients in both the post-RDT/pre-AMS and post-RDT/AMS groups were more likely to have an ID consult within 24 hours of blood culture draw.
Table 1.
Baseline Clinical Characteristics by Group
| Characteristic | Pre-RDT Pre-AMS (n = 237) | Post-RDT Pre-AMS (n = 308) | Post-RDT Post-AMS (n = 287) | P Value |
|---|---|---|---|---|
| Age, median (SD), y | 57.1 (15) | 56.2 (16.1) | 54.1 (16.6) | .08 |
| Female, No. (%) | 92 (38.7) | 116 (37.5) | 120 (41.8) | .57 |
| Charlson Comorbidity Index, median (IQR) | 2 (1–4) | 2 (1–4) | 2 (1–4) | .43 |
| Immune compromise, No. (%) | 18 (7.6) | 15 (4.8) | 13 (4.5) | .25 |
| Prior ESBL (1 y), No. (%) | 9 (3.8) | 25 (8.2) | 23 (8.0) | .09 |
| Prior CRE (1 y), No. (%) | 10 (4.2) | 10 (3.2) | 1 (0.4) | .01 |
| Prior antibiotics (90 d), No. (%) | 23 (9.7) | 54 (17.5) | 43 (14.9) | .03 |
| ID consult within 24 h, No. (%) | 84 (50.3)a | 208 (67.8) | 240 (83.6) | <.0001 |
| ICU at time of BSI, No. (%) | 93 (39.2) | 91 (29.6) | 115 (40.1) | .01 |
| Polymicrobial BSI, No. (%) | 15 (6.3) | 30 (9.7) | 30 (10.5) | .24 |
| Target organism isolatesb | ||||
| Acinetobacter spp. | 11 (4.6) | 22 (7.1) | 14 (4.9) | <.0001 |
| Citrobacter spp. | 0 (0) | 3 (1) | 4 (1.4) | |
| Enterobacter spp. | 31 (13.1) | 43 (13.9) | 45 (16.7) | |
| Escherichia coli | 95 (40.1) | 118 (38.3) | 100 (34.8) | |
| Klebsiella oxytoca | 4 (1.7) | 5 (1.6) | 9 (2.9) | |
| Klebsiella pneumoniae | 63 (26.6) | 76 (24.6) | 73 (23.7) | |
| Pseudomonas aeruginosa | 34 (14.3) | 34 (11) | 41 (14.3) | |
| Proteus spp. | 7 (2.9) | 15 (4.9) | 15 (5.2) | |
| Resistance marker detected | ||||
| CTX-M | -- | 30 (9.7) | 28 (9.8) | .67 |
| CTX-M & KPC | -- | 2 (0.6) | 0 (0) | |
| KPC | -- | 4 (1.3) | 2 (0.7) | |
| OXA | -- | 2 (0.6) | 1 (0.3) | |
| Source BSI, No. (%) | ||||
| Bone/joint | 4 (1.7) | 1 (0.3) | 9 (3.1) | .03 |
| Endovascular | 20 (8.4) | 33 (10.7) | 33 (11.5) | |
| Skin/soft tissue | 22 (9.2) | 20 (6.5) | 25 (8.7) | |
| Respiratory | 33 (13.9) | 27 (8.7) | 36 (12.5) | |
| Intra-abdominal | 40 (16.8) | 82 (26.5) | 52 (18.1) | |
| Urinary | 76 (31.2) | 91 (29.6) | 73 (25.4) | |
| Unknown | 39 (18.8) | 53 (17.7) | 59 (20.4) |
Abbreviations: AMS, antimicrobial stewardship; BSI, bloodstream infection; CRE, carbapenem-resistant Enterobacteriaceae; ESBL, extended-spectrum beta-lactamase; IQR, interquartile range; RDT, rapid diagnostic test.
aID consult, missing n = 71.
bPolymicrobial infections included.
Verigene-BC GN missed RDT-target GN organisms in 31 cases, 17 (5.5%) in the post-RDT/pre-AMS group vs 14 (4.9%) in the post-RDT/AMS group (P = .87). The most commonly missed organisms included K. pneumoniae (10, 32.3%), P. aeruginosa (6, 19.4%), Acinetobacter spp. (5, 16.1%), and Enterobacter spp. (3, 9.7%). The majority of missed on-panel organisms occurred in polymicrobial BSIs (28, 90.3%). Despite the presence of missed GN organisms, all patients were placed on in vitro active therapy, and 87.1% (27) were placed on optimal therapy.
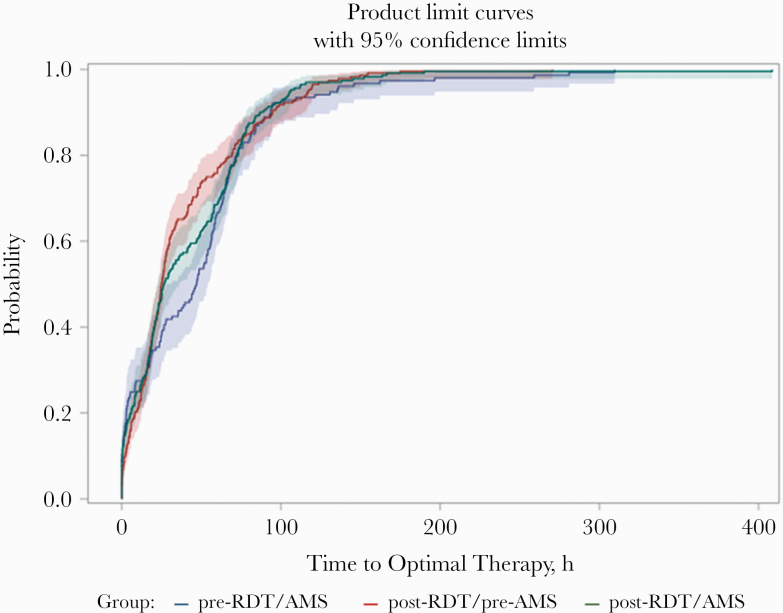
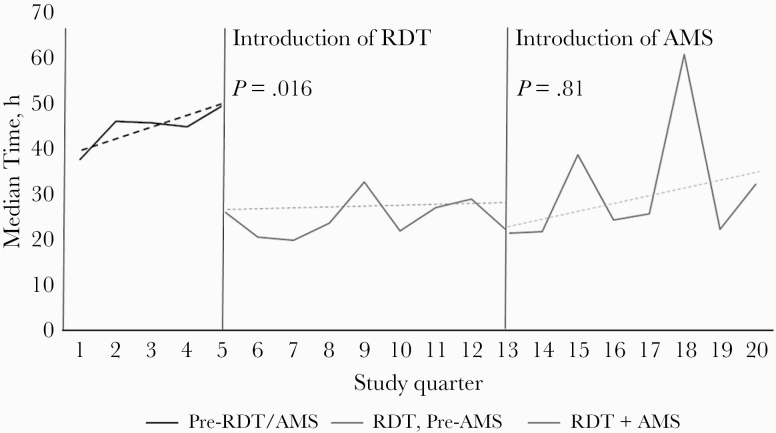
The median time to optimal antibiotic therapy (IQR), in hours, decreased in both post-RDT groups (47 [7.9–67.7] hours pre-RDT/pre-AMS vs 24.9 [12.4–55.2] hours post-RDT/pre-AMS vs 26.5 [10.3–66.5] hours post-RDT/AMS; P = .09 log-rank test) (Figure 2). Through interrupted time-series analysis, there were no significant differences in trends per quarter for time to optimal antibiotic therapy (Figure 3). The median time to optimal therapy did significantly decrease with the introduction of RDT (P = .016) but not with AMS in addition to RDT (P = .81). With the pre-RDT/AMS group as the comparator, the unadjusted hazard ratio for time to optimal therapy in the post-RDT/pre-AMS group was 1.31 (95% CI, 1.02–1.68), and in the post-RDT/post-AMS group it was 1.21 (95% CI, 0.95–1.54).
Figure 2.
Kaplan-Meier cumulative probability of receipt of optimal therapy by group. Abbreviations: AMS, antimicrobial stewardship; RDT, rapid diagnostic test.
Figure 3.
Interrupted time series analysis of trends in time to optimal therapy by group. Abbreviations: AMS, antimicrobial stewardship; RDT, rapid diagnostic test.
Statistical interaction was present between the exposure group and ID consult within 24 hours of blood culture draw, and therefore the results were stratified by presence of ID consult. Additionally, univariable Cox regression demonstrated potential confounding by source of BSI and admission to the ICU at the time of blood culture draw. Admission to the ICU and source of infection remained independently associated with time to optimal therapy after multivariable Cox regression analysis within each stratum of ID consult (Table 2). With pre-RDT/AMS as the reference group, there was no difference in time to optimal therapy in the post-RDT/pre-AMS group or post-RDT/AMS group among patients in the non–ID consult stratum. Within the ID consult stratum, controlling for source and ICU stay, both post-RDT/pre-AMS (adjusted hazard ratio [aHR], 1.34; 95% CI, 1.04–1.72) and post-RDT/AMS (aHR, 1.28; 95% CI, 1.01–1.64) had improved time to optimal therapy.
Table 2.
Multivariable Cox Proportional Hazards Model for Time to Optimal Therapy
| Variable | Unadjusted HR (95% CI) | Adjusted HR (95% CI) ID = Yes (n = 455) | Adjusted HR (95% CI) ID = No (n = 162) |
|---|---|---|---|
| Post-RDT/pre-AMS (ref = pre-RDT/AMS) | 1.31 (1.02–1.68) | 1.34 (1.04–1.72) | 0.93 (0.63–1.37) |
| Post-RDT/AMS (ref = pre-RDT/AMS) | 1.21 (0.95–1.54) | 1.28 (1.01–1.64) | 0.84 (0.54–1.29) |
| ICU at GN BSI (ref = No ICU) | 0.92 (0.76–1.11) | 0.95 (0.78–1.16) | 1.01 (0.7–1.45) |
| Source of BSI = urinary (ref = nonurinary) | 1.24 (1–1.5) | 1.28 (1.03–1.58) | 1.64 (1.17–2.29) |
Abbreviations: AMS, antimicrobial stewardship; BSI, bloodstream infection; HR, hazard ratio; ICU, intensive care unit; ID, infectious diseases consult; IQR, interquartile range; RDT, rapid diagnostic test.
The proportion of patients placed on in vitro active therapy during inpatient treatment of their GN BSI was similar among all groups (99.5% pre-RDT/AMS vs 98.4% post-RDT/pre-AMS vs 100% post-RDT/AMS; P = .055). The proportion of patients placed on optimal therapy increased with both introduction of RDT and AMS intervention (66.5% vs 78.9% vs 83.2%; P < .0001). Among those placed on optimal therapy, antibiotic escalation occurred most frequently in the post-RDT/pre-AMS group (15.3% vs 39.1% vs 13.1%; P < .0001). Time to antibiotic escalation (IQR) significantly decreased with the introduction of RDT (48.4 [17.6–66.5] hours pre-RDT/AMS vs 20.4 [14.9–30.2] hours post-RDT/pre-AMS vs 21.9 [16.5–35.9] hours post-RDT/AMS; P = .02). Antibiotic de-escalation occurred in 45.2% pre-RDT/AMS vs 31.7% post-RDT/pre-AMS vs 39.1% post-RDT/AMS (P = .018). Time to antibiotic de-escalation (IQR) did not significantly change with the introduction of RDT or AMS intervention (60.9 [47.6–83.6] hours pre-RDT/AMS vs 65.3 [26.2–89.5] hours post-RDT/pre-AMS vs 66.7 [51.7–81.6] hours post-RDT/AMS; P = .47).
Among related clinical outcomes, overall length of stay (IQR), in days, was not significantly different between groups (16.9 [6.4–32.5] vs 15.9 [7.8–29.8] vs 18.9 [7.2–35.9]; P = .7). Importantly, post-BSI length of stay (IQR) also did not significantly differ 9.5 [5.1–18.8] vs 9.8 [5.4–20] vs 11.3 [6–21.1]; P = .17). All-cause inpatient mortality was lower in the post-RDT/AMS group (15.9% vs 14.9% vs 3.8%; P < .0001).
Phenotypic resistance to advanced-generation cephalosporins, piperacillin-tazobactam, or carbapenems was seen in 124 (15.6%) Enterobacterales isolated from blood cultures. Among those patient samples, 93 were available for additional molecular testing. A total of 75 (79.8%) isolates had at least 1 beta-lactamase resistance gene identified by PCR, with blaCTX-M being the most common (Table 3). A total of 59 isolates had both Verigene BC-GN and PCR data available for comparison. Resistance secondary to blaCTX-M was present by Verigene BC-GN in 40 isolates. This was confirmed by PCR in 36 (90%), while 3 more were identified to harbor blaCTX-M by PCR but not Verigene BC-GN. This resulted in an agreement between Verigene BC-GN and PCR for blaCTX-M of 72.3% (95% CI, 57%–82%). Among these 59 isolates, 5 had blaKPC present by Verigene BC-GN, which was confirmed in 4 isolates by PCR. One organism, which also had blaCTX-M identified by both Verigene BC-GN and PCR, also carried blaKPC that was not identified by Verigene BC-GN. This resulted in an agreement between Verigene BC-GN and PCR of 78.1% (95% CI, 65.1%–86.3%). Only 4 phenotypically resistant organisms were negative for all tested resistance genes.
Table 3.
Additional Molecular Analysis of Phenotypically Resistant Enterobacterales
| Resistance Marker Detected | Total (n = 93), No. (%) | Pre-RDT Pre-AMS (n = 34), No. (%) | Post-RDT Pre-AMS (n = 36), No. (%) | Post-RDT Post-AMS (n = 23), No. (%) |
|---|---|---|---|---|
| Molecular analysisa | ||||
| TEM | 13 (13.9) | 7 (20.6) | 6 (16.7) | 0 (0) |
| SHV | 12 (12.9) | 8 (23.5) | 1 (2.8) | 3 (13) |
| CTX-M | 51 (54.8) | 11 (32.4) | 22 (61.1) | 18 (78.3) |
| KPC | 7 (7.5) | 2 (5.9) | 4 (11.1) | 1 (4.4) |
Abbreviations: AMS, antimicrobial stewardship; RDT, rapid diagnostic test.
aTwo isolates had both TEM and SHV identified; 2 isolates have both CTX-M and KPC identified.
DISCUSSION
This study evaluated the impact of RDT availability with and without active AMS intervention on time to optimal antibiotic therapy in GN BSI. In the groups with RDT availability, time to optimal therapy significantly decreased. Interestingly, the impact of RDT was only significantly associated with improved time to optimal therapy in those with an ID consult, which remained significant after controlling for confounding variables. Time to optimal therapy was not significantly impacted by the introduction of the AMS intervention, although the proportion of patients eventually placed on optimal therapy increased. The observed lack of additional impact of AMS on time to optimal therapy was likely due to the high proportion of patients seen by ID. In-patient all-cause mortality was also significantly lower in the post-RDT/post-AMS group, although confirmation of a causal association is beyond the scope of the current study.
Numerous retrospective studies have demonstrated the ability to appropriately de-escalate antibiotics and improve patient clinical outcomes in gram-positive BSI, in particular with AMS intervention [33–36]. Studies evaluating the impact of RDT on GN BSI, however, are more limited [16–18, 37]. In a quasi-experimental study, Rivard et al. examined the impact of concurrent Verigene BC-GN and AMS intervention on >800 patients with GN-BSI [17]. Proportionally, antibiotic switch occurred in a similar amount of patients, but the median time to switch significantly decreased with the introduction of RDT/AMS intervention, from 44 to 28.6 hours. This patient population is similar to the current study; however, there were limited data on relevant confounders or the percentage of patients with ID consult. Although the exposure was a combination of RDT and AMS intervention, the primary outcome demonstrated similar results to the current study.
A novel feature of our study is the focus on the presence of ID consult even in the setting of an AMS. The importance of active AMS intervention on RDT results to improve clinical outcomes in BSIs has been demonstrated in previous literature [16]. The exact mechanisms have not been fully elucidated but likely center on the timely attention of those with advanced ID training. In a cross-sectional survey of non-ID physicians at the University of Nebraska Medical Center, Donner et al. evaluated non-ID provider confidence and comprehension interpreting and acting upon microbiology results [38]. Among the 156 respondents, 81.6% reported adjusting antibiotic therapy based on traditional microbiology, while only 60% reported adjusting based on RDT results. Additionally, correctness on knowledge-based questions ranged from 50% to 86%, with common errors surrounding interpretation of Enterobacterales and antibiotic de-escalation. Consultation with ID-trained individuals has been shown to improve clinical outcomes in BSI, with most studies focused on the management of S. aureus BSI [39–41]. Recently, Burnham et al. evaluated the impact of ID consultation across multidrug-resistant infections [42]. ID consult in multidrug-resistant Enterobacterales was associated with significantly deceased risk of 30-day mortality, lending support to the current findings.
Potential reasons for the scarcity of data to support use of RDTs in GN BSI include the increased diversity of pathogenic organisms, the complexity and multifactorial nature of antibiotic resistance, and the potential downstream clinical consequences of missed organism identification and/or phenotypic resistance [43, 44]. Overall, in vitro studies have demonstrated high sensitivity and specificity for organisms and resistance determinants; however, these studies typically occur in monomicrobial blood samples with on-panel targets without mention of phenotypic resistance [13, 14, 45]. In a previous study, Pogue et al. demonstrated a high degree of positive agreement between phenotypic resistance and genetics resistance with on-panel organisms with the exception of non-lactose-fermenting organisms [43]. For instance, we previously confirmed a high level of agreement between phenotypic advanced-generation cephalosporin resistance and the presence of CTX-M, lending confidence to our algorithm recommendation to recommend de-escalation in Enterobacterales without detection of resistance determinant [26]. In the current study, PCR confirmed that blaCTX-M was the most common driver for third-generation cephalosporin resistance at our institution. There were, however, discrepancies in detection between Verigene BC-GN and PCR that resulted in an agreement of 72.3%, but this must be interpreted within the limitation of a small sample of clinical isolates tested. Additionally, the differences in detection could be secondary to differences in analytic techniques and need to be further investigated [25, 46]. The application of RDTs that provide phenotypic susceptibility information, either in place of or in addition to genetic resistance testing, although beyond the scope of this paper, is an area of ongoing research [47].
There are notable limitations to the study. Given the retrospective nature, review of decision-making regarding antibiotic therapy can only be evaluated based on information contained in the EMR. Additionally, due to incomplete data before 2015, a proportion of pre-RDT/AMS patients were missing data on presence of ID consult. A second notable limitation is the lack of consensus-based definition of optimal antibiotic therapy [48]. The current definition of optimal antibiotic therapy is similar to the definitions used in previous investigations and was done with an algorithmic approach, but a certain level of subjectivity must be considered in this assessment. Lastly, external generalization of these findings may be limited, as there was a high proportion of patients who had an ID consult at the time of gram-negative BSI, likely due to the extensive ID services available at UMMC. Previous studies of RDT that included GN BSI reported a much smaller proportion of patients seen by an ID specialist [17, 18]. This is significant, as ID consult was an effect measure modifier. In institutions where ID presence is limited, AMS intervention may have a higher impact than that currently demonstrated, as these ID consults may be serving as an extension of AMS activities with respect to responding to RDT results.
In conclusion, the introduction of RDT in GN BSI resulted in a significant decrease in time to optimal antibiotic therapy, by a median of ~22 hours from blood culture draw. Additionally, the overall proportion of patients placed on optimal antibiotic therapy increased. Infectious diseases consultation was a significant interaction, highlighting the importance of having ID-trained individuals, even outside of AMS, review RDT results in a timely fashion. More experience is needed on the impact of antibiotic de-escalation and overall clinical outcomes.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The authors thank Nora Loughry and Sanjay Chainani for assistance with data collection and validation.
Author contributions. K.C.C., E.L.H., J.K.J., and S.L. contributed to the conception/design of the work, acquisition, analysis, and interpretation of results, and drafting and revising the final manuscript. S.H. contributed to the acquisition, analysis, and interpretation of results and drafting and revising the final manuscript.
Financial support. This study was funded by Making a Difference in Infectious Diseases (MAD-ID).
Potential conflicts of interest. K.C.C. has served as a speaker for Luminex Corporation and GenMark Diagnostics. J.K.J. and K.C.C. have received study supplies from BioFire Diagnostics and GenMark Diagnostics. J.K.J. has served as a speaker for GenMark Diagnostics. E.L.H. and S.L. report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 2013; 19:501–9. [DOI] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention. The biggest antibiotic-resistant threats in the U.S. Centers for Disease Control and Prevention. 2018 Available at: https://www.cdc.gov/drugresistance/biggest_threats.html. Accessed 24 February 2019.
- 3. Marquet K, Liesenborgs A, Bergs J, et al. Incidence and outcome of inappropriate in-hospital empiric antibiotics for severe infection: a systematic review and meta-analysis. Crit Care 2015; 19:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lodise TP, Zhao Q, Fahrbach K, et al. A systematic review of the association between delayed appropriate therapy and mortality among patients hospitalized with infections due to Klebsiella pneumoniae or Escherichia coli: how long is too long? BMC Infect Dis 2018; 18:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ibrahim EH, Sherman G, Ward S, et al. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 2000; 118:146–55. [DOI] [PubMed] [Google Scholar]
- 6. Zaragoza R, Artero A, Camarena JJ, et al. The influence of inadequate empirical antimicrobial treatment on patients with bloodstream infections in an intensive care unit. Clin Microbiol Infect 2003; 9:412–8. [DOI] [PubMed] [Google Scholar]
- 7. Kang CI, Kim SH, Park WB, et al. Bloodstream infections caused by antibiotic-resistant gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother 2005; 49:760–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zilberberg MD, Shorr AF, Micek ST, et al. Multi-drug resistance, inappropriate initial antibiotic therapy and mortality in gram-negative severe sepsis and septic shock: a retrospective cohort study. Crit Care 2014; 18:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teshome BF, Vouri SM, Hampton N, et al. Duration of exposure to antipseudomonal β-lactam antibiotics in the critically Ill and development of new resistance. Pharmacotherapy 2019; 39(3):261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62:e51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tamma PD, Avdic E, Keenan JF, et al. What is the more effective antibiotic stewardship intervention: preprescription authorization or postprescription review with feedback? Clin Infect Dis 2017; 64:537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mancini N, Infurnari L, Ghidoli N, et al. Potential impact of a microarray-based nucleic acid assay for rapid detection of gram-negative bacteria and resistance markers in positive blood cultures. J Clin Microbiol 2014; 52:1242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ledeboer NA, Lopansri BK, Dhiman N, et al. Identification of gram-negative bacteria and genetic resistance determinants from positive blood culture broths by use of the verigene gram-negative blood culture multiplex microarray-based molecular assay. J Clin Microbiol 2015; 53:2460–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walker T, Dumadag S, Lee CJ, et al. Clinical impact of laboratory implementation of verigene BC-GN microarray-based assay for detection of gram-negative bacteria in positive blood cultures. J Clin Microbiol 2016; 54:1789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Avdic E, Carroll KC. The role of the microbiology laboratory in antimicrobial stewardship programs. Infect Dis Clin North Am 2014; 28:215–35. [DOI] [PubMed] [Google Scholar]
- 16. Timbrook TT, Morton JB, McConeghy KW, et al. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64:15–23. [DOI] [PubMed] [Google Scholar]
- 17. Rivard KR, Athans V, Lam SW, et al. Impact of antimicrobial stewardship and rapid microarray testing on patients with gram-negative bacteremia. Eur J Clin Microbiol Infect Dis 2017; 36:1879–87. [DOI] [PubMed] [Google Scholar]
- 18. Bookstaver PB, Nimmich EB, Smith TJ, et al. Cumulative effect of an antimicrobial stewardship and rapid diagnostic testing bundle on early streamlining of antimicrobial therapy in gram-negative bloodstream infections. Antimicrob Agents Chemother 2017; 61:e00189-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Porter AM, Bland CM, Young HN, et al. Comparison of pharmacist-directed management of multiplex PCR blood culture results with conventional microbiology methods on effective and optimal therapy within a community hospital. Antimicrob Agents Chemother 2018; 63:e01575-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayakawa K, Mezaki K, Kobayakawa M, et al. Impact of rapid identification of positive blood cultures using the verigene system on antibiotic prescriptions: a prospective study of community-onset bacteremia in a tertiary hospital in Japan. PLoS One 2017; 12:e0181548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kapadia SN, Abramson EL, Carter EJ, et al. The expanding role of antimicrobial stewardship programs in hospitals in the United States: lessons learned from a multisite qualitative study. Jt Comm J Qual Patient Saf 2018; 44:68–74. [DOI] [PubMed] [Google Scholar]
- 22. Joint Commission. Antimicrobial stewardship. Available at: http://www.jointcommission.org/topics/hai_antimicrobial_stewardship.aspx. Accessed 25 February 2019.
- 23.Clinical & Laboratory Standards Institute. Clinical & Laboratory Standards Instiute (CLSI) M100. Available at: https://clsi.org/standards/products/free-resources/access-our-free-resources/. Accessed 7 April 2019.
- 24. The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters, version 9.0. Available at: http://www.eucast.org/clinical_breakpoints/. Accessed 2 August 2019.
- 25. Luminex Corporation. Verigene® gram-negative blood culture test. Available at: https://www.luminexcorp.com/clinical/infectious-disease/verigene-bloodstream-infection-tests/gram-negative-blood-culture/. Accessed 3 March 2018.
- 26. Claeys KC, Schlaffer KE, Heil EL, et al. Validation of an antimicrobial stewardship-driven verigene blood-culture gram-negative treatment algorithm to improve appropriateness of antibiotics. Open Forum Infect Dis 2018; 5:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Augustine MR, Testerman TL, Justo JA, et al. Clinical risk score for prediction of extended-spectrum β-lactamase-producing enterobacteriaceae in bloodstream isolates. Infect Control Hosp Epidemiol 2017; 38:266–72. [DOI] [PubMed] [Google Scholar]
- 28. Nüesch-Inderbinen MT, Hächler H, Kayser FH. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur J Clin Microbiol Infect Dis 1996; 15:398–402. [DOI] [PubMed] [Google Scholar]
- 29. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 2011; 70:119–23. [DOI] [PubMed] [Google Scholar]
- 30. Sedighi M, Halajzadeh M, Ramazanzadeh R, et al. Molecular detection of β-lactamase and integron genes in clinical strains of Klebsiella pneumoniae by multiplex polymerase chain reaction. Rev Soc Bras Med Trop 2017; 50:321–8. [DOI] [PubMed] [Google Scholar]
- 31. Rasheed JK, Jay C, Metchock B, et al. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother 1997; 41:647–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pagani L, Dell’Amico E, Migliavacca R, et al. Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in Northern Italy. J Clin Microbiol 2003; 41:4264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neuner EA, Pallotta AM, Lam SW, et al. Experience with rapid microarray-based diagnostic technology and antimicrobial stewardship for patients with gram-positive bacteremia. Infect Control Hosp Epidemiol 2016; 37:1361–6. [DOI] [PubMed] [Google Scholar]
- 34. Aitken SL, Hemmige VS, Koo HL, et al. Real-world performance of a microarray-based rapid diagnostic for gram-positive bloodstream infections and potential utility for antimicrobial stewardship. Diagn Microbiol Infect Dis 2015; 81:4–8. [DOI] [PubMed] [Google Scholar]
- 35. MacVane SH, Raux BR, Smith TT. Evaluation of rapid polymerase chain reaction-based organism identification of gram-positive cocci for patients with a single positive blood culture. Eur J Clin Microbiol Infect Dis 2019; 38:1471–9. [DOI] [PubMed] [Google Scholar]
- 36. Avdic E, Wang R, Li DX, et al. Sustained impact of a rapid microarray-based assay with antimicrobial stewardship interventions on optimizing therapy in patients with gram-positive bacteraemia. J Antimicrob Chemother 2017; 72:3191–8. [DOI] [PubMed] [Google Scholar]
- 37. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction–based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61:1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Donner LM, Campbell WS, Lyden E, Van Schooneveld TC. Assessment of rapid-blood-culture-identification result interpretation and antibiotic prescribing practices. J Clin Microbiol 2017; 55:1496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wenzler E, Wang F, Goff DA, et al. An automated, pharmacist-driven initiative improves quality of care for Staphylococcus aureus bacteremia. Clin Infect Dis 2017; 65:194–200. [DOI] [PubMed] [Google Scholar]
- 40. Smith JR, Frens JJ, Snider CB, Claeys KC. Impact of a pharmacist-driven care package on Staphylococcus aureus bacteremia management in a large community healthcare network: a propensity score-matched, quasi-experimental study. Diagn Microbiol Infect Dis 2018; 90:50–4. [DOI] [PubMed] [Google Scholar]
- 41. Vogel M, Schmitz RP, Hagel S, et al. Infectious disease consultation for Staphylococcus aureus bacteremia—a systematic review and meta-analysis. J Infect 2016; 72:19–28. [DOI] [PubMed] [Google Scholar]
- 42. Burnham JP, Kollef MH. Treatment of severe skin and soft tissue infections: a review. Curr Opin Infect Dis 2018; 31:113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pogue JM, Heil EL, Lephart P, et al. An antibiotic stewardship program blueprint for optimizing verigene BC-GN within an institution: a tale of two cities. Antimicrob Agents Chemother 2018; 62:e02538-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Humphries RM, Spafford K, MacVane S. Reply to Pogue and Heil, “The clinical impact of a negative molecular β-lactamase gene test for Enterobacteriaceae : let’s not let perfect be the enemy of really good.” J Clin Microbiol 2020; 58:e02114-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ward C, Stocker K, Begum J, et al. Performance evaluation of the Verigene® (Nanosphere) and FilmArray® (BioFire®) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis 2015; 34:487–96. [DOI] [PubMed] [Google Scholar]
- 46. Weile J, Knabbe C. Current applications and future trends of molecular diagnostics in clinical bacteriology. Anal Bioanal Chem 2009; 394:731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dare RK, Lusardi K, Pearson C, et al. Clinical impact of accelerate pheno rapid blood culture detection system in bacteremic patients. Clin Infect Dis. In press. [DOI] [PubMed] [Google Scholar]
- 48. Spivak ES, Cosgrove SE, Srinivasan A. Measuring appropriate antimicrobial use: attempts at opening the black box. Clin Infect Dis 2016; 63:1639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.